Give us the tools and we will finish the job
-Winston Churchill
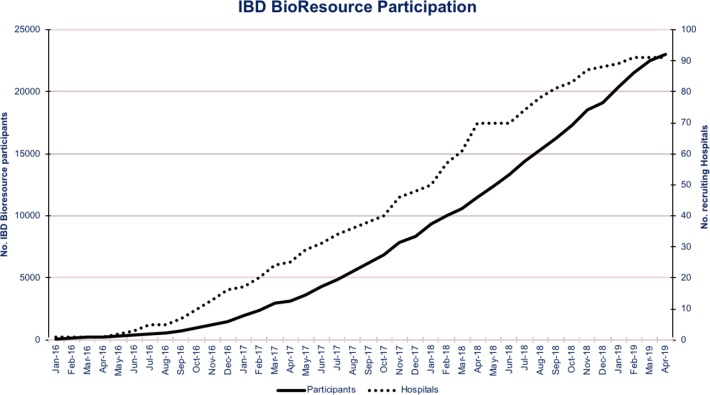
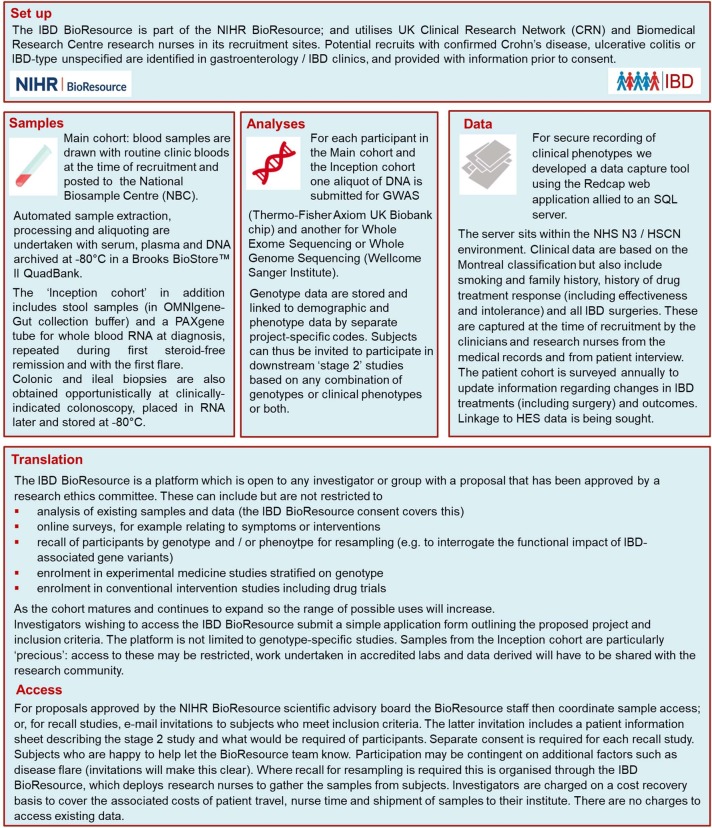
An alliance of clinicians, academics, research nurses, funders, coordinators, programmers and, most importantly, patients has come together in the UK to deliver a powerful new platform to accelerate Crohn’s and Colitis research—the inflammatory bowel disease (IBD) BioResource. As part of the National Institute for Health Research (NIHR) BioResource for translational research, 25 000 patients in over 90 hospitals UK-wide have signed up since we launched in January 2016 (figure 1). All have detailed phenotypes databased including Montreal classification,1 treatment response history (updated annually), surgical history and comorbidities (see IBD BioResource panel descriptive, Clinical data collection sheet and Health and Lifestyle questionnaire). Serum, plasma and DNA samples are banked; and genome-wide genetic profiling undertaken. Participants’ data and samples can be studied, and they themselves surveyed or recalled for resampling or downstream studies (see figure 2). Critically, such studies can be led by any UK or overseas investigator whether from the worlds of clinical research, pharmacovigilance, science or industry.
Figure 1.
IBD BioResource recruitment in over 90 hospitals in the UK. IBD, inflammatory bowel disease.
Figure 2.
How the IBD BioResource works. HES, Hospital Episode Statistics; HSCN, Health Social Care Network; IBD, inflammatory bowel disease; NHS, National Health Service.
What is the IBD BioResource for
A key motivation is to leverage recent genetics advances, and by understanding the functional impact of IBD-associated gene variants accelerate translation of the new knowledge for clinical benefit. Beyond this, it is increasingly evident that the IBD BioResource can facilitate a wide spectrum of research. This might include anything from mining existing data or samples or surveying the cohort regarding outcomes of newly licensed treatments, to pharmacogenetic research. It could also be used to expedite recruitment to intervention studies, including experimental medicine and conventional drug trials.
Gene discovery in Crohn’s disease and ulcerative colitis has placed these conditions at the forefront of the field of common disease genetics. As the UK IBD Genetics Consortium we have, through national and international collaboration, helped deliver a number of landmark studies in IBD.2–14 More than 240 confirmed IBD susceptibility loci have been identified to date. New druggable pathways continue to be identified and new pathogenic insights continue to accrue,4 particularly from those loci where the causal variants have been identified and functionally characterised. Examples of the latter include the association between IBD and the R381Q variant in the interleukin 23 receptor and between CD and the T300A variant in the autophagy gene ATG16L1.15–17
For most IBD risk loci, however, the causal genes and causal variants await identification. There are instances of hard-fought progress, with dissection of individual loci allowing new genes to be characterised and new biological insights gained18 19; and using innovative genetic ‘fine mapping’ techniques it has been possible to refine ~20% of risk loci to a single variant.2 However, functional characterisation of the latter is awaited, and is made all the more challenging by the fact that most are non-coding (regulatory). Furthermore, more than 50% map to genomic regions lacking homology with any known functional motif. The prize may be new biological insight not just about IBD pathogenesis but potentially also new understanding of the regulation of gene transcription, but no number of DNA samples can deliver this. Without recourse to mouse models or genetically manipulated cell lines, with all their problems, investigators will be able to use the IBD BioResource to access and resample cohorts of patients homozygous for risk and wild-type alleles at any locus. Already, despite only having recently reached maturity, such ‘stage 2’ recall-by-genotype studies are in progress. These are interrogating the functional impact of cytokine polymorphisms, non-coding RNA’s and HLA variants implicated in our genome wide association studies (GWAS), to better understand how these contribute to IBD pathogenesis and to specific subphenotypes.
Critical to the success of the IBD BioResource will be the willingness of patients to participate in annual surveys (to gather long term data re treatment outcomes etc) and stage 2 studies. It is early to be drawing conclusions, but in stage 2 studies to date 30%–60% of eligible participants have agreed to help. Maintaining engagement with patients and potential stage 2 users is clearly a key role of the team of four IBD BioResource coordinators and one data manager. As well as running the annual surveys, they maintain the website (www.ibdbioresource.nihr.ac.uk), circulate newsletters and promote activities through social media (Twitter and LinkedIn). Over the next 5 years, it is anticipated that longitudinal data from surveys will be substantially augmented by access to NHS digital and potential collaborations with the IBD Registry and Health Data Research UK, to minimise the risk of participants being lost to follow-up.
In addition to its recall/resample functionality, the IBD BioResource will also be used as the substrate for a new suite of genetics analyses. There is still more mileage in GWAS for risk variants: as the IBD dataset expands so the number of loci detected will increase in linear proportion, and new insights gained.20 Beyond GWAS increasing attention is turning to exome and whole genome sequencing, particularly to ascertain low frequency risk variants of potentially greater effect size than are typically identified by GWAS.3 These may contribute to the ‘missing heritability’ of IBD and other common diseases.21 Two limiting factors to date have been sequencing costs and the need for very large sample sizes to detect rare variant associations with statistical confidence. Both are becoming tractable. All IBD BioResource participants will undergo 50x exome sequencing or 20x genome sequencing and the data will be meta-analysed with other datasets in the international IBD genetics consortium in well powered studies; and the results made available to the research community.
Translational aspects
As well as identifying susceptibility genes, there is increasing interest in identifying biomarkers for clinically relevant outcomes such as prognosis and treatment response, the aim being to deliver on the promise of personalised medicine. This requires detailed phenotypes on large sample sets—something the IBD BioResource has, with 95% completion of core data fields on the first 24 000 participants. Lee et al previously used GWAS to identify loci associated with Crohn’s disease prognosis22; and blood monocytes from genotype-selected NIHR BioResource subjects to interrogate the function of a FOXO3 polymorphism.23 The expanding IBD BioResource dataset should identify more markers associated with disease course and enable development of progressively more accurate ‘polygenic scores’ for prognosis. The latter use the power of genome-wide data (rather than just genome-wide significant hits), potentially combined with environmental modifyers such as smoking, to substantially improve predictive accuracy.24 This could be used at the time of IBD diagnosis to identify individuals destined for a complex disease course with multiple flares, perhaps warranting early and aggressive therapy.
Regarding treatment outcomes, pharmacogenetic analyses have recently identified a number of clinically important associations. Thus, NUDT15 polymorphisms are associated with thiopurine-induced leucopenia both in Asians and Europeans25 26; and human leukocyte antigen (HLA) DRB1*0701 is associated with risk of thiopurine pancreatitis.27 Other work lead by Tariq Ahmad in Exeter, including samples from the IBD BioResource, has identified strong association between HLA DQA1-05 (found in 40% of people of European origin) and immunogenicity to anti tumour necrosis factor (TNF) therapy.28 Studies to demonstrate the utility of testing for these in the IBD clinic is a near-term objective for IBD BioResource investigators.
Future pharmacogenetic studies will also benefit from the scale of the IBD BioResource. By recruiting from hospital clinics, it is biased towards more severely affected patients. Thus >13 000 have been treated with thiopurines and >9000 with anti-TNF therapy, all with detailed treatment outcomes recorded. Through the IBD BioResource, all participants can be recontacted or recalled if more detailed information or more samples are required. Even relatively recently introduced therapies are strongly represented, for example, over 1400 participants have received vedolizumab—and through an annual ‘treatment update’ survey, we expect to see this number climbing rapidly. We thus have a clear opportunity to better understand how these treatments are being used in routine practice, and for further pharmacogenetic analyses, for example, of treatment response. There may also be a role for the IBD BioResource in pharmacovigilance for new therapies.
Supporting intervention studies and drug trials
Can the IBD BioResource directly support intervention studies, including drug trials? We believe so. Later this year, it will be used to recruit to the IBD BOOST study, surveying 12 000 patients and then recruiting 1180 with self-reported symptoms of fatigue, pain and/or urgency for different interventions. There is also potential utility for drug trials recruitment. There is widespread recognition that drug development takes too long, is too costly and potentially high risk.29 With the recent expansion of licensed IBD therapies industry may see the cost of developing a new drug, particularly one that ends up as fourth or fifth line therapy, as too great—and might shelve some of the potentially valuable new treatments.30 Different strategies are required, potentially including experimental and precision medicine to investigate subgroups of patients who have distinct pathogenic pathways and in whom drugs which target those pathways may demonstrate high efficacy (hence warranting earlier use in such biomarker-defined subsets). The IBD BioResource, with its detailed genome-wide data, is ideally positioned to support such studies.
At a more general level, the IBD BioResource is well placed to accelerate recruitment to conventional phase 3 studies—widely regarded as a major bottle-neck in drug development. This should be of great interest to the pharma industry and clinical research organisations. Potential participants meeting inclusion criteria for a particular study (identified from the IBD BioResource database of phenotypes and drug histories) and under a trial site hospital can be provided with trial details and notified that if their IBD is flaring and they are interested they should contact the research nurse at their site. Many patients are only too willing to participate in research but a major block is giving them the opportunity.31 Inherent to the problem is that most drug trial recruitment happens in clinic, but requires the clinician meeting the patient in flare to know about the study, be aware of inclusion criteria and to match these up to the patient in front of them. This presupposes an alignment of knowledge, time and motivation in busy clinicians, and all too often one or more of these is lacking. Having a large cohort of research-engaged IBD BioResource participants who have consented to screening of their medical records and can be contacted directly regarding research should help circumvent this bottleneck.
The future
A key goal over the next 5 years is to recruit an inception cohort of 1000 individuals newly diagnosed with IBD who will undergo more detailed sampling (including stool, biopsy tissue and whole blood for RNA—unconfounded by the effects of drug treatment) and longitudinal follow-up. In due course, this will facilitate and expand existing research into the determinants, predictors and biomarkers of disease course and treatment response.32 33
The IBD BioResource has more than 90 hospitals participating UK-wide, and is now recruiting at ~1000 patients per month (figure 1). With 25 000 highly characterised patients already signed up, it is very definitely ‘open for business’ and now able to support and expedite a wide range of studies. What began as a resource to enable post-GWAS studies has become a wider multidimensional platform ready for use by the global IBD research community. Its job is to accelerate IBD research across all domains, improve treatments and outcomes, and perhaps one day ‘finish the job’ by achieving a cure. We are keen to see this tool used!
To apply to use the IBD BioResource please visit
https://BioResource.nihr.ac.uk/researchers/researchers/application-process/
Enquiries re its use to ibd@bioresource.nihr.ac.uk
Acknowledgments
We thank NIHR BioResource volunteers for their participation, and gratefully acknowledge NIHR BioResource centres, NHS Trusts and staff for their contribution. In addition to the staff at the NIHR BioResource, we would like to thank the IBD BioResource administrators, coordinators and data manager.
Footnotes
Funding: The IBD BioResource is part of the NIHR BioResource for Translational Research and is partner funded by the Medical Research Council (RG74456), the NIHR, the Wellcome Trust (RG84515), Open Targets (RG84515) and Crohn’s and Colitis UK (RG82018).
Disclaimer: The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health.
Competing interests: None declared.
Provenance and peer review: Commissioned; externally peer reviewed.
Collaborators: Achuth Shenoy; Ailsa Hart; Ajay Muddu; Ajay Verma; AlanLobo; Alan Wiles; Albert Davies; Alexandra Kent; Alison Simmons; Andi Li; AndrewCole; Anjan Dhar; Anne Phillips; Annette Woods; Anton Gunasekera; ArabisOglesby; Babur Javaid; Bijay Baburajan; Byron Theron; Carl Anderson; CatherineThorbinson; Cathryn Edwards; Cathryn Preston; Charles Murray; Charlie Lees; ChiragOza; Chris MacDonald; Christian Selinger; Christopher Lamb; Christopher Mathew;Chuka Nwokolo; Colin Rees; Conor Lahiff; Dan Sharpstone; Daniel Gavin; DavidHobday; Deepak Kejariwal;Deepthy Francis; Deven Vani; Ehab Abdelmalek; EmmaWesley; Francisco Porras-Perez;Fraser Cummings; Ganesh Sivagi; Gareth Parkes; GeorgeMacfaul; Gordon Moran; Helen Jane Dallal; Helen Steed; Jack Satsangi; JamesLee;James Lindsay; Jamie Barbour; Jeff Butterworth; Jeremy Sanderson, JimmyLimdi; John Beckly; John deCaestecker; John Gordon; John Mansfield; JohnMcLaughlin; John Ramage; John Saunders; Jonathan Landy; Joseph Collum; JoyWilkins; Jude Tidbury; Juliette Loehry; Kath Phillis; Katherine Smith; KatieClark; Khalid Elamin; Klaartje Bel Kok; Konrad Koss; Laetitia Pele; LauraHancock; Leena Sinha; Leonie Grellier; Luke Jostins; Lynn O’Donohoe; MarkJarvis; Mark Tremelling; Martyn Carter; Matthew Brookes; Matthew Johnson; MelissaSmith; Michael Sprakes; Michelle Baker-Moffat; Mina Hanna; Monica Bose; MoniraRahman; Natalie Prescott; Nick Kennedy; Nikolaos Koukias; Paul Banim; PaulSmith; Peter Irving; Phil Roberts; Rachel Simpkins; Rasha Shawky; RichardAppleby; Richard Keld; Richard Pollok; Richard Shenderey; Roisin Bevan; RonitDas;Ruth Penn; Salil Singh; Shaji Sebastian; Simon McLaughlin; Simon Panter; SreedharSubramanian; Stamatia Chatzinikolaou; Stephen Foley; Stephen Lewis; StuartBloom; Subramaniam Ramakrishnan; Sunil Samuel; Sunny Nair; Tariq Ahmad; TheresaTindall; Timi Patani; Udi Shmueli; Vinod Patel.
Patient consent for publication: Not required.
Contributor Information
On behalf of the IBD BioResource Investigators:
Ehab Abdelmalek, Richard Appleby, Bijay Baburajan, Michelle Baker-moffat, Paul Banim, Jamie Barbour, John Beckly, Roisin Bevan, Stuart Bloom, Monica Bose, Matthew Brookes, Jeff Butterworth, Carl Anderson, Martyn Carter, Stamatia Chatzinikolaou, Katie Clark, Andrew Cole, Fraser Cummings, Helen Jane Dallal, Albert Davies, John Decaestecker, Anjan Dhar, Cathryn Edwards, Khalid Elamin, Stephen Foley, Daniel Gavin, John Gordon, Leonie Grellier, Anton Gunasekera, Grace Hamill, Laura Hancock, Mina Hanna, Ailsa Hart, David Hobday, Peter Irving, Mark Jarvis, Babur Javaid, Matthew Johnson, Richard Keld, Nick Kennedy, Nikolaos Koukias, Alexandra Kent, Klaartje Bel Kok, Konrad Koss, Conor Lahiff, Jonathan Landy, Charlie Lees, Stephen Lewis, Andi Li, Jimmy Limdi, James Lindsay, Alan Lobo, Juliette Loehry, Chris Macdonald, George Macfaul, John Mansfield, Simon Mclaughlin, John Mclaughlin, Lin Mear, Gordon Moran, Ajay Muddu, Charles Murray, Sunny Nair, Chuka Nwokolo, Lynn O’donohoe, Arabis Oglesby, Chirag Oza, Simon Panter, Gareth Parkes, Timi Patani, Vinod Patel, Ruth Penn, Anne Phillips, Kath Phillis, Richard Pollock, Natalie Prescott, Cathryn Preston, Monira Rahman, John Ramage, Subramaniam Ramakrishnan, Colin Rees, Phil Roberts, Sunil Samuel, John Saunders, Shaji Sebastian, Christian Selinger, Dan Sharpstone, Richard Shenderey, Achuth Shenoy, Udi Shmueli, Alison Simmons, Salil Singh, Leena Sinha, Ganesh Sivagi, Katherine Smith, Paul Smith, Melissa Smith, Michael Sprakes, Helen Steed, Sreedhar Subramanian, Tariq Ahmad, Byron Theron, Jude Tilbury, Theresa Tindall, Mark Tremelling, Deven Vani, Ajay Verma, Emma Wesley, Alan Wiles, Joy Wilkins, Annette Woods, Rachel Simpkins, Catherine Thorbinson, Deepthy Francis, Rasha Shawky, Laetitia Pele, Jack Satsangi, Luke Jostins, Christopher Lamb, Christopher Mathew, Jeremy Sanderson, and James Lee
Collaborators: On behalf of the IBD BioResource Investigators
References
- 1. Satsangi J, Silverberg MS, Vermeire S, et al. The Montreal classification of inflammatory bowel disease: controversies, consensus, and implications. Gut 2006;55:749–53. 10.1136/gut.2005.082909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Huang H, Fang M, Jostins L, et al. Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature 2017;547:173–8. 10.1038/nature22969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Luo Y, de Lange KM, Jostins L, et al. Exploring the genetic architecture of inflammatory bowel disease by whole-genome sequencing identifies association at ADCY7. Nat Genet 2017;49:186–92. 10.1038/ng.3761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. de Lange KM, Moutsianas L, Lee JC, et al. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat Genet 2017;49:256–61. 10.1038/ng.3760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cleynen I, Boucher G, Jostins L, et al. Inherited determinants of Crohn’s disease and ulcerative colitis phenotypes: a genetic association study. Lancet 2016387:156–67. 10.1016/S0140-6736(15)00465-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu JZ, van Sommeren S, Huang H, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet 2015;47:979–86. 10.1038/ng.3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jostins L, Ripke S, Weersma RK, et al. Host-microbe interactions shape genetic risk for inflammatory bowel disease. Nature 2012;491:119–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Franke A, McGovern DP, Barrett JC, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet 2010;42:1118–25. 10.1038/ng.717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Anderson CA, Boucher G, Lees CW, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet 2011;43:246–52. 10.1038/ng.764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Barrett JC, Lee JC, Lees CW, et al. Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nat Genet 2009;41:1330–4. 10.1038/ng.483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Barrett JC, Hansoul S, Nicolae DL, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet 2008;40:955–62. 10.1038/ng.175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fisher SA, Tremelling M, Anderson CA, et al. Genetic determinants of ulcerative colitis include the ECM1 locus and five loci implicated in Crohn’s disease. Nat Genet 2008;40:710–2. 10.1038/ng.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Parkes M, Barrett JC, Prescott NJ, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn’s disease susceptibility. Nat Genet 2007;39:830–2. 10.1038/ng2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007;447:661–78. 10.1038/nature05911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sarin R, Wu X, Abraham C. Inflammatory disease protective R381Q IL23 receptor polymorphism results in decreased primary CD4+ and CD8+ human T-cell functional responses. Proc Natl Acad Sci U S A 2011;108:9560–5. 10.1073/pnas.1017854108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lassen KG, Kuballa P, Conway KL, et al. Atg16L1 T300A variant decreases selective autophagy resulting in altered cytokine signaling and decreased antibacterial defense. Proc Natl Acad Sci U S A 2014;111:7741–6. 10.1073/pnas.1407001111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Murthy A, Li Y, Peng I, et al. A Crohn’s disease variant in Atg16l1 enhances its degradation by caspase 3. Nature 2014;506:456–62. 10.1038/nature13044 [DOI] [PubMed] [Google Scholar]
- 18. Cader MZ, Boroviak K, Zhang Q, et al. C13orf31 (FAMIN) is a central regulator of immunometabolic function. Nat Immunol 2016;17:1046–56. 10.1038/ni.3532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mohanan V, Nakata T, Desch AN, et al. C1orf106 is a colitis risk gene that regulates stability of epithelial adherens junctions. Science 2018;359:1161–6. 10.1126/science.aan0814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Parkes M, Cortes A, van Heel DA, et al. Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat Rev Genet 2013;14:661–73. 10.1038/nrg3502 [DOI] [PubMed] [Google Scholar]
- 21. Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature 2009;461:747–53. 10.1038/nature08494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee JC, Biasci D, Roberts R, et al. Genome-wide association study identifies distinct genetic contributions to prognosis and susceptibility in Crohn’s disease. Nat Genet 2017;49:262–8. 10.1038/ng.3755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee JC, Espéli M, Anderson CA, et al. Human SNP links differential outcomes in inflammatory and infectious disease to a FOXO3-regulated pathway. Cell 2013;155:57–69. 10.1016/j.cell.2013.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dudbridge F, Pashayan N, Yang J. Predictive accuracy of combined genetic and environmental risk scores. Genet Epidemiol 2018;42:4–19. 10.1002/gepi.22092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang SK, Hong M, Baek J, et al. A common missense variant in NUDT15 confers susceptibility to thiopurine-induced leukopenia. Nat Genet 2014;46:1017–20. 10.1038/ng.3060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Walker GJ, Harrison JW, Heap GA, et al. Association of Genetic Variants in NUDT15 With Thiopurine-Induced Myelosuppression in Patients With Inflammatory Bowel Disease. JAMA 2019;321:773–85. 10.1001/jama.2019.0709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Heap GA, Weedon MN, Bewshea CM, et al. HLA-DQA1-HLA-DRB1 variants confer susceptibility to pancreatitis induced by thiopurine immunosuppressants. Nat Genet 2014;46:1131–4. 10.1038/ng.3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sazonovs A, Kennedy N, Moutsianas L, et al. HLA-DQA1*05 is associated with the development of antibodies to anti-TNF therapy. BioRxiv 2018. 10.1101/410035 [DOI] [Google Scholar]
- 29. Kaitin KI. Deconstructing the drug development process: the new face of innovation. Clin Pharmacol Ther 2010;87:356–61. 10.1038/clpt.2009.293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Argollo M, Fiorino G, Hindryckx P, et al. Novel therapeutic targets for inflammatory bowel disease. J Autoimmun 2017;85:103–16. 10.1016/j.jaut.2017.07.004 [DOI] [PubMed] [Google Scholar]
- 31. Moorcraft SY, Marriott C, Peckitt C, et al. Patients' willingness to participate in clinical trials and their views on aspects of cancer research: results of a prospective patient survey. Trials 2016;17:17 10.1186/s13063-015-1105-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Biasci D, Lee JC, Noor NM, et al. A blood-based prognostic biomarker in IBD. Gut 2019;68:1386–95. 10.1136/gutjnl-2019-318343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Parkes M, Noor NM, Dowling F, et al. PRedicting Outcomes For Crohn’s dIsease using a moLecular biomarkEr (PROFILE): protocol for a multicentre, randomised, biomarker-stratified trial. BMJ Open 2018;8:e026767 10.1136/bmjopen-2018-026767 [DOI] [PMC free article] [PubMed] [Google Scholar]