Abstract
DNA methyltransferases are ubiquitous enzymes conserved in bacteria, plants and opisthokonta. These enzymes, which methylate cytosines, are involved in numerous biological processes, notably development. In mammals and higher plants, methylation patterns established and maintained by the cytosine DNA methyltransferases (DMTs) are essential to zygotic development. In fungi, some members of an extensively conserved fungal-specific DNA methyltransferase class are both mediators of the Repeat Induced Point mutation (RIP) genome defense system and key players of sexual reproduction. Yet, no DNA methyltransferase activity of these purified RID (RIP deficient) proteins could be detected in vitro. These observations led us to explore how RID-like DNA methyltransferase encoding genes would play a role during sexual development of fungi showing very little genomic DNA methylation, if any. To do so, we used the model ascomycete fungus Podospora anserina. We identified the PaRid gene, encoding a RID-like DNA methyltransferase and constructed knocked-out ΔPaRid defective mutants. Crosses involving P. anserina ΔPaRid mutants are sterile. Our results show that, although gametes are readily formed and fertilization occurs in a ΔPaRid background, sexual development is blocked just before the individualization of the dikaryotic cells leading to meiocytes. Complementation of ΔPaRid mutants with ectopic alleles of PaRid, including GFP-tagged, point-mutated and chimeric alleles, demonstrated that the catalytic motif of the putative PaRid methyltransferase is essential to ensure proper sexual development and that the expression of PaRid is spatially and temporally restricted. A transcriptomic analysis performed on mutant crosses revealed an overlap of the PaRid-controlled genetic network with the well-known mating-types gene developmental pathway common to an important group of fungi, the Pezizomycotina.
Author summary
Sexual reproduction is considered to be essential for long-term persistence of eukaryotic species. Sexual reproduction is controlled by strict mechanisms governing which haploids can fuse (mating) and which developmental paths the resulting zygote will follow. In mammals, differential genomic DNA methylation patterns of parental gametes, known as ‘DNA methylation imprints’ are essential to zygotic development, while in plants, global genomic demethylation often results in female-sterility. Although animal and fungi are evolutionary related, little is known about epigenetic regulation of gene expression and development in multicellular fungi. Here, we report on a gene of the model fungus Podospora anserina, encoding a protein called PaRid that share several features with known DNA methyltransferases. We showed that expression of the catalytically functional version of the PaRid protein is required in the maternal parental strain to form zygotes. By establishing the transcriptional profile of PaRid mutant strain, we identified a set of PaRid target genes. Half of them are also targets of a mating-type transcription factor known to be a major regulator of sexual development. So far, there was no other example of identified RID targets shared with a well-known developmental pathway that is common to an important group of fungi, the Pezizomycotina.
Introduction
A covalently modified DNA base, the 5-methylcytosine (5-meC) is common in genomes of organisms as diverse as bacteria, fungi, plants and animals. In eukaryotes, when present, this epigenetic modification is associated with down-regulation of gene expression and suppression of transposon activity [1]. Patterns of cytosine methylation are established and maintained through subsequent DNA replication cycles by the cytosine DNA methyltransferases (DMTs), a class of enzymes conserved from bacteria to mammals. Five families of eukaryotic DMTs can be distinguished [2,3]: 1) the “maintenance” DMT family which includes the mammalian DNMT1 [4], the plant MET1 [5] and the fungal DIM-2 enzymes [6]; 2) the “de novo” DMT family which includes the mammalian DNMT3A and DNMT3B [7] and the plant Domains Rearranged Methyltransferase DRM2 [8]; 3) the flowering plant-specific “maintenance” chromomethylase (CMT) family, which includes the Arabidopsis thaliana CMT2 and CMT3 enzymes [9–11]; 4) the fungal-specific DMT-like family which includes the Ascobolus immersus Masc1 [12] and the Neurospora crassa RID proteins [13]; 5) the putative CG-specific “maintenance” DMTs of the structurally divergent DNMT5 family [14,15]. DMTs from the first three families have been shown to methylate cytosines in vitro, while no such activity has ever be demonstrated either for the fungal DMT-like protein [12,13] nor for the DMTs of the DNMT5 family [14]. A sixth family, typified by Dnmt2 which is now known to be a tRNA methyltransferase, was therefore discarded from the bona fide DMT group [16]. Surprisingly, genes encoding putative DMTs can also be found in genome of species actually having very few, if any, 5-meC (e.g., Dictyostelium discoideum [17], Drosophila melanogaster [18] and Aspergillus nidulans [19]). These DMT-like proteins might be endowed with a still undetermined function. By contrast, most of the ascomycetous yeasts, including the model organism Saccharomyces cerevisiae and the human pathogen Candida albicans, lack genes encoding putative DMT-like enzymes. A notable exception includes the fission yeast Schizosaccharomyces pombe genome that contains a Dnmt2-like homolog. To date, the presence of 5-mC in the ascomycetous yeast genomes remains controversial [20,21].
In mammals and higher plants, methylation of cytosines is mostly associated with stable and inheritable repression of gene transcription. Therefore, it plays an essential role in genome stability and developmental programs such as imprinting and X chromosome inactivation [22]. In mice, loss of function of DNMT1 results in embryonic lethality [23] whereas flowering plants that present mutation in MET1 display a spectrum of viable developmental abnormalities [24,25]. In fungi, however, alteration of 5-meC content results in less contrasted outcomes. In the model ascomycete N. crassa, inactivation of the dim-2 gene, which encodes an enzyme from the “maintenance” DMT family, results in complete loss of DNA methylation. Remarkably, the dim-2 mutants behave as wild-type strains with respect to their vegetative life cycle and their sexual reproduction cycle [6]. These observations indicate that although N. crassa displays substantial cytosine methylation in some genomic compartments, this epigenetic modification is dispensable for all developmental processes observed in laboratory conditions. RID (for RIP Deficient), the second putative DMT of N. crassa belongs to the fungal-specific DMT-like family. The RID protein plays an essential role in RIP (Repeat-Induced point mutation), a genome defense mechanism conserved among Pezizomycotina fungi [13,26,27]. As defined in N. crassa by Selker and colleagues, RIP occurs in haploid parental nuclei after fertilization but before karyogamy. Repetitive DNA sequences originally described as longer than 400 bp are detected during the RIP process and are subsequently subjected to extensive conversion of cytosine to thymine (C-to-T) [28,29]. Over the extent of the RIP targeted repeats, the remaining cytosines are heavily methylated. Furthermore, in N. crassa, recent findings showed that linker regions located in between RIPed repeats are subjected to RID-independent RIP [30]. This cytosine to thymine mutagenic process is mediated by DIM-2 and relies on heterochromatin-related pathway. In this fungus, the absence of RID abolishes RIP on newly formed repeats without causing additional defect, such mutants displaying wild-type vegetative growth and sexual cycle.
The analogous MIP (Methylation Induced Premeiotically) process discovered in A. immersus leads to cytosine methylation only, within repeats [31]. Disruption of Masc1, which encodes a member of the fungal-specific DMT-like family, results in abolition of the de novo methylation of repeats but also in severely impaired sexual development [12]. Indeed, when Masc1 is absent in both parental stains, the resulting crosses are arrested at an early stage of sexual reproduction and no dikaryotic cells are formed. Similar defects of sexual development have been observed when RID-like DmtA and TrRid knockout mutants were constructed in A. nidulans [19] and Trichoderma reesei [32], respectively (Wan-Chen Li and Ting-Fang Wang, personal communication). This suggests that, unlike RID of N. crassa, several members of the fungal-specific DMT-like family could be involved in both genome defense and development [33], as for Masc1, which appears as the founding prototype of this family of putative DMTs.
Unlike A. nidulans and T. reesei, the model ascomycete P. anserina does not produce asexual conidia and therefore relies only on sexual reproduction to propagate, before the colony dies of senescence. Besides, this fungus displays an active RIP process prior to the formation of the zygotes, but the RIPed targets are not methylated [34,35]. In this context, we explored the function of PaRid, a DMT homologue closely related to the N. crassa RID protein. In the present study, we showed that the PaRid fungal-specific putative DMT protein is essential to ensure proper sexual development and that a catalytically dead version of this protein looses this function. Moreover, we demonstrated that PaRid is required in the maternal parental strain. Indeed, in the absence of PaRid, even if the wild-type allele is present in the nuclei coming from the male gamete, the sexual development is blocked before the dikaryon formation. By establishing the transcriptional profile of mutant crosses, we also identified a set of PaRid target genes. A substantial subset of these genes was previously identified as targets of FPR1, a mating-type transcription factor known to be a major regulator of fertilization and subsequent sexual development.
Results
PaRid is essential to ensure proper sexual development
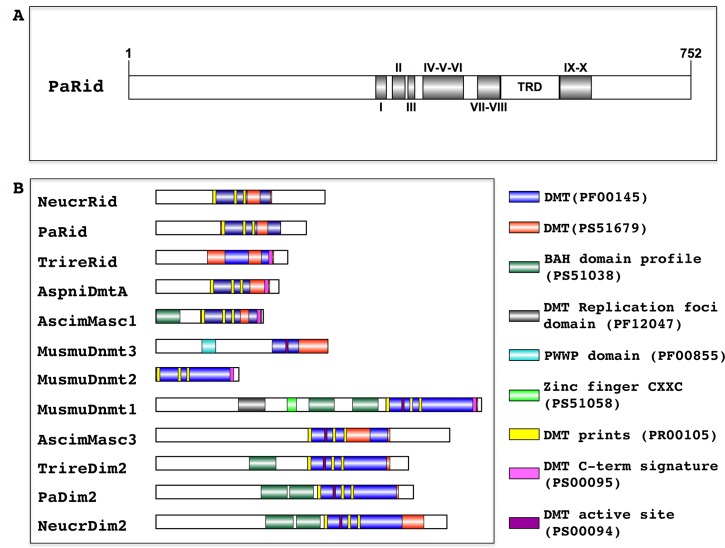
The P. anserina genome contains a single gene, namely PaRid (Pa_1_19440) [36], which encodes a 752 amino acid protein closely related to fungal-specific DMT-like proteins (Fig 1A). In addition of conserved DMT domain motifs (I to X) and expected Target Recognition Domain variable region (TRD), PaRid shows typical signatures of cytosine methyltransferases (InterPro domains PF00145, PS51679, PR00105) (Fig 1B). This protein belongs to the same phylogenetic group (DMT-like fungal-specific family) as the previously described N. crassa RID protein (41% identity) [19], A. immersus Masc1 protein (33% identity) [12] and A. nidulans DmtA (31% identity) (Fig 2). As already pointed out for Masc1/Rid DMT-like, these proteins, including PaRid, have peculiar features [33]. They show a non-canonical EQT (glutamate-glutamine-threonine) triad in motif VI instead of the ENV (glutamate-asparagine-valine) triad shared by all other eukaryotic C5-cytosine methyltransferases (Fig 3) as well as a remarkably short TRD domain (80 amino acids).
Fig 1. Structural analysis of DNA methyltransferase proteins.
(A) Domain architecture of P. anserina putative DNA methyltransferase PaRid. The catalytic domains contain 10 conserved motifs (I—X) and a target recognition domain (TRD) located between the motifs VIII and IX. The amino acid length is indicated. (B) Domain architecture of DNA methyltransferase proteins (DMT). The functional domain analysis was performed using InterProScan and visualized using IBS. Mus musculus: MusmuDnmt1: NP_001300940.1, MusmuDnmt2: NP_034197.3, MusmuDnmt3A: NP_031898.1; Ascobolus immersus, AscimMasc1 (AAC49849.1), AscimMasc3; Aspergillus nidulans AspniDmtA: XP_664242.1, Neurospora crassa NeucrRid: AAM27408.1, NeucrDim2: XP_959891.1; Podospora anserina PaDim2: Pa_5_9100; PaRid: Pa_1_19440; Trichoderma reesei TrireDim2 XP_006964860.1; TrireRid: AEM66210.1. Cytosine-specific DNA methyltransferase domains: PF00145, PR00105, PS51679, PS00095, PS00094; protein-DNA interaction domains: bromo-associated homology (BAH) domain PS51038, Replication foci targeting sequence (RFTS) PF12047, Zinc finger motif (CXXC) PS51058, PWWP domain (PF00855). Because its sequence is too divergent, PaDnmt5 (Pa_4_2960) was not included.
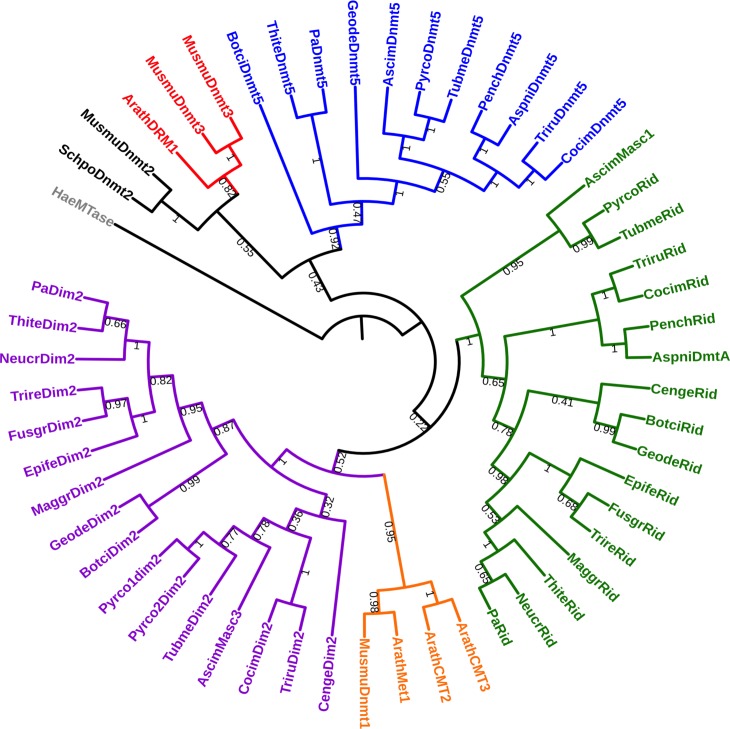
Fig 2. Phylogenetic analysis of mammal, plant and fungal DNA methyltransferases.
The maximum likelihood tree resolved five groups i) the Dnmt1/Met1/CMT group (orange), ii) the Dnmt2, group (black) iii) the Dnmt3/DRM group (red) iv) the fungal Dim2-like group (purple) and v) the fungal-specific Rid-like group (green), vi) the DNMT5 group (blue). Haemophilus aegyptius (HaeMTase: WP_006996493.1) Arabidopsis thaliana (ArathMet1: NP_199727.1; ArathCMT2: NP_193637.2; ArathCMT3: NP_177135.1; ArathDRM1: NP_197042.2; ArathDRM2: NP_196966.2), Ascobolus immersus (AscimMasc3: CE37440_11164; AscimMasc1: AAC49849.1; AscimDnmt5: RPA73956.1), Aspergillus nidulans (AspniDmtA: XP_664242.1, AspniDnmt5: XP_663680.1), Botrytis cinerea (BotciDim2: XP_024553164.1; BotciRid: XP_024550989.1, BotciDnmt5: XP_024550790.1), Cenococcum geophilum (CengeDim2: OCK96497.1; CengeRid: OCK89234.1), Coccidioides immitis (CocimDim2: XP_001247991.2; CocimRid: XP_001239116.2; CocimDnmt5: XP_001247253.2) Epichloe festucae (EpifeDim2: annotated in this study; EpifeRid: AGF87103.1), Fusarium graminearum (FusgrDim2: EYB34029.1; FusgrRid: XP_011320094.1) Pseudogymnoascus destructans (GeodeDim2: XP_024321957.1; GeodeRid: XP_024328520.1; GeodeDnmt5: XP_024320712.1) Magnaporthe grisea (MaggrDim2: XP_003718076.1; MaggrRid: XP_003720946.1) Mus musculus (MusmuDnmt1: NP_001300940.1; MusmuDnmt3A: NP_031898.1; MusmuDnmt3B: NP_001003961.2, MusmuDnmt2: NP_034197.3), Neurospora crassa (NeucrDim2: XP_959891.1; NeucrRid: AAM27408.1), Penicillium chrysogenum (PenchRid: XP_002563814.1; PenchDnmt5: XP_002561360.1), Podospora anserina (PaDim2: Pa_5_9100; PaRid: Pa_1_19440; PaDnmt5: Pa_4_2960), Pyronema confluens (Pyrco1dim2: PCON_02009m.01; Pyrco2dim2: PCON_01959m.01, PircoRid: PCON_06255m.01; PyrcoDnmt5: CCX08765.1), Schizosaccharomyces pombe (SchpoDnmt2: NP_595687.1), Thielavia terrestris (ThiteDim2: XP_003654318.1; ThiteRid: XP_003651414.1; ThiteDnmt5: XP_003650845.1), Trichoderma reesei (TrireDim2 XP_006964860.1; TrireRid: AEM66210.1), Trichophyton rubrum (TriruDim2: XP_003239082.1; TriruRid: XP_003239287.1; TriruDnmt5: XP_003236242.1), Tuber melanosporum (TubmeDim2: XP_002837027.1; TubmeRid: XP_002842459.1; TubmeDnmt5: XP_002837747.1).
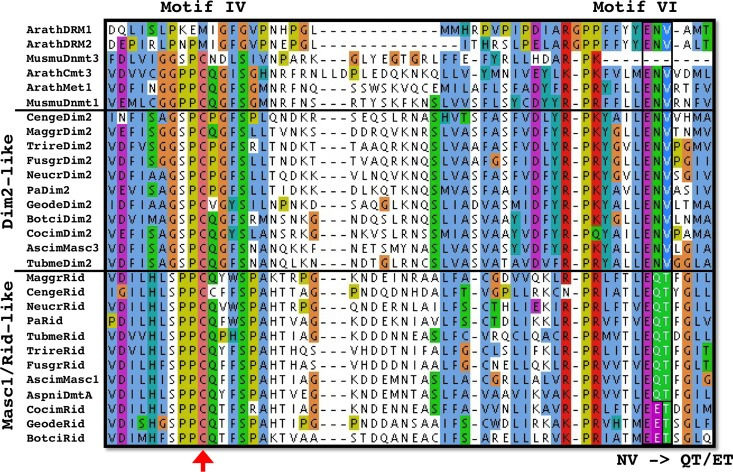
Fig 3. Evolution of the catalytic motifs IV and VI of the fungal Rid-like DNA methyltranferases.
A key catalytic step is the nucleophilic attack of the DNA methyltransferases on the sixth carbon of the target cytosine. This attack is made by the cysteine residue (red arrow) of the conserved PCQ triad (motif IV). This reaction is catalyzed by protonation of the N3 position of the cytosine by the glutamate residue of the conserved ENV triad (motif VI). In the Masc1/Rid-like group of enzymes, the ENV triad is replaced by either the EQT triad (e.g. N. crassa Rid, A. immersus Masc1, P. anserina PaRid, etc.) or the EET triad (e.g. B. cinerea Rid, P. destructans Rid, etc.). Arath: Arabidopsis thaliana, Musmu: Mus musculus, Cenge: Cenococcum geophilum, Maggr: Magnaporthe grisea, Trire: Trichoderma reesei, Fusgr: Fusarium graminearum, Neucr: Neurospora crassa, Pa: Podospora anserina, Geode: Pseudogymnoascus destructans, Botci: Botrytis cinerea, Cocim: Coccidioides immitis. Ascim: Ascobolus immersus, Tubme: Tuber melanosporum, Aspni: Aspergillus nidulans. See above for accession numbers of the corresponding proteins.
An EST database generated from vegetatively grown P. anserina showed no expression of PaRid [36]. However, the PaRid expression profile extracted from a microarray transcriptional time-course analysis performed during P. anserina’s sexual development (Bidard and Berteaux-Lecellier, GEO accession no. GSE104632) showed a peak at T12 (12 hours post fertilization) followed by a decrease at T30 (30 hours post fertilization) (S1A Fig). Besides RT-PCR experiments (S1B Fig) indicated that PaRid transcripts could be detected up to T96 (96 hours post fertilization). Searching the available P. anserina RNA-seq data [37], we did not find any evidence of non-coding RNA at the PaRid locus (S1C Fig), as described for sense-antisense DmtA/tmdA transcripts found in A. nidulans [19].
To gain insight about a potential function of PaRid during the life cycle of P. anserina, we deleted the corresponding gene. Replacement of the PaRid wild-type allele with the hygromycin-B resistance marker generated the ΔPaRid null allele (PaRid::hph) (Materials and methods and S2 Fig). The ΔPaRid mutant strains displayed wild-type phenotypes with respect to vegetative growth and vegetative developmental programs (i.e. germination, mycelium pigmentation, aerial hyphae production, branching, anastomosis, longevity, stress resistance, See S2 Table, S4 and S5 Figs), indicating that the PaRid gene is dispensable for the vegetative phase of P. anserina’s life cycle.
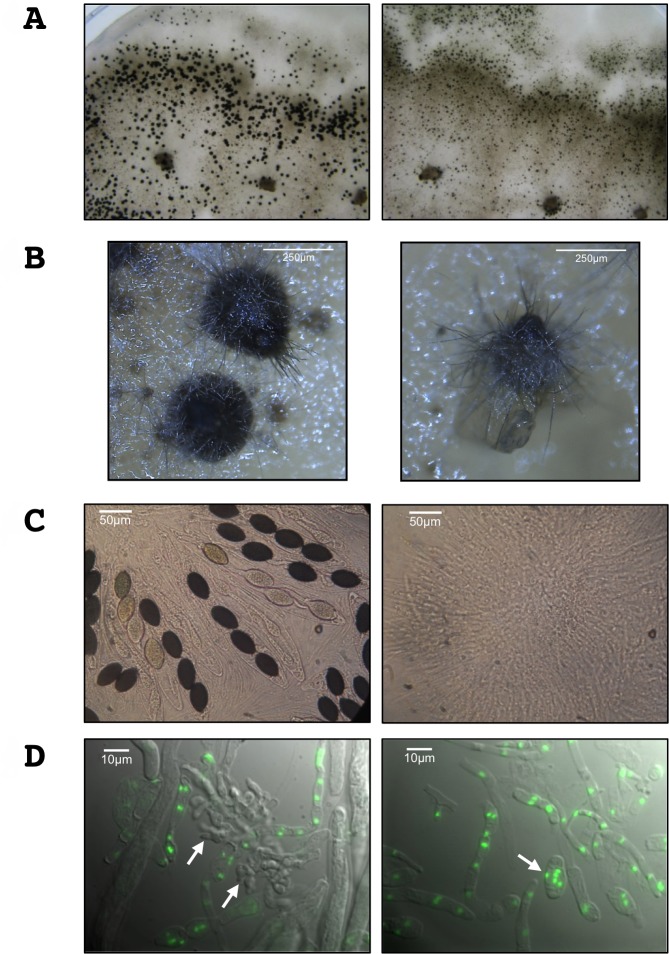
We then investigated the ability of the ΔPaRid mutant strains to perform sexual reproduction (see S3 Fig for description). During the first 30 hours post-fertilization, the sexual development of homozygous ΔPaRid crosses is indistinguishable from that of wild-type crosses. In homozygous ΔPaRid crosses, the fruiting bodies are formed normally, both in timing (24 hours post-fertilization) and in number (4333 in average per cross +/- 368, N = 5), as compared to homozygous wild-type crosses (4477 in average per cross +/- 458, N = 5) (S3 Table, Fig 4A). However, while the fructifications originating from wild-type crosses continued to develop up to 45 hours post-fertilization, those originating from homozygous ΔPaRid crosses stopped maturing around 30 hours after fertilization, forming distinctive micro-perithecia only. Although these micro-perithecia never grew into fully mature fruiting bodies, they displayed no visible morphological defects (Fig 4B). Importantly, they were fully pigmented and harbored typical necks and ostioles. As a consequence of this early developmental arrest, while one fully matured perithecium would produce hundreds of asci after 96 hours post fertilization, the ΔPaRid micro-perithecia are barren (Fig 4C).
Fig 4. PaRid is essential to complete fruiting body development and to produce ascospores.
(A) Homozygous crosses of wild type S strains (left panel) and of ΔPaRid strains (right panel) on M2 medium after 5 days at 27°C. Each dark dot is one fruiting body resulting from one event of fertilization. The homozygous ΔPaRid cross forms reduced-size fruiting bodies only (right panel). (B) Close up of fruiting bodies (perithecia) originating from either a wild-type genetic background (left panel) or a ΔPaRid genetic background (right panel). Scale bar: 250 μm. (C) After 4 days of growth at 27°C, the wild type fruiting bodies start to produce ascospores (left panel) while the mutant micro-perithecia are barren (right panel). Scale bar: 50 μm. (D) Fluorescence microscopy pictures of 48h-old fruiting body content from homozygous crosses of wild type S strains (left panel) and of ΔPaRid strains (right panel), performed on M2 medium at 27°C. The nuclei are visualized thanks to histone H1-GFP fusion protein. Croziers are readily formed inside the wild type perithecia (left panel, white arrows) while no crozier but large plurinucleate ascogonial cells only are seen inside the ΔPaRid perithecia (right panel, white arrow). Scale bar: 10 μm.
Furthermore, heterozygous orientated crosses showed that when the wild-type PaRid allele was present in the female gamete genome (i.e. ascogonia) and the ΔPaRid null allele was present in the male gamete genome (i.e. spermatia), the fruiting-body development was complete and resulted in the production of asci with ascospores (S6B Fig). The progeny isolated from this cross showed the expected 1:1 segregation of the ΔPaRid allele. On the contrary, when the ΔPaRid null allele was present in the female gamete genome and the wild-type PaRid allele was present in the male gamete genome, the fruiting body development was blocked and the resulting micro-perithecia are barren (S6B Fig). Altogether, these results indicate that (1) ΔPaRid mutants formed both male and female gametes and that these gametes are able to fuse since (2) fertilization occurred as efficiently in homozygous ΔPaRid mutant crosses than in wild-type crosses, yielding a similar number of fruiting bodies per crosses (3) the wild-type PaRid allele must be present in the maternal haploid genome for completion of sexual development. Notably, because reciprocal heterozygous orientated crosses with mat− and mat+ wild-type strains behaved similarly, the observed ΔPaRid phenotype was not mating-type dependent.
PaRid does not contribute to the development of maternal tissues
Fruiting bodies in Pezizomycotina are integrated structure made of two major kinds of tissues. The outer part, also called envelope or peridium, is exclusively made of maternal tissue whereas the inner part, the zygotic tissue is issued from fusion of the two parental haploid gametes [38]. To check whether the ΔPaRid sterility resulted from a peridium defect or from a developmental defect of the zygotic tissue, we set up trikaryon crosses involving the Δmat strain [39,40]. Because the Δmat strain lacks the genes required for fertilization, it does not participate either as male or female in sexual reproduction. However, the Δmat mycelium is able to provide maternal hyphae required to build fruiting bodies. Consequently, the Δmat strain can only complement mutants defective for the formation of the envelope but cannot complement zygotic tissue dysfunction. We observed that the Δmat; mat+ ΔPaRid; mat− ΔPaRid trikaryons yielded micro-perithecia only (S6C Fig), equivalent in size and shape to the micro-perithecia generated by the mat+ ΔPaRid; mat− ΔPaRid dikaryons. These results indicated a defect of zygotic tissues in ΔPaRid mutants. Furthermore, grafting two-day-old ΔPaRid micro-perithecia onto a wild-type mycelium did not result in further development of the fruiting bodies and did not restore ascus production either (S4 Table), which disproves the hypothesis of a metabolic deficiency or a nutrient shortage being the cause of the observed ΔPaRid mutant strain defects [41]. Conversely, two-day-old (not yet mature) wild-type perithecia grafted onto ΔPaRid mycelia, continued to develop into fully mature perithecia and expelled the usual amount of ascospores confirming that the ΔPaRid mutant mycelium is able to provide all required nutrients. It is to note that wild-type perithecia do not develop when grafted on wild-type mycelium grown on water agar medium, or PaMpk1, PaMKK1, PaASK1 and IDC1 mutant mycelia grown on regular M2 medium [41], which are all conditions in which the mycelium is unable to provide enough nutrients for fruiting-body development.
In the absence of PaRid in female gametes, sexual development is blocked before the dikaryon formation
As mentioned above, in P. anserina, fertilization results in formation of a plurinucleate ascogonium located inside the fruiting bodies. This ascogonium gives rise to dikaryotic cells that differentiate specialized cells (croziers) in which karyogamy leads to the formation of zygotes (S3 Fig). The diploid cells immediately enter meiosis to yield ascospores. In the homozygous ΔPaRid crosses, dissection of micro-perithecium contents (N = 5) performed 48 hours post-fertilization showed plurinucleate ascogonial cells, but an absence of dikaryotic cells. This observation indicated that the sexual development was blocked before the formation of the dikaryotic cells (Fig 4D). Control experiment performed in the same conditions on perithecium contents collected from wild-type crosses (N = 5), typically showed 20 to 30 dikaryotic cells per perithecium (Fig 4D). As mentioned above, heterozygous crosses where the ΔPaRid null allele was present in the female gamete genome and the wild-type PaRid allele in the male gamete genome resulted in the formation of micro-perithecia. When their content was dissected (N = 5), no dikaryotic cells were observed as in the homozygous ΔPaRid crosses. By contrast, heterozygous crosses where the wild-type PaRid allele was present in the female gamete genome and the ΔPaRid null allele in the male gamete genome resulted in the formation of fully mature perithecia that contain dikaryotic cells (N = 5). Altogether, these results show that a wild-type PaRid allele must be present in the haploid genome of P. anserina’s female gametes but not in the haploid genome of P. anserina’s male gametes for the dikaryotic cells to form.
Nevertheless, when homozygous ΔPaRid crosses are incubated from three to four weeks in the culture room (as opposed to the 96 hours post-fertilization needed for wild-type crosses to yield offspring), few asci were produced. A total of three asci were collected from 20 independent homozygous ΔPaRid crosses showing micro-perithecia only, whereas tens of thousands can typically be recovered from a single wild-type cross. These asci reflect a leaky phenotype of the PaRid mutant (see Discussion section). Each of these 12 ΔPaRid dikaryotic ascospores displayed wild-type shape, germinated efficiently and generated bona fide ΔPaRid mutant strains.
Complementation of the ΔPaRid mutants with ectopic alleles
To verify that the PaRid deletion was responsible for the sexual development arrest when absent from the female gamete haploid genome, we transformed a ΔPaRid strain with a PaRid-HA allele (see Materials and Methods section). Among the phleomycin-resistant transformants that were recovered (N = 109), 84% showed a complete restoration of fertility (Table 1). Moreover, when the expression of PaRid was driven by the highly and constitutively active AS4 promoter (AS4-PaRid-HA allele) [42], 67% of the phleomycin-resistant transformants (N = 78) completed sexual development and produced ascospores (Table 1). Among these complemented transformants, no noticeable additional vegetative or reproductive phenotypes were observed (S4 Fig).
Table 1. Complementation experiments.
| Alleles | Fertile | Sterile |
|---|---|---|
| PaRid-HA | 92 | 17 |
| PaRid-GFP-HA | 20 | 43 |
| AS4-PaRid-HA | 52 | 26 |
| AS4-PaRid-GFP-HA | 61 | 34 |
| PaRidC403S-HA | 0 | 89 |
| AS4-PaRidC403S-HA | 0 | 55 |
A collection of alleles was introduced in a ΔPaRid strain and subsequent transformants where crossed to a ΔPaRid strain of compatible mating type. Number of fertile versus sterile transformants is indicated in the table. When none of the transformants showed a restored fertility, PCR amplifications were performed to check the presence of full-length ectopic alleles. Those from which we cannot amplify the corresponding fragment were discarded. For each transformation experiments, expression of the HA-tagged proteins was confirmed by western blot on selected transformants (see S7 Fig). See Materials and methods section for details on allele features.
To investigate whether the putative enzymatic function of the PaRid protein was essential to P. anserina’s sexual development, we constructed the PaRidC403S-HA and the AS4-PaRidC403S-HA point-mutated alleles, which both encode a catalytically-dead PaRid protein [2]. To perform the methylation transfer, this cysteine residue of the conserved PCQ triad (proline-cysteine-glutamine) located in motif IV forms a covalent bond with the cytosines that will be modified (Fig 3). This cysteine residue is invariant in all eukaryotic C5-cytosine methyltransferases and its substitution results in loss of activity in mammalian DNA methyltransferases Dnmt3A and Dnmt3B [43]. After transformation of a knockout ΔPaRid strain, independent phleomycin-resistant transformants were recovered (N = 89 for the PaRidC403S-HA allele and N = 55 for the AS4-PaRidC403S-HA allele). Although they presented at least one full-length copy of either the PaRidC403S-HA allele or the AS4-PaRidC403S-HA, none of these transformants showed any complementation of the ΔPaRid sterility (Table 1). For selected transformants, we confirmed that the corresponding point-mutated protein PaRidC403S was readily expressed (S7 Fig). Again, these transformants could not be distinguished from ΔPaRid mutants when crossed to either wild-type strains or ΔPaRid mutant strains.
PaRid cellular localization
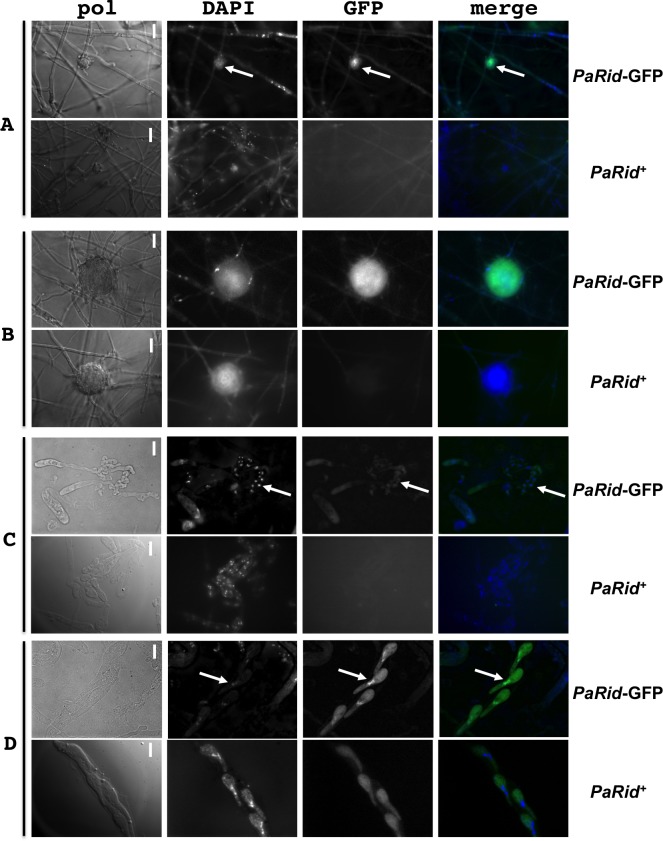
To investigate the subcellular localization of PaRid, we expressed two GFP-tagged chimeric versions of this protein (Table 1 and Fig 5). The PaRid-GFP-HA allele was driven by its native promoter, while the AS4-PaRid-GFP-HA allele was driven by the highly and constitutively active AS4 promoter. After transformation of a ΔPaRid strain, independent phleomycin-resistant transformants were recovered (N = 63 for the PaRid-GFP-HA allele and N = 95 for the AS4-PaRid-GFP-HA allele). Since 32% of the phleomycin-resistant strains obtained after transformation with the PaRid-GFP-HA allele and 64% of those obtained after transformation with the AS4-PaRid-GFP-HA allele showed a complete restoration of the ΔPaRid fertility defect (Table 1), both tagged-alleles were proven to be expressed and to encode functional proteins. Fluorescence under the native promoter was too weak to be monitored, therefore contructs with PaRid under the control of the AS4 promoter were subsequently analyzed. All the complemented AS4-PaRid-GFP-HA transformants had a wild-type phenotype (i.e., displayed no spurious phenotypes that could be linked to a putative constitutive overexpression of PaRid). The PaRid-GFP fluorescence was observed in the female gametes (ascogonia and protoperithecia, Fig 5A and 5B) but neither in mycelium nor in the male gametes (spermatia). This expression pattern is in line with both the ΔPaRid maternal sterility and the absence of PaRid ESTs in vegetative mycelium. Surprisingly, fluorescence could no longer be observed in croziers (formed around 30 hours post-fertilization) (Fig 5C), but signal resumed during ascospore formation (from 42 to 96 hours post-fertilization) (Fig 5D). In ascospores, PaRid-GFP was nuclear since the cytoplasmic signal can be attributed to self-fluorescence (Fig 5D). Seeing some GFP signal during sexual development, from ascogonia to ascospores, further supports the detection of PaRid transcripts by RT-PCR up to 96 hours post-fertilization (S1A and S1B Fig), whereas in the microarray experiments, the ratio of ascogenous tissues versus vegetative tissues (perithecium envelope) might be low, thereby masking PaRid’s expression profile.
Fig 5. Expression and subcellular localization of PaRid during P. anserina life cycle.
PaRid-GFP expression was assayed from a ΔPaRid:AS4-PaRid-GFP-HA strain showing wild type phenotypes (PaRid-GFP). As a control, self-fluorescence was assayed from a wild-type strain (No GFP-tagged protein, PaRid+). No GFP signal can be observed in mycelium, however, a significant and specific GFP signal can be found in ascogonia (A, white arrows are pointing at ascogonia) and protoperithecia (B). Surprisingly, no GFP signal is observed in the croziers (C, white arrows are pointing at croziers) but a strong signal can be noticed in the mature ascospores (D, white arrows are pointing at the nuclei of one ascospore). Since self-fluorescence can be detected in the cytoplasm of ascospores, but not in the nuclei, the PaRid-GFP protein localization is nuclear. From left to right: bright-field (pol), DAPI staining, GFP channel and merge of the two latest. Scale bar: 1,5 μm (A and B), 5 μm (C and D).
Transcriptomic reprograming in response to ΔPaRid developmental arrest
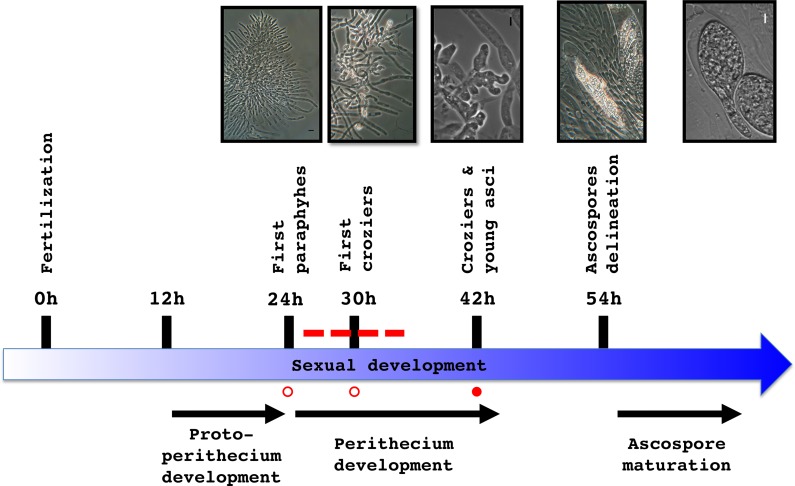
We explored the transcriptional profile of the ΔPaRid micro-perithecia using P. anserina’s microarrays, representing 10,556 predicted coding sequences (CDS) [44]. To this end, ΔPaRid mat+ female gametes were fertilized by wild-type mat- male gametes and micro-perithecia were collected 42 hours post-fertilization (T42). This time point was chosen to unravel the broadest set of differentially expressed genes, given that 1) micro-perithecia originating from ΔPaRid crosses do not form croziers and thus might stop to develop around 30 hours (T30) post-fertilization, 2) in wild-type crosses, croziers are formed from 30 hours (T30) to 42 hours post-fertilization (T42) afterward karyogamy can proceed (Fig 6). To identify the differentially expressed CDS (DE CDS), we compared the transcriptional profile of the ΔPaRid micro-perithecia to that of the wild-type perithecia at both 24 (T24) and 30 (T30) hours post-fertilization (Bidard and Berteaux-Lecellier, GEO accession no. GSE104632), hypothesizing that these two time points likely correspond to the window of time that immediately precedes and/or spans the ΔPaRid developmental arrest. Doing so, we identified 451 CDS which expression was either down-regulated (217 CDS) or up-regulated (234 CDS) by a fold change (FC) ≥ 2 (S5 Table) at both T24 and T30 time points. This set of DE CDS represented 4.4% of P. anserina’s predicted CDS. Actually, none of the CDS had opposite patterns of differential expression at T24 and T30.
Fig 6. Transcriptomic analysis experimental design.
Schematic developmental time course from fertilization to ascospore maturation of wild-type crosses. Total RNA was extracted from wild-type perithecia at T24 and T30 (open circles) and from ΔPaRid micro-perithecia at T42 (solid circle); Dotted line: time frame during which the ΔPaRid developmental blockage might occur. Light microphotographs of the upper panel illustrate the various developmental steps indicated along the time course.
Functional annotation of the complete set of up- and down-regulated genes
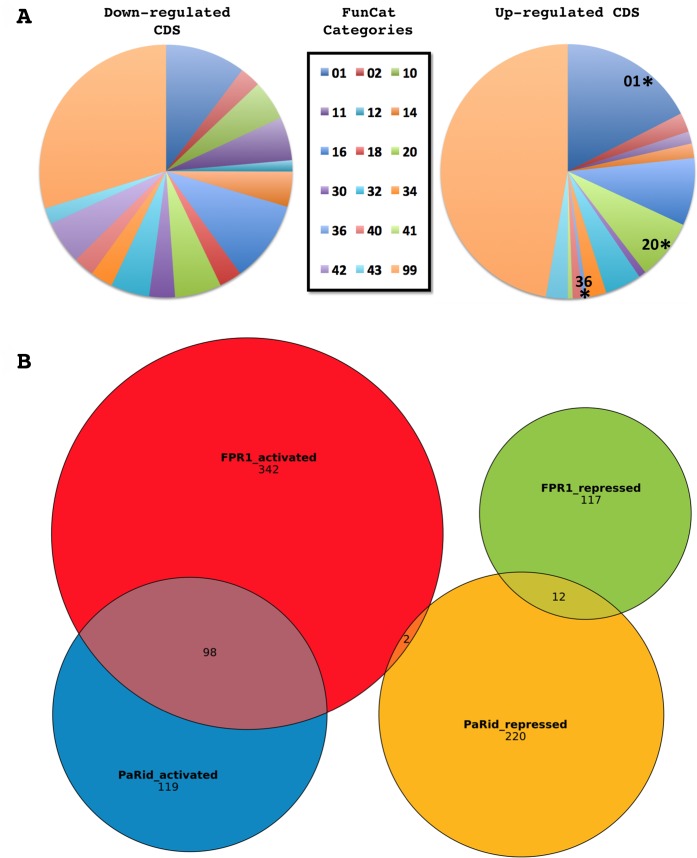
CDS with no predicted function or domain were not over-represented in neither down-regulated (72 out of 217) nor up-regulated (80 out of 234) sets of DE CDS when compared to the total of 3553 such CDS in the genome (S5 Table). The fraction of DE CDS showing N. crassa or S. cerevisiae orthologs was 69.6% and 32.2% respectively in the down-regulated set and 50.9% and 11.9% respectively in the up-regulated set (S5 Table). These observations show that DE CDS having an ortholog either in N. crassa or in S. cerevisiae were significantly less abundant in the up-regulated set than in the down-regulated set (p-value = 0.0455 and 3.10*10−5, respectively). This bias existed also when the up-regulated CDS having an ortholog either in N. crassa or in S. cerevisiae where compared to those of the complete P. anserina’s set of CDS (p-value = 0.0317 and 9.38*10−8, respectively), since among the 10,556 annotated CDS of P. anserina, 6873 are N. crassa’s orthologs (65,1%) and 3312 are S. cerevisiae’s orthogs (31,4%) [36]. This might indicate that the up-regulated set appears more divergent and species specific than the down-regulated set which belongs preferentially to the conserved fungal genome core. We then performed a FunCat analysis [45] to further characterize the function of the DE CDS (Fig 7A, Table 2). Approximately two-third of them belonged to the “Unclassified” category (category number 99) either in the up- or down-regulated sets. Among the 72 classified CDS of the down-regulated set, no FunCat categories were enriched (See Table 2). By contrast, the up-regulated set was enriched in CDS that fell in the “Metabolism” (category number 01) and “Cellular transport” categories (category number 20).
Fig 7. Transcriptomic analysis of differentially expressed CDS in response to ΔPaRid developmental arrest.
(A) Functional categories in the down- and up-regulated CDS sets. Legend of pie charts corresponds to the FunCat categories, see Table 2 for details. Stars mark significantly enriched functional categories (p-value < 0.05). (B) Venn diagram of PaRid and FPR1 targets.
Table 2. Main functional category analysis (FunCat).
| FUNCTIONAL CATEGORY | Number of genes | Enrichment P-VALUE |
|---|---|---|
| DOWN regulated genes | ||
| 01 Metabolism | 50 | 0.6015477193364 |
| 02 Energy | 13 | 0.1175222171120 |
| 10 Cell cycle and DNA processing | 25 | 0.2907628248754 |
| 11 Transcription | 27 | 0.2549629641085 |
| 12 Protein synthesis | 7 | 0.7780707022438 |
| 14 Protein fate | 22 | 0.8422873544643 |
| 16 Protein with binding function or co-factor requirement | 51 | 0.8512335723436 |
| 18 Protein activity regulation | 14 | 0.1205020546264 |
| 20 Cellular transport | 29 | 0.6204933767605 |
| 30 Cellular communication | 16 | 0.1342730657708 |
| 32 Cell rescue, defense and virulence | 24 | 0.3026236390720 |
| 34 Interaction with the cellular environment | 14 | 0.3090089528220 |
| 40 Cell fate | 13 | 0.1673709514827 |
| 42 Biogenesis of cellular components | 27 | 0.0669108316950 |
| 43 Cell type differentiation | 10 | 0.7303867300457 |
| 99 Unclassified | 145 | 0.9252711123620 |
| UP regulated genes | ||
| 01 Metabolism * | 57 | 1.251208625*10−09 |
| 02 Energy | 8 | 0.0605014394295 |
| 11 Transcription | 5 | 0.9863844124318 |
| 14 Protein fate | 6 | 0.9874423656325 |
| 16 Protein with binding function or co-factor requirement | 28 | 0.5725800327713 |
| 20 Cellular transport * | 25 | 0.0071875703621 |
| 30 Cellular communication | 3 | 0.8848848194025 |
| 32 Cell rescue, defense and virulence | 15 | 0.0951283621522 |
| 34 Interaction with the cellular environment | 8 | 0.2011467605014 |
| 36 Systemic interaction with the environment * | 2 | 0.0002724146627 |
| 40 Cell fate | 4 | 0.6104680132063 |
| 41 Systemic development | 2 | 0.0761279956280 |
| 43 Cell type differentiation | 9 | 0.1081833860122 |
| 99 Unclassified | 154 | 0.8954838244443 |
* Functional category significantly enriched, i.e. corresponding p-value < 0.05
Most of the down-regulated DE CDS are involved in developmental pathways
The down-regulated ΔPaRid CDS set was enriched in transcription factors (TFs, FunCat sub-category 11.02.03.04.01 transcriptional activators, p-value = 1.02*10−4). Notably, out of the 17 TFs identified (S6 Table, S8 Fig), 11 were also FPR1 targets. Furthermore, Pa_1_16860 and Pa_3_1720 orthologs in N. crassa [46,47] and A. nidulans [48,49] influence sexual development (S6 Table). On the same note, the N. crassa ortholog of Pa_1_22930 plays a significant role in sexual development [47]. Yet, we did not identify in this down-regulated CDS set, the P. anserina’s orthologs of most of the N. crassa TFs that have been found to be differentially expressed during sexual development [46,50–53].
We also noticed three enzymes related to NAD/NADP oxidoreduction and belonging to the developmental class: a NADPH dehydrogenase (Pa_6_6330), a NADP-dependent oxidoreductase (Pa_5_11750) and a glycerol-3-phosphate dehydrogenase [NAD+] (Pa_1_6190), as well as proteins involved in cellular signal transduction by regulating the phosphorylation status of the intracellular inositol trisphosphate messenger, including PaIPK2 [54] (Pa_5_1490, Pa_6_9890, Pa_1_18990). Interestingly, inositol phosphates are required for both fruiting body number and proper development in Sordariales [55].
In this down-regulated set, we also identified widely conserved CDS regulating cell division: Ras GTPase-activating proteins (Pa_1_10960 and Pa_6_7140), Rho-GTPase-activating protein (Pa_7_10800), cell division control protein (Pa_3_3430), replicating licensing factor (Pa_4_8520) and serine/threonine-protein kinases (Pa_1_9100 and Pa_7_9140). In line with active DNA replication, we also spotted some enzymes or cofactors involved in the nucleotide metabolism, such as a 3',5'-cyclic-nucleotide phosphodiesterase (Pa_7_2860), a dimethyladenosine transferase (Pa_2_13220) and two guanine nucleotide-binding protein alpha-2 subunits (Pa_2_10260, Pa_5_11490). The reduced expression of genes involved in promoting cell division was clearly in line with the growth arrest of the ΔPaRid micro-perithecia. The developmental category may be more informative to uncover some CDS acting downstream of the PaRid network. In this category, we identified the PEX3 peroxisomal biogenesis factor (Pa_7_8080). The crucial role of β-oxidation and the glyoxylate cycle during sexual development has already been documented [56]. We also found a VelvetA-like-1 protein (VeA, Pa_3_6550) [57]. Present in many fungi, the Velvet protein complex seems to have expanded its conserved role in developmental programs to more specific roles related to each organism’s needs [58,59]. Likewise, the IDC1 gene that was found down-regulated in this study is required for both cell fusion and development of the envelope of the fruiting bodies. The list of the down-regulated CDS includes a putative protein presenting a fascilin (FAS1) domain. Such extracellular domain is thought to function as a cell adhesion domain [60] and therefore might play a key role in the development of multicellular structures. A further connection to cell shape dynamics and cytoskeleton was found with the down-regulation of an annexin protein (Pa_6_1130). This calcium-dependent phospholipid-binding protein family has been linked with membrane scaffolding, organization and trafficking of vesicles, endocytosis, exocytosis, signal transduction, DNA replication, etc. The N. crassa homolog of this annexin (NCU04421) was shown to be up-regulated during asexual sporulation [61].
The gene expression regulation class mostly contained general regulating factors as CDS encoding chromatin remodeling proteins. As down-regulated chromatin remodeling factors, we identified two histone-lysine N-methyltransferases, Pa_7_3820 homologous to Set-9/KMT5, which methylates the lysine 20 of histone H4 [62] and Pa_3_3820 homologous to Set-4, one acetyltransferase (Pa_3_10520), one ATP-dependent RNA helicase (Pa_4_8200) and one SNF2 family ATP-dependent chromatin-remodeling factor (Pa_7_9570). Some CDS encoding DNA/RNA processing factors were also found down-regulated in the ΔPaRid mutant micro-perithecia. As such, we identified a putative cruciform DNA recognition protein (Pa_2_440), an ATPase involved in DNA repair (Pa_6_4260), a SIK1-like RNA-binding protein (Pa_5_12950) and the telomere length regulator protein Rif1 (Pa_1_3890). In mammals as in yeast, Rif1 is required for checkpoint-mediated cell cycle arrest in response to DNA damage [63].
Analysis of the up-regulated CDS set uncovers enrichment in the “Metabolism” and “Cellular transport” functional categories
Half of the 26 most up-regulated CDS were putative proteins of unknown function devoid of conserved domains (S5 Table), included the one showing the highest FC (Pa_4_5390). Two CDS encoded enzymes involved in the metabolism of amino acids (Pa_4_140 and Pa_1_5740), two were encoding transporters (Pa_3_420 and Pa_6_11600) and three CDS were involved in secondary metabolism (Pa_4_4580, Pa_5_11000 and Pa_5_720, see below).
More importantly, CDS belonging to primary metabolism were enriched in this up-regulated set (p-value = 2.35*10−9). For instance among the CAZymes, 11 glycoside hydrolases (GH) were found in this set (enrichment p-value = 0.0186). The predicted secondary metabolite clusters identified in this study are mostly composed of CDS encoding proteins of unknown function. Nonetheless, some of them contain CDS encoding putative secondary metabolism related functions: HC-toxin synthetases (Pa_3_11193, Pa_3_11220), cytochrome P450 proteins (Pa_3_2900, Pa_4_4580, Pa_4_4570, Pa_1_9520, Pa_6_7810), polyketide synthase (Pa_5_11000), multidrug efflux systems (Pa_3_11220, Pa_4_3775), trichodiene oxygenase (Pa_3_5540) and an O-methylsterigmatocystin oxidoreductase (Pa_2_7080). If they are not the result of cellular stresses induced in the ΔPaRid mutants, these secondary metabolites could act as secondary messengers during P. anserina sexual development. CDS encoding transporters were also over-represented in the up-regulated CDS set (p-value = 0.0071, see Table 2), as if arrest of perithecium development would generate cellular flux. Surprisingly, we also identified eight CDS encoding HET domain-containing proteins (Pa_2_4570, Pa_2_9350, Pa_2_8040, Pa_3_2610, Pa_5_1080, Pa_5_7650, Pa_6_1970, Pa_6_6730) (enrichment p-value = 0.0390). The HET gene family is known to prevent cell fusion in filamentous fungi by inducing cell death program when genetically different nuclei cohabit in a common cytoplasm [64]. Formation of heterokaryotic mycelia and potential incompatibility responses to non-self are vegetative processes that might be repressed through the PaRid network.
Comparison of PaRid regulated genes and mating-type target genes shows a significant overlap
We compared the ΔPaRid DE CDS to the mating-type target genes identified previously [65]. P. anserina has a typical heterothallic mating-type structure with two idiomorphs. The MAT1-1 idiomorph (mat-) is characterized by the MAT1-1-1 gene, which encodes the FMR1 MATα-HMG domain containing protein, while the MAT1-2 idiomorph (mat+) is composed of the MAT1-2-1 gene, which encodes the FPR1 MATA_HMG domain containing protein (reviewed in [66]). Both proteins are transcription factors essential for fertilization in heterothallic Pezizomycotina and development of the fruiting bodies (reviewed in [38]). Microarray comparisons of wild-type mat+ versus fpr1- mutant strains, and wild-type mat- versus fmr1- mutant strains revealed 571 and 232 target genes of FPR1 and FMR1, respectively [65]. The authors have determined that among the FPR1 target genes, 442 are activated and 129 are repressed. Similarly, among the FMR1 target genes, 151 are activated and 81 are repressed. Comparing these activated and repressed mating-type target genes with the genes down- and up-regulated in the ΔPaRid mutant strain showed a significant overlap (Fig 7B, S5 Table). The 217 PaRid activated CDS contained 98 FPR1 activated targets, which was clearly indicative of a strong enrichment (p-value = 5.40*10−46). FPR1 acts also as a repressor: accordingly, we found 14 FPR1 repressed targets among the 234 PaRid repressed CDS, which did not correspond to any enrichment (p-value > 0.05) (Fig 7B, S5 Table). Strikingly, we noticed that there is no CDS that would be both activated by PaRid and repressed by FPR1, while only two CDS were repressed by PaRid and activated by FPR1 (Fig 7B, S5 Table). This observation indicates a strong congruence of the regulatory pathways of PaRid and mating-type gene FPR1. Conversely, only 17 FMR1 targets were identified in the 541 PaRid regulated CDS. This low number of FMR1 targets (mat- idiomorph) is consistent with the ΔPaRid mat+ mutant strain being used as the female partner in the microarray experiments (Table 3) and our above observation that PaRid is dispensable in the male partner (here mat-). Out of the 17 FMR1 targets, five were found in the ΔPaRid down-regulated CDS set whereas 12 were found in the ΔPaRid up-regulated CDS set (S5 Table).
Table 3. Crosses used in transcriptome analysis.
| Denomination | Crosses | Female strain | Male strain | Sampling time |
|---|---|---|---|---|
| Wild type cross | S mat+ x S mat- | S mat+ | S mat- | T24 and T30 |
| ΔPaRid cross | S mat+ ΔPaRid x S mat- | S mat+ ΔPaRid | S mat- | T42 |
Discussion
Sexual reproduction is considered to be essential for long-term persistence of eukaryotic species [67]. Only a few asexual lineages are known to persist over a long period of time without sex [68], most eukaryotes engaging sexual reproduction at some point in their life cycle. Studies have shown that sex reduces the accumulation of deleterious mutations compared to asexual reproduction [69] but also provide a faster adaptive response, by bringing together favorable gene combinations [70,71]. In multicellular eukaryotes, sexual reproduction is controlled by strict mechanisms governing which haploids can fuse (mating) and which developmental paths the resulting zygote will follow. These strict mechanisms are both genetically and epigenetically determined. Among the epigenetic modifications that control gene expression, DNA methylation reprogramming allows cells to shape their identity by launching and maintaining differential transcriptional programs in each cell type. In mammals, specific differential genomic DNA methylation patterns of parental gametes, known as ‘DNA methylation imprints’ are not essential to karyogamy but to zygotic development [72] while global genomic demethylation in A. thaliana results in male-fertile but female-sterile plants [24,25]. It has been hypothesized that the regulation of gene expression by DNA methylation during the development of higher eukaryotes may have been acquired from ancestral mechanisms of genome defense against invasive repetitive elements such as transposons. Some of the multicellular fungi are endowed with a homology-based genome defense system (RIP or RIP-like) exhibiting epigenetic features in addition to a functional link to sexual development [12,19]. Nonetheless, very little is known about epigenetic regulation of gene expression and development in multicellular fungi, although animal and fungi are each other’s closest relatives. In this study, we took advantage of a model ascomycete that displays ongoing RIP but no DNA methylation [15], to explore the role of PaRid, a DMT-like protein, part of the Masc1/RID family, conserved in fungi [12,13].
PaRid is involved in the formation of croziers
We report that PaRid plays a central role in the mid-time course of sexual development of P. anserina. Homozygous ΔPaRid crosses, as heterozygous ones, can perform fertilization, which depends on the recognition of male cells by female organs. Therefore PaRid is not involved in the early steps of sexual development. However, the PaRid wild-type allele must be expressed in the female gametes for further perithecium development. The most conspicuous developmental step after fertilization is the formation of the dikaryotic cells, which is preceded by the division of parental nuclei in a syncytium (ascogonial plurinucleate cell). If PaRid is absent from the maternal lineage, the perithecia stop to grow prematurely (micro-perithecia phenotype) and no croziers emerge from the biparental plurinucleate ascogonial cells that they contain. Furthermore providing surrogate maternal tissue either by grafting the mutant micro-perithecia to a wild-type mycelium or by introducing a Δmat partner to the ΔPaRid mat+; ΔPaRid mat− dikaryon did not rescue the maturation defect of the fruiting bodies, as previously shown for some of the P. anserina female sterile mutants [40,73–75]. This result demonstrates that the ΔPaRid mutant sterility is not due to a perithecium envelope building failure, nor to an improper feeding of the maturing fruiting body, but to the lack of ascogenous tissue. To date, among the P. anserina sterile mutants that have been studied, these macroscopic (micro-perithecia) and microscopic (no dikaryotic cells) phenotypes are both found only in the ΔSmr1 mutant [76]. SMR1, MAT1-1-2 in the standard nomenclature [77], is a conserved protein of unknown molecular function, which is required for the initial development of the dikaryotic stage of both Gibberella zeae [78] and Sordaria macrospora [79]. A noticeable difference between PaRid and SMR1/MAT1-1-2 is that the former must be present in the maternal lineage (mat+ or mat-), while the latter is present only in the mat- nucleus (male or female). Genetic observations support the idea that SMR1 diffuses from mat- to mat+ nuclei inside the fruiting body, even if mat- nuclei come from a male gamete [80]. In contrast, our observations based on the tagging of PaRid with the GFP indicate that this gene is not expressed in the male gamete, and complementation experiments suggest that PaRid is not expressed in nuclei coming from male gametes inside the perithecia. It has been proposed that SMR1 releases the developmental arrest following inter-nuclear recognition between two compatible nuclei in the plurinucleate ascogonial cell and could consequently trigger crozier formation [81]. Besides, in contrast to the ΔPaRid and ΔSmr1 deletions, the previously identified P. anserina mutants affecting the zygotic lineage (Smr2, Cro1 and Ami1), generate either uninucleate croziers [34,35,76] or plurinucleate croziers [82]. Consequently, because the corresponding mutants do indeed form these early hook-shaped structures, PaRid probably acts at earlier developmental stages than Smr2, Cro1 and Ami1, in line with its expression pattern. Taken together, these data suggest that SMR1 releases the developmental arrest following inter-nuclear recognition by diffusing from the mat- to mat+ nucleus, while PaRid is a maternal gene that is required for further development, especially the formation of the croziers.
PaRid is an activator of the mid-time course of P. anserina’s sexual development
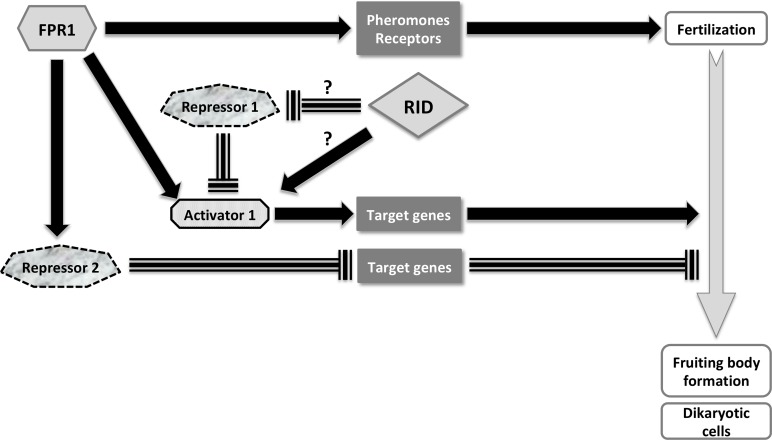
In an effort to uncover the PaRid regulatory network and identify potential co-factors, we performed a transcriptomic analysis of mutant crosses. Strikingly, this study established that almost half of the CDS activated by PaRid (45.16% of the down-regulated set) are also activated by the FPR1 transcription factor [65]. In agreement with the fertilization ability of ΔPaRid, the mat+ prepropheromone gene and the mat+ pheromone receptor gene are not included in the set of genes down-regulated in ΔPaRid background. Consequently, we concluded that 1) PaRid is a key actor of the middle steps of P. anserina sexual development; 2) PaRid along with FPR1 is involved in the developmental pathway that follows fertilization and leads to formation of dikaryotic cells; 3) PaRid, as FPR1, is an activator of this pathway. Furthermore, the expression of fpr1 is not deregulated in the ΔPaRid mutant background, and conversely the expression of PaRid is not deregulated in the fpr1- mutant background [65]. This result suggests that PaRid acts neither upstream nor downstream of FPR1, but rather as an independent branched pathway mediated by a cascade of repressors and activators, which is the more parsimonious model to explain the overlap of PaRid and FPR1 target genes (Fig 8). The PaRid-regulated CDS set includes many FPR1 targets (n = 112) and few FMR1 targets (n = 17). This observation further supports the maternal effect of the PaRid deletion, as we have used a mat+ (FPR1) strain as a maternal strain in our experiments. Determination of mating-type gene targets was performed at the stage of competence for fertilization [65], a stage belonging to the very early steps of sexual development of perithecia. As the mating-type gene targets are likely to vary according to the developmental stage [81], their determination in the middle steps of fruiting-body development would be more appropriate to the comparison with those of PaRid and might reveal even more extensive overlap. MAT1-2-1 (FPR1) and MAT1-1-1 (FMR1) are ubiquitous in Pezizomycotina (reviewed in [66]). We propose that the overlap of mating-type targets genes and PaRid-regulated genes is present in all fungi in which the inactivation of Rid results in sexual development defect. At present, Rid linked sexual defect were described in A. immersus [12] and A. nidulans [19]. Unfortunately, neither mating type target genes nor Rid regulated genes were available in these fungi to test our proposal.
Fig 8. Schematic representation of PaRid & FPR1 developmental pathways during sexual development.
FPR1, a MATα-HMG transcription factor is essential to fertilization and development of fruiting bodies. This mating type protein can act either as an activator or as a repressor. This study established that PaRid shares part of the FPR1 positive regulatory circuit, which is at work after fertilization to build the fructification and to form the dikaryotic cells. As the methyltransferase activity is required for PaRid function, we hypothesized that PaRid might repress a repressor or alternatively might activate an activator of the FPR1 regulatory circuit [118]. Solid black arrow is indicative of activation. Dashed T-line is indicative of repression. Solid grey arrow represents the sexual development time line.
However, this list of DE CDS did not draw a clear picture of a potential epigenetic regulation process, if any, in which PaRid could be involved, nor the nature of the putative repressor immediately downstream of it (Repressor 1 in Fig 8). Notably, neither the constitutive heterochromatin factors (the putative H3K9me3 methylase: Pa_6_990, the ortholog of the heterochromatin protein 1: Pa_4_7200, the components of the DCDC complex of N. crassa [83]: Pa_6_250, Pa_7_5460, Pa_2_10970, Pa_3_6830) nor the facultative heterochromatin factors (Pa_1_6940 encoding the putative H3K27 methylase and the associated subunits of the PRC2 complex: Pa_3_4080, Pa_2_11390) are differentially expressed. Genes involved in the STRIPAK complex [84] were not differentially expressed either. In consequences, the nature of Repressor 1 cannot be hypothesized yet (Fig 8). Nonetheless, this study identified a set of TFs that might be candidate to enter the specific PaRid genetic network, since eight out of 17 TFs of the ΔPaRid down-regulated CDS set are not FPR1 targets. Additional functional studies on a selected subset of specifically DE CDS identified in this study are required to explore the PaRid downstream pathway(s), which are essential to sexual development.
By contrast, to the down-regulated set, the up-regulated one only contains a handful of developmentally relevant CDS. The functional annotation of the up-regulated set points toward CDS involved in physiological responses to the cellular stress following micro-perithecium developmental arrest.
One toolkit, different outcomes
The ΔPaRid mutant phenotype resembles the ones observed in A. immersus [12] and in A. nidulans [19], where development of croziers and of the subsequent ascospores are never observed in Masc1 mutants or dmtA mutants, respectively. Remarkably, sexual development of homozygous T. reesei rid mutant crosses deleted for the rid homolog (TrRid) allows the formation of dikaryotic cells but is blocked during karyogamy (Wan-Chen Li and Ting-Fang Wang, personal communication). On the contrary, neither barren perithecia nor fertility defect are observed in N. crassa crosses homozygous for the rid null allele [13], although this fungus shows heavy DNA methylation of repeats subjected to RIP. The function of rid orthologs has also been addressed in fungal species that reproduce asexually (Aspergillus flavus [85], Cryphonectria parasitica [86], Metarhizium robertsii [87]). Interestingly, in these species, in addition to a decrease of DNA methylation contents, the absence of DMT-like fungal specific Masc1/RID proteins results in a large palette of phenotypes including reduction of clonal dispersion (conidiation and sclerotial production), defects of mycelium morphology, decrease in secondary metabolite production and/or virulence toward plant hosts. In Magnaporthe grisea, a pathogenic fungus that can reproduce through both sexual and asexual reproduction, deletion of the rid ortholog also has a negative impact on conidiation and mycelium morphology but not on virulence [74].
Altogether, these observations are puzzling. It has led us to consider why such a gene, known to be involved in RIP-like genome defense systems and conserved both in terms of presence and sequence identity, would play an essential role during the sexual development in organisms showing RIP mutations at low level, and very little genomic DNA methylation, if any [15]. One can wonder if RIP, as a genome defense mechanism operating after fertilization but before karyogamy, is not by itself a checkpoint required for the development of dikaryotic hyphae in P. anserina, A. nidulans and A. immersus, and for karyogamy in T. reesei. In this hypothesis, in the absence of the ‘genomic quality control’ of the two haploid parental nuclei performed by RIP before every single karyogamy, the sexual development might stop. In this study, it was not possible to test whether PaRid is also required for RIP since homozygous ΔPaRid mutant crosses were barren, except for rare sporadic asci (only 3 recovered to date) and RIP frequency is low in P. anserina [34]. Nevertheless, all of the 12 ascospores that we recovered from these mutant crosses were indistinguishable from those of wild-type: their shape and pigmentation were standard, they germinated efficiently and subsequent genetic analyses showed that they did not result from uniparental lineages. Consequently, if RIP functions as a checkpoint, it does not prevent viability of progeny that circumvents the function of PaRid. In N. crassa, after karyogamy, genome integrity is further checked through the meiotic silencing of unpaired DNA process (MSUD) [89]. MSUD proceeds during meiosis to reversibly silence genes contained in DNA segments that are not paired with their homolog during meiotic homolog pairing. Because evolution often scrambles chromosomal micro-syntheny, the MSUD prevents interspecific crosses. If deletion of PaRid results in RIP deficient strains, then inactivation of genome defense system acting on haploid parental nuclei has clearly different outcomes, since it would promote repeat spreading.
Ancestral dual roles for DMT-like fungal specific Masc1/RID?
Alternatively, PaRid might play a dual role, possibly relying on two independent enzymatic functions: one as the central effector of RIP and the other as a positive regulator of gene expression during the early steps of sexual development. Our results show that the PaRid function relies on its methyltransferase activity, and that this enzymatic activity has to be present in the female partner. Ascogonia, the female gametes, are small structures of specialized hyphae that differentiate within the mycelium [38]. As such, they represented an extremely low fraction of the so-called “vegetative” tissue when four-day-old mycelium was collected to extract total RNA. This might be why no PaRid mRNAs were detected at this stage while genetic data further supported by cytoplasmic observations indicate that PaRid is specifically present in the female gametes. Besides, since driving its expression from a constitutive promoter did not result in any unusual phenotypes, regulation of PaRid expression, both in terms of timing and cell-type specificity, might proceed through post-transcriptional and/or post-translational mechanisms. Although we did not detect any evidence of antisense PaRid transcript as described in A. nidulans [19], regulation of expression through non-coding RNA (ncRNA) is widely conserved in mammals, Drosophila, plants and S. pombe [90,91]. Therefore, we cannot rule out that the PaRid pattern of expression is also regulated by ncRNA. Alternatively, the PaRid half-life might be tightly regulated by the ubiquitin proteasome system (UPS) [92–95]. Again, this hypothesis remains to be further explored.
Masc1/RID fungal-specific DMTs-like form a quite ancient group of proteins, whose initial DNA-methyltransferase function may have evolved differently in distinct lineages. The predicted structures of RID homologues, although conserved, are not identical [96]. In particular, the Masc1/DmtA/TrRid/PaRid proteins include a compact catalytic domain with a short C-terminal extension (at most 133 amino acids for P. anserina) when compared to that of N. crassa (260 amino acids). Mutant analyses revealed that the Masc1/DmtA/TrRid/PaRid proteins fulfill both sexual developmental functions and genome defense (either RIP is still active or traces of RIP are present as relics in the genomes), whereas the N. crassa RID protein plays a more specialized role, limited to its genome defense function. Taken these observations altogether, one can wonder whether the long C-terminal extension of N. crassa RID is responsible for its restricted function. In addition, despite a structural and functional conservation of the Masc1/DmtA/TrRid/PaRid group of proteins, some species-specific co-factors would be required for enzymatic catalysis. To date, none of them has been identified, although it has been suggested that the DIM-2 ortholog of M. grisea, MoDIM-2, mediates the MoRID de novo methylation [88,97].
Finally, since the Masc1/DmtA/TrRid/PaRid group appears to have a non-canonical motif VI structure when compared to all other prokaryotic and eukaryotic C5-cytosine methyltransferases [2], it is possible that this class of enzymes has acquired exclusive catalytic and/or substrate properties [33]. Although they do not share the features of Dnmt2, the mammalian tRNA cytosine methyltransferase, one can hypothesize that some of these fungal DMT-like enzymes could methylate RNA substrate(s). However, to date, only N6-methyladenosine (m6A), the most prevalent modification of mRNA in eukaryotes, has been linked to developmental functions. For example, the budding yeast N6-methyladenosine IME4 controls the entry of diploid cells into meiosis [98] while lack of the A. thaliana ortholog MTA leads to embryonic lethality [99].
Further functional studies on the fungal DMT-like proteins should also help understand whether the critical PaRid-related developmental program is a conserved feature of Pezizomycotina, secondarily lost by N. crassa during fungal evolution.
Materials and methods
Strains and culture conditions
The strains used in this study derived from the « S » wild-type strain that was used for sequencing [36,100]. Standard culture conditions, media and genetic methods for P. anserina have been described [41,101] and the most recent protocols can be accessed at http://podospora.i2bc.paris-saclay.fr. Construction of the Δmus51::su8-1 strain lacking the mus-51 subunit of the complex involved in end-joining of broken DNA fragments was described previously [102]. In this strain, DNA integration mainly proceeds through homologous recombination. Mycelium growth is performed on M2 minimal medium. Ascospores do not germinate on M2 but on a specific G medium. The methods used for nucleic acid extraction and manipulation have been described [103,104]. Transformation of P. anserina protoplasts was carried out as described previously [105].
Identification and deletion of the P. anserina PaRid gene
The PaRid gene was identified by searching the complete genome of P. anserina with tblastn [106], using RID (NCU02034.7) [13] as query. One CDS Pa_1_19440 (accession number CDP24336) resembling this query with significant score was retrieved. To confirm gene annotation, PaRid transcripts were amplified by RT-PCR experiments performed on total RNA extracted from developing perithecia (female partner mat+/male partner mat-) at either 2 days (T48) or 4 days (T96) post-fertilization using primers 5s-PaRid/3s-PaRid (See S1 Table). Sequencing of the PCR products did not identify any intron in the PaRid ORF, thus confirming the annotation (see S1B Fig).
Deletion was performed on a Δmus51::su8-1 strain as described in [74] and verified by Southern blot as described in [107]. Because PaRid is genetically linked to the mating-type locus, no recombination occurs between the two loci and therefore the gene deletion has been performed in the two mating-type backgrounds to get sexually compatible ΔPaRid mutants. For each deletion, several independent transformants have been checked. One mutant of each mating type has been then selected for complementation and further analyses.
Allele construction
Two plasmids were mainly used in this study: the pAKS106 plasmid which derived from the pBCPhleo plasmid [108] and the pAKS120 plasmid which derived from pAKS106. pAKS106 contains an HA tag sequence followed by the ribP2 terminator at the unique ClaI and ApaI sites [109]. The pAKS120 was generated by cloning the promoter of the AS4 gene, which is highly and constitutively expressed throughout the life cycle [42], at the unique NotI site. Thus, recombinant C-terminus HA-tagged proteins can be produced from pAKS106 and pAKS120, the expression of the corresponding fusion gene being either under the control of its own promoter (pAKS106) or under the control of the constitutive highly expressed AS4 promoter (pAKS120). Because our allele construction strategy required high-fidelity DNA synthesis, all PCR amplifications were performed using the Phusion High Fidelity DNA polymerase (Thermo Scientific), according to the manufacturer’s protocol and the resulting alleles were systematically sequenced to ensure that no mutation was introduced during PCR amplification (see S1 Table for primers sequences) and that, when relevant, the HA-tag or GFP sequences were in the appropriate reading frame.
Constructing the PaRid-HA and AS4-PaRid-HA alleles and their GFP-tagged version
To complement the ΔPaRid::hph mutant strain, the wild-type PaRid allele was cloned into the pAKS106 plasmid. To this end, the wild-type PaRid allele was PCR amplified from the GA0AB186AD09 plasmid [36], using the primers PaRIDHAFXbaI and PaRIDHAREcoRI (see S1 Table). The 2914-bp amplicon, which corresponds to 653-bp promoter sequence followed by the complete PaRid CDS, minus the stop codon, was digested with the XbaI/EcoRI restriction enzymes and cloned into the pAKS106 plasmid, also hydrolyzed with the XbaI/EcoRI restriction enzymes to obtain the PaRid-HA allele. In order to put the PaRid allele under the control of a highly and constitutively expressed promoter, we then constructed the AS4-PaRid-HA allele. To do so, the wild-type PaRid allele was PCR amplified from the GA0AB186AD09 plasmid [36], using the primers PaRIDAS4XbaI and PaRIDHAREcoRI (see S1 Table). This pair of primers was designed to amplify the complete PaRid ORF, minus the stop codon. The resulting 2263-bp amplicon was digested with the XbaI/EcoRI restriction enzymes and cloned into the pAKS120 plasmid, also hydrolyzed with the XbaI/EcoRI restriction enzymes.
To tag the PaRid protein with the Green Fluorescent Protein (GFP), we PCR amplified the ORF of the eGFP minus the stop codon with primers RIDGFPFEcoRI and RIDGFPRClaI (see S1 Table), using the pEGFP-1 plasmid (Clontech, Mountain View, CA) as template. The resulting 713-bp fragment was digested with EcoRI and ClaI and cloned in frame with the PaRid allele either into the PaRid-HA plasmid or the AS4-PaRid-HA plasmid (see above), previously digested with the same restriction enzymes. This yielded the chimeric PaRid-GFP-HA and AS4-PaRid-GFP-HA alleles.
Constructing the site-directed mutant PaRidC403S-HA
The PaRidC403S allele was constructed by in vitro site-directed mutagenesis. The GA0AB186AD09 plasmid harboring the wild-type PaRid allele was amplified by inverse PCR, using the divergent overlapping primers RIDmut1 and RIDmut2. The RIDmut1 primer leads to G-to-C substitution at position 1208 of the PaRid sequence (see S1 Table, marked in bold italics), that matches the converse C-to-G substitution located in the RIDmut2 primer (see S1 Table, marked in bold italics). The PCR amplicons were digested with the DpnI enzyme (Fermentas), self-ligated and then transformed into E. coli competent cells. The PaRid allele was sequenced from several plasmids extracted from independent chloramphenicol resistant E. coli colonies. One allele showing no other mutation but the directed TGT (cysteine) to TCT (serine) transvertion was selected as the PaRidC403S allele. The PaRidC403S allele was then PCR amplified using the primers PaRIDHAFXbaI and PaRIDHAREcoRI. The 2914-bp amplicon, which corresponds to 653-bp promoter sequence followed by the complete PaRid ORF, at the exception of the stop codon, were digested with the XbaI/EcoRI restriction enzymes and cloned into the pAKS106 plasmid, also hydrolysed with the XbaI/EcoRI restriction enzymes. This resulted in the PaRidC403S-HA allele. Then, to get the PaRid allele under the control of a highly and constitutively expressed promoter, we constructed the AS4-PaRidC403S-HA allele. To do so, the PaRidC403S allele was PCR amplified from the pAKS106-PaRidC403S-HA, using the primers PaRIDAS4XbaI and PaRIDHAREcoRI (see S1 Table). This pair of primers was designed to amplify the complete PaRid ORF minus the stop codon. The resulting 2263-bp amplicon was digested with the XbaI/EcoRI restriction enzymes and cloned into the pAKS120 plasmid, also hydrolyzed with the XbaI/EcoRI restriction enzymes. This yielded the pAKS120-AS4-PaRidC403S-HA plasmid. All the PCR-amplified fragments were sequenced. All the strains used in this study are listed in S7 Table.
Cytology & microscopy analysis
Perithecia were harvested from 12 to 96 h after fertilization. Tissue fixation was performed as in [110]. Pictures were taken with a Leica DMIRE 2 microscope coupled with a 10-MHz Cool SNAPHQ charge-coupled device camera (Roper Instruments), with a z-optical spacing of 0.5 mm. The GFP filter was the GFP-3035B from Semrock (Ex: 472nm/30, dichroïc: 495nm, Em: 520nm/35). The Metamorph software (Universal Imaging Corp.) was used to acquire z-series. Images were processed using the ImageJ software (NIH, Bethesda).
Phenotypic analyses
Spermatium counting was performed as follows: each strain was grown on M2 medium at 27°C for 21 days. To collect spermatia, cultures were washed with 1.5 mL of 0.05% Tween 20 in sterile water. Numeration proceeded through Malassez counting chamber. Grafting was assayed as in [41]. Western blot analyses were performed on perithecia grown for two days, as in [109]. We used the anti-HA high affinity monoclonal antibody from mouse to recognize the HA-peptide (clone HA-7, ref H3663, Sigma-Aldrich). Detection was performed using Chromogenic Western Blot Kit (Anti-Mouse, EnzoLife, ENZ-KIT182), molecular scale was given by the Prestained Protein Molecular Weight Marker (Thermo Scientific, 11583100)
Phylogenetic analysis
Orthologous genes were identified using FUNGIpath [111] and MycoCosm portal [112] and manually verified by reciprocal Best Hits Blast analysis. Sequences were aligned using MUSCLE (http://www.ebi.ac.uk/Tools/msa/muscle/) and trimmed using Jalview to remove non-informative regions (i.e. poorly aligned regions and/or gaps containing regions). Trees were constructed with PhyML 3.0 software with default parameters and 100 bootstrapped data set [113]. The tree was visualized with the iTOL version 4.3 (http://itol.embl.de/). Functional annotation was performed using InterPro 71.0 (http://www.ebi.ac.uk/interpro/search/sequence-search), Panther Classification System version 14.0 (http://www.pantherdb.org/panther/), PFAM 32.0 (http://pfam.xfam.org/), Prosite (http://prosite.expasy.org/). Proteins were drawn using IBS v1.0.2 software (http://ibs.biocuckoo.org/)
RNA preparation for microarray
The male partner was grown for 6 days at 27°C on M2 medium. Then, spermatia were collected by washing the resulting mycelium with 1.5 mL of H2O per Petri dish (104 spermatia / ml). The female partner strains were grown for 4 days at 27°C on M2 covered with cheesecloth (Sefar Nitex 03-48/31 Heiden). The sexual development time-course experiment started when female partners were fertilized by spermatia (1.5*104 spermatia / cross). This time point was referred to as T0 (0 hour). By scraping independent crosses, samples of 20 to 100 mg of growing perithecia were harvested 24 hours (T24) and 30 hours (T30) after fertilization from wild-type crosses and 42 hours (T42) after fertilization from ΔPaRid crosses and immediately flash-frozen in liquid nitrogen. For each time point, we collected three biological replicates originating from three independent crosses. The frozen samples materials were grinded using a Mikro-dismembrator (Sartorius, Goettingen, Germany). Total P. anserina RNAs were extracted using the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) with DNase treatment. The quality and quantity of total RNA was assessed using a Nanodrop spectrophotometer (Nanodrop Technologies, Wilmington, USA) and the Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, USA). This protocol was performed on two genetically distinct crosses as specified in Table 3.
Labelling and microarray hybridization
Gene expression microarrays for P. anserina consisted of a custom 4×44 K platform (AMADID 018343, Agilent, Santa Clara, USA) containing 10,556 probes on each array with each probe in four replicates. Microarray hybridization experiments including target preparation, hybridization and washing were performed as described in [44]. The experimental samples were labeled with the Cy5 dye and reference sample with the Cy3 dye. The reference sample is a mixture of RNAs extracted from different growth conditions.
Microarray data acquisition
Microarrays were scanned using the Agilent Array Scanner at 5μm/pixel resolution with the eXtended Dynamic Range (XDR) function. Array quality and flagging were performed as described [114]. Raw data pre-processing was performed with Feature Extraction (v9.5.3) software (Agilent technologies), with the GE2-v4_95_Feb07 default protocol. Microarray data reported in this paper have been deposited in the Gene Expression Omnibus database under the accession no. GSE104632.
Statistical analyses: Microarray data normalization, differential analysis and functional enrichment
Raw data normalization and differential analysis were done using the MAnGO software [115], and a moderate t-test with adjustment of p-values [116] was computed to measure the significance with each expression difference. Differential analyses have been performed between the wild-type cross at 24 (T24) and 30 (T30) hours after fertilization and the mutant cross. Fold change values (FC) were calculated from the ratio between normalized intensities at each time of the time course. Differentially expressed (DE) coding sequences (CDS) in the ΔPaRid cross were defined as those whose adjusted P-values are inferior to 0,001 and the absolute value of FC higher than 2. The resulting list was called DE CDS specific of ΔPaRid cross. Functions of genes identified as up- or down-regulated in transcriptomics data were explored using FunCat functional categories (Level 1, as described in [45]). To assess whether any of the functions were observed in up- or down-regulated lists of genes at a frequency greater than that expected by chance, p-values were calculated using hypergeometric distribution as described in [117].
Supporting information
(A) Average expression profiles (y-axis) of PaRid (Pa_1_19440) during sexual development (x-axis, hours). (B) Amplification of PaRid transcripts (2296 bp) by RT-PCR. MW: GeneRuler DNA Ladder Mix (Thermo Fisher Scientific), RT T48, RT T96: RT-PCR performed on RNA extracted from 2 days or 4 days post fertilization developing perithecia, gDNA: genomic DNA, NRT: PCR performed on RNA extracted from 2 days or 4 days post fertilization developing perithecia, Neg: No RNA. See (Materials and methods section for details). (C) Coverage of RNA-seq mapped reads at the PaRid locus [37]. RNA-seq experiments were performed on RNAs extracted from non-germinated ascospores (Ascospores), eight hours germinating ascospores (Germinated ascospores), 1-day- or 4-day-old mycelia, 2 days or 4 days post fertilization developing perithecia.
(TIF)
Schematic representations of the endogenous and disrupted loci are given in. Replacement by homologous recombination of the wild type PaRid allele by the disrupted ΔPaRid allele results in the substitution of a 3.2 kb PstI fragment by a 2.3 kb PstI fragment as revealed by hybridization of the 5’UTR digoxygenin-labeled probe (S2A) and in the substitution of a 2.8 kb PstI fragment by a 4.0 kb PstI fragment as revealed by hybridization of the 3’UTR digoxygenin-labeled probe (S2A).
(TIF)
Major steps of P. anserina’s life cycle as shown by a schematic representation (upper panel) and the corresponding light microphotographs (lower panel). P. anserina’s life cycle begins with the germination of an ascospore (A) that gives rise to a haploid mycelium (B). After three days of growth, both male gametes (spermatia, B, top) and female gametes (ascogonia, B, bottom) are formed. Because most of the ascospores carry two different and sexually compatible nuclei (mat+ and mat- mating types) P. anserina strains are self-fertile (pseudo-homothallism). Before fertilization occurs, ascogonia can mature into protoperithecia by recruiting protective maternal hyphae to shelter the ascogonial cell. A pheromone/receptor signaling system allows the ascogonia to recognize and fuse with spermatia of compatible mating type (heterothallism). Fertilization initiates the development of the fruiting body (perithecia, C) in which the dikaryotic mat+/mat- fertilized ascogonium forms. Further development leads to a three-celled hook-shaped structure called the crozier (D). The two parental nuclei in the middle cell of the crozier fuse (karyogamy, schematic representation D) to form a diploid nucleus, which then immediately undergoes meiosis. The four resulting haploid nuclei undergo mitosis. In most cases, ascospores are formed around 2 non-sister nuclei within the developing ascus. On rare occasions, two ascospores are formed around only one haploid nucleus each, leading to a five-ascospore ascus (E, photograph). Scale bar: 10 μm in (A-D); 200 μm in (E).
(TIF)
Strains were grown on M2 minimal medium for 6 days at 27°C. S: Wild-type strain. See Materials and Methods section for details on the complemented ΔPaRid:AS4:PaRid:HA strain.
(TIF)
(A) Cryosensitivity assay: growth rate at two sub-optimal temperatures compared with optimal temperature. (B) Thermosensitivity assay: growth rate at 37°C compared with optimal temperature. (C) Longevity assay. For each genotype, growth was assayed on 3 independent cultures after 4 days of growth at the indicated temperatures. Each experiment was performed three times. For each genotype, longevity was measured on three independent cultures, issued from three individual ascospores, as described in [42].
(TIF)
(A) Morphological comparison of perithecia obtained from wild-type crosses (PaRid), ΔPaRid crosses (ΔPaRid) and ΔSmr1 crosses (ΔSmr1). Size and morphology of the ΔPaRid and ΔSmr1 perithecia are alike. Scale bar: 250 μm. (B) Perithecia obtained in the indicated trikaryons on M2 medium after 5 days at 27°C. The Δmat; PaRid+ mat-; PaRid+ mat+ trikaryons form typical fully developed perithecia (left panels). By contrast, only blocked micro-perithecia are formed by the Δmat; ΔPaRid mat-; ΔPaRid mat+ trikaryons (right panels). (C) Heterozygous orientated crosses PaRid+ mat- × ΔPaRid mat+ after 5 days at 27°C. When the wild-type PaRid+ allele is present in the female gametes genome and the mutant ΔPaRid allele is present in the male gamete genome fully developed perithecia are formed, conversely when the mutant ΔPaRid allele is present in the female gametes and the wild-type PaRid+ allele is present in the male gamete, only blocked micro-perithecia are formed. Left panel, scale bar: 1 mm, right panel, scale bar: 500 μm.
(TIF)
Western-blot was performed as described in Material and Methods section and probed with an anti-HA antibody that specifically detects the HA-tagged proteins. The PaRid protein (83 kDa) was barely detectable when the expression of the ectopic alleles was driven from its native promoter (ΔPaRid:PaRid, ΔPaRid:PaRidC403S). However, it was readily produced when the expression of the ectopic alleles was under the control of a strong and constitutive promoter (ΔPaRid:AS4-PaRid, ΔPaRid:AS4-PaRidC403S). Complementation of the fertility defect was obtained by insertion of the wild type PaRid-HA allele or by insertion of the AS4-PaRid-HA only. Negative controls (No PaRid-HA tagged): PaRid+ wild-type strains mat+ (lane 1) & mat- (lane 3), ΔPaRid mutant strain (lane 4), ΔPaRid:PaRid+-GFP strain (no HA tag, GFP only, complemented strain, lane 7). ΔPaRid:PaRid+ = ΔPaRid mutant strain harboring an ectopic wild type PaRid+-HA allele (complemented strain, lane 5), ΔPaRid:AS4-PaRid = ΔPaRid mutant strain harboring an ectopic AS4-PaRid+-HA allele (complemented strain, lane 6), ΔPaRid:AS4-PaRidC403S = ΔPaRid mutant strain harboring an ectopic AS4-PaRidC403S-HA allele (non-complemented strain, lane 8). ΔPaRid:PaRidC403S = ΔPaRid mutant strain harboring an ectopic catalytically dead PaRidC403S-HA allele (two non-complemented strains, lane 9 & 10), MW: Prestained Protein Molecular Weight Marker (Thermo Scientific, lane 2). See the material and method section for details on the alleles construction and features.
(TIF)
If four out of 17 have orthologs in A. nidulans, A. immersus and N. crassa (Pa_1_17860, Pa_1_22930, Pa_2_740 and Pa_2_5020), Pa_4_1960 is the only TF of the set showing no orthologs into the genomes of these three species. Although Pa_1_18880 and Pa_7510 display orthologs either in N. crassa or in N. crassa and A. nidulans, their phylogenetic positions were ambiguous, suggesting some species specialization. See S6 Table for protein names. Only bootstrap values > 0.5 are indicated on the corresponding branches.
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
DE DOWN: down-regulated CDS. DE UP: up-regulated CDS. DOWN FC max: list of CDS with maximum fold change higher than -5. UP FC max: list of CDS with maximum fold change higher than +5. A: target of mating-type transcription factors, activated; R: target of mating-type transcription factors, repressed; 0: CDS controlled neither by FPR1 nor FMR1. d = the corresponding gene was previously deleted [65].
(XLS)
A = ascospores, P = perithecia. d = the corresponding gene was previously deleted [65].
(XLS)
(DOCX)
Acknowledgments
We acknowledge the technical assistance of Sylvie François. We are thankful to Philippe Silar for providing resources to start the project as well as for fruitful scientific discussion and to Ting-Fang Wang for sharing unpublished data and stimulating scientific discussion. We thank Nadia Ponts for performing the DNA methylation assays, Gaëlle Lelandais for performing statistics and Gwenaël Ruprich-Robert for providing biological samples. We thank Cécile Fairhead, Tatiana Giraud, Marc-Henri Lebrun, Nadia Ponts and Annie Sainsard-Chanet for critical reading of the manuscript and fruitful discussions.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
PG, FC and FM were supported by grants from UMR8621, UMR9198 and Agence Nationale de la Recherche Grant ANR- 05-Blan-O385-02. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012. May 29;13(7):484–92. 10.1038/nrg3230 [DOI] [PubMed] [Google Scholar]
- 2.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. 10.1146/annurev.biochem.74.010904.153721 [DOI] [PubMed] [Google Scholar]
- 3.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010. March;11(3):204–20. 10.1038/nrg2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bestor TH. Cloning of a mammalian DNA methyltransferase. Gene. 1988. December 25;74(1):9–12. 10.1016/0378-1119(88)90238-7 [DOI] [PubMed] [Google Scholar]
- 5.Finnegan EJ, Dennis ES. Isolation and identification by sequence homology of a putative cytosine methyltransferase from Arabidopsis thaliana. Nucleic Acids Res. 1993. May 25;21(10):2383–8. 10.1093/nar/21.10.2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kouzminova E, Selker EU. dim-2 encodes a DNA methyltransferase responsible for all known cytosine methylation in Neurospora. EMBO J. 2001. August 1;20(15):4309–23. 10.1093/emboj/20.15.4309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet. 1998. July;19(3):219–20. 10.1038/890 [DOI] [PubMed] [Google Scholar]
- 8.Cao X, Springer NM, Muszynski MG, Phillips RL, Kaeppler S, Jacobsen SE. Conserved plant genes with similarity to mammalian de novo DNA methyltransferases. Proc Natl Acad Sci U S A. 2000. April 25;97(9):4979–84. 10.1073/pnas.97.9.4979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henikoff S, Comai L. A DNA methyltransferase homolog with a chromodomain exists in multiple polymorphic forms in Arabidopsis. Genetics. 1998. May;149(1):307–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lindroth AM, Cao X, Jackson JP, Zilberman D, McCallum CM, Henikoff S, et al. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science. 2001. June 15;292(5524):2077–80. 10.1126/science.1059745 [DOI] [PubMed] [Google Scholar]
- 11.Bartee L, Malagnac F, Bender J. Arabidopsis cmt3 chromomethylase mutations block non-CG methylation and silencing of an endogenous gene. Genes Dev. 2001. July 15;15(14):1753–8. 10.1101/gad.905701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malagnac F, Wendel B, Goyon C, Faugeron G, Zickler D, Rossignol JL, et al. A gene essential for de novo methylation and development in Ascobolus reveals a novel type of eukaryotic DNA methyltransferase structure. Cell. 1997. October 17;91(2):281–90. 10.1016/s0092-8674(00)80410-9 [DOI] [PubMed] [Google Scholar]
- 13.Freitag M, Williams RL, Kothe GO, Selker EU. A cytosine methyltransferase homologue is essential for repeat-induced point mutation in Neurospora crassa. Proc Natl Acad Sci U S A. 2002. June 25;99(13):8802–7. 10.1073/pnas.132212899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huff JT, Zilberman D. Dnmt1-independent CG methylation contributes to nucleosome positioning in diverse eukaryotes. Cell. 2014. March 13;156(6):1286–97. 10.1016/j.cell.2014.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bewick AJ, Hofmeister BT, Powers RA, Mondo SJ, Grigoriev IV, James TY, et al. Diversity of cytosine methylation across the fungal tree of life. Nat Ecol Evol. 2019. February 18; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh C-L, Zhang X, et al. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006. January 20;311(5759):395–8. 10.1126/science.1120976 [DOI] [PubMed] [Google Scholar]
- 17.Katoh M, Curk T, Xu Q, Zupan B, Kuspa A, Shaulsky G. Developmentally regulated DNA methylation in Dictyostelium discoideum. Eukaryot Cell. 2006. January;5(1):18–25. 10.1128/EC.5.1.18-25.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lyko F, Whittaker AJ, Orr-Weaver TL, Jaenisch R. The putative Drosophila methyltransferase gene dDnmt2 is contained in a transposon-like element and is expressed specifically in ovaries. Mech Dev. 2000. July;95(1–2):215–7. [DOI] [PubMed] [Google Scholar]
- 19.Lee DW, Freitag M, Selker EU, Aramayo R. A cytosine methyltransferase homologue is essential for sexual development in Aspergillus nidulans. PloS One. 2008;3(6):e2531 10.1371/journal.pone.0002531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mishra PK, Baum M, Carbon J. DNA methylation regulates phenotype-dependent transcriptional activity in Candida albicans. Proc Natl Acad Sci U S A. 2011. July 19;108(29):11965–70. 10.1073/pnas.1109631108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang Y, Gao X-D, Wang Y, Yuan B-F, Feng Y-Q. Widespread existence of cytosine methylation in yeast DNA measured by gas chromatography/mass spectrometry. Anal Chem. 2012. August 21;84(16):7249–55. 10.1021/ac301727c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Panning B, Jaenisch R. DNA hypomethylation can activate Xist expression and silence X-linked genes. Genes Dev. 1996. August 15;10(16):1991–2002. 10.1101/gad.10.16.1991 [DOI] [PubMed] [Google Scholar]
- 23.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992. June 12;69(6):915–26. 10.1016/0092-8674(92)90611-f [DOI] [PubMed] [Google Scholar]
- 24.Ronemus MJ, Galbiati M, Ticknor C, Chen J, Dellaporta SL. Demethylation-induced developmental pleiotropy in Arabidopsis. Science. 1996. August 2;273(5275):654–7. 10.1126/science.273.5275.654 [DOI] [PubMed] [Google Scholar]
- 25.Finnegan EJ, Peacock WJ, Dennis ES. Reduced DNA methylation in Arabidopsis thaliana results in abnormal plant development. Proc Natl Acad Sci U S A. 1996. August 6;93(16):8449–54. 10.1073/pnas.93.16.8449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Selker EU, Stevens JN. Signal for DNA methylation associated with tandem duplication in Neurospora crassa. Mol Cell Biol. 1987. March;7(3):1032–8. 10.1128/mcb.7.3.1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hane Williams, Taranto Solomon, Oliver. Repeat-Induced Point Mutation in Fungi: A Fungal-Specific Endogenous Mutagenesis Process In: Genetic Transformation Systems in Fungi. Verlag: Springer; 2015. p. 55–68. [Google Scholar]
- 28.Cambareri EB, Jensen BC, Schabtach E, Selker EU. Repeat-induced G-C to A-T mutations in Neurospora. Science. 1989. June 30;244(4912):1571–5. 10.1126/science.2544994 [DOI] [PubMed] [Google Scholar]
- 29.Gladyshev E, Kleckner N. Direct recognition of homology between double helices of DNA in Neurospora crassa. Nat Commun. 2014. April 3;5:3509 10.1038/ncomms4509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gladyshev E, Kleckner N. DNA sequence homology induces cytosine-to-thymine mutation by a heterochromatin-related pathway in Neurospora. Nat Genet. 2017. June;49(6):887–94. 10.1038/ng.3857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rhounim L, Rossignol JL, Faugeron G. Epimutation of repeated genes in Ascobolus immersus. EMBO J. 1992. December;11(12):4451–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li W-C, Chen C-L, Wang T-F. Repeat-induced point (RIP) mutation in the industrial workhorse fungus Trichoderma reesei. Appl Microbiol Biotechnol. 2018. January 8; [DOI] [PubMed] [Google Scholar]
- 33.Gladyshev E. Repeat-Induced Point Mutation and Other Genome Defense Mechanisms in Fungi. Microbiol Spectr. 2017. July;5(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bouhouche K, Zickler D, Debuchy R, Arnaise S. Altering a gene involved in nuclear distribution increases the repeat-induced point mutation process in the fungus Podospora anserina. Genetics. 2004. May;167(1):151–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Graïa F, Berteaux-Lecellier V, Zickler D, Picard M. ami1, an orthologue of the Aspergillus nidulans apsA gene, is involved in nuclear migration events throughout the life cycle of Podospora anserina. Genetics. 2000. June;155(2):633–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Espagne E, Lespinet O, Malagnac F, Da Silva C, Jaillon O, Porcel BM, et al. The genome sequence of the model ascomycete fungus Podospora anserina. Genome Biol. 2008;9(5):R77 10.1186/gb-2008-9-5-r77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silar P, Dauget J-M, Gautier V, Grognet P, Chablat M, Hermann-Le Denmat S, et al. A gene graveyard in the genome of the fungus Podospora comata. Mol Genet Genomics MGG. 2018. October 4; [DOI] [PubMed] [Google Scholar]
- 38.Peraza-Reyes L, Malagnac F. Sexual Development in Fungi Springer; 2016. 524 p. (The Mycota I, Growth, Differentiation and Sexuality). [Google Scholar]
- 39.Coppin E, Arnaise S, Contamine V, Picard M. Deletion of the mating-type sequences in Podospora anserina abolishes mating without affecting vegetative functions and sexual differentiation. Mol Gen Genet MGG. 1993. November;241(3–4):409–14. 10.1007/bf00284694 [DOI] [PubMed] [Google Scholar]
- 40.Jamet-Vierny C, Debuchy R, Prigent M, Silar P. IDC1, a pezizomycotina-specific gene that belongs to the PaMpk1 MAP kinase transduction cascade of the filamentous fungus Podospora anserina. Fungal Genet Biol FG B. 2007. December;44(12):1219–30. 10.1016/j.fgb.2007.04.005 [DOI] [PubMed] [Google Scholar]
- 41.Silar P. Grafting as a method for studying development in the filamentous fungus Podospora anserina. Fungal Biol. 2011. August;115(8):793–802. 10.1016/j.funbio.2011.06.005 [DOI] [PubMed] [Google Scholar]
- 42.Silar P, Picard M. Increased longevity of EF-1 alpha high-fidelity mutants in Podospora anserina. J Mol Biol. 1994. January 7;235(1):231–6. 10.1016/s0022-2836(05)80029-4 [DOI] [PubMed] [Google Scholar]
- 43.Bestor TH, Verdine GL. DNA methyltransferases. Curr Opin Cell Biol. 1994. June;6(3):380–9. [DOI] [PubMed] [Google Scholar]
- 44.Bidard F, Imbeaud S, Reymond N, Lespinet O, Silar P, Clave C, et al. A general framework for optimization of probes for gene expression microarray and its application to the fungus Podospora anserina. 2010;3(1):171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruepp A, Zollner A, Maier D, Albermann K, Hani J, Mokrejs M, et al. The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 2004;32(18):5539–45. 10.1093/nar/gkh894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Colot HV, Park G, Turner GE, Ringelberg C, Crew CM, Litvinkova L, et al. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci U S A. 2006. July 5;103(27):10352–7. 10.1073/pnas.0601456103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carrillo AJ, Schacht P, Cabrera IE, Blahut J, Prudhomme L, Dietrich S, et al. Functional Profiling of Transcription Factor Genes in Neurospora crassa. G3 Genes Genomes Genet. 2017. September 1;7(9):2945–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hagiwara D, Kondo A, Fujioka T, Abe K. Functional analysis of C2H2 zinc finger transcription factor CrzA involved in calcium signaling in Aspergillus nidulans. Curr Genet. 2008. December;54(6):325–38. 10.1007/s00294-008-0220-z [DOI] [PubMed] [Google Scholar]
- 49.Han KH, Han KY, Yu JH, Chae KS, Jahng KY, Han DM. The nsdD gene encodes a putative GATA-type transcription factor necessary for sexual development of Aspergillus nidulans. Mol Microbiol. 2001. July;41(2):299–309. 10.1046/j.1365-2958.2001.02472.x [DOI] [PubMed] [Google Scholar]
- 50.Aramayo R, Metzenberg RL. Meiotic transvection in fungi. Cell. 1996. July 12;86(1):103–13. 10.1016/s0092-8674(00)80081-1 [DOI] [PubMed] [Google Scholar]
- 51.Feng B, Haas H, Marzluf GA. ASD4, a new GATA factor of Neurospora crassa, displays sequence-specific DNA binding and functions in ascus and ascospore development. Biochemistry. 2000. September 12;39(36):11065–73. 10.1021/bi000886j [DOI] [PubMed] [Google Scholar]
- 52.Li D, Bobrowicz P, Wilkinson HH, Ebbole DJ. A mitogen-activated protein kinase pathway essential for mating and contributing to vegetative growth in Neurospora crassa. Genetics. 2005. July;170(3):1091–104. 10.1534/genetics.104.036772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McCluskey K, Wiest AE, Grigoriev IV, Lipzen A, Martin J, Schackwitz W, et al. Rediscovery by Whole Genome Sequencing: Classical Mutations and Genome Polymorphisms in Neurospora crassa. G3 GenesGenomesGenetics. 2011. September 1;1(4):303–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xie N, Ruprich-Robert G, Chapeland-Leclerc F, Coppin E, Lalucque H, Brun S, et al. Inositol-phosphate signaling as mediator for growth and sexual reproduction in Podospora anserina. Dev Biol. 2017. 01;429(1):285–305. 10.1016/j.ydbio.2017.06.017 [DOI] [PubMed] [Google Scholar]
- 55.Teichert I, Nowrousian M, Pöggeler S, Kück U. Chapter Four—The Filamentous Fungus Sordaria macrospora as a Genetic Model to Study Fruiting Body Development In: Friedmann T, Dunlap JC, Goodwin SF, editors. Advances in Genetics [Internet]. Academic Press; 2014. [cited 2019 Feb 10]. p. 199–244. Available from: http://www.sciencedirect.com/science/article/pii/B9780128001493000044 10.1016/B978-0-12-800149-3.00004-4 [DOI] [PubMed] [Google Scholar]
- 56.Peraza Reyes L, Berteaux-Lecellier V. Peroxisomes and sexual development in fungi. Front Physiol [Internet]. 2013. [cited 2018 Jul 14];4 Available from: https://www.frontiersin.org/articles/10.3389/fphys.2013.00244/full [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li S, Myung K, Guse D, Donkin B, Proctor RH, Grayburn WS, et al. FvVE1 regulates filamentous growth, the ratio of microconidia to macroconidia and cell wall formation in Fusarium verticillioides. Mol Microbiol. 2006. December;62(5):1418–32. 10.1111/j.1365-2958.2006.05447.x [DOI] [PubMed] [Google Scholar]
- 58.Calvo AM, Bok J, Brooks W, Keller NP. veA is required for toxin and sclerotial production in Aspergillus parasiticus. Appl Environ Microbiol. 2004. August;70(8):4733–9. 10.1128/AEM.70.8.4733-4739.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Karimi Aghcheh R, Németh Z, Atanasova L, Fekete E, Paholcsek M, Sándor E, et al. The VELVET A orthologue VEL1 of Trichoderma reesei regulates fungal development and is essential for cellulase gene expression. PloS One. 2014;9(11):e112799 10.1371/journal.pone.0112799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim J-E, Kim S-J, Lee B-H, Park R-W, Kim K-S, Kim I-S. Identification of Motifs for Cell Adhesion within the Repeated Domains of Transforming Growth Factor-β-induced Gene,βig-h3. J Biol Chem. 2000. June 10;275(40):30907–15. 10.1074/jbc.M002752200 [DOI] [PubMed] [Google Scholar]
- 61.Greenwald CJ, Kasuga T, Glass NL, Shaw BD, Ebbole DJ, Wilkinson HH. Temporal and spatial regulation of gene expression during asexual development of Neurospora crassa. Genetics. 2010. December;186(4):1217–30. 10.1534/genetics.110.121780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang Y, Reddy B, Thompson J, Wang H, Noma K, Yates JR, et al. Regulation of Set9-mediated H4K20 methylation by a PWWP domain protein. Mol Cell. 2009. February 27;33(4):428–37. 10.1016/j.molcel.2009.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Harari Y, Rubinstein L, Kupiec M. An anti-checkpoint activity for rif1. PLoS Genet. 2011. Dec;7(12):e1002421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Glass NL, Kaneko I. Fatal Attraction: Nonself Recognition and Heterokaryon Incompatibility in Filamentous Fungi. Eukaryot Cell. 2003. February;2(1):1–8. 10.1128/EC.2.1.1-8.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bidard F, Aït Benkhali J, Coppin E, Imbeaud S, Grognet P, Delacroix H, et al. Genome-wide gene expression profiling of fertilization competent mycelium in opposite mating types in the heterothallic fungus Podospora anserina. PloS One. 2011;6(6):e21476 10.1371/journal.pone.0021476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dyer PS, Inderbitzin P, Debuchy R. Mating-Type Structure, Function, Regulation and Evolution in the Pezizomycotina In: Growth, Differentiation and Sexuality. Third. Springer International Publishing; 2016. p. 351–85. (Mycota). [Google Scholar]
- 67.Goddard MR, Godfray HCJ, Burt A. Sex increases the efficacy of natural selection in experimental yeast populations. Nature. 2005. March 31;434(7033):636–40. 10.1038/nature03405 [DOI] [PubMed] [Google Scholar]
- 68.Judson OP, Normark BB. Ancient asexual scandals. Trends Ecol Evol. 1996. February;11(2):41–6. [DOI] [PubMed] [Google Scholar]
- 69.Bruggeman J, Debets AJM, Wijngaarden PJ, deVisser JAGM, Hoekstra RF. Sex slows down the accumulation of deleterious mutations in the homothallic fungus Aspergillus nidulans. Genetics. 2003. June;164(2):479–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Coleman JJ, Rounsley SD, Rodriguez-Carres M, Kuo A, Wasmann CC, Grimwood J, et al. The genome of Nectria haematococca: contribution of supernumerary chromosomes to gene expansion. PLoS Genet. 2009. August;5(8):e1000618 10.1371/journal.pgen.1000618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Becks L, Agrawal AF. The evolution of sex is favoured during adaptation to new environments. PLoS Biol. 2012;10(5):e1001317 10.1371/journal.pbio.1001317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999. October 29;99(3):247–57. 10.1016/s0092-8674(00)81656-6 [DOI] [PubMed] [Google Scholar]
- 73.Malagnac F, Lalucque H, Lepère G, Silar P. Two NADPH oxidase isoforms are required for sexual reproduction and ascospore germination in the filamentous fungus Podospora anserina. Fungal Genet Biol FG B. 2004. November;41(11):982–97. 10.1016/j.fgb.2004.07.008 [DOI] [PubMed] [Google Scholar]
- 74.Lalucque H, Malagnac F, Brun S, Kicka S, Silar P. A non-Mendelian MAPK-generated hereditary unit controlled by a second MAPK pathway in Podospora anserina. Genetics. 2012. June;191(2):419–33. 10.1534/genetics.112.139469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ait Benkhali J, Coppin E, Brun S, Peraza-Reyes L, Martin T, Dixelius C, et al. A network of HMG-box transcription factors regulates sexual cycle in the fungus Podospora anserina. PLoS Genet. 2013;9(7):e1003642 10.1371/journal.pgen.1003642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Arnaise S, Zickler D, Le Bilcot S, Poisier C, Debuchy R. Mutations in mating-type genes of the heterothallic fungus Podospora anserina lead to self-fertility. Genetics. 2001. October;159(2):545–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Turgeon BG, Yoder OC. Proposed nomenclature for mating type genes of filamentous ascomycetes. Fungal Genet Biol FG B. 2000. October;31(1):1–5. 10.1006/fgbi.2000.1227 [DOI] [PubMed] [Google Scholar]
- 78.Zheng Q, Hou R, Juanyu null, Zhang null, Ma J, Wu Z, et al. The MAT locus genes play different roles in sexual reproduction and pathogenesis in Fusarium graminearum. PloS One. 2013;8(6):e66980 10.1371/journal.pone.0066980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Klix V, Nowrousian M, Ringelberg C, Loros JJ, Dunlap JC, Pöggeler S. Functional characterization of MAT1-1-specific mating-type genes in the homothallic ascomycete Sordaria macrospora provides new insights into essential and nonessential sexual regulators. Eukaryot Cell. 2010. June;9(6):894–905. 10.1128/EC.00019-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Arnaise S, Debuchy R, Picard M. What is a bona fide mating-type gene? Internuclear complementation of mat mutants in Podospora anserina. Mol Gen Genet MGG. 1997. September;256(2):169–78. 10.1007/pl00008611 [DOI] [PubMed] [Google Scholar]
- 81.Turgeon BG, Debuchy R. Cochliobolus and Podospora: mechanism of sex determination and the evolution of reproductive lifestyle. In: Sex in Fungi, Molecular Determination and Evolutionary Implications. 2007. [Google Scholar]
- 82.Berteaux-Lecellier V, Zickler D, Debuchy R, Panvier-Adoutte A, Thompson-Coffe C, Picard M. A homologue of the yeast SHE4 gene is essential for the transition between the syncytial and cellular stages during sexual reproduction of the fungus Podospora anserina. EMBO J. 1998. August 10;17(5):1248–58. 10.1093/emboj/17.5.1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lewis ZA, Adhvaryu KK, Honda S, Shiver AL, Knip M, Sack R, et al. DNA methylation and normal chromosome behavior in Neurospora depend on five components of a histone methyltransferase complex, DCDC. PLoS Genet. 2010;6(11):e1001196 10.1371/journal.pgen.1001196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kück U, Beier AM, Teichert I. The composition and function of the striatin-interacting phosphatases and kinases (STRIPAK) complex in fungi. Fungal Genet Biol FG B. 2016. May;90:31–8. 10.1016/j.fgb.2015.10.001 [DOI] [PubMed] [Google Scholar]
- 85.Yang K, Liang L, Ran F, Liu Y, Li Z, Lan H, et al. The DmtA methyltransferase contributes to Aspergillus flavus conidiation, sclerotial production, aflatoxin biosynthesis and virulence. Sci Rep. 2016. March 16;6:23259 10.1038/srep23259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.So K-K, Ko Y-H, Chun J, Bal J, Jeon J, Kim J-M, et al. Global DNA Methylation in the Chestnut Blight Fungus Cryphonectria parasitica and Genome-Wide Changes in DNA Methylation Accompanied with Sectorization. Front Plant Sci [Internet]. 2018. February 2 [cited 2018 Oct 7];9 Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5801561/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang Y, Wang T, Qiao L, Zhu J, Fan J, Zhang T, et al. DNA methyltransferases contribute to the fungal development, stress tolerance and virulence of the entomopathogenic fungus Metarhizium robertsii. Appl Microbiol Biotechnol. 2017. May;101(10):4215–26. 10.1007/s00253-017-8197-5 [DOI] [PubMed] [Google Scholar]
- 88.Jeon J, Choi J, Lee G-W, Park S-Y, Huh A, Dean RA, et al. Genome-wide profiling of DNA methylation provides insights into epigenetic regulation of fungal development in a plant pathogenic fungus, Magnaporthe oryzae. Sci Rep. 2015. February 24;5:8567 10.1038/srep08567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shiu PK, Raju NB, Zickler D, Metzenberg RL. Meiotic silencing by unpaired DNA. Cell. 2001. December 28;107(7):905–16. 10.1016/s0092-8674(01)00609-2 [DOI] [PubMed] [Google Scholar]
- 90.Keller C, Bühler M. Chromatin-associated ncRNA activities. Chromosome Res. 2013;21(6–7):627–41. 10.1007/s10577-013-9390-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Taylor DH, Chu ET-J, Spektor R, Soloway PD. Long Non-Coding RNA Regulation of Reproduction and Development. Mol Reprod Dev. 2015. December;82(12):932–56. 10.1002/mrd.22581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ezhkova E, Tansey WP. Proteasomal ATPases link ubiquitylation of histone H2B to methylation of histone H3. Mol Cell. 2004. February 13;13(3):435–42. [DOI] [PubMed] [Google Scholar]
- 93.Köhler A, Schneider M, Cabal GG, Nehrbass U, Hurt E. Yeast Ataxin-7 links histone deubiquitination with gene gating and mRNA export. Nat Cell Biol. 2008. June;10(6):707–15. 10.1038/ncb1733 [DOI] [PubMed] [Google Scholar]
- 94.van der Lee R, Lang B, Kruse K, Gsponer J, Sánchez de Groot N, Huynen MA, et al. Intrinsically Disordered Segments Affect Protein Half-Life in the Cell and during Evolution. Cell Rep. 2014. September 15;8(6):1832–44. 10.1016/j.celrep.2014.07.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Collins GA, Goldberg AL. The Logic of the 26S Proteasome. Cell. 2017. May 18;169(5):792–806. 10.1016/j.cell.2017.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Amselem J, Lebrun M-H, Quesneville H. Whole genome comparative analysis of transposable elements provides new insight into mechanisms of their inactivation in fungal genomes. BMC Genomics. 2015. December;16(1):141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ikeda K, Van Vu B, Kadotani N, Tanaka M, Murata T, Shiina K, et al. Is the fungus Magnaporthe losing DNA methylation? Genetics. 2013. November;195(3):845–55. 10.1534/genetics.113.155978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Clancy MJ, Shambaugh ME, Timpte CS, Bokar JA. Induction of sporulation in Saccharomyces cerevisiae leads to the formation of N6-methyladenosine in mRNA: a potential mechanism for the activity of the IME4 gene. Nucleic Acids Res. 2002. October 15;30(20):4509–18. 10.1093/nar/gkf573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhong S, Li H, Bodi Z, Button J, Vespa L, Herzog M, et al. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell. 2008. May;20(5):1278–88. 10.1105/tpc.108.058883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Grognet P, Bidard F, Kuchly C, Tong LCH, Coppin E, Benkhali JA, et al. Maintaining two mating types: structure of the mating type locus and its role in heterokaryosis in Podospora anserina. Genetics. 2014. May;197(1):421–32. 10.1534/genetics.113.159988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rizet G, Engelmann C. Contribution à l’étude génétique d’un Ascomycète tétrasporé: Podospora anserina. Rev Cytol Biol Veg. 1949;11:201–304. [Google Scholar]
- 102.Lambou K, Malagnac F, Barbisan C, Tharreau D, Lebrun M-H, Silar P. The crucial role of the Pls1 tetraspanin during ascospore germination in Podospora anserina provides an example of the convergent evolution of morphogenetic processes in fungal plant pathogens and saprobes. Eukaryot Cell. 2008. October;7(10):1809–18. 10.1128/EC.00149-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, et al. Current protocols in molecular biology. Wiley Interscience; New York; 1987. [Google Scholar]
- 104.Lecellier G, Silar P. Rapid methods for nucleic acids extraction from Petri dish-grown mycelia. Curr Genet. 1994. February;25(2):122–3. [DOI] [PubMed] [Google Scholar]
- 105.Brygoo Y, Debuchy R. Transformation by integration in Podospora anserina. I. Methodology and phenomenology. Mol Gen Genet. 1985;200:128–31. [Google Scholar]
- 106.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990. October 5;215(3):403–10. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 107.Grognet P, Lalucque H, Silar P. The PaAlr1 magnesium transporter is required for ascospore development in Podospora anserina. Fungal Biol. 2012. October;116(10):1111–8. 10.1016/j.funbio.2012.08.004 [DOI] [PubMed] [Google Scholar]
- 108.Silar P. Two new easy-to-use vectors for transformations. Fungal Genet Newsl. 1995;42:73. [Google Scholar]
- 109.Kicka S, Bonnet C, Sobering AK, Ganesan LP, Silar P. A mitotically inheritable unit containing a MAP kinase module. Proc Natl Acad Sci U S A. 2006. September 5;103(36):13445–50. 10.1073/pnas.0603693103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Berteaux-Lecellier V, Picard M, Thompson-Coffe C, Zickler D, Panvier-Adoutte A, Simonet JM. A nonmammalian homolog of the PAF1 gene (Zellweger syndrome) discovered as a gene involved in caryogamy in the fungus Podospora anserina. Cell. 1995. June 30;81(7):1043–51. 10.1016/s0092-8674(05)80009-1 [DOI] [PubMed] [Google Scholar]
- 111.Grossetête S, Labedan B, Lespinet O. FUNGIpath: a tool to assess fungal metabolic pathways predicted by orthology. BMC Genomics. 2010. February 1;11:81 10.1186/1471-2164-11-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Grigoriev IV, Nikitin R, Haridas S, Kuo A, Ohm R, Otillar R, et al. MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res. 2014. January;42(Database issue):D699–704. 10.1093/nar/gkt1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010. May;59(3):307–21. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- 114.Bidard F, Clavé C, Saupe SJ. The transcriptional response to nonself in the fungus Podospora anserina. G3 Bethesda Md. 2013. June 21;3(6):1015–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Marisa L, Ichanté J-L, Reymond N, Aggerbeck L, Delacroix H, Mucchielli-Giorgi M-H. MAnGO: an interactive R-based tool for two-colour microarray analysis. Bioinforma Oxf Engl. 2007. September 1;23(17):2339–41. [DOI] [PubMed] [Google Scholar]
- 116.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Stat Methodol. 1995;57:289–300. [Google Scholar]
- 117.Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, et al. GO::TermFinder—open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinforma Oxf Engl. 2004. December 12;20(18):3710–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Harris CJ, Scheibe M, Wongpalee SP, Liu W, Cornett EM, Vaughan RM, et al. A DNA methylation reader complex that enhances gene transcription. Science. 2018. 07;362(6419):1182–6. 10.1126/science.aar7854 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Average expression profiles (y-axis) of PaRid (Pa_1_19440) during sexual development (x-axis, hours). (B) Amplification of PaRid transcripts (2296 bp) by RT-PCR. MW: GeneRuler DNA Ladder Mix (Thermo Fisher Scientific), RT T48, RT T96: RT-PCR performed on RNA extracted from 2 days or 4 days post fertilization developing perithecia, gDNA: genomic DNA, NRT: PCR performed on RNA extracted from 2 days or 4 days post fertilization developing perithecia, Neg: No RNA. See (Materials and methods section for details). (C) Coverage of RNA-seq mapped reads at the PaRid locus [37]. RNA-seq experiments were performed on RNAs extracted from non-germinated ascospores (Ascospores), eight hours germinating ascospores (Germinated ascospores), 1-day- or 4-day-old mycelia, 2 days or 4 days post fertilization developing perithecia.
(TIF)
Schematic representations of the endogenous and disrupted loci are given in. Replacement by homologous recombination of the wild type PaRid allele by the disrupted ΔPaRid allele results in the substitution of a 3.2 kb PstI fragment by a 2.3 kb PstI fragment as revealed by hybridization of the 5’UTR digoxygenin-labeled probe (S2A) and in the substitution of a 2.8 kb PstI fragment by a 4.0 kb PstI fragment as revealed by hybridization of the 3’UTR digoxygenin-labeled probe (S2A).
(TIF)
Major steps of P. anserina’s life cycle as shown by a schematic representation (upper panel) and the corresponding light microphotographs (lower panel). P. anserina’s life cycle begins with the germination of an ascospore (A) that gives rise to a haploid mycelium (B). After three days of growth, both male gametes (spermatia, B, top) and female gametes (ascogonia, B, bottom) are formed. Because most of the ascospores carry two different and sexually compatible nuclei (mat+ and mat- mating types) P. anserina strains are self-fertile (pseudo-homothallism). Before fertilization occurs, ascogonia can mature into protoperithecia by recruiting protective maternal hyphae to shelter the ascogonial cell. A pheromone/receptor signaling system allows the ascogonia to recognize and fuse with spermatia of compatible mating type (heterothallism). Fertilization initiates the development of the fruiting body (perithecia, C) in which the dikaryotic mat+/mat- fertilized ascogonium forms. Further development leads to a three-celled hook-shaped structure called the crozier (D). The two parental nuclei in the middle cell of the crozier fuse (karyogamy, schematic representation D) to form a diploid nucleus, which then immediately undergoes meiosis. The four resulting haploid nuclei undergo mitosis. In most cases, ascospores are formed around 2 non-sister nuclei within the developing ascus. On rare occasions, two ascospores are formed around only one haploid nucleus each, leading to a five-ascospore ascus (E, photograph). Scale bar: 10 μm in (A-D); 200 μm in (E).
(TIF)
Strains were grown on M2 minimal medium for 6 days at 27°C. S: Wild-type strain. See Materials and Methods section for details on the complemented ΔPaRid:AS4:PaRid:HA strain.
(TIF)
(A) Cryosensitivity assay: growth rate at two sub-optimal temperatures compared with optimal temperature. (B) Thermosensitivity assay: growth rate at 37°C compared with optimal temperature. (C) Longevity assay. For each genotype, growth was assayed on 3 independent cultures after 4 days of growth at the indicated temperatures. Each experiment was performed three times. For each genotype, longevity was measured on three independent cultures, issued from three individual ascospores, as described in [42].
(TIF)
(A) Morphological comparison of perithecia obtained from wild-type crosses (PaRid), ΔPaRid crosses (ΔPaRid) and ΔSmr1 crosses (ΔSmr1). Size and morphology of the ΔPaRid and ΔSmr1 perithecia are alike. Scale bar: 250 μm. (B) Perithecia obtained in the indicated trikaryons on M2 medium after 5 days at 27°C. The Δmat; PaRid+ mat-; PaRid+ mat+ trikaryons form typical fully developed perithecia (left panels). By contrast, only blocked micro-perithecia are formed by the Δmat; ΔPaRid mat-; ΔPaRid mat+ trikaryons (right panels). (C) Heterozygous orientated crosses PaRid+ mat- × ΔPaRid mat+ after 5 days at 27°C. When the wild-type PaRid+ allele is present in the female gametes genome and the mutant ΔPaRid allele is present in the male gamete genome fully developed perithecia are formed, conversely when the mutant ΔPaRid allele is present in the female gametes and the wild-type PaRid+ allele is present in the male gamete, only blocked micro-perithecia are formed. Left panel, scale bar: 1 mm, right panel, scale bar: 500 μm.
(TIF)
Western-blot was performed as described in Material and Methods section and probed with an anti-HA antibody that specifically detects the HA-tagged proteins. The PaRid protein (83 kDa) was barely detectable when the expression of the ectopic alleles was driven from its native promoter (ΔPaRid:PaRid, ΔPaRid:PaRidC403S). However, it was readily produced when the expression of the ectopic alleles was under the control of a strong and constitutive promoter (ΔPaRid:AS4-PaRid, ΔPaRid:AS4-PaRidC403S). Complementation of the fertility defect was obtained by insertion of the wild type PaRid-HA allele or by insertion of the AS4-PaRid-HA only. Negative controls (No PaRid-HA tagged): PaRid+ wild-type strains mat+ (lane 1) & mat- (lane 3), ΔPaRid mutant strain (lane 4), ΔPaRid:PaRid+-GFP strain (no HA tag, GFP only, complemented strain, lane 7). ΔPaRid:PaRid+ = ΔPaRid mutant strain harboring an ectopic wild type PaRid+-HA allele (complemented strain, lane 5), ΔPaRid:AS4-PaRid = ΔPaRid mutant strain harboring an ectopic AS4-PaRid+-HA allele (complemented strain, lane 6), ΔPaRid:AS4-PaRidC403S = ΔPaRid mutant strain harboring an ectopic AS4-PaRidC403S-HA allele (non-complemented strain, lane 8). ΔPaRid:PaRidC403S = ΔPaRid mutant strain harboring an ectopic catalytically dead PaRidC403S-HA allele (two non-complemented strains, lane 9 & 10), MW: Prestained Protein Molecular Weight Marker (Thermo Scientific, lane 2). See the material and method section for details on the alleles construction and features.
(TIF)
If four out of 17 have orthologs in A. nidulans, A. immersus and N. crassa (Pa_1_17860, Pa_1_22930, Pa_2_740 and Pa_2_5020), Pa_4_1960 is the only TF of the set showing no orthologs into the genomes of these three species. Although Pa_1_18880 and Pa_7510 display orthologs either in N. crassa or in N. crassa and A. nidulans, their phylogenetic positions were ambiguous, suggesting some species specialization. See S6 Table for protein names. Only bootstrap values > 0.5 are indicated on the corresponding branches.
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
DE DOWN: down-regulated CDS. DE UP: up-regulated CDS. DOWN FC max: list of CDS with maximum fold change higher than -5. UP FC max: list of CDS with maximum fold change higher than +5. A: target of mating-type transcription factors, activated; R: target of mating-type transcription factors, repressed; 0: CDS controlled neither by FPR1 nor FMR1. d = the corresponding gene was previously deleted [65].
(XLS)
A = ascospores, P = perithecia. d = the corresponding gene was previously deleted [65].
(XLS)
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.