Abstract
In this work, we developed novel core-shell nanoparticle systems with magnetic core and polymer shell via atom transfer radical polymerization for use as high affinity nanoadsorbents for organic contaminants in water and wastewater treatment. Polyphenolic-based moieties, curcumin multiacrylate (CMA) and quercetin multiacrylate (QMA), were incorporated into poly(ethylene glycol) (PEG) based polymeric shells to create high affinity binding sites for the capture of polychlorinated biphenyls (PCBs) as a model pollutant. The resulting magnetic nanoparticles (MNPs) were characterized by Fourier transform infrared spectroscopy (FTIR), thermogravimetric analysis (TGA), transmission electron microscopy (TEM), X-ray diffraction (XRD), dynamic light scattering (DLS), and UV-visible spectroscopy. The affinity of these novel materials for PCB 126 was evaluated and fitted to the nonlinear Langmuir model to determine binding affinities (KD). The KD values obtained were: PEG MNPs (8.42 nM) < IO MNPs (8.23nM) < QMA MNPs (5.88 nM) < CMA MNPs (2.72 nM), demonstrating that the presence of polyphenolic-based moieties enhanced PCB 126 binding affinity, which is hypothesized to be a result of π – π stacking interactions. These values are lower that KD values for activated carbon, providing strong evidence that these novel core-shell nanoparticles have a promising application as nanoadsorbents for specific organic contaminants offering a cost effective alternative to current remediation approaches.
Keywords: Core-shell nanoparticles, Iron oxide, Environmental remediation
1. Introduction
Water is the most essential natural resource for human life, yet only 0.03% of the total available water on earth can be utilized for human consumption, and over 1 billion people lack access to safe drinking water [1,2]. The spread of a wide range of environmental contaminants in surface water has become a worldwide problem, affecting human health and the ecological environment [3].
Polychlorinated biphenyls (PCBs) are some of the most persistent, ubiquitous, and bio-accumulated pollutants in the environment, even though their production was banned in 1979 in the United States and in 2001 by the Stockholm Convention on Persistent Organic Pollutants [4–6]. PCBs have poor aqueous solubility and low volatility, which makes their extraction from the environment especially challenging. Because of environmental cycling, PCBs have been distributed worldwide [4]. Current remediation techniques for persistent organic pollutants, such as PCBs, involve dredging and subsequent deposition in landfills, or complete degradation through incineration or chemical dehalogenation techniques [7]. However, it has been shown that these techniques could result in harmful byproducts when insufficient temperatures are reached during incineration, can require organic solvents that are often more toxic than the pollutants being remediated, and could contribute to the pollutant’s ubiquitous nature through air, water and slurry transport processes in the landfill’s surrounding environment [8,9].
Significant advances have been made in wastewater treatment and water remediation. Oxidation, photocatalytic degradation, membrane filtration, ion exchange, adsorption/separation processes and bioremediation all show promising results [10–12]. Nevertheless, their application has been limited due to several factors, of which the most important are efficiency [3], energy requirements [13] and economic cost [14]. In contrast, adsorption is a useful strategy because of its ease of application, low cost and rich sorbent variety. The unique properties of sorbent materials such as porosity, large surface area, mechanical strength, tunable shapes and morphologies and a variety of functional groups present on their surface are being exploited for a range of industrial applications (e.g., heavy metal separation from water) [15,16]. Furthermore, nanoadsorbents have a very high specific surface area and associated sorption sites, provide very short diffusion paths, allow tunable surface chemistry [17], and have been successfully used in environmental applications with promising performance in pollutant mitigation and/or removal.
Among nanoadsorbents, magnetic nanoparticles (MNPs) have gained interest as promising alternatives to current water treatment techniques that can meet the stringent water quality standards at lower costs and higher efficiencies [18–20]. Iron oxide magnetic nanoparticles (IO MNPs), such as magnetite (Fe3O4) or maghemite (Ƴ-Fe2O3), possess superparamagnetic properties, when small enough. Because of this feature, an external magnetic field will rapidly aggregate the IO MNPs together, and once the magnetic field is removed, their magnetization decreases to zero, resulting in them being re-dispersed [21,22]. Therefore, IO MNPs in combination with an external magnetic field can be used as a separation tool for organic contaminants from aqueous or slurry matrices, without requiring centrifugation or filtration steps even when dealing with raw environmental samples. IO MNPs can also generate heat in response to exposure to an alternating magnetic field (AMF), which can cause local modification of its properties, such as thermal treatment or binding properties. Additionally, these IO MNPs can be regenerated without the need of harmful solvents or the generation of secondary byproducts [23–25]. Most importantly, IO MNPs can be easily synthesized with readily available materials and low-cost methods, making them ideal for large-scale operations.
Furthermore, the surface of the IO MNPs can be easily modified to incorporate a variety of materials, such as organic molecules, polymers, surfactants, oligonucleotides, among others, that improve the stability of the MNPs in solution and help prevent their aggregation, as well as providing additional functionalities for tailored applications. The incorporation of the IO MNPs into core-shell structures has been widely exploited because of its versatility in shell materials that can provide desired functionality, while the magnetic core functions as the means for magnetic separation. In order to obtain the desired functionalities, there are several strategies that have been used either as ‘grafting to’ or ‘grafting from’ the MNP surface. Of particular interest are methods which involve surface-initiated atom transfer radical polymerization (ATRP), a ‘grafting from’ approach widely used today [26–29]. ATRP is a controlled “living” radical polymerization which allows for the synthesis of core-shell nanoparticles with tunable thickness. The magnetic properties of these nanocomposites have enabled their use in environmental applications for capture and/or separation where they can be easily decanted out of solution [26,30,31].
To obtain larger adsorption capacities for a specific compound, various functional monomers or crosslinkers can be incorporated in the ATRP reaction, which will modify the chemical composition of the adsorbent via its shell. Plant derived polyphenols, such as quercetin and curcumin, are a well-known class of naturally occurring antioxidants rich in aromatic and phenolic moieties. The prevalence of these types of functionalities have been observed in computational analysis of the monoclonal antibody S2B1, which possesses high selectivity and nanomolar binding affinities for coplanar, non-ortho-chlorinated PCB congeners. The sterically constrained deep binding pocket present in this antibody presents aromatic residues of tyrosine and arginine, where pi-cation interactions with the center of the PCB molecule take place [32]. These π-π stacking interactions between PCB and aromatic residues have also been observed in other antibodies, as well as in water-sediment interactions where humin and humic matter act as PCB sinks [33–37]. Therefore, by incorporating plant derived polyphenols into core-shell magnetic nanoparticles, their aromatic and phenolic moieties will improve the adsorption behavior for organic contaminants such as PCBs.
In this work, core-shell magnetic nanoparticles were prepared using ATRP to coat IO MNPs with a PEG-based polymer shell crosslinked with acrylated plant derived polyphenols. Two different polyphenols, curcumin and quercetin, were acrylated and incorporated into the core-shell magnetic nanoparticles to enhance their adsorption capacity for PCBs. The functionalized nanoparticle systems were characterized for size, shell coating percent, response to a static magnetic field and stability. The binding isotherm for a model contaminant, PCB 126, was studied, and the binding constants for the fours systems synthesized were evaluated using the Langmuir adsorption model.
2. Experimental details
2.1. Materials
Iron (III) chloride hexahydrate (FeCl3 • 6 H2O); iron chloride tetrahydrate (FeCl2 • 4 H2O); 2-bromo2-methyl propionic acid (BMPA); 2,2’ bipyridine (Bpy); copper(I) bromide (CuBr); copper powder (< 425 micron), triethyl amine (TEA), acryloyl chloride, and potassium carbonate (K2CO3) were obtained from Sigma Aldrich (St Louis, MO). Ammonium hydroxide (NH4OH) was purchased from EMD Chemicals (Gibbstown, NJ). Poly(ethylene glycol) 400 dimethacrylate (PEG400DMA) was obtained from Polysciences INC. (Warrington, PA). Curcumin was purchased from Chem-Impex International, Inc. (Bensenville, IL) and quercetin was purchased from Cayman Chemicals (Ann Arbor, MI). 3,3′,4,4′,5-Pentachlorobiphenyl (PCB-126) in isooctane was purchased from Accustandard (New Haven, CT). All solvents (isooctane, ethanol HPLC grade, tetrahydrofuran (THF); dichloromethane (DCM), and acetonitrile (ACN)) were obtained from Fisher Scientific (Hannover Park, IL). All materials were used as received.
2.2. Curcumin multiacrylate synthesis and purification
Curcumin multiacrylate (CMA) was prepared by reacting curcumin with acryloyl chloride according to the protocol described by Patil et al. [38,39]. Briefly, curcumin was dissolved in THF at a concentration of 50 mg/mL. Both acryloyl chloride and TEA were added at a 3:1 ratio with respect to curcumin. The reaction mixture was purged with nitrogen for 20 min and allowed to react overnight. The reaction mixture was then filtered to remove the byproduct salts formed. The THF was evaporated and the remaining solid was re-dissolved in DCM. This solution was then purified by washing three times with K2CO3 0.1 M to remove any unreacted acryloyl chloride, and again thrice with HCl 0.1 M) to remove unreacted TEA. Finally the DCM was evaporated to obtain CMA.
2.3. Quercetin multiacrylate synthesis and purification
Quercetin multiacrylate (QMA) was prepared by the reaction of quercetin with acryloyl chloride according to the method described by Gupta et al. [40]. Briefly, quercetin was dissolved in anhydrous THF at a concentration off 100 mg/mL. Acryloyl chloride and K2CO3 were both added at a 6:1 ratio with respect to quercetin. The reaction vessel was purged with nitrogen for 20 min and allowed to react overnight. The reaction mixture was then filtered to remove the byproduct salts formed. The THF was evaporated and the remaining solid was re-dissolved in DCM. This solution was then purified by washing three times with K2CO3 0.1 M to remove unreacted acryloyl chloride. Finally the DCM was evaporated to obtain QMA.
2.4. Iron oxide nanoparticle synthesis
Iron oxide magnetic nanoparticles (IO MNPs) were synthesized via a one-pot co-precipitation method [22]. A 2:1 molar ratio of FeCl3 • 6 H2O and FeCl2 • 4 H2O, respectively, were dissolved in 40 mL of deionized (DI) water and combined in a sealed 3-neck flask under vigorous stirring and nitrogen flow to achieve an inert synthesis environment. The solution was heated to 85 °C and, at this point, 5 mL of NH4OH (30.0% v/v) was injected dropwise into the vessel. The reaction was carried out for 1 h at this temperature. The nanoparticles were then magnetically decanted and washed three times with DI water. Finally, the particles were re-suspended in 45 mL of DI water and dialyzed against water for 24 h (100 kDA molecular weight cutoff).
2.5. Surface initiated polymerization
The core-shell nanoparticles were prepared by minor modifications of the previously reported method by Wydra et al. [41]. Briefly, the uncoated nanoparticles and the BMPA initiator were mixed at a 1:4 molar ratio in a 75–25 ethanol – DI water solution. The mixture was stirred for 24 h at room temperature. The particles were then washed three times with ethanol. The initiator coated particles (BMPA MNPs) were then suspended in ethanol for the ATRP reaction. The amount of catalyst used was determined based on a macromere ratio. The ratios used were 1:0.04 for Bpy and 1:0.01 for CuBr. Additionally, 2–3 crystals of Cu(0) were combined with the catalyst in 5 mL of ethanol. The catalyst solution and particles were then placed in a 3-neck flask under nitrogen bubbling and heated to 50 °C. The acrylated polyphenol (CMA or QMA), was mixed with 8 mmol of the macromere in a 90:10 molar ratio, and injected into the reaction vessel once it reached a temperature of 50 °C. The reaction was carried out for 24 h. After this, the particles were magnetically decanted and washed three times with ethanol, five times with a 50–50% (v/v) ACN/DCM solution, and twice with a 50–50% (v/v) ethanol/DI water solution. Finally, the particles were re-suspended in DI water.
2.6. Particle characterization
2.6.1. Fourier transform infrared (FTIR) spectra
Attenuated total reflectance FTIR (ATR-FTIR) was used to determine the surface functionalization with a Varian Inc. 7000e spectrometer. Dried samples were placed on the diamond ATR crystal and the spectrum was obtained between 700 and 4000 cm−1 using 32 scans.
2.6.2. Thermogravimetric analysis (TGA)
TGA was used to quantify the mass percent of the coating on the particle systems using a Netzsch Instruments STA 449A system. Approximately 5 mg of the dry sample was heated at a rate of 5 °C/ minute until a temperature of 120 °C under constant nitrogen flow. The system was kept isothermal for 20 min to vaporize residual water. The sample continued to be heated at 5 °C/minute until a temperature of 600 °C. The presented mass loss values are normalized to the mass after the isothermal heating at 120 °C.
2.6.3. Transmission electron microscopy (TEM)
TEM images of the samples were obtained using a JOEL 2010F at an accelerating voltage of 200 keV. The nanoparticles were diluted to a 1 mg/mL concentration in DI water and then dried on lacey carbon TEM grids prior to analysis.
2.6.4. X-ray diffraction (XRD)
The X-ray patterns of the nanoparticles were obtained using a Siemens D-500 X-ray spectrometer with a CuKα radiation source (λ = 1.54 Å) at 40 kV and 30 mA scanning from 5° to 65°, at a scan rate of 1°/minute. The XRD patterns were used to estimate the particle’s crystal domain using the Scherrer equation [42]:
| (1) |
where τ is the mean size of the ordered crystalline domains, K is a dimensionless shape factor with a value close to unity (for iron oxide, K = 0.8396), λ is the X-ray wavelength, β is the line broadening at half the maximum intensity (FWHM) after subtracting the instrumental line broadening, and θ is the Bragg angle, in radians (17.72°). Additionally, we use the XRD patterns to confirm the magnetic crystal structure of the iron oxide nanoparticles before and after coating.
2.6.5. Dynamic light scattering (DLS)
DLS measurements were obtained using a Malvern Zetasizer, Nano ZS90 instrument. The nanoparticle solutions were diluted to 200 μg/mL and probe sonicated for 10 min prior to analysis.
2.6.6. Ultraviolet (UV)-visible spectroscopy
The stability of the nanoparticles was analyzed using a Cary Win 50 probe UV-visible spectrophotometer. The magnetic nanoparticles were diluted to 200 μg/mL in DI water, and probe sonicated for 10 min. The samples were then placed in a quartz cuvette and their change in absorbance was read at 540 nm for a period of 12 h.
2.7. PCB 126 binding studies
The binding capacity of the MNPs to PCB 126 was conducted under equilibrium conditions, as determined by previous kinetic studies. All experiments were carried out using 0.1 mg of the core-shell nanoparticles (CMA MNPs, QMA MNPs, PEG MNPs, and IO MNPs), suspended in a 99:1 DI water to ethanol solvent in 3 mL borosilicate glass vials.
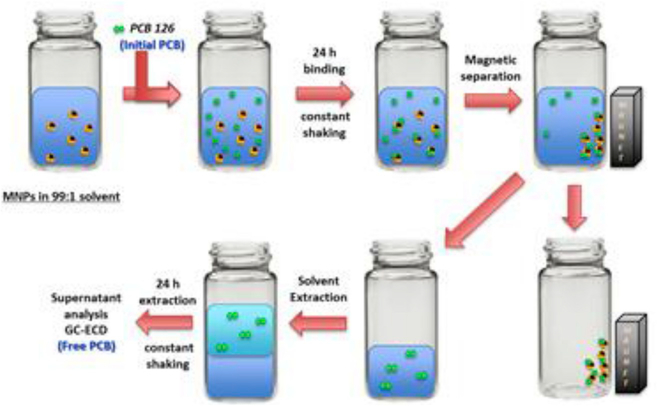
Different PCB 126 stocks of varying concentrations were freshly prepared in ethanol. Binding experiments were carried out in batch conditions. The samples were spiked with the PCB 126 stock solutions to obtain the initial concentrations ranging from 0.003 to 0.1 ppm, all while maintaining a solvent ratio of 99:1 of DI water to ethanol. The samples were initially sonicated for 10 min to ensure a well dispersed sample and then subjected to orbital shaking for 24 h at 200 rpm and room temperature conditions, in order to evaluate the equilibrium binding. At the end of the binding study, the MNP suspension was separated by exposure to a static magnet for ~10 min, as seen in Fig. 1. The supernatant containing the unbound PCB 126 was placed into a new borosilicate glass vial and a 1:1 liquid extraction using isooctane was performed for 24 h. Finally the organic phase, rich in PCB 126, was collected using a Hamilton syringed and transferred to a glass chromatography vial for analysis. At this point each sample was spiked with the internal standard, 5’-fluoro-3,3’,4,4’,5-pentachlorobiphenyl (F-PCB 126) The PCB 126 concentrations before and after binding were determined using an Agilent 6890N gas chromatograph coupled to electron capture detection (GC-ECD), equipped with an Agilent HP-5MS UI column (30×0.25×0.25). All binding studies were carried out in triplicates.
Fig. 1.
Schematic representation of the binding studies conducted with PCB 126 in a 99:1 DI water ethanol solvent.
The equilibrium adsorption of PCB 126 was evaluated according to the Langmuir isotherm model. This model assumes a monolayer adsorption on a homogeneous surface where all existing binding sites are energetically equivalent. These sites are all identical, and once a site is filled, no interactions occur between the adsorbed molecules [43]. The Langmuir model is represented by the following equation:
| (2) |
Where qe (mg/g) represents the adsorption capacity at equilibrium, Ce (mg/L) is the equilibrium concentration of the adsorbate, KD (L/mg) is the adsorption coefficient of the adsorbant related to the energy of adsorption, and Bmax (mg/g) is the maximum binding capacity of the adsorbant.
3. Results and discussion
Core-shell magnetic nanoparticles were prepared via surface initiated atom transfer radical polymerization. Two acrylated polyphenols, curcumin multiacrylate and quercetin multiacrylate, were selected as functional crosslinkers due to their unique properties and structure similarity to PCB binding domains in antibodies and humin matter. The reaction process followed a 3 step process. First, the uncoated nanoparticles were synthesized using the co-precipitation method, where Fe (III) and Fe (II) salts were dissolved in DI water in a 2:1 ratio and heated to 85 °C, at which NH4OH was added to precipitate the iron oxide magnetic nanoparticles. In the second step, the uncoated nanoparticles, suspended in ethanol, were mixed with BMPA in a 1:4 molar ratio for 24 h at room temperature. Finally, the BMPA-coated nanoparticles were reacted with PEG400DMA and the acrylated polyphenol in an inert environment, using bipyridine and copper salts as catalyst, to obtain core-shell magnetic nanoparticles.
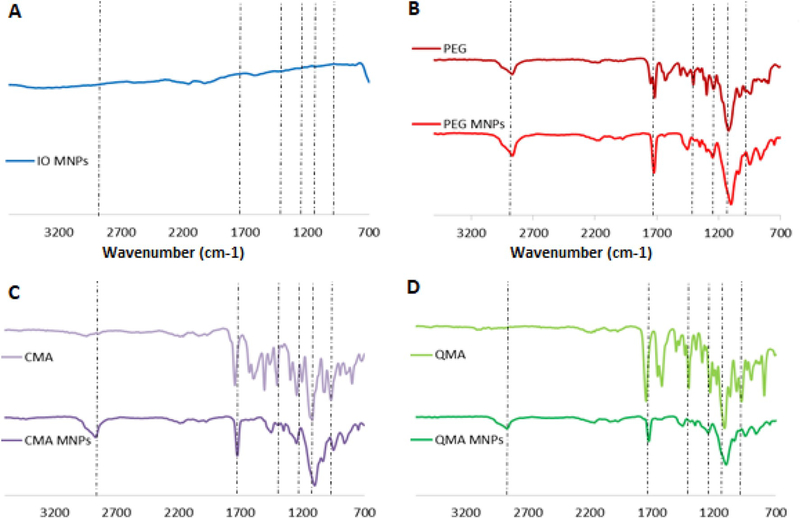
FTIR analysis confirms the successful ATRP reaction. The spectrum in Fig. 2 demonstrates the incorporation of the polyphenol-based moieties, QMA and CMA, and the PEG400DMA. The presence of peaks at ~1750 cm−1 and ~1100 cm−1 in all the synthesized core-shell MNPs correspond to the carbonyl band (C=O) stretching and ether band (CO-C) stretching from the PEG400DMA. For the CMA and QMA core-shell systems, the appearance of additional peaks is seen, confirming the incorporation of the polyphenols onto the coating. In the CMA MNPs spectra, the presence of three peaks between 1604 cm−1 and 1400 cm−1 are attributed to the symmetric ring vibrations of the benzene rings present in CMA. Furthermore, less intense peaks 1026 cm−1 and 964.4 cm−1 correspond to the enol (C-O-C) peak, and the benzoate C-H vibrations of the aromatic rings respectively. Similarly, the QMA MNPs spectrum exhibits the presence of a broad peak at 1600 cm−1 and two shorter peaks at 1432 cm−1 and 1404 cm−1 that correspond to the aromatic ring vibrations of the benzene rings present in QMA. Additionally, the enol group peak of QMA is observed at 1122 cm−1.
Fig. 2.
FTIR spectra of the synthesized core-shell magnetic nanoparticles.
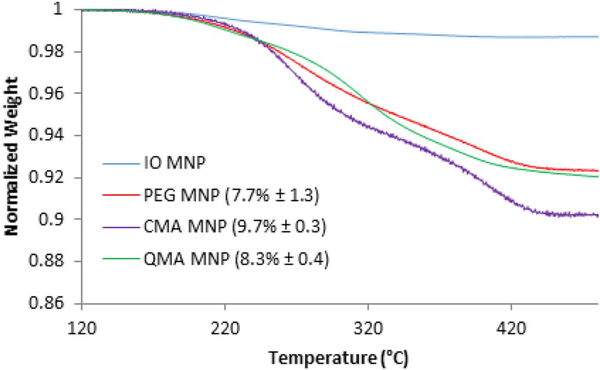
To further characterize the coating on the core-shell MNPs, quantification of this coating was conducted using TGA, as shown in Fig. 3. Minimal weight loss was observed for the uncoated iron oxide nanoparticles. However, a significant weight loss of 9.7%, 8.3% and 3.2% was observed for the CMA, QMA and PEG coated magnetic nanoparticle systems, respectively, suggesting the successful ATRP reaction being conducted on the surface of the magnetic nanoparticles.
Fig. 3.
Mass loss profile with increasing temperature of the synthesized core-shell magnetic nanoparticles.
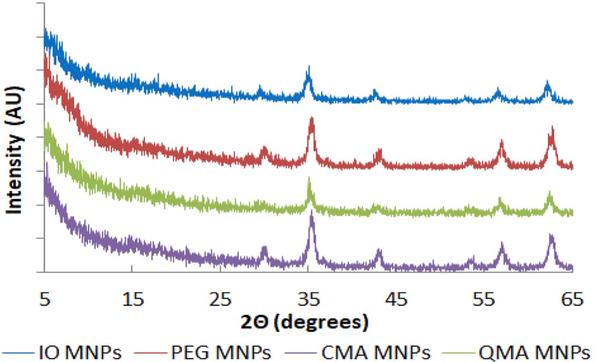
The core-shell MNPs exhibit a tendency to be attracted to a static magnet, as can be seen in Fig. 4. The black aqueous dispersion of MNPs is rapidly magnetically decanted, leaving a transparent solution after exposure to a nearby magnet. This indicates that the core of the MNPs remains superparamagnetic after the ATRP synthesis. Additionally, the XRD patterns of the iron oxide core-shell MNPs synthesized are in agreement with the JCPDS card (19–0629) associated with magnetite. Similarly, the broad diffraction lines in the XRD patterns suggest the nano-crystallite nature of the magnetite particles [48,49]. The sharp peaks present in the diffractograms in Fig. 5 indicate the formation of a crystalline magnetite structure. The highest intensity peak seen for the 35.5° (2θ) corresponds to the (3 1 1) reflection plane of the iron oxide crystalline structure, which was used in the Scherrer equation to calculate the crystallite size of the core-shell MNPs. The calculated crystallite size from the XRD spectra is depicted in Table 1.
Fig. 4.
Suspended solution of CMA MNPs and capture of CMA MNPs in a static magnetic field (right).
Fig. 5.
XRD patterns of the synthesized core-shell magnetic nanoparticles.
Table 1.
Size analysis from XRD diffractograms using the Scherrer equation and hydrodynamic size analysis via dynamic light scattering of the synthesized core-shell MNPs (mean ± std dev. for three independent batches and three samples from each batch).
| MNP system | XRD crystal size (nm) | Hydrodynamic size (nm)* | PDI |
|---|---|---|---|
| IO MNPs | 13.4 ± 0.9 | 126.5 ± 0.9 | 0.12 ± 0.02 |
| BMPA MNPs | 10.8 ± 0.7 | 141.0 ± 0.3 | 0.14 ± 0.05 |
| PEG MNPs | 12.9 ± 1.6 | 222.7 ± 10.6 | 0.18 ± 0.10 |
| CMA MNPs | 9.5 ± 1.2 | 254.6 ± 19.4 | 0.15 ± 0.05 |
| QMA MNPs | 9.0 ± 1.4 | 232.8 ± 9.6 | 0.20 ± 0.03 |
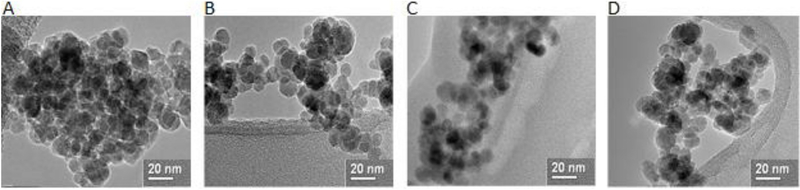
From the TEM images of the core-shell nanoparticles in Fig. 6. it can be seen that the core iron oxide nanoparticle size ranges between 8 and 12 nm. This size is in accordance with values previously reported by our lab group [21,41,46]. As seen in Table 1, these values are similar to those obtained for the crystal size using the Scherrer equation.
Fig. 6.
TEM images of (a) iron oxide magnetic nanoparticles, (b) PEG coated magnetic nanoparticles, (c) CMA coated nanoparticles and (d) QMA coated magneticnanoparticles.
The hydrodynamic diameter of the core-shell MNPs was determined via dynamic light scattering (DLS) and reported as Z-average, with the variability in particle size within the batches being quantified by the polydispersity index (PDI), as presented in Table 1. The coated MNPs demonstrated a slightly larger aggregate size than the uncoated nanoparticles. It was observed that the hydrodynamic size of the uncoated particles is significantly larger than the size reported from the TEM (Fig. 6) and XRD analysis (Table 1). This is due to the agglomeration of the iron oxide particles in the dispersed state, and it suggests that the core-shell systems are most likely small agglomerates of IO MNPs at the core which are encapsulated within the PEG400DMA-polyphenol-based coatings.
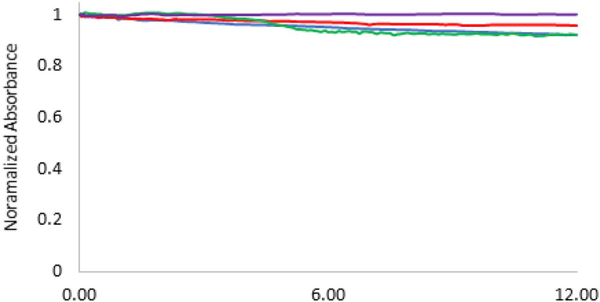
In order to maximize the pollutant binding capacity of the core-shell MNPs in aqueous environments, their stability in solution is very important as further agglomeration could cause the nanoparticles to fall out of solution and limit the available surface for adsorption to occur. Thus, the stability of the core-shell systems in DI water was analyzed for a period of 12 h, after probe sonication for 10 min. All the synthesized systems demonstrated good stability over the period of time studied, as seen in Fig. 7.
Fig. 7.
Normalized absorbance (at 540 nm) of the MNPs in DI water for 12h using UV-visible spectroscopy.
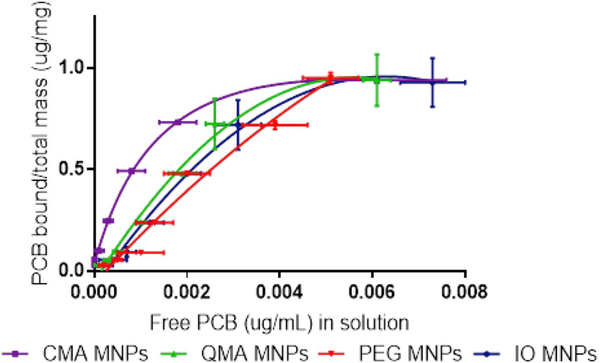
The binding capacity of the nanoparticles for PCB 126 was studied under equilibrium conditions and constant shaking at room temperature. Seven different PCB 126 concentrations were used at a loading of 0.1 mg/mL of the nanoparticles to obtain a binding isotherm. The equilibrium time of 24 h was determined from previous kinetic studies where the contact time varied from 30 min to 1 week. The adsorption isotherm for PCB 126 onto the IO MNPs, PEG MNPs, CMA MNPs and QMA MNPs is shown in Fig. 8. It can be seen that for all systems the amount of PCB 126 adsorbed increases as the free concentration of PCB increased, until an adsorption plateau was reached. This behavior matches what is commonly seen nanoparticles used in the adsorption or organic contaminants such as PCBs [47] and dyes [48]. The CMA MNPs bind more PCB at lower free adsorbate concentrations, and as the plateau is reached, it behaves very similarly to the other three systems. The Langmuir model provides a good fit for the experimental data (R2 > 0.95), and thus can be used to describe the adsorption behavior of the MNP systems. The use of the Langmuir model suggests that the adsorption of PCB 126 onto the MNP systems occurs through monolayer adsorption where there is little to no interaction between the adsorbed PCB molecules. This can be explained due to the planar nature of PCB 126. Previous studies have demonstrated that planar molecules, such as PCB 126, can more closely approach the sorption surface of adsorbent, allowing for a favorable π-cloud interaction between the aromatic groups present in the adsorbent and those in the sorbate molecules [49,50]. The maximum adsorption capacity (Bmax) and Langmuir adsorption coefficients (KD) for each system were calculated and are presented in Table 2.
Fig. 8.
Adsorption isotherms for PCB 126 of the core-shell systems at room temperature. PCB 126 initial concentrations from 0.003 to 0.1ppm fitted using the Langmuir model.
Table 2.
Langmuir binding constants for the binding isotherms of PCB 126 for the four nanoparticle systems synthesized (n= 12, except for PEG MNPs where n= 15).
| MNP system | Bmax (mg/g) | 95% CI | KD (nM) | 95% CI | R2 |
|---|---|---|---|---|---|
| IO MNPs | 0.99 | 0.98 to 1.01 | 8.23 | 6.10 to 6.37 | 0.963 |
| PEG MNPs | 1.91 | 0.98 to 2.75 | 8.42 | 6.54 to 14.24 | 0.980 |
| CMA MNPs | 1.06 | 1.02 to 1.09 | 2.72 | 2.50 to 3.00 | 0.993 |
| QMA MNPs | 1.06 | 1.02 to 1.10 | 5.88 | 5.58 to 6.24 | 0.956 |
The binding isotherms were obtained by running four independent studies with newly synthesized materials and preparing three independent samples for each concentration in each of these studies. Although there is some variability between each batch, the amount of PCB bound per total mass at the lower end of the binding isotherm for CMA MNPs is significantly higher than the other curves, based on the confidence intervals, indicating a higher affinity for PCB 126. However, because of this batch to batch variability, there is no significant difference in the behavior of the other three systems (IO MNPs, PEG MNPs and QMA MNPs). This behavior is further confirmed when looking at the scatter plots with confidence intervals for each individual initial concentration, where the confidence intervals indicate that the CMA MNPs have a significantly higher affinity than the other systems (see supportive information S1 – S7). For each initial concentration level, from the confidence intervals, differences between the systems can be observed. For example, when the initial PCB concentration level is 0.003 ppm, the estimate difference between CMA MNPs and IO MNPs is of 0.0022 with p-values less than 0.0001.
The maximum binding capacity of all the magnetic nanomaterials is relatively the same for all of the systems and close to 1 mg/g, with the only exception being the PEG MNPs which is closer to 2 mg/g. These values are all much lower than those normally reported for other carbonaceous materials, specifically activated carbon, which normally present values of maximum loading of higher orders of magnitude [49,51]. However, the Langmuir adsorption coefficients obtained for the IO MNPs, PEG MNPs, CMA MNPs and QMA MNPs are 8.23 nM, 8.42 nM, 2.72 nM and 5.88 nM, respectively. These values are lower than the reported KD of 15.2 nM for activated carbon made of coconut shell binding specifically to PCB 126 [52], showing promising adsorption capacities for our newly synthesized materials to outcompete activated carbon, which is the gold standard in environmental remediation/biding of organic contaminants. Additionally, the KD values obtained for our acrylated polyphenol containing core-shell MNPs are very close to what is reported for specific binding of PCB 126 by the monoclonal antibody S2B1 (2.5 ± 0.01 nM) [32], further demonstrating the high affinity of these materials for PCB 126.
More closely examining the KD values in Table 2, it is seen that their affinity for PCB 126 is as follows: PEG MNPs < IO MNPs < QMA MNPs < CMA MNPs. This order demonstrates that the presence of the acrylated polyphenols, CMA and QMA, as crosslinkers enhances the binding affinity for PCB 126. This can be explained because of their aromatic rich nature which provides sites for π – π stacking interactions between the nanoparticle surface and the PCB in solution. In contrast, the PEG MNPs present a lower affinity for PCB 126 than the IO MNPs. This was expected as the hydrophilic nature of the PEG400DMA is expected to hamper the adsorption of the hydrophobic PCB 126 onto the nanoparticle surface [53]. Furthermore, this emphasizes the important role that the aromatic rich acrylated polyphenols have in enhancing PCB 126 binding by, not only allowing for π – π interactions with the adsorbate, but also increasing the hydrophobic nature of the nanomaterial. These results show the great promise for our magnetic nanomaterials to be used as remediation alternatives for harmful contaminants in the environment.
4. Conclusions
This study reports the successful synthesis of novel core-shell magnetic nanoparticles using ATRP to coat iron oxide nanoparticles with a PEG-based polymer shell with and without acrylated plant derived polyphenols as additional functional crosslinkers. Curcumin multiacrylate and quercetin multiacrylate were incorporated to enhance pollutant binding capacity of the core-shell nanoparticles. Equilibrium binding studies were conducted at seven different PCB concentrations, and binding isotherms for each MNP system synthesized were obtained. The Langmuir model was used to obtain biding coefficients and the maximum binding capacity of the nanoparticles. It was seen that the maximum binding capacity of these materials was lower than what is reported for carbonaceous materials. However, it was demonstrated that these materials possess higher binding affinity coefficients for PCB 126 than activated carbon, which is the gold standard for organic pollutant adsorption. Furthermore, we were able to demonstrate the binding enhancement for PCB 126 by incorporating only 10 mol% of acrylated naturally occurring polyphenols, curcumin and quercetin, and obtaining binding capacities similar to those observed for antibodies. These materials can be further optimized to enhance the binding capacity by modifying the loading of the polyphenol, and can be further explored as capture agents for other organic contaminants in the environment. Overall, we have obtained novel nanomaterials that can bind PCB 126 in aqueous media with high affinities, and are feasible alternatives for environmental remediation of harmful organic contaminants.
Supplementary Material
HIGHLIGHTS.
Polyphenol containing magnetic core-shell nanoparticles were synthesized via ATRP.
CMA and QMA incorporation increases affinity for PCB 126 binding in aquous media.
Novel nanomaterials have higher affinity for PCB 126 than activated carbon.
Acknowledgements
The authors would like to thank Dr. Andrew Morris and Dr. Sony Soman for their help in method development for GC-ECD analysis and allowing us to use their facilities at the University of Kentucky’s small molecule mass spectrometry core laboratory. This project was supported by the grant number P42ES007380 the National Institute of Environmental Health Sciences. The content of this paper is solely the responsibility of the authors and does not necessarily represent the view of the National Institute of Environmental Health Sciences.
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.matchemphys.2018.10.045.
References
- [1].UNEP. Natural solutions for water security Water and Biodiversity. International Day for Biological Diversity – Convention on Biological Diversity; May 22, 2013. [Google Scholar]
- [2].Mittal H, Mishra SB, Carbohydr. Polym 101 (2014) 1255. [DOI] [PubMed] [Google Scholar]
- [3].Xu P, et al. , Sci. Total Environ 424 (2012) 1. [DOI] [PubMed] [Google Scholar]
- [4].Dang VD, Walters DM, Lee CM, Am CM, J. Environ. Sci 8 (2012) 11. [Google Scholar]
- [5].Howell N, et al. , Chemosphere 70 (2008) 593. [DOI] [PubMed] [Google Scholar]
- [6].Joes KC, de Voogt P, Environ. Pollut 100 (1999) 209–221. [DOI] [PubMed] [Google Scholar]
- [7].NCI, Table 2. Food Sources of the Total Omega 6 Fatty Acids (18:2 + 20:4), Listed in Descending Order by Percentages of Their Contribution to Intake, Based on Data from the National Health and Nutrition Examination Survey 2005–2006, NIH, 2013, http://appliedresearch.cancer.gov/diet/foodsources/fatty_acids/table2.html. [Google Scholar]
- [8].Petriello MC, Newsome BJ, Dziubla TD, Hilt JZ, Bhattacharyya D, Hennig B B, Sci. Total Environ 491–492 (2014) 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ham SY, Kim YJ, Lee DH, Chemosphere 106 (2008) 226. [DOI] [PubMed] [Google Scholar]
- [10].Zhu J, et al. , RSC Adv. 2 (2012) 4844. [Google Scholar]
- [11].Argawal S, et al. , J. Environ. Eng 133 (2007) 1075. [Google Scholar]
- [12].Gomes HI, Dias-Ferreira C, Ribeiro AB, Sci. Total Environ 444 (2013) 237. [DOI] [PubMed] [Google Scholar]
- [13].Hilal-Mert E, et al. , React. Funct. Polym 73 (2013) 175. [Google Scholar]
- [14].Gollavelli G, Chang C, Ling Y, ACS Sustain. Chem. Eng 1 (2013) 462. [Google Scholar]
- [15].Abd Ali L, Wan Ibrahim W, Sulaiman A, Kamboh M, Sanagi M, Talanta 148 (2016) 191. [DOI] [PubMed] [Google Scholar]
- [16].Abdelwahab MS, Fathallah EM, Chem. Eng. J 223 (2013) 318. [Google Scholar]
- [17].Qu X, Alvarez PJJ, Li Q, Water Res. 47 (2013) 3931. [DOI] [PubMed] [Google Scholar]
- [18].Masciangioli T, Zhang WX, Environ. Sci. Technol 37 (2003) 102A. [DOI] [PubMed] [Google Scholar]
- [19].Qu X, Brame J, Li Q, Alvarez PJJ, Acc. Chem. Res 46 (2013) 834. [DOI] [PubMed] [Google Scholar]
- [20].Zhao Z, Lan J, Li G, Jiang G, Iron-based magnetic nanomaterials on wastewater treatment, in: Reisner DE, Pradeep T (Eds.), Aquananotechnology: Global Prospects, CRC Press, Baton Rouge, 2015, p. 265. [Google Scholar]
- [21].Frimpong R, Dou J, Pechan M, Hilt JZ, Magn J. Magn Mater. 322 (2010) 326. [Google Scholar]
- [22].Zhou Q, Li J, Wang M, Zhao D, Crit. Rev. Environ. Sci. Technol 8 (2016) 7833–826. [Google Scholar]
- [23].Huang YK, Keller AA AA, Water Res. 80 (2015) 159. [DOI] [PubMed] [Google Scholar]
- [24].Huang Y, Keller AA, Sustain. Chem Eng 1 (2013) 731. [Google Scholar]
- [25].Geng Z, Lin Y, Yu X, Shen Q, Ma L, Li Z, Pan N, Wang X, J. Mater. Chem 22 (2012) 3527. [Google Scholar]
- [26].Vestal CR, Zhang ZJ, J. Am. Chem. Soc 124 (2002) 14312. [DOI] [PubMed] [Google Scholar]
- [27].Zhuo Y, Wang S, Ding B, Yang Z, Chem. Eng. J 138 (2008) 578. [Google Scholar]
- [28].Wang Y, Teng X, Wang JS, Yang H, Nano Lett. 3 (2003) 789. [Google Scholar]
- [29].Mittal H, Mishra SB, Carbohydr. Polym 101 (2014) 1255. [DOI] [PubMed] [Google Scholar]
- [30].Choi H, Argarwal S, Al-Abed SR, Environ Sci Tecnol 43 (2009) 488. [DOI] [PubMed] [Google Scholar]
- [31].Meenach SA, Hilt JZ, Anderson KW, Acta Biomater. 6 (2010) 1039. [DOI] [PubMed] [Google Scholar]
- [32].Pellequer JL, Chen SW, Keum YS, Karu AE, Li QX, Roberts VA, J. Mol. Recogn 18 (2005) 282. [DOI] [PubMed] [Google Scholar]
- [33].Lin C, Chinnappan R, Acharya K, Pellequer JL, Jankowiak R, Biophys. Chem 154 (2011) 35. [DOI] [PubMed] [Google Scholar]
- [34].Inui H, et al. , J. Agric. Food Chem 60 (2012) 1605. [DOI] [PubMed] [Google Scholar]
- [35].Gobas F, Zhang X, Chemosphere 25 (1992) 1961. [Google Scholar]
- [36].Sharma B, Gardner KH, Melton J, Hawkins A, Tracey G, Environ. Eng. Sci 26 (2009) 1371. [Google Scholar]
- [37].Palchoudhury S, Lead JR, Environ. Sci. Technol 48 (2014) 14558. [DOI] [PubMed] [Google Scholar]
- [38].Patil V, Dziubla TD, Kalika DS, Polymer 75 (2015) 88. [Google Scholar]
- [39].Patil V V, Gutierrez AM, Sunkara M, Morris AJ, Hilt JZ, Kalika DS, Dziubla TD, J. Nat. Prod 80 (2017) 1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Gupta P, Authimoolam S, Hilt JZ, Dziubla TD, Acta Biomater. 27 (2015) 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Wydra R, Kruse A, Bae Y, Anderson KW, Hilt JZ, Mater. Sci. Eng. C 33 (2013) 4660. [DOI] [PubMed] [Google Scholar]
- [42].Andrade AL, Souza DM, Pereira MC, Fabris JD, Domingues RZ, Quim. Nova 33 (2012) 524. [Google Scholar]
- [43].Langmuir I, AIChE J 38 (1916) 121. [Google Scholar]
- [46].Hauser A, Mathias R, Anderson KW, Hilt JZ, Mater. Chem. Phys 160 (2015) 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Wang M, Liu P, Wang Y, Zhou D, Ma C, Zhang D, Zhan J, J. Colloid Interface Sci 447 (2015) 1. [DOI] [PubMed] [Google Scholar]
- [48].Sharma G, Naushad M, Kumar A, Rana S, Sharma S, Bhatnagar A, Stadler FJ, Ghfar AA, Khan MR, Process Saff Environ 109 (2017) 301. [Google Scholar]
- [49].Liu L, Fokkink R, Koelmans AA, Environ. Toxicol. Chem 35 (2015) 1650. [DOI] [PubMed] [Google Scholar]
- [50].Jonker MTO, Keolmans AA, Environ. Sci. Technol 26 (2002) 3725. [DOI] [PubMed] [Google Scholar]
- [51].Kleineidam S, Schüth C, Grathwohl P P, Environ. Sci. Technol 36 (2002) 304. [DOI] [PubMed] [Google Scholar]
- [52].Koelmans AA, Meulman B, Meijer T, Jonker MTO MTO, Environ. Sci. Technol 43 (2009) 736. [DOI] [PubMed] [Google Scholar]
- [53].Phatthanakittiphong T, Tae Seo G. Nanomater. 6 (2016) 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.