Abstract
Purpose
The present study aims to investigate the role of ELF3-AS1 in oral squamous cell carcinoma (OSCC).
Patients and methods
A total of 112 patients with OSCC were admitted in Guangdong Provincial Stomatological Hospital from March 2016 to March 2019. RT-qPCR, cells and transient transfections, cell proliferation rate measurements and Western blots were carried out to analyze the samples.
Results
In the present study, we showed that ELF3-AS1 and glucose transporter 1 (GLUT1) were both upregulated in OSCC tissues, and those two factors were positively correlated. In OSCC cells, ELF3-AS1 overexpression resulted in upregulation, while ELF3-AS1 siRNA silencing caused downregulated expression of GLUT1 and glucose uptake. ELF3-AS1 and GLUT1 overexpression resulted in increased rate of OSCC cells, while ELF3-AS1 and GLUT1 siRNA silencing resulted in decreased proliferation rate of OSCC cells. In addition, GLUT1 siRNA silencing attenuated the effects of ELF3-AS1 overexpression.
Conclusion
Therefore, ELF3-AS1 promotes the proliferation of OSCC cells by reprogramming glucose metabolism.
Keywords: oral squamous cell carcinoma, lncRNA ELF3-AS1, GLUT1, glucose uptake
Introduction
Malignances developed from head and neck account for about 5% of all malignancies, and more than 90% of all head and neck cancer cases are oral squamous cell carcinoma (OSCC).1 OSCC in most cases is diagnosed at advanced stages due to the following reasons: 1) OSCC lack obvious symptoms at early stages2 and 2) with cancer development, OSCC will show similar symptoms to other oral lesions, such as denture-related traumatic ulcer.3 OSCC at late stages lack effective therapeutic approaches, and prognosis is generally poor.4,5 In spite of efforts made on OSCC treatment, the overall 5-year survival rate of OSCC is still below 50%.5 Therefore, novel therapeutic approaches are always needed.
Although the tumorigenesis of OSCC is closely associated with many risk factors, such as smoking, alcohol abuse, areca consumption and HPV infection,6 genetic alterations are considered as the major players in new aspects of the tumorigenesis and progression of OSCC.7,8 In spite of the lack of protein-coding capacity, long (>200 nt) noncoding RNAs participate in diverse biological processes through its roles in gene expression regulation.9 Altered expression of lncRNAs is frequently observed during cancer development, and regulation of certain key lncRNAs may affect cancer development by directly regulating the expression of oncogenes or tumor suppressors.10,11 A considerable number of differentially expressed lncRNA has been identified in OSCC.12 Most of the differentially expressed lncRNAs are more tissue-specific than protein-coding genes and interact with cancer-related pathways to promote or suppress the progression of OSCC.13 In a recent study, Guo et al reported a novel lncRNA named ELF3-AS1 as an oncogene in bladder cancer.14 This lncRNA attracted our attention because preliminary microarray data also showed its upregulation in OSCC and its positive correlation with glucose transporter 1 (GLUT1), which is a key player in glucose metabolism.15 The present study was therefore carried out to investigate the role of ELF3-AS1 in OSCC.
Materials and methods
Research patients
A total of 112 patients with OSCC were admitted in Guangdong Provincial Stomatological Hospital from March 2016 to March 2019. This study selected 60 out of those 112 patients. Inclusion criteria are as follows: 1) newly diagnosed cases; 2) confirmed by histopathological examinations; 3) no therapies were initiated before this study. Exclusion criteria are as follows: 1) any therapies were performed within 100 days before this study; 2) any other obvious clinical disorders were observed. The 60 OSCC patients included 11, 21, 18 and 10 cases at American Joint Committee on Cancer stages I–IV,16 respectively. According to tumor location, there were 18 cases of upper location, 24 cases of middle location and 18 cases of lower location. According to tumor grade, there were 19, 24 and 17 cases at grades I, II and IV, respectively. All patients were informed with the details of experimental designed and this study was approved by the aforementioned hospital committee before the admission of patients.
All patients were diagnosed through histopathological biopsy. During biopsy, OSCC (cancer) tissues and noncancer tissues within 3 cm around tumors were obtained from each patient. All tissues were confirmed by at least 3 experienced pathologists. Weight of biopsies ranged from 0.06 to 0.11 g.
Cells and transient transfections
This study included two OSCC cell lines, SCC090 and SCC25 (ATCC, USA). Cell culture medium was Eagle’s Minimum Essential Medium (ATCC® 30–2003™, USA) containing 10% fetal bovine serum (FBS, Sigma-Aldrich, USA), 100 U/mL penicillin and 100 μg/mL streptomycin. Cells were cultivated under conditions of 5% CO2 and 37°C. Cell subculture was performed by 0.05% trypsin digestion followed by cell suspension preparation. ELF3-AS1 (Accession: NR_146472.1) and GLUT1 (Accession: KR711970.1) expression vector were constructed using pcDNA3 vector by Sangon (Shanghai, China). ELF3-AS1 (5ʹ- GCCUCUUUGUGGCUGAAUCUC-3ʹ) and GLUT1 siRNA (5ʹ-GCUCAGCAGCGUGGGCCACAG-3ʹ) as well as negative control siRNA (5ʹ-UCUCUGAUUGUAACUGGGAUA-3ʹ) were designed and synthesized by GenePharma (Shanghai, China). SCC090 and SCC25 were collected for transient transfections when cell confluence reached 70–90%. Lipofectamine 2000 (Invitrogen, CA, USA) was used to transfect 10 nM ELF3-AS1 or GLUT1 expression vector, or 10 nM empty pcDNA3 vector (negative control, NC), or 40 nM ELF3-AS1 or GLUT1 siRNA, or 40 nM negative control siRNA (negative control, NC) into 105 cells. Control group (C) of this study included cells with no transfections. Subsequent experiments were performed using cells harvested at 24 hrs after transfections.
RT-qPCR
Ribozol reagent (VWR Life Science, USA) was mixed with 105 cells or 0.03 g tissue (ground in liquid nitrogen before use) to perform total RNA extractions. Following DNase I digestion, AMV Reverse Transcriptase XL (Clontech, USA) and Luna® Universal One-Step RT-qPCR Kit (NEB, USA) were used to perform reverse transcriptions and prepare qPCR reaction mixtures. With 18S rRNA or GAPDH as endogenous control, expressions of ELF3-AS1 and GLUT1 mRNA were detected and expression levels were normalized based on g2−∆∆CT method. Primer sequences were as follows: 5ʹ-TGAAGTCATCACGAACCGC-3ʹ (forward) and 5ʹ-GGAGCCCCAAGTTAATGCG-3ʹ (reverse) for ELF3-AS1; 5ʹ-AGGTGATCGAGGAGTTCTA-3ʹ (forward) and 5ʹ-TCAAAGGACTTGCCCAGTTT-3ʹ (reverse) for GLUT1; 5ʹ-CCAGGGCTGCTTTTAACTCT-3ʹ (forward) and 5ʹ-GGACTC CACGACGTACTCA-3ʹ (reverse) for GAPDH; 5ʹ-CTACCACATCCAAGGAAGCA-3ʹ (forward) and 5ʹ-TTTTTCGTCACTACCTCCCCG-3ʹ (reverse) for human 18S rRNA. Three replicates were set for each experiment.
Glucose uptake assay
Before glucose uptake assay, Krebs–Ringe–-HEPES (KRH) buffer was prepared. KRH contains 25 mM HEPES (pH 7.4), 1.3 mM CaCl2, 120 mM NaCl, 1.3 mM KH2PO4, 1.2 mM MgSO4 and 5 mM KCl. SCC090 and SCC25 cells were collected at 24 hrs posttranscriptions, and 5×105cells were washed with KRH buffer. After washing, cells were mixed with fresh KRH buffer supplementing with 1 μCi of [3H]-2-deoxyglucose (Perkin Elmer Life Sciences). Cells were cultivated for 30 mins at 37°C, following by adding 3 volumes of ice-cold KRH to halt glucose uptake. Finally, a liquid scintillation spectrometry was used to measure radioactivity. Glucose uptake was represented by disintegrations per minute.
Cell proliferation rate measurement
SCC090 and SCC25 cells (3×104 cells collected at 24 hrs after transfections) were mixed with 1 mL Eagle’s Minimum Essential Medium (10% FBS) to prepare single-cell suspensions. Cells were cultivated in 96-well plates with 0.1 mL cell suspension per well, and cell culture conditions were 5% CO2 and 37°C. Cells were collected at 24, 48, 72 and 96 hrs after the beginning of cell culture. CCK-8 solution (10 μL, Sigma-Aldrich, USA) was added into each well at 2 hrs before cell collection. Finally, OD (450 nm) values were measured using SmartReader 96 Microplate Absorbance Reader for 96 Well Plate (Alkali Scientific, USA) to reflect cell proliferation.
Western blot
SCC090 and SCC25 cells (2×105 cells collected at 24 hrs after transfections) were mixed with 1.5 mL RIPA Buffer (Sangon, Shanghai, China) to extract total protein. Protein samples were denatured and electrophoresis was then performed using 10% SDS-PAGE gel. Following gel transfer (PVDF membrane) and blocking (2 hrs in 5% nonfat milk at room temperature), GLUT1 (1: 1200, ab15309, Abcam) or GAPDH (1: 1200, ab8245, Abcam) primary antibodies (rabbit polyclonal, overnight at 4°C), and IgG-HRP (1: 1000, MBS435036, MyBioSource) secondary antibody (goat anti-rabbit, 2 hrs at room temperature) were used to incubate with the membranes sequentially. ECL (Sigma-Aldrich, USA) was used to develop signals and signals were normalized using Image J v1.46.
Statistical analysis
All data presented in this paper were mean values, which were calculated using data from 3 biological replicates. Differences between two types of tissues were analyzed by performing paired test. Differences among different cell treatment groups were explored using ANOVA (one-way) and Tukey test. Correlations were analyzed by linear regression. p<0.05 was statistically significant.
Results
ELF3-AS1 and GLUT1 were upregulated in OSCC
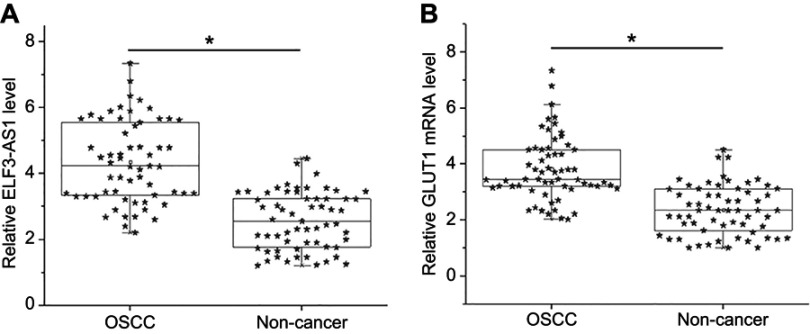
ELF3-AS1 and GLUT1 mRNA in two types of tissues were detected by performing RT-qPCR. Expression data were compared between two types of tissues by performing paired t-test. It was observed that expression levels of ELF3-AS1 (Figure 1A) and GLUT1 mRNA (Figure 1B) were both significantly higher in OSCC tissues comparing to noncancer tissues (p<0.05).
Figure 1.
ELF3-AS1 and GLUT1 were upregulated in OSCC. RT-qPCR analyzed by paired t test showed that expression levels of ELF3-AS1 (A) and GLUT1 mRNA (B) were both significantly higher in OSCC tissues comparing to noncancer tissues. (*p<0.05).
Abbreviations: OSCC, oral squamous cell carcinoma; ELF3-AS1, E74 like ETS transcription factor 3-antisense RNA 1; GLUT1, glucose transporter 1.
ELF3-AS1 and GLUT1 were positively correlated in OSCC
Correlation between ELF3-AS1 and GLUT1 was analyzed by performing linear regression. The results showed that, in OSCC tissues, ELF3-AS1 and GLUT1 were positively and significantly correlated (R square=0.7633, p<0.0001; Figure 2A). However, ELF3-AS1 and GLUT1 were not significantly correlated in noncancer tissues (R square=7.009e-005, p=0.9494; Figure 2B).
Figure 2.
ELF3-AS1 and GLUT1 were positively correlated in OSCC. Linear regression showed that ELF3-AS1 and GLUT1 were positively and significantly correlated in OSCC tissues (A), but not in noncancer tissues (B).
ELF3-AS1 positively regulated GLUT1 expression and glucose uptake in OSCC cells
CC090 and SCC25 cells were transfected with ELF3-AS1 expression vector and ELF3-AS1 siRNA. Compared to C and NC two controls, ELF3-AS1 expression was significantly altered at 24 hrs after the transfection of ELF3-AS1 expression vector or ELF3-AS1 siRNA (Figure 3A). Moreover, compared to two controls, ELF3-AS1 overexpression resulted in upregulated (Figure 3B), while ELF3-AS1 siRNA silencing caused downregulated (Figure 3C) expression of GLUT1 and glucose uptake (p<0.05).
Figure 3.
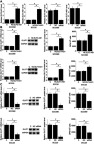
ELF3-AS1 positively regulated GLUT1 expression and glucose uptake in OSCC cells. After transfection of ELF3-AS1 expression vector and ELF3-AS1 siRNA, ELF3-AS1 expression was significantly altered at 24 h comparing to C and NC groups (A). Moreover, ELF3-AS1 overexpression resulted in upregulated (B), while ELF3-AS1 siRNA silencing caused downregulated (C) expression of GLUT1 and glucose uptake (*p<0.05).
Abbreviations: ELF3-AS1, E74 like ETS transcription factor 3-antisense RNA 1; GLUT1, glucose transporter 1.
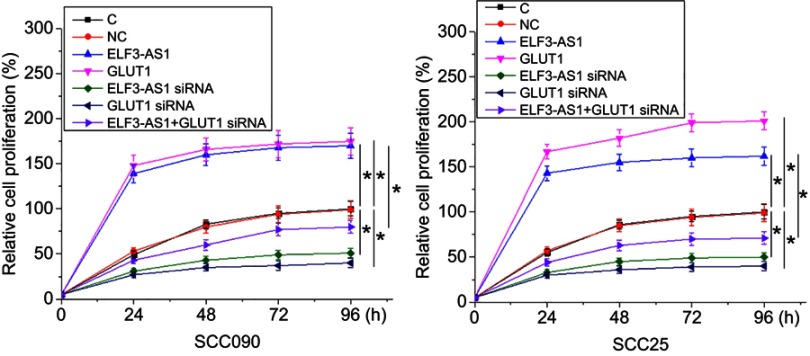
ELF3-AS1 positively regulated OSCC cell proliferation through GLUT1
GLUT1 expression vector and ELF3-AS1 siRNA were also transfected into SCC090 and SCC25 cells, and transfections were confirmed by RT-qPCR (data not shown). Cell proliferation data were compared among different cell transfection groups by performing ANOVA (one-way) and Tukey test. Compared to two controls (C and NC), ELF3-AS1 and GLUT1 overexpression resulted in a significantly increased proliferation rate of OSCC cells (p<0.05). In contrast, the proliferation rate of OSCC cells was significantly decreased after ELF3-AS1 and GLUT1 siRNA silencing (p<0.05). In addition, GLUT1 siRNA silencing attenuated the effects of ELF3-AS1 overexpression (Figure 4, p<0.05).
Figure 4.
ELF3-AS1 positively regulated OSCC cell proliferation through GLUT1. Cell proliferation data analyzed by ANOVA (one-way) and Tukey test showed that ELF3-AS1 and GLUT1 overexpression resulted in increased proliferation rate of OSCC cells. However, ELF3-AS1 and GLUT1 siRNA silencing resulted in decreased proliferation rate of OSCC cells. In addition, GLUT1 siRNA silencing attenuated the effects of ELF3-AS1 overexpression. *p<0.05.
Abbreviations: ELF3-AS1, E74 like ETS transcription factor 3-antisense RNA 1; GLUT1, glucose transporter 1.
Discussion
The expression pattern and functionality of ELF3-AS1 have been investigated in the present study. We observed that ELF3-AS1 was upregulated in OSCC and may promote the proliferation of OSCC cells by upregulating GLUT1, which is a key player in glucose transport.15
Due to the rapid growth and proliferation nature of cancer cells, altered glucose metabolism in cancer is necessary to provide sufficient energy to support the activities of cancer cells.17 GLUT1 is a key player in glucose metabolism by mediating the transport of glucose across plasma membranes and to be metabolized in cells.18 Therefore, GLUT1 is usually overexpressed in cancer cells compared to normal cells.19 Consistently, our study observed the upregulated GLUT1 expression in OSCC. In addition, GLUT1 positively regulated the proliferation of OSCC cells. Those data further confirmed the oncogenic role of GLUT1 in OSCC.
Recent studies have identified a big number of differentially expressed lncRNAs in OSCC.12,13 These lncRNAs may interact with one or multiple signaling pathways to participate in the pathogenesis of OSCC.12,13 Previous studies have shown that lncRNAs are also critical players in glucose metabolism in cancer cells. LncRNAs regulate multiple downstream targets involved in glucose metabolism, such as glucose transporters (GLUT1 and GLUT4), enzymes (pyruvate carboxylase, G6P and so on), oncogenes (c-Myc) and so on.20 In liver cancer, lncRNA NBR2 regulates the expression of GLUT1 to modulate the sensitivity of cancer cell to phenformin.21 In another study, GLUT1 was proven to be involved in the HOTAIR-induced glycolysis in many types of cancer.22 In the present study, we showed that ELF3-AS1 positively regulated the expression of GLUT1 mRNA and protein, as well as glucose uptake in OSCC cells. LncRNAs can regulate the expression of genes at posttranscriptional or translational levels.9 Therefore, ELF3-AS1 may affect the stability of GLUT1 to affect glucose uptake in OSCC cells. However, the molecular mechanism is still hardly known. More experimental studies are still needed.
Conclusion
In conclusion, ELF3-AS1 was upregulated in OSCC, and ELF3-AS1 may positively regulate GLUT1 to promote the proliferation of OSCC cells.
Acknowledgment
We received financial support from Guangdong Province Science and Technology Innovation Strategy Special Fund Project (2018KJYZ014).
Ethics approval and informed consent
The research has been carried out in accordance with the World Medical Association Declaration of Helsinki, ethical approval was obtained from the Ethics Committee of Guangdong Provincial Stomatological Hospital and that all subjects provided written informed consent in this work.
Author contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332 [DOI] [PubMed] [Google Scholar]
- 2.Singh T, Schenberg M. Delayed diagnosis of oral squamous cell carcinoma following dental treatment. Ann R Coll Surg Engl. 2013;95(5):369–373. doi: 10.1308/003588413X13629960045599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Valente VB, Takamiya AS, Ferreira LL, et al. Oral squamous cell carcinoma misdiagnosed as a denture-related traumatic ulcer: a clinical report. J Prosthet Dent. 2016;115(3):259–262. doi: 10.1016/j.prosdent.2015.08.024 [DOI] [PubMed] [Google Scholar]
- 4.Wang B, Zhang S, Yue K, Wang XD. The recurrence and survival of oral squamous cell carcinoma: a report of 275 cases. Chin J Cancer. 2013;32(11):614–618. doi: 10.5732/cjc.012.10219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Massano J, Regateiro FS, Januario G, Ferreira A. Oral squamous cell carcinoma: review of prognostic and predictive factors. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2006;102(1):67–76. doi: 10.1016/j.tripleo.2005.07.038 [DOI] [PubMed] [Google Scholar]
- 6.Santos HB, dos Santos TK, Paz AR, et al. Clinical findings and risk factors to oral squamous cell carcinoma in young patients: a 12-year retrospective analysis. Med Oral Patol Oral Cir Bucal. 2016;21(2):e151–e156. doi: 10.4317/medoral.20770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lau HK, Wu ER, Chen MK, et al. Effect of genetic variation in microRNA binding site in WNT1-inducible signaling pathway protein 1 gene on oral squamous cell carcinoma susceptibility. PLoS One. 2017;12(4):e0176246. doi: 10.1371/journal.pone.0176246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Z, Li G, Wei S, et al. Genetic variants in selected pre-microRNA genes and the risk of squamous cell carcinoma of the head and neck. Cancer. 2010;116(20):4753–4760. doi: 10.1002/cncr.25323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engreitz JM, Haines JE, Perez EM, et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016;539(7629):452–455. doi: 10.1038/nature20149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang G, Lu X, Yuan L. LncRNA: a link between RNA and cancer. Biochim Biophys Acta. 2014;1839(11):1097–1109. doi: 10.1016/j.bbagrm.2014.08.012 [DOI] [PubMed] [Google Scholar]
- 11.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29(4):452–463. doi: 10.1016/j.ccell.2016.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pentenero M, Bowers LM, Jayasinghe R, et al. World workshop on oral medicine VII: clinical evidence of differential expression of lncRNAs in oral squamous cell carcinoma: a scoping review. Oral Dis. 2019;25(Suppl 1):88–101. doi: 10.1111/odi.13076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pentenero M, Bowers L, Jayasinghe R, et al. World workshop on oral medicine VII: functional pathways involving differentially expressed lncRNAs in oral squamous cell carcinoma. Oral Dis. 2019;25(Suppl 1):79–87. doi: 10.1111/odi.13051 [DOI] [PubMed] [Google Scholar]
- 14.Guo Y, Chen D, Su X, Chen J, Li Y. The lncRNA ELF3-AS1 promotes bladder cancer progression by interaction with kruppel-like factor 8. Biochem Biophys Res Commun. 2019;508(3):762–768. doi: 10.1016/j.bbrc.2018.11.183 [DOI] [PubMed] [Google Scholar]
- 15.Wu N, Zheng B, Shaywitz A, et al. AMPK-dependent degradation of TXNIP upon energy stress leads to enhanced glucose uptake via GLUT1. Mol Cell. 2013;49(6):1167–1175. doi: 10.1016/j.molcel.2013.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mascitti M, Rubini C, De Michele F, et al. American joint committee on cancer staging system 7th edition versus 8th edition: any improvement for patients with squamous cell carcinoma of the tongue? Oral Surg Oral Med Oral Pathol Oral Radiol. 2018;126(5):415–423. doi: 10.1016/j.oooo.2018.07.052 [DOI] [PubMed] [Google Scholar]
- 17.Annibaldi A, Widmann C. Glucose metabolism in cancer cells. Curr Opin Clin Nutr Metab Care. 2010;13(4):466–470. doi: 10.1097/MCO.0b013e32833a5577 [DOI] [PubMed] [Google Scholar]
- 18.Olson AL, Pessin JE. Structure, function, and regulation of the mammalian facilitative glucose transporter gene family. Annu Rev Nutr. 1996;16:235–256. doi: 10.1146/annurev.nu.16.070196.001315 [DOI] [PubMed] [Google Scholar]
- 19.Szablewski L. Expression of glucose transporters in cancers. Biochim Biophys Acta. 2013;1835(2):164–169. doi: 10.1016/j.bbcan.2012.12.004 [DOI] [PubMed] [Google Scholar]
- 20.Fan C, Tang Y, Wang J, et al. Role of long non-coding RNAs in glucose metabolism in cancer. Mol Cancer. 2017;16(1):130. doi: 10.1186/s12943-017-0699-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu X, Gan B. lncRNA NBR2 modulates cancer cell sensitivity to phenformin through GLUT1. Cell Cycle. 2016;15(24):3471–3481. doi: 10.1080/15384101.2016.1249545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei S, Fan Q, Yang L, et al. Promotion of glycolysis by HOTAIR through GLUT1 upregulation via mTOR signaling. Oncol Rep. 2017;38(3):1902–1908. doi: 10.3892/or.2017.5840 [DOI] [PubMed] [Google Scholar]