SUMMARY
Despite the recognition nearly a century ago that the human microbiome plays a clinically relevant role in drug disposition, mechanistic insights and translational applications are still limited. Here, we highlight the recent re-emergence of “pharmacomicrobiomics”, which seeks to understand how inter-individual variations in the microbiome shape drug efficacy and side effect profiles. Multiple bacterial species, genes, and enzymes have already been implicated in the direct biotransformation of drugs, both from targeted “case studies” and from systematic computational and experimental analyses. Indirect mechanisms are also at play; for example, microbial interactions with the host immune system can have broad impacts on immunomodulatory drugs. Finally, we discuss multiple emerging strategies for the precise manipulation of complex microbial communities to improve treatment outcomes. In the coming years, we anticipate a shift towards a more comprehensive view of precision medicine that encompasses our human and microbial genomes and their combined metabolic activities.
Keywords: Pharmacomicrobiomics, microbiome, precision medicine, biotransformation, immunology, immunotherapy, bacteriophages, genome editing, pharmacokinetics, pharmacodynamics
In Brief
Here, Lam et al. review the reciprocal interactions between drugs and the human microbiome. They discuss how inter-individual microbiome variations shape drug efficacy, the direct biotransformation of drugs by microbiota, and indirect microbiome-dependent mechanisms at work. They also examine microbiome engineering and how that could be leveraged for microbiome-based therapeutics.
Why do drugs work well for one patient while failing to have an effect or inducing severe side effects on the next? The answer is complicated and multifaceted. Multiple decades of progress in pharmacogenetics and pharmacogenomics (Thorn et al., 2013), have revealed the fundamental mechanisms through which drugs are metabolized by hepatocytes, enterocytes, and other cell types throughout the body and the importance of drug transporters in mediating the absorption, distribution, and elimination of drugs and other foreign compounds (xenobiotics). These studies have also helped illuminate drug mechanism of action and helped to identify novel drug targets. Robust associations between human genotype and drug outcomes has enabled diagnostic tests that inform drug selection and dosing (Thorn et al., 2013).
In comparison, the pharmacomicrobiomics field is still in the dark ages. Although there is a long and rich history of research on antibiotic resistance mechanisms in pathogenic bacteria (Lewis, 2013) we still know very little about how drugs interact with our resident microbial communities and the mechanisms through which the microbiome shapes drug pharmacokinetics and pharmacodynamics. Several questions remain unanswered: How many drugs are influenced by the microbiome? What are the microbial species, genes, and enzymes involved in the direct biotransformation of drugs? Do some drugs act in part by shaping the microbiome? How does microbial colonization alter host pathways for drug metabolism, absorption, or even mechanism of action? Given the complexity of the microbiome, is it possible to develop targeted approaches for controlling microbial metabolic activity and/or their interactions with host tissues?
Here, we summarize and discuss the recent studies that have ventured into a new frontier by revealing mechanistic insights into the reciprocal interactions between drugs and the human microbiome. Our focus in this review is on host-targeted (i.e., non-antibiotic) drugs. Antibiotics are discussed elsewhere in this Special Issue of Cell Host & Microbe. While much of the current literature focuses on the distal gut microbiome, we also highlight studies that have begun to apply these concepts to microbes in other body habitats, including the reproductive tract and even within solid tumors. Finally, we discuss exciting new developments in engineering the microbiome and how that might be leveraged to design microbiome-based therapeutics. These innovative new approaches have broad relevance beyond microbial drug metabolism, opening up new opportunities to target components of the microbiome or even to harness the microbiome for new medicines. Together, these studies provide a proof-of-principle that a mechanistic understanding of the role of the microbiome in pharmacology is possible and offer strong support for the hypothesis that the microbiome represents a neglected yet clinically relevant contributor to drug response. They also emphasize the utility of an interdisciplinary approach that borrows concepts and methods from biochemistry, chemical biology, computer science, genetics, and genomics, among others.
From case studies to systematic analyses of microbial drug metabolism
The vast evolutionary and metabolic diversity found within host-associated microbial communities can be overwhelming. A well-studied example comes from genes involved in carbohydrate metabolism: whereas the human genome encodes around two dozen distinct enzymes, the suite of carbohydrate-active enzyme families encoded by the microbiome numbers in the hundreds (Turnbaugh et al., 2010). The combined arsenal of distinct microbial enzymes involved in the metabolism of endogenous and exogenous small molecules remains to be determined.
One indication of the complexity of this enzymatic machinery comes from the growing list of known drugs that can be metabolized by the human gut microbiome (Spanogiannopoulos et al., 2016). These compounds vary in structure, molecular weight, polarity, and solubility making it highly likely that a large set of enzymes underlie their biotransformation. One prominent example is the bacterial hydrolysis of glucuronic acid residues from anti-cancer and anti-inflammatory drugs (Pellock and Redinbo, 2017). where a remarkable degree of diversity in bacterial β-glucuronidase structure translates to differences in substrate scope (Pollet et al., 2017). In total, 279 unique proteins, which could be binned into 6 structural categories with varying ability to accommodate small molecules versus macromolecules like polysaccharides, were discovered. Importantly, automated annotation pipelines will often mask this fine-scale diversity, which in this case would greatly underestimate the disparate strategies gut bacteria use for scavenging sugar from diverse chemical scaffolds.
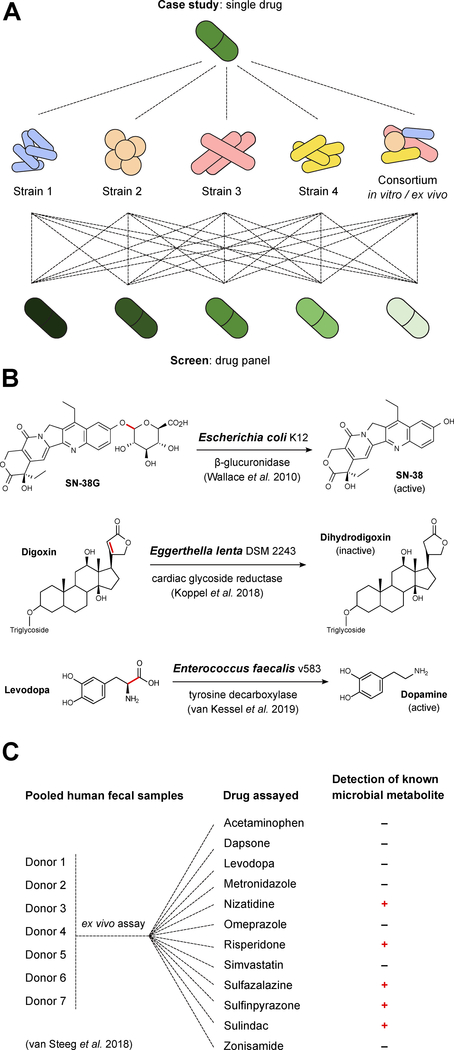
Due to our limited understanding of the microbial enzymes involved in non-antibiotic drug biotransformation, multiple researchers have chosen to pursue in-depth mechanistic analyses of single drugs of interest. These “case studies” (Figure 1B) have helped to provide fundamental insights that are now being leveraged to design more systematic analyses of drug metabolism by complex host-associated microbial communities.
Figure 1. Drug metabolism: from case studies to screens.
(A) Case studies and screens are complementary approaches. Detailed case studies enable the discovery and mechanistic analysis of the microbial species, genes, and enzymes involved in drug biotransformations, whereas screens enable more high-throughput testing of drug metabolism by individual strains or bacterial consortia. (B) Case studies have identified bacterial strains involved in the transformation of drugs or their metabolites; examples include digoxin (Eggerthella lenta DSM 2243), levodopa (Enterococcus faecalis v583), and SN-38 (Escherichia coli K12). (C) Ex vivo screens can be applied to human fecal samples to identify drugs subject to microbial metabolism by a particular individual or individuals. A recent study used ex vivo incubations of a pooled fecal sample from 7 donors with a panel of 12 drugs and identified metabolites for nizatidine, risperidone, sulfasalazine, sulfinpyrazone, and sulindac.
We recently discovered and characterized an enzyme (Cgr2, cardiac glycoside reductase 2) that is sufficient for the reductive inactivation of the cardiac drug digoxin by select strains of the gut Actinobacterium Eggerthella lenta (Haiser et al., 2013; Koppel et al., 2018). Due to the lack of genetics tools for E. lenta, we used comparative genomics to identify a single genomic locus--an 8 gene cgr-associated gene cluster--predictive of digoxin reduction. Transcriptomics (RNA-seq) narrowed this list down to 2 candidate genes that were significantly upregulated in the presence of digoxin, whereas heterologous expression and in vitro biochemical characterization confirmed that a single enzyme (Cgr2) is sufficient for digoxin reduction. Surprisingly, this enzyme appears to be specific for cardenolides (toxic plant compounds like digoxin), raising questions as to whether Cgr2 evolved in environmental contexts that have more routine exposure to plant toxins. Analysis of homologous reductases across bacterial genomes and human gut microbiomes highlighted the vast enzymatic diversity and need for a systematic functional characterization of these novel reductase sub-families (Koppel et al., 2018).
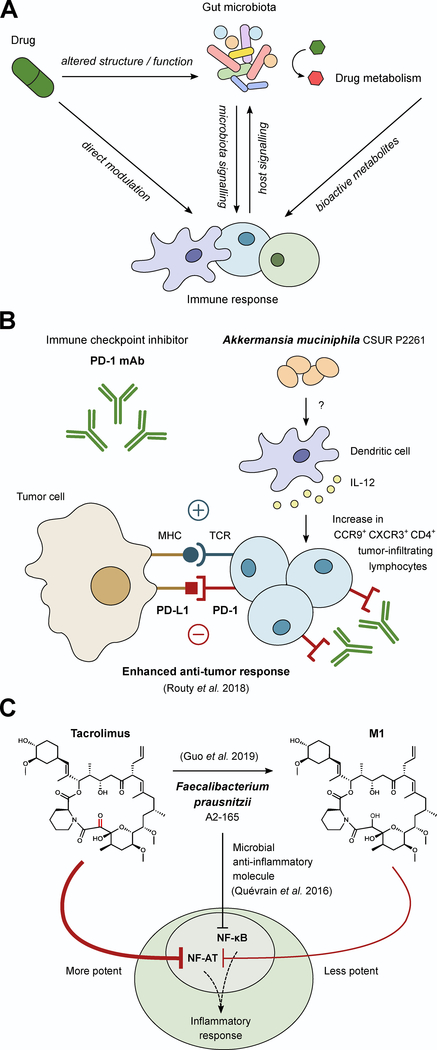
A more recent example of gut bacterial drug inactivation comes from studies of the immunosuppressant tacrolimus, which shows high and unexplained variability in its efficacy for organ transplantation patients. An analysis of 19 kidney transplant recipients revealed that patients requiring dose escalation were enriched for the gut bacterium Faecalibacterium prausnitzii relative to stable dose controls (Lee et al., 2015). This finding was surprising given the recent discovery of specific anti-inflammatory proteins expressed by F. prausnitzii (Quévrain et al., 2016), which might imply that elevated levels of this bacterium would be beneficial for drug efficacy. This paradox was recently explained through the discovery that F. prausnitzii and other gut Firmicutes are capable of metabolizing tacrolimus to an inactive metabolite unique to bacteria and detectable in stool samples from patients (Guo et al., 2019). Thus, gut bacterial strains can have opposing impacts relevant to disease; strain-level variations and/or environmental factors may tip the scales between the extent of bacterial drug biotransformation versus host immunomodulation for each patient.
Importantly, the impact of host-associated bacteria on drug metabolism is generalizable to other body habitats (Klatt et al., 2017) and even diseased tissue (Geller et al., 2017). Comparisons of the vaginal microbiotas of patients given the anti-retroviral drug tenofovir revealed enhanced drug efficacy in patients dominated by Lactobacillus relative to women with high abundance of Gardnerella vaginalis (Klatt et al., 2017). Measurements of drug levels in patients and following in vitro incubations with G. vaginalis suggested that drug inactivation by bacteria is responsible for the reduced efficacy. Interestingly, a recent study of pancreatic ductal adenocarcinoma arrived at a similar conclusion but with the opposite approach (Geller et al., 2017). Bacterial contamination of cancer cell lines led to resistance to the anti-cancer drug gemcitabine, which was attributed to the long isoform of the bacterial enzyme cytidine deaminase. Intratumoral injections of cytidine deaminase-expressing gut bacterial isolates impaired the treatment of disease in mouse models and 76% of the analyzed patient cancer samples had detectable levels of bacteria, mainly Gammaproteobacteria.
It is also possible for bacteria to interfere with drug bioavailability through the peripheral activation of compounds prior to reaching their target tissue. A recent study revealed that a tyrosine decarboxylase conserved in Enterococcus faecalis is necessary and sufficient to convert the Parkinson’s Disease medication levodopa (L-DOPA) to dopamine (van Kessel et al., 2019). Data from rodent models and patient cohorts supported the key role of E. faecalis in determining drug bioavailability (van Kessel et al., 2019), prompting ongoing efforts to identify small molecule inhibitors that are selective for the bacterial enzyme but do not impact host decarboxylation.
Taken together, these and other recent case studies (Zimmermann et al., 2019) highlight that bacterial drug metabolism is a general mechanism through which the microbiome in the gastrointestinal and reproductive tracts, and perhaps even within diseased tissue, alters drug response. However, it is difficult to predict the overall impact of direct microbial drug metabolism from these studies. To address this major gap in our knowledge, multiple research groups have begun developing complementary computational and experimental methods to systematically study the metabolism of drugs by host-associated microorganisms (Figure 1A).
We have made some early progress towards establishing a computational tool for predicting microbial drug metabolism (Mallory et al., 2018). Our approach leverages recent advances in comparing and characterizing chemical transformations using continuous vector representations. By applying this approach to the MetaCyc database (Caspi et al., 2018) we were able to systematically cluster reactions. Analysis of a “gold standard” set of known drug-metabolite transformations revealed matches with similar substructure modifications, motivating ongoing work to expand this method to assess the full set of FDA-approved compounds and their potential sensitivity to gut bacterial metabolism.
A major bottleneck in validating these predictions is the lack of robust platforms for the systematic quantification of large drug panels and their microbial metabolites. Recently, two research groups (Chankhamjon et al., 2019; van de Steeg et al., 2018) (Figure 1C) incubated human stool samples with drugs under laboratory conditions (t=24 hours in both studies) and subsequently subjected the samples to HPLC-MS analysis of the drugs and drug metabolites. The hit rate varied between studies, with 5/12 (41%) of the tested drugs metabolized in the first study (van de Steeg et al., 2018) and 57/438 (13%) of the tested drugs metabolized in the second study (Chankhamjon et al., 2019). Combined with prior literature-based surveys (Spanogiannopoulos et al., 2016), this brings the total list of drugs sensitive to in vitro and/or ex vivo microbial drug metabolism to 108 (96 of which are not considered antibiotics). This number is undoubtedly an underestimate given the focus on drugs that were already known to be metabolized in the former study (van de Steeg et al., 2018) and the use of only a single stool sample in the latter (Chankhamjon et al., 2019). More work is necessary to determine the predictive power of these or other laboratory tests for in vivo drug metabolism, analogous to the well-established in vitro to in vivo extrapolation (IVIVE) methods used for mammalian drug metabolism. Importantly, even if some of these reactions are not physiologically relevant, they could still provide important insights into the diverse biotransformations catalyzed by human gut bacteria and a starting point for mechanistic dissection.
Collateral impacts of non-antibiotic drugs on the human microbiome
Recent large-scale surveys of the human gut microbiome have revealed that environmental, rather than genetic, factors explain more of the observed variation in community structure (Falony et al., 2016; Rothschild et al., 2018). Remarkably, medication use explained the most variation in a cohort of >1000 Belgian individuals (Falony et al., 2016) outweighing even the reported variation in dietary intake. An obvious caveat to these cross-sectional studies is that it is not possible to determine the reason for the strong association between drugs and microbial community structure given the confounding impact of host pathophysiology, treatment-associated shifts in dietary supplements and other lifestyle factors, or other socioeconomic factors.
One approach, albeit artificial, for testing the direct impact of drugs on human gut microbial communities is to use the ex vivo incubation of human stool samples with panels of drugs. To this end, we performed a small screen of 8 antibiotics and 6 non-antibiotic drugs against the gut microbiomes of 3 unrelated individuals (Maurice et al., 2013). Incubations were performed for 4 hours immediately upon sample collection to minimize changes in bacterial physiology and community structure in response to storage and/or more prolonged incubations. As expected, the antibiotics consistently induced bacterial cell damage across these complex microbial communities. None of the tested non-antibiotic drugs had a significant impact on bacterial physiology or community-wide growth; however, we were able to identify changes in the expression of putative genes for drug tolerance mechanisms and drug metabolism (Maurice et al., 2013).
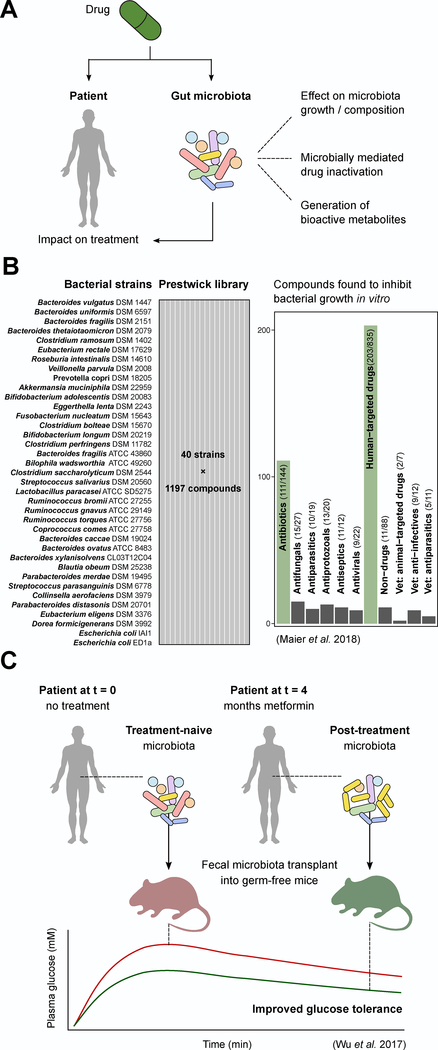
A more recent study described a large-scale screen of >1,000 drugs against 38 gut bacterial species (Maier et al., 2018) (Figure 2B). Surprisingly, 24% of the tested non-antibiotic drugs inhibited the growth of at least one bacterial strain at what was estimated to be a physiologically relevant concentration. However, it remains unclear how this antimicrobial activity would change given different drug formulations, cotherapies, or other host and environmental factors. More importantly, the threshold used to assign inhibitory activity was relatively modest (25% growth decrease in mono-culture relative to controls), consistent with our prior work (Maurice et al., 2013) suggesting that non-antibiotic drugs may act primarily by shaping microbial community gene expression, cellular physiology, and metabolic activity in the absence of full growth inhibition or cell lysis.
Figure 2. Off-target effects of drugs on the gut microbiota and their consequences for disease treatment.
(A) Drugs can interact with the microbiota in a number of ways to potentially impact the treatment of disease, including affecting microbial growth and composition, being subject to inactivation and thus leading to reduced availability, or being converted to bioactive metabolites that can exert their own effects including toxicity. (B) A large screen of 40 bacterial strains against the 1,197-compound Prestwick library has demonstrated that multiple host-targeted (non-antibiotic) drugs can impact members of the human gut microbiota in vitro, with as-yet unknown effects in vivo or during the course of therapy. (C) Drug-induced changes to the microbiota during the course of therapy may contribute to or influence drug efficacy on the host. A recent study with metformin found that post-treatment (after 4 months of treatment) stool transplanted into germ-free mice improved glucose tolerance compared to transplant of treatment-naive stool for 2 of 3 patients tested.
Assuming that some of the direct impact of non-antibiotic drugs on the microbiome is observed in patients, the next question is the degree to which drug-induced changes in the microbiome have downstream consequences related to mechanism-of-action and/or side effect profiles. The first proof-of-principle that drug-induced shifts in the gut microbiome are relevant to disease outcomes comes from studies of the type 2 diabetes medication metformin (Wu et al., 2017). A double-blind randomized control trial revealed a significant association between metformin use and shifts in the human gut microbiome. Transplantation of the stool microbiomes of patients pre- and post-metformin into germ-free mice was sufficient to improve glucose tolerance, providing causal evidence that changes in the gut microbiome over the course of therapy are relevant to disease (Figure 2C). These results appear to be mediated, at least in part, by the direct effect of metformin on gut bacterial gene expression (Wu et al., 2017); however, more work is necessary to dissect the mechanisms through which metformin shapes the microbiome and how the altered microbiome signals to regulate host glucose homeostasis.
One of the major consequences of altering gut microbial community structure is to change the immune system, which is responsive to a diverse array of microbial factors. We recently tested the hypothesis that changes to the human gut microbiome in response to non-antibiotic drugs could potentiate drug response by altering immune status (Nayak et al., 2019). A combination of in vitro and ex vivo experiments revealed that methotrexate (widely used for rheumatoid arthritis, RA) significantly alters gut bacterial growth and transcriptional activity, with a bias towards inhibiting the growth of Bacteroidetes over the other abundant members of the gut microbiome. Dose-dependent methotrexate-induced changes in the gut microbiome were also observed in “humanized” mice--formerly germ-free animals colonized with human stool samples from patients and healthy controls. These changes were robust to delivery method and dietary folate supplementation, of which methotrexate is an analog. Remarkably, longitudinal analyses of RA patients revealed that the degree to which the microbiome was altered by methotrexate was positively associated with drug efficacy whereas transplantation of pre- and post-treatment stool samples from one donor into germ-free mice provided initial evidence that microbial shifts may contribute to drug response by decreasing host pro-inflammatory responses such as activated T cells and Th17 cells (Nayak et al., 2019).
As with the direct metabolism of drugs by host-associated microorganisms, it is likely that these two initial case studies are just the first of many drug-microbiome interactions with consequences for treatment outcomes. Importantly, the emerging area of toxicomicrobiomics will be able to leverage the broader findings within the microbiome field, as researchers across many disease areas begin to better understand the mechanistic links between the component species, genes, enzymes, and metabolites within the human microbiome and host pathways relevant to the treatment of disease (Figure 2A).
Microbial manipulation of immunomodulatory drug response
A well-studied and important area of microbiome research is the intimate link between the microbiome and the host immune system. While much of this research is focused on the etiology or predisposition to disease, it has recently become clear that these host-microbiome interactions are also critically important for response to immunomodulatory drugs.
Oncology has led the way in this area, in particular three independent landmark studies each linked differences in the gut microbiome to the efficacy of cancer immunotherapeutic PD-1 blockade (Gopalakrishnan et al., 2018; Matson et al., 2018; Routy et al., 2018), despite different cohorts and disease contexts. Two studies focused on skin cancer (melanoma) (Gopalakrishnan et al., 2018; Matson et al., 2018) whereas the third studied a combination of lung cancer (non-small-cell lung carcinoma) and kidney cancer (renal cell carcinoma) (Routy et al., 2018). Analysis of the gut microbiomes of responders and non-responders revealed multiple bacteria associated with improved drug efficacy, including Akkermansia muciniphila (Routy et al., 2018), Bifidobacterium longum (Matson et al., 2018), and F. prausnitzii (Gopalakrishnan et al., 2018). Transplantation of intact human gut microbial communities (Gopalakrishnan et al., 2018; Matson et al., 2018; Routy et al., 2018) or even specific strains of A. muciniphila (Routy et al., 2018) into mouse models of disease led to enhanced systemic and anticancer immunity where A. muciniphila gavage combined with anti-PD1 therapy resulted in an IL-12 dependent accumulation of CCR9+ CXCR3+ CD4+ tumor infiltrating lymphocytes and corresponding decreases in tumor size (Figure 3B). Together, these studies emphasize the ability for changes in gut microbial community structure to impact drug efficacy even for organs far removed from the gastrointestinal tract: i.e. skin, lung, and kidney.
Figure 3. Microbial masters of immunomodulatory drugs.
(A) Due to the complex interplay between the immune system and the microbiota there are many potential ways microbes could alter the activity of immunomodulatory drugs. This includes immune manipulation by the microbiota, drug-induced changes in microbial community structure and function, drug-induced changes in immune function that feedback to changes in the microbiome, and the direct microbial biotransformation of drugs, all of which could impact the activity of immunomodulatory drugs. (B) During anti-PD-1 therapy, the inhibitory signal of PD-L1 from tumor cells is blocked, allowing for enhanced anti-tumor response. However, the efficacy of anti-PD-1 therapy depends on sufficient numbers of tumor infiltrating lymphocytes (TILs). The composition of the microbiota can influence the quantity of TILs as demonstrated in recent work in which colonization with A. muciniphila increases CCR9+ CXCR3+ CD4+ TILs in an IL-12-dependent manner during anti-PD-1 treatment in mice. (C) F. prausnitzii has been associated with anti-inflammatory responses through the inhibition of NF-κB signaling via a microbial anti-inflammatory molecule. Despite this association, patients on the immunosuppressant drug tacrolimus with elevated levels of F. prausnitzii required higher drug doses because F. prausnitzii metabolizes tacrolimus into a less active metabolite (M1), providing an example of a bacterium that can have opposing impacts on therapy.
Multiple key questions remain: why did each study reveal different bacterial biomarkers of drug response? Is this a reflection of differences in cohorts, cancer types, methodology, and/or other factors? Are these host-microbiome interactions unique to immune checkpoint blockade or more reflective of the broader mechanisms through which the microbiome impacts host immunity? Consistent with the latter hypothesis, F. prausnitzii has been implicated in inflammatory bowel disease (Sokol et al., 2008), A. muciniphila in metabolic syndrome (Everard et al., 2013), and B. longum in autoimmune disease (Srutkova et al., 2015). Even more perplexingly, why do these three bacterial species that are typically associated with decreased inflammation correlate with improved immunotherapy response, which is by definition enhanced immune activity? Perhaps this is reflective of the context-dependent way in which these host-microbiome interactions play out or due to strain-level differences in the microbial factors that alter host immune function.
Outside of oncology, several studies have investigated the impact of microbial metabolism on anti-inflammatory drugs. A classic example is the activation of drugs by gut bacterial metabolism, including the reduction of prodrugs such as sulfasalazine into their active metabolites (Spanogiannopoulos et al., 2016). More recently, gut bacterial metabolism has been implicated in the gastrointestinal toxicity of non-steroidal anti-inflammatory drugs (NSAIDs). A two-dimensional small intestinal cell culture assay linked NSAID-induced cytotoxicity to the uncoupling of mitochondrial oxidative phosphorylation, increased reactive oxygen species, and increased intestinal permeability (Bhatt et al., 2018). Studies in mice suggest that bacterial β-glucuronidases are responsible for this toxicity due to their ability to re-activate NSAIDs in the gut lumen following Phase II glucuronidation in the liver (LoGuidice et al., 2012). Of note, structural and biochemical analyses of diverse gut bacterial β-glucuronidases revealed that efficiency of metabolism of the NSAID diclofenac glucuronide cannot be readily predicted from a commonly used model substrate or the structure of the active site (Biernat et al., 2019).
The gut microbiome may also play a role in coordinating the response to biologic therapies by shaping immune function (Ananthakrishnan et al., 2017). The distal gut microbiomes of inflammatory bowel disease (Crohn’s and ulcerative colitis) patients receiving anti-integrin therapy (vedolizumab) were tracked prior to therapy. Crohn’s disease patients that achieved remission exhibit higher baseline diversity, enrichments for Roseburia inulinivorans and Burkholderiales sp., and enrichments for branched chain amino acid synthesis pathways. A neural network algorithm could accurately predict drug response. Interestingly, this approach did not generalize to ulcerative colitis; however, it did perform well on a small cohort of patients given anti-TNF. More work is necessary to expand this work to larger cohorts across independent patient cohorts, institutions, and countries to determine the degree to which the microbiome can accurately predict the response to immunomodulatory drugs.
The mechanisms underlying these microbiome biomarkers of drug response still remain to be determined; are these effects mediated by microbial immune manipulation, by microbial metabolism of immunomodulatory drugs, or a more complex cross-talk between the microbiota, immune system and drugs (Figure 3A)? Given the observation that multiple immunomodulatory drugs alter the growth of gut bacteria (Maier et al., 2018) and the broad impacts of different bacteria on immune responses (Hooper et al., 2012) one could imagine a scenario where these alterations act in synergy to promote a beneficial response; for example, A. muciniphila and anti-PD1 immunotherapy (Figure 3B) or alternatively act in opposing manners negating a response such as F. prausnitzii and tacrolimus (Figure 3C). Approaches such as gnotobiotic studies and more sophisticated systems for the in vitro co-culture of microorganisms with host tissues (Yissachar et al., 2017) could aid in deconvoluting these complex interactions.
Towards precise modification of the gut microbiota
Building a complete encyclopedia of drug-microbiome interactions will help us to understand the myriad ways in which host-associated microorganisms can interact with molecules inside the body leading to downstream effects on the host. The next logical question is whether and how we can translate this knowledge to improve the treatment of disease. The idea of manipulating the gut microbiota is certainly not new; however, current strategies used in the clinic lack precision--they may effect profound changes to the microbiome where smaller, defined changes are desired. For example, antibiotics are often prescribed for infections typically caused by a single organism but can have far-reaching and long-lasting impacts on the microbiota (Dethlefsen and Relman, 2010). In contrast, the more recent development of highly selective antibacterials, for example targeting only staphylococcal species (Yao et al., 2016) or Clostridium difficile (Thorpe et al., 2018), appears to limit collateral damage to the microbiota when compared to broad-spectrum antibiotics.
Fecal microbiota transplantation (FMT) is another strategy that introduces large changes by transferring a sample from a healthy donor into the gut of a patient with a depleted or less diverse microbiota, as in the case of recurrent C. difficile infection (Khanna et al., 2017; Weingarden et al., 2015). Stool samples from different donors or even the same donor across time may vary substantially, impacting the set of microorganisms being transferred. Here too, efforts such as those to develop a defined bacterial cocktail for use in treating C. difficile infection or other gastrointestinal infections or disorders (Munoz et al., 2016; Petrof et al., 2013) signifies progress towards increased standardization, enabling improvements in precision and control as well as the ability to assess possible limitations of a given therapeutic. Advances in microbiome-targeted therapeutics and the current challenges in the field have been reviewed previously (Mimee et al. 2016). Here, we focus on several exciting examples of recent work describing novel strategies for precise manipulation that either leverage or directly target the gut microbiota, and we group these strategies into four broad categories (Figure 4A): (1) inhibiting metabolic activities carried out by gut bacterial enzymes; (2) removing specific bacterial species or strains; (3) introducing or engrafting engineered strains into the gut; and (4) directly genetically modifying bacterial cells that are present within the gastrointestinal tract.
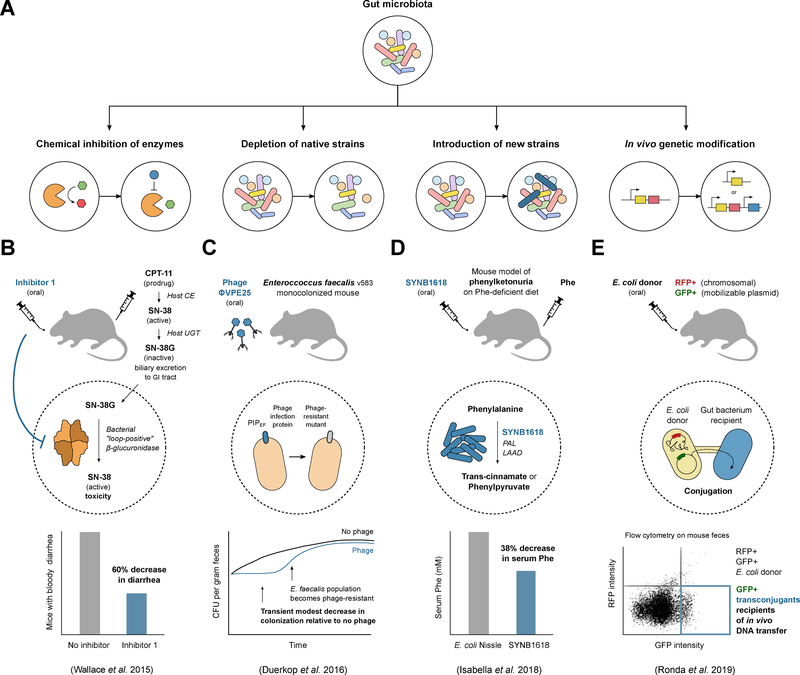
Figure 4. Strategies for precision engineering of host-associated microbial communities.
(A) Methods to manipulate either the membership or activities of the gut microbiota can be broadly grouped into four categories: inhibiting microbiome-encoded enzymes, depleting native members of the microbiota, adding new members that confer beneficial functions, or directly genetically modifying the existing microbiota through the addition or removal of genes. (B) The prodrug CPT-11 is activated by host carboxylesterases (CE) to active SN-38 and subsequently inactivated by host UDP-glucuronosyltransferase in the liver to inactive SN-38G via addition of a glucuronide moiety. SN-38G undergoes biliary excretion to the GI tract, where bacterial β-glucuronidases can liberate the glucuronide, leading to reactivation of SN-38 and toxicity in the form of diarrhea. Treating mice with Inhibitor 1 against β-glucuronidases is effective in alleviating CPT-11-induced bloody diarrhea, but the inhibitor is only active against a subset of β-glucuronidase enzymes that have a specific loop structure (“loop-positive”). (C) Phage ΦVPE25 lytic against Enterococcus faecalis v583 were administered orally to mice that were monocolonized with the same strain. Colonization by E. faecalis was found to be transiently reduced relative to mice given a no-phage control, but phage exposure eventually led to the gut bacterial population becoming phage-resistant, with E. faecalis isolates from mouse stool confirmed to have mutations in the membrane Phage Infection Protein (PIP) that confers resistance to ΦVPE25. (D) Engineered for the treatment of phenylketonuria, SYNB1618 is a derivative of E. coli Nissle carrying enzymes for the breakdown of phenylalanine (Phe): Phe is converted to trans-cinnamate by phenylalanine ammonia lyase (PAL) or to phenylpyruvate by L-amino acid deaminase (LAAD). A mouse model of phenylketonuria given SYNB1618 orally and challenged with an injection of Phe showed decreased levels of Phe in the serum relative to mice given the control E. coli Nissle. (E) An emerging strategy for manipulating the microbiome is to introduce genetic constructs directly into organisms colonizing the host. A recent study used bacterial conjugation from an orally introduced E. coli donor to transfer a GFP marker to recipient gut bacteria. The E. coli donor was engineered to carry RFP on the chromosome and GFP on a mobilizable plasmid, allowing GFP-only transconjugant bacteria to be distinguished from the donor by flow cytometry on mouse fecal samples.
Microbial enzymes catalyzing undesirable reactions in the gut can be viewed as “druggable” targets, toward which selective and non-lethal enzyme inhibitors may be developed (Wallace and Redinbo, 2013). Extensive work has been performed to develop inhibitors against gut bacterial β-glucuronidases with the goal of reducing the gastrointestinal toxicity of drugs used for cancer, inflammation, and other indications (Biernat et al., 2019; Wallace et al., 2015). To date, β-glucuronidase crystal structures have been solved for members of the Firmicutes, Bacteroidetes, and Proteobacterium phyla. Remarkably, although the overall structure of these phylogenetically disparate homologs is conserved, the analyzed enzymes differ in their catalytic efficiency, substrate specificity, and sensitivity to inhibition. For example, an inhibitor against β-glucuronidases was found to improve drug-induced toxicity in mice from bacterial reactivation of the chemotherapeutic CPT-11, but was found to be effective only for β-glucuronidases with a loop structure (Wallace et al., 2015) (Figure 4B). These studies provide an important proof-of-principle that a detailed molecular understanding of the key microbial biotransformations can enable the selective inhibition of members of widespread protein families.
With the recent elucidation of multiple additional microbial genes responsible for drug metabolism, the approach used for β-glucuronidases is now poised to be expanded to disrupt a broader suite of microbial enzymes that impact drug disposition and side effect profiles. For example, the drug L-DOPA is meant to be converted to dopamine after crossing the blood-brain barrier; however, the conversion to dopamine can occur before L-DOPA enters the central nervous system, through the action of (1) human DOPA decarboxylase present in cells of the peripheral nervous system or (2) by bacterial tyrosine decarboxylases found in the small intestine, the site of L-DOPA absorption (van Kessel et al., 2019). Inhibitors of human DOPA decarboxylase, such as carbidopa, can be co-administered with L-DOPA to reduce its decarboxylation, but importantly, these inhibitors are inactive against bacterial decarboxylases (van Kessel et al., 2019). The cardiac drug digoxin is a promising target for inhibition because its reduction to dihydrodigoxin is catalyzed by an enzyme that to date has only been found in one bacterial species, E. lenta (Koppel et al., 2018). An obvious disadvantage to this approach is that each of these new inhibitor compounds would require its own drug development cycle as an adjuvant therapy, a process that is both time-consuming and expensive.
One alternative may be to develop a broad-spectrum inhibitor of a class of enzymes. Exciting progress towards this comes in the form of activity-based probes, which are selectively reactive molecules that act as substrates to become covalently bound to an enzyme, enabling quantification of enzyme activity within a complex mixture. Fluorescently tagged activity-based probes can be used to label live cells, and this technique has been applied in combination with fluorescence-activated cell sorting (FACS) to identify active members of the gut microbiota with β-glucuronidase activity (Whidbey et al., 2018), which coupled to structural information (Wallace et al., 2015) could be used to design broad spectrum inhibitors.
Alternatively, it may be possible to disrupt an entire class of enzymes through interfering with essential cofactors. For example, tungsten has been used to disrupt molybdenum-cofactor-dependent enzymes that are used by members of the family Enterobacteriaceae in anaerobic respiration. Using mouse models of colitis, oral tungsten treatment was found to reduce inflammation by preventing the expansion of E. coli and other Enterobacteriaceae in the gut (Zhu et al., 2018). Although tungsten has the potential to cause off-target effects due to the generality of the molybdenum cofactor, tungsten did not inhibit in vitro growth of Bacteroides and Clostridium strains, did not impact butyrate production by the microbiota in vivo, nor did it appear to affect the Enterobacteriacae in the absence of inflammation.
Another approach to microbiome modification is to selectively deplete strains with undesirable activities, such as those that act on drugs to form toxic metabolites. One attractive avenue is to utilize existing bacterial viruses (bacteriophage or phage) to selectively remove bacterial members from the gut, akin to the use of phage therapy against pathogens (Merril et al., 2003; Kortright et al., 2019). Some argue that phages and their bacterial hosts have evolved over time to have an ultimately balanced relationship, allowing each to exist in nature, making phage-based therapeutics targeted at the gut microbiota challenging and complete eradication of species or strains unlikely. Consistent with this idea, when phage isolated from wastewater against the opportunistic pathogen E. faecalis were administered via the drinking water to mice monocolonized with E. faecalis, there was a transient and modest reduction in bacterial abundance (compared to control mice) followed by a rise in colonization level due to the population becoming phage-resistant; isolates of E. faecalis from mouse feces were confirmed to carry mutations in the membrane phage infection protein that confer resistance to phage (Duerkop et al., 2016) (Figure 4C). On the other hand, there is some evidence supporting the idea that phages can modulate bacterial populations in the gut. When germ-free mice colonized with a defined bacterial community were given either a mixture of virus-like particles purified from feces (Reyes et al., 2013) or a small set of lytic phages each targeting a different bacterium (Hsu et al., 2018), the abundance of targeted members decreased and phage abundance correspondingly increased. This indicates that phage are capable of knocking down levels of their bacterial targets in vivo, at least transiently, and that cascading effects on bacterial populations can occur when decreased abundance of one member promotes or inhibits others. In another study indicating that phage may be underappreciated in therapy, fecal filtrate transplantation (containing no bacteria, in contrast with FMT) successfully treated 5 patients with chronic relapsing C. difficile infection (Ott et al., 2017). These studies suggest that despite the existence of bacterial defense mechanisms against phage predation, phages may be exploited for modifying the gut microbiota under some circumstances.
Instead of using phages in their natural form, another avenue is to engineer them to be better or more specific bacterial killers than nature has evolved (Kilcher and Loessner, 2018). For example, the use of phage to deliver a CRISPR-Cas system for sequence-specific killing of pathogens in an animal host has been demonstrated: targeting the eae gene that encodes a virulence factor in enterohemorrhagic E. coli using a Galleria mellonella larvae infection model (Citorik et al., 2014) and targeting a kanamycin resistance gene present in only one of two strains of Staphylococcus aureus using a mouse skin infection model (Bikard et al., 2014). It will be interesting to see if this strategy can be extended to members of the gut microbiota for selective removal of strains encoding specific genes, (e.g., drug metabolism genes), particularly for bacteria that are present at high abundance in the gastrointestinal tract.
In contrast to removing strains, there are efforts to introduce engineered strains into the host as live bacterial therapeutics, an application that is similar to the use of probiotics that are meant to confer beneficial functions. Two recent studies described engineering Escherichia coli to express genes that could complement absent host functions in human metabolic diseases caused by genetic mutations. The first study focused on the disease phenylketonuria (PKU), a condition caused by a defect in the human gene encoding phenylalanine hydroxylase, leading to decreased metabolism of the amino acid phenylalanine, the accumulation of which can cause severe mental disability. A strain called SYNB1618, derived from E. coli Nissle, was constructed to express proteins involved in phenylalanine degradation, inducible both aerobically and under anaerobic conditions, i.e., in the gut (Isabella et al., 2018). Using a mouse model of PKU, mice that were injected with phenylalanine and orally dosed with SYNB1618 were found to have an average 38% decrease in blood phenylalanine levels compared to mice dosed with the control strain Nissle (Figure 4D). A similar strategy was used in the second study to address host enzyme defects leading to the condition hyperammonemia, in which ammonia levels are elevated in the blood: another derivative of Nissle, SYNB1020, was engineered for high expression of enzymes for the biosynthesis of arginine from ammonia (Kurtz et al., 2019). First, the repressor of the arginine biosynthesis operon was deleted and second, the first enzyme in the pathway was replaced with a feedback-insensitive mutant and placed under the control of promoter induced in anaerobic conditions. Together, these two modifications enabled higher ammonia consumption and arginine production than Nissle, and SYNB1020 was shown to lower levels of ammonia in the blood using two different mouse models of hyperammonemia. These applications demonstrate the use of engineered strains for increasing intestinal amino acid degradation and biosynthesis but could be naturally extended to other activities that provide a health benefit to the host.
Both “synbiotic” strains, SYNB1618 and SYNB1020, were designed to have auxotrophies (diaminopimelic acid and thymidine, respectively) with the rationale that this would facilitate biocontainment because the strains would be restricted in their ability to replicate both inside and outside of the host where exogenous sources of these nutrients required for growth are low (Isabella et al., 2018; Kurtz et al., 2019). On the other hand, if one wanted stable maintenance in the host, an introduced strain would have to be competitive against those members of the native microbiota that occupy a similar niche. Indeed, one challenge for the development of novel microbiome-targeted therapeutics is to understand engraftment. For example, the SER-109 therapeutic developed by Seres, containing spores prepared from batches of healthy donor stool samples, showed promising results in a Phase 1 trial for patients with recurrent C. difficile infection, achieving the primary end point of absence of diarrhea and C. difficile-positive stool during the 8 weeks following treatment (Khanna et al., 2016); however, SER-109 performed worse than anticipated in Phase 2, potentially due in part to challenges in achieving adequate engraftment (Henn et al., 2017; Ratner, 2016).
Rather than relying on the inherent ability of “wild” strains to colonize the gut, engraftment can be viewed as yet another characteristic that is amenable to engineering or manipulation. This was recently demonstrated by two different groups using a similar strategy for engraftment: by providing mice with a diet containing the polysaccharide porphyran (found in seaweed), it was possible to engraft species that are naturally and exclusively able to use this energy source, such as Bacteroides plebeius (Kearney et al., 2018) or Bacteroides ovatus (Shepherd et al., 2018), to high levels in the gut. Colonization levels were shown to be tunable by controlling the amount of porphyran in the diet; furthermore, the operon conferring the ability to use porphyran was engineered into two additional species for engraftment, Bacteroides stercoris and Bacteroides thetaiotaomicron (Shepherd et al., 2018). A caveat of this strategy is that it would likely be confined to nutrients that have limited utilization by native members of the microbiota. For example, the higher prevalence of porphyran utilization genes in the gut microbiomes of the Japanese population (Hehemann et al., 2010) may make porphyran-mediated engraftment less feasible as a general strategy among this group.
Other efforts to modify the gut microbiota indicate the movement away from a strategy of introducing engineered strains to that of directly genetically editing bacteria that are actively colonizing the gastrointestinal tract. Such an approach has been called in vivo or in situ engineering, referring to bacterial genetic modification events that occur in the host rather than in the lab. This approach was recently demonstrated using conjugation from an E. coli donor to transfer a “genetic payload” to recipient cells in the gut, detectable by fluorescent protein expression (GFP) and selectable by antibiotic resistance (Ronda et al., 2019) (Figure 4E). The payload was delivered via libraries of plasmids carrying origins of replication for maintenance in diverse bacteria or carried on transposable elements for insertion in recipient bacterial genomes. This is a powerful method to introduce genetic constructs into the gut microbiota, although in terms of control, strategies for biocontainment may be more challenging than for single engineered strains, particularly in the case of mobilizable and broad-host range replicative plasmids that can undergo secondary transfer events should recipient cells have endogenous DNA mobilization systems. GFP-positive transconjugants were only detected in stool up to a few days after oral delivery of an E. coli donor that was not well-suited to colonizing the gut, but a few transconjugant strains (Proteus mirabilis and Escherichia fergusonii, isolated from feces post-transfer) were shown to be able to mediate secondary transfer, and when used as donor strains themselves for conjugation, were found to lengthen GFP-positive transconjugant persistence in the stool (Ronda et al., 2019).
In vivo microbiome engineering is an exciting prospect, and these as well as strain engineering efforts will be supported by the development of new genetic tools for manipulation of diverse gut bacterial species (Heap et al., 2010; Lim et al., 2017; Mimee et al., 2015). These and other genetic tools and approaches will enable further progress towards precision modification of the gut microbiota as well as molecular mechanistic investigation of the impacts that bacteria have on host health.
So much to do, so little time
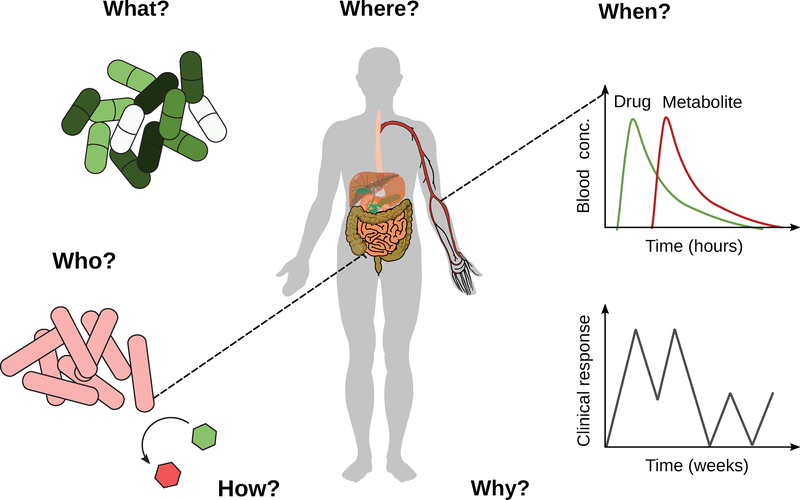
Despite the remarkable progress in the emerging field of pharmacomicrobiomics, our main takeaway so far is that the gap in our scientific knowledge is immense, especially given the broad implications of the studies discussed herein across multiple disease areas and traditional scientific disciplines. Moving forward, it will be important to begin to address the general questions of who, what, when, where, why, and how (Figure 5), with the goal of developing a fundamental set of principles (or rules) through which the microbiome impacts the treatment of disease that can be used to design more precise interventions.
Figure 5. Known unknowns: who, what, when, where, why, and how.
Fundamental gaps in our knowledge related to the role of the microbiome in pharmacology and toxicology.
Who While we now have a better sense for the key microbial species involved in drug response, far more systematic efforts are necessary. Better tools are needed to predict which members of complex human gut microbiotas are involved in drug pharmacokinetics and pharmacodynamics together with improved methods for quantifying their absolute abundance and activity in situ. As discussed above, the continued development of chemical probes promises to enable the culture-independent identification of microorganisms involved in specific reactions of interest, as recently demonstrated for bacteria that express β-glucuronidases (Whidbey et al., 2018).
What/How The current set of 108 drugs directly metabolized by human gut bacteria provide a strong foundation for a deep dive into the underlying genetic, biochemical, and enzymatic mechanisms responsible. Just as these mechanistic studies have provided trends and guiding principles for host drug metabolism (Thorn et al., 2013), a similar commitment to careful mechanistic studies of the microbiome will likely be informative. In parallel, “multi-omic” datasets that couple metagenomics, metatranscriptomics, metaproteomics, and metabolomics will help to assess the relevance of the identified mechanisms in the appropriate physiological context while also providing new hypotheses for in-depth dissection.
Where A major gap in our knowledge about the microbiome relates to the physical niche in which host-microbiome interactions and microbial biotransformations occur. Pioneering work has begun to co-localize microbial species and metabolites (Garg et al., 2017), paving the way towards similar studies that account for the precise location along the length and width of the gastrointestinal tract and other body habitats. Contrary to the long-standing dogma that the microbiome can only impact small molecules in the distal gut, recent work on L-DOPA has provided evidence that the microbiome can even impact the first-pass metabolism of drugs that are rapidly absorbed within the small intestine (van Kessel et al., 2019).
When It is now clear that the microbiome and its interactions with host pathophysiology are incredibly dynamic, requiring a more concerted effort to determine the time scales at which key processes are carried out. In the context of pharmacology, it is critical to determine the acute impact of the microbiome on pharmacokinetics (minutes to hours) while also considering the dynamic nature of the microbiome over the course of therapy (days to years). The same questions are important to consider when assessing the lasting effects of non-antibiotic drugs on the gut microbiota after treatment is completed. This area of study will provide opportunities to re-examine the mechanisms responsible for changes in drug response over time; for example, the widespread phenomenon of resistance to cancer chemotherapies.
Why The final and perhaps most perplexing question relates to the evolution of the direct and indirect role of the microbiome in pharmacology. By definition, most drugs are xenobiotics, i.e. they are not normally found within the human body. How does an enzyme evolve and get maintained in a microbial genome in the absence of its substrate? A common explanation is promiscuous enzymatic activity, wherein enzymes meant to target endogenous substrates can also accommodate xenobiotics; however, for some enzymes (like Cgr2) the endogenous substrate(s) remain elusive (Koppel et al., 2018). Alternatively, these drug biotransformations may have evolved in a similar context to antibiotic resistance mechanisms, perhaps through ancient microbial interactions and/or exposure to plant secondary metabolites. On the surface, interactions with the immune system seem more obvious; however, it is perhaps surprising that multiple prevalent bacteria efficiently boost immune response, which one might expect to lead to their eradication.
If you’ve read this far, we would like to extend a warm invitation to join in these efforts. The challenges are immense and our knowledge so limited that it will take a community of collaborative scientists across multiple traditional disciplines to maintain forward momentum. Without a dramatic expansion in our research efforts, it seems likely that the dream of therapies based upon a deep knowledge base and diverse toolkit for the precise manipulation of the human microbiome will remain just that.
Box 1. Glossary of terms.
Drug disposition: the combination of absorption, distribution, metabolism, and elimination of a drug.
Microbiome: often used interchangeably with microbiota. Used here to refer to the aggregate genetic material of a microbiota.
Microbiota: the combined set of microorganisms found with a given human body habitat.
Pharmacogenetics: the study of how human genetic polymorphisms impact drugs.
Pharmacogenomics: the study of how the human genome impacts drugs.
Pharmacomicrobiomics: the study of how the microbiome impacts drugs.
Pharmacokinetics: the study of how drugs move throughout the body.
Pharmacodynamics: the study of drugs and their mechanism of action.
Toxicomicrobiomics: an emerging area of study that considers the broad impacts of toxic compounds on cells across all domains of life.
Xenobiotics: compounds foreign to a living organism such as drugs or environmental pollutants.
Acknowledgments
We thank the other members of the Turnbaugh lab for pivotal discussions and critical feedback on the manuscript. This work was supported by the National Institutes of Health (R01HL122593; R21CA227232; 5T32AI060537). PJT is a Chan Zuckerberg Biohub investigator and a Nadia’s Gift Foundation Innovator supported, in part, by the Damon Runyon Cancer Research Foundation (DRR-42–16) and the Searle Scholars Program (SSP-2016–1352). Fellowship support was provided by the Canadian Institutes of Health Research (KNL) and the Microbial Pathogenesis and Host Defense Training Grant (5T32AI060537).
Footnotes
Declaration of Interests
Dr. Turnbaugh is on the scientific advisory boards for Kaleido, Seres, SNIPRbiome, uBiome, and WholeBiome; there is no direct overlap between the current work and these consulting duties. All other authors have no relevant declarations.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ananthakrishnan AN, Luo C, Yajnik V, Khalili H, Garber JJ, Stevens BW, Cleland T, and Xavier RJ (2017). Gut microbiome function predicts response to anti-integrin biologic therapy in inflammatory bowel diseases. Cell Host Microbe 21, 603–610.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt AP, Gunasekara DB, Speer J, Reed MI, Peña AN, Midkiff BR, Magness ST, Bultman SJ, Allbritton NL, and Redinbo MR (2018). Nonsteroidal anti-inflammatory drug-induced leaky gut modeled using polarized monolayers of primary human intestinal epithelial cells. ACS Infect Dis 4, 46–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biernat KA, Pellock SJ, Bhatt AP, Bivins MM, Walton WG, Tran BNT, Wei L, Snider MC, Cesmat AP, Tripathy A, et al. (2019). Structure, function, and inhibition of drug reactivating human gut microbial β-glucuronidases. Sci. Rep 9, 825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikard D, Euler CW, Jiang W, Nussenzweig PM, Goldberg GW, Duportet X, Fischetti VA, and Marraffini LA (2014). Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat. Biotechnol 32, 1146–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi R, Billington R, Fulcher CA, Keseler IM, Kothari A, Krummenacker M, Latendresse M, Midford PE, Ong Q, Ong WK, et al. (2018). The MetaCyc database of metabolic pathways and enzymes. Nucleic Acids Res. 46, D633–D639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chankhamjon P, Javdan B, Lopez J, Hull R, Chatterjee S, and Donia MS (2019). Systematic mapping of drug metabolism by the human gut microbiome. Biorxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citorik RJ, Mimee M, and Lu TK (2014). Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nat. Biotechnol 32, 1141–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, and Relman DA (2010). Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc. Natl. Acad. Sci. U. S. A 108, 4554–4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerkop BA, Huo W, Bhardwaj P, Palmer KL, and Hooper LV (2016). Molecularbasis for lytic bacteriophage resistance in enterococci. mBio 7, e01304–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, et al. (2013). Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. U. S. A 110, 9066–9071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falony G, Joossens M, Vieira-Silva S, Wang J, Darzi Y, Faust K, Kurilshikov A, Bonder MJ, Valles-Colomer M, Vandeputte D, et al. (2016). Population-level analysis of gut microbiome variation. Science 352, 560–564. [DOI] [PubMed] [Google Scholar]
- Garg N, Wang M, Hyde E, da Silva RR, Melnik AV, Protsyuk I, Bouslimani A, Lim YW, Wong R, Humphrey G, et al. (2017). Three-dimensional microbiome and metabolome cartography of a diseased human lung. Cell Host Microbe 22, 705–716.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geller LT, Barzily-Rokni M, Danino T, Jonas OH, Shental N, Nejman D, Gavert N, Zwang Y, Cooper ZA, Shee K, et al. (2017). Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science 357, 1156–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, Prieto PA, Vicente D, Hoffman K, Wei SC, et al. (2018). Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 359, 97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Crnkovic CM, Won K-J, Yang X, Lee JR, Orjala J, Lee H, and Jeong H (2019). Commensal gut bacteria convert the immunosuppressant tacrolimus to less potent metabolites. Drug Metab. Dispos 47, 194–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiser HJ, Gootenberg DB, Chatman K, Sirasani G, Balskus EP, and Turnbaugh PJ (2013). Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science 341, 295–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heap JT, Kuehne SA, Ehsaan M, Cartman ST, Cooksley CM, Scott JC, and Minton NP (2010). The ClosTron: Mutagenesis in Clostridium refined and streamlined. J. Microbiol. Methods 80, 49–55. [DOI] [PubMed] [Google Scholar]
- Hehemann J-H, Correc G, Barbeyron T, Helbert W, Czjzek M, and Michel G (2010). Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature 464, 908–912. [DOI] [PubMed] [Google Scholar]
- Henn M, Ford C, O’Brien E, Wortman J, Simmons S, Diao L, Litcofsky K, Bernardo P, Aunins J, Cook D, et al. (2017). Gastrointestinal tract microbiome dynamics following treatment with ser-109, an investigational oral microbiome therapeutic to reduce the recurrence of Clostridium difficile Infection (CDI) In Open Forum Infectious Diseases, (Oxford University Press US; ), S389–S390. [Google Scholar]
- Hooper LV, Littman DR, and Macpherson AJ (2012). Interactions between the microbiota and the immune system. Science 336, 1268–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu BB, Gibson TE, Yeliseyev V, Liu Q, Bry L, Silver PA, and Gerber GK (2019). Dynamic modulation of the gut microbiota and metabolome by bacteriophages in a mouse model. Cell Host Microbe 25, 803–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isabella VM, Ha BN, Castillo MJ, Lubkowicz DJ, Rowe SE, Millet YA, Anderson CL, Li N, Fisher AB, West KA, et al. (2018). Development of a synthetic live bacterial therapeutic for the human metabolic disease phenylketonuria. Nat. Biotechnol 36, 857–864. [DOI] [PubMed] [Google Scholar]
- Kearney SM, Gibbons SM, Erdman SE, and Alm EJ (2018). Orthogonal dietary niche enables reversible engraftment of a gut bacterial commensal. Cell Rep. 24, 1842–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kessel SP, Frye AK, El-Gendy AO, Castejon M, Keshavarzian A, van Dijk G, and El Aidy S (2019). Gut bacterial tyrosine decarboxylases restrict levels of levodopa in the treatment of Parkinson’s disease. Nat. Commun 10, 310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna S, Pardi DS, Kelly CR, Kraft CS, Dhere T, Henn MR, Lombardo M-J, Vulic M, Ohsumi T, Winkler J, et al. (2016). A novel microbiome therapeutic increases gut microbial diversity and prevents recurrent Clostridium difficile infection. J. Infect. Dis 214, 173–181. [DOI] [PubMed] [Google Scholar]
- Khanna S, Vazquez-Baeza Y, González A, Weiss S, Schmidt B, Muñiz-Pedrogo DA, Rainey JF, Kammer P, Nelson H, Sadowsky M, et al. (2017). Changes in microbial ecology after fecal microbiota transplantation for recurrent C. difficile infection affected by underlying inflammatory bowel disease. Microbiome 5, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilcher S, and Loessner MJ (2018). Engineering bacteriophages as versatile biologics. Trends Microbiol. 27, 355–367. [DOI] [PubMed] [Google Scholar]
- Klatt NR, Cheu R, Birse K, Zevin AS, Perner M, Noël-Romas L, Grobler A, Westmacott G, Xie IY, Butler J, et al. (2017). Vaginal bacteria modify HIV tenofovir microbicide efficacy in African women. Science 356, 938–945. [DOI] [PubMed] [Google Scholar]
- Koppel N, Bisanz JE, Pandelia M-E, Turnbaugh PJ, and Balskus EP (2018). Discovery and characterization of a prevalent human gut bacterial enzyme sufficient for the inactivation of a family of plant toxins. Elife 7, e33953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortright KE, Chan BK, Koff JL, and Turner PE (2019). Phage Therapy: A Renewed Approach to Combat Antibiotic-Resistant Bacteria. Cell Host Microbe 25, 219–232 [DOI] [PubMed] [Google Scholar]
- Kurtz CB, Millet YA, Puurunen MK, Perreault M, Charbonneau MR, Isabella VM, Kotula JW, Antipov E, Dagon Y, Denney WS, et al. (2019). An engineered E.coli Nissle improves hyperammonemia and survival in mice and shows dose-dependent exposure in healthy humans. Sci. Transl. Med 11, eaau7975. [DOI] [PubMed] [Google Scholar]
- Lee JR, Muthukumar T, Dadhania D, Taur Y, Jenq RR, Toussaint NC, Ling L, Pamer E, and Suthanthiran M (2015). Gut microbiota and tacrolimus dosing in kidney transplantation. PLoS One 10, e0122399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis K (2013). Platforms for antibiotic discovery. Nat. Rev. Drug Discov 12, 371–387. [DOI] [PubMed] [Google Scholar]
- Lim B, Zimmermann M, Barry NA, and Goodman AL (2017). Engineered regulatory systems modulate gene expression of human commensals in the gut. Cell 169, 547–558.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoGuidice A, Wallace BD, Bendel L, Redinbo MR, and Boelsterli UA (2012). Pharmacologic targeting of bacterial β-glucuronidase alleviates nonsteroidal anti-inflammatory drug-induced enteropathy in mice. J. Pharmacol. Exp. Ther 341, 447–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier L, Pruteanu M, Kuhn M, Zeller G, Telzerow A, Anderson EE, Brochado AR, Fernandez KC, Dose H, Mori H, et al. (2018). Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 555, 623–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory EK, Acharya A, Rensi SE, Turnbaugh PJ, Bright RA, and Altman RB (2018). Chemical reaction vector embeddings: towards predicting drug metabolism in the human gut microbiome. Pac. Symp. Biocomput 23, 56–67. [PMC free article] [PubMed] [Google Scholar]
- Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre M-L, Luke JJ, and Gajewski TF (2018). The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 359, 104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurice CF, Haiser HJ, and Turnbaugh PJ (2013). Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell 152, 39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merril CR, Scholl D, and Adhya SL (2003). The prospect for bacteriophage therapy in Western medicine. Nat. Rev. Drug Discov 2, 489–497. [DOI] [PubMed] [Google Scholar]
- Mimee M, Citorik RJ, Lu TK (2016) Microbiome therapeutics - Advances and challenges. Adv Drug Deliv Rev 105, 44–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimee M, Tucker AC, Voigt CA, and Lu TK (2015). Programming a human commensal bacterium, Bacteroides thetaiotaomicron, to sense and respond to stimuli in the murine gut microbiota. Cell Systems 1, 62–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz S, Guzman-Rodriguez M, Sun J, Zhang Y-G, Noordhof C, He S-M, Allen-Vercoe E, Claud EC, and Petrof EO (2016). Rebooting the microbiome. Gut Microbes 7, 353–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak RR, Alexander M, Stapleton-Grey K, Ubeda C, Scher JU, and Turnbaugh PJ (2019). Perturbation of the human gut microbiome by a non-antibiotic drug contributes to the resolution of autoimmune disease. Biorxiv. [Google Scholar]
- Ott SJ, Waetzig GH, Rehman A, Moltzau-Anderson J, Bharti R, Grasis JA, Cassidy L, Tholey A, Fickenscher H, Seegert D, et al. (2017). Efficacy of sterile fecal filtrate transfer for treating patients with Clostridium difficile infection. Gastroenterology 152, 799–811.e7. [DOI] [PubMed] [Google Scholar]
- Pellock SJ, and Redinbo MR (2017). Glucuronides in the gut: Sugar-driven symbioses between microbe and host. J. Biol. Chem 292, 8569–8576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrof EO, Gloor GB, Vanner SJ, Weese SJ, Carter D, Daigneault MC, Brown EM, Schroeter K, and Allen-Vercoe E (2013). Stool substitute transplant therapy for the eradication of Clostridium difficile infection: “RePOOPulating” the gut. Microbiome 1, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollet RM, D’Agostino EH, Walton WG, Xu Y, Little MS, Biernat KA, Pellock SJ, Patterson LM, Creekmore BC, Isenberg HN, et al. (2017). An atlas of β-glucuronidases in the human intestinal microbiome. Structure 25, 967–977.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quévrain E, Maubert MA, Michon C, Chain F, Marquant R, Tailhades J, Miquel S, Carlier L, Bermudez-Humaran LG, Pigneur B, et al. (2016). Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn’s disease. Gut 65, 415–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratner M (2016). Seres’s pioneering microbiome drug fails mid-stage trial. Nat. Biotechnol 34, 1004–1005. [DOI] [PubMed] [Google Scholar]
- Reyes A, Wu M, McNulty NP, Rohwer FL, and Gordon JI (2013). Gnotobiotic mouse model of phage-bacterial host dynamics in the human gut. Proceedings of the National Academy of Sciences 110, 20236–20241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronda C, Chen SP, Cabral V, Yaung SJ, and Wang HH (2019). Metagenomic engineering of the mammalian gut microbiome in situ. Nat. Methods 16, 167–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D, Costea PI, Godneva A, Kalka IN, Bar N, et al. (2018). Environment dominates over host genetics in shaping human gut microbiota. Nature 555, 210–215. [DOI] [PubMed] [Google Scholar]
- Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillere R, Fluckiger A, Messaoudene M, Rauber C, Roberti MP, et al. (2018). Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359, 91–97. [DOI] [PubMed] [Google Scholar]
- Shepherd ES, DeLoache WC, Pruss KM, Whitaker WR, and Sonnenburg JL (2018). An exclusive metabolic niche enables strain engraftment in the gut microbiota. Nature 557, 434–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux J-J, Blugeon S, Bridonneau C, Furet J-P, Corthier G, et al. (2008). Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. U. S. A 105, 16731–16736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanogiannopoulos P, Bess EN, Carmody RN, and Turnbaugh PJ (2016). The microbial pharmacists within us: a metagenomic view of xenobiotic metabolism. Nat. Rev. Microbiol 14, 273–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srutkova D, Schwarzer M, Hudcovic T, Zakostelska Z, Drab V, Spanova A, Rittich B, Kozakova H, and Schabussova I (2015). Bifidobacterium longum CCM 7952 Promotes Epithelial Barrier Function and Prevents Acute DSS-Induced Colitis in Strictly Strain-Specific Manner. PLoS One 10, e0134050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Steeg E, Schuren FHJ, Obach RS, van Woudenbergh C, Walker GS, Heerikhuisen M, Nooijen IHG, and Vaes WHJ (2018). An Ex Vivo Fermentation Screening Platform to Study Drug Metabolism by Human Gut Microbiota. Drug Metab. Dispos 46, 1596–1607. [DOI] [PubMed] [Google Scholar]
- Thorn CF, Klein TE, and Altman RB (2013). PharmGKB: the Pharmacogenomics Knowledge Base. Methods Mol. Biol 1015, 311–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe CM, Kane AV, Chang J, Tai A, Vickers RJ, and Snydman DR (2018). Enhanced preservation of the human intestinal microbiota by ridinilazole, a novel Clostridium difficile-targeting antibacterial, compared to vancomycin. PLoS One 13, e0199810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Henrissat B, and Gordon JI (2010). Viewing the human microbiome through three-dimensional glasses: integrating structural and functional studies to better define the properties of myriad carbohydrate-active enzymes. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun 66, 1261–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace BD, and Redinbo MR (2013). The human microbiome is a source of therapeutic drug targets. Curr. Opin. Chem. Biol 17, 379–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace BD, Roberts AB, Pollet RM, Ingle JD, Biernat KA, Pellock SJ, Venkatesh MK, Guthrie L, O’Neal SK, Robinson SJ, et al. (2015). Structure and Inhibition of Microbiome β-Glucuronidases Essential to the Alleviation of Cancer Drug Toxicity. Chem. Biol 22, 1238–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingarden A, González A, Vázquez-Baeza Y, Weiss S, Humphry G, Berg-Lyons D, Knights D, Unno T, Bobr A, Kang J, et al. (2015). Dynamic changes in short- and long-term bacterial composition following fecal microbiota transplantation for recurrent Clostridium difficile infection. Microbiome 3, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whidbey C, Sadler NC, Nair RN, Volk RF, DeLeon AJ, Bramer LM, Fansler SJ, Hansen JR, Shukla AK, Jansson JK, et al. (2018). A probe-enabled approach for the selective isolation and characterization of functionally active subpopulations in the gut microbiome. J. Am. Chem. Soc 41, 42–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Esteve E, Tremaroli V, Khan MT, Caesar R, Mannerås-Holm L, Ståhlman M, Olsson LM, Serino M, Planas-Félix M, et al. (2017). Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat. Med 23, 850–858. [DOI] [PubMed] [Google Scholar]
- Yao J, Carter RA, Vuagniaux G, Barbier M, Rosch JW, and Rock CO (2016). A pathogen-selective antibiotic minimizes disturbance to the microbiome. Antimicrob. Agents Chemother. 60, 4264–4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yissachar N, Zhou Y, Ung L, Lai NY, Mohan JF, Ehrlicher A, Weitz DA, Kasper DL, Chiu IM, Mathis D, et al. (2017). An intestinal organ culture system uncovers a role for the nervous system in microbe-immune crosstalk. Cell 168, 1135–1148.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Winter MG, Byndloss MX, Spiga L, Duerkop BA, Hughes ER, Büttner L, de Lima Romão E, Behrendt CL, Lopez CA, et al. (2018). Precision editing of the gut microbiota ameliorates colitis. Nature 553, 208–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann M, Zimmermann-Kogadeeva M, Wegmann R, and Goodman AL (2019). Separating host and microbiome contributions to drug pharmacokinetics and toxicity. Science 363, eaat9931. [DOI] [PMC free article] [PubMed] [Google Scholar]