Abstract
Microbial communities can perform a variety of behaviors that are useful in both therapeutic and industrial settings. Engineered communities that differ in composition from naturally-occurring communities offer a unique opportunity for improving upon existing community functions and expanding the range of microbial community applications. This has prompted recent advances in various community design approaches including artificial selection procedures, reduction from existing communities, combinatorial evaluation of potential microbial combinations, and model-based in silico community optimization. Computational methods in particular offer a likely avenue towards improved synthetic community development going forward. This review introduces each class of design approach and surveys their recent applications and notable innovations, closing with a discussion of existing design challenges and potential opportunities for advancement.
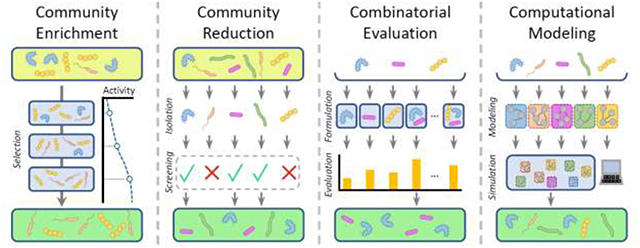
Graphical abstract
Introduction
Microbial communities have long been recognized for the important impact they have on human health, agriculture, and industry. In the context of health, specifically, recent studies have highlighted the key role that the human microbiome – the ensemble of microorganisms that live in and on the human body – plays in human physiology, immunity, development, and nutrition. For example, microbially-produced butyrate contributes to colonic health via its function as an important energy source [1] and anti-inflammatory agent [2]. Proper immune system development also relies on the gut microbiome, which modulates aspects such as lymphoid structure development and T cell differentiation [3,4]. Beyond human health, microbial activity is important to a range of industrial applications including microbially-mediated denitrification in wastewater treatment [5,6] and biofuel production [7,8]. These microbial influences also extend to agriculture, where bacteria support plant access to nitrogen [9,10] and phosphorus [11].
These myriad important functions suggest that microbial communities can serve as a prime target for medical, agricultural, and industrial advancement and have sparked recent interest in developing microbiome-based biotechnologies and therapeutics [12,13]. Specifically, microbiome engineering – the manipulation of naturally occurring microbial communities or the construction of synthetic communities to produce a specific function – is a promising tool for improving and innovating upon various clinical and industrial applications. Such attempts to engineer a given community can be done by various approaches. One simple microbiome engineering technique is the modulation of environmental conditions to effect changes in community function. This approach is frequently applied to optimize bioreactor communities, which are used for the production and degradation of various compounds. Indeed, previous studies have shown that bioreactor operators can improve community function by changing substrate composition [14–16], aeration [17], pH [14,18], and temperature [19]. Another simple approach is to modify a community by adding beneficial species or removing undesirable ones. Antibiotics are perhaps the most prevalent example of this approach in therapeutics, acting as a tool for removing pathogenic species. Probiotics, on the other hand, are an example of an additive tool, aiming to improve gut community function by introducing beneficial species [13,20,21].
Importantly, however, for microbiome engineering to realize its full potential, more sophisticated techniques must be employed to provide researchers with greater control when engineering microbial communities. This is especially important for the construction of synthetic communities that consist of a human-developed mixture of species. In the context of microbiome-based therapy, such synthetic communities could enable more precise modulation of the microbiome and bypass the negative side effects of antibiotics usage [22,23] or potential inconsistency of probiotic engraftment [24,25]. Synthetic communities can also be useful for industrial applications, where the novel coupling of microbial metabolisms can lead to the improved production or degradation of economically important compounds [26]. More generally, synthetic communities provide a more flexible and powerful engineering approach, allowing researchers to engineer the community as a whole, rather than being limited to the perturbation of an existing community. However, one key challenge in developing such synthetic communities is the identification of novel species compositions that optimize, or at least improve upon, desired functions. This task, which can be thought of as ‘rational design’ for community engineering, is far from trivial and various techniques have been developed and applied to address this need.
In this review, we survey a variety of approaches for designing synthetic compositions with targeted functions, highlighting recent methodological innovations and applications. Importantly, we focus on methods for designing synthetic compositions (i.e., new mixtures of existing microbial species), rather than the use of genetically engineered microbes in synthetic compositions, which has been reviewed elsewhere [27–30]. We begin with approaches that rely on selective pressures and adaptation of community composition to reach synthetic compositions with optimized functions. Next, we cover methods that use microbial isolates from existing communities to formulate reduced, well-defined compositions that recapitulate the desired functions of the source communities. We then describe techniques that evaluate possible synthetic compositions in a combinatorial manner to identify desirable compositions. Finally, we describe the recently expanding range of computational tools that identify candidate compositions predicted to optimize a desired function. We conclude by discussing the challenges and potential opportunities that are still present in the field of synthetic community composition design. Since therapeutic community design studies are currently limited, we illustrate the application of certain design approaches using examples from industrial settings.
Community enrichment towards synthetic compositions
Naturally occurring microbial communities can carry out an amazing variety of functions, many of which could be harnessed towards clinical, industrial, and environmental applications. For example, the human gut microbiome can perform multiple metabolic processes that are crucial for the host, including harvesting energy from the diet [31], synthesizing important vitamins [32], and resisting pathogen colonization via competitive exclusion [33]. Indeed the transplantation of fecal microbiota from healthy donors has proven effective at treating several gastrointestinal disorders [13,34–36]. Similarly, different soil communities can degrade various pollutants, such as diesel fuel [37] and polycyclic aromatic hydrocarbons [38], or prevent non-biodegradable pollutants, such as uranium [39], from contaminating water supplies by catalyzing their conversion to insoluble forms. However, the efficiency at which naturally occurring communities perform these functions may not be sufficient for industrial settings, necessitating an optimization procedure that can build upon such communities and ultimately produce synthetic communities with enhanced capabilities. A common approach to achieve this goal is through enrichment – a community design methodology that aims to reach a community composition with optimal desired capabilities by subjecting an existing community to environmental conditions that favor species that can perform the target function. To date, enrichment has primarily been used for biotechnological application, including microbial fuel cells (MFCs), biopolymer production, and biohydrogen production, which will be described below.
MFCs have become a prime target of microbial community engineering due to both the promise of efficient microbially-mediated electricity generation and the wide range of substrates that they can utilize. This biotechnology was originally inspired by marine sediment communities that reduce various elements for energy in a manner that can be exploited to generate electricity [40–43]. Recent MFC applications can now utilize a wide range of substrates (depending on the specific microbial community employed) including glucose [44,45], acetate [44], lactate [46], cellulose [47,48], and ammonium [49]. Indeed, MFCs can even consume various industrial waste products, enabling the coupling of electricity generation with waste degradation [50–53]. Though naturally occurring communities can already achieve these tasks, MFCs seeded with such communities often undergo compositional changes and exhibit gradual improvement in efficiency over time as the community adapts to operating in the MFC environment. Such changes were shown to include enrichment for species potentially related to current generation [54] and degradation of the supplied substrate [55], and the observed changes in community composition during extended operation of an MFC were demonstrated to be linked to concurrent increases in MFC efficiency [56].
Notably, while MFC communities experience inherent and appropriate selective pressures, other biotechnologies may require the application of artificial selection procedures to optimize community function. For example, to increase the yield and efficiency of microbial communities grown and harvested for biopolymers used in biodegradable plastics, researchers have applied an artificial feast-famine cycle [57,58]. This cycle selects for communities that store energy (in the form of biopolymers) more efficiently during the feast phase so that energy is available during the famine phase. This procedure can be further enhanced by introducing phosphate limitation, which can ensure that biopolymer production is advantageous while also reducing the growth of sub-communities that do not contribute to production [59]. Recent work has also demonstrated that photosynthetic communities can be enriched for biopolymer production without a famine step when oxygen is limited [60].
Artificial selection procedures have also improved microbial community hydrogen production, though in this context, artificial section is often applied as a pretreatment rather than as part of post-enrichment operating conditions. Such pretreatments include heat shock, acidic or basic incubation, freeze drying, and chloroform treatment [61]. Each of these pretreatments aims to enrich for hydrogen-producing species in the original community while excluding hydrogen consumers. Interestingly, though, the efficacy of different pretreatments is inconsistent across different studies, likely due to difference in the set of species present in the initial community [62]. Consequently, the discovery of new promising hydrogen-producing communities necessitates the re-evaluation of these enrichment procedures to identify the best pretreatment [63].
Community reduction from existing compositions
Some microbial community applications may impose specific restrictions on the species that can be present in the synthetic community. For example, microbiome therapeutics must meet various regulatory guidelines [64], and be devoid of pathogenic species so as to avoid inadvertently infecting the recipient [12]. However, it may not be possible to fully satisfy such restrictions using enrichment approaches due to the relatively broad and unspecified nature of environmental selection. For example, applying environmental conditions that inhibit the growth of pathogens may simultaneously negatively impact the growth of desirable species. This challenge can be addressed by a complementary design approach, referred to in this review as community reduction, wherein individual members of some initial community are isolated and characterized to rationally determine whether they should be used in the synthetic community. While some community members may be lost during the isolation step [65], this approach provides better control over community composition and enables a more principled selection of desirable species and the explicit exclusion of undesirable ones.
This design paradigm has been used, for example, to reconstruct synthetic communities for treating Clostridium difficile infection (CDI). CDI is a gastrointestinal infection where C. difficile, a spore-forming, antibiotic-resistant enteric pathogen, dominates the gut microbiome, causing inflammation and diarrhea [66]. Previous clinical studies have demonstrated that CDI can be effectively treated with fecal microbiota transplantation (FMT) from a healthy donor [35]. This makes CDI a prime target for synthetic FMTs that could potentially recapitulate the same beneficial effects using a simplified, well-defined community composition. Indeed, an early study used a mixture of ten previously identified and isolated intestinal species to formulate a reduced synthetic CDI treatment composition [67]. In this study, all five patients treated with the synthetic composition exhibited marked improvement, similar to that observed in a patient treated with a donor stool sample. Surprisingly, one of these five patients was previously treated unsuccessfully with a donor stool transplant, suggesting that reduced synthetic FMT compositions may not be strictly inferior in efficacy to traditional FMTs. In a more recent study, a synthetic FMT composition was designed by isolating as many individual species as possible from a single donor’s stool [68]. These isolates were screened for pathogens, and the remaining nonpathogenic isolates were mixed to form a synthetic community. Both patients treated with this synthetic community composition responded well to treatment, and longitudinal sampling revealed notable engraftment of the species in the synthetic composition, though this declined over time.
The community reduction approach has also shown success when used for non-CDI therapeutic applications. For example, rather than targeting C. difficile, Caballero et al. investigated the role that gut species play in resisting Vancomycin-resistant Enterococcus (VRE) colonization in mice [69]. As part of this larger study, the authors isolated ampicillin-resistant strains from mouse stool and examined which strains could confer VRE resistance. From these experiments, they identified a four-strain synthetic composition that both resisted VRE colonization and ameliorated pre-established VRE colonization. In another example, Atarashi et al. set out to design a synthetic composition that would induce Treg cells in the mouse colon [70]. The authors isolated species from a human donor’s chloroform-treated stool and found that a synthetic community composed of 17 isolates induced Treg cells in germ-free mice to a similar extent as the original chloroform-treated stool. The same group, as part of a larger study of intestinal Th17 cell induction, also used community reduction to successfully identify a synthetic composition of 20 human gut strains with notable Th17 cell induction in mice [71]. These examples illustrate the potential of community reduction to recapitulate various important gut microbiome functions, making it a promising tool for future microbiome therapeutics. One important caveat to note, however, is that community reduction inherently cannot design synthetic compositions with novel functions, thus restricting its wider applicability.
Combinatorial evaluation of potential compositions
One of the unique benefits of synthetic communities is that they can include combinations of species that never co-occur in naturally occurring communities, potentially facilitating a wider range of metabolic capacities. Such synthetic communities may therefore be able to perform certain functions better than existing communities (or communities obtained via enrichment or reduction) or even perform entirely new functions. This is of particular interest for industrial applications such as the production of biofuel and other biological compounds [26]. Indeed, exploration of novel metabolic coupling in synthetic communities has already proved successful, demonstrating potential applications for the production of various resources including hydrogen [72], acetic acid [73–75], and lactic acid [76,77], as well as the degradation of undesirable substances including polycyclic aromatic hydrocarbons [78] and cellulose [79,80]. To go beyond a simple trial and error exploration approach for identifying such beneficial combinations, researchers can employ a more comprehensive process, referred to in this review as combinatorial evaluation, the systematic enumeration, construction, and evaluation of possible combinations of a set of species to identify the best-performing composition. The set of species used could, for example, consist of candidate species that are believed to contribute to the desired function.
When the number of species to consider is small, combinatorial evaluation can be performed in its ideal form, constructing and assessing all possible combinations of the species of interest. For example, to optimize the biodegradation of dyes in textile wastewater, researchers isolated three species from a textile wastewater plant and evaluated the degradation capabilities of all combinations of these three species [81]. In fact, due to the relatively small number of species considered, they were also able to evaluate additional compositions that varied in the relative abundances of each species.
Importantly, however, as the number of candidate species grows, the number of potential compositions grows exponentially, quickly rendering the evaluation of all possible combinations impossible. This setting calls for techniques that can drastically reduce the number of evaluated compositions. One such technique is fractional factorial design (FFD). In general terms, FFD aims to estimate the effects of, and potentially the interactions between, particular components of a system on a specified output [82]. These effect and interaction measurements then provide a basis for mathematically identifying an optimal parameterization of these components. In the context of microbial community design, FFD reduces the required number of evaluated compositions by carefully selecting a subset of potential community compositions that can isolate specific species effects or interactions of interest. One important caveat is that FFD achieves this reduction in evaluated compositions by assuming negligible effects of higher-order interactions. However, if later evidence suggests that one or more higher-order interactions have important contributions, a technique called foldover design can be applied to efficiently determine those specific interaction effects based on the findings of the original FFD experiment [82].
Microbial community function optimization via FFD has historically focused on factors external to the community. For example, various studies have used FFD to examine the impact of environmental factors such as substrate composition [15,37,83–85], pH [83,84,86], temperature [83], and heavy metal presence [86] on specific community functions. More recent efforts, however, have used FFD, or FFD-like techniques, to investigate the potential of individual species effect estimation. A recent study, for example, has used random gut community subsets to estimate individual microbial contributions to host phenotypes [87]. Though these compositions were randomly selected (rather than specifically constructed to most efficiently separate individual contributions), the results of this study suggest that FFD could be applied in a similar manner for synthetic composition optimization by treating species as the components of interest. Indeed, one group has already applied FFD to develop wastewater treatment communities [88,89]. In this pair of studies, the authors used FFD to estimate the contributions of both individual species and interspecies interactions to total organic carbon (TOC) degradation [88] and substrate utilization rate [89]. They then employed this information to develop synthetic compositions with improved biodegradation capacities. Interestingly, both studies found that the optimized three- and four-strain communities performed better than a baseline mixture of all six strains evaluated, demonstrating the utility of FFD in synthetic community design.
Another technique for efficiently evaluating potential compositions is the definition of microbial consortia that will be treated as single units when enumerating possible species combinations (i.e., each combination will either include or exclude all species in a given consortium). This technique is particularly useful when a microbial consortium has previously demonstrated a desirable emergent function. For example, one group observed that a consortium of marine species, named the NPMC, could efficiently fix CO2 [90]. They later treated this consortium as a single candidate community member when using a combinatorial evaluation approach to develop a synthetic community for CO2 fixation [91]. Importantly, in addition to reducing the pool of available species to six candidate community members (one of which was the NPMC), this approach also allowed the researchers to include species that could not be isolated from the NPMC in the final community. In a separate study, the same group designed a synthetic community for lignocelluloytic enzyme activity by considering a synthetic consortium previously designed for cellulolytic activity [80] alongside several fungal strains [92]. These studies highlight the benefits of this approach, allowing researchers to evaluate designed communities with higher complexity without drastically increasing the number of evaluated compositions.
Computational model-based design of synthetic compositions
The design paradigms described above rely on various approaches for characterizing and assessing candidate community compositions, with techniques like FFD and consortium inclusion allowing researchers to reduce the set of compositions ultimately evaluated. Importantly, however, such approaches may still entail evaluating many compositions that a priori might be expected to perform the desired function poorly based on existing knowledge of microbial ecology, genomics, and metabolism. Indeed, databases such as NCBI [93] and IMG [94] provide access to an ever increasing number of sequenced microbial genomes, which when coupled with various gene annotation databases, such as KEGG [95] and MetaCyc [96], can be used to infer the functional capacities of individual microbial species. Design methodologies that could harness such information to pinpoint community compositions that are likely to successfully and efficiently perform the desired function might dramatically reduce the time and labor needed for experimental evaluation. As a simple example, a collection of previously sequenced and genomically-annotated microbial species could be searched to identify species whose genomes confer the capacity to perform a certain function, even when these species have never been experimentally tested for that function in the lab. In this review, however, we will focus on more sophisticated computational methods for designing synthetic community compositions, primarily highlighting methods that aim to model community-level metabolism and to identify synthetic compositions that are predicted to perform the desired function well. We refer to this approach as computational model-based design.
Microbial community metabolism can be modeled with varying levels of complexity and using a variety of modeling frameworks [97,98]. A relatively simple form of metabolic modeling, often referred to as network-based or topology-based modeling, represents each species as a directed network where nodes denote metabolites and edges connect substrates to products, reflecting the set of metabolic reactions the modeled species can catalyze. With this framework, community metabolism can be modeled as a collection of such networks, where outputs from one network can be used as input for another. A recently introduced design algorithm, termed CoMiDA [99], has utilized this modeling framework to identify a minimal set of microbial species that collectively provide the enzymatic capacity required to synthesize a set of desired products from a predefined set of available substrates. To achieve this, the CoMiDA algorithm integrates a graph-theoretic representation of network flow with the set cover problem to consider all possible metabolic paths from substrates to products and to detect the minimal set of species that can catalyze these reactions. The obtained solution can provide a starting point for further synthetic community experimentation and development. Another design algorithm, termed MultiPus [100], utilizes a similar framework but aims to minimize the number of reactions and inter-microbial transfers, rather than the number of species.
While network-based models are easy to construct and analyze, they generally only account for the potential metabolic capacity of each species, rather than for the way each species will behave in a given environment. Accordingly, communities designed by CoMiDA or MultiPus are indeed guaranteed to have the metabolic potential to carry out the desired function, but may not actually perform this function in reality. Instead, the accurate estimation of microbiome behavior requires a detailed model of microbial metabolism, one that can predict the specific activity of each species, the flux through each reaction, the uptake and secretion rate of environmentally available metabolites, and the growth rate of each species in a given environment. One such modeling framework utilizes constraint-based models and flux balance analysis (FBA) [101,102]. Such models can predict the steady state metabolic activity of a given species by identifying a set of metabolic fluxes that maximize microbial growth while adhering to a set of thermodynamic constraints [103,104]. Recent years have witnessed an explosion of studies that aim to extend constraints-based modeling from single species models to community models that can predict community-level metabolism, species interactions, and community dynamics [105–108]. Building on these effort, a recent design method, termed FLYCOP [109], has utilized a previously introduced community modeling framework to evaluate synthetic composition function in silico. Importantly, the underlying modeling framework accounts for community dynamics and spatial community organization, as well as metabolism-mediated species-interactions via changes to the shared environment [107]. Using this framework, FLYCOP explores potential synthetic compositions using a stochastic search procedure and identifies an optimized composition. Interestingly, FLYCOP is not restricted to optimizing metabolic activity, and can also consider a community’s growth over time. This allowed the authors to identify an initial synthetic composition of four cross-feeding strains that optimized community stability. This ability to optimize community stability could be extremely important for therapeutics, where treatment may require the community to function for a prolonged period.
Another modeling approach that can be useful for community design efforts, especially when stability is an important consideration, aims to model community ecological dynamics rather than community metabolism. These models capture how the abundance of each community member impacts the abundances of others over time [110,111]. Such models can be especially useful when interactions between species may not be mediated via metabolism or when detailed metabolic models are not available. In one example, a group optimized a synthetic composition for both a non-metabolic function and community stability through a combination of ecological modeling and experimental characterization of individual microbial activity [112]. In this case, the authors aimed to develop a community for Treg induction in the mouse colon that would persist over time. To achieve this, the authors first created a model of community induction effectiveness for a set of Clostridia strains using data on Treg induction contributions. They then simulated the community’s ecological dynamics using a previously published ecological model for those strains [110], and used their induction model to estimate Treg induction over time. This enabled them to predict each potential composition’s inductive effect and stability simultaneously. Such integration of different modeling framework may be a promising avenue for future expansion and improvement of computational design capabilities.
Synthetic composition design challenges and opportunities
The previous sections have surveyed recent applications of, and advances in, synthetic composition design. Importantly, however, there are still many daunting challenges, as well as exciting opportunities, for future development in this field. Clearly, each design approach described above has its own strengths and weaknesses that make it more suitable for certain applications and less appropriate for others and that impose a specific set of challenges and opportunities (Table 1). Enrichment can often work without detailed knowledge of individual species, instead relying on understanding the desired biological process in order to create a selection procedure. However, each novel application may require a completely new selection procedure, which could be challenging to develop [113]. Additionally, the time required for optimization can be extensive, with some experiments showing continued improvement over the course of months [58,114]. The community reduction and combinatorial evaluation approaches similarly avoid the need for detailed mechanistic data while also incorporating greater control over the specific species used. Unfortunately, due to our inability to culture a large fraction of microbial species [65], community reduction can suffer from a failure to isolate a set of species sufficient to recapitulate the original community’s function. Additionally, community reduction does not inherently optimize a synthetic composition’s function, but rather only identifies a well-defined synthetic composition with the desired function. Combinatorial evaluation, on the other hand, can suffer from tractability issues regarding the number of compositions to evaluate, as described above. Finally, computational model-based design can drastically decrease the time and labor needed to identify optimal, or near-optimal compositions, but it requires detailed and thorough mechanistic and/or ecological data about the species being considered. Understanding these differences between approaches, as well as considering the available time, labor, and knowledge resources, will help future designers select the approach best suited for their specific application.
Table 1.
Summary of synthetic community design methods
| Description | Requirements | Benefits | Downsides | Example usage | Design approach |
|---|---|---|---|---|---|
| Environmental conditions are modified to promote the growth of species that perform a desired function |
|
|
|
Fradinho et al., 2016 [60] Korkakaki et al., 2017 [59] Kumari et al., 2017 [63] |
Enrichment |
| Species are isolated from an existing community, screened to keep desirable species and/or exclude undesirable species, and then reconstituted into a simplified community that recapitulates the source community’s function |
|
|
|
Petrof et al., 2013 [68] Atarashi et al., 2015 [71] Caballero et al, 2017 [69] |
Community reduction |
| All possible combinations of a set of candidate species are evaluated for their performance of a desired function and the best-performing composition is selected |
|
|
|
Hu et al., 2016 [91] Hu et al., 2017 [92] | Combinatorial evaluation |
| Mechanistic models of microbial functional capacities and activities and/or ecological models of community dynamics are used to evaluate potential community compositions in silico, identifying one or more optimized compositions for further experimental validation |
|
|
|
Eng and Borenstein, 2016 [99] Julien-Laferriere et al., 2016 [100] Garcia-Jimenez et al., 2018 [109] Stein et al., 2018 [112] |
Computational model-based design |
While each design technique offers unique benefits, the power and flexibility of computational model-based design, combined with the recent expansion of available genomic, metabolic, and other mechanistic data, render such computational design efforts an especially promising route toward rapid advancement in synthetic community design. For example, methods that can concurrently optimize multiple community functions could enable synthetic therapeutic communities to simultaneously treat diverse health concerns, such as metabolic deficiencies and pathogenic infections, while also ensuring community stability. There are however substantial obstacles that must be overcome for model-based design to reach its full potential. One key challenge is the inability of many currently modeling frameworks to directly incorporate knowledge of non-metabolic microbial interactions into models of community function, which recent evidence suggests can be important factors in shaping the human gut microbiome [115]. Ecological models that are based on observed community dynamics may only partially capture the outcomes of such interactions, and it is likely that the nuanced effects of key interaction mediators are ignored [116]. The potentially important functional impact of higher-order interactions (i.e., interactions involving multiple species in the community) [117–119] poses another challenge for computational methods, specifically when such methods evaluate only a subset of possible compositions. This calls for more sophisticated methods that efficiently search the space of potential compositions while adequately accounting for higher-order interactions, which would be especially advantageous.
Perhaps the most promising avenue for advancement in synthetic design might entail the integration of multiple design approaches such that the weaknesses of one approach are addressed by the incorporation of a complementary strategy. Indeed, there is already evidence that communities designed using one method can be further improved via orthogonal design techniques. The computationally-designed synthetic Treg induction community described above [112] was developed from a Treg induction community originally designed using community reduction [70]. In this case, community reduction identified a set of culturable species that could form a synthetic Treg induction community and computational design optimized the composition to improve its function. Pairing enrichment with combinatorial evaluation or model-based design could also offer potentially fruitful composite approaches. Specifically, the enrichment procedure could begin with compositions identified and pre-optimized by other design techniques, rather than environmentally sampled or randomly constructed initial communities. Such method integration could potentially reduce the time required to select for an optimal composition since the initial community is hopefully closer in composition to the final community that enrichment would achieve. Additionally, this could help augment combinatorial evaluation or computational design, which may result in sub-optimal communities due to insufficient coverage of evaluated compositions or insufficient mechanistic and ecological knowledge respectively. Such innovative combinations of design approaches could enhance or enable the development of synthetic compositions that may have previously been challenging due to the various limitations of each individual approach.
Conclusions
In this review, we have surveyed various methods for designing synthetic microbial communities, highlighting their utility in formulating and optimizing communities for a wide variety of applications. These methods range from experimentally driven techniques, including enrichment, community reduction, and combinatorial evaluation, to computational approaches that employ mechanistic models of microbial function and ecological models of community dynamics. We have described various successful applications of these methods to both industrial and therapeutic synthetic community design, noting interesting and important observations made during the design process. Perhaps the most important of these observations is that optimized communities are often smaller and less complex than naturally occurring communities. Indeed, as in some cases mentioned above, simpler synthetic communities with fewer strains have achieved better performance than their more complex counterparts. This suggests that design methods will continue to play an important role in identifying the particular subcommunities that best achieve specific functions. Given this, we believe that continued advancement in all classes of design approaches will greatly expand the uses of, and improve the efficacy of, synthetic microbial communities.
Highlights.
Synthetic microbial communities can be engineered for targeted functions
Artificial selection has been employed to improve existing communities
Several methods have been used to identify new compositions with desired functions
Recent progress in computational model-based design has enabled novel design tools
Computational and integrative approaches are promising avenues for advancement
Acknowledgements
We thank the members of the Borenstein Lab for helpful feedback. This work was supported by the NIH New Innovator Award DP2AT00780201 and by the NIH grant 1R01GM124312–01 to EB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Donohoe DR, Garge N, Zhang X, Sun W, O’Connell TM, Bunger MK, Bultman SJ: The Microbiome and Butyrate Regulate Energy Metabolism and Autophagy in the Mammalian Colon. Cell Metab 2011, 13:517–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Canani RB, Costanzo MDi, Leone L, Pedata M, Meli R, Calignano A: Potential beneficial effects of butyrate in intestinal and extraintestinal diseases. World J Gastroenterol 2011, 17:1519–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hooper LV, Littman DR, Macpherson AJ: Interactions between the microbiota and the immune system. Science 2012, 336:1268–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kabat AM, Srinivasan N, Maloy KJ: Modulation of immune development and function by intestinal microbiota. Trends Immunol 2014, 35:507–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pitt WW, Hancher CW, Patton BD: Biological reduction of nitrates in wastewaters from nuclear processing using a fluidized-bed bioreactor. Nucl Chem Waste Manag 1981, 2:57–70. [Google Scholar]
- 6.Daims H, Nielsen JL, Nielsen PH, Schleifer KH, Wagner M: In situ characterization of Nitrospira-like nitrite-oxidizing bacteria active in wastewater treatment plants. Appl Environ Microbiol 2001, 67:5273–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abate C, Callieri D, Rodríguez E, Garro O: Ethanol production by a mixed culture of flocculent strains of Zymomonas mobilis and Saccharomyces sp. Appl Microbiol Biotechnol 1996, 45:580–583. [DOI] [PubMed] [Google Scholar]
- 8.Mamma D, Koullas D, Fountoukidis G, Kekos D, Macris BJ, Koukios E: Bioethanol from sweet sorghum: Simultaneous saccharification and fermentation of carbohydrates by a mixed microbial culture. Process Biochem 1996, 31:377–381. [Google Scholar]
- 9.Franche C, Lindström K, Elmerich C: Nitrogen-fixing bacteria associated with leguminous and non-leguminous plants. Plant Soil 2009, 321:35–59. [Google Scholar]
- 10.Rovira AD: Rhizosphere research - 85 years of progress and frustration In The Rhizosphere and Plant Growth. Edited by Keister DL, Cregan PB. Springer; Netherlands; 1991:3–13. [Google Scholar]
- 11.Afzal A: Rhizobium and Phosphate Solubilizing Bacteria Improve the Yield and Phosphorus Uptake in Wheat (Triticum aestivum). Int J Agri Biol 2008, 10:85–88. [Google Scholar]
- 12.••.Mimee M, Citorik RJ, Lu TK: Microbiome therapeutics — Advances and challenges. Adv Drug Deliv Rev 2016, 105:44–54.This review details known mechanisms for gut microbiome effects on the host and describes previous work in microbiome therapeutics development, including probiotics and microbiota transplants. This information provides a valuable background for understanding how microbiome therapeutics can improve host health, what approaches have been successful, and where the field is heading.
- 13.Marchesi JR, Adams DH, Fava F, Hermes GDA, Hirschfield GM, Hold G, Quraishi MN, Kinross J, Smidt H, Tuohy KM, et al. : The gut microbiota and host health: a new clinical frontier. Gut 2016, 65:330–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cui M, Yuan Z, Zhi X, Shen J: Optimization of biohydrogen production from beer lees using anaerobic mixed bacteria. Int J Hydrogen Energy 2009, 34:7971–7978. [Google Scholar]
- 15.Pholchan MK, Baptista J de C, Davenport RJ, Curtis TP: Systematic study of the effect of operating variables on reactor performance and microbial diversity in laboratory-scale activated sludge reactors. Water Res 2010, 44:1341–1352. [DOI] [PubMed] [Google Scholar]
- 16.Bassin JP, Kleerebezem R, Rosado AS, van Loosdrecht MCM, Dezotti M: Effect of Different Operational Conditions on Biofilm Development, Nitrification, and Nitrifying Microbial Population in Moving-Bed Biofilm Reactors. Environ Sci Technol 2012, 46:1546–1555. [DOI] [PubMed] [Google Scholar]
- 17.Simova ED, Frengova GI, Beshkova DM: Effect of Aeration on the Production of Carotenoid Pigments by Rhodotorula rubra-lactobacillus casei Subsp. casei Co-Cultures in Whey Ultrafiltrate. Z Naturforsch 2003, 58:225–229. [DOI] [PubMed] [Google Scholar]
- 18.Fang HHP, Liu H: Effect of pH on hydrogen production from glucose by a mixed culture. Bioresour Technol 2002, 82:87–93. [DOI] [PubMed] [Google Scholar]
- 19.Zoetemeyer RJ, Arnoldy P, Cohen A, Boelhouwer C: Influence of temperature on the anaerobic acidification of glucose in a mixed culture forming part of a two-stage digestion process. Water Res 1982, 16:313–321. [Google Scholar]
- 20.Walsh CJ, Guinane CM, O’Toole PW, Cotter PD: Beneficial modulation of the gut microbiota. FEBS Lett 2014, 588:4120–4130. [DOI] [PubMed] [Google Scholar]
- 21.Derrien M, van Hylckama Vlieg JET: Fate, activity, and impact of ingested bacteria within the human gut microbiota. Trends Microbiol 2015, 23:354–66. [DOI] [PubMed] [Google Scholar]
- 22.Bartlett JG: Narrative Review: The New Epidemic of Clostridium difficile –Associated Enteric Disease. Ann Intern Med 2006, 145:758. [DOI] [PubMed] [Google Scholar]
- 23.Theriot CM, Koenigsknecht MJ, Carlson PE, Hatton GE, Nelson AM, Li B, Huffnagle GB, Z Li J, Young VB: Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat Commun 2014, 5:3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zmora N, Zilberman-Schapira G, Suez J, Mor U, Dori-Bachash M, Bashiardes S, Kotler E, Zur M, Regev-Lehavi D, Brik RB-Z, et al. : Personalized Gut Mucosal Colonization Resistance to Empiric Probiotics Is Associated with Unique Host and Microbiome Features. Cell 2018, 174:1388–1405.e21. [DOI] [PubMed] [Google Scholar]
- 25.Maldonado-Gómez MX, Martínez I, Bottacini F, O’Callaghan A, Ventura M, van Sinderen D, Hillmann B, Vangay P, Knights D, Hutkins RW, et al. : Stable Engraftment of Bifidobacterium longum AH1206 in the Human Gut Depends on Individualized Features of the Resident Microbiome. Cell Host Microbe 2016, 20:515–526. [DOI] [PubMed] [Google Scholar]
- 26.Bader J, Mast-Gerlach E, Popović MK, Bajpai R, Stahl U: Relevance of microbial coculture fermentations in biotechnology. J Appl Microbiol 2010, 109:371–87. [DOI] [PubMed] [Google Scholar]
- 27.Mee MT, Wang HH, Lo TM, Nguyen HX, Ling H, Leong SS, Poh CL, Chang MW, Knip M, Hyoty H, et al. : Engineering ecosystems and synthetic ecologies. Mol Biosyst 2012, 8:2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pandhal J, Noirel J: Synthetic microbial ecosystems for biotechnology. Biotechnol Lett 2014, 36:1141–1151. [DOI] [PubMed] [Google Scholar]
- 29.•.Johns NI, Blazejewski T, Gomes AL, Wang HH: Principles for designing synthetic microbial communities. Curr Opin Microbiol 2016, 31:146–153.This review discusses the various tools and strategies available for designing synthetic communities via genome engineering, describing techniques for creating beneficial metabolic interactions between species, the use of FBA models to evaluate potential genomic manipulations, and possible approaches to ensure community stability through engineered interactions. This serves as an informative introduction to an alternative suite of methods for synthetic community design that could not be covered in the scope of this review.
- 30.Zerfaß C, Chen J, Soyer OS: Engineering microbial communities using thermodynamic principles and electrical interfaces. Curr Opin Biotechnol 2018, 50:121–127. [DOI] [PubMed] [Google Scholar]
- 31.Backhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI: The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci 2004, 101:15718–15723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guarner F, Malagelada J-R: Gut flora in health and disease. Lancet 2003, 361:512–519. [DOI] [PubMed] [Google Scholar]
- 33.Buffie CG, Pamer EG: Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol 2013, 13:790–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kelly CR, Kahn S, Kashyap P, Laine L, Rubin D, Atreja A, Moore T, Wu G: Update on Fecal Microbiota Transplantation 2015: Indications, Methodologies, Mechanisms, and Outlook. Gastroenterology 2015, 149:223–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rossen NG, MacDonald JK, de Vries EM, D’Haens GR, de Vos WM, Zoetendal EG, Ponsioen CY: Fecal microbiota transplantation as novel therapy in gastroenterology: A systematic review. World J Gastroenterol 2015, 21:5359–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aroniadis OC, Brandt LJ: Fecal microbiota transplantation: past, present and future. Curr Opin Gastroenterol 2013, 29:79–84. [DOI] [PubMed] [Google Scholar]
- 37.Molina-Barahona L, Rodríguez-Vázquez R, Hernández-Velasco M, Vega-Jarquín C, Zapata-Pérez O, Mendoza-Cantú A, Albores A: Diesel removal from contaminated soils by biostimulation and supplementation with crop residues. Appl Soil Ecol 2004, 27:165–175. [Google Scholar]
- 38.Frutos FJG, Escolano O, García S, Babín M, Fernández MD: Bioventing remediation and ecotoxicity evaluation of phenanthrene-contaminated soil. J Hazard Mater 2010, 183:806–813. [DOI] [PubMed] [Google Scholar]
- 39.Williams KH, Long PE, Davis JA, Wilkins MJ, N’Guessan AL, Steefel CI, Yang L, Newcomer D, Spane FA, Kerkhof LJ, et al. : Acetate Availability and its Influence on Sustainable Bioremediation of Uranium-Contaminated Groundwater. Geomicrobiol J 2011, 28:519–539. [Google Scholar]
- 40.Reimers CE, Tender LM, Fertig S, Wang W: Harvesting Energy from the Marine Sediment−Water Interface. Environ Sci Technol 2001, 35:192–195. [DOI] [PubMed] [Google Scholar]
- 41.Du Z, Li H, Gu T: A state of the art review on microbial fuel cells: A promising technology for wastewater treatment and bioenergy. Biotechnol Adv 2007, 25:464–482. [DOI] [PubMed] [Google Scholar]
- 42.Lovley DR: Microbial fuel cells: novel microbial physiologies and engineering approaches. Curr Opin Biotechnol 2006, 17:327–332. [DOI] [PubMed] [Google Scholar]
- 43.Logan BE, Hamelers B, Rozendal R, Shroder U, Keller J, Freguia S, Aelterman P, Verstraete W, Rabaey K: Microbial Fuel Cells: Methodology and Technology. Environ Sci Technol 2006, 40:5181–5192. [DOI] [PubMed] [Google Scholar]
- 44.Zhang E, Xu W, Diao G, Shuang C: Electricity generation from acetate and glucose by sedimentary bacterium attached to electrode in microbial-anode fuel cells. J Power Sources 2006, 161:820–825. [Google Scholar]
- 45.Chaudhuri SK, Lovley DR: Electricity generation by direct oxidation of glucose in mediatorless microbial fuel cells. Nat Biotechnol 2003, 21:1229–1232. [DOI] [PubMed] [Google Scholar]
- 46.Kim B-H, Kim H-J, Hyun M-S, Park D-H: Direct electrode reaction of Fe (III)-reducing bacterium, Shewanella putrefaciens. J Microbiol Biotechnol 1999, 9:127–131. [Google Scholar]
- 47.Ren Z, Thomas E Ward A, Regan JM: Electricity Production from Cellulose in a Microbial Fuel Cell Using a Defined Binary Culture. Environ Sci Technol 2007, 41:4781–4786. [DOI] [PubMed] [Google Scholar]
- 48.Seto A, Saito Y, Matsushige M, Kobayashi H, Sasaki Y, Tonouchi N, Tsuchida T, Yoshinaga F, Ueda K, Beppu T: Effective cellulose production by a coculture of Gluconacetobacter xylinus and Lactobacillus mali. Appl Microbiol Biotechnol 2006, 73:915–921. [DOI] [PubMed] [Google Scholar]
- 49.He Z, Kan J, Wang Y, Huang Y, Mansfeld F, Nealson KH: Electricity Production Coupled to Ammonium in a Microbial Fuel Cell. Environ Sci Technol 2009, 43:3391–3397. [DOI] [PubMed] [Google Scholar]
- 50.Kargi F, Eker S: Electricity generation with simultaneous wastewater treatment by a microbial fuel cell (MFC) with Cu and Cu–Au electrodes. J Chem Technol Biotechnol 2007, 82:658–662. [Google Scholar]
- 51.Moon H, Chang IS, Kim BH: Continuous electricity production from artificial wastewater using a mediator-less microbial fuel cell. Bioresour Technol 2006, 97:621–627. [DOI] [PubMed] [Google Scholar]
- 52.Clauwaert P, Rabaey K, Aelterman P, De Schamphelaire L, Pham TH, Boeckx P, Nico Boon A, Willy Verstraete: Biological Denitrification in Microbial Fuel Cells. Environ Sci Technol 2007, 41:3354–3360. [DOI] [PubMed] [Google Scholar]
- 53.Oh S, Logan BE: Hydrogen and electricity production from a food processing wastewater using fermentation and microbial fuel cell technologies. Water Res 2005, 39:4673–4682. [DOI] [PubMed] [Google Scholar]
- 54.Bond DR, Holmes DE, Tender LM, Lovley DR: Electrode-reducing microorganisms that harvest energy from marine sediments. Science 2002, 295:483–5. [DOI] [PubMed] [Google Scholar]
- 55.Lu L, Huggins T, Jin S, Zuo Y, Ren ZJ: Microbial Metabolism and Community Structure in Response to Bioelectrochemically Enhanced Remediation of Petroleum Hydrocarbon-Contaminated Soil. Environ Sci Technol 2014, 48:4021–4029. [DOI] [PubMed] [Google Scholar]
- 56.Rabaey K, Boon N, Siciliano SD, Verhaege M, Verstraete W: Biofuel cells select for microbial consortia that self-mediate electron transfer. Appl Environ Microbiol 2004, 70:5373–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reis MAM, Serafim LS, Lemos PC, Ramos AM, Aguiar FR, Van Loosdrecht MCM: Production of polyhydroxyalkanoates by mixed microbial cultures. Bioprocess Biosyst Eng 2003, 25:377–385. [DOI] [PubMed] [Google Scholar]
- 58.Moralejo-Gárate H, Mar’atusalihat E, Kleerebezem R, van Loosdrecht MCM: Microbial community engineering for biopolymer production from glycerol. Appl Microbiol Biotechnol 2011, 92:631–9. [DOI] [PubMed] [Google Scholar]
- 59.Korkakaki E, van Loosdrecht MCM, Kleerebezem R: Impact of phosphate limitation on PHA production in a feast-famine process. Water Res 2017, 126:472–480. [DOI] [PubMed] [Google Scholar]
- 60.•.Fradinho JC, Reis MAM, Oehmen A: Beyond feast and famine: Selecting a PHA accumulating photosynthetic mixed culture in a permanent feast regime. Water Res 2016, 105:421–428.This work evaluates a novel enrichment procedure for obtaining biopolymer-producing communities, observing that a constant supply of acetate under oxygen-limited conditions and high light availability can lead to photosynthetic communities with increased biopolymer productivity. Both the explanation of the rationale behind the enrichment procedure and the analysis of different modifications provide a useful example of enrichment method development.
- 61.Wang J, Wan W: Comparison of different pretreatment methods for enriching hydrogen-producing bacteria from digested sludge. Int J Hydrogen Energy 2008, 33:2934–2941. [Google Scholar]
- 62.Wang J, Wan W: Factors influencing fermentative hydrogen production: A review. Int J Hydrogen Energy 2009, 34:799–811. [Google Scholar]
- 63.Kumari S, Das D: Improvement of biohydrogen production using acidogenic culture. Int J Hydrogen Energy 2017, 42:4083–4094. [Google Scholar]
- 64.••.Petrof EO, Khoruts A: From stool transplants to next-generation microbiota therapeutics. Gastroenterology 2014, 146:1573–1582.This review covers the history of FMTs, details the technical and health concerns associated with FMT processing and application, and describes previous research in developing synthetic FMT communities. As synthetic FMTs appear to be a promisingly versatile and powerful tool in microbiome therapeutics, this discussion of FMT successes, challenges, and regulatory considerations offers useful context for future developments in the field.
- 65.Pham VHT, Kim J: Cultivation of unculturable soil bacteria. Trends Biotechnol 2012, 30:475–484. [DOI] [PubMed] [Google Scholar]
- 66.Leffler DA, Lamont JT: Clostridium difficile Infection. N Engl J Med 2015, 372:1539–1548. [DOI] [PubMed] [Google Scholar]
- 67.Tvede M, Rask-Madsen J: BACTERIOTHERAPY FOR CHRONIC RELAPSING CLOSTRIDIUM DIFFICILE DIARRHOEA IN SIX PATIENTS. Lancet 1989, 333:1156–1160. [DOI] [PubMed] [Google Scholar]
- 68.••.Petrof EO, Gloor GB, Vanner SJ, Weese SJ, Carter D, Daigneault MC, Brown EM, Schroeter K, Allen-Vercoe E: Stool substitute transplant therapy for the eradication of Clostridium difficile infection: “RePOOPulating” the gut. Microbiome 2013, 1:3.In this study, researchers developed a synthetic FMT community by isolating microbial strains from donor stool, screening for pathogens, and then combining the remaining isolates into a therapeutic community that showed promising results in treating CDI. This serves as a prime example of community reduction for therapeutics design and highlights key benefits of the approach, including pathogen exclusion and the use of culturable species that can be maintained in the lab for repeated therapeutic formulation on demand.
- 69.Caballero S, Kim S, Carter RA, Leiner IM, Sušac B, Miller L, Kim GJ, Ling L, Pamer EG: Cooperating Commensals Restore Colonization Resistance to Vancomycin-Resistant Enterococcus faecium. Cell Host Microbe 2017, 21:592–602.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, et al. : T reg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 2013, 500. [DOI] [PubMed] [Google Scholar]
- 71.Atarashi K, Tanoue T, Ando M, Kamada N, Nagano Y, Narushima S, Suda W, Imaoka A, Setoyama H, Nagamori T, et al. : Th17 Cell Induction by Adhesion of Microbes to Intestinal Epithelial Cells. Cell 2015, 163:367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Asada Y, Tokumoto M, Aihara Y, Oku M, Ishimi K, Wakayama T, Miyake J, Tomiyama M, Kohno H: Hydrogen production by co-cultures of Lactobacillus and a photosynthetic bacterium, Rhodobacter sphaeroides RV. Int J Hydrogen Energy 2006, 31:1509–1513. [Google Scholar]
- 73.Collet C, Gaudard O, Péringer P, Schwitzguébel J-P: Acetate production from lactose by Clostridium thermolacticum and hydrogen-scavenging microorganisms in continuous culture—Effect of hydrogen partial pressure. J Biotechnol 2005, 118:328–338. [DOI] [PubMed] [Google Scholar]
- 74.Kondo T, Kondo M: Efficient production of acetic acid from glucose in a mixed culture of Zymomonas mobilis and Acetobacter sp. J Ferment Bioeng 1996, 81:42–46. [Google Scholar]
- 75.Talabardon M, Schwitzguebel J-P, Peringer P, Yang S-T: Acetic Acid Production from Lactose by an Anaerobic Thermophilic Coculture Immobilized in a Fibrous-Bed Bioreactor. Biotechnol Prog 2000, 16:1008–1017. [DOI] [PubMed] [Google Scholar]
- 76.Taniguchi M, Tokunaga T, Horiuchi K, Hoshino K, Sakai K, Tanaka T: Production of l-lactic acid from a mixture of xylose and glucose by co-cultivation of lactic acid bacteria. Appl Microbiol Biotechnol 2004, 66:160–165. [DOI] [PubMed] [Google Scholar]
- 77.Roble ND, Ogbonna JC, Tanaka H: l-Lactic acid production from raw cassava starch in a circulating loop bioreactor with cells immobilized in loofa (Luffa cylindrica). Biotechnol Lett 2003, 25:1093–1098. [DOI] [PubMed] [Google Scholar]
- 78.Boonchan S, Britz ML, Stanley GA: Degradation and mineralization of high-molecular-weight polycyclic aromatic hydrocarbons by defined fungal-bacterial cocultures. Appl Environ Microbiol 2000, 66:1007–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Haruta S, Cui Z, Huang Z, Li M, Ishii M, Igarashi Y: Construction of a stable microbial community with high cellulose-degradation ability. Appl Microbiol Biotechnol 2002, 59:529–534. [DOI] [PubMed] [Google Scholar]
- 80.Poszytek K, Ciezkowska M, Sklodowska A, Drewniak L: Microbial Consortium with High Cellulolytic Activity (MCHCA) for Enhanced Biogas Production. Front Microbiol 2016, 7:324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ayed L, Achour S, Khelifi E, Cheref A, Bakhrouf A: Use of active consortia of constructed ternary bacterial cultures via mixture design for Congo Red decolorization enhancement. Chem Eng J 2010, 162:495–502. [Google Scholar]
- 82.Gunst RF, Mason RL: Fractional factorial design. Wiley Interdiscip Rev Comput Stat 2009, 1:234–244. [Google Scholar]
- 83.Prakasham RS, Rao CS, Rao RS, Lakshmi GS, Sarma PN: l-asparaginase production by isolated Staphylococcus sp. ? 6A: design of experiment considering interaction effect for process parameter optimization. J Appl Microbiol 2007, 102:1382–1391. [DOI] [PubMed] [Google Scholar]
- 84.Skonieczny MT, Yargeau V: Biohydrogen production from wastewater by Clostridium beijerinckii: Effect of pH and substrate concentration. Int J Hydrogen Energy 2009, 34:3288–3294. [Google Scholar]
- 85.Jiménez J, Guardia-Puebla Y, Romero-Romero O, Cisneros-Ortiz ME, Guerra G, Morgan-Sagastume JM, Noyola A: Methanogenic activity optimization using the response surface methodology, during the anaerobic co-digestion of agriculture and industrial wastes. Microbial community diversity. Biomass and Bioenergy 2014, 71:84–97. [Google Scholar]
- 86.Kikot P, Viera M, Mignone C, Donati E: Study of the effect of pH and dissolved heavy metals on the growth of sulfate-reducing bacteria by a fractional factorial design. Hydrometallurgy 2010, 104:494–500. [Google Scholar]
- 87.•.Faith JJ, Ahern PP, Ridaura VK, Cheng J, Gordon JI: Identifying gut microbe-host phenotype relationships using combinatorial communities in gnotobiotic mice. Sci Transl Med 2014, 6:220ra11.By evaluating a collection of randomly generated subsets of a 17-strain mouse gut community, the authors estimated various individual microbial impacts on host phenotypes including intestinal metabolite concentrations, body fat, and T_reg induction. This approach is similar to FFD and demonstrates how FFD-like approaches could be used for purposes such as informing synthetic community design.
- 88.Chen Y, Lin C-J, Jones G, Fu S, Zhan H: Enhancing biodegradation of wastewater by microbial consortia with fractional factorial design. J Hazard Mater 2009, 171:948–953. [DOI] [PubMed] [Google Scholar]
- 89.Chen Y, Lin C-J, Jones G, Fu S, Zhan H: Application of statistical design for the optimization of microbial community of synthetic domestic wastewater. Biodegradation 2011, 22:205–213. [DOI] [PubMed] [Google Scholar]
- 90.Hu J, Wang L, Zhang S, Le Y, Fu X: Feasibility of a Two-Step Culture Method to Improve the CO2-Fixing Efficiency of Nonphotosynthetic Microbial Community and Simultaneously Decrease the Spontaneous Oxidative Precipitates from Mixed Electron Donors. Appl Biochem Biotechnol 2014, 173:2307–2320. [DOI] [PubMed] [Google Scholar]
- 91.•.Hu J, Xue Y, Li J, Wang L, Zhang S, Wang Y-N, Gao M-T: Characterization of a designed synthetic autotrophic–heterotrophic consortia for fixing CO 2 without light. RSC Adv 2016, 6:78161–78169.The authors developed a carbon fixation community using a combinatorial evaluation approach to consider a previously-described consortium of marine microbes along with several individual isolates from that consortium. This work provides an example of a scenario where a reduced community cannot adequately recapitulate the original community’s function while also demonstrating the utility of sub-consortia in combinatorial evaluation.
- 92.Hu J, Xue Y, Guo H, Gao M, Li J, Zhang S, Tsang YF: Design and composition of synthetic fungal-bacterial microbial consortia that improve lignocellulolytic enzyme activity. Bioresour Technol 2017, 227:247–255. [DOI] [PubMed] [Google Scholar]
- 93.Agarwala R, Barrett T, Beck J, Benson DA, Bollin C, Bolton E, Bourexis D, Brister JR, Bryant SH, Canese K, et al. : Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 2018, 46:D8–D13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen I-MA, Markowitz VM, Chu K, Palaniappan K, Szeto E, Pillay M, Ratner A, Huang J, Andersen E, Huntemann M, et al. : IMG/M: integrated genome and metagenome comparative data analysis system. Nucleic Acids Res 2017, 45:D507–D516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kanehisa M: KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 2000, 28:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Caspi R, Billington R, Fulcher CA, Keseler IM, Kothari A, Krummenacker M, Latendresse M, Midford PE, Ong Q, Ong WK, et al. : The MetaCyc database of metabolic pathways and enzymes. Nucleic Acids Res 2018, 46:D633–D639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Greenblum S, Chiu H, Levy R, Carr R, Borenstein E: Towards a predictive systems-level model of the human microbiome: progress, challenges, and opportunities. Curr Opin Biotechnol 2013, 24:810–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Borenstein E: Computational systems biology and in silico modeling of the human microbiome. Brief Bioinform 2012, 13:769–80. [DOI] [PubMed] [Google Scholar]
- 99.•.Eng A, Borenstein E: An algorithm for designing minimal microbial communities with desired metabolic capacities. Bioinformatics 2016, 32:2008–2016.This work introduces an algorithm for identifying a minimal set of species with the collective capacity to catalyze the conversion of a set of substrates to desired products using an integer linear programming formulation of a joint set cover-network flow problem. This represents the first computational algorithm developed specifically to design synthetic microbial communities.
- 100.Julien-Laferrière A, Bulteau L, Parrot D, Marchetti-Spaccamela A, Stougie L, Vinga S, Mary A, Sagot M-F, Ro D-K, Bernstein HC, et al. : A Combinatorial Algorithm for Microbial Consortia Synthetic Design. Sci Rep 2016, 6:29182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Heirendt L, Arreckx S, Pfau T, Mendoza SN, Richelle A, Heinken A, Haraldsdóttir HS, Wachowiak J, Keating SM, Vlasov V, et al. : Creation and analysis of biochemical constraint-based models: the COBRA Toolbox v3.0. arXiv Prepr 2017, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.••.Magnúsdóttir S, Heinken A, Kutt L, Ravcheev DA, Bauer E, Noronha A, Greenhalgh K, Jäger C, Baginska J, Wilmes P, et al. : Generation of genome-scale metabolic reconstructions for 773 members of the human gut microbiota. Nat Biotechnol 2016, 35:81–89.The authors used a semi-automated pipeline to generate in silico models of metabolism for individual gut microbes and demonstrated their utility for identifying sufficient growth media, characterizing inter-microbial interactions, and analyzing 16S rRNA and metagenomic data. The resulting metabolic models have been made available and serve as a valuable resource for microbiome modeling and synthetic community design.
- 103.Kauffman KJ, Prakash P, Edwards JS: Advances in flux balance analysis. Curr Opin Biotechnol 2003, 14:491–496. [DOI] [PubMed] [Google Scholar]
- 104.Orth JD, Thiele I, Palsson BØ: What is flux balance analysis? Nat Biotechnol 2010, 28:245–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chiu H-C, Levy R, Borenstein E: Emergent biosynthetic capacity in simple microbial communities. PLoS Comput Biol 2014, 10:e1003695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chan SHJ, Simons MN, Maranas CD, Segal E, Henry C, Kupiec M, von Mering C: SteadyCom: Predicting microbial abundances while ensuring community stability. PLOS Comput Biol 2017, 13:e1005539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Harcombe WR, Riehl WJ, Dukovski I, Granger BR, Betts A, Lang AH, Bonilla G, Kar A, Leiby N, Mehta P, et al. : Metabolic resource allocation in individual microbes determines ecosystem interactions and spatial dynamics. Cell Rep 2014, 7:1104–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zomorrodi AR, Islam MM, Maranas CD: d-OptCom: Dynamic multi-level and multi-objective metabolic modeling of microbial communities. ACS Synth Biol 2014, 3:247–57. [DOI] [PubMed] [Google Scholar]
- 109.••.García-Jiménez B, García JL, Nogales J: FLYCOP: metabolic modeling-based analysis and engineering microbial communities. Bioinformatics 2018, 34:i954–i963.This study introduced a computational framework for designing synthetic communities that can optimize for both community metabolism and temporal dynamics using a community-level dynamic FBA model and applied it to various design tasks. FLYCOP represents the first FBA-based approach for designing synthetic communities.
- 110.•.Bucci V, Tzen B, Li N, Simmons M, Tanoue T, Bogart E, Deng L, Yeliseyev V, Delaney ML, Liu Q, et al. : MDSINE: Microbial Dynamical Systems INference Engine for microbiome time-series analyses. Genome Biol 2016, 17:121.This paper presents a toolbox for both inferring various properties of microbial community dynamics from time-series data and utilizing such data to predict interactions between species as well extrapolate future community dynamics, supporting toolbox performance with the results of multiple validation experiments. This toolbox offers a variety of important functionalities that can augment and enable various model-based design approaches.
- 111.Bucci V, Xavier JB: Towards Predictive Models of the Human Gut Microbiome. J Mol Biol 2014, 426:3907–3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.••.Stein RR, Tanoue T, Szabady RL, Bhattarai SK, Olle B, Norman JM, Suda W, Oshima K, Hattori M, Gerber GK, et al. : Computer-guided design of optimal microbial consortia for immune system modulation. Elife 2018, 7:e30916.Using a collection of T_reg-inducing Clostridia strains, the authors developed a combined functional and ecological model to predict the stability and induction capabilities of different strain combinations. They validated predicted stability and induction differences between synthetic compositions in an experimental setting and also used this model to optimize a synthetic T_reg induction community. This work demonstrates a promising general framework for optimizing non-metabolic functions and integrating models of community dynamics.
- 113.Kleerebezem R, van Loosdrecht MCM: Mixed culture biotechnology for bioenergy production. Curr Opin Biotechnol 2007, 18:207–12. [DOI] [PubMed] [Google Scholar]
- 114.Johnson K, Jiang Y, Kleerebezem R, Muyzer G, van Loosdrecht MCM: Enrichment of a mixed bacterial culture with a high polyhydroxyalkanoate storage capacity. Biomacromolecules 2009, 10:670–6. [DOI] [PubMed] [Google Scholar]
- 115.Verster AJ, Ross BD, Radey MC, Bao Y, Goodman AL, Mougous JD, Borenstein E: The Landscape of Type VI Secretion across Human Gut Microbiomes Reveals Its Role in Community Composition. Cell Host Microbe 2017, 22:411–419.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.•.Momeni B, Xie L, Shou W: Lotka-Volterra pairwise modeling fails to capture diverse pairwise microbial interactions. Elife 2017, 6:e25051.The authors used mechanistic simulations of two-species microbial communities to investigate the ability of pairwise ecological models to predict community dynamics when interactions were facilitated by one or more mediator compounds, finding that Lotka-Volterra models can fail to adequately capture multi-mediator interactions. This work demonstrates the importance of mechanistic models, including those that describe non-metabolic interactions, for computational design methods to accurately evaluate potential synthetic compositions.
- 117.Werner EE, Peacor SD: A REVIEW OF TRAIT‐MEDIATED INDIRECT INTERACTIONS IN ECOLOGICAL COMMUNITIES. Ecology 2003, 84:1083–1100. [Google Scholar]
- 118.Gould AL, Zhang V, Lamberti L, Jones EW, Obadia B, Gavryushkin A, Korasidis N, Carlson JM, Beerenwinkel N, Ludington WB: High-dimensional microbiome interactions shape host fitness. bioRxiv 2018, doi: 10.1101/232959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sanchez-Gorostiaga A, Bajić D, Osborne ML, Poyatos JF, Sanchez A: High-order interactions dominate the functional landscape of microbial consortia. bioRxiv 2018, doi: 10.1101/333534. [DOI] [PMC free article] [PubMed] [Google Scholar]


