While it has long been recognized and documented in epidemiologic studies that early life stress (ELS) can predispose individuals to higher all-cause mortality at potentially alarming rates (Dube, Felitti, Dong, Giles, & Anda, 2003; Felitti et al., 1998; Marmot et al., 1991), and that early life protective factors, including social/relational factors, can prevent or reverse the consequences of ELS (Farrell, Simpson, Carlson, Englund, & Sung, 2016), our ability to identify and systematically test possible mechanisms is necessarily compromised in human studies by ethical and practical constraints. A successful animal model of a complex human phenomenon provides an elegance that is rooted in its specificity and built with rigorous attention to mechanism. In addition to insights provided by natural experiments and deliberate intervention in human work, scientific advances have benefited from at least three important preclinical models that have provided specific manipulations of early postnatal life experiences (differences in licking and grooming; maternal separation/deprivation; caregiver maltreatment) followed by careful study of their behavioral, physical, cognitive, physiologic, neural and epigenetic consequences. Because the behavioral (anxiety, depression), physical (growth), cognitive (deficits in memory), physiologic (stress-system), and neural (prefrontal cortex, hippocampal, amygdala volume) changes seen in preclinical models mirror those often seen in children and adults who have experienced ELS, much attention has been given to considering whether the core mechanisms identified in preclinical models can explain (and potentially be used to reverse) negative outcomes in humans. In particular, the promise of epigenetic mechanisms (most prominently DNA methylation) as perhaps the critical and reversible process that embeds experience within the body and brain to influence physical and mental health, and which may even be transmittable across generations, has been an increasing target for empirical and theoretical work. This promise, so beautifully articulated by early pioneers in this work (Champagne, 2010; Zhang & Meaney, 2010), has drawn human researchers to collect or use banked tissue samples for characterization of DNA methylation.
This important step forward opens the possibility for meaningful cross-species dialog; specifically, where does human data match or contradict the preclinical models, how might preclinical models be further manipulated to probe these differences, and how might human studies better measure meaningful analogs to the preclinical work? In the interest of advancing this goal, we provide a classification structure to identify aspects of preclinical and human studies that should correspond for best comparison (based on available data) and highlight places where this match could be increased. This is critical because we cannot fully appreciate similarities or differences across species (and their implications for theory and intervention) without careful alignment and adjustment to methods in preclinical and human studies as more data become available. In fact, it has been the case that key papers (e.g. McGowan et al., 2009) have investigated epigenetic markers identified in one preclinical model (e.g. NR3C1 in hippocampus; licking and grooming model) when in fact the human sample (human suicide victims with and without child abuse histories) better matches a different preclinical model (caregiver maltreatment) which would have suggested another epigenetic target (e.g. BDNF in PFC). Of course, this has sometimes happened due to timing (the licking and grooming model was available first); however, this should not preclude future work from taking these similarities and differences into account. Further, the opportunities afforded by the development of the maternal separation/deprivation model (Daniels et al., 2009; Murgatroyd et al., 2009) and the caregiver maltreatment model (Roth et al., 2009, Blaze et al., 2013) have not been fully realized. Serendipitously, some studies have revealed parallels across models that are dissimilar, which brings to light the key question of how specific these processes in fact are, and whether that specificity holds across species.
To further this conversation and spur research that takes advantage of these opportunities, we organize the existing literature by providing a classification structure for the match between characteristics of the human studies and the three most prominent preclinical models of early postnatal stress. Specifically we evaluate the match between 63 human studies and each of three identified animal models of early life stress (licking/grooming, separation, maltreatment) on six key variables (timing of ELS, timing of epigenetic sampling, type of ELS, the degree to which sex of participants was addressed, tissue source for the epigenetic sample(s), and target DNA methylation loci examined). Our intention with this classification scheme is to systematically identify where there is translational relevance and some next steps for this line of research to increase translation and replicability to accelerate scientific advance.
Method
Model selection.
We began with the licking and grooming model (e.g. Weaver et al., 2004; Champagne et al., 2006), arguably the most prominent model examining how early experience may change developmental trajectories as mediated via DNA methylation. Because many human studies that cite the licking and grooming model focus on adversity (vs. positive caregiving following mild challenge), we next included two prominent models of early life adversity, the maternal separation/deprivation model (Daniels et al., 2009; Murgatroyd et al., 2009) and the caregiver maltreatment model (Roth et al., 2009, Blaze et al., 2013). Our intention was to allow a thorough evaluation of translational relevance by allowing a contrast with three animal models that differ in the type of ELS. Please see Table 1 for details on each model, particularly with respect to the match variables evaluated in the classification scheme.
Table 1.
Core features of the three selected preclinical models of postnatal early life stress.
| Model | High/Low LG-ABN | Maternal Separation/Deprivation | Caregiver Maltreatment |
|---|---|---|---|
| Description | Typically derived from a strain of animals previously observed to engaged in different base-rates of licking and grooming (LG) of pups; sometimes includes handling of pups (and thus short dam-pup separations) to induce differential LG behavior | Longer separations of pups and dams, ranging from a few hours per day, (often repeated for 9–12 days), | Stress-induced maternal adverse behaviors (dropping, stepping on, dragging and neglecting) |
| Seminal Epigenetic Study/Studies | Weaver et al., 2004; Champagne, et al., 2006 | Daniels et al., 2009; Murgatroyd et al., 2009 | Roth et al., 2009; Blaze et al., 2013 |
| Additional Seminal Studies of Model | Liu et al., 1997; Caldji et al., 1998; Champagne, et al., 2001 | Plotsky & Meaney, 1993 | Roth & Sullivan, 2005; Doherty et al., 2017 |
| Species | Rat | Rat (Daniels); Mice (Murgatroyd) | Rat |
| Strain/Genotype | Long-Evans hooded rat | Sprague-Dawley rat; C57BL/6J C57BL/6N and DBA/2J mice | Long-Evans rat |
| Sex of Caregiver | Mother | Mother | Mother |
| Sex of Offspring | Not specified; often only male in studies of NR3C1; only female in studies of ERα | Both studied, sex differences widespread | Both - sex differences found in epigenetics, sex differences in behavior |
| Timing of Early Experience | First postnatal week | PD1–10 3hr/day, Murgatroyd; PD2–14 3hr/day, Daniels; PD2–13 4hr/day, Chen; PD9 24hrs, Kember | PD1–7; 30 min/day exposure to maltreating caregiver |
| Physiology Measured | HPA-response to restraint stress; decreased levels of GR mRNA, decreased ability to downregulate CHR and ACTH release | Higher circulating and stress responsive corticosterone | Not measured |
| Behavior Observed | More “anxious” behaviors (e.g. less open field exploration) in low LGs, e.g. Francis et al., 1999; decreased hippocampal dependent memory in low LGs, e.g. Liu et al., 2000, Bredy et al., 2003 Less LG of own pups | More “anxious” behaviors (less time in open field) in separated rats, less approach to novel objects; poorer memory, anhedonia | Females- more “anxious” prior to birth, mistreat own offspring, less adaptive coping in forced swim, less approach to novel objects; Males- deficits in ability to extinguish fear memory |
| Age/Stage of Epigenetic Sampling | E20/PD1: ns PD6, PD21, PD90: p<.001 | PD21; “various ages” | PD8, PD30, PD90 |
| Tissue/location | Brain/hippocampus | Brain/hippocampus | Brain/PFC hippocampus |
| Epigenetic change measured | NR3C1, 17 promoter methylation, decreased H3–K9 acetylation and NGFI-A binding in low LG; 100+ kilobase pairs Chr18, McGowan et al., 2011 ERα | demethylation of AVP, No change to NR3C1 17, Daniels et al., 2009 & Murgatroyd et al., 2009 | BDNF, decreased expression in PFC but not hippocampus and increased BDNF methylation at exon IV and IX, CpG11 for adults maltreated as infants; increase in BDNF methylation in PFC and hippocampus in offspring of maltreated mothers |
Bold indicates findings from the seminal epigenetic work cited, italics denotes important additions or clarifications from prior or subsequent studies
Article selection process.
A systematic search was performed to verify the selected rodent preclinical models of early postnatal stress and to identify the body of existing human studies. We selected the three key rodent models as those most consistently studied with respect to postnatal caregiving influences on epigenetics, and varying in their degree of severity. For each, the seminal article (or articles) describing the epigenetic results was used as a root. To allow further classification of the specifics of the model, prior or subsequent seminal work was also referenced. See Table 1 for abstracted core components of each model based on this approach. Notably, each model includes offspring outcomes, and sometimes, for female animals, there is effort to understand subsequent mothering (and its epigenetic mechanisms). We also note that related work in non-human primates has been documented. Although beyond the scope of this review these models offer further tests of cross-species differentiation and replication of core mechanisms and can provide important insight (Kundakovic & Champagne, 2015). Finally, in all three animal models of ELS, DNA methylation was consistently examined, though other complementary epigenetic processes have been studied as impacted by ELS (histone modification, Xie et al., 2013; chromatin remodeling, Weaver et al., 2017). Therefore, we focused our systematic review on DNA methylation specifically.
Next, the following advanced search queries were run in the U.S. National Library of Medicine “medline” database:
methylation AND (DNA OR epigenetic) AND (“early life” OR “maternal behavior”) NOT autism NOT schizophrenia NOT ethanol NOT alcohol NOT cancer NOT metastable NOT diabetes NOT lead NOT nutrition NOT asthma NOT allergy NOT allergic NOT pollution NOT pollutants [no date restrictions, through July 2, 2018]. This resulted in 340 hits.
methylation AND (DNA OR epigenetic) AND (“early life” OR “maternal”) AND human NOT infection NOT substance NOT autism NOT schizophrenia NOT ethanol NOT alcohol NOT cancer NOT metastable NOT diabetes NOT lead NOT nutrition NOT asthma NOT allergy NOT allergic NOT pollution NOT pollutants [no date restrictions, through July 2, 2018] This resulted in 1198 hits.
Articles citing Champagne et al., 2006, Daniels et al., 2009, Murgatroyd et al., 2009, Roth et al., 2009 [no date restrictions, through July 2, 2018]. This resulted in 176, 27, 293, and 310 citing articles, respectively.
Articles citing Weaver et al., since the last systematic review (Tureki & Meaney, 2016 included through July 2014). This resulted in 440 hits (August 1, 2014 through July 2, 2018), as well as 26 of the 27 human articles selected by Turecki & Meaney, 2016 (one was subsequently retracted).
Entries from these searches were examined, and empirical articles with human data that examined epigenetic markers of some type of postnatal caregiver-related ELS or caregiving were retained (a total of 63 articles). This included studies of infants or children whose mothers had been previously stressed or had suffered from mental illness (during or before pregnancy and in the infants’ early postnatal life), and those examining data from adults with ELS exposures involving caregivers (prior maltreatment, caregiver mental illness, and socioeconomic position). Samples of mothers with limited information on childhood adversity and no methylation data from children were excluded, and one study of child methylation as the result of stress exposure during grandmaternal gestation was also excluded because of limited information on subsequent adversity in the mother or child’s life. One study that constrained epigenome-wide association study (EWAS) results to those CpGs associated with an adult outcome (BMI) was also excluded. Similarly, studies that sampled only placental tissue or cord blood were excluded as the focus of this paper was on postnatal epigenetic differences as related to ELS experiences (though this would be a fruitful area for future work to examine parallels with preclinical models of prenatal stress).
Selection of match variables.
Both human and animal models suggest that in addition to the type of ELS, the targeted epigenetic location/process and the species under study, investigators should consider the age/developmental stage both at the time of the early stress and at the time of epigenetic sampling, the sex of the individual, and the origin of the epigenetic sample (e.g. brain, blood, buccal, saliva). The rationale for including each of these, including a description of cross-species translation limitations, is provided in the following paragraphs.
Type of ELS.
Early stress in humans can come in many forms which may not always be independent. For example, neglect is common in children exposed to abuse (Cicchetti & Handley, 2017), and high levels of parental stress can reduce the quality of parenting behaviors in both rodents and humans (Doherty, Blaze, Keller & Roth, 2017, Wray, 2015). The selected preclinical models include differences in parental care (levels of licking and grooming, sometimes augmented by brief handling stress), maternal separation (separations of hours and up to a day of dams and pups shortly after birth, which is species atypical for length of time a dam is out of the nest), and maltreatment (neglectful and abusive parenting following maternal resource restriction and novelty stress). These models have each suggested a targeted epigenetic locus that may be responsible for long term offspring outcomes. The licking and grooming model (LG) heavily implicates the glucocorticoid receptor gene NR3C1, the maternal separation model (SEP) reports epigenetic differences in the gene responsible for synthesis of the hormone vasopressin, AVP, and the maltreatment model (MALTX) implicates the gene coding for synthesis of the protein brain-derived neurotropic factor, BDNF.
Timing of ELS.
The appropriate timing for comparable human epigenetic programming to that seen in animal models is unknown. In many cases, ELS paradigms used in rodents apply the ELS within the first and certainly by the end of the second postnatal week. This may correspond to roughly 6 months of stress experience in the human infant, though developmental age/stage comparisons across species are difficult (Sengupta, 2013). In the rat, weaning can happen around postnatal day 21, while human infants are often weaned between 9–12 months, although there is considerable variability (Canadian Pediatric Society, 2004). Both rodents and humans reach sexual maturity before social maturity. Human studies suggest that methylation differences continue to be settling through at least 5 years of age, and of course both human and animal work has demonstrated the dynamic nature of the epigenome over the course of development and its exquisite sensitivity to experiences outside of sensitive periods of development (Bale, 2014, Gitik et al., 2018, Kanherkar, Bhatia-Dey, & Csoka, 2014).
Age/Developmental Stage for Epigenetic Sampling.
In many cases, this variable has been systematically examined in the key animal models, with animals being sacrificed and sampled immediately post ELS, and at many developmental time points into adulthood. Human studies vary in the timing of the epigenetic sampling from as early as right at birth (or even prenatally) all the way through senescence. Without longitudinal work, understanding when differences should be expected and importantly when change is most likely will be elusive.
Sex.
In all three preclinical models, sex differences have been documented and can be quite important. In fact, sexually-dimorphic phenotypes in rats result in part from methylation differences that naturally occur (McCarthy & Nugent, 2015, Nugent, et al., 2015, Kolodkin, & Auger, (2011) and follow differences in preferential maternal licking and grooming and anogenital stimulation of male offspring (Kosten & Nielsen, 2014). A little discussed fact is that many rodent studies utilize litters that were culled to include only male animals. It would be interesting to address potential sex differences in ordinary parenting behaviors following ELS. Resultant hypotheses could be tested for relevance in human work.
Tissue Type & Location.
This cross-species limitation of tissue source is probably the best recognized, as in most cases, animal models use brain tissue and in most cases, human studies use peripheral samples. However, it is certainly possible to routinely assess peripheral epigenetics in animal models and sometimes possible to use stored blood or buccal samples for human populations that later have post-mortem brain tissue available. Human samples also commonly use blood drawn under different or unspecified conditions, and from different components (e.g. venous, cord blood, placenta). Creating careful correlations across tissue type would go a long way toward advancing crosstalk, and in the few studies that have done this work, both reasonable correlations (Smith et al., 2015) and discrepancies (Armstrong, Lesseur, Conradt, Lester & Marsit, 2014) have so far been documented.
Determination of degree of correspondence.
Information on each of the possible 6 match variables was extracted (see Table 2) and a percent match to each animal model was estimated. In particular, for each preclinical model, the study was given a summed score from 0–6 as to whether they matched on each of the following: (1) timing of ELS (1=neonatal and less than 6 postnatal months; .5=includes less than 2 years; 0=prenatal and over age 2 or across all of childhood), and (2) the offspring age at time of epigenetic sampling (1=matched if postnatal year up to age 30 years, .5 if postnatal year 31–50; 0=over age 50 because rodent models typically extend to postnatal day (PND) 90 or occasionally PND 180, but have not assess aged rats). Specific to each animal model we also coded degree of match to (3) ELS type (1=positive maternal care for LG model, maternal separation for SEP model, and abuse/neglect for MALTX model; .5 for partial match (e.g. maternal depression was coded as a partial match for both the LG and the SEP models given research on the association between maternal depression and both parenting and child outcomes) and 0 for no match); (4) offspring sex (1=matched on sex tested by the model{male for LG}, or both sexes included and paper explicitly tested for sex differences; 0=not matched); (5) tissue source examined in each model (brain, buccal, saliva, blood) and source for brain tissue (hippocampal vs. PFC) of the epigenetic sample analyzed (1=source and location match e.g. hippocampus for the LG and SEP models, and both hippocampus and PFC for the MALTX model; .5 for only hippocampus in the MALTX model; 0=not matched, (0 was coded for all sample types except brain tissue because all three animal models focus on brain tissue); and, (6) whether they assessed the model-appropriate target epigenetic location directly or discussed its analysis from an epigenome-wide (EWAS) scan (NR3C1 for LG, AVP for SEP, and BDNF for MALTX; 1=matched or EWAS, 0=not matched).
Table 2.
Included human empirical articles with ELS and epigenetic information (n=63).
| 1st Author | Year | PMID | ELS type | ELS age | ELS Data Source | Sex | Racial/Ethnic Group | Sample Location | EPI target | EPI age/stage |
|---|---|---|---|---|---|---|---|---|---|---|
| DNA methylation determined from brain tissue (predominantly hippocampal) | ||||||||||
| Labonte B | 2012 | PMID:22752237 | CH abuse | CH | PM Proxy | M | FrCan Ca | Quebec Suicide Bank | EWAS promoters | adult PM |
| Labonte B | 2012 | PMID:22444201 | CH abuse | CH | PM Proxy | M | FrCan Ca | Quebec Suicide Bank | GR | adult PM |
| McGowan PO | 2009 | PMID:19234457 | CH abuse | CH | PM Proxy | M | FrCan Ca | Quebec Suicide Bank | NR3C1 promoter | adult PM |
| Nemoda Z | 2015 | PMID:25849984 | MatDep | CH | PM Proxy | M | FrCan Ca | Quebec Suicide Bank | 450K | adult PM |
| Suderman M | 2012 | PMID:23045659 | CH abuse | CH | PM Proxy | M | FrCan Ca | Quebec Suicide Bank | 6.5M base pairs | adult PM |
| DNA methylation determined from buccal tissue | ||||||||||
| Braithwaite EC | 2015 | PMID:25875334 | MatDep | postnatal | EPDS | F/M | British Ca | UK | NR3C1, BDNF | 2 PNM |
| Conradt E | 2016 | PMID:26822444 | MatDep/MatSen | 0–4 PNM | CESD/coded | F/M | CaAm/AfAm | Rhode Island | NR3C1, 11-B-HSD2 | 4 PNM |
| Essex MJ | 2013 | PMID:21883162 | parental stress | 05 PNY | rep | F/M | CaAm | Wisconsin | 27K | 15 PNY |
| Giarraputo J | 2017 | PMID:27653086 | preterm/medical risk | neonatal | rec | F/M | CaAm, multi, Hisp | Rhode Island NICU | NR3C1 | neonatal |
| Kumsta R | 2016 | PMID:27271856 | OR | 0–43 PNM | rec | F/M | Romanian | England | 450K | 15 PNY |
| Lapp HE | 2018 | PMID:29475055 | CH adversities | 0–18yrs | ACE Q | F/M | CaAm, AfAm | Boston | MT-ND6 | adults |
| Lester BM | 2015 | PMID:26585459 | preterm | neonatal | rec | F/M | CaAm, multi, Hisp | Rhode Island | NR3C1,HDS11B2 | neonatal |
| Moore SR | 2017 | PMID:29162165 | tact | 4X 5 PNW | diary | F/M | Ca, Asian | Vancouver | NR3C1,OPRM1,OXTR, BDNF, EWAS | 4.5 PNY |
| Non AL | 2016 | PMID:27218411 | OR | CH | rec | F/M | Er | Bucharest | SLC6A4; FKBP5 | 12 PNY |
| DNA methylation determined from saliva sample | ||||||||||
| Cicchetti D | 2017 | PMID:29162187 | CH maltx | CH <9 PNY | rec | F/M | AfAm/CaAm | Upstate NY | NR3C1, 450K | 9.37 PNY |
| Efstathopoulos P | 2018 | PMID:29921868 | SES/PeerPrb | CH | rep | F/M | Swedish | S. Sweden | NR3C1 | 13–14 PNY |
| King L | 2017 | PMID:28918249 | MatDep | pre/perinatal | EPDS | F/M | Ca | Vancouver | OXT, IGR btwn OXT/AVP | 2.9 PNY |
| Melas PA | 2013 | PMID:23449091 | CH adversities | CH | rep | F/M | Er | Stockholm | MAOA, NR3C1 | adults |
| Murgatroyd C | 2015 | PMID:25942041 | MatDep/tact | 5 & 9 PNW | EPDS | F/M | British Ca | Wirral Penninsula, UK | NR3C1 | 14 PNM |
| Parade SH | 2017 | PMID:29162169 | CH maltx | 0–5 PNY | rec | F/M | Hisp, CaAm, AfAm, other | Rhode Island | HTR2A (Serotonin) | 3–5 PNY |
| Parade SH | 2016 | PMID:26822445 | CH maltx | 0–5 PNY | rec | F/M | Hisp, CaAm, AfAm, other | Rhode Island | NR3C1 | 3–5 PNY |
| Parent J | 2017 | PMID:29162170 | CH maltx | 0–5 PNY | rec | F/M | Hisp, CaAm, AfAm, other | Rhode Island | NR3C1 | 2x, 3–5 PNY |
| Tyrka AR | 2015 | PMID:25997773 | CH maltx | 0–5 PNY | rec | F/M | Hisp, CaAm, AfAm, other | Rhode Island | NR3C1 | 3–5 PNY |
| Weder N | 2014 | PMID:24655651 | CH maltx | 5–14 PNY | rec/int | F/M | Hisp, AfAm, CaAm, multi | Connecticut | 450K | <6MO removal |
| DNA methylation determined from blood sample (variation in procedure) | ||||||||||
| Borghol N | 2012 | PMID:22422449 | CH SES | CH | rep | M | British Ca | 1958 UK Birth Cohort | 20K scan | 45 PNY |
| Bustamante AC | 2016 | PMID:27475889 | CH Trauma | CH | CTQ | F/M | AfAm/CaAm | Detroit | NR3C1 | 49.6 PNY |
| Cao-Lei L | 2014 | PMID:25710121 | prenatal hardship | prenatal | rep | F/M | FrCan Ca | Quebec Proj Ice Storm | 450K | 8 & 15 PNY |
| Cao-Lei L | 2015 | PMID:25238154 | prenatal stress | prenatal | rep | F/M | FrCan Ca | Quebec Proj Ice Storm | 450K | 15 PNY |
| Dalle Molle R | 2012 | PMID:23168995 | parental care | CH | PBI | F/M | not specified | PROTAIA project | BDNF | adolescents |
| Duman EA | 2015 | PMID:25995833 | CH Trauma | CH | CTQ | M | CaAm | Upstate NY | SLC6A4 | 18–77 PNY |
| Farrell C | 2018 | PMID:29793048 | CH Trauma | CH | CTQ | F/M | Irish | Dublin | NR3C1, FKBP5 | 18–45 PNY |
| Gouin JP | 2017 | PMID:28785027 | CH SES & abuse | CH | ret rep | F/M | FrCan Ca | Quebec | OXTR | 27 PNY |
| Houtepen LC | 2016 | PMID:26997371 | CH trauma | CH | CTQ | F/M | Er | Netherlands | 450K | adults |
| Janusek LW | 2017 | PMID:27765646 | CH SES & truama | CH | ret rep | M | AfAm | Chicago | IL6 promoter | 18–25 PNY |
| Kantake M | 2014 | PMID:25023132 | MatSep | neonatal | NICU sep | F/M | Japanese | Japan | NR3C1 | PND 4 |
| Kantake M | 2018 | PMID:29796117 | IUGR | pre/neonatal | rec | F/M | Japanese | Japan | NR3C1 | 0–2 PNM |
| Khulan B | 2014 | PMID:25247593 | MatSep | CH | rec | M | Er | Helsinki Birth Cohort | 450K | 61.5 PNY |
| Lam LL | 2012 | PMID:23045638 | CH SES | CH | occupation | F/M | Ca, Asian | Vancouver | 14K array | 24–45 PNY |
| Levine ME | 2015 | PMID:25658624 | CH SES & truama | CH | ret rep | F/M | CaAm | Health & Retirement Study | IL1B IL8 PTGS2 | 51–95PNY |
| Martin-Blanco | 2014 | PMID:25048180 | CH Trauma | CH | CTQ | F/M | CaAm | Hospitalized | NR3C1 | 29 PNY |
| Marzi SJ | 2018 | PMID:29325449 | CH Victimization | CH | multiple | F/M | not specified | Britian (E_Risk Study) | 450K | 18 PNY |
| Naumova OY | 2016 | PMID:22123582 | accept/reject | CH | mat rep | F/M | AfAm | US urban | EWAS | 17–29.5 PNY |
| Naumova OY | 2012 | PMID:26822446 | OR | CH | rec | F/M | Slavic | Russia | 27K | 7–10 PNY |
| Needham BL | 2015 | PMID:26295359 | MatEd | CH | ret rep | F/M | CaAm, AfAm, Hisp | MESA study | 450K | 55–94 PNY |
| Peng H | 2018 | PMID:29781947 | CH Trauma | CH | ETI | F/M | not specified | national, THS & MMS | NR3C1,BDNF, SLC6A4, MAOA/B | 20–60 PNY |
| Perroud N | 2016 | PMID:26350166 | CH maltx | CH | CTQ | F/M | Er | Switzerland | 5HT3AR | 30–45 mean PNY |
| Perroud N | 2011 | PMID:22832351 | CH maltx | CH | CTQ | F/M | Er | France/Sweden; hosp. | NR3C1 | adults |
| Perroud N | 2014 | PMID:24690014 | genocide | prenatal | site | F | Tutsi | Rwanda | NR3C1 &NR3C2 | 17–18 PNY |
| Prados J | 2015 | PMID:25612291 | CH maltx | CH | ret rep | F/M | Er | Switzerland | 450K | 32–42 mean PNY |
| Provenzi L | 2017 | PMID:28959218 | Matsen | 3 PNM | coded | F/M | Er | Milan | SLC6A4 | at NICU discharge |
| Radtke KM | 2015 | PMID:26080088 | CH maltx | CH | Int | F/M | Varied | Germany | 450K | 11–21PNY |
| Radtke KM | 2011 | PMID:22832523 | Mat IPV | CH | rep | F/M | Varied | Germany | NR3C1 | 10–19 PNY |
| Romens SE | 2015 | PMID:25056599 | CH maltx | CH | rec | F/M | CaAm, AfAm | Wisconsin | NR3C1 | 11–14 PNY |
| Smith JA | 2017 | PMID:28678593 | neighborhood | CH | rep/census | F/M | CaAm, AfAm, Hisp | MESA study | 18 genes, NR3C1, AVP, BDNF | 70 PNY |
| Steiger H | 2013 | PMID:23417893 | CH abuse | CH | int | F | Ca | Candian; psychiatric | NR3C1 | 17–48 PNY |
| Suderman M | 2014 | PMID:24618023 | CH abuse | CH | ret rep | M | Br Ca | 1958 UK Birth Cohort | 20K scan | 45 PNY |
| Tehranifar P | 2013 | PMID:23196856 | CH maltx | CH | rep | F | AfAm, Hisp, CaAm | NY Women’s Birth Cohort | Sat2, Alu, Line-1 | 38–46 PNY |
| Tyrka AR | 2012 | PMID:22295073 | CH abuse, PL | CH | CTQ | F/M | CaAm | Rhode Island | NR3C1 | 18–65 PNY |
| Tyrka AR | 2016 | PMID:27378548 | CH abuse, PL | CH | CTQ | F/M | not specified | Rhode Island | NR3C1 | not specified |
| Unternaehrer E | 2015 | PMID:26061800 | maternal care | CH | PBI | F/M | Er | Switzerland | BDNF, OXTR | 22–33 PNY |
| van der Knaap LJ | 2014 | PMID:24713862 | CH adversity | CH | youth rep | F/M | Er | Dutch, TRAILS | NR3C1 | 14–18 |
| Wankerl M | 2014 | PMID:24937096 | prenatal stress, maltx | CH | rep | F/M | Er | Germany | SERT | young adults |
| Yehuda R | 2014 | PMID:24832930 | parental PTSD | CH | offspring rep | F/M | Jewish | New York | NR3C1 | 47–58 PNY |
ELS Abbreviations: CH=childhood, IUGR=intrauterine growth restriction, Maltx=maltreatment, Mat+maternal Dep=depression, Ed=education, IPV = intimate partner violence OR= orphange, PL=parental loss,
PeerPbs = peer problems, Sen=sensitivity, Sep= separation, SES = socioeconomic status, tact=tactile
Timing Abbreviations: CH=childhood, PNM=postnatal month, PNY=postnatal year, PNW=postnatal week
ELS Data Source Abbreviations: CESD=Center for Epidemiological Studies Depression Scale Revised, CTQ=Childhood Trauma Questionnaire, EPDS=Ediburgh Postnatal Depression Scale, ETI = Early Trauma Inventory,
Int=interview, PBI=Parental Bonding Instrument, PM proxy=postmortem proxy interview with next-of-kin, rec= medical record, rep=reported, ret=retrospective, sep=separation
Race/Ethnicity Abbreviations: Br=British, Ca=Caucasian, CaAm=Caucsian-American, AfAm=African American, Er=European, FrCan=French Canadian, Hisp=Hispanic
race listed if at least 20% of sample
Results
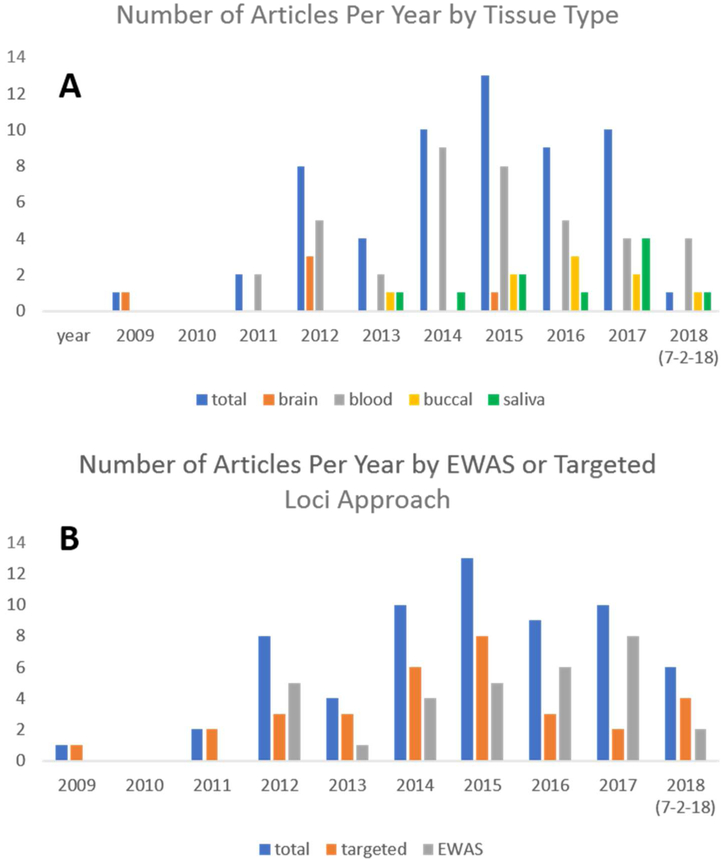
Of the 2,784 articles that were identified from the search strategies described above (this number includes duplicates that were surfaced by more than one strategy), 212 were evaluated in close detail and 63 met final criteria and are included in Table 2. We include here only empirical work, however it is interesting to note that most of the 2,784 records were commentaries and review papers, an indication of the great interest in this topic across many disciplines (biology, psychology, psychiatry, neuroscience, medicine, education etc.). The body of human work is also quite impressive, especially given the difficulty inherent in this type of study. As can be seen in Figure 1a, human studies have been consistently published since 2012, with shifting tissue type preferences (particularly increasing use of saliva samples). Targeted loci approaches continue to be common, but EWAS approaches are increasingly utilized (see Figure 1b). However, it is clear that more human work is needed, particularly more work that builds from the preclinical work and that evaluates the wealth of available specific and testable hypotheses with longitudinal or intervention studies.
Figure 1.
Publication trends for included articles (n=63). Panel (a) indicates tissue type by publication year and panel (b) indicates EWAS versus targeted loci by year.
We organize the results according to tissue type studied. Because the animal work all uses brain tissue, in all cases excepting the five postmortem brain papers there is a mismatch between the human and animal work. Although use of human brain tissue is clearly restricted (and presents its own limitations), because epigenetic signals are critically involved in cell-type specificity we elect to organize our review to highlight that immediately upon departing from a match in tissue type a degree of translational specificity is lost and should temper interpretation. Within each section (divided by tissue type), we then summarize the degree of match on other key variables (type and timing of ELS, epigenetic target). Our intention is to illuminate places where the match between human and animal work could be increased to further theoretical understanding and practical implications. Table 3 provides detail on the classification determination made for each of the 63 studies for each model and variable assessed, which could have ranged from 0% (matched on 0/6 variables) to 100% (matched on 6/6 variables).
Table 3.
Model fit ratings and summaries for all included articles (n=63)
| 1st Author | Year | ALL MODELS | LG MODEL | SEP MODEL | MALTX MODEL | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ELS Age | EPI Age | ELS Type | Sex | EPI source & Loc | EPI Target | LG Match Score | LG % Fit | ELS Type | Sex | EPI source & Loc | EPI Target | SEP Match Score | SEP % fit | ELS Type | Sex | EPI source & Loc | EPI Target | MALTX Match Score | MALTX %flt | ||
| DNA methylation determined from brain tissue (predominantly hippocampal) | |||||||||||||||||||||
| Labonte B | 2012 | 0.0 | 0.5 | 0.0 | 1.0 | 1.0 | 1.0 | 3.5 | 0.58 | 0.0 | 0.0 | 1.0 | 1.0 | 2.5 | 0.42 | 1.0 | 0.0 | 0.5 | 1.0 | 3.0 | 0.50 |
| Labonte B | 2012 | 0.0 | 0.5 | 0.0 | 1.0 | 1.0 | 1.0 | 3.5 | 0.58 | 0.0 | 0.0 | 1.0 | 0.0 | 1.5 | 0.25 | 1.0 | 0.0 | 0.5 | 0.0 | 2.0 | 0.33 |
| McGowan PO | 2009 | 0.0 | 0.5 | 0.0 | 1.0 | 1.0 | 1.0 | 3.5 | 0.58 | 0.0 | 0.0 | 1.0 | 0.0 | 1.5 | 0.25 | 1.0 | 0.0 | 1.0 | 0.0 | 2.5 | 0.42 |
| Nemoda Z | 2015 | 0.0 | 0.5 | 0.5 | 1.0 | 1.0 | 1.0 | 4.0 | 0.67 | 0.5 | 0.0 | 1.0 | 1.0 | 3.0 | 0.50 | 0.0 | 0.0 | 0.5 | 1.0 | 2.0 | 0.33 |
| Suderman M | 2012 | 0.0 | 0.5 | 0.0 | 1.0 | 1.0 | 1.0 | 3.5 | 0.58 | 0.0 | 0.0 | 1.0 | 1.0 | 2.5 | 0.42 | 1.0 | 0.0 | 0.5 | 1.0 | 3.0 | 0.50 |
| DNA methylation determined from buccal tissue | |||||||||||||||||||||
| Braithwaite EC | 2015 | 1.0 | 1.0 | 0.5 | 1.0 | 0.0 | 1.0 | 4.5 | 0.75 | 0.5 | 1.0 | 0.0 | 0.0 | 3.5 | 0.58 | 0.0 | 1.0 | 0.0 | 1.0 | 4.0 | 0.67 |
| Conradt E | 2016 | 1.0 | 1.0 | 1.0 | 1.0 | 0.0 | 1.0 | 5.0 | 0.83 | 0.5 | 1.0 | 0.0 | 0.0 | 3.5 | 0.58 | 0.0 | 1.0 | 0.0 | 0.0 | 3.0 | 0.50 |
| Essex MJ | 2013 | 0.5 | 1.0 | 0.5 | 1.0 | 0.0 | 1.0 | 4.0 | 0.67 | 0.5 | 1.0 | 0.0 | 1.0 | 4.0 | 0.67 | 0.0 | 1.0 | 0.0 | 1.0 | 3.5 | 0.58 |
| Giarraputo J | 2017 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 3.0 | 0.50 | 1.0 | 0.0 | 0.0 | 0.0 | 3.0 | 0.50 | 0.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.33 |
| Kumsta R | 2016 | 0.5 | 0.5 | 0.0 | 1.0 | 0.0 | 1.0 | 3.0 | 0.50 | 1.0 | 1.0 | 0.0 | 1.0 | 4.0 | 0.67 | 0.5 | 1.0 | 0.0 | 1.0 | 3.5 | 0.58 |
| Lapp HE | 2018 | 0.0 | 0.5 | 0.0 | 1.0 | 0.0 | 0.0 | 1.5 | 0.25 | 0.0 | 1.0 | 0.0 | 0.0 | 1.5 | 0.25 | 1.0 | 1.0 | 0.0 | 0.0 | 2.5 | 0.42 |
| Lester BM | 2015 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 3.0 | 0.50 | 1.0 | 0.0 | 0.0 | 0.0 | 3.0 | 0.50 | 0.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.33 |
| Moore SR | 2017 | 1.0 | 1.0 | 1.0 | 1.0 | 0.0 | 1.0 | 5.0 | 0.83 | 0.0 | 1.0 | 0.0 | 0.0 | 3.0 | 0.50 | 0.0 | 1.0 | 0.0 | 1.0 | 4.0 | 0.67 |
| Non AL | 2016 | 0.5 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 2.5 | 0.42 | 1.0 | 1.0 | 0.0 | 0.0 | 3.5 | 0.58 | 0.5 | 1.0 | 0.0 | 0.0 | 3.0 | 0.50 |
| DNA methylation determined from saliva sample | |||||||||||||||||||||
| Cicchetti D | 2017 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 1.0 | 3.0 | 0.50 | 0.0 | 1.0 | 0.0 | 1.0 | 3.0 | 0.50 | 1.0 | 1.0 | 0.0 | 1.0 | 4.0 | 0.67 |
| Efstathopoulos P | 2018 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 1.0 | 3.0 | 0.50 | 0.0 | 1.0 | 0.0 | 0.0 | 2.0 | 0.33 | 0.0 | 1.0 | 0.0 | 0.0 | 2.0 | 0.33 |
| King L | 2017 | 1.0 | 1.0 | 0.5 | 1.0 | 0.0 | 0.0 | 3.5 | 0.58 | 0.5 | 1.0 | 0.0 | 0.5 | 4.0 | 0.67 | 0.0 | 1.0 | 0.0 | 0.0 | 3.0 | 0.50 |
| Melas PA | 2013 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 2.0 | 0.33 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.17 | 0.5 | 1.0 | 0.0 | 0.0 | 1.5 | 0.25 |
| Murgatroyd C | 2015 | 1.0 | 1.0 | 1.0 | 0.0 | 0.0 | 1.0 | 4.0 | 0.67 | 0.5 | 1.0 | 0.0 | 0.0 | 3.5 | 0.58 | 0.0 | 1.0 | 0.0 | 0.0 | 3.0 | 0.50 |
| Parade SH | 2017 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 2.0 | 0.33 | 0.0 | 1.0 | 0.0 | 0.0 | 2.0 | 0.33 | 1.0 | 1.0 | 0.0 | 0.0 | 3.0 | 0.50 |
| Parade SH | 2016 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 1.0 | 3.0 | 0.50 | 0.0 | 1.0 | 0.0 | 0.0 | 2.0 | 0.33 | 1.0 | 1.0 | 0.0 | 0.0 | 3.0 | 0.50 |
| Parent J | 2017 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 1.0 | 3.0 | 0.50 | 0.0 | 1.0 | 0.0 | 0.0 | 2.0 | 0.33 | 1.0 | 1.0 | 0.0 | 0.0 | 3.0 | 0.50 |
| Tyrka AR | 2015 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 1.0 | 3.0 | 0.50 | 0.0 | 1.0 | 0.0 | 0.0 | 2.0 | 0.33 | 1.0 | 1.0 | 0.0 | 0.0 | 3.0 | 0.50 |
| Weder N | 2014 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 1.0 | 3.0 | 0.50 | 0.0 | 1.0 | 0.0 | 1.0 | 3.0 | 0.50 | 1.0 | 1.0 | 0.0 | 1.0 | 4.0 | 0.67 |
| DNA methylation determined from blood sample (variation in procedure) | |||||||||||||||||||||
| Borghol N | 2012 | 0.0 | 0.5 | 0.0 | 1.0 | 0.0 | 1.0 | 2.5 | 0.42 | 1.0 | 0.0 | 0.0 | 1.0 | 2.5 | 0.42 | 0.0 | 0.0 | 0.0 | 1.0 | 1.5 | 0.25 |
| Bustamante AC | 2016 | 0.0 | 0.5 | 0.0 | 1.0 | 0.0 | 1.0 | 2.5 | 0.42 | 0.0 | 1.0 | 0.0 | 0.0 | 1.5 | 0.25 | 1.0 | 1.0 | 0.0 | 0.0 | 2.5 | 0.42 |
| Cao-Lei L | 2014 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 2.0 | 0.33 | 0.0 | 0.0 | 0.0 | 1.0 | 2.0 | 0.33 | 0.0 | 0.0 | 0.0 | 1.0 | 2.0 | 0.33 |
| Cao-Lei L | 2015 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 1.0 | 3.0 | 0.50 | 0.0 | 1.0 | 0.0 | 1.0 | 3.0 | 0.50 | 0.0 | 1.0 | 0.0 | 1.0 | 3.0 | 0.50 |
| Dalle Molle R | 2012 | 0.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.33 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.17 | 0.0 | 0.0 | 0.0 | 1.0 | 2.0 | 0.33 |
| Duman EA | 2015 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 2.0 | 0.33 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.17 | 1.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.33 |
| Farrell C | 2018 | 0.0 | 0.5 | 0.0 | 0.0 | 0.0 | 1.0 | 1.5 | 0.25 | 0.0 | 0.0 | 0.0 | 0.0 | 0.5 | 0.08 | 1.0 | 0.0 | 0.0 | 0.0 | 1.5 | 0.25 |
| Gouin JP | 2017 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 2.0 | 0.33 | 1.0 | 1.0 | 0.0 | 0.0 | 3.0 | 0.50 | 1.0 | 1.0 | 0.0 | 0.0 | 3.0 | 0.50 |
| Houtepen LC | 2016 | 0.0 | 0.5 | 0.0 | 1.0 | 0.0 | 1.0 | 2.5 | 0.42 | 0.0 | 1.0 | 0.0 | 1.0 | 2.5 | 0.42 | 1.0 | 1.0 | 0.0 | 1.0 | 3.5 | 0.58 |
| Janusek LW | 2017 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 2.0 | 0.33 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.17 | 1.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.33 |
| Kantake M | 2014 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 3.0 | 0.50 | 1.0 | 0.0 | 0.0 | 0.0 | 3.0 | 0.50 | 0.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.33 |
| Kantake M | 2018 | 1.0 | 1.0 | 0.0 | 1.0 | 0.0 | 1.0 | 4.0 | 0.67 | 0.0 | 1.0 | 0.0 | 0.0 | 3.0 | 0.50 | 0.0 | 1.0 | 0.0 | 0.0 | 3.0 | 0.50 |
| Khulan B | 2014 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 2.0 | 0.33 | 1.0 | 0.0 | 0.0 | 1.0 | 2.0 | 0.33 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | 0.17 |
| Lam LL | 2012 | 0.0 | 0.5 | 0.0 | 1.0 | 0.0 | 1.0 | 2.5 | 0.42 | 1.0 | 1.0 | 0.0 | 1.0 | 3.5 | 0.58 | 0.0 | 1.0 | 0.0 | 1.0 | 2.5 | 0.42 |
| Levine ME | 2015 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.17 | 1.0 | 1.0 | 0.0 | 0.0 | 2.0 | 0.33 | 1.0 | 1.0 | 0.0 | 0.0 | 2.0 | 0.33 |
| Martin-Blanco | 2014 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 1.0 | 3.0 | 0.50 | 0.0 | 1.0 | 0.0 | 0.0 | 2.0 | 0.33 | 1.0 | 1.0 | 0.0 | 0.0 | 3.0 | 0.50 |
| Marzi SJ | 2018 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 1.0 | 3.0 | 0.50 | 0.0 | 1.0 | 0.0 | 1.0 | 3.0 | 0.50 | 1.0 | 1.0 | 0.0 | 1.0 | 4.0 | 0.67 |
| Naumova OY | 2016 | 0.0 | 1.0 | 1.0 | 1.0 | 0.0 | 1.0 | 4.0 | 0.67 | 0.0 | 1.0 | 0.0 | 1.0 | 3.0 | 0.50 | 0.5 | 1.0 | 0.0 | 1.0 | 3.5 | 0.58 |
| Naumova OY | 2012 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 1.0 | 3.0 | 0.50 | 1.0 | 1.0 | 0.0 | 1.0 | 4.0 | 0.67 | 0.0 | 1.0 | 0.0 | 1.0 | 3.0 | 0.50 |
| Needham BL | 2015 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 2.0 | 0.33 | 0.0 | 1.0 | 0.0 | 1.0 | 2.0 | 0.33 | 0.0 | 1.0 | 0.0 | 1.0 | 2.0 | 0.33 |
| Peng H | 2018 | 0.0 | 0.5 | 0.0 | 1.0 | 0.0 | 1.0 | 2.5 | 0.42 | 0.0 | 1.0 | 0.0 | 0.0 | 1.5 | 0.25 | 1.0 | 1.0 | 0.0 | 1.0 | 3.5 | 0.58 |
| Perroud N | 2016 | 0.0 | 0.5 | 0.0 | 1.0 | 0.0 | 0.0 | 1.5 | 0.25 | 0.0 | 1.0 | 0.0 | 0.0 | 1.5 | 0.25 | 1.0 | 1.0 | 0.0 | 0.0 | 2.5 | 0.42 |
| Perroud N | 2011 | 0.0 | 0.5 | 0.0 | 1.0 | 0.0 | 1.0 | 2.5 | 0.42 | 0.0 | 1.0 | 0.0 | 0.0 | 1.5 | 0.25 | 1.0 | 1.0 | 0.0 | 0.0 | 2.5 | 0.42 |
| Perroud N | 2014 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 1.0 | 3.0 | 0.50 | 0.0 | 1.0 | 0.0 | 0.0 | 2.0 | 0.33 | 0.0 | 1.0 | 0.0 | 0.0 | 2.0 | 0.33 |
| Prados J | 2015 | 0.0 | 0.5 | 0.0 | 1.0 | 0.0 | 1.0 | 2.5 | 0.42 | 0.0 | 1.0 | 0.0 | 1.0 | 2.5 | 0.42 | 1.0 | 1.0 | 0.0 | 1.0 | 3.5 | 0.58 |
| Provenzi L | 2017 | 1.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 3.0 | 0.50 | 0.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.33 | 0.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.33 |
| Radtke KM | 2015 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 1.0 | 3.0 | 0.50 | 0.0 | 1.0 | 0.0 | 1.0 | 3.0 | 0.50 | 1.0 | 1.0 | 0.0 | 1.0 | 4.0 | 0.67 |
| Radtke KM | 2011 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 2.0 | 0.33 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.17 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.17 |
| Romens SE | 2015 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 2.0 | 0.33 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.17 | 1.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.33 |
| Smith JA | 2017 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 2.0 | 0.33 | 0.0 | 1.0 | 0.0 | 1.0 | 2.0 | 0.33 | 0.0 | 1.0 | 0.0 | 1.0 | 2.0 | 0.33 |
| Steiger H | 2013 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 2.0 | 0.33 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.17 | 1.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.33 |
| Suderman M | 2014 | 0.0 | 0.5 | 0.0 | 1.0 | 0.0 | 1.0 | 2.5 | 0.42 | 0.0 | 0.0 | 0.0 | 1.0 | 1.5 | 0.25 | 1.0 | 0.0 | 0.0 | 1.0 | 2.5 | 0.42 |
| Tehranifar P | 2013 | 0.0 | 0.5 | 0.0 | 0.0 | 0.0 | 0.0 | 0.5 | 0.08 | 0.0 | 0.0 | 0.0 | 0.0 | 0.5 | 0.08 | 1.0 | 0.0 | 0.0 | 0.0 | 1.5 | 0.25 |
| Tyrka AR | 2012 | 0.0 | 1.0 | 0.5 | 1.0 | 0.0 | 1.0 | 3.5 | 0.58 | 0.0 | 1.0 | 0.0 | 0.0 | 2.0 | 0.33 | 1.0 | 1.0 | 0.0 | 0.0 | 3.0 | 0.50 |
| Tyrka AR | 2016 | 0.0 | 0.5 | 0.0 | 1.0 | 0.0 | 1.0 | 2.5 | 0.42 | 0.0 | 1.0 | 0.0 | 0.0 | 1.5 | 0.25 | 1.0 | 1.0 | 0.0 | 0.0 | 2.5 | 0.42 |
| Unternaehrer E | 2015 | 0.0 | 1.0 | 1.0 | 1.0 | 0.0 | 0.0 | 3.0 | 0.50 | 0.0 | 1.0 | 0.0 | 0.0 | 2.0 | 0.33 | 0.0 | 1.0 | 0.0 | 1.0 | 3.0 | 0.50 |
| van der Knaap LJ | 2014 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 1.0 | 3.0 | 0.50 | 0.0 | 1.0 | 0.0 | 0.0 | 2.0 | 0.33 | 0.5 | 1.0 | 0.0 | 0.0 | 2.5 | 0.42 |
| Wankerl M | 2014 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 2.0 | 0.33 | 0.0 | 1.0 | 0.0 | 0.0 | 2.0 | 0.33 | 1.0 | 1.0 | 0.0 | 0.0 | 3.0 | 0.50 |
| Yehuda R | 2014 | 0.0 | 0.5 | 0.0 | 1.0 | 0.0 | 1.0 | 2.5 | 0.42 | 0.5 | 1.0 | 0.0 | 0.0 | 2.0 | 0.33 | 0.0 | 1.0 | 0.0 | 0.0 | 1.5 | 0.25 |
| Average Model Fit: Match Score, % Match | 2.8 | 0.46 | 2.3 | 0.38 | 2.6 | 0.44 | |||||||||||||||
Note. For each empirical paper, the match between the preclinical models and the study procedures is provided, coded as described in the method section. In the first unshaded vertical section, the match between ELS age and EPI age is provided as a single number with reference to all three preclinical models, because the comparison preclinical models do not differ from each other on these factors as we have defined them. In the next three vertical sections, we provide the match between each of the three models and the empirical paper with respect to ELS type, sex of the sampled individual, epigenetic tissue source and location (EPI Source & Loc; for example, brain vs. blood, and within brain, hippocampal vs. prefrontal), epigenetic target (EPI Target, for example NR3C1), the match score, which is a sum of their match on each of the six match criteria, and the resultant percent match between that empirical paper and each preclinical model.
Postmortem brain tissue.
Five papers met the search criteria and used post-mortem human brain tissue (top row of Tables 2 and 3, Labonte et al., 2012a 2012b, McGowan, et al., 2009, Nemoda et al., 2015, Suderman et al., 2012). Upon classification, it appears there may be substantial overlap in the subjects used in all five papers, which all use hippocampal brain tissue from the Quebec Suicide Brain Bank from Caucasian males of French Canadian decent and rely on postmortem proxy interviews to establish early histories of abuse, neglect or maternal depression. However, sample sizes range from 12–25 suicide completers with early life adversity, suggesting the samples are at most only partially overlapping in exact tissue samples. Although there are other studies evaluating epigenetic markers within human brain tissue (Puglia, Lillard, Morris, & Connelley, 2015, Wockner et al., 2014), none of those that emerged from our search also include measures of ELS. Overall, these five studies of the Quebec Suicide Brain Bank fit 1.5–4 of the six evaluated study match criteria, resulting in a 25–67% match rating. Therefore, even when the studies were matched on tissue type, they were not matched on most other criteria (due in part to limitations inherent in postmortem studies), limiting translational specificity. We note, however, that by examining DNA methylation locations specific to studies of maltreatment (or by using EWAS) specificity could be increased.
Buccal cells.
Nine papers met the search criteria and used buccal cells (second section of Tables 2 and 3). Three of these examined samples with ELS occurring from months to years in duration and with broad measures of adversity (duration in institutional care, Adverse Childhood Experiences (ACE score)) three evaluated the effects of maternal depression or parental stress early in life (2) or in infancy/preschool (1), two similar samples evaluated risk status in preterm infants recruited from a neonatal intensive care unit in Rhode Island, and the final evaluated infant tactile stimulation at 5 postnatal weeks. Because animal models examine targeted exposures to ELS, studies with broad ELS exposure windows and downstream DNA methylation assessments up to many years later lack translational specificity. Two studies used a EWAS approach, one with the 27K beadchip and one with the 450K beadchip; five examined NR3C1 and four of these also looked at other candidate loci (2 including BDNF), one looked at SLC6A4 and FKBP5, and one looked at MT-ND6. Thus, studies ranged in their specificity regarding epigenetic target loci. Overall, these studies fit 1.5–5 of the six evaluated study match criteria, resulting in a 25–83% match rating. The highest rating was achieved only for the two studies that examined postnatal positive maternal caregiving behaviors and otherwise matched the LG model versus using the LG model to understand models of early adversity. More work examining positive caregiving is badly needed to fully examine the cross-species replication of the LG model, and when separation or maltreatment is the human ELS, inclusion of AVP and BDNF (and ideally via EWAS) would increase translational specificity.
Saliva samples.
Ten papers met the search criteria and used saliva (third section of tables 2 and 3). Nine examined samples with ELS occurring from months to years in duration (four of which were from the same project, although sample sizes vary across papers), and one evaluated prenatal and perinatal stress. Seven studies examined NR3C1 explicitly; one of these also looked at the MAOA gene and one also used an EWAS approach. One study used an EWAS approach, with the 450K beadchip. One study focused exclusively on OXT-related genes, and the final study on the serotonin transporter gene (HTR2A). Similarly to studies with buccal cells, EWAS improves the opportunity for translational match across models, as does time-limited ELS exposure and epigenetic measurement closely following ELS. Overall, these studies fit 1.5–4 of the six evaluated study match criteria, resulting in 25–67% match rating.
Blood samples.
The majority of human studies (39) used blood (bottom row of tables 2 and 3), typically whole blood, though there is an increasing focus on accounting for cell type which is not common in the early work. Of these, in 21 the ELS included childhood maltreatment (abuse, neglect, or “trauma”, which included both family upheaval and abuse). In six papers, the primary ELS was exposure to low socioeconomic status, sometimes measured as maternal education, parental occupation, and in one case as neighborhood disadvantage, and SES was included in an additional three papers also examining maltreatment. In four studies, parental/maternal care differences was the ELS/experience measured. In three studies the ELS was maternal separation or orphanage care, for two it included prenatal stress exposure, and one each intrauterine growth restriction, prenatal genocide exposure, parental PTSD, and maternal intimate partner violence exposure. Some type of EWAS was common (n=13, 33%) with number of loci increasing as more sophisticated beadchips became available. However, none published to date use the currently recommended 850K beadchip. Sixteen (41%) also specifically discuss NR3C1. Of the remaining ten, four targeted serotonin genes, two BDNF (one of these also included OXT), one OXT, and the remaining three less common targets in ELS research. Overall, these studies fit .5–4 of the six evaluated study match criteria, resulting in 8–67% match rating. This lower match is largely driven by the fact that so many studies examined severe adversity and focused on NR3C1.
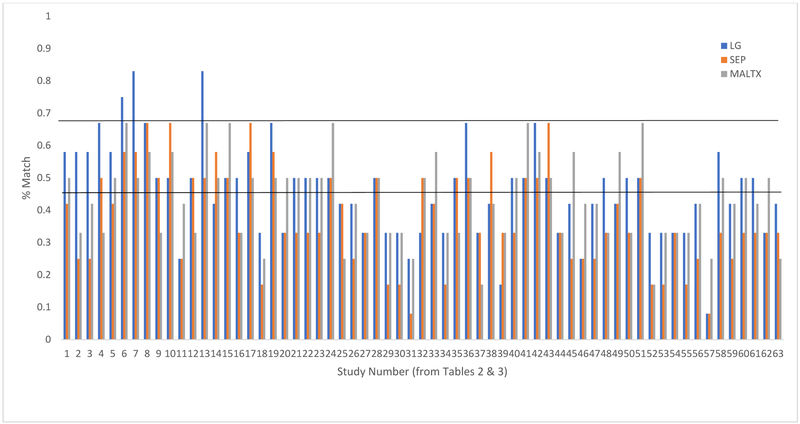
Across tissue types, match between human studies and each preclinical model was less than 50% on average, (LG=46%, SEP=38%, MALTX=44%), with a range of 8% to 83% (see Figure 2 for an illustration of these results). Rarely does the match exceed 50% (22%, 14% and 10% of the time, for each preclinical model respectively), and in only two cases did the match exceed 75%. See Table 3 for the fit across models.
Figure 2.
Model fit between human studies of ELS and epigenetics with three animal models. Note. For each empirical paper, a bar indicates the match with each of three animal models as described in Table 3.
Discussion
Several interesting results emerged from this systematic classification. Our review highlighted the degree to which human studies have missed opportunities for translational relevance, with an overreliance on the licking/grooming model even in recent work examining maltreatment (Tyrka et al., 2015) or separation (Kantake et al., 2014). This overreliance is further overly dependent on the original Weaver et al., 2004 study which only examined methylation of NR3C1 (and subsequent histone acetylation and NGFI-A binding) versus taking into account later data suggesting that widespread methylation across chromosome 18 may be a consequence of differences in maternal licking and grooming in the first week of life (McGowan, et al., 2011). In most cases when a candidate locus was used and there was a model mismatch it occurred when NR3C1 methylation status was assessed in participants with maltreatment history. However, the opposite mismatch also occurred, for example BDNF assessed in participants as a function of positive parenting, (Unternaehrer, et al., 2016). Immediate steps using existing EWAS data could increase specificity by testing for AVP and/or BDNF as appropriate for the ELS experienced, as well as allowing emergence of novel loci.
Systematic attention to timing of ELS has been grossly underspecified in most human studies, despite clear evidence that timing matters in both preclinical models (where in fact ELS must sometimes occur in the first postnatal week to impact epigenetic signatures) and human studies (where consequence of ELS extend at least to stress experienced in the first several postnatal years). Relatedly, the timing of the epigenetic sample is very important, and an understanding of when and how methylation patterns change across species is badly needed. There is some evidence, for example, that in rodents at PND1, no individual differences in methylation can be detected (Weaver et al., 2004), while in humans a number of studies have documented individual epigenetic differences in cord blood (Oberlander et al., 2008). Twin studies also reveal rapid changes in early life (Martino et al., 2013). This is a particularly important area for future work, as the promise of prevention lies in knowing when methylation differences occur so as to intervene in a timely fashion. Simple cross-species age-equivalents are clearly inappropriate given these data (stress experienced for seven days early in the life of a rat for example, would be generously extended to the equivalent of 6 or 9 postnatal human months). Both animal and human studies could contribute to this question by including multiple sampling time points of both ELS and DNA methylation, and in animal work by examining DNA methylation changes association with experimental variation of the timing and duration of ELS.
The human literature is predominantly cross-sectional. Longitudinal studies are a common tool in human developmental studies and could be exploited here to identify when differences emerge and when (and how) they are or can be ameliorated. Indeed, only one paper in this review measured change in DNA methylation (Parent et al., 2017; saliva sample), and the results are opposite to the interpretation usually made from similar single time point data (decreasing and lower time 2 methylation for the maltreated children vs. non-maltreated children at NR3C1 exons 1D and 1F). The human studies also nearly entirely lack attempts at intervention, pharmacologically, as is done in animal models, but also behaviorally – despite the literally hundreds of commentaries and reviews that include reference to this possibility (e.g. Heim & Binder, 2012; Szyf & Bick, 2013, Tureki & Meaney, 2016).
Epigenome-wide association studies are underrepresented in both animal and human work, despite a long history with many of these same genes suggesting that a candidate approach is perhaps inefficient or ineffective for predicting complex behaviors or disease. If both animal and human work systematically included EWAS approaches (and considered making this information publicly available) progress could be accelerated. Indeed, again referring back to the literature on candidate genes, repositories with very large samples may be needed to detect small but possibly important differences, suggesting that systematic efforts to collect data (even in free-standing projects) that could be deposited collectively may be an important step to consider now.
Differences in tissue type could also be more readily understood if multiple tissue types were collected in human and animal work. Indeed, if animal models were to systematically evaluate the correlation between brain and blood DNA methylation across the epigenome, (particularly with attention to blood cell type), match ratings would go up for over half of the published work and our ability to determine cross-species replication would be quickly improved. Although more difficult, replication analyses across tissues within human studies have also been published (for a blood-brain example, see Houtepen, 2016).
Relatedly, genotypes of human participants under study are far too infrequently considered. This is problematic in two ways – some genotypes are more susceptible to developmental stress and may be more readily promoted or silenced via epigenetic processes. Most work also focuses on DNA methylation (see Mitchell, Schneper & Notterman, 2016 for an exceptionally clear description of the biology of methylation), and our search criteria restricted our review to evaluation of the DNA methylation literature. Other important epigenetic processes are likely at play (see for example, recent work on the epigenetics of telomeres, Blaze, Asok & Roth, 2015), and efforts to align the human and preclinical work on these processes is also needed.
Sex differences are also largely ignored despite the fact that all three preclinical models have robust sex differences. Increased specification and systematic expansion in breadth is critical to realize the promise of this new mechanism for improving human health and well-being. Existing human data sets could systematically examine sex differences to readily contribute to this question. Because of power, repositories may be needed to fully appreciate individual differences, including sex. Human work would also benefit from greater attention to diversity with regard to race and ethnicity, perhaps even considering modeling of ancestral genetic differences (e.g. Parent, et al., 2017).
Within animal models of ELS, comparison across species may also be fruitful. While there is some evidence for similarity across species, especially among rodents, and on occasion remarkably across species as divergent as fish (McGhee & Bell, 2014), sometimes patterns are confusingly divergent within a species (Long-Evans vs. Sprague-Dawley rats; Jawahar, Murgatroyd, Harrison & Baune, 2015) or more similar between rodents and humans (e.g. Dolle Molle et al., 2012) but not with other primates (Kinnally, et al., 2011). Careful attention to species when making comparisons would aid in specificity and help to work toward an understanding of underlying differences.
Finally, although beyond the scope of this review, similar important parallels could be drawn with preclinical models of prenatal epigenetic changes following stress and the large human literature on epigenetics in placental, cord blood and maternal blood at birth. For example, the highly cited paper by Oberlander and colleagues on NR3C1 methylation in cord blood of newborns who experienced exposure to maternal prenatal depression (Oberlander et al., 2008) could be evaluated for its match to preclinical models of prenatal stress and epigenetic modification vs. to postnatal epigenetic models We did not include such work, in part because data comparing tissues from the same infants suggests methylation patterns in placenta, cord blood, and saliva may be weakly or negatively correlated (Armstrong et al., 2014; Ollikainen et al., 2010). A review of studies of placental and cord blood would, however, be particularly helpful for untangling age parallels between human and rodent newborns with regard to timing of prenatal and postnatal stress and methylation changes.
In conclusion, immediate steps with existing data sets could increase match specificity by examining the putative loci from samples with EWAS data. New or existing blood samples from well-studied animal models could be evaluated to clearly establish correlations between blood cell type methylation and brain region methylation in prominent genes. Animal models could also utilize EWAS technology much more frequently as is now common in human work. With regard to new study design, greater specificity in timing of both ELS exposure and epigenetic sampling, and including more than one epigenetic sample (across tissues and within tissues across time) would be of great benefit. Indeed, the great interest in DNA methylation as patterning that is influenced by experience and then relatively stable across time could and should be systematically evaluated across time, tissue, and loci. This systematic review suggests that we are underutilizing the power of three well-established ELS animal models due to insufficient matching between methods in the models and in human work. As is central to the tradition of ISDP, we encourage these steps, especially in this subfield which has been the focus of the work of so many ISDP members across decades.
Contributor Information
Sarah Watamura, University of Denver, Psychology.
Tania Roth, University of Delaware, Psychology.
References
- Armstrong DA, Lesseur C, Conradt E, Lester BM, & Marsit CJ (2014). Global and gene-specific DNA methylation across multiple tissues in early infancy: implications for children’s health research. The FASEB Journal, 28(5), 2088–2097. 10.1096/fj.13-238402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bale TL (2014). Lifetime stress experience: transgenerational epigenetics and germ cell programming. Dialogues in clinical neuroscience, 16(3), 297–305. PMC4214173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaze J, Asok A, & Roth TL (2015). The long-term impact of adverse caregiving environments on epigenetic modifications and telomeres. Frontiers in Behavioral Neuroscience, 9, 79 10.3389/fnbeh.2015.00079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghol N, Suderman M, McArdle W, Racine A, Hallett M, Pembrey M, … & Szyf M (2011). Associations with early-life socio-economic position in adult DNA methylation. International journal of epidemiology, 41(1), 62–74. doi: 10.1093/ije/dyr147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braithwaite EC, Kundakovic M, Ramchandani PG, Murphy SE, & Champagne FA (2015). Maternal prenatal depressive symptoms predict infant NR3C1 1F and BDNF IV DNA methylation. Epigenetics, 10(5), 408–417. doi.org/ 10.1080/15592294.2015.1039221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante AC, Aiello AE, Galea S, Ratanatharathorn A, Noronha C, Wildman DE, & Uddin M (2016). Glucocorticoid receptor DNA methylation, childhood maltreatment and major depression. Journal of affective disorders, 206, 181–188. doi: 10.1016/j.jad.2016.07.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldji C, Tannenbaum B, Sharma S, Francis D, Plotsky PM, & Meaney MJ (1998). Maternal care during infancy regulates the development of neural systems mediating the expression of fearfulness in the rat. Proceedings of the National Academy of Sciences, 95(9), 5335–5340. PMCID: PMC20261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canadian Pediatric Society (2004). Weaning from the breast. Paediatrics & Child Health, 9(4), 249–253. doi: 10.1093/pch/9.4.249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao-Lei L, Elgbeili G, Massart R, Laplante DP, Szyf M, & King S (2015). Pregnant women’s cognitive appraisal of a natural disaster affects DNA methylation in their children 13 years later: Project Ice Storm. Translational psychiatry, 5(2), e515. doi: 10.1038/tp.2015.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao-Lei L, Massart R, Suderman MJ, Machnes Z, Elgbeili G, Laplante DP, … & King S (2014). DNA methylation signatures triggered by prenatal maternal stress exposure to a natural disaster: Project Ice Storm. PloS ONE, 9(9), e107653. doi: 10.1371/journal.pone.0107653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champagne FA, Weaver IC, Diorio J, Dymov S, Szyf M, & Meaney MJ (2006). Maternal care associated with methylation of the estrogen receptor-α1b promoter and estrogen receptor-α expression in the medial preoptic area of female offspring. Endocrinology, 147(6), 2909–2915. doi: 10.1210/en.2005-1119 [DOI] [PubMed] [Google Scholar]
- Champagne F, Diorio J, Sharma S, & Meaney MJ (2001). Naturally occurring variations in maternal behavior in the rat are associated with differences in estrogen-inducible central oxytocin receptors. Proceedings of the National Academy of Sciences, 98(22), 12736–12741. doi: 10.1073/pnas.221224598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicchetti D, & Handley ED (2017). Methylation of the glucocorticoid receptor gene, nuclear receptor subfamily 3, group C, member 1 (NR3C1), in maltreated and nonmaltreated children: Associations with behavioral undercontrol, emotional lability/negativity, and externalizing and internalizing symptoms. Development and Psychopathology, 29(5), 1795–1806. doi: 10.1017/S0954579417001407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conradt E, Hawes K, Guerin D, Armstrong DA, Marsit CJ, Tronick E, & Lester BM (2016). The contributions of maternal sensitivity and maternal depressive symptoms to epigenetic processes and neuroendocrine functioning. Child Development, 87(1), 73–85. doi: 10.1111/cdev.12483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalle Molle R, Portella AK, Goldani MZ, Kapczinski FP, Leistner-Segala S, Salum GA, … & Silveira PP (2012). Associations between parenting behavior and anxiety in a rodent model and a clinical sample: relationship to peripheral BDNF levels. Translational Psychiatry, 2(11), e195. doi: 10.1038/tp.2012.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels WMU, Fairbairn LR, Van Tilburg G, McEvoy CRE, Zigmond MJ, Russell VA, & Stein DJ (2009). Maternal separation alters nerve growth factor and corticosterone levels but not the DNA methylation status of the exon 1 7 glucocorticoid receptor promoter region. Metabolic Brain Disease, 24(4), 615 10.1007/s11011-009-9163-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty TS, Blaze J, Keller SM, & Roth TL (2017). Phenotypic outcomes in adolescence and adulthood in the scarcity-adversity model of low nesting resources outside the home cage. Developmental Psychobiology, 59(6), 703–714. 10.1002/dev.21547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube SR, Felitti VJ, Dong M, Giles WH, & Anda RF (2003). The impact of adverse childhood experiences on health problems: Evidence from four birth cohorts dating back to 1990. Preventive Medicine, 37, 268–277. doi: 10.1016/S0091-7435(03)00123-3 [DOI] [PubMed] [Google Scholar]
- Duman EA, & Canli T (2015). Influence of life stress, 5-HTTLPR genotype, and SLC6A4 methylation on gene expression and stress response in healthy Caucasian males. Biology of Mood & Anxiety Disorders, 5(1), 2. doi: 10.1186/s13587-015-0017-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efstathopoulos P, Andersson F, Melas PA, Yang LL, Villaescusa JC, Rȕegg J, … & Lavebratt C (2018). NR3C1 hypermethylation in depressed and bullied adolescents. Translational Psychiatry, 8(1), 121 DOI 10.1038/s41398-018-0169-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essex MJ, Thomas Boyce W, Hertzman C, Lam LL, Armstrong JM, Neumann S, & Kobor MS (2013). Epigenetic vestiges of early developmental adversity: childhood stress exposure and DNA methylation in adolescence. Child Development, 84(1), 58–75. doi: 10.1111/j.1467-8624.2011.01641.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell C, Doolin K, O’Leary N, Jairaj C, Roddy D, Tozzi L, … & Szyf M (2018). DNA methylation differences at the glucocorticoid receptor gene in depression are related to functional alterations in hypothalamic–pituitary–adrenal axis activity and to early life emotional abuse. Psychiatry research, 265, 341–348. doi.org/ 10.1016/j.psychres.2018.04.064 [DOI] [PubMed] [Google Scholar]
- Felitti VJ, Anda RF, Nordenberg D, Williamson DF, Spitz AM, Edwards V,…Marks JS (1998). Relationship of childhood abuse and household dysfunction to many of the leading causes of death in adults. American Journal of Preventive Medicine, 14(4), 245–258. doi: 10.1016/S0749-3797(98)00017-8 [DOI] [PubMed] [Google Scholar]
- Giarraputo J, DeLoach J, Padbury J, Uzun A, Marsit C, Hawes K, & Lester B (2016). Medical morbidities and DNA methylation of NR3C1 in preterm infants. Pediatric Research, 81(1–1), 68. doi: 10.1038/pr.2016.185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitik M, Holliday ED, Leung M, Yuan Q, Logue SF, Tikkanen R, … & Gould TJ (2018). Choline ameliorates adult learning deficits and reverses epigenetic modification of chromatin remodeling factors related to adolescent nicotine exposure. Neurobiology of learning and memory. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouin JP, Zhou QQ, Booij L, Boivin M, Côté SM, Hébert M, … & Vitaro F (2017). Associations among oxytocin receptor gene (OXTR) DNA methylation in adulthood, exposure to early life adversity, and childhood trajectories of anxiousness. Scientific Reports, 7(1), 7446. doi: 10.1038/s41598-017-07950-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim C, & Binder EB (2012). Current research trends in early life stress and depression: Review of human studies on sensitive periods, gene–environment interactions, and epigenetics. Experimental Neurology, 233(1), 102–111. doi: 10.1016/S0749-3797(98)00017-8 [DOI] [PubMed] [Google Scholar]
- Houtepen LC, Vinkers CH, Carrillo-Roa T, Hiemstra M, Van Lier PA, Meeus W,… & Schalkwyk LC (2016). Genome-wide DNA methylation levels and altered cortisol stress reactivity following childhood trauma in humans. Nature Communications, 7, 10967. doi: 10.1038/ncomms10967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janusek LW, Tell D, Gaylord-Harden N, & Mathews HL (2017). Relationship of childhood adversity and neighborhood violence to a proinflammatory phenotype in emerging adult African American men: an epigenetic link. Brain, Behavior, and Immunity, 60, 126–135. .doi.org/ 10.1016/j.bbi.2016.10.006 [DOI] [PubMed] [Google Scholar]
- Jawahar MC, Murgatroyd C, Harrison EL, & Baune BT (2015). Epigenetic alterations following early postnatal stress: a review on novel aetiological mechanisms of common psychiatric disorders. Clinical Epigenetics, 7(1), 122. doi: 10.1186/s13148-015-0156-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanherkar RR, Bhatia-Dey N, & Csoka AB (2014). Epigenetics across the human lifespan. Frontiers in Cell and Developmental Biology, 2, 49. doi: 10.3389/fcell.2014.00049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantake M, Ohkawa N, Iwasaki T, Ikeda N, Awaji A, Saito N, … & Shimizu T (2018). Postnatal relative adrenal insufficiency results in methylation of the glucocorticoid receptor gene in preterm infants: a retrospective cohort study. Clinical Epigenetics, 10(1), 66. doi.org/ 10.1186/s13148-018-0497-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantake M, Yoshitake H, Ishikawa H, Araki Y, & Shimizu T (2014). Postnatal epigenetic modification of glucocorticoid receptor gene in preterm infants: a prospective cohort study. BMJ Open, 4(7), e005318. doi: 10.1136/bmjopen-2014-005318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khulan B, Manning JR, Dunbar DR, Seckl JR, Raikkonen K, Eriksson JG, & Drake AJ (2014). Epigenomic profiling of men exposed to early-life stress reveals DNA methylation differences in association with current mental state. Translational Psychiatry, 4(9), e448. doi: 10.1038/tp.2014.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King L, Robins S, Chen G, Yerko V, Zhou Y, Nagy C, … & Zelkowitz P (2017). Perinatal depression and DNA methylation of oxytocin-related genes: a study of mothers and their children. Hormones and Behavior, 96, 84–94. doi: 10.1016/j.yhbeh.2017.09.006 [DOI] [PubMed] [Google Scholar]
- Kinnally EL, Feinberg C, Kim D, Ferguson K, Leibel R, Coplan JD, & Mann JJ (2011). DNA methylation as a risk factor in the effects of early life stress. Brain, Behavior, and Immunity, 25(8), 1548–1553. doi: 10.1016/j.bbi.2011.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodkin MH, & Auger AP (2011). Sex difference in the expression of DNA methyltransferase 3a in the rat amygdala during development. Journal of Neuroendocrinology, 23(7), 577–583. doi: 10.1111/j.1365-2826.2011.02147.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumsta R, Marzi SJ, Viana J, Dempster EL, Crawford B, Rutter M, … & Sonuga-Barke EJ (2016). Severe psychosocial deprivation in early childhood is associated with increased DNA methylation across a region spanning the transcription start site of CYP2E1. Translational Psychiatry, 6(6), e830. doi: 10.1038/tp.2016.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundakovic M, & Champagne FA (2015). Early-life experience, epigenetics, and the developing brain. Neuropsychopharmacology, 40(1), 141. doi: 10.1038/npp.2014.140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labonté B, Suderman M, Maussion G, Navaro L, Yerko V, Mahar I, … & Turecki G (2012). Genome-wide epigenetic regulation by early-life trauma. Archives of General Psychiatry, 69(7), 722–731. doi: 10.1001/archgenpsychiatry.2011.2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labonté B, Yerko V, Gross J, Mechawar N, Meaney MJ, Szyf M, & Turecki G (2012). Differential glucocorticoid receptor exon 1B, 1C, and 1H expression and methylation in suicide completers with a history of childhood abuse. Biological Psychiatry, 72(1), 41–48. doi: 10.1016/j.biopsych.2012.01.034 [DOI] [PubMed] [Google Scholar]
- Lam LL, Emberly E, Fraser HB, Neumann SM, Chen E, Miller GE, & Kobor MS (2012). Factors underlying variable DNA methylation in a human community cohort. Proceedings of the National Academy of Sciences, 109(Supplement 2), 17253–17260. doi/ 10.1073/pnas.1121249109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapp HE, Ahmed S, Moore CL, & Hunter RG (2018). Toxic stress history and hypothalamic-pituitary-adrenal axis function in a social stress task: Genetic and epigenetic factors. Neurotoxicology and Teratology. doi.org/ 10.1016/j.ntt.2018.01.011 [DOI] [PubMed] [Google Scholar]
- Lester BM, Marsit CJ, Giarraputo J, Hawes K, LaGasse LL, & Padbury JF (2015). Neurobehavior related to epigenetic differences in preterm infants. Epigenomics, 7(7), 1123–1136. doi: 10.2217/epi.15.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine ME, Cole SW, Weir DR, & Crimmins EM (2015). Childhood and later life stressors and increased inflammatory gene expression at older ages. Social Science & Medicine, 130, 16–22. doi: 10.1016/j.socscimed.2015.01.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Diorio J, Tannenbaum B, Caldji C, Francis D, Freedman A, … & Meaney MJ (1997). Maternal care, hippocampal glucocorticoid receptors, and hypothalamic-pituitary-adrenal responses to stress. Science, 277(5332), 1659–1662. [DOI] [PubMed] [Google Scholar]
- Marmot MG, Stansfeld S, Patel C, North F, Head J, White I, … & Smith GD (1991). Health inequalities among British civil servants: the Whitehall II study. The Lancet, 337(8754), 1387–1393. [DOI] [PubMed] [Google Scholar]
- Martín-Blanco A, Ferrer M, Soler J, Salazar J, Vega D, Andión O, … & Pérez V (2014). Association between methylation of the glucocorticoid receptor gene, childhood maltreatment, and clinical severity in borderline personality disorder. Journal of Psychiatric Research, 57, 34–40. doi: 10.1016/j.jpsychires.2014.06.011 [DOI] [PubMed] [Google Scholar]
- Martino D, Loke YJ, Gordon L, Ollikainen M, Cruickshank MN, Saffery R, & Craig JM (2013). Longitudinal, genome-scale analysis of DNA methylation in twins from birth to 18 months of age reveals rapid epigenetic change in early life and pair-specific effects of discordance. Genome Biology, 14(5), R42. doi: 10.1186/gb-2013-14-5-r42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzi SJ, Sugden K, Arseneault L, Belsky DW, Burrage J, Corcoran DL, … & Odgers CL (2018). Analysis of DNA methylation in young people: limited evidence for an association between victimization stress and epigenetic variation in blood. American Journal of Psychiatry, 175(6), 517–529. doi: 10.1176/appi.ajp.2017.17060693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy MM, & Nugent BM (2015). At the frontier of epigenetics of brain sex differences. Frontiers in Behavioral Neuroscience, 9, 221. doi: 10.3389/fnbeh.2015.00221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee KE, & Bell AM (2014). Paternal care in a fish: epigenetics and fitness enhancing effects on offspring anxiety. Proceedings of the Royal Society B, 281(1794), 20141146. doi: 10.1098/rspb.2014.1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan PO, Sasaki A, D’alessio AC, Dymov S, Labonté B, Szyf M, … & Meaney MJ (2009). Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nature Neuroscience, 12(3), 342. doi: 10.1038/nn.2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan PO, Suderman M, Sasaki A, Huang TC, Hallett M, Meaney MJ, & Szyf M (2011). Broad epigenetic signature of maternal care in the brain of adult rats. PloS ONE, 6(2), e14739. doi: 10.1371/journal.pone.0014739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melas PA, Wei Y, Wong CC, Sjöholm LK, Åberg E, Mill J, … & Lavebratt C (2013). Genetic and epigenetic associations of MAOA and NR3C1 with depression and childhood adversities. International Journal of Neuropsychopharmacology, 16(7), 1513–1528. doi: 10.1017/S1461145713000102 [DOI] [PubMed] [Google Scholar]
- Mitchell C, Schneper LM, & Notterman DA (2015). DNA methylation, early life environment, and health outcomes. Pediatric Research, 79(1–2), 212. doi: 10.1038/pr.2015.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore SR, McEwen LM, Quirt J, Morin A, Mah SM, Barr RG, … & Kobor MS (2017). Epigenetic correlates of neonatal contact in humans. Development and Psychopathology, 29(5), 1517–1538. doi: 10.1017/S0954579417001213 [DOI] [PubMed] [Google Scholar]
- Murgatroyd C, Patchev AV, Wu Y, Micale V, Bockmühl Y, Fischer D, … & Spengler D (2009). Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nature Neuroscience, 12(12), 1559. doi: 10.1038/nn.2436 [DOI] [PubMed] [Google Scholar]
- Murgatroyd C, Quinn JP, Sharp HM, Pickles A, & Hill J (2015). Effects of prenatal and postnatal depression, and maternal stroking, at the glucocorticoid receptorgene. Translational Psychiatry, 5(5), e560. doi: 10.1038/tp.2014.140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumova OY, Hein S, Suderman M, Barbot B, Lee M, Raefski A, … & Grigorenko EL (2016). Epigenetic patterns modulate the connection between developmental dynamics of parenting and offspring psychosocial adjustment. Child Development, 87(1), 98–110. doi: 10.1111/cdev.12485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumova OY, Lee M, Koposov R, Szyf M, Dozier M, & Grigorenko EL (2012). Differential patterns of whole-genome DNA methylation in institutionalized children and children raised by their biological parents. Development and Psychopathology, 24(1), 143–155. doi: 10.1017/S0954579411000605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needham BL, Smith JA, Zhao W, Wang X, Mukherjee B, Kardia SL, … & Diez Roux AV (2015). Life course socioeconomic status and DNA methylation in genes related to stress reactivity and inflammation: The multi-ethnic study of atherosclerosis. Epigenetics, 10(10), 958–969. doi: 10.1080/15592294.2015.1085139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoda Z, Massart R, Suderman M, Hallett M, Li T, Coote M, … & Steiner M (2015). Maternal depression is associated with DNA methylation changes in cord blood T lymphocytes and adult hippocampi. Translational Psychiatry, 5(4), e545. doi: 10.1038/tp.2015.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Non AL, Hollister BM, Humphreys KL, Childebayeva A, Esteves K, Zeanah CH, … & Drury SS (2016). DNA methylation at stress-related genes is associated with exposure to early life institutionalization. American Journal of Physical Anthropology, 161(1), 84–93. doi: 10.1002/ajpa.23010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent BM, Wright CL, Shetty AC, Hodes GE, Lenz KM, Mahurkar A, … & McCarthy MM (2015). Brain feminization requires active repression of masculinization via DNA methylation. Nature Neuroscience, 18(5), 690. doi: 10.1038/nn.3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberlander TF, Weinberg J, Papsdorf M, Grunau R, Misri S, & Devlin AM (2008). Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses. Epigenetics, 3(2), 97–106. PMID:18536531 [DOI] [PubMed] [Google Scholar]
- Ollikainen M, Smith KR, Joo EJH, Ng HK, Andronikos R, Novakovic B, … & Craig JM (2010). DNA methylation analysis of multiple tissues from newborn twins reveals both genetic and intrauterine components to variation in the human neonatal epigenome. Human Molecular Genetics, 19(21), 4176–4188. doi: 10.1093/hmg/ddq336 [DOI] [PubMed] [Google Scholar]
- Parade SH, Ridout KK, Seifer R, Armstrong DA, Marsit CJ, McWilliams MA, & Tyrka AR (2016). Methylation of the glucocorticoid receptor gene promoter in preschoolers: Links with internalizing behavior problems. Child Development, 87(1), 86–97. doi: 10.1111/cdev.12484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parade SH, Novick AM, Parent J, Seifer R, Klaver SJ, Marsit CJ, … & Tyrka AR (2017). Stress exposure and psychopathology alter methylation of the serotonin receptor 2A (HTR2A) gene in preschoolers. Development and psychopathology, 29(5), 1619–1626. doi: 10.1017/S0954579417001274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent J, Parade SH, Laumann LE, Ridout KK, Yang BZ, Marsit CJ, … & Tyrka AR (2017). Dynamic stress-related epigenetic regulation of the glucocorticoid receptor gene promoter during early development: the role of child maltreatment. Development and Psychopathology, 29(5), 1635–1648. doi: 10.1017/S0954579417001298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Zhu Y, Strachan E, Fowler E, Bacus T, Roy-Byrne P, … & Zhao J (2018). Childhood Trauma, DNA Methylation of Stress-related Genes, and Depression: Findings from Two Monozygotic Twin Studies. Psychosomatic Medicine. 10.1097/PSY.0000000000000604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perroud N, Paoloni-Giacobino A, Prada P, Olié E, Salzmann A, Nicastro R, … & Huguelet P (2011). Increased methylation of glucocorticoid receptor gene (NR3C1) in adults with a history of childhood maltreatment: a link with the severity and type of trauma. Translational Psychiatry, 1(12), e59. doi: 10.1038/tp.2011.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perroud N, Rutembesa E, Paoloni-Giacobino A, Mutabaruka J, Mutesa L, Stenz L, … & Karege F (2014). The Tutsi genocide and transgenerational transmission of maternal stress: epigenetics and biology of the HPA axis. The World Journal of Biological Psychiatry, 15(4), 334–345. doi.org/ 10.3109/15622975.2013.866693 [DOI] [PubMed] [Google Scholar]
- Perroud N, Zewdie S, Stenz L, Adouan W, Bavamian S, Prada P, … & Paoloni-Giacobino A (2016). Methylation of serotonin receptor 3A in ADHD, borderline personality, and bipolar disorders: link with severity of the disorders and childhood maltreatment. Depression and Anxiety, 33(1), 45–55. doi: 10.1002/da.22406 [DOI] [PubMed] [Google Scholar]
- Plotsky PM, & Meaney MJ (1993). Early, postnatal experience alters hypothalamic corticotropin-releasing factor (CRF) mRNA, median eminence CRF content and stress-induced release in adult rats. Molecular Brain Research, 18(3), 195–200. doi: 10.1016/0169-328X(93)90189-V [DOI] [PubMed] [Google Scholar]
- Prados J, Stenz L, Courtet P, Prada P, Nicastro R, Adouan W, … & Perroud N (2015). Borderline personality disorder and childhood maltreatment: A genome-wide methylation analysis. Genes, Brain and Behavior, 14(2), 177–188. doi: 10.1111/gbb.12197 [DOI] [PubMed] [Google Scholar]
- Provenzi L, Fumagalli M, Giorda R, Morandi F, Sirgiovanni I, Pozzoli U, … & Montirosso R (2017). Maternal sensitivity Buffers the association between slc6a4 Methylation and socio-emotional stress response in 3-Month-Old Full Term, but not very Preterm infants. Frontiers in Psychiatry, 8, 171. doi: 10.3389/fpsyt.2017.00171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radtke KM, Schauer M, Gunter HM, Ruf-Leuschner M, Sill J, Meyer A, & Elbert T (2015). Epigenetic modifications of the glucocorticoid receptor gene are associated with the vulnerability to psychopathology in childhood maltreatment. Translational Psychiatry, 5(5), e571. doi: 10.1038/tp.2015.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romens SE, McDonald J, Svaren J, & Pollak SD (2015). Associations between early life stress and gene methylation in children. Child Development, 86(1), 303–309. doi: 10.1111/cdev.12270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth TL, & Sullivan RM (2005). Memory of early maltreatment: neonatal behavioral and neural correlates of maternal maltreatment within the context of classical conditioning. Biological Psychiatry, 57(8), 823–831. doi: 10.1016/j.biopsych.2005.01.032 [DOI] [PubMed] [Google Scholar]
- Roth TL, Lubin FD, Funk AJ, & Sweatt JD (2009). Lasting epigenetic influence of early-life adversity on the BDNF gene. Biological Psychiatry, 65(9), 760–769. doi: 10.1016/j.biopsych.2008.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta P (2013). The laboratory rat: relating its age with human’s. International Journal of Preventive Medicine, 4(6), 624–30. PMCID:PMC3733029 [PMC free article] [PubMed] [Google Scholar]
- Smith AK, Kilaru V, Klengel T, Mercer KB, Bradley B, Conneely KN, … & Binder EB (2015). DNA extracted from saliva for methylation studies of psychiatric traits: evidence tissue specificity and relatedness to brain. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics, 168(1), 36–44. doi: 10.1002/ajmg.b.32278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JA, Zhao W, Wang X, Ratliff SM, Mukherjee B, Kardia SL, … & Needham BL (2017). Neighborhood characteristics influence DNA methylation of genes involved in stress response and inflammation: The Multi-Ethnic Study of Atherosclerosis. Epigenetics, 12(8), 662–673. doi.org/ 10.1080/15592294.2017.1341026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiger H, Labonté B, Groleau P, Turecki G, & Israel M (2013). Methylation of the glucocorticoid receptor gene promoter in bulimic women: associations with borderline personality disorder, suicidality, and exposure to childhood abuse. International Journal of Eating Disorders, 46(3), 246–255. doi: 10.1002/eat.22113 [DOI] [PubMed] [Google Scholar]
- Suderman M, Borghol N, Pappas JJ, Pereira SMP, Pembrey M, Hertzman C, … & Szyf M (2014). Childhood abuse is associated with methylation of multiple loci in adult DNA. BMC Medical Genomics, 7(1), 13. doi: 10.1186/1755-8794-7-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suderman M, McGowan PO, Sasaki A, Huang TC, Hallett MT, Meaney MJ, … & Szyf M (2012). Conserved epigenetic sensitivity to early life experience in the rat and human hippocampus. Proceedings of the National Academy of Sciences, 109(Supplement 2), 17266–17272. doi/ 10.1073/pnas.1121260109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyf M, & Bick J (2013). DNA methylation: a mechanism for embedding early life experiences in the genome. Child Development, 84(1), 49–57. doi: 10.1111/j.1467-8624.2012.01793.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tehranifar P, Wu HC, Fan X, Flom JD, Ferris JS, Cho YH, … & Terry MB (2013). Early life socioeconomic factors and genomic DNA methylation in mid-life. Epigenetics, 8(1), 23–27. doi.org/ 10.4161/epi.22989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turecki G, & Meaney MJ (2016). Effects of the social environment and stress on glucocorticoid receptor gene methylation: a systematic review. Biological Psychiatry, 79(2), 87–96. doi: 10.1016/j.biopsych.2014.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrka AR, Parade SH, Eslinger NM, Marsit CJ, Lesseur C, Armstrong DA, … & Seifer R (2015). Methylation of exons 1 D, 1 F, and 1 H of the glucocorticoid receptor gene promoter and exposure to adversity in preschool-aged children. Development and Psychopathology, 27(2), 577–585. doi: 10.1017/S0954579415000176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrka AR, Parade SH, Welch ES, Ridout KK, Price LH, Marsit C, … & Carpenter LL (2016). Methylation of the leukocyte glucocorticoid receptor gene promoter in adults: associations with early adversity and depressive, anxiety and substance-use disorders. Translational Psychiatry, 6(7), e848. doi: 10.1038/tp.2016.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrka AR, Price LH, Marsit C, Walters OC, & Carpenter LL (2012). Childhood adversity and epigenetic modulation of the leukocyte glucocorticoid receptor: preliminary findings in healthy adults. PloS ONE, 7(1), e30148. doi: 10.1371/journal.pone.0030148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unternaehrer E, Meyer AH, Burkhardt SC, Dempster E, Staehli S, Theill N, … & Meinlschmidt G (2015). Childhood maternal care is associated with DNA methylation of the genes for brain-derived neurotrophic factor (BDNF) and oxytocin receptor (OXTR) in peripheral blood cells in adult men and women. Stress, 18(4), 451–461. doi.org/ 10.3109/10253890.2015.1038992 [DOI] [PubMed] [Google Scholar]
- van der Knaap LJ, Oldehinkel AJ, Verhulst FC, van Oort FV, & Riese H (2015). Glucocorticoid receptor gene methylation and HPA-axis regulation in adolescents. The TRAILS study. Psychoneuroendocrinology, 58, 46–50. doi: 10.1038/tp.2014.22 [DOI] [PubMed] [Google Scholar]
- Wankerl M, Miller R, Kirschbaum C, Hennig J, Stalder T, & Alexander N (2014). Effects of genetic and early environmental risk factors for depression on serotonin transporter expression and methylation profiles. Translational psychiatry, 4(6), e402. doi: 10.1038/tp.2014.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver IC, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, Seckl JR, … & Meaney MJ (2004). Epigenetic programming by maternal behavior. Nature Neuroscience, 7(8), 847. doi: 10.1038/nn1276 [DOI] [PubMed] [Google Scholar]
- Weaver IC, Korgan AC, Lee K, Wheeler RV, Hundert AS, & Goguen D (2017). Stress and the emerging roles of chromatin remodeling in signal integration and stable transmission of reversible phenotypes. Frontiers in Behavioral Neuroscience, 11, 41. doi: 10.3389/fnbeh.2017.00041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weder N, Zhang H, Jensen K, Yang BZ, Simen A, Jackowski A, … & O’Loughlin K (2014). Child abuse, depression, and methylation in genes involved with stress, neural plasticity, and brain circuitry. Journal of the American Academy of Child & Adolescent Psychiatry, 53(4), 417–424. doi: 10.1016/j.jaac.2013.12.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wockner LF, Noble EP, Lawford BR, Young RM, Morris CP, Whitehall VL, & Voisey J (2014). Genome-wide DNA methylation analysis of human brain tissue from schizophrenia patients. Translational Psychiatry, 4(1), e339 10.1038/tp.2013.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehuda R, Daskalakis NP, Lehrner A, Desarnaud F, Bader HN, Makotkine I, … & Meaney MJ (2014). Influences of maternal and paternal PTSD on epigenetic regulation of the glucocorticoid receptor gene in Holocaust survivor offspring. American Journal of Psychiatry, 171(8), 872–880. doi: 10.1176/appi.ajp.2014.13121571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, An Q, Goldberg J, Quyyumi AA, & Vaccarino V (2015). Promoter methylation of glucocorticoid receptor gene is associated with subclinical atherosclerosis: a monozygotic twin study. Atherosclerosis, 242(1), 71–76. [DOI] [PMC free article] [PubMed] [Google Scholar]