Abstract
In this paper, we propose a 3D multi-atlas-based prostate segmentation method for MR images, which utilizes patch-based label fusion strategy. The atlases with the most similar appearance are selected to serve as the best subjects in the label fusion. A local patch-based atlas fusion is performed using voxel weighting based on anatomical signature. This segmentation technique was validated with a clinical study of 13 patients and its accuracy was assessed using the physicians’ manual segmentations (gold standard). Dice volumetric overlapping was used to quantify the difference between the automatic and manual segmentation. In summary, we have developed a new prostate MR segmentation approach based on nonlocal patch-based label fusion, demonstrated its clinical feasibility, and validated its accuracy with manual segmentations.
Keywords: Prostate segmentation, MRI, multi-atlas, label fusion, prostate cancer
1. INTRODUCTION
Prostate cancer is the second leading cause of cancer death for USA male populations [1]. Prostate MRI image segmentation has been an area of intense research due to the increased use of MRI as a modality for the clinical workup of prostate cancer, e.g. diagnosis and treatment planning [2–4]. Segmentation is useful for various tasks: to accurately localize prostate boundaries for radiotherapy [5], perform volume estimation to track disease progression [6], to initialize multi-modal registration algorithms [7, 8], and to obtain the region of interest for computer-aided detection of prostate cancer [9]. Currently, physicians’ manual segmentation is the gold standard in the clinic. However, manual segmentation of the prostate is time consuming and often subject to inter- and intra-observer variation.
Accurate prostate segmentation for MRI data can be challenging due to the image noise, inter-patient anatomical differences, and the similar intensities of the prostate and surrounding tissues (e.g., the bladder). To overcome these challenges, several segmentation techniques have been proposed. Deformable models (DMs) have been particularly popular, especially active shape models (ASMs) and level-sets. For example, a DM framework was proposed for 3D prostate segmentation from T2-MRI, in which the DM evolution was controlled by voxel intensity and a statistical shape model [10]. Zhu et al. [11] proposed a hybrid 2D/3D ASM-based methodology for 3D MRI prostate segmentation. Ghose et al. [12] proposed a similar approach that aligned T2-MRI data, and then used an active appearance model (AAM), an extension of ASM, guided by appearance and shape information to segment the prostate. Also, Gao et al. [13] aligned MR images before segmenting the prostate using a level-set guided by appearance information and a learned shape prior. In addition, McClure et al. [14] utilized a nonnegative matrix factorization (NMF) feature fusion method to create a more robust model for guiding the evolution of a 3D level-set deformable model for MRI DWI prostate segmentation. A probabilistic anatomical atlas was used by Martin et al. [15] to constrain a DM-based framework for segmenting the prostate from 3D T2-MR images. In [16] a multi-feature landmark-free active appearance model was presented for segmenting 2-D medical images. These methods yield good results but cannot handle medical images in a 3D manner. Statistical-based techniques have also been utilized for segmenting prostates from MRI data. For example, a probabilistic graph-cut-based framework for 3D T2-MRI prostate segmentation based on a probabilistic atlas was proposed in [17]. Firjany et al. [18] proposed a Markov random field (MRF) image model for 2D dynamic contrast enhanced (DCE)-MRI prostate segmentation that combined a graph cut approach with a prior shape model of the prostate and the visual appearance of the prostate image, modeled using a linear combination of discrete Gaussians (LCDG). This method was later extended in [19] to allow for 3D prostate segmentation from DCE-MRI data. A maximum a posteriori (MAP)-based framework that performed automated 3D MRI prostate segmentation using an MRF model and statistical shape information was proposed in [20]. Also, an atlas-based segmentation approach was presented to extract the prostate from MR images based on averaging the best atlases that match the image to be segmented [21]. In [22] another technique was proposed to use an automated atlas approach to segment the prostate region based on a selective and iterative method for performance level estimation (SIMPLE) based alignment technique. In addition to DMs and statistical-based techniques, several other methods have been proposed to segment the prostate from MR images. A semi-automated edge detection technique in [23] was proposed for MRI prostate segmentation based on a static wavelet transform [24] to locate the prostate edges. Also, a semi-automated approach is proposed to use a prostate shape prior to detect the contour in each slice and then refined them to form a 3D prostate surface [25]. Additionally, random walk classification was used for MRI prostate segmentation in [26].
In this study we propose to integrate similar appearance-specific atlases and patch-based voxel weighting into label propagation framework to automatically segment prostate from MR images. This approach has 2 distinctive strengths: 1) Instead of performing the fusion of nonlinearly deformed template structures, the proposed method achieves the labeling of each voxel individually by comparing its surrounding patch with patches in training subjects in which the labels of the central voxels are known. When the patch under study resembles a patch in the training subjects, their central voxels are considered to belong to the same structure, and this training patch is used to estimate the final label. 2) Contrary to classical majority voting schemes that give the same weight to all the samples, the nonlocal means scheme enables the robust distinction of the most similar samples according to their local patch-based anatomical features. Finally, a patch-based weighting is used to perform a pixel-based aggregation of the labels ensuring the independency of the votes.
2. METHODS
Figure 1 shows a schematic flow chart of the automated multi-atlas segmentation of the prostate for MR images using patch-based label fusion. After an initial preprocessing for all images, all of the atlases are non-rigidly registered to a target image. The resulting transformation is then used to propagate the anatomical structure labels of the atlas into the space of the target image. The top L similar atlases are further chosen by measuring intensity difference in the region of interest around the prostate. Finally, using voxel weighting based on patch-based anatomical signature, the label that the majority of all warped labels predict for each voxel is used for the final segmentation of the target image. The three major steps are briefly described below.
Figure 1.
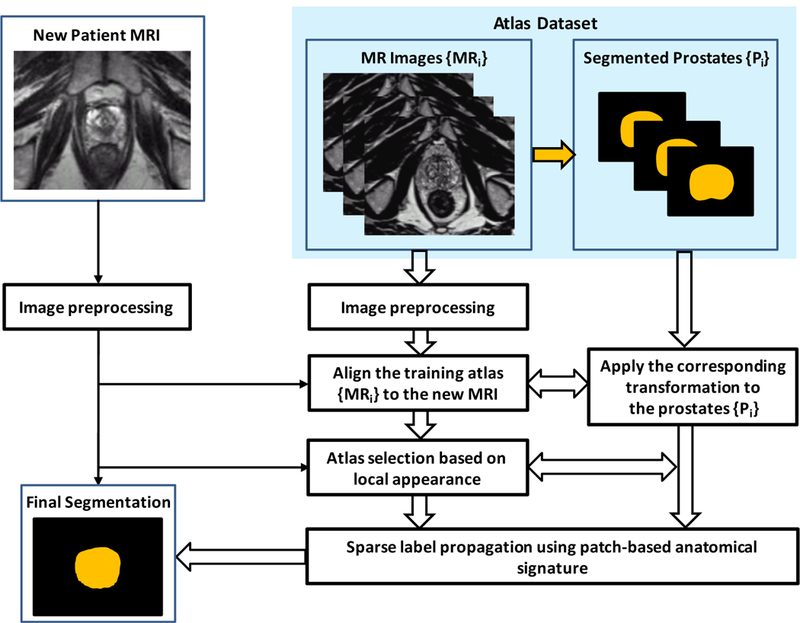
Schematic flow chart of the proposed algorithm for 3D MR prostate segmentation.
2.1. Image Preprocessing and Registration
Before segmentation, pre-processing is performed for all MR images in the dataset, which includes reducing noise, inhomogeneity correction and inter-subject intensity normalization [24]. The same processing will be performed for a new patient’s images that will be segmented. Such pre-processing steps are designed to improve the accuracy of the registration and segmentation. During the alignment processing of the training set, we first select one MR image as a template, and align other MR images to the template image. And we use the corresponding transformation obtained from training image alignment to align the segmented prostates (binary mask) to the template prostate. Since the segmented prostate of each training image is available, in order to optimize the alignment of training set we again align each training image to the template image by registering the binary segmentation prostates to the template prostate. When a newly acquired MR image comes, all aligned training images in the training set are registered to this new image. The deformable registration is used to obtain the spatial deformation field between the new MR image and training images. The same transformations are applied to the segmented prostates in the training set.
2.2. Atlas Selection
Due to the significant appearance difference of the prostate in both shape and intensity, the aligned atlas dataset may contain redundant atlases which could affect the segmentation accuracy. Therefore, an atlas selection should be performed to identify these similar atlases for label fusion. In order to deal with this problem we compare the intensity difference around the rectal region and rank the atlases in the aligned dataset. Here we use the normalized sum of the squared difference (NSSD) across the initialization mask instead of the normalized mutual information (NMI) over the image [27, 28]. This strategy is chosen because NSSD is sensitive to the variations in contrast and luminance. The NSSD intensity differences between each atlas and target image are defined over a region of interest to measure local image appearance base on the L2 norm. Within the ranked database, the top L atlases with the most similar appearance (smaller NSSDs) around the prostate region are selected for final multi-atlas segmentation.
2.3. Patch-Based Label Fusion
One of the most popular multi-atlases based image segmentation methods is the nonlocal mean label propagation strategy [29], and it can be summarized as follows. Given N aligned training images and their segmentation ground labels {(Ii, Gi), i = 1, …, N}, for a new treatment image Inew, each voxel in Inew is correlated to each voxel in Ii with a graph weight . Then, we can estimate the corresponding label for the voxel in Inew by performing label propagation from the atlases as below
| (1) |
where Ω denotes the image domain, and G denotes the prostate probability map of Inew estimated by multi-atlases based labeling. if voxel belongs to the prostate region in Ii, and otherwise. Through its corresponding graph weight , we can represent the contribution of each candidate voxel in the training image Ii during label propagation, and we can propagate its corresponding anatomical label in the ground truth segmentation image to the reference voxel in the target image with a weight . Therefore, the core problem to perform label propagation is how to define the graph weight with respect to each candidate voxel , which reflects the contribution of during label fusion.
In [29], is determined based on the intensity patch difference between the reference voxel and candidate voxel . Here, the graph weight is given by,
| (2) |
where τ is the smoothing parameter, and Σ is a diagonal matrix which represents the variance in each feature dimension. denotes the neighborhood of voxel in image Ii, and it is defined as the M × M × M sub-volume centered at . denotes the features of image I at voxel . , with and denote the patch-based signature at of Inew and at of Ii, respectively.
Due to the noise and anatomical complexity of prostate MR images, patch-based representation using voxel intensities alone may not be able to effectively distinguish prostate and non-prostate voxels. We use patch-based anatomical features as signature for each voxel to characterize the prostate image appearance. Three types of images features, namely, the Gabor wavelet (GW) feature, the histogram of gradient (HOG) feature, and the local binary pattern (LBP) feature, are extracted from a small image patch centered at each voxel of each aligned training image. Gabor and HOG features can provide complementary anatomical information to each other, and LBP can capture texture information from the input image [30]. However, the graph weight in Eq. (2) may not be able to effectively identify the most representative candidate voxels in atlases to estimate the prostate probability of a reference voxel, especially when the reference voxel is located near the prostate boundary, which is the most difficult region to segment correctly. So we propose to enforce the sparsity constraint in the conventional label propagation framework to resolve this issue. More specifically, the sparse graph weights based on least absolute shrinkage and selection operator (LASSO) [31] are estimated to reconstruct the patch-based signature of each voxel by signature of neighboring voxels in the training images. To do this, firstly signatures , can be organized as columns in a matrix Θ. Then, the corresponding sparse coefficient vector of voxel can be estimated by solving the following optimization problem:
| (3) |
The optimal solution of this equation is denoted as . Here we can set the graph weight to the corresponding element in with respect to in image Ii. Then, we can use the prostate probability map G estimated by Eq. (1) to localize the prostate in the new treatment image. Through enforcing the sparsity constraint, candidate voxels assigned with large graph weights are all from the prostate regions, while candidate voxels from non-prostate regions are mostly assigned with zero or very small graph weights.
3. EXPERIMENTS AND RESULTS
The proposed segmentation method was tested with MR images of 13 prostate-cancer patients. All MR data were acquired using a Siemens MR scanner and an external-body surface coil. Each 3D T2W MR data sets consisted of 256×256×176 voxels to cover the whole pelvis and the voxel size was 1.0×1.0×1.0 mm3. All prostate glands were contoured in MR images by an experienced physician. We used leave-one-out cross-validation method to evaluate the proposed segmentation algorithm. In other words, we used the 12 training images and segmented prostates as the training set and applied the proposed method to process the remaining subject. Our prostate segmentations are compared with the physicians’ manual contours. The Dice volume overlap was calculated for quantitative evaluation. Figure 2 is an example that shows the proposed segmentation method works well for 3D MR images of the prostate and achieved similar results as compared to the manual segmentation. We successfully performed the segmentation method for all enrolled patients. Figure 3 shows the Dice volume overlaps between our automatic and manual segmentations for all patients. Overall, the averaged Dice volume overlap was 89.5±2.6%, which demonstrated the accuracy of the proposed segmentation method.
Figure 2.
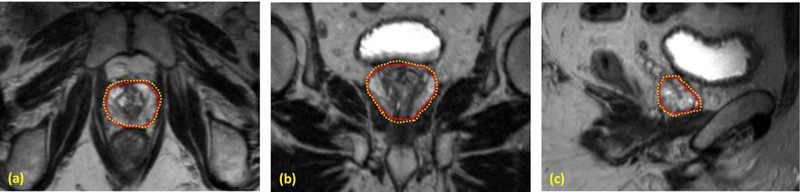
Comparison between the automatic and manual segmentations. (a) Axial, (b) coronal and (c) sagittal T2 MR images of the prostate. The automatic prostate segmentation is shown is yellow dashed line and the manual segmentation is shown in red dotted line.
Figure 3.
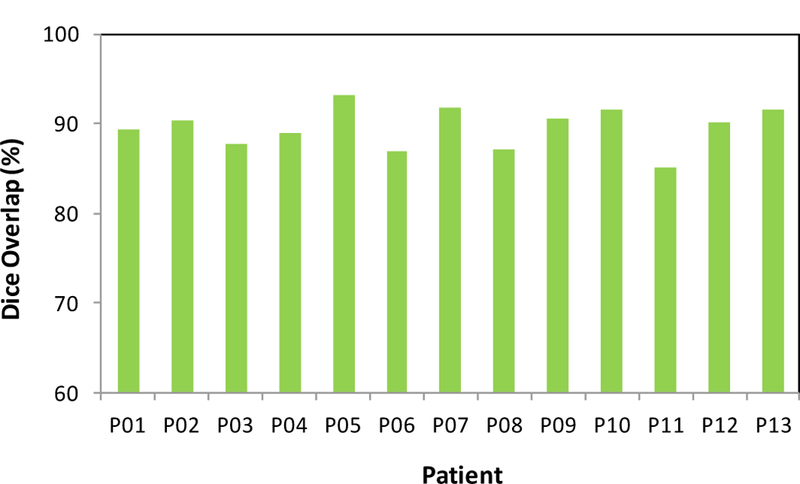
Dice volume overlaps between the automatic and manual segmentations.
4. DISCUSSION AND CONCLUSION
We report a novel 3D MR prostate segmentation method based on the patch-based label fusion framework. The atlases with the most similar global appearance are selected to serve as the best subjects in the label fusion. A local patch-based atlas fusion is performed using voxel weighting based on anatomical signature. In this study, we have demonstrated its clinical feasibility, and validated its accuracy with manual segmentations (gold standard). This segmentation technique could be a useful tool in image-guided interventions for prostate-cancer diagnosis and treatment.
ACKNOWLEDGEMENTS
This research is supported in part by the Department of Defense (DoD) Prostate Cancer Research Program (PCRP) Award W81XWH-13-1-0269 and Winship Cancer Institute.
REFERENCES
- [1].[Prostate Cancer Foundation, http://www.prostatecancerfoundation.org], (2008).
- [2].Hoeks CMA, Hambrock T, Yakar D et al. , “Transition Zone Prostate Cancer: Detection and Localization with 3-T Multiparametric MR Imaging,” Radiology, 266(1), 207–217 (2013). [DOI] [PubMed] [Google Scholar]
- [3].Yang X, and Fei B, “3D prostate segmentation of ultrasound images combining longitudinal image registration and machine learning” Proc. SPIE 8316, 83162O (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Yang X, Schuster D, Master V et al. , “Automatic 3D segmentation of ultrasound images using atlas registration and statistical texture prior,” Proc. SPIE 7964, 796432, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Yang XF, Rossi P, Ogunleye T et al. , “Prostate CT segmentation method based on nonrigid registration in ultrasound-guided CT-based HDR prostate brachytherapy,” Medical Physics, 41(11), (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Litjens G, Toth R, van de Ven W et al. , “Evaluation of prostate segmentation algorithms for MRI: The PROMISE12 challenge,” Medical Image Analysis, 18(2), 359–373 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Yang X, Rossi P, Mao H et al. , “A New MR-TRUS Registration for Ultrasound-Guided Prostate Interventions,” Proc. SPIE 94151Y-94151Y-9, (2015). [DOI] [PMC free article] [PubMed]
- [8].Yang X, Rossi P, Ogunleye T et al. , “A New 3D Neurovascular Bundles (NVB) Segmentation Method based on MR-TRUS Deformable Registration,” Proc. SPIE 941319–941319-7, (2015). [DOI] [PMC free article] [PubMed]
- [9].Tiwari P, Kurhanewicz J, and Madabhushi A, “Multi-kernel graph embedding for detection, Gleason grading of prostate cancer via MRI/MRS,” Medical Image Analysis, 17(2), 219–235 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Allen PD, Graham J, Williamson DC et al. , “Differential segmentation of the prostate in MR images using combined 3D shape modelling and voxel classification” 410–413.
- [11].Zhu Y, Williams S, and Zwiggelaar R, “A hybrid ASM approach for sparse volumetric data segmentation,” Pattern Recognition and Image Analysis, 17(2), 252–258. [Google Scholar]
- [12].Ghose S, Oliver A, Martí R et al. , “A hybrid framework of multiple active appearance models and global registration for 3D prostate segmentation in MRI” 8314, 83140S-83140S-9. [Google Scholar]
- [13].Gao Y, Sandhu R, Fichtinger G et al. , “A coupled global registration and segmentation framework with application to magnetic resonance prostate imagery,” IEEE Trans Med Imaging, 29(10), 1781–94 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].McClure P, Khalifa F, Soliman A et al. , “A Novel NMF Guided Level-set for DWI Prostate Segmentation,” Journal of Computer Science & Systems Biology, 7, 209–2016 (2014). [Google Scholar]
- [15].Martin S, Troccaz J, and Daanenc V, “Automated segmentation of the prostate in 3D MR images using a probabilistic atlas and a spatially constrained deformable model,” Medical Physics, 37(4), 1579–90 (2010). [DOI] [PubMed] [Google Scholar]
- [16].Toth R, and Madabhushi A, “Multifeature Landmark-Free Active Appearance Models: Application to Prostate MRI Segmentation,” Ieee Transactions on Medical Imaging, 31(8), 1638–1650 (2012). [DOI] [PubMed] [Google Scholar]
- [17].Ghose S, Mitra J, Oliver A et al. , “Graph cut energy minimization in a probabilistic learning framework for 3D prostate segmentation in MRI” 125–128.
- [18].Firjany A, Elnakib A, El-Baz A et al. , [Novel Stochastic Framework for Accurate Segmentation of Prostate in Dynamic Contrast Enhanced MRI] Springer Berlin Heidelberg, Berlin, Heidelberg: (2010). [Google Scholar]
- [19].Firjani A, Elnakib A, Khalifa F et al. , “A new 3D automatic segmentation framework for accurate segmentation of prostate from DCE-MRI” 1476–1479.
- [20].Makni N, Puech P, Lopes R et al. , “Combining a deformable model and a probabilistic framework for an automatic 3D segmentation of prostate on MRI,” Int J Comput Assist Radiol Surg, 4(2), 181–8 (2009). [DOI] [PubMed] [Google Scholar]
- [21].Klein S, van der Heide UA, Lips IM et al. , “Automatic segmentation of the prostate in 3D MR images by atlas matching using localized mutual information,” Medical Physics, 35(4), 1407–1417 (2008). [DOI] [PubMed] [Google Scholar]
- [22].Dowling JA, Fripp J, Chandra S et al. , [Fast Automatic Multi-atlas Segmentation of the Prostate from 3D MR Images] Springer Berlin Heidelberg, Berlin, Heidelberg: (2011). [Google Scholar]
- [23].Flores-Tapia D, Thomas G, Venugopal N et al. , “Semi automatic MRI prostate segmentation based on wavelet multiscale products” 3020–3023. [DOI] [PubMed]
- [24].Yang XF, and Fei BW, “A wavelet multiscale denoising algorithm for magnetic resonance (MR) images,” Measurement Science & Technology, 22(2), (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Vikal S, Haker S, Tempany C et al. , “Prostate contouring in MRI guided biopsy,” Proc SPIE Int Soc Opt Eng, 7259, 72594A (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Khurd P, Grady L, Gajera K et al. , [Facilitating 3D Spectroscopic Imaging through Automatic Prostate Localization in MR Images Using Random Walker Segmentation Initialized via Boosted Classifiers] Springer Berlin Heidelberg, Berlin, Heidelberg: (2011). [Google Scholar]
- [27].Aljabar P, Heckemann RA, Hammers A et al. , “Multi-atlas based segmentation of brain images: Atlas selection and its effect on accuracy,” Neuroimage, 46(3), 726–738 (2009). [DOI] [PubMed] [Google Scholar]
- [28].Yang XF, Wu N, Cheng GH et al. , “Automated Segmentation of the Parotid Gland Based on Atlas Registration and Machine Learning: A Longitudinal MRI Study in Head-and-Neck Radiation Therapy,” International Journal of Radiation Oncology Biology Physics, 90(5), 1225–1233 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Rousseau F, Habas PA, and Studholme C, “A Supervised Patch-Based Approach for Human Brain Labeling,” Ieee Transactions on Medical Imaging, 30(10), 1852–1862 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Yang X, Rossi P, Jani A et al. , “BEST IN PHYSICS (IMAGING): 3D Prostate Segmentation in Ultrasound Images Using Patch-Based Anatomical Feature,” Medical Physics, 42(6), 3685–3685 (2015). [Google Scholar]
- [31].Aseervatham S, Antoniadis A, Gaussier E et al. , “A sparse version of the ridge logistic regression for large-scale text categorization,” Pattern Recognition Letters, 32(2), 101–106 (2011). [Google Scholar]


