Abstract
Targeted panel, whole exome, or whole genome DNA sequencing using next-generation sequencing (NGS) allows for extensive high-throughput investigation of molecular machines/systems such as the LINC complex. This includes the identification of genetic variants in humans that cause disease, as is the case for some genes encoding LINC complex proteins. The relatively low cost and high speed of the sequencing process results in large datasets at various stages of analysis and interpretation. For those not intimately familiar with the process, interpretation of the data might prove challenging. This review lays out the most important and most commonly used materials and methods of NGS. It also discusses data analysis and potential pitfalls one might encounter because of peculiarities of the laboratory methodology or data analysis pipelines.
Keywords: DNA sequencing, DNA sequence analysis, LINC complex, Mutation, Next-generation sequencing, Polymorphism, Sequence variants
1. Introduction
Mutations in genes encoding LINC complex proteins have been linked to human disease. Mutations in SYNE1 encoding nesprin-1 cause autosomal recessive cerebellar ataxia, either pure or with associated features such as motor neuron involvement [1–4]. SYNE1 mutations also cause autosomal recessive arthrogryposis multiplex congenita, a disorder characterized by congenital joint contractures and reduced fetal movements [5–7]. Linkage of SYNE1 mutations to these autosomal recessive diseases is robust, as the pathogenic alleles clearly segregate with affected individuals in several families. Similarly, homozygosity for a protein truncating mutation in SYNE4, which encodes nesprin-4 expressed in the hair cells of the inner ear, has been shown to segregate with progressive high-frequency hearing loss in two families of Iraqi-Jewish ancestry [8]. An autosomal dominantly inherited point mutation in SYNE2 leading to an amino acid substation in nesprin-2β1 has also been shown to segregate among first-degree relatives with an Emery-Dreifuss muscular dystrophy-like phenotype [9].
There have been other reports of mutations in genes encoding LINC complex proteins leading to disease where segregation within families has not been demonstrated. Autosomal dominant sequence variations in SYNE1 have been reported in individuals with Emery-Dreifuss muscular dystrophy-like phenotypes [9–11]. Sequence variations in SUN1 and SUN2 have also been reported in individuals with Emery-Dreifuss muscular dystrophy-like phenotypes [12]. Functional abnormalities in cells expressing the protein variants and in the case of Synel genetically modified mice suggest that these sequence variants could be pathogenic [9–15]. Furthermore, mutations in EMD and LMNA, respectively, encoding the LINC complex-associated proteins emerin and A-type lam-ins that bind to SUNs, also cause Emery-Dreifuss muscular dystrophy [16, 17].
The advent of next-generation sequencing (NGS) has allowed for the analysis of large panels of genes and even whole exomes in disease gene discovery research as well as in clinical practice [18, 19]. NGS using the predominant Illumina technology, which is a highly parallelized version of Sanger sequencing generating short (up to 300 bp) reads, involves library preparation, target capture, and the sequencing process proper followed by data processing and analysis. The initial step in library generation is DNA fragmentation. Unless whole genome sequencing is performed (in which case the genomic DNA library is directly sequenced), various PCR-based or hybridization-based methodologies are used to capture the genomic regions of interest to be sequenced. Subsequently, adaptors are ligated to the DNA fragments that allow attachment of the individual library molecules to a solid surface for amplification (cluster generation) and sequencing by synthesis through annealing of sequencing primers followed by template-dependent extension. Mixing of multiple samples in a shared sequencing process is made possible by individual specific molecular tags also introduced via the adaptor molecules. If cost is an important consideration, multiple adaptor-ligated individually tagged libraries can be used for capture, although that might compromise efficiency of the process. Cluster generation is a solid phase amplification step that results in hundreds of millions of clusters each consisting of thousands of clones of the individual library molecules densely scattered on a glass slide, called the flowcell. During the sequencing process, the fluorescent signal corresponding to the incorporating nucleotides in the individual clusters is electronically converted to hundreds of millions of individual DNA sequences corresponding to the DNA molecule clones in individual clusters. Sequence data must then be aligned so variants relative to the reference can be identified and evaluated for their potential role in pathogenesis.
As more researchers utilize this technology and as more data becomes available from its use in routine clinical practice, care must be taken in concluding that sequence variants cause disease. This applies to genes encoding LINC complex proteins [3, 7]. Determining the pathogenicity of sequence variants, especially with-out precise phenotypic descriptions and sequences of family members, requires review of the literature and available databases, careful consideration of population allele frequency, and variant data from other individuals or other families that have the same variant. Analysis programs can also be used to determine how a variant potentially affects protein structure or expression. Complementary analyses such as repeat expansion testing, methylation testing, transcriptome analysis, and copy number assessment can provide additional information. Ultimately, bench experimentation may be required to confirm that a rare variant uncovered by NGS is functionally disruptive and potentially pathogenic. For example, when whole exome sequencing identified a missense mutation in LMNA reported in the literature to abolish prelamin A processing in vitro, we performed cell biological experiments on the patient’s fibroblast to confirm there was accumulation of the unprocessed pathogenic protein [20].
We review the materials and methods used for Illumina NGS and the identification of disease-causing variants. The scope of this chapter does not allow for a detailed description of the entire NGS process. Rather, we provide a general overview for non-geneticists who study molecular machines/systems such as the LINC complex and how alterations in their components may cause human disease.
2. Materials
Several different kits, reagents, and devices are commercially available for library preparation, target capture, and sequencing. With regard to sequencers, Illumina has emerged as the unequivocal leader. We describe some of the instruments, reagents, and kits we use for NGS.
2.1. Library Preparation, Target Capture, and NGS Equipment
Sonicator for DNA fragmentation. We recommend the Covaris S2 System Sonicator from VWR or its derivatives that can handle multiple samples simultaneously. Reproducible fragment size and size distribution of the library is essential requirement for NGS sequencing. The Covaris sonicator can perform this task in a highly reproducible manner without direct contact of the instrument with the sample. The multiplexing, automated versions can handle eight samples at a time, making this tedious and time-consuming process somewhat less of a challenge.
Hardware for DNA quantitation and library quality assessment. We use the Qubit Fluorometer from Invitrogen (Q32857) to obtain highly accurate measurements of DNA concentration before fragmentation. This is absolutely essential for NGS sequencing. Besides concentration, the size and size distribution of the sonicated DNA fragments is also critical. This is best assessed using the Fragment Analyzer, the Advanced Analytical Quantitation, or the Bioanalyzer from Agilent. Quantitation of the library with the successfully attached adaptors is best done using real-time PCR with CFX96 Real-Time System, BioRad, or equivalent. Precise assessment of the quality and quantity of library generated is essential for efficient clustering and representative mixing of libraries if more than one sample is sequenced at a time. Within the recommended cluster density range, sequence yield correlates directly with the cluster density obtained on the flowcell.
Hardware for DNA capture. Standard PCR machines with heated lids are used for hybridization-based capture (Agilent Sureselect reagents or their equivalents).
Sequencers. Illumina is the leader in the manufacturing of NGS instruments. Most laboratories currently use the models HiSeq 1500 or 2500 and 3000 or 4000. The numbers refer to whether the machine can run a single (1500; 3000) or two flowcells (2500; 4000) at the same time or whether cluster generation on the flowcell is randomly spaced (1500, 2500) or patterned (3000, 4000). Larger laboratories use customized and serially linked versions of these instruments (HiSeq XTen and XFive) to sequence exclusively genomes. The names of these instruments reflect their price in millions of US dollars and are out of reach of most academic research, hospital-based, or even private laboratories. New technology on the horizon is the NovaSeq machine that is predicted to drive down the cost of whole genome sequencing during its production cycle within the next few years from approximately $1000 to $300 dollars. Other NGS machines, such as Life Technologies’ Proton machine, use a different chemistry and a pH-based incorporation detection system that is less accurate around homopolymer regions [21]. This limits its usefulness for discovery of novel variants on a genomic scale. The platform provided by Pacific Biosystems allows for sequencing of individual DNA molecules over 10,000 base pairs but has a high error rate and has a limited throughput. Large genome centers use it as a corollary instrument, but it is rarely seen in the clinical molecular laboratory environment [22]. We therefore focus on generation and analysis of data obtained using the Illumina instrument product line.
2.2. Kits and Custom Reagents
SureSelect Exome V6 Capture Library from Agilent (5190–8865); one per sample.
TruSeq Custom Amplicon kit for 96 samples from Illumina (FC-130–1001); one per 96 sample.
SureSelectXT Reagent kit for 96 samples from Agilent (G9641B); one per sample.
Dynabeads MyOne Streptavidin T1 from Thermo Fisher (65602); one per sample.
Herculase II Fusion DNA Polymerase from Agilent (600677); one per sample.
Library Quant Kit (Illumina Universal) from Kapa Biosystems (KK4824); three per sample.
AgenCourt AMPure XP from Beckman Coulter (A63882); one bottle.
Qubit dsDNA Broad Range Assay Kit from Life Tech (Q32853); one kit.
-
Qubit dsDNA High Sensitivity assay kit Life Tech (Q32854); one kit.
Details regarding the use of these kits and reagents are provided in the manufacturers’ instructions and Illumina library preparation and sequencing protocols. In the Subheading 3, we address some important considerations relating to their use.
2.3. Computational Hardware (Recommended Minimum)
Network attached storage (NAS) capable of storing 20 tera-bytes of data.
Linux virtual machine: 4 processors and 32 GB RAM, running CentOS.
Windows Workstation: two 12 core Intel E5–2690v3 processors and 128 GB RAM.
Windows Server 2012: R2 Standard 64-bit.
2.4. Computational Software
CentOS Linux operating system.
NextGENe v2.4.02 from Softgenetics.
bcl2fastQ Conversion Software v1.8.4 from Illumina.
Variant annotation and filtering software: Golden Helix SNP or Variation Suite.
Genome MaNaGer™; current availability is limited to data reanalysis by MNG Laboratories (fee for service).
3. Methods
3.1. Library Preparation and Selection of Targeted Regions
The cost and computational complexity of whole genome sequencing makes it impractical for most laboratories. The alternative is to enrich and select genomic regions to sequence. Methodology such as targeted PCR or hybridization-based capture can be used to select relatively small targeted regions, such as specific genes, or more expansive regions, such as whole exomes. Selection of the best approach is based on the scenario, test volume, laboratory setup, and affordability.
Long-range PCR amplification is a necessity for thorough assessment of ambiguously mapping regions of the genome. Primers flanking ambiguously mapping regions should be used to avoid artifacts due to divergent variation in highly similar genomic regions. A list of such problematic regions can be found in Mandelker et al. [23]. An example is mitochondrial genome sequencing, which is performed optimally on a single amplicon of the mitochondrial genome, removing the possibility of artifacts due to sequencing of mitochondrial pseudogenes located in the nuclear genome. Long- range PCR is not easily scalable, and thus most laboratories resign to the increased false negative and false positive rates in these regions due to ambiguous mapping and unpredictable representation percentage of specific alleles. This is a serious issue, since these sequences represent about 2% of all exomic coding regions.
Multiplex PCR approaches are best suited for sequencing of relatively small (less than a megabase) noncontiguous genomic regions such as specific exons of genes. This approach allows fast, high-volume testing even with limited starting material available. Targeted screens for carriers of mutations in a specific gene are a good application for this method. We have found TruSeq Amplicon reagents by Illumina to be well suited for most applications. Limitation of this method is that it cannot be used to identify large deletions, even if the precise position of the deletion is known. Since the amplified regions from a specific target region are all the same size, the experiments should be designed to take into account the danger of duplicate reads that arise if a low number of template DNA molecules are used as starting material.
Hybridization-based selection of regions of interest is recommended if the region to be sequenced is greater than one mega-base, although it also works for smaller regions. Kits containing oligonucleotide baits (RNA or DNA) synthesized using various technologies are commercially available, such as the Agilent SureSelectXT or equivalents from other manufacturers such as Illumina or IDT. Some allow or encourage capturing multiple libraries simultaneously with a single capture reagent. The smaller the region of interest, the more one can save on sequencing cost using a single capture reagent for a large number of combined libraries. Using individual capture for each sample, however, allows greater reproducibility between experiments and thus allows obtaining copy number information from the sequencing data with great reliability. This approach requires at least 100 ng DNA to perform. Agilent Sureselect reagents perform well for both custom and off-the-shelf (e.g., exome) panels. The flexibility of this platform is important if the panel of targeted genes changes over time. Since hybridization capture uses randomly fragmented DNA as an input, duplicate reads are easier to identify. The ratio of forward and reverse reads over specific nucleotides is also much better balanced than with multiplexed PCR-based methods.
Transcriptome analysis can be thought of as another approach to focus on a subset of genomic regions without the need to specifically amplify or capture by hybridization the regions of interest. The cellular transcriptional machinery essentially does the work for you. All that needs to be done is removal of the high abundance structural RNAs using a hybridization-based approach. Transcriptomes provide an integrated output of the actual living state of the cell/tissue which could be very difficult or impossible to establish from analysis of even whole genome sequencing. Transcriptome analysis is also invaluable to assess the effects of splice site variants and even regulatory mutations that are outside the scope of most capture-based targeted amplification schemes. This method is essential in cancer genomics, and in that case, generally the tumor is available for “tissue-specific” transcript evaluation.
3.2. DNA Sequencing and Data Acquisition
Illumina technology is based on synthesis of a new DNA molecule complementary to a template strand. The main difference from the Sanger method is that the sequencing reaction is massively parallelized, meaning that results can be recorded from hundreds of millions of template DNA molecules simultaneously. Another important difference is that this technology generates relatively short (100–150 base pair) paired-end reads compared to the 500–1000 base pair reads that can be generated by traditional Sanger methodology. This is a significant limitation when it comes to precise mapping of variants in non-unique sequences within the genome. In addition, identification and sizing of repeat expansions are also limited to a size of approximately 100 base pairs. However, detection of variants in a subset of the DNA molecules interrogated is more sensitive than what can be achieved using Sanger methodology. This is because each original interrogated DNA molecule generates an independent sequence and, depending on the depth of sequencing, many dozens or hundreds of molecules are investigated for each region of interest. Detection of a mutation at less than 1% representation, as can occur in patients with mitochondrial heteroplasmy, chimerism, or mosaicism, requires additional indexing [24]. Following the initial couple of cycles, the sequencer gives a quantity and quality estimate of the reads that will be obtained from the run. This is an important step that allows decision to be made whether the run should be continued or aborted potentially preventing completion of a very expensive failed experiment.
3.3. Raw Data Quality Assessment
The overall quality of the sequences obtained is largely dependent on four components: (1) the expertise of the technologist, (2) the quality and quantity of the starting material, (3) the quality of the capture, and (4) the sequencing reagents and the reliability and precision of the sequencing instrument itself. The most reliable way to assess the performance of the instrument is through the use of an internal control library generated of the phage Phi X 174 (PhiX Control v3 (catalog # FC-110–3001)). When this control library is mixed in with the sample(s), it will yield sequence and sequence quality information independent of the quality of the sample library. The machine aligns the phage-derived sequence to an internal reference, providing information about the error rate associated with the run and the overall quality of the sequences obtained. An average Phred quality score of ≥Q30 for ≥90% of the reads indicates a successful run. Setting a lower cutoff for quality depends on particular circumstances and specific limitations in sample quality and quantity. If there is a significant difference between the quality of the reads obtained from the Phi X 174 control and the sample library, there is a set of troubleshooting steps in the Illumina instruments’ users’ manuals to identify the source of the problem. The projected total yield (in gigabases) from a run is one metric provided by the machine that allows predictions about the number of reads that the run will generate. This is variable depending on the specific instrument used and the success of the clustering but allows generation of a very good estimate about the depth of coverage to be expected for the specific region of interest. The Illumina instruments users’ manuals also provide a plethora of other metrics that allow troubleshooting of the sequence generation process.
3.4. Demultiplexing
Most NGS runs, depending on the size of the targeted regions, contain multiple samples in a single flowcell lane. Sample-specific reads are sorted from the mixed data using demultiplexing. Demultiplexing is a process whereby the sample specific indexes (short DNA sequences, usually 6-mers), introduced into the sample during the library preparation process, are read and used for sorting the reads into individual bins corresponding to the samples sequenced. The indexes are read in an independent priming reaction (Read2) and are kept in association with the forward (Read1) and reverse (Read3) reads obtained from the same cluster. This association allows assignment of the reads from specific clusters to specific samples. Selection of compatible indices for a specific run is a crucial process that has to be carefully supervised to avoid misassignment of reads to the wrong sample (or to no sample at all). In some cases, 96 or more indices can be used in the same batch. However, for most large panels and exomes, the number is more likely to be up to 20 or up to 6 samples, respectively. At the end of the process, “FastQ” files are generated which contain the individual sequences obtained from a sample with a quality score (Q1–40) assigned to each nucleotide.
3.5. Sequence Alignment and Mutation Calling
Sequence alignment and mutation calling pipelines such the Genome Analysis Toolkit (GATK; Broad Institute) have been described in detail and also have recommendations available (Best Practices) to guide the user in setting them up either in the laboratory or in the “cloud” [25–27]. Setting up these pipelines requires significant computational hardware and an experienced bioinformatics staff, which is often beyond the resources of smaller laboratories [28]. For smaller laboratories, we recommend soft-ware packages will well developed graphical user interfaces such as NextGENe produced by Softgenetics. These can be run on PCs and sometimes on MACs and provide a primary data analysis capability with some basic annotations. Knowing exactly what the soft-ware is doing or not doing, as well as being able to adapt it to the task at hand, is of huge importance. From this perspective, open-source software is preferred to software packages with inaccessible, unmodifiable code. That said, the algorithms used by most pipelines, open-source or commercial, use Burrows-Wheeler aligner and the Genome Analysis Toolkit pipeline as described in the Best Practices guidelines for mutation calling [26, 27]. The input into these pipelines is FastQ files (individual reads with quality scores for each base called) generated by the sequencer. The output of the alignment software is a Binary Alignment/Map (BAM) file that contains the genomic coordinates corresponding to the beginning and end of each read. The information relating to the variants called is summarized in a variant call format (vcf) file ordered according to specific chromosomal positions. Information in the BAM files also allows for the visualization of the aligned reads and the variants called and can be used for assessment of coverage depth throughout the region of interest. These files can be conveniently viewed using the integrated genome viewer (reference) generated by the Broad Institute.
It is important to understand the significance of mean or average coverage depth and what that means for the specificity and sensitivity of the testing. These are the metrics generally provided on clinical reports of panels/whole exome or genome sequencing and can be used to compare the products of various laboratories. Average coverage in itself provides no information about poorly covered regions and their size. We recommend the service provider gives three coverage statistics: the percent of region of interest (ROI) covered 5-, 10-, and 30-fold. Thirtyfold coverage is generally considered the desirable minimum coverage to avoid false positive and false negative calls for constitutional samples for unique genomic regions. Tenfold coverage is generally considered to be sufficient to provide a good indication for the absence of a mutation at a specific position; if the variant does not show up even once out of ten reads, the sample is almost certainly wild type at that site. Fivefold coverage is helpful to pick up homozygous or apparently homozygous variants in poorly covered regions, thus decreasing the false negative rate. In our experience, all variants that make it into a report need to be confirmed using an alternative method (such as targeted PCR). This is especially true for variants with less than 30-fold coverage. However, the most significant cause of false negative results is due to limitations in the size and content of the ROI. One has to verify the completeness of the inclusion of regions with known pathogenic variants in the capture reagent used. Not all ROIs/capture reagents are created equal. An exome capture reagent from one company can represent half of the ROI of the exome capture reagent of another company, and they might both omit a significant percentage of hard to capture regions with pathogenic variants.
3.6. Data Interpretation and Reporting
The vcf files listing the position and nature of the identified variants in a tab delimited text file format generated from most NGS platforms are too large and too poorly annotated for human inter-pretation. When faced with the hundreds of thousands to millions of variant calls in partial, whole exome or genome datasets, respectively, interpretation cannot be done without a database providing variant annotation and filtering. There are many publicly available databases with indications of pathogenicity of previously identified variants, such as ClinVar (National Center for Biotechnology Information), HGMD (Institute of Medical Genetics in Cardiff), OMIM (Johns Hopkins), and COSMIC (Sanger Institute). Others have data on allele frequency in the general population, such as 1000 Genomes Project (The International Genome Sample Resource), Exome Variant Server (University of Washington), ExAC (Broad Institute), and gnomAD (Broad Institute). However, having an internal dataset reference, generated with the same methodology and thus containing the same set of artifacts, is extremely helpful to provide accurate classification and reporting of variants [29]. Many off-the-shelf software packages for variant interpretation emphasize phenotype-based filtering as one of the early steps in the process. In our experience, this is not recommended, since medical and family histories for the patients are often scarce or nonexistent. Therefore, filtering should use phenotype information only as one of the last steps to avoid discarding unexpected pathogenic variants. This may be just as important for targeted panels as for interpretation of a whole exome. For this reason, most large centers have developed their own analysis pipe-line/database that can be updated regularly and allow the filtering algorithm to be controlled depending on the particular dataset. The list of variants requiring expert human review can be significantly shortened if informative family members are sequenced along with the proband. Most commonly these are the parents, but if they are not available, having healthy unaffected first-degree relatives and affected distant relatives can be of great help to predict the pathogenicity of a given rare variant not previously described in public databases.
The MNG Genome MaNaGer filtering strategy for variants is summarized in Fig. 1. Briefly, it classifies variants first into four categories using reference and reportable range filters:
Fig. 1.
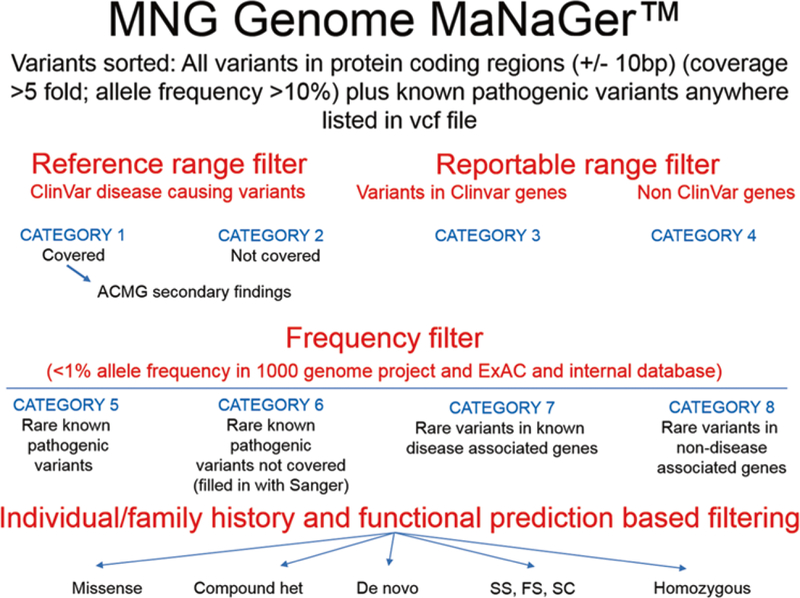
The MNG Genome MaNaGer strategy for variant filtering and annotation. See text for details. American College of Medical Genetics and Genomics (ACMG) secondary findings are those unrelated to the indication for ordering the sequencing but of medical value for patient care. Other abbreviations used in figure: het heterozygous, SS splice site, FS frameshift, SC stop codon
Category 1: variants with a clear pathogenic or likely pathogenic annotation in ClinVar that are present in the patient
Category 2: variants with a clear pathogenic or likely pathogenic annotation in ClinVar that are not covered in the patient
Category 3: all variants in disease-associated genes in ClinVar and other scientific literature (all category 1 variants are also present in this list)
Category 4: all variants in genes that have not been associated with disease yet
With the exception of the first two categories, these files are too large to be reviewed without further filtering. In a following step, we therefore use a frequency filter to generate four additional categories that contain the variants in categories 1–4 that are not present in any known population at greater than 1% allele frequency:
Category 5: rare known pathogenic variants
Category 6: rare known pathogenic variants not covered (filled in using targeted method such as PCR followed by Sanger sequencing)
Category 7: rare variants in known disease-associated genes
Category 8: rare variants in non-disease associated genes
The lists in categories 5–8 are more manageable for human review. We have developed a specific order to review them based on information obtained from individual and family histories as well as functional predictions (the most important five subcategories are listed in Fig. 1):
Homozygous variants
Disruptive variants; splice site, frameshift, stop codon variants
Variants unique to proband sample
Variants unique to case (trio)
De novo variants; not present in parents
Compound heterozygous changes
Variants that cause an amino acid change (missense)
Variants that after review (preferably) by three independent reviewers are judged by any one of the reviewers as of interest are further discussed in a grand rounds-like forum to determine if they should be further considered or reported based on American College of Medical Genetics and Genomics criteria.
The American College of Medical Genetics and Genomics has developed specific guidelines for the classification of variants as “pathogenic,” “likely pathogenic,” “unknown significance,” “likely benign,” and benign [30]. These guidelines are based on medical knowledge about disease frequencies, modes of inheritance, in silico prediction of the disruptive nature (nonsense, frameshift, splice site, predicted damaging/disruptive) of the variants as well as data available about specific genomic regions where known pathogenic variants cluster. The guidelines also take into account the presence or absence of the variants in healthy or affected parents (whether inherited or de novo). We use these guidelines in our daily clinical routine conservatively, recognizing that there is a danger to use many weak lines of evidence for declaring a variant pathogenic or likely pathogenic [30,31].
Reporting variants in the clinical setting carries great responsibility. The same is true for publishing them in the biomedical literature. The public, physicians and non-physician scientists vary greatly in their ability to rationally evaluate the significance of a reported variant. What is reported to a physician and patient or in the literature, even if properly qualified as a variant of unknown significance, may be perceived as a cause of the disease. This becomes a more significant problem as the number of the genes sequenced increases, while knowledge about the genes’ functions lags behind. In such instances, it is critical to obtain parental DNA to assess whether the mutation is present in unaffected parents or it arose de novo. Every clinical report should contain a sentence stating that such testing needs to be performed before the significance of the reported find can be properly established. This is even true in situations when variants carry a pathogenic or likely pathogenic label based on out-dated or insufficient evidence. Notably, such information is lacking for some rare variants of genes encoding LINC complex proteins that have been claimed to cause disease [10–13].
Although estimates vary, greater than 10% of variants labeled as pathogenic in ClinVar are rare ethnicity-specific variants with no clinical significance. There have been many efforts to increase data sharing among laboratories to assist with correct interpretation but much remains to be done. [32]. Until a time when the number of sequenced genomes allows a better phenotype-genotype correlation, sequencing trios (proband and parents), rather than probands alone, should be used for clinical and research studies. Reporting should be performed cautiously and in a conservative manner, both as to the number of variants reported and assessment of their clinical significance.
Another important concept generally applied throughout medicine but somewhat neglected in clinical genetics is comprehensive rather than method-specific testing. Many successful NGS companies emphasize the low cost of their platform but fail to emphasize that NGS—even whole exome sequencing—can only provide answers to the patient’s clinical problem in about 30% of cases. Combining tests for other genetic alterations or their manifestations, such as repeat expansions, methylation/imprinting disorders, copy number changes, chromosomal rearrangements, and transcript processing defects, and offering a synthesis of their results in a single report is something that needs to become the norm rather than the exception. Finally, despite comprehensive genetic testing, bench research using cultured cells or model organisms/animals is sometimes necessary to determine if a newly discovered rare variant actually affects protein function or expression that can underlie pathology.
3.7. Applying Information from Basic Research to Clinical Variant Interpretation
Unfortunately, only a fraction of variants discovered in clinical practice make it to basic scientists, and there is often an unnecessary delay in the transfer and review of information. This handicap can be overcome by forming strong collaborations between clinical molecular diagnosticians and basic scientists to develop and apply predictive screening tools and functional verification methodology for newly discovered variants. Of utmost importance is establishment of a system that facilitates or automates mapping human phenotype-associated mutations onto the functional models of not only individual genes and proteins but also multi-molecular complexes, such as the LINC complex. Clearly there are some encouraging examples of such efforts, but we believe this area requires much more attention and resources [33].
Another prerequisite for discovering new disease-gene associations is developing better structural and functional interaction networks connecting genes and encoded proteins. A systems approach focusing on disruption of molecular assemblies and pathways rather than individual proteins is a highly promising yet not fully exploited area of modern molecular diagnostic practice. Incorporating transcriptome analysis with both transcript level and processing assessment into routine genetic testing is already a reality in some laboratories. Furthermore, combining genetic data with proteomic data to identify protein levels and modify-cations using mass spectrometry might further enhance an under-standing of the metabolic homeostasis of cells and tissues [34]. This is particularly true for molecular machines/systems such as the LINC complex, in which different protein isoforms may be expressed at different levels in various cell types or tissues.
4. Conclusions
The field of genomic scale molecular diagnosis is rapidly expanding. Genomic datasets from individuals of diverse genetic back-grounds are accumulating at an ever-increasing rate. Analysis and processing of these data and deposition of the frequency and effect prediction information for individual variants gained from it will make an increasing portion of data analysis amenable to automation. That said, there will continue to be a need for basic scientists giving “personalized” attention to newly identified variants, including those in protein components of the LINC complex.
Acknowledgments
H.J.W. is supported by National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under award numbers R01AR048997 and R01AR068636. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Disclosures
P.L.N. is an employee of MNG Laboratories and owns equity in the company. H.J.W. serves on the scientific advisory board of MNG Laboratories and receives fees for this service.
References
- 1.Gros-Louis F, Dupre N, Dion P et al. (2007) Mutations in SYNE1 lead to a newly discovered form of autosomal recessive cerebellar ataxia. Nat Genet 39:80–85 [DOI] [PubMed] [Google Scholar]
- 2.Noreau A, Bourassa CV, Szuto A et al. (2013) SYNE1 mutations in autosomal recessive cerebellar ataxia. JAMA Neurol 70:1296–1231 [DOI] [PubMed] [Google Scholar]
- 3.Synofzik M, Smets K, Mallaret M et al. (2016) SYNE1 ataxia is a common recessive ataxia with major non-cerebellar features: a large multi-centre study. Brain 2016(139):1378–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mademan I, Harmuth F, Giordano I et al. (2016) Multisystemic SYNE1 ataxia: confirming the high frequency and extending the mutational and phenotypic spectrum. Brain 139:e46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Attali R, Warwar N, Israel A et al. (2009) Mutation of SYNE-1, encoding an essential component of the nuclear lamina, is responsible for autosomal recessive arthrogryposis. Hum Mol Genet 18:3462–3469 [DOI] [PubMed] [Google Scholar]
- 6.Laquérriere A, Maluenda J, Camus A et al. (2014) Mutations in CNTNAP1 and ADCY6 are responsible for severe arthrogryposis multi-plex congenita with axoglial defects. Hum Mol Genet 23:2279–2289 [DOI] [PubMed] [Google Scholar]
- 7.Baumann M, Steichen-Gersdorf E, Krabichler B et al. (2017) Homozygous SYNE1 mutation causes congenital onset of muscular weakness with distal arthrogryposis: a genotype-phenotype correlation. Eur J Hum Genet 25:262–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horn HF, Brownstein Z, Lenz DR et al. (2013) The LINC complex is essential for hearing. J Clin Invest 123:740–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Q, Bethmann C, Worth NF et al. (2007) Nesprin-1 and −2 are involved in the pathogenesis of Emery Dreifuss muscular dystrophy and are critical for nuclear envelope integrity. Hum Mol Genet 16:2816–2833 [DOI] [PubMed] [Google Scholar]
- 10.Puckelwartz MJ, Kessler EJ, Kim G (2010) Nesprin-1 mutations in human and murine cardiomyopathy. J Mol Cell Cardiol 48:600–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou C, Li C, Zhou B et al. (2017) Novel nesprin-1 mutations associated with dilated cardiomyopathy cause nuclear envelope disruption and defects in myogenesis. Hum Mol Genet 26:2258–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meinke P, Mattioli E, Haque F et al. (2014) Muscular dystrophy-associated SUN1 and SUN2 variants disrupt nuclear-cytoskeletal connections and myonuclear organization. PLoS Genet 10:e1004605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Puckelwartz MJ, Kessler E, Zhang Y et al. (2009) Disruption of nesprin-1 produces an Emery Dreifuss muscular dystrophy-like phenotype in mice. Disruption of nesprin-1 produces an Emery Dreifuss muscular dystrophy-like phenotype in mice. Hum Mol Genet 18:607–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J, Felder A, Liu Y et al. (2010) Nesprin 1 is critical for nuclear positioning and anchor-age. Hum Mol Genet 19:329–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stroud MJ, Feng W, Zhang J et al. (2017) Nesprin-1 a2 is essential for mouse postnatal viability and nuclear positioning in skeletal muscle. J Cell Biol 216:1915–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bione S, Maestrini E, Rivella S et al. (1994) Identification of a novel X-linked gene responsible for Emery-Dreifuss muscular dystrophy. Nat Genet 8:323–327 [DOI] [PubMed] [Google Scholar]
- 17.Bonne G, Di Barletta MR, Varnous S et al. (1999) Mutations in the gene encoding lamin A/C cause autosomal dominant Emery-Dreifuss muscular dystrophy. Nat Genet 21:285–288 [DOI] [PubMed] [Google Scholar]
- 18.Bamshad MJ, Ng SB, Bigham AW et al. (2011) Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet 12:745–755 [DOI] [PubMed] [Google Scholar]
- 19.Nagy PL, Mansukhani M (2015) The role of clinical genomic testing in diagnosis and discovery of pathogenic mutations. Expert Rev Mol Diagn 15:1101–1105 [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Lichter-Konecki U, Anyane-Yeboa K et al. (2016) A mutation abolishing the ZMPSTE24 cleavage site in prelamin A causes a progeroid disorder. J Cell Sci 129:1975–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boland JF, Chung CC, Roberson D et al. (2013) The new sequencer on the block: comparison of Life Technology’s Proton sequencer to an Illumina HiSeq for whole-exome sequencing. Hum Genet 132:1153–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.English AC, Salerno WJ, Hampton OA et al. (2014)Assessing structural variation in a personal genome-towards a human reference diploid genome. BMC Genomics 16:286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mandelker D, Amr SS, Pugh T et al. (2014) Comprehensive diagnostic testing for stereo-cilin: an approach for analyzing medically important genes with high homology. J Mol Diagn 16:639–647 [DOI] [PubMed] [Google Scholar]
- 24.Kennedy SR, Schmitt MW, Fox EJ et al. (2014) Detecting ultralow-frequency mutations by duplex sequencing. Nature Protoc 9:2586–5606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKenna A, Hanna M, Banks E et al. (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DePristo MA, Banks E, Poplin R et al. (2011) A framework for variation discovery and geno-typing using next-generation DNA sequencing data. Nat Genet 43:491–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van der Auwera GA, Carneiro MO et al. (2013) From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 43:11.10.1–11.1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsai EA, Shakbatyan R, Evans J et al. (2016) Bioinformatics workflow for clinical whole genome sequencing at Partners HealthCare Personalized Medicine. J Pers Med 6:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang K, Li M, Hakonarson H (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38:e164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richards S, Aziz N, Bale S et al. (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17:405–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Green RC, Berg JS, Grody WW et al. (2013) ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med 15:565–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Topol EJ (2015) The big medical data miss: challenges in establishing an open medical resource. Nat Rev Genet 16:253–254 [DOI] [PubMed] [Google Scholar]
- 33.Luu TD, Rusu AM, Walter V et al. (2012) MSV3d: database of human MisSense Variants mapped to 3D protein structure. Database (Oxford) 2012:bas018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alfaro JA, Sinha A, Kislinger T et al. (2014) Onco-proteogenomics: cancer proteomics joins forces with genomics. Nat Methods 11:1107–1113 [DOI] [PubMed] [Google Scholar]


