Abstract
Purpose
MicroRNA-195 is dysregulated in different kinds of cancers and plays a pivotal role in tumorigenesis. It may function as a prognostic biomarker for cancers. However, the results from articles were not consistent. This study was designed to validate the prognostic value of microRNA-195 in human tumors.
Methods
We conducted a detailed search on PubMed until December 31, 2018. The quality of these publications was assessed on the basis of a list of key reviews presented by PRISMA statement. The pooled hazard ratios (HR) and pooled odds ratios (OR) of each 95% confidence interval (95% CI) were calculated to assess the effect.
Results
This meta-analysis included 12 studies involving 940 cancer patients to assess the prognostic value of miR-195 in different solid tumors. The results showed that patients with high expression of miR-195 had favorable tumor-node-metastasis (late vs early: pooled OR =0.16, 95% CI: 0.11–0.22, P<0.001), lymph node metastasis (pooled OR =0.25, 95% CI: 0.18–0.35, P<0.001) and distant metastasis (pooled OR =0.26, 95% CI: 0.13–0.52, P<0.001). At the same time, high levels of miR-195 expression were closely correlated with better overall survival (pooled HR =0.46, 95% CI: 0.36–0.58, P<0.001).
Conclusion
Elevated microRNA-195 may serve as a potential biomarker to predict a favorable prognosis for various cancer types in China.
Keywords: biomarker, prognosis, microRNA-195, expression, meta-analysis
Introduction
According to estimates by the International Agency for Research on Cancer (IARC), with the rapid growth and aging of the global population due to changes in fertility and life expectancy, by 2018, approximately 18.1 million new cancer cases occurred worldwide and 9.6 million patients die of cancer although cancer mortality has declined for several consecutive years.1,2 If cancer could be diagnosed earlier and appropriate treatment for cancer can be given in time, the patients may have more opportunities for recovery. However, the diagnosis and treatment of cancer is limited. Thus, it is imperative to find a new biomarker to prognosis.
MicroRNA (miRNA), a small non-coding RNA molecule (usually 21–23 nucleotides in length), is an evolutionarily conserved non-coding RNA molecule that passes through the 3ʹ-untranslation (UTR) region of the target messenger RNA (mRNA).3 The complementary sequence of 3ʹ-UTR is involved in post-transcriptional gene regulation. Emerging studies suggest that miRNAs can regulate gene expression after transcription by degrading target mRNA or inhibiting protein translation. Through high-throughput profiling, miRNA dysregulation has been widely observed at different stages of cancer. Up-regulation of specific miRNA leads to inhibition of tumor suppressor gene expression. On the contrary, down-regulation of specific miRNA leads to increased oncogene expression. Both of these conditions have a malignant effect on cell proliferation, differentiation and apoptosis, and contribute to tumor growth and progression.4 Therefore, miRNAs are considered to be diagnostic, prognostic and therapeutic biomarkers for almost all cancers.5
MicroRNA-195 (miRNA-195 or miR-195, Gene ID: 406971) gene, originating in intron 7, is located on human chromosome 17p13.1, which is a highly conserved miRNA in a variety of diseases such as cancer, heart failure and schizophrenia.6,7 We used miRBase (http://mirbase.org/) to predict miR-195 stem ring structure, which indicated that miR-195 hairpins produced “lead” miR-195-5p and sister “passenger” miR-195-3p. Interestingly, the predominantly expressed form in recent studies is miR-195-5p.8 MiR-195 has been reported to be involved in the progression of cancer by regulating key genes and signaling pathways, thus significantly affecting the carcinogenicity of various cancers,9 which includes non-small cell lung cancer,10 prostate cancer,11 gastric cancer,12 colorectal cancer,13 hepatocellular cancer,14 esophageal squamous cell carcinoma15 and cervical cancer.16 Meanwhile, CDK6, Bcl-2 and WEE1 are target genes of miR-195, which are involved in miR-195-mediated cell cycle and apoptosis effects.17–19 So far, numerous studies have shown that the clinicopathological features and the overall survival of patients in various cancers are closely related to the dysregulation of miR-195. However, the results in several studies were inconsistent and conflicting. To more accurately assess the relationship between the expression level of miR-195 and the clinical prognosis of cancer patients, we conducted a meta-analysis of published studies.
Methods
Search strategy
Published studies were searched in PubMed, updated before December 31, 2018. We used the following keywords: “tumor” or “tumour” or “cancer” or “carcinoma” or “malignant tumor” or “neoplasm” and “miRNA-195” or “miR-195” or “microRNA-195”. Articles published in full-text in English were required.
Data extraction
In order to ensure the accuracy of data extraction, the two authors (CY and WN) extracted the valuable data that were screened out according to the inclusion criteria and exclusion criteria, respectively. The following information was collected and compiled from the inclusive studies: name of first author; year of publication; country; miR-195 detection method; follow-up time; cut-off value (method of distinguishing high and low expression groups); sample size (overall number of patients); quantity of lymph node metastasis (LNM) or distant metastasis (DM) patients; the total number of patients with different tumor node metastasis (TNM) stage in high and low miR-195 expression level groups. When the author provided univariate and multivariate analysis, we preferred the second method. Any controversial information was settled by discussion among the authors.
Quality assessment
The quality of the relevant studies was evaluated using the Newcastle-Ottawa Scale (NOS).20 The NOS score ranges from 0 to 9. The score is greater than or equal to 6 for high quality.
Criteria of inclusion and exclusion
Criteria conforming to the inclusion are as follows: (1) analysis of the prognostic role of miR-195 in cancer patients; (2) grouping patients according to the high and low expression levels of miR-195; (3) studying the relationship between miR-195 expression and clinicopathological parameters; (4) providing HR for available data access to 95% CI.
Exclusion criteria: (1) reviews, case reports, letters, conference abstracts or studies on animal experiments; (2) data duplication or overlap studies; (3) sample size of study less than 30 items.
Statistical methods
Meta-analysis was carried out conforming to the PRISMA statement and used Stata (version 12.0, Stata Corp, College Station, USA) software.21,22 The relationship between the expression level of miR-195 and clinical outcomes was assessed by odds ratio (OR), hazard ratio (HR) and each corresponding 95% CI. Cochran’s Q test and Higgins I-squared statistic method were exclusively used for assessing the heterogeneity of the included trials.22 If the Q test does not show significant heterogeneity in qualitative studies, we prefer to use a fixed effect model, ie, I2<50% or PQ>0.05. However, when I2>50% or PQ<0.05 is considered to be significant heterogeneity, a random effects model will be performed. Sensitivity analysis was applied to test the origin of heterogeneity and the stability of the results. If the HR with 95% CI of the OS was not directly written in the text, we obtained the HR and 95% CI from Kaplan-Meier curve of the OS by using the Engauge Digitizer 4.1 software and excel of Tierney et al.23,24 Publication bias was evaluated through funnel plot by Egger’s and Begg’s test and P<0.05 means obvious statistically significance.25
Results
Involved studies’ features
The article retrieval strategy was illustrated in Figure 1. A total of 271 articles were searched and the data were obtained from the PubMed database. Among them, because the data we need are incomplete, we excluded 250 publications. After we reviewed the title and abstract, we selected 21 publications for further full-text screening. From the left 14 publications, one was uncorrected proofed version and one was studied in Pakistan. For the representativeness of the population we included, 12 studies from China with total 940 patients were included to perform meta-analysis. All included literature described the relationship between miR-195 and tumor prognosis. Of them, Chen et al pointed out miR-195-5p was the target molecule while other 12 studies did not specifically analyze the effect of sister chain 5p or 3p on tumor prognosis. The cancer types included renal cell carcinoma,26 esophageal squamous cell carcinoma,15 colorectal cancer,13 clear cell renal cell carcinoma,27 laryngeal squamous cell carcinoma,28 hepatocellular carcinoma,14 oral squamous cell carcinoma,29 cervical cancer,16 prostate cancer,11 osteosarcoma,36 non-small cell lung cancer37 and pancreatic cancer.30 All included studies used qRT-PCR to detect the expression level of miR-195. There were two cut-off values to distinguish high and low expression groups (mean level and median value). All characteristics and differences of each study are reflected in Table 1.
Figure 1.
The flow chart of meta-analysis.
Table 1.
Main characteristics of the studies included in the meta-analysis
| Study | Year | Country | Cancer type | MicroRNA-195 exprssion | Detection method | HR statistic | HR (95% CI) for OS high/low | Survival analysis | NOS | Cut-off value | Follow-up (months) | Sample size | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| High | Low | ||||||||||||||||||
| Total | TNM for advaced stage | LNM | DM | Total | TNM for advaced stage | LNM | DM | ||||||||||||
| Wang et al13 | 2012 | China | CRC | 24 | 3 | 4 | - | 61 | 33 | 34 | - | qRT-PCR | Date in paper | 0.41 (0.19–0.89) | Multivariate | 8 | Median value | 60 | 85 |
| Sun et al15 | 2014 | China | ESCC | 46 | 5 | 10 | - | 52 | 35 | 30 | - | qRT-PCR | Date in paper | 0.17 (0.08–0.79) | Multivariate | 9 | Median value | 60 | 98 |
| Han et al36 | 2015 | China | OS | 50 | 29 | - | 18 | 57 | 46 | - | 34 | qRT-PCR | Survival curve | 0.44 (0.24–0.83) | Univariate | 9 | Mean level | 80 | 107 |
| Su et al37 | 2016 | China | NSCLC | 50 | 10 | 24 | - | 50 | 33 | 39 | - | qRT-PCR | Survival curve | 0.51 (0.28–0.93) | Univariate | 9 | Median value | 60 | 100 |
| Sun et al27 | 2016 | China | ccRCC | 13 | 3 | 4 | - | 17 | 3 | 12 | - | qRT-PCR | - | - | - | 6 | Mean level | - | 30 |
| Zhou et al16 | 2016 | China | CC | 25 | - | 10 | - | 25 | - | 19 | - | qRT-PCR | - | - | - | 6 | Mean level | - | 50 |
| Chen et al26 | 2017 | China | RCC | 33 | 8 | 4 | - | 34 | 20 | 14 | - | qRT-PCR | Survival curve | 0.45 (0.14–1.45) | Univariate | 8 | Mean level | 60 | 67 |
| Shuang et al28 | 2017 | China | LSCC | 61 | 39 | 15 | - | 61 | 60 | 26 | - | qRT-PCR | Date in paper | 0.36 (0.13–0.96) | Multivariate | 9 | Mean level | 120 | 122 |
| Yu et al14 | 2017 | China | HCC | 42 | 6 | - | - | 88 | 45 | - | - | qRT-PCR | Survival curve | 0.51 (0.31–0.84) | Univariate | 9 | Mean level | 60 | 130 |
| Wang et al29 | 2017 | China | OSCC | 19 | 1 | - | - | 21 | 6 | - | - | qRT-PCR | - | - | - | 6 | Mean level | - | 40 |
| Zhou et al30 | 2017 | China | PC | 20 | 5 | 4 | - | 22 | 14 | 12 | - | qRT-PCR | Survival curve | 0.56 (0.26–1.21) | Univariate | 8 | Median value | 50 | 42 |
| Cai et al11 | 2018 | China | PCa | 42 | - | - | 2 | 27 | - | - | 11 | qRT-PCR | Date in paper | 0.51 (0.27–0.95) | Multivariate | 6 | Mean level | 200 | 69 |
Notes: The dashes represent no data; TNM for advanced stage: III+IV or III+IV or IIb+III+IV (PC, OS); DM: distant metastasis (OS, PCa).
Abbreviations: TNM, tumor node metastasis; DM, distant metastasis; LNM, lymph node metastasis; NOS, Newcastle-Ottawa Scale; CRC, colorectal cancer; ESCC, esophageal squamous cell carcinoma; OS, osteosarcoma; NSCLC, non-small cell lung cancer; ccRCC, clear cell renal cell carcinoma; CC, cervical cancer; RCC, renal cell cancer; LSCC, laryngeal squamous cell carcinoma; HCC, hepatocellular carcinoma; OSCC, oral squamous cell carcinoma; PC, pancreatic cancer; PCa, prostate cancer.
Study characteristics of association between miR-195 expression and TNM stage (advanced stage vs early stage: III/IV vs I/II or IIb/III/IV vs IIa/I/II)
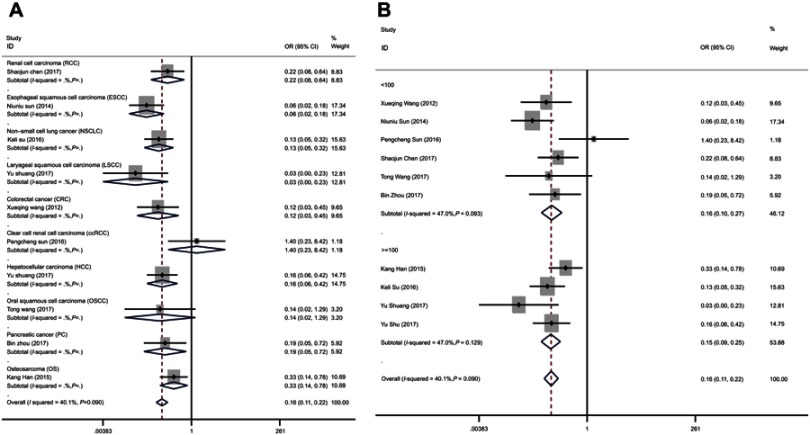
Ten researches containing 770 patients were enrolled to assess the prognostic value of miR-195 expression in cancer patients (Figure 2A). Because no heterogeneity was found in 10 reports (I2=40.1%, PQ =0.09), the pooled OR and its 95% CI were analyzed using a fixed effects model. We found that high expression level of miR-195 suggested a good phase of TNM (pooled OR =0.20, 95% CI: 0.11–0.36, P<0.001). Figure 2A shows the results we analyzed in the Stata software. We found that the increased miR-195 expression was related with earlier TNM stage in esophageal squamous cell carcinoma (OR =0.06, 95% CI: 0.02–0.180, P<0.001), non-small cell lung cancer (OR =0.129, 95% CI: 0.05–0.32, P<0.001), renal cell carcinoma (OR =0.22, 95% CI: 0.08–0.64, P<0.001), hepatocellular carcinoma (OR =0.16, 95% CI: 0.06–0.42, P<0.001), pancreatic cancer (OR =0.19, 95% CI: 0.05–0.72, P=0.015), laryngeal squamous cell carcinoma (OR =0.03, 95% CI: 0.004–0.23, P=0.001), colorectal cancer (OR =0.121, 95% CI: 0.03–0.45, P=0.002), osteosarcoma (OR =0.33, 95% CI: 0.0.14–0.75, P=0.012), but not related with clear cell renal cell carcinoma (OR =1.40, 95% CI: 0.23–8.42, P=0.71) and oral squamous cell carcinoma (OR =0.14, 95% CI: 0.02–1.29, P=0.082). In addition, we performed the subgroup analysis for sample size (Figure 2B). The sample size of the enrolled studies was from 30 to 130. The sample size of 100 was selected as the boundary. We found a significant association between the TNM stage and the miR-195 expression level in the subgroup of sample size ≥100 (pooled OR =0.15, 95% CI: 0.09–0.25, P<0.001), with no heterogeneity (I2=47.0%, PQ=0.129). In the subgroup of sample size <100, the heterogeneity is not obvious (I2=47.0%, PQ =0.093) and the result is also significant (pooled OR =0.16, 95% CI: 0.10–0.0.27, P<0.001). Therefore, the sample size does not affect the result.
Figure 2.
Forest plots of the subgroup analyses evaluating ORs of miR-195 for TNM by the factors of (A) cancer type; (B) sample size.
Abbreviation: OR, overall survival.
Association between miR-195 expression and lymph node metastasis (LNM)
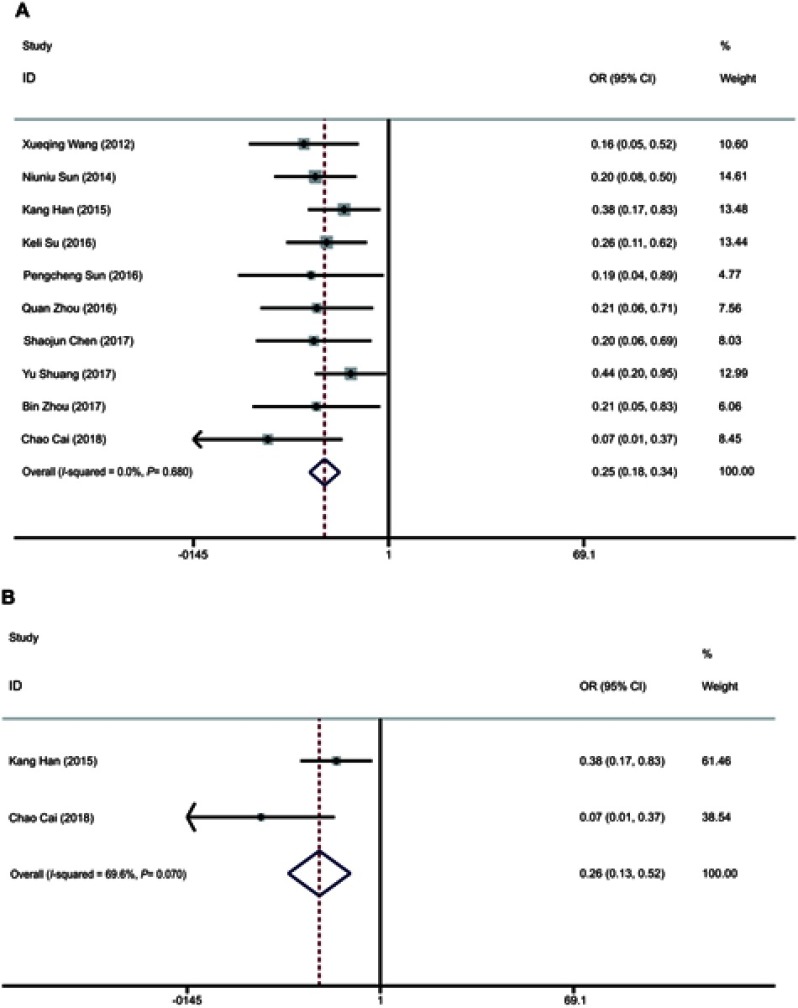
A total of 594 patients with LNM were enrolled in 8 studies (Figure 3A). We compared the high and low expression of miR-195 by using a fixed effect model (I2=0.0%, PQ =0.680). The combined result showed that patients with low expression of miR-195 had a tendency to metastasize to lymph nodes (pooled OR =0.25, 95% CI: 0.18–0.35, P<0.001).
Figure 3.
Forest plots evaluating ORs of miR-195 for (A) LNM and (B) DM.
Abbreviations: OR, overall survial; LNM, lymph node metastasis; DM, distant metastasis.
Association between miR-195 expression and distant metastasis (DM)
Two eligible studies with 176 patients were used to detect the relationship between miR-195 expression levels and DM (Figure 3B). The heterogeneity was found in the studies (I2=69.6%, PQ=0.07), with the pooled OR =0.26 (95% CI: 0.13–0.52, P<0.001 in a random-effect model). Therefore, the results indicated that patients with high levels of miR-195 expression in the tumor have a lower risk of DM. However, more samples were needed to be included to further verify this result.
Association between miR-195 expression and overall survival (OS)
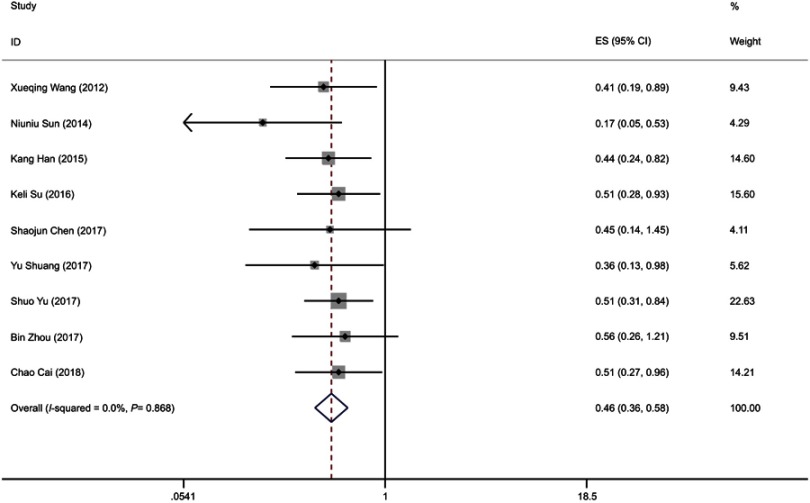
A total of 9 studies of 853 cancer patients were enrolled to detect the correlation between overall survival (OS) and miR-195 in cancers (Figure 4). Since the OS group did not detect heterogeneity (I2=0.0%, PQ=0.868), this analysis used a fixed model. There is a strong correlation between high expression levels of miR-195 and better OS in cancer patients (pooled HR =0.46, 95% CI: 0.36–0.58, P<0.001) (Figure 4). However, the relationship is not significant in pancreatic cancer (HR =0.560, 95% CI: 0.26–1.21, P=0.139) and renal cell carcinoma (HR =0.450, 95% CI: 0.14–1.45, P=0.181).
Figure 4.
Forest plot of pooled HRs of elevated miR-195 expression for OS.
Abbreviations: HR, hazard ratio; OS, overall survival.
Sensitivity analysis
To assess the stability and reliability of this meta-analysis, we performed sensitivity analyses on the TNM, LNM and OS groups, respectively, and omit each study sequentially. In the TNM group, the results showed that there was no significant impact on individual research which affected the pooled OR (Figure 5A). In addition, sensitivity analysis in LNM (Figure 5B) or OS (Figure 5C) group suggested same results after we excluded each study sequentially.
Figure 5.

Sensitivity analysis. (A) TNM. (B) LNM. (C) OS.
Abbreviations: TNM, tumor-node-metastasis; LNM, lymph node metastasis; OS, overall survival.
Publication bias
Figure 6 shows the funnel plot of the article reported in the current meta-analysis. All studies enrolled in the TNM (Figure 6A), LNM (Figure 6B) and OS (Figure 6C) group with an even distribution around the horizon line showed no obvious publication bias. Egger’s test and Begg’s test were carried out on the publication bias to test the symmetry of the funnel plot. Finally, significance of publication bias in LNM (P=0.008 for Egger’s test, P=0.049 for Begg’s test) and marginal significance of publication bias in OS (P=0.051 for Egger’s test, P=0.076 for Begg’s test) was obtained. There was no significant publication bias in TNM (P=0.451 for Egger’s test, P=0.929 for Begg’s test).
Figure 6.

Funnel plot analysis of potential publication bias (Begg’s test). (A) TNM. (B) LNM. (C) OS.
Abbreviations: TNM, tumor-node-metastasis; LNM, lymph node metastasis; OS, overall survival.
Discussion
Cancer-associated miRNAs act by binding to the 3ʹ-untranslation region (UTR) of specific mRNAs and play a significant and multifaceted role in the development and progression of tumors.31 A large number of miRNAs have been found to be associated with clinical features of a variety of cancers. For example, high expression of miR-21 has been confirmed to correlate with worse overall survival significantly in carcinomas of the digestive system.32 Increased expression of miR-31 is reported to be associated with poor overall survival and cancer-specific survival in patients with general cancers, which suggests that miR-31 may be a useful clinical prognostic biomarker.33 Therefore, it is of great significance to study the expression levels of miRNAs in cancers.
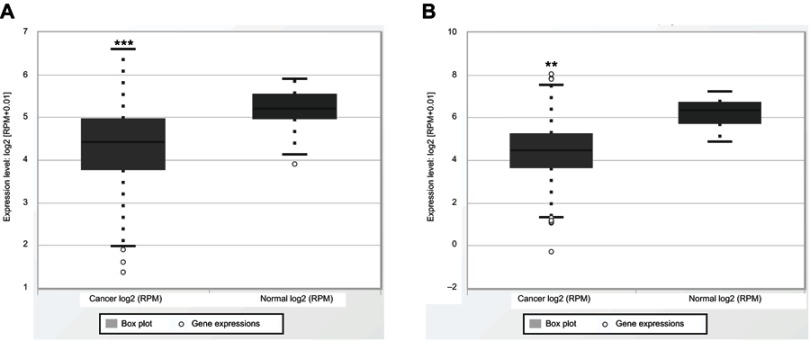
It is reported that miR-195 in the microenvironment plays a significant and versatile role in the occurrence and development of malignant tumors by affecting the survival and proliferation of tumor cells, promoting the spread of tumors and reducing the response to anticancer drugs.7 A certain amount of studies have investigated the prognostic value of miR-195 expression level in various cancer types but shows inconsistent results. For instance, Yang et al found that up-regulation of miR-195 can promote hepatoma cell apoptosis and decrease CDK2 expression by upregulating the expression of large tumor suppressor kinase 2 (LATS2) and p53.34 Whereas, Wang et al reported that miR-195 contributes to the promotion of HCC lung metastasis through negative regulation of FGF2 and VEGFA.35 Besides, we validate with data available on the miR-195 expression level and cancer data sets from The Cancer Genome Atlas (TCGA) database by boxplots. Osteosarcoma, cervical cancer, laryngeal squamous cell carcinoma and oral squamous cell carcinoma could not be acquired in the database. We found the expression of miR-195 in cancer tissues was lower than that in adjacent tissues in most of the tumors we included except for colorectal cancer and clear cell renal cell carcinoma. We picked the most representative types of esophageal squamous cell carcinoma and hepatocellular carcinoma in Figure 7A and B and other included tumors were shown in the supplementary materials (Figure S1A–F). The exact mechanism between miR-195 and different kinds of cancer patients remains unclear. Therefore, we conducted the meta-analysis from published studies to obtain a more accurate estimate of the prognostic value of miR-195 in cancer patients.
Figure 7.
Validation with data available on the expression level of miR-195 and cancer data sets from TCGA database. (A) Esophageal squamous cell carcinoma. (B) Hepatocellular carcinoma. **P<0.01; ***P<0.001.
Abbreviation: TCGA, The Cancer Genome Atlas.
In this study, we analyzed the relationship between a specific microRNA and cancer patient based on the results of 940 cancer patients in 12 independent studies. For all we know, this is the first study to perform a comprehensive meta-analysis to detect the effects of the dysregulation of miR-195 and prognosis associated with cancer patients, which indicates that elevated miR-195 significantly predicted longer overall survival time of cancer patients. Detailed analysis found that the higher the expression level of miR-195, the lower the risk of LNM and DM in tumor patients, and the more favorable the TNM stage in cancer patients. Although included studies applied different cut-off values to distinguish the high and low expression of miR-195, it does not affect the consistency of our final results. As for publication bias in OS and LNM group, we considered that the population in our enrolled studies were all Chinese, which may result in marginal significance of publication bias in HR for OS and significance of publication bias in OR for LNM. Besides, the reasons for potential publication bias may be ascribed to publications that tended to show positive results as well as relatively strict inclusion criteria that we made. But these interference factors did not affect the overall conclusion of the study.
Nevertheless, few limits shall be claimed in this analysis. Firstly, our analysis only absorbed 12 studies with a total of 940 patients and the included studies were mainly based on Chinese patients so that we ignored the subgroup analysis according to age, sex, depth of tumor invasion and tumor size, which are factors that cancer progression is associated with. Secondly, the HRs of 4 studies were extracted from survival curves, which were less accurate. Thirdly, the definition of each cut-off value is inconsistent. Therefore, it is imperative to establish a standard, uniform cut-off value. In addition, in order to achieve more meaningful results, a survey involving uniform disconnection is required. Fourthly, since only research of Chen et al26 in the included articles specifically described miR-195-5p while other 11 articles did not specify which sister chain make sense, we can only discuss the overall effect of miR-195. Finally, we only enrolled literature published in English, which limited the population and ethnicity involved.
Conclusion
To conclude, the results indicated that elevated miR-195 may be a good prognostic biomarker for the clinical outcome of human cancer in Chinese. However, the exact functional role of miR-195 remains to be further studied through larger sample studies, which may validate and enhance the prognostic value of miR-195 in future and global cancer patients.
Supplementary material
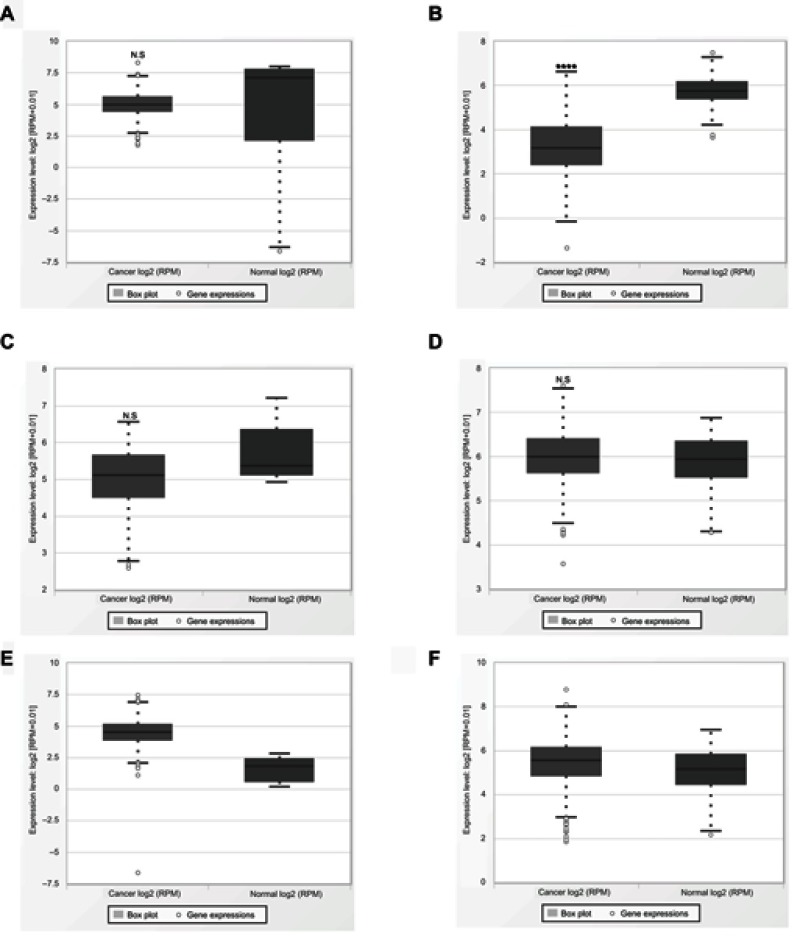
Validation with data available on the expression level of miR-195 and cancer data sets from TCGA database. (A) Non-small cell lung cancer. (B) Renal cell cancer. (C) Pancreatic cancer. (D) Prostate cancer. (E) Colorectal cancer. (F) Clear cell renal cell carcinoma.
Abbreviation: TCGA, The Cancer Genome Atlas.
Acknowledgments
We thank the authors of all the included studies. This study was supported by the Project of Standard Diagnosis and Treatment of Key Disease of Jiangsu Province (BE2015722), the Project of the Peak of the Six Talents of Jiangsu Province (WSN-018), and the Scientific Research Foundation for Health of Jiangsu Province (H201408).
Disclosure
The authors reported no conflicts of interest in this work.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Cronin KA, Lake AJ, Scott S, et al. Annual report to the nation on the status of cancer, part I: national cancer statistics. Cancer Am Cancer Soc. 2018;124:2785–2800. doi: 10.1002/cncr.31551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kong YW, Ferland-McCollough D, Jackson TJ, Bushell M. microRNAs in cancer management. Lancet Oncol. 2012;13:e249–e258. doi: 10.1016/S1470-2045(12)70140-7 [DOI] [PubMed] [Google Scholar]
- 4.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028 [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flavin RJ, Smyth PC, Laios A, et al. Potentially important microRNA cluster on chromosome 17p13.1 in primary peritoneal carcinoma. Mod Pathol. 2009;22:197–205. doi: 10.1038/modpathol.2008.135 [DOI] [PubMed] [Google Scholar]
- 7.He JF, Luo YM, Wan XH, Jiang D. Biogenesis of MiRNA-195 and its role in biogenesis, the cell cycle, and apoptosis. J Biochem Mol Toxicol. 2011;25:404–408. doi: 10.1002/jbt.20396 [DOI] [PubMed] [Google Scholar]
- 8.Yu W, Liang X, Li X, et al. MicroRNA-195: a review of its role in cancers. Oncotargets Ther. 2018;11:7109–7123. doi: 10.2147/OTT.S183600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan B, Manley J, Lee J, Singh SR. The emerging roles of microRNAs in cancer metabolism. Cancer Lett. 2015;356:301–308. doi: 10.1016/j.canlet.2014.10.011 [DOI] [PubMed] [Google Scholar]
- 10.Liu B, Qu J, Xu F, et al. MiR-195 suppresses non-small cell lung cancer by targeting CHEK1. Oncotarget. 2015;6:9445–9456. doi: 10.18632/oncotarget.3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu C, Guan H, Wang Y, et al. miR-195 inhibits EMT by targeting FGF2 in prostate cancer cells. PLoS One. 2015;10:e144073. doi: 10.1371/journal.pone.0144073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen YH, Xie ZB, Yue AM, et al. Expression level of microRNA-195 in the serum of patients with gastric cancer and its relationship with the clinicopathological staging of the cancer. Eur Rev Med Pharmacol Sci. 2016;20:1283–1287. [PubMed] [Google Scholar]
- 13.Wang X, Wang J, Ma H, Zhang J, Zhou X. Downregulation of miR-195 correlates with lymph node metastasis and poor prognosis in colorectal cancer. Med Oncol. 2012;29:919–927. doi: 10.1007/s12032-011-9880-5 [DOI] [PubMed] [Google Scholar]
- 14.Yu S, Jing L, Yin XR, et al. MiR-195 suppresses the metastasis and epithelial-mesenchymal transition of hepatocellular carcinoma by inhibiting YAP. Oncotarget. 2017;8:99757–99771. doi: 10.18632/oncotarget.20909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun N, Ye L, Chang T, Li X, Li X. microRNA-195-Cdc42 axis acts as a prognostic factor of esophageal squamous cell carcinoma. Int J Clin Exp Pathol. 2014;7:6871–6879. [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou Q, Han LR, Zhou YX, Li Y. MiR-195 suppresses cervical cancer migration and invasion through targeting Smad3. Int J Gynecol Cancer. 2016;26:817–824. doi: 10.1097/IGC.0000000000000686 [DOI] [PubMed] [Google Scholar]
- 17.Deng Z, Wang Y, Fang X, et al. Research on miRNA-195 and target gene CDK6 in oral verrucous carcinoma. Cancer Gene Ther. 2017;24:282–288. doi: 10.1038/cgt.2017.18 [DOI] [PubMed] [Google Scholar]
- 18.Zhu J, Ye Q, Chang L, et al. Upregulation of miR-195 enhances the radiosensitivity of breast cancer cells through the inhibition of BCL-2. Int J Clin Exp Med. 2015;8:9142–9148. [PMC free article] [PubMed] [Google Scholar]
- 19.Bhattacharya A, Schmitz U, Wolkenhauer O, et al. Regulation of cell cycle checkpoint kinase WEE1 by miR-195 in malignant melanoma. Oncogene. 2013;32:3175–3183. doi: 10.1038/onc.2012.324 [DOI] [PubMed] [Google Scholar]
- 20.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25:603–605. doi: 10.1007/s10654-010-9491-z [DOI] [PubMed] [Google Scholar]
- 21.McInnes M, Moher D, Thombs BD, et al. Preferred reporting items for a systematic review and meta-analysis of diagnostic test accuracy studies: the PRISMA-DTA statement. JAMA. 2018;319:388–396. doi: 10.1001/jama.2017.19163 [DOI] [PubMed] [Google Scholar]
- 22.Chaimani A, Mavridis D, Salanti G. A hands-on practical tutorial on performing meta-analysis with Stata. Evid Based Ment Health. 2014;17:111–116. doi: 10.1136/eb-2014-101967 [DOI] [PubMed] [Google Scholar]
- 23.DerSimonian R, Laird N. Meta-analysis in clinical trials revisited. Contemp Clin Trials. 2015;45:139–145. doi: 10.1016/j.cct.2015.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8. doi: 10.1186/1745-6215-8-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Egger M, Davey SG, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen S, Wang L, Yao X, et al. miR-195-5p is critical in REG gamma-mediated regulation of wnt/beta-catenin pathway in renal cell carcinoma. Oncotarget. 2017;8:63986–64000. doi: 10.18632/oncotarget.19256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun P, Wang L, Lu Y, et al. MicroRNA-195 targets VEGFR2 and has a tumor suppressive role in ACHN cells via PI3K/Akt and Raf/MEK/ERK signaling pathways. Int J Oncol. 2016;49:1155–1163. doi: 10.3892/ijo.2016.3608 [DOI] [PubMed] [Google Scholar]
- 28.Shuang Y, Li C, Zhou X, Huang YW, Zhang L. Expression of miR-195 in laryngeal squamous cell carcinoma and its effect on proliferation and apoptosis of Hep-2. Eur Rev Med Pharmacol Sci. 2017;21:3232–3238. [PubMed] [Google Scholar]
- 29.Wang T, Ren Y, Liu R, et al. miR-195-5p suppresses the proliferation, migration, and invasion of oral squamous cell carcinoma by targeting TRIM14. Biomed Res Int. 2017;2017:7378148. doi: 10.1155/2017/7378148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou B, Sun C, Hu X, et al. MicroRNA-195 suppresses the progression of pancreatic cancer by targeting DCLK1. Cell Physiol Biochem. 2017;44:1867–1881. doi: 10.1159/000485876 [DOI] [PubMed] [Google Scholar]
- 31.Dong H, Lei J, Ding L, et al. MicroRNA: function, detection, and bioanalysis. Chem Rev. 2013;113:6207–6233. 10.1021/acs.analchem.8b03246 [DOI] [PubMed] [Google Scholar]
- 32.Zhou X, Wang X, Huang Z, et al. Prognostic value of miR-21 in various cancers: an updating meta-analysis. PLoS One. 2014;9:e102413. doi: 10.1371/journal.pone.0102413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang S, Hu J, Zhang D, et al. Prognostic role of microRNA-31 in various cancers: a meta-analysis. Tumor Biol. 2014;35:11639–11645. doi: 10.1007/s13277-014-2492-x [DOI] [PubMed] [Google Scholar]
- 34.Yang X, Yu J, Yin J, et al. MiR-195 regulates cell apoptosis of human hepatocellular carcinoma cells by targeting LATS2. Pharmazie. 2012. Jul;67(7):645-51. [PubMed] [Google Scholar]
- 35.Wang M, Zhang J, Tong L, et al. MiR-195 is a key negative regulator of hepatocellular carcinoma metastasis by targeting FGF2 and VEGFA. Int J Clin Exp Patho. 2015;8:14110–14120. [PMC free article] [PubMed] [Google Scholar]
- 36.Han K, Chen X, Bian,N, et al. MicroRNA profiling identifies MiR-195 suppresses osteosarcoma cell metastasis by targeting CCND1. Oncotarget 2015;6:8875–8889. doi: 10.18632/oncotarget.3560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Su K, Zhang T, Wang Y, et al. Diagnostic and prognostic value of plasma microRNA-195 in patients with non-small cell lung cancer. World J Surg Oncol 2016;14:224. doi: 10.1186/s12957-016-0980-8 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Validation with data available on the expression level of miR-195 and cancer data sets from TCGA database. (A) Non-small cell lung cancer. (B) Renal cell cancer. (C) Pancreatic cancer. (D) Prostate cancer. (E) Colorectal cancer. (F) Clear cell renal cell carcinoma.
Abbreviation: TCGA, The Cancer Genome Atlas.