Abstract
Interventional pharmacology is one of medicine’s most potent weapons against disease. These drugs, however, can result in damaging side effects and must be closely monitored. Pharmacovigilance is the field of science that monitors, detects, and prevents adverse drug effects. Safety efforts begin during the development process, using in vivo and in vitro studies, continue through clinical trials, and extend to post-marketing surveillance of ADRs in real-world populations. Future toxicity and safety challenges, including increased polypharmacy and patient diversity, stress the limits of these traditional tools. Massive amounts of newly available data present an opportunity for using artificial intelligence and machine learning to improve drug safety science. Here, we explore recent advances as applied to pre-clinical drug safety and post-marketing surveillance with a specific focus on machine and deep learning approaches.
Keywords: Pharmacovigilance, Machine Learning, Deep Learning, Adverse Drug Reactions
The challenge of keeping drugs safe
Drug safety is a major challenge in bring new drugs to market. Unexpected toxicities are a major source of attrition during clinical trials and post-marketing safety concerns cause unnecessary morbidity and mortality. Adverse events (AEs), or adverse drug reactions (ADRs) when causality is demonstrated, are unexpected effects occurring from a normal dosage of the drug. Between 2008 and 2017, the Food and Drug Administration (FDA) approved 321 novel drugsi. Over the same period of time, the FDA Adverse Event Reporting System (FAERS)ii recorded more than 10 million AE reports, among which 5.8 million were serious reports and 1.1 million were AEs related to death. AEs burden our health system causing 2 million hospital stays each year and lengthening visits by 1.7 to 4.6 daysi. The economic, social, and health burden of toxicity and safety assessment is an essential and pressing public health concern.
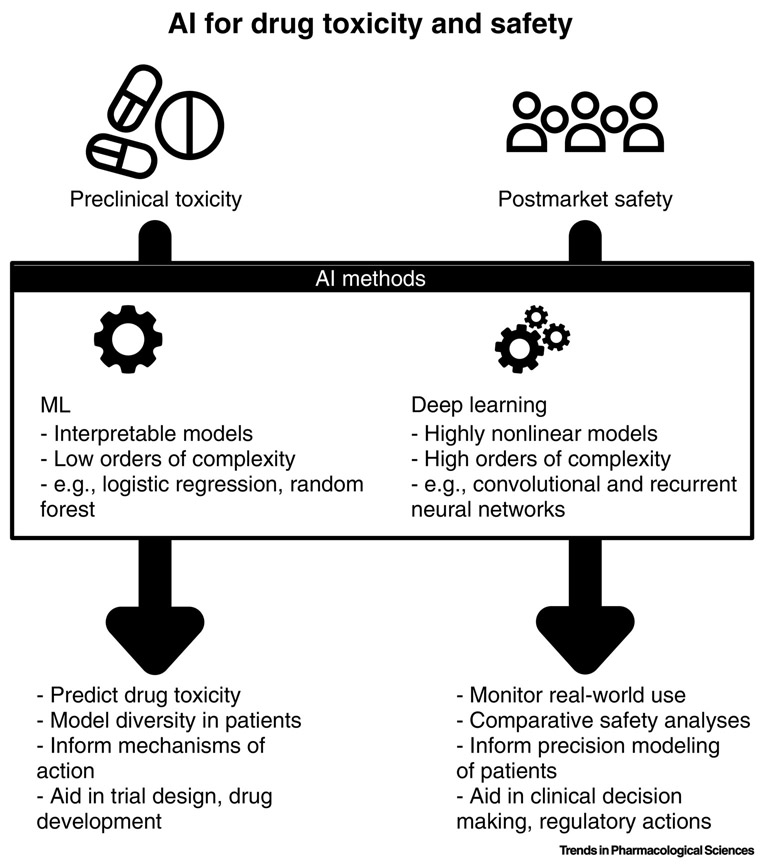
There are two complementary systems to address drug safety (Figure 1). Before a drug is approved, clinical trials ensure that this drug is safe and effective for its intended use. Once a drug is marketed, drugs are monitored through AE reports to ensure a drug’s safety information is up to date, a process called pharmacovigilance (PV). However, neither of these processes are error proof as clinical trials suffer from structural limitations. For example, it is impossible to test for all potential synergistic effects or to conduct trials on populations large enough to detect rare AEs. Until recently, women and the elderly were considered special sub-groups for clinical trials. These trials have focused on designing drugs for the average patient [1] even at a time when there are increasing calls for precision medicine to enable the “right drug at the right dose to the right patient”[2]. Once drugs are approved, it is the purview of programs to monitor drug safety. These agencies use databases of spontaneously collected AE reports to flag leads and perform confirmatory follow up analyses. However, these spontaneous reports are known to suffer from biases such as underreporting which is especially troublesome for rare events and drug-drug interactions (DDIs)[3]. The research community has turned to statistical and computational approaches to address these limitations and supplement its PV toolbox [4, 5].
Figure 1:
Artificial intelligence and machine learning present an opportunity for improving drug safety. These algorithms enable a data-driven approach to toxicity and safety assessments that can identify patterns that otherwise would be overlooked. Traditional machine learning, including methods like logistic regression, random forests, and support vector machines can produce interpretable models with relatively low complexity. These methods are desirable when the goal is to understand how the predictors affect the incidence or risk of an adverse event. A new class of methods, called deep neural networks – and often referred to as “artificial intelligence” – allows for more complex models to be built at the cost of requiring significantly more data. The benefit of using these algorithms is that they can automatically identify non-linear patterns in the data without requiring much manual intervention. Common examples that have been used in drug safety research include convolutional and recurrent neural networks. In both cases, these models have been used to in pre-clinical drug toxicity study, to model patient diversity, and to facilitate lead selection and trial design, and in post-marking surveillance to conduct comparative effectiveness research, identify drug-drug interactions, and to aid in clinical decision making. AI-assisted drug safety and toxicity science remains a nascent and growing field that requires further research to evaluate its potential clinical impact.
Over the past decade, we have seen two phenomena occur: (1) the explosion of freely accessible databases of medical, chemical, and pharmacological knowledge, along with the rapid adoption of electronic health record (EHR) systems stimulated by the Health Information Technology for Economic and Clinical Health (HITECH) Actiii; and (2) the development of novel computational methods in the realm of machine learning (ML) (see Glossary) and deep learning (DL) a popular re-branding of neural networks - catalyzed by exponential increase in compute power and data availability. Below, we explore the recent literature leveraging artificial intelligence (AI) methods, both ML and DL, on novel data sources for pre-clinical and in post-marketing surveillance for PV (Figure 1). We further encourage the reader to reference the following introductory reviews for more details on ML [6, 7, 8] and DL[9, 10].
Pre-clinical Drug Safety
AI techniques have been shown to play an important role in pre-market drug safety, especially in the area of toxicity evaluation. Drug toxicity determination is a main step in drug design and involves identifying the AEs of chemicals on humans, plants, animals, and the environment[11]. Pre-clinical evaluations are a necessity for preventing toxic drugs from reaching clinical trials. Despite this, high toxicity is still a major contributor to drug failure accounting for two-thirds of post-market drug withdrawals[12] and for one-fifth of failures during clinical trials[13]. Thus, accurate toxicity estimates are necessary for ensuring drug safety, and can help reduce the cost and development time of bringing new drugs to market. Animal studies have historically been the most conventional approach taken to assess toxicity[11, 12, 13]. However, these studies are constrained by cost, time, and ethical considerations. Numerous computational, in silico, approaches have demonstrated utility in estimating the toxicity of drug candidates. These approaches predict toxicity by evaluating various features of the drug and include target-based predictions and Quantitative Structure-Activity Relationships (QSAR). Below, we focus largely on ML and DL approaches in the area of QSAR and discuss DL techniques in toxicity prediction and assessment.
Quantitative Structure-Activity Relationships (QSAR)
QSAR is a method that establishes quantitative relationships between chemical or structural characteristics and pharmacological activity[14]. QSAR methods have been used to model numerous drug safety endpoints including drug lethal dose 50% (LD50)values, skin/eye irritation, and tissue-specific toxicity[15]. Specifically, a QSAR model can analyze the relationship between several predictors (e.g. molecular properties) and a response (e.g. biological activities such as binding affinity)[16]. Good models will be highly predictive and fairly easy to interpret. There are several types of ML approaches that have been used for QSAR modeling.
Regression
Early QSAR techniques relied on multivariate linear regression to assess the chemical properties of drug candidates[17]. These approaches are sensitive to high data dimensionality and feature correlation which may result in overfitting, and limited interpretability. Modern regression-based approaches incorporate feature selection techniques to address these concerns. One such technique is the use of a penalized regression model. L1 regularization, which is used in least absolute shrinkage and selection operator (LASSO), aims to prevent overfitting by reducing the number of features and only selecting subsets that are most relevant to the QSAR model prediction[18]. L2 regularization, which is used in ridge regression (RR), aims to alleviate collinearity by reducing the effective number of features used in the model. Recently, Algamal et al.[16] proposed a weight adjustment to the adaptive LASSO aimed to improve the selection of correlated descriptors. This approach demonstrated potential when used to develop a QSAR prediction of the anti-cancer potency of various imadazo[4,5-b]pyridine derivatives. The authors also proposed applying L1-norm regularization in the selection of significant descriptors for anti-hepatitis C virus activity of thiourea derivatives[19]. While regression-based approaches have demonstrated utility in QSAR prediction, assumptions of linearity, which are inherent in regression, as well as issues of dimensionality affect most QSAR modelling tasks. Currently, the most common alternatives are support vector machines (SVM) and ensemble approaches, such as random forest, due to their high predictive accuracy, robustness, and ease of interpretation[20].
Support Vector Machines
Support Vector Machines (SVM) is an approach that aims to find a hyperplane in an n dimensional space (n is defined by the number of features) that discriminatively classifies the data. For example, if there are 2 input features, the hyperplane is a line. With 3 features, the hyperplane is a 2-dimensional plane[21]. Support vectors are data points used to build the SVM. These data points are located near the hyperplane and influence the orientation and the position of this hyperplane. These support vectors are used to maximize the margin of the classifier. SVM performs classification by using a kernel function to map vectors into a higher dimensional feature space.
In a recent QSAR modeling of histone deacetylase 1 (HDAC1) inhibitors, SVM exhibited the best performance in predicting activity value when compared with naive Bayes, k-nearest neighbor (k-NN), and random forest algorithms[22]. Specifically, when SVM was used in conjunction with a Chemistry Development Kit (CDK)iv[23] molecular fingerprint for HDAC1 activity prediction with 5-fold cross validation of a training set, it achieved an area under the receiver operating curve (AUC)=0.91, with 97% sensitivity, and 50% specificity. The training set consisted of over 2,300 human HDAC1 inhibitors extracted from the Binding Databasev, BindingDB[24]. This model also performed well (AUC=0.89, with 95% sensitivity, and 75% specificity) when validated using an external set of 413 ChEMBLvicompounds[25].
Another recent study by Nehoei et al.[26] used a genetic algorithm (GA) [23] variable selection approach with SVM to develop QSAR models for the prediction of vascular endothelial growth factor receptor 2 (VEGFR-2) inhibition by aminopyrimidine-5-carbaldehyde oxime derivatives. More recently, Algamal et al.[27] applied an L1-norm SVM approach to build a QSAR classification model for neuraminidase inhibitors of influenza A virus (H1N1).
Ensemble learning
Ensemble methods combine several ML models into a robust predictive model. In doing so, they have improved predictive performance when compared with a single model, and are often less susceptible to bias and overfitting. Random forest is an ensemble learning algorithm that can handle class imbalances and avoid overfitting, two common challenges in QSAR modeling. Ng et al.[28] used a decision forest model to predict estrogen receptor binding, using a 3,308 chemical training set from the FDA’s Estrogenic Activity Databasevii. Models showed good performance with an internal accuracy of 92% and an external validation ranging from 70-89% accuracy. More recently, Lee et al.[29] demonstrated the use of random forest in ligand-based QSAR modelling using ChEMBL bioactivity data. Training of the 1,121 developed QSAR models showed an overall AUC = 0.97 using 5-fold cross validation. Testing on an external validation set showed an accuracy of 0.89.
To further highlight the impact of class imbalances in QSAR modeling, Grenet et al.[30] showed that many commonly used ML modeling approaches under-performed when using data from ToxCastviii, an US Environmental Protection Agency (EPA) managed public dataset containing chemical structure and bioactivity data. These data are imbalanced, and highly enriched for inactive compounds that are negative for toxicity in in vitro assays and for whom a half maximal activity concentration (AC50) could not be measured. When SVM, random forest, linear discriminant analysis (LDA), and neural networks were used on the ToxCast data, the AUC ranged between 0.6 and 0.73 across the methods, well below the expected performance. In response, the authors developed a stacked generalization approach, an ensemble method that consists of training a learning algorithm by combining the predictions of other algorithms. This stacked approached showed better performance with more models reaching an AUC>0.80 than any of the single QSAR classifiers.
Software
QSAR can also be used to predict target-based activities like toxicity. The field of target-driven toxicity prediction is heavily infiltrated with proprietary tools. Many of these use classical ML algorithms but refine the type of data used to calculate predictions. TargeTox[31] ix and PrOCTOR[32] x are two examples of recent open-source toxicity prediction tools.
TargeTox leverages protein target data with a network-based approach and gradient boosting to identify potentially toxic drugs. This approach builds protein networks using a distance metric following the assumption that neighboring biological entities share functional roles, thus hypothesizing that toxicity responses can be isolated to specific network regions. The built networks, pharmacological, and functional impact data from the public datasets, like DrugBankxi[33] and ChEMBL[25] (Table 1), comprise the model features. A gradient boosting classifier is then applied to develop a quantitative toxicity prediction score for each drug. While the authors specifically discuss a gradient boosting ensemble approach, any classifier with a regularization function and the capacity to handle nonlinear relationships can be applied. TargeTox has multiple model variants based on which distance metric is used for network calculation. The top performing approach uses a diffusion state distance (DSD) with a subset of reference points to calculate distance to the closest protein bound by a drug candidate. This method achieved an AUC of .743 with a sensitivity of 0.75 and specificity of 0.658 when trained and tested on data from DrugBank[33] and ClinicalTrails.govxii with 5-fold cross validation. The novelty of TargeTox is its ability to generate protein network data as well as combine other pharmacological and functional features into a ML classifier for toxicity prediction.
TABLE 1:
Open-source Databases with molecular, or pharmacological information
| Name | Description | References | Resource |
|---|---|---|---|
| DrugBank | a bioinformatics and chemoinformatics resource about drug data and drug target | [33,49] | xi |
| ChEMBL | a manually curated database of bioactive molecules with drug-like properties | [25,52] | vi |
| SIDER | a database on marketed medicines and their recorded adverse drug reactions | [50,51] | xvi |
| ChEBI | a freely available dictionary of molecular entities focused on “small” chemical compounds | [53] | xx |
| PubChem | an open chemistry database at the National Institutes of Health (NIH) | [54] | xxi |
| Reactome | a curated and peer-reviewed pathway database | [55] | xxii |
| KEGG | a database resource for understanding high-level functions and utilities of the biological system, the organism and the ecosystem, from molecular-level information | [56] | xxiii |
PrOCTOR is a target-based toxicity prediction software that in addition to network information, also incorporates chemical properties into its scoring. To develop a PrOCTOR score, the algorithm combines chemical structure properties of the drug candidate (e.g. molecular weight, polar surface area, quantitative estimate of drug-likeness (QED)) along with protein target information (e.g. network connectivity, tissue-specific expression). Drug target data is extracted from public datasets including DrugBank[33], GTExxiii[34], and ExACxiv[35]. Compared to TargeTox, the PrOCTOR model includes many more features with a total of 48 variables (34 target-based, 10 structure, and 4 drug-likeness) per drug compound model. A random forest classifier is used on the 48-feature model to develop a PrOCTOR score which assesses the likelihood of toxicity. This model constructs 50 decision trees using a subset of the features and uses the tree consensus to predict outcome. PrOCTOR showed high performance (AUC=0.83) and accuracy (ACC = 0.75) with high sensitivity (0.75) and specificity (0.74) when trained on a set of 784 FDA drugs with 10-fold cross validation. Further, when tested on a set of FDA-approved drugs, the algorithm scored 3 drugs with known toxicity events, docetaxel, bortezomib, and rosiglitazone, with the worst score. PrOCTOR’s ability to leverage multiple types of target and structure-based features for toxicity prediction places sets it above many other target-based algorithms.
Deep Learning
DL is an extension of ANNs which uses a hierarchy of ANNs to learn useful features from raw data. A Merck-sponsored Kaggle competition in 2012 introduced DL to the field of drug discovery. The winning team used DL on a set of diverse QSAR data sets to predict activity values for various compounds[20]. Recently, numerous studies of toxicity modelling have used DL approaches.
To assess hepatotoxicity, Xu et al. used DL to build drug-induced liver injury (DILI) prediction models with chemical structure data[36]. The authors used a recurrent neural network to construct the models. The best model was trained on 475 drugs and predicted an external validation set of about 200 drugs with an accuracy of 86.9%, sensitivity of 82.5%, and specificity of 92.9%. This model outperformed previously reported DILI prediction models.
Deep convolutional neural networks (CNNs) are a class of DL networks that learn representations of raw images from pixel information as a hierarchy of images from which features can be extracted and used to classify complex patterns[37]. CNNs have been used to predict toxicity from images of cells pre-treated with a set of drugs [37]. This approach was able to effectively predict a broad spectrum of toxicity mechanisms from different drugs, nuclear stains, and cell lines. Tong et al. also used a CNN strategy in a protein structure analysis task[38]. Specifically, a 3D CNN approach was used to analyze amino acid microenviroments and predict effects of mutations on protein structure. No prior knowledge or feature assumptions were required for this prediction task. And, the approach demonstrated a two-fold increase in accuracy prediction compared with models that require hand-selected features. Other DL approaches that have been used to assess drug toxicity include autoencoders[39], generative adversarial networks (GANs)[40] and long short-term memory (LSTM)[41], among others.
Post-marketing surveillance
In 1962, it was revealed that thousands of babies were born with malformed limbs because thalidomide, a mild sleeping pill, had no contraindications for pregnant women to whom it was often prescribed off-label [42]. The WHO ”Programme for International Drug Monitoring”xv was created following this disaster. Since 1978, the Uppsala Monitoring Centre (UMC) in Sweden is the global coordinator for PV in collaboration with the WHO, and counts 134 full member countries with national agencies supporting patient safety and drug AE reporting systems. These initiatives are the proof that safety assessment in clinical trials has its limit, and that drug safety needs to be actively monitored during the entire market life of drugs.
In the United States, the FDA maintains FAERS, a database containing adverse event reports, medication error reports and product quality complaints resulting in AEs. These self-reported Individual Case Safety Reports (ICSRs) have been a major data source for post-marketing drug safety mining. The classical methods to evaluate causality include the Naranjo algorithm[43], the Venulet algorithm[44], and the World Health Organization-Upsala Monitoring Centre (WHO-UMC) system for standardized case causality assessment, among others[45].
Naturally, spontaneous reporting systems (SRS) like FAERS enabled data mining methods to identify statistical associations between drugs and AEs [46, 47]. But with known short-comings, such as confounding biases and under-reporting[48], the attention has shifted to other data sources and advanced computational methods that could replace or complement existing resources. Below we discuss the approaches and data sources that can support post-marketing PV along with the associated AI-driven methods needed to extract information and learn from it.
System Pharmacology
System pharmacology is the study of drug action using principals from systems biology, considering the effect of drug on the entire system rather than a single target or metabolizing enzyme. This approach promises to explain unexpected drug effects that may result from complex interactions of targets and pathways. Application of systems pharmacology to adverse drug events differs from its use in drug discovery in that it’s focused on off-target effects and clinical observations of adverse reactions. In addition, it is one of the most data-rich approach to drug safety for in silico ADR mining. A variety of open databases are available and have been listed in Table 1. As a consequence of the rich data sources available, investigators in system pharmacology for adverse drug effects (ADEs) now have methods of choice involving network approaches and the ability to integrate multiple types of features. Lorberbaum et al.[57] proposed the modular assembly of drug safety subnetworks (MADSS) where they generated protein networks using knowledge bases that were pruned with literature mining, genome-wide association study (GWAS) data, assigned phenotype target with DrugBank and ChEMBL, and finally, trained random forest models on network metrics to predict new drugs causing AEs. Raja et al.[58] focused on DDIs by mining the literature to integrate drug-gene interactions (DGIs) at different scales and trained random forest models to predict DDIs with a gold standard corpus, focusing specifically on cutaneous diseases. Sornalakshmi et al.[59] trained SVM models on similarity measures such as 2D molecular structure similarity, 3D pharmacologic similarity, interaction profile fingerprint (IPF) similarity, target similarity and ADE similarity from DrugBank and SIDERxvito predict drug pairs likely to interact with each other based on a literature-based gold standard. Xu et al. [36] used various pharmaceutical compound datasets and neural networks to encode these drugs using the undirected graph recursive neural networks (UG-RNN) introduced by Lusci et al. [60] and classified them between DILI positive and DILI negative compounds. Herrero et al.[61] used pharmacokinetic (PK) and pharmacodynamic (PD) properties from DrugBank and other sources, along with drug-enzymes relationship data to build neural networks supervised models taking Lexicompxvii and Vidal compendiaxviii as ground truth for DDI labels.
All of the above approaches heavily rely on molecular features directly linked to these drugs along with phenotypic evidence of their effects, and the source datasets are usually open. However, the clinical information used are often limited to specific outcomes, missing on the longitudinal patient medical history, and all the other clinical covariates. In contrast, EHRs are closed datasets that have been used to compensate the episodic aspect of SRS and provide observational data captured during medical encounters.
EHR mining
Stimulated by the HITECH Act, the rapid and widespread adoption of EHRs that we have witnessed this past decade has also enabled researchers to tap into these rich and noisy sources of clinical data for PV. EHR data stand out in their challenging heterogeneity: these temporal data sources include categorical data such as diagnostic, procedure and medication codes, but also continuous laboratory tests and measurement values, along with large volumes of semi-structured and unstructured medical notes and reports.
Structured EHR data
Structured data such as diagnoses, procedures, medications and laboratory tests present the advantage of requiring the least pre-processing for ML approaches. Zhao et al.[62] studied nine different weighting strategies regarding how to use drugs, diagnoses and measurements as features in supervised learning algorithms for ADR prediction.
Bayesian methods have been popular in modeling medical outcomes. Benefiting from a knowledge-rich domain, a variety of Bayesian approach have been used for adverse event prediction by including prior medical knowledge. Bekker et al.[63] used Bayesian network representations to model the effect of drugs on the progression of multiple co-morbidities using prescriptions and diagnoses from primary care data. Moghaddass et al.[64] proposed to generalize the self-controlled case series (SCCS) with a multivariate hierarchical Bayesian model to leverage latent factor analysis (LFA) and bring more interpretability regarding the effects of transient multi-drug exposures on a collection of health outcomes. As an alternative, Morel et al.[65] proposed another multivariate SCCS method based on convolution of step functions with point drug exposures to estimate the effect of longitudinal features. Kuang et al. directly followed up with that multiple SCCS and presented a baseline regularization to take into account individual-specific, time dependent occurrence rate of AEs. More recently, they have also presented a version of that model for drug repurposing[66].
Rather than predicting discrete outcomes, it is also possible to model drug responses by predicting dynamic time series of observational data to select the best treatment courses. Xu et al. [67] estimated individualized treatment response (ITR) curves with Bayesian non-parametrics (BNP). They modeled creatinine time series response of treatments used in managing kidney function and demonstrated a gain in accuracy compared to baseline models. Structured data have the advantage of being easily pre-processed for ML and DL algorithms, but they also have to rely on mappings, data structures and terminologies that could impede reproducibility. More importantly, because they are primarily designed for billing purposes, EHR databases present a number of biases, including confounding bias and selection bias[68], and do not provide the whole picture of their patients’ care trajectory.
Clinical notes and biomedical corpora
Beyond structured data, which present both standardization and mapping challenges and may not be readily accessible, biomedical and clinical corpora represent a valuable resource. Clinical corpora enable the learning of medical language model that embed prior knowledge and help with the pragmatics of context-specific use of certain words. Natural Language Processing (NLP) methods are essential in this area to extract concepts and apply ML or learn embedding representations of these documents to directly make predictions. Abacha et al.[69] developed a hybrid feature-based and kernel-based system for DDI detection and classification applied to the DDI-Extraction-2013 corpus. That corpus contained 1017 medical texts, including abstracts from MEDLINE and documents describing DDIs from the DrugBank database, annotated with DDIs and pharmacological substances for the supervision of the learning task. Mower et al.[70] built embedding of semantic predications (ESP), by extracting concept-relationship-concept triples from the literature with the SemRep NLP systemxix. They trained a kNN model with the Exploring and Understanding ADRs by Integrative Mining of Clinical Records and Biomedical Knowledge (EU-ADR), and the Observational Medical Outcomes Partnership (OMOP) datasets, two national networks that have defined common data models, as ground truth and showed good generalization performances to predict binary ADE outcomes. Kim et al.[71] opted for a naive Bayes classifier to predict the likelihood of ADR in textual data from expert opinion on ADR case reports from the Korean Adverse Event Reporting System database.
NLP has dramatically benefited from advances in DL in the recent years to build better language models, with the development of word embeddings[72], sequence to sequence (seq2seq) learning[73], and more recently attention mechanisms [74, 75, 76]. The clinical domain has always been a challenging field of application for NLP, and these novel methods have been promptly applied to PV problems[77]. Language models can be trained using the biomedical literature and then applied to clinical notes, as demonstrated by Dev et al.[78], where the authors used MEDLINE to learn a better representation of concepts found in narrative logs for classification of ADEs. A recurrent theme in NLP for the detection of drug to AE relationship prediction is the need to first detect the concept (i.e., Named Entity Recognition, (NER)), and then perform the learning task. These two tasks as showed in the studies previously mentioned can both be conducted with neural networks. With its natural properties of connections and weights, DL enables multi-task learning (MTL), an approach that consists of sharing the weights of the neural networks between multiple tasks to improve overall performances. Zhang et al.[79] used this technique to jointly learn NER in texts for AE cases and ADE classification between serious and non-serious effects. Similarly, Li et al. [80] applied biLSTM networks on the Medication, Indication and Adverse Drug Events (MADE) 1.0 challenge for NER with a conditional random field network, and for relations extraction with an attention mechanism. More recently and using the same dataset, Yang et al. [81] developed a similar LSTM model for NER but extracted relations between concepts and ADEs by comparing SVM and random forests.
These NLP techniques have also been used with data collected on social media and in online health communities. Some of the reviews covering covering these applications are[82, 59, 83]. There is evidence that users on social media disseminate information comparable to ICSRs and ADEs can be classified from these high-noise data sources[84]. Post-marketing PV has been conducted using Twitter data [85, 86] with embedding techniques and biLSTM deep classifiers that outperform conditional random field methods, discussion forums[87], or more domain-specific health social networking sites[88].
Concluding Remarks
The availability of publicly accessible data, adoption of EHR systems, and development of novel ML and DL approaches has transformed the field of PV. This article focuses specifically on advances in AI techniques in the context of pre-clinical drug safety and post-marketing surveillance. We encourage the reader to reference systemic reviews on PV[21, 89, 90, 91, 92] which present a more complete picture landscape of PV.
In the recent years, we have observed a growing integration of multiscale data, from molecular databases to clinical datasets, in conjunction with a democratization of DL models to leverage these different data types. Neural nets have been used so far mostly for NLP applications in PV, but they have integrated the most recent state-of-the-art concepts such as attention mechanisms and multi tasks learning. Their applications are starting to be used beyond that scope, both in chemoinformatics and with clinical observational data. We noted that most of the approaches in the recent years that aim at predicting ADEs have been using annotated datasets. This almost exclusive use of supervised models has its limits, as the prediction of novel and unknown drug effects cannot rely on labeled data.
This is only the dawn of AI, and numerous questions remain such as how to address class imbalances in supervised modeling tasks, and how to incorporate unsupervised approaches in PV studies (Outstanding Questions). Techniques such as GANs hold promise in addressing some of these concerns. For example, novel unsupervised approaches using GANs that can generate in silco molecules with desired chemical properties are starting to emerge[93, 94], showing great promise for drug safety.
OUTSTANDING QUESTIONS.
Could the discussed methods address the class imbalance issues that are prevalent in QSAR modeling, or are more advanced approaches needed?
What are the limitations to using unsupervised methods that would address the scarcity of annotated datasets?
How can deep generative models change the game of pre-clinical drug safety?
While academic research has witnessed a drastic increase in the use of ML and DL, the community will begin to see these approaches entering into practice at a growing rate. For example, the FDA recently released plans for a new regulatory framework to promote the development of safe medical devices using AI algorithms. We expect that this will extend to drug development and safety in the future. Appropriate regulatory frameworks will need to be established to control for the risk of false positives. Overall, the risk of implementing AI approaches for PV is low and the opportunity high as it may have a positive impact on healthcare.
HIGHLIGHTS.
The expansion of publicly available resources and the adoption of electronic health records (EHR) has enabled the use of artificial intelligence (AI) methods for pharmacovigilance.
Pre-clinical quantitative structure-activity relationship (QSAR) is largely moving toward ensemble machine learning methods and deep learning approaches.
Post-marketing pharmacovigilance relies on a variety of data sources such as molecular, chemoinformatic and clinical databases, as well as social media and biomedical literature.
Deep learning-powered Natural Language Processing (NLP) methods including word embeddings and attention mechanisms are the techniques of choice to extract drug-adverse event (AE) relationships in text data.
Acknowledgments:
The authors are supported by NIH NIGMS R01GM107145.
GLOSSARY
- Attention mechanism
a process that allows to look at elements in a sequence as a whole and learn a distribution that weights their contextual importance
- AC50
Concentration for half-maximal activity, as derived from the Hill equation. It is a common potency measured employed in toxicity testing
- Area under the Receiver Operating Curve (AUC)
Area between the curve and the x-axis. Heuristic used to evaluate the performance of classification models with AUC =1 indicating perfect classification.
- Bayesian methods
Bayesian and Frequentist are two different approaches to defining probabilities. The Bayesian approach is to see as a representation of uncertainty, while frequentists see probabilities as a long-term frequency of an event.
- Bayesian non-parametrics (BNP)
a class of methods where the complexity of a model is defined by the data
- Convolutional neural networks (ConvNet or CNN)
were pioneered by LeCun et al. in 1990 in their first application for handwritten digit recognition. They consist of training filters that performs convolutional products with the input data and learn more and more high level features. ConvNets have to learn comparatively less weights than fully connected neural networks, and are particularly efficient for computer vision applications.
- Data dimensionality
Refers to the number of attributes present in a dataset
- Deep learning (DL)
Deep learning (DL) is a sub-field of machine learning where the algorithms learn abstractions of the input features that they use to make the predictions. These algorithms are characterized by a higher capacity than classic machine learning techniques (i.e., have higher degrees of freedom). It is in essence the trade-off of DL: what we gain in capacity and automated feature engineering, we lose it in a higher dimension space of parameters that is more complex and time-consuming to explore.
- Diffusion state distance (DSD)
Distance metric based on properties of graph diffusion designed to capture distinctions between annotations in protein-protein interactions
- Embedding of semantic predications (ESP)
a method to generate semantic vector representations of biomedical terms inspired by the skipgram-with-negative-sampling (SGNS). SGNS is an embedding method that uses neural networks to associate terms to their context in a corpus
- Genetic algorithm (GA)
GA is a stochastic variable selection method that solves optimization problems by applying Darwinian hypotheses of evolution
- Gradient boosting classifier
An approach used for classification and regression that builds a predictive model using a combination of individually weaker prediction models
- k-fold cross validation
A resampling procedure in which the data is split into k groups in order to estimate and assess model performance
- k-nearest neighbor (k-NN)
Algorithm used for classification in which the data are separated into several classes to help predict the classification of a new data point
- Latent factor analysis (LFA)
A statistical method used to describe variability in observed and correlated variables in term of unobserved variables called latent factors.
- LD50 (Lethal dose 50%)
Amount of an administered substance that kills 50% of a test sample
- Linear discriminant analysis (LDA)
Statistical, machine learning technique which seeks to find a linear combinate of features that separates two or more classes.
- Machine learning (ML)
a field of artificial intelligence in which algorithms are trained to perform tasks and make predictions by learning directly from the data, without being explicit programmed. ML methods can broadly be classified into two classes based on how the data learn to make predictions: supervised and unsupervised learning. In supervised learning, an algorithm is used to learn the mapping between input variables and an output, such as a label. The goal is for the algorithm to learn to predict a correct output when a new input is provided. In unsupervised learning, there are no assigned labels to the input training data. Here, the machine’s goal is to learn representations of the input data that can be used for tasks such as predicting future inputs, and decision making, without an output
- Naïve bayes
set of supervised learning algorithm based on Bayes theorem with the assumption of conditional independence between feature pairs
- Named Entity Recognition (NER)
a method that identified tokens in unstructured text and map them to concepts or categories in terminologies
- Natural Language Processing (NLP)
Subfield of computer science that aims at manipulating and making sense of natural language data
- Random forest
An ensemble learning approach in classification and regression which constructs decision trees during training and produces the class (classification) or mean predication (regression) of individual trees.
- Regression
Statistical approach which seeks to find relationships between dependent variables and one or more independent variables. Multivariate regression estimates a single regression model with more than one outcome variable.
- Self-controlled case series (SCCS)
A method in epidemiological study design where the subjects are their own control.
- Undirected Graph Recursive Neural Networks (UG-RNN)
In chemoinformatics, a model where, for a molecule of N atoms, a multilayer perceptron (MLP) recursively crawls through N different representations of this molecule. The vectors generated as a result are then averaged to compute a prediction for the molecule
- Word embeddings
In Natural Language Processing (NLP), one of the main challenges is finding a good representation for the vocabulary the corpus covers. While some methods simply encode tokens in binary vectors with a sparse representation, word embeddings learn a representation that takes into account the token's context
Footnotes
Conflict of interest: The authors do not have any conflicts of interest to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
xviii): http://www.vidal-dis.com/
xxiii): https://www.genome.jp/kegg/
References
- [1].Tannenbaum C, Day D, et al. , Age and sex in drug development and testing for adults, Pharmacological research 121 (2017) 83–93. [DOI] [PubMed] [Google Scholar]
- [2].Collins FS, Varmus H, A new initiative on precision medicine, New England Journal of Medicine 372 (9) (2015) 793–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Marengoni A, Onder G, Guidelines, polypharmacy, and drug-drug interactions in patients with multimorbidity, BMJ: British Medical Journal (Online) 350. [DOI] [PubMed] [Google Scholar]
- [4].Tatonetti NP, Patrick PY, Daneshjou R, Altman RB, Datadriven prediction of drug effects and interactions, Science translational medicine 4 (125) (2012) 125ra31–125ra31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Tatonetti NP, Fernald GH, Altman RB, A novel signal detection algorithm for identifying hidden drug-drug interactions in adverse event reports, Journal of the American Medical Informatics Association 19 (1) (2011) 79–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Rajkomar A, Dean J, Kohane IS, Machine learning in medicine., The New England journal of medicine 380 14 (2019) 1347–1358. [DOI] [PubMed] [Google Scholar]
- [7].Ba,stanlar Y, Ozuysal M, Introduction to machine learning, in: miR-Nomics: MicroRNA Biology and Computational Analysis, Springer, 2014, pp. 105–128. [DOI] [PubMed] [Google Scholar]
- [8].Handelman G, Kok H, Chandra R, Razavi A, Lee M, Asadi H, ed octor: machine learning and the future of medicine, Journal of internal medicine 284 (6) (2018) 603–619. [DOI] [PubMed] [Google Scholar]
- [9].Miotto R, Wang F, Wang S, Jiang X, Dudley JT, Deep learning for healthcare:review, opportunities and challenges, Briefings in bioinformatics 19 (6) (2017) 1236–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Xiao C, Choi E, Sun J, Opportunities and challenges in developing deep learning models using electronic health records data: a systematic review, Journal of the American Medical Informatics Association 25 (10) (2018) 1419–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Raies AB, Bajic VB, In silico toxicology: computational methods for the prediction of chemical toxicity, Wiley Interdisciplinary Reviews: Computational Molecular Science 6 (2) (2016) 147–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Onakpoya IJ, Heneghan CJ, Aronson JK, Worldwide withdrawal of medicinal products because of adverse drug reactions: a systematic review and analysis, Critical reviews in toxicology 46 (6) (2016) 477–489. [DOI] [PubMed] [Google Scholar]
- [13].Segall MD, Barber C, Addressing toxicity risk when designing and selecting compounds in early drug discovery, Drug discovery today 19 (5) (2014) 688–693. [DOI] [PubMed] [Google Scholar]
- [14].Roy K, Kar S, Das RN, Understanding the basics of QSAR for applications in pharmaceutical sciences and risk assessment, Academic press, 2015. [Google Scholar]
- [15].Patlewicz G, Fitzpatrick JM, Current and future perspectives on the development, evaluation, and application of in silico approaches for predicting toxicity, Chemical research in toxicology 29 (4) (2016) 438–451. [DOI] [PubMed] [Google Scholar]
- [16].Algamal ZY, Lee MH, Al-Fakih AM, Aziz M, High-dimensional qsar prediction of anticancer potency of imidazo [4, 5-b] pyridine derivatives using adjusted adaptive lasso, Journal of Chemometrics 29 (10) (2015) 547–556. [Google Scholar]
- [17].Luco JM, Ferretti FH, Qsar based on multiple linear regression and pls methods for the anti-hiv activity of a large group of hept derivatives, Journal of chemical information and computer sciences 37 (2) (1997) 392–401. [DOI] [PubMed] [Google Scholar]
- [18].Tibshirani R, Regression shrinkage and selection via the lasso, Journal of the Royal Statistical Society: Series B (Methodological) 58 (1) (1996) 267–288. [Google Scholar]
- [19].Algamal Z, Lee M, A new adaptive l1-norm for optimal descriptor selection of high-dimensional qsar classification model for anti-hepatitis c virus activity of thiourea derivatives, SAR and QSAR in Environmental Research 28 (1) (2017) 75–90. [DOI] [PubMed] [Google Scholar]
- [20].Ma J, Sheridan RP, Liaw A, Dahl GE, Svetnik V, Deep neural nets as a method for quantitative structure-activity relationships, Journal of chemical information and modeling 55 (2) (2015) 263–274. [DOI] [PubMed] [Google Scholar]
- [21].Lo Y-C, Rensi SE, Torng W, Altman RB, Machine learning in chemoinformatics and drug discovery, Drug discovery today. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Shi J, Zhao G, Wei Y, Computational qsar model combined molecular descriptors and fingerprints to predict hdac1 inhibitors, médecine/sciences 34 [2018] 52–58. [DOI] [PubMed] [Google Scholar]
- [23].Willighagen EL, Mayfield JW, Alvarsson J, Berg A, Carlsson L, Jeliazkova N, Kuhn S, Pluskal T, Rojas-Chertó M, Spjuth O, et al. , The chemistry development kit (cdk) v2. 0: atom typing, depiction, molecular formulas, and substructure searching, Journal of cheminformatics 9 [1] [2017] 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Liu T, Lin Y, Wen X, Jorissen RN, Gilson MK, Bindingdb: a webaccessible database of experimentally determined protein–ligand binding affinities, Nucleic acids research 35 (suppl 1] [2006] D198–D201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gaulton A, Bellis LJ, Bento AP, Chambers J, Davies M, Hersey A, Light Y, McGlinchey S, Michalovich D, Al-Lazikani B, et al. , Chembl: a large-scale bioactivity database for drug discovery, Nucleic acids research 40 (Dl) [2011] D1100–D1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Nekoei M, Mohammadhosseini M, Pourbasheer E, Qsar study of vegfr2 inhibitors by using genetic algorithm-multiple linear regressions (gamlr) and genetic algorithm-support vector machine (ga-svm): a comparative approach, Medicinal Chemistry Research 24 [7] [2015] 3037–3046. [Google Scholar]
- [27].Algamal Z, Qasim M, Ali H, A qsar classification model for neuraminidase inhibitors of influenza a viruses (h1n1) based on weighted penalized support vector machine, SAR and QSAR in Environmental Research 28 [5] [2017] 415–426. [DOI] [PubMed] [Google Scholar]
- [28].Ng HW, Doughty SW, Luo H, Ye H, Ge W, Tong W, Hong H, Development and validation of decision forest model for estrogen receptor binding prediction of chemicals using large data sets, Chemical research in toxicology 28 [12] [2015] 2343–2351. [DOI] [PubMed] [Google Scholar]
- [29].Lee K, Lee M, Kim D, Utilizing random forest qsar models with optimized parameters for target identification and its application to targetfishing server, BMC bioinformatics 18 (16) (2017) 567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Grenet I, Merlo K, Comet JP, Tertiaux R, Rouquié D, Dayan F, Stacked generalization with applicability domain outperforms simple qsar on in vitro toxicological data, Journal of Chemical Information and Modeling. [DOI] [PubMed] [Google Scholar]
- [31].Lysenko A, Sharma A, Boroevich KA, Tsunoda T, An integrative machine learning approach for prediction of toxicity-related drug safety, Life science alliance 1 (6) (2018) e201800098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Gayvert KM, Madhukar NS, Elemento O, A data-driven approach to predicting successes and failures of clinical trials, Cell chemical biology 23 (10) (2016) 1294–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Law V, Knox C, Djoumbou Y, Jewison T, Guo AC, Liu Y, Maciejewski A, Arndt D, Wilson M, Neveu V, et al. , Drugbank 4.0: shedding new light on drug metabolism, Nucleic acids research 42 (D1) (2013) D1091–D1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].G. Consortium, et al. , The genotype-tissue expression (gtex) pilot analysis: multitissue gene regulation in humans, Science 348 (6235) (2015) 648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Karczewski KJ, Weisburd B, Thomas B, Solomonson M, Ruderfer DM, Kavanagh D, Hamamsy T, Lek M, Samocha KE, Cummings BB, et al. , The exac browser: displaying reference data information from over 60 000 exomes, Nucleic acids research 45 (D1) (2016) D840–D845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Xu Y, Dai Z, Chen F, Gao S, Pei J, Lai L, Deep learning for drug-induced liver injury, Journal of chemical information and modeling 55 (10) (2015) 2085–2093. [DOI] [PubMed] [Google Scholar]
- [37].Jimenez-Carretero D, Abrishami V, Fernández-de Manuel L, Palacios I, Quílez-Alvarez A, Díez-Sánchez A, del Pozo MA, Mon-toya MC, Tox (r) cnn: Deep learning-based nuclei profiling tool for drug toxicity screening, PLoS computational biology 14 (11) (2018) e1006238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Torng W, Altman RB, 3d deep convolutional neural networks for amino acid environment similarity analysis, BMC bioinformatics 18 (1) (2017) 302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Hu Q, Feng M, Lai L, Pei J, Prediction of drug-likeness using deep autoencoder neural networks, Frontiers in genetics 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kadurin A, Nikolenko S, Khrabrov K, Aliper A, Zhavoronkov A, drugan: an advanced generative adversarial autoencoder model for de novo generation of new molecules with desired molecular properties in silico, Molecular pharmaceutics 14 (9) (2017) 3098–3104. [DOI] [PubMed] [Google Scholar]
- [41].Altae-Tran H, Ramsundar B, Pappu AS, Pande V, Low data drug discovery with one-shot learning, ACS central science 3 (4) (2017) 283–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ridings JE, The thalidomide disaster, lessons from the past, in: Teratogenicity Testing, Springer, 2013, pp. 575–586. [DOI] [PubMed] [Google Scholar]
- [43].Naranjo CA, Busto U, Sellers EM, Sandor P, Ruiz I, Roberts E, Janecek E, Domecq C, Greenblatt D, A method for estimating the probability of adverse drug reactions, Clinical Pharmacology & Therapeutics 30 (2) (1981) 239–245. [DOI] [PubMed] [Google Scholar]
- [44].Venulet J, Ciucci A, Berneker G, Updating of a method for causality assessment of adverse drug reactions., International journal of clinical pharmacology, therapy, and toxicology 24 (10) (1986) 559–568. [PubMed] [Google Scholar]
- [45].Agbabiaka TB, Savović J, Ernst E, Methods for causality assessment of adverse drug reactions, Drug safety 31 (1) (2008) 21–37. [DOI] [PubMed] [Google Scholar]
- [46].Sakaeda T, Tamon A, Kadoyama K, Okuno Y, Data mining of the public version of the fda adverse event reporting system, International journal of medical sciences 10 (7) (2013) 796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Banda JM, Evans L, Vanguri RS, Tatonetti NP, Ryan PB, Shah NH, A curated and standardized adverse drug event resource to accelerate drug safety research, Scientific data 3 (2016) 160026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hazell L, Shakir SA, Under-reporting of adverse drug reactions, Drug safety 29 (5) (2006) 385–396. [DOI] [PubMed] [Google Scholar]
- [49].Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, Sajed T, Johnson D, Li C, Sayeeda Z, et al. , Drugbank 5.0: a major update to the drugbank database for 2018, Nucleic acids research 46 (D1) (2017) D1074–D1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Kuhn M, Campillos M, Letunic I, Jensen LJ, Bork P, A side effect resource to capture phenotypic effects of drugs, Molecular systems biology 6(1) (2010) 343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Kuhn M, Letunic I, Jensen LJ, Bork P, The sider database of drugs and side effects, Nucleic acids research 44 (D1) (2015) D1075–D1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Gaulton A, Hersey A, Nowotka M, Bento AP, Chambers J, Mendez D, Mutowo P, Atkinson F, Beilis LJ, Cibrián-Uhalte E, et al. , The chembl database in 2017, Nucleic acids research 45 (D1) (2016) D945–D954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Hastings J, Owen G, Dekker A, Ennis M, Kale N, Muthukrishnan V, Turner S, Swainston N, Mendes P, Steinbeck C, Chebi in 2016: Improved services and an expanding collection of metabolites, Nucleic acids research 44 (D1) (2015) D1214–D1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, Li Q, Shoemaker BA, Thiessen PA, Yu B, et al. , Pubchem 2019 update: improved access to chemical data, Nucleic acids research 47 (D1) (2018) D1102–D1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Joshi-Tope G, Gillespie M, Vastrik I, D’Eustachio P, Schmidt E, de Bono B, Jassal B, Gopinath G, Wu G, Matthews L, et al. , Reactome: a knowledgebase of biological pathways, Nucleic acids research 33 (suppl 1) (2005) D428–D432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K, Kegg: new perspectives on genomes, pathways, diseases and drugs, Nucleic acids research 45 (D1) (2016) D353–D361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Lorberbaum T, Nasir M, Keiser MJ, Vilar S, Hripcsak G, Tatonetti NP, Systems pharmacology augments drug safety surveillance, Clinical Pharmacology & Therapeutics 97 (2) (2015) 151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Raja K, Patrick M, Elder JT, Tsoi LC, Machine learning workflow to enhance predictions of adverse drug reactions (adrs) through druggene interactions: Application to drugs for cutaneous diseases, Scientific reports 7 (1) (2017) 3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Sornalakshmi K, Vadivu G, Sujatha G, Hemavathi D, A survey on using social media data analytics for pharmacovigilance, Research Journal of Pharmacy and Technology 10 (10) (2017) 3474–3478. [Google Scholar]
- [60].Lusci A, Pollastri G, Baldi P, Deep architectures and deep learning in chemoinformatics: the prediction of aqueous solubility for drug-like molecules, Journal of chemical information and modeling 53 (7) (2013) 1563–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Herrero-Zazo M, Lille M, Barlow DJ, Application of machine learning in knowledge discovery for pharmaceutical drug-drug interactions., in: KDWeb, 2016. [Google Scholar]
- [62].Zhao J, Temporal weighting of clinical events in electronic health records for pharmacovigilance, in: Bioinformatics and Biomedicine (BIBM), 2015 IEEE International Conference on, IEEE, 2015, pp. 375–381. [Google Scholar]
- [63].Bekker J, Hommersom A, Lappenschaar M, Davis J, Measuring adverse drug effects on multimorbity using tractable bayesian networks, arXiv preprint arXiv: 1612.03055. [Google Scholar]
- [64].Moghaddass R, Rudin C, Madigan D, The factorized self-controlled case series method: An approach for estimating the effects of many drugs on many outcomes, Journal of Machine Learning Research 17 (185) (2016) 1–24. [Google Scholar]
- [65].Morel M, Bacry E, Gaïffas S, Guilloux A, Leroy F, Convsccs: convolutional self-controlled case series model for lagged adverse event detection, arXiv preprint arXiv: 1712.08243. [DOI] [PubMed] [Google Scholar]
- [66].Kuang Z, Bao Y, Thomson J, Caldwell M, Peissig P, Stewart R, Willett R, Page D, A machine-learning-based drug repurposing approach using baseline regularization, in: Computational Methods for Drug Repurposing, Springer, 2019, pp. 255–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Xu Y, Xu Y, Saria S, A bayesian nonparametric approach for estimating individualized treatment-response curves, in: Machine Learning for Healthcare Conference, 2016, pp. 282–300. [Google Scholar]
- [68].Haneuse S, Daniels M, A general framework for considering selection bias in ehr-based studies: what data are observed and why?, eGEMs 4 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Abacha AB, Chowdhury MFM, Karanasiou A, Mrabet Y, Lavelli A, Zweigenbaum P, Text mining for pharmacovigilance: Using machine learning for drug name recognition and drug-drug interaction extraction and classification, Journal of biomedical informatics 58 [2015] 122–132. [DOI] [PubMed] [Google Scholar]
- [70].Mower J, Subramanian D, Cohen T, Learning predictive models of drug side-effect relationships from distributed representations of literature-derived semantic predications, Journal of the American Medical Informatics Association 25 [10] [2018] 1339–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Kim HH, Rhew KY, A machine learning approach to classification of case reports on adverse drug reactions using text mining of expert opinions, in: Advances in Computer Science and Ubiquitous Computing, Springer, 2017, pp. 1072–1077. [Google Scholar]
- [72].Mikolov T, Chen K, Corrado G, Dean J, Efficient estimation of word representations in vector space, arXiv preprint arXiv:1301.3781. [Google Scholar]
- [73].Sutskever I, Vinyals O, Le QV, Sequence to sequence learning with neural networks, in: Advances in neural information processing systems, 2014, pp. 3104–3112. [Google Scholar]
- [74].Luong M-T, Pham H, Manning CD, Effective approaches to attention-based neural machine translation, arXiv preprint arXiv: 1508.04025. [Google Scholar]
- [75].Britz D, Goldie A, Luong M-T, Le Q, Massive exploration of neural machine translation architectures, arXiv preprint arXiv:1703.03906. [Google Scholar]
- [76].Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, Kaiser L, Polosukhin I, Attention is all you need, in: Advances in Neural Information Processing Systems, 2017, pp. 5998–6008. [Google Scholar]
- [77].Luo Y, Thompson WK, Herr TM, Zeng Z, Berendsen MA, Jonnalagadda SR, Carson MB, Starren J, Natural language processing for ehr-based pharmacovigilance: a structured review, Drug safety 40 (11) (2017) 1075–1089. [DOI] [PubMed] [Google Scholar]
- [78].Dev S, Zhang S, Voyles J, Rao AS, Automated classification of adverse events in pharmacovigilance, in: Bioinformatics and Biomedicine (BIBM), 2017 IEEE International Conference on, IEEE, 2017, pp. 1562–1566. [Google Scholar]
- [79].Zhang S, Dev S, Voyles J, Rao AS, Attention-based multi-task learning in pharmacovigilance, in: 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), IEEE, 2018, pp. 2324–22328. [Google Scholar]
- [80].Li F, Liu W, Yu H, Extraction of information related to adverse drug events from electronic health record notes: Design of an end-to-end model based on deep learning, JMIR medical informatics 6 (4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Yang X, Bian J, Gong Y, Hogan WR, Wu Y, Madex: A system for detecting medications, adverse drug events, and their relations from clinical notes, Drug safety (2019) 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Sarker A, Ginn R, Nikfarjam A, OConnor K, Smith K, Jayaraman S, Upadhaya T, Gonzalez G, Utilizing social media data for pharmacovigilance: a review, Journal of biomedical informatics 54 (2015) 202–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Tricco AC, Zarin W, Lillie E, Jeblee S, Warren R, Khan PA, Robson R, Hirst G, Straus SE, et al. , Utility of social media and crowd-intelligence data for pharmacovigilance: a scoping review, BMC medical informatics and decision making 18 (1) (2018) 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Comfort S, Perera S, Hudson Z, Dorrell D, Meireis S, Nagarajan M, Ramakrishnan C, Fine J, Sorting through the safety data haystack: Using machine learning to identify individual case safety reports in socialdigital media, Drug safety 41 (6) (2018) 579–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Calix RA, Gupta R, Gupta M, Jiang K, Deep gramulator: Improving precision in the classification of personal health-experience tweets with deep learning, in: Bioinformatics and Biomedicine (BIBM], 2017 IEEE International Conference on, IEEE, 2017, pp. 1154–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Cocos A, Fiks AG, Masino AJ, Deep learning for pharmacovigilance: recurrent neural network architectures for labeling adverse drug reactions in twitter posts, Journal of the American Medical Informatics Association 24 (4] (2017] 813–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Liu J, Wang G, Pharmacovigilance from social media: An improved random subspace method for identifying adverse drug events, International Journal of Medical Informatics 117 (2018] 33–43. [DOI] [PubMed] [Google Scholar]
- [88].Nikfarjam A, Sarker A, Oconnor K, Ginn R, Gonzalez G, Pharmacovigilance from social media: mining adverse drug reaction mentions using sequence labeling with word embedding cluster features, Journal of the American Medical Informatics Association 22 (3] (2015] 671–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Wu Z, Li W, Liu G, Tang Y, Network-based methods for prediction of drug-target interactions, Frontiers in pharmacology 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Dana D, Gadhiya S, St Surin L, Li D, Naaz F, Ali Q, Paka L, Yamin M, Narayan M, Goldberg I, et al. , Deep learning in drug discovery and medicine; scratching the surface, Molecules 23 (9] (2018] 2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Hammann F, Schöning V, Drewe J, Prediction of clinically relevant drug-induced liver injury from structure using machine learning, Journal of Applied Toxicology. [DOI] [PubMed] [Google Scholar]
- [92].Zhang L, Zhang H, Ai H, Hu H, Li S, Zhao J, Liu H, Applications of machine learning methods in drug toxicity prediction, Current topics in medicinal chemistry 18 (12] (2018] 987–997. [DOI] [PubMed] [Google Scholar]
- [93].Benhenda M, Chemgan challenge for drug discovery: can ai reproduce natural chemical diversity?, arXiv preprint arXiv:1708.08227. [Google Scholar]
- [94].Preuer K, Renz P, Unterthiner T, Hochreiter S, Klambauer G, Fréchet chemblnet distance: A metric for generative models for molecules, arXiv preprint arXiv: 1803.09518. [DOI] [PubMed] [Google Scholar]
- [95].LeCun Y, Boser BE, Denker JS, Henderson D, Howard RE, Hubbard WE, Jackel LD, Handwritten digit recognition with a back-propagation network, in: NIPS, 1989. [Google Scholar]