Abstract
Studies measuring dichlorodiphenyltrichloroethane (DDT) exposure during key windows susceptibility including the intrauterine period suggest that DDT exposure is associated with breast cancer risk. We hypothesized that prenatal DDT exposure is associated with DNA methylation. Using prospective data from 316 daughters in the Child Health and Development Study, we examined the association between prenatal exposure to DDTs and DNA methylation in blood collected in midlife (mean age: 49 years). To identify differentially methylated regions (DMRs) associated with markers of DDTs (p,p’-DDT and the primary metabolite of p,p’-DDT, p,p’-DDE, and o,p’-DDT, the primary constituents of technical DDT), we measured methylation in 30 genes important to breast cancer. We observed DDT DMRs in three genes, CCDC85A, CYP1A1 and ZFPM2, each of which has been previously implicated in pubertal development and breast cancer susceptibility. These findings suggest prenatal DDT exposure may have life-long consequence through alteration in genes relevant to breast cancer.
Keywords: children cohort, DDT, DNA methylation, prenatal exposure, organochlorine, white blood cells, windows of susceptibility
Introduction
The pesticide dichlorodiphenyl-trichloroethane (p,p’-DDT, referred to generically as the pesticide, DDT), its primary metabolite (dichlorodiphenyldichloroethylene, p,p’-DDE) and an isomer that was a contaminant of commercial DDT (orthoparaDDT or o,p’-DDT) are endocrine disrupting chemicals (EDCs).(1) DDT was used widely in the U. S. beginning in 1945, peaking in 1959 and was banned in 1972.(2) Evidence from both in vivo and in vitro assays demonstrates that DDT can impact estrogen signaling through the ligand binding domain of ERα and ERβ.(3–8) DDT and o,p´-DDT, the most estrogenic components, also support the growth of breast tumors in animal models.(9, 10) Thus, DDT exposure may affect breast cancer risk both as a direct carcinogen and through alterations in hormonal signaling.(10, 11)
The mammary gland undergoes several developmental changes during the intrauterine period, puberty, childbearing and menopause which may be more susceptible windows for exogenous or endogenous carcinogenic influences.(12–15) In a prospective study of pregnant women in the Child Health and Development Studies (CHDS), Cohn and colleagues reported that high levels of serum p,p’-DDT were associated with a 5-fold increase in risk of breast cancer in women exposed to DDT prior to age 14 years, while there was no association in women who were not exposed before age 14.(16) Furthermore, CHDS daughters who had high in utero exposures to o,p’-DDT had a nearly four-fold increased risk of breast cancer after 54-years of follow-up.(17) These findings support the hypothesis that environmental exposures during windows of susceptibility when the breast is developing may increase the risk of breast cancer.(18) Women highly exposed to DDT in utero include women born in the 1960s, before DDT was banned. These women are just now approaching the age of increased risk for breast cancer. Thus the potential association of prenatal DDT exposure with breast cancer is relevant to the search for midlife biomarkers of risk.
Epigenetic biomarkers, such as DNA methylation, can change the activity of a DNA segment without changing the underlying DNA sequence and are essential to developmental processes and genomic imprinting.(19–21) White blood cells (WBCs) are common and readily accessible sources of DNA to determine methylation differences related to exposures.(22–25) Measuring methylation in repetitive elements in WBC DNA, we previously found overall methylation levels were lower in women with breast cancer compared with their unaffected sister controls.(26, 27) Increasing evidence also suggests that many breast cancer susceptibility genes may be altered through epigenetic alterations. For example, silencing of BRCA1 by methylation has been observed in breast cancer tissues,(28–31) and in WBCs.(32–34)
Fetal development is a critical time period when most of the epigenetic landscape is established,(21) and environmental exposures during this lifestage in addition to other vulnerable windows of susceptibility may alter epigenetic alternations.(35–37) For example, many epigenome-wide association studies reported maternal cigarette smoking during pregnancy was associated with altered DNA methylation in specific CpG sites in infants, children and adolescents.(38–42) Using information from the New York Women’s Birth Cohort, we previously reported that there are persistent DNA methylation changes in these specific CpG sites in midlife associated with prenatal smoking exposure.(43) These observations suggest that maternal smoking during pregnancy is associated with offspring DNA methylation in WBC across the lifecourse. Animal studies have demonstrated that EDCs may affect long-term health outcomes through epigenetic mechanisms (reviewed in (44)). Some limited evidence exists for DDT exposure and DNA methylation alterations in humans.(45–48)
In this report we test the hypothesis that DDT-associated changes in DNA methylation at midlife could account for DDT associations with breast cancer risk we observed in the Child Health and Development Studies cohort (CHDS). We investigated this hypothesis by examining the relation of in utero DDT exposure to methylation of breast cancer-associated genes at midlife in CHDS daughters.
Materials and methods
Study participants
Female offspring who were born into the Oakland, California based CHDS pregnancy cohort from 1959–1967 and who participated in the “Three Generations of Breast Cancer (3Gs)” or the Health Disparities Study (DISPAR), in adulthood at ages 44–54, between 2010 and 2013, formed the basis for this study. Details of the 3Gs and DISPAR study recruitment and response are described elsewhere.(49, 50) In order to be eligible for the current study, Prenatal Environmental Determinants of Intergenerational Risk (PEDIGREE), participants were further required to complete a home visit and/or provide a bio-specimen (saliva or blood sample) and to have an existing or pending mammogram within a year of recruitment. Using these criteria, two groups were invited to participate: daughters whose mothers had been diagnosed with breast cancer (n=231) and daughters whose mothers had not been known to have a breast cancer diagnosis as of the time of recruitment (n=281). Authorization to collect mammography was received from 491 (96%) and mammograms were successfully collected for 397 (81%). For this study, WBC DNA with consent was available for 335 (84%) participants. We further required available data on all study variables (organochlorine measures, age, race/ethnicity and body mass index) to achieve our final analysis sample (n=316). The study was approved by the Institutional Review Boards of Columbia University and the Public Health Institute (Oakland, California).
Exposure constructs.
We measured DDT using maternal serum samples collected from each mother 1–3 days after she gave birth.(16) Specifically, we measured p,p´-DDT, the active ingredient of DDT; o,p´-DDT, a low concentration contaminant; and p,p´-DDE, the most abundant p,p´-DDT metabolite (for details see(16, 51)).
Blood-based Biomarkers in Midlife.
We profiled DNA methylation levels in 30 candidate genes selected based on published genetic or epigenetic association with at least one or more of the following categories: (i) genes that are associated with breast cancer identified in genome-wide association studies (GWAS), or mutations related to breast cancer risk,(52, 53) (ii) genes related to age at menarche from GWAS,(54) (iii) genes related to growth and development from GWAS(55) or EWAS(56) (iv) genes involved in DNA recombination and repair (http://sciencepark.mdanderson.org/labs/wood/dna_repair_genes.html), and (v) selected candidate CpG sites from a previous study of adolescents that showed evidence of methylation changes between girls with and without a breast cancer family history in our exploratory genome-wide DNA methylation profiling from pilot work in 48 girls,(57) but outside the above selection criteria (Supplementary Table 1). For each locus, we selected the bis-seq primer locations based on the chromatin states defined by Ernst et al.(58) and available in the UCSC human genome browser.(59) We focused on active promoters and enhancers, insulators, and poised chromatin, since these regions are implicated in gene regulation in cancer (60–62) and are often enriched in disease‐associated DMRs.(63–65) In addition, we covered a few loci with repressed chromatin or transcription associated chromatin states. When indicated, annotation of GWAS peaks was performed using the NHGRI-EBI catalog (53) and literature searches for smaller-scale genetic association studies.
DNA extraction and bisulfite treatment
We extracted genomic DNA from whole blood samples by a salting out procedure; lysing cells with SDS in a nuclei lysis buffer and treating with RNase A (final 133 μg/mL) and RNase T1 (final 20 units/mL) to remove RNA. We coprecipitated proteins with NaCl (330 μL of saturated NaCl added per 1mL solution) by centrifugation and recovered genomic DNA from the supernatant by precipitation with 100% ethanol, washed it in 70% ethanol, and dissolved the DNA in the Tris-EDTA buffer. We bisulfite-converted genomic DNA (500 ng) using the EpiTect Bisulfite Kit (Qiagen, Germantown, MD), as per the manufacturer’s instructions and resuspended the DNA in 20uL of distilled water with storage at −20°C. We performed all DNA methylation assays blinded to DDT exposure data.
Targeted bisulfite DNA sequencing
We examined DNA methylation at the 30 selected loci using targeted massive parallel bisseq on genomic DNA from the WBC samples, as described.(66) Oligonucleotide primers were designed in MethPrimer(67) (Supplemental Table 1). Bisulfite-converted DNA was amplified and barcoded by PCR on a Fluidigm AccessArray high throughput PCR machine, followed by Nextgen (Illumina MiSeq) sequencing, as described.(66) After trimming for adaptors, low-quality bases (Phred score<30) and sequenced DNA fragments (reads) with a length <40 bp with TrimGalore (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/), the reads were aligned to the human genome (GRCh37 build) using Bismark aligner.(68) We used the Bismark extractor for methylation calling and since low coverage and low PCR complexity can lead to less representative estimates of DNA methylation levels, we filtered out values where the coverage was less than 100 reads and the complexity for each amplicon (estimated by the number of DNA methylation patterns represented at least 10 times) was less than the median complexity score of the amplicon across samples +/−1 standard deviation (SD). To rule out technical outliers due to low PCR complexity, which can occur due to amplification of limiting amounts of staring DNA, we applied a filtering algorithm based on the complexity of the sequenced DNA fragments. The complexity score was estimated by the number of DNA methylation patterns represented at least 10 times. Values where the complexity score was less than the mean score of the given amplicon across samples minus 1 SD were filtered out. Post complexity QC filtering showed lower but still high variance in methylation level. Therefore, all analyses presented in this paper have been performed on the filtered data. We used Bismark software to determine the percentage of methylation at each CpGs and then calculated the mean percentage methylation in each amplicon/gene by averaging the percentages of methylation across all CpGs in the amplicon/gene.
Statistical methods
We calculated the percentage of methylation at each CpG position by dividing the total number of methylated reads by the total number of reads (sum of methylated and unmethylated reads). While methylation differences at single CpG sites might have biological relevance, alteration of regulatory DNA elements often induces methylation changes affecting multiple contiguous CpGs spanning up to several kb of DNA. Each amplicon/gene contained a different number of CpG sites (Supplemental Table 1). We averaged percentages of methylation at each CpG across all CpGs in the amplicon/gene and use that value as an indication of percentage of methylation for each amplicon/gene for further data analysis. To assess the association between DNA methylation and in utero DDT exposure, we compared percentage of methylation for each amplicon/gene across different levels of markers of DDT exposure in maternal serum using the Kruskal-Wallis test. The rejection of the null hypothesis concludes that there is no difference in percentage of DNA methylation of the gene among the levels of DDT exposure. We divided each daughter participant into different DDT exposure groups using the cutpoint values of each markers of DDTs from our previous study:(17) for p,p´-DDT (low exposure, < 8.09 μg/L; median exposure, 8.09–13.90 μg/L; high exposure, > 13.90 μg/L), o,p´-DDT (low exposure, ≤ 0.42 μg/L; median exposure, 0.43–0.72 μg/L; high exposure, > 0.72 μg/L) and p,p´-DDE (low exposure, ≤ 35.23 μg/L; median exposure, > 35.23–58.49 μg/L; high exposure, > 58.49 μg/L). We used these cuptoint values as our previous study showed an association of prenatal DDT exposure with breast cancer and thus might have biological relevance. Both age and race/ethnicity were associated with DDT exposure,(16, 49) and DNA methylation.(69, 70) For each DMR, we carried out a multivariable analysis adjusting for age and race/ethnicity using linear regression models with percentage of DNA methylation as the outcome, and categorical variables for markers of DDT exposure. We also tested interaction with family history of breast cancer. All analyses were performed with SAS software 9.4 (SAS Institute, Cary, NC) and R.3.10.
Results
Table 1 presents the distributions of selected characteristics and levels of prenatal DDT exposure of participants. The mean age of daughter participants when blood was drawn was 49.3 years (SD=2.0). The mean levels of prenatal DDT exposure were 12.4 (SD=7.7, μg/L) for p,p’-DDT, 47.0 (SD=20.4, μg/L) for p,p’-DDE and 0.51 (SD=0.44, μg/L) for o,p’-DDT. Overall, the percentages of daughter participants with high prenatal DDT exposure were 30.4% for p,p’-DDT, 21.8% for p,p’-DDE and 21.8% for o,p’-DDT. Supplemental Table 2 present the spearman correlation of prenatal DDT exposure with age and race/ethnicity. Consist with previous studies, (16, 49), prenatal DDT exposure were associated with age and race/ethnicity.
Table 1.
Demographic characteristics and in utero DDT exposure of daughters in the Child Health and Development Studies Pregnancy Cohort (N=316)
| Variable | Mean, SD |
|---|---|
| Age | 49.3 (2.0) |
| BMI | 28.0 (7.3) |
| p-p’-DDT, μg/L | 12.4 (7.7) |
| p-p’-DDE, μg/L | 47.0 (20.4) |
| o-p’-DDT, μg/L | 0.51 (0.44) |
| Race/Ethnicity of mother | No, % |
| White | 226 (71.5) |
| Black | 49 (15.5) |
| Hispanic | 12 (3.8) |
| Asian | 14 (4.4) |
| Other | 15 (4.8) |
| p-p’-DDT | |
| Low (<8.09, μg/L) | 112 (35.4) |
| Median (8.09–13.90, μg/L) | 108 (34.2) |
| High (>13.90, μg/L) | 96 (30.4) |
| p-p’-DDE | |
| Low (≤35.23, μg/L) | 108 (34.2) |
| Median (>35.23–58.49, μg/L) | 139 (44.0) |
| High (>58.49, μg/L) | 69 (21.8) |
| o-p’-DDT | |
| Low (≤0.42, μg/L) | 166 (52.5) |
| Median (0.43–0.72, μg/L) | 81 (25.6) |
| High (>0.72, μg/L) | 69 (21.8) |
| Mother has breast cancer | |
| No | 173 (54.8) |
| Yes | 143 (45.2) |
SD: standard deviation
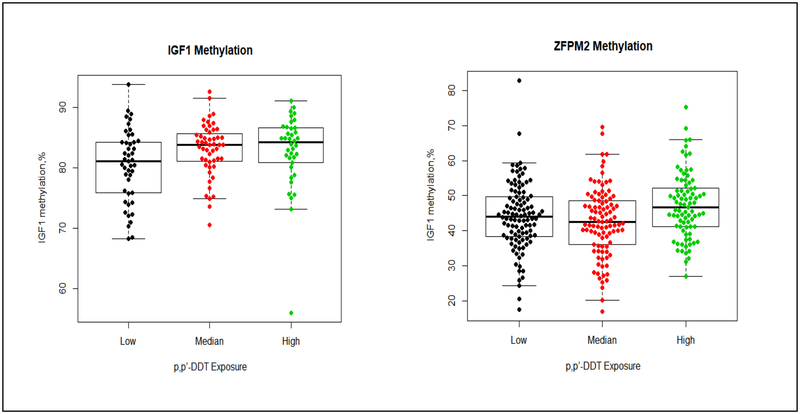
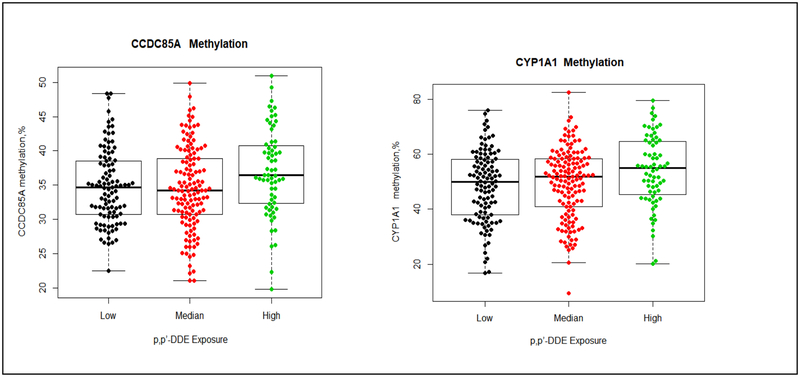
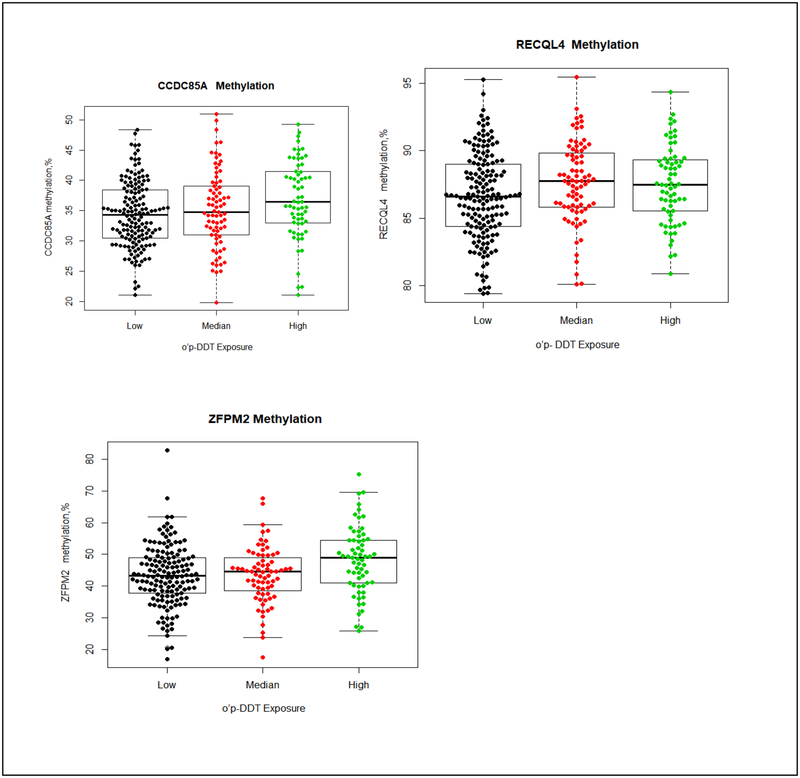
The distribution of methylation in the 30 genes by prenatal DDT exposure are summarized in Tables 2–4. Using the Kruskal-Wallis test to compared values of percentage of methylation across different DDTs exposure groups without adjusting any covariate, we observed higher prenatal p,p’-DDT exposure was associated with higher methylation in IGF1 and ZFPM2. Methylation in IGF1 was 3.2% higher in daughter participants with higher p,p’-DDT than participants with lower exposure (81.1±8.4%, 83.8±4.9%, and 84.2±5.8% for <8.09, 8.09–13.90 and >13.90, μg/L of p,p’-DDT, p=0.03). The median of percentage of methylation levels in ZFPM2 were 44.1± 11.2%, 42.5 ±12.5% and 46.6 ±11.0% (p=0.009) for <8.09, 8.09–13.90 and >13.90, μg/L of p,p’-DDT (Table 2 and Figure 1). Higher methylation in CCDC85A, and CYP1A1 were associated with higher prenatal p,p’-DDE exposure; methylation in CCDC85A, and CYP1A1 were 1.7%, and 5.0% higher in daughter participants with the highest p,p’-DDE than participants with lowest exposure (Table 3 and Figure 2). Methylation in CCDC85A, RECQL4 and ZFPM2 were associated with intrauterine o,p’-DDT exposure (Table 4 and Figure 3). The difference in methylation between highest and lowest o,p’-DDT groups ranged from 0.9% for RECQL4 to 5.6% for ZFPM2.
Table 2.
Distribution of methylation levels in 30 genes by p-p’-DDT exposure of daughters in the Child Health and Development Studies Pregnancy Cohort
| p-p’-DDT <8.09, μg/L | p-p’-DDT: 8.09–13.90, μg/L | p-p’-DDT >13.90, μg/L | |||||
|---|---|---|---|---|---|---|---|
| Gene | Median, % | IQR | Median, % | IQR | Median, % | IQR | P.value |
| ADRB1 | 24.4 | 9.0 | 24.5 | 9.4 | 24.3 | 8.7 | 0.76 |
| ARHGEF7 | 44.7 | 10.3 | 46.5 | 8.3 | 44.4 | 9.4 | 0.61 |
| BRCA1 | 81.2 | 11.2 | 82.4 | 12.5 | 83.1 | 11.0 | 0.47 |
| CCDC85A | 34.7 | 7.7 | 34.5 | 9.9 | 35.7 | 8.2 | 0.57 |
| CCNL1 | 0.2 | 0.6 | 0.5 | 1.1 | 0.2 | 0.5 | 0.05 |
| CDH1 | 7.4 | 2.1 | 7.1 | 2.4 | 7.7 | 2.9 | 0.07 |
| CELF4 | 56.6 | 10.7 | 55.6 | 11.9 | 54.7 | 12.3 | 0.60 |
| chr11 | 7.7 | 4.1 | 8.2 | 4.3 | 8.4 | 5.0 | 0.26 |
| chr12 | 78.5 | 4.9 | 78.3 | 5.4 | 78.4 | 4.8 | 0.60 |
| chr16 | 81.7 | 5.4 | 82.3 | 6.2 | 81.4 | 5.8 | 0.73 |
| chr4 | 81.9 | 8.6 | 81.2 | 8.9 | 81.8 | 9.0 | 0.44 |
| CYP1A1 | 51.2 | 20.4 | 50.8 | 17.5 | 53.1 | 16.2 | 0.09 |
| DLGAP2 | 97.1 | 1.1 | 97.1 | 1.2 | 96.9 | 1.1 | 0.24 |
| ESR1 | 54.2 | 20.3 | 55.1 | 19.7 | 55.2 | 21.3 | 0.69 |
| GAB2 | 96.8 | 2.6 | 97.2 | 2.8 | 97.0 | 3.5 | 0.30 |
| GNA12 | 38.4 | 17.9 | 35.0 | 25.9 | 38.8 | 23.5 | 0.47 |
| IGF1 | 81.1 | 8.4 | 83.8 | 4.9 | 84.2 | 5.8 | 0.03 |
| MCHR2 | 13.3 | 3.8 | 13.3 | 3.5 | 12.9 | 4.4 | 0.85 |
| OBSCN | 92.4 | 1.8 | 92.8 | 1.9 | 92.4 | 2.0 | 0.09 |
| PCDHGB1 | 39.5 | 7.8 | 41.3 | 7.7 | 39.7 | 8.3 | 0.15 |
| PEX14 | 66.7 | 7.4 | 66.2 | 6.2 | 65.8 | 8.2 | 0.67 |
| RAD51L1 | 98.2 | 2.0 | 98.2 | 1.2 | 98.0 | 1.5 | 0.44 |
| RECQL4 | 86.7 | 4.6 | 87.2 | 4.3 | 87.5 | 3.8 | 0.25 |
| SLC39A14 | 87.3 | 10.1 | 87.2 | 9.8 | 87.3 | 12.1 | 0.63 |
| SLC6A3 | 73.8 | 4.7 | 73.8 | 6.5 | 74.6 | 5.2 | 0.41 |
| TCF7L2 | 52.4 | 14.2 | 49.9 | 16.2 | 51.4 | 15.3 | 0.44 |
| TERT | 96.6 | 3.0 | 96.9 | 2.4 | 96.7 | 2.9 | 0.56 |
| XRCC3 | 79.7 | 3.3 | 79.5 | 2.8 | 79.9 | 4.1 | 0.86 |
| ZFPM2 | 44.1 | 11.2 | 42.5 | 12.5 | 46.6 | 11.0 | 0.0090 |
| ZNF483 | 35.7 | 4.3 | 36.4 | 3.1 | 35.4 | 3.2 | 0.56 |
p value of Kruskal-Wallis test
IQR: The interquartile range
Table 4.
Distribution of methylation levels in 30 genes by o,p’-DDT exposure of daughters in the Child Health and Development Studies Pregnancy Cohort
| o,p’-DDT ≤0.42, μg/L | o,p’-DDT: 0.43–0.72, μg/L | o,p’-DDT >0.72, μg/L | |||||
|---|---|---|---|---|---|---|---|
| Gene | Median | IQR | Median | IQR | Median | IQR | P.value |
| ADRB1 | 24.3 | 8.9 | 24.8 | 8.6 | 25.1 | 11.6 | 0.43 |
| ARHGEF7 | 45.5 | 8.6 | 44.4 | 9.3 | 44.6 | 9.2 | 0.90 |
| BRCA1 | 81.9 | 12.1 | 81.2 | 12.8 | 82.3 | 10.5 | 0.97 |
| CCDC85A | 34.3 | 8.0 | 34.8 | 8.0 | 36.4 | 8.5 | 0.01 |
| CCNL1 | 0.3 | 0.8 | 0.3 | 0.7 | 0.3 | 0.6 | 0.90 |
| CDH1 | 7.4 | 2.2 | 7.1 | 3.0 | 7.9 | 2.9 | 0.05 |
| CELF4 | 56.5 | 10.0 | 56.3 | 12.6 | 55.0 | 12.4 | 0.40 |
| chr11 | 8.0 | 4.4 | 8.2 | 4.7 | 7.5 | 5.8 | 0.90 |
| chr12 | 78.6 | 5.0 | 78.1 | 5.2 | 78.1 | 4.9 | 0.95 |
| chr16 | 81.6 | 5.5 | 82.3 | 6.2 | 81.9 | 5.8 | 0.17 |
| chr4 | 81.4 | 8.6 | 81.5 | 9.6 | 82.1 | 8.3 | 0.88 |
| CYP1A1 | 50.3 | 19.7 | 51.9 | 16.4 | 54.0 | 14.5 | 0.05 |
| DLGAP2 | 97.1 | 1.0 | 97.1 | 1.2 | 97.0 | 1.3 | 0.75 |
| ESR1 | 55.0 | 19.2 | 54.9 | 22.1 | 55.2 | 21.1 | 0.98 |
| GAB2 | 97.0 | 2.7 | 96.9 | 2.7 | 97.1 | 3.5 | 0.96 |
| GNA12 | 36.0 | 20.8 | 37.8 | 23.8 | 42.4 | 26.8 | 0.35 |
| IGF1 | 83.2 | 5.5 | 83.4 | 7.2 | 83.1 | 9.5 | 0.92 |
| MCHR2 | 13.2 | 3.5 | 12.7 | 4.5 | 13.4 | 3.9 | 0.37 |
| OBSCN | 92.7 | 1.8 | 92.5 | 1.7 | 91.9 | 2.1 | 0.05 |
| PCDHGB1 | 40.1 | 7.7 | 40.2 | 7.9 | 40.5 | 9.4 | 0.66 |
| PEX14 | 65.6 | 7.2 | 67.8 | 7.5 | 66.5 | 8.1 | 0.37 |
| RAD51L1 | 98.1 | 1.8 | 98.3 | 1.3 | 98.1 | 1.8 | 0.33 |
| RECQL4 | 86.6 | 4.6 | 87.8 | 4.0 | 87.5 | 3.9 | 0.03 |
| SLC39A14 | 87.2 | 8.6 | 87.6 | 10.1 | 87.2 | 15.9 | 0.45 |
| SLC6A3 | 73.5 | 5.2 | 74.5 | 6.2 | 74.5 | 5.1 | 0.13 |
| TCF7L2 | 52.5 | 13.0 | 49.6 | 15.8 | 51.4 | 16.2 | 0.28 |
| TERT | 96.7 | 2.8 | 96.6 | 2.8 | 96.8 | 2.8 | 0.96 |
| XRCC3 | 79.7 | 3.2 | 79.7 | 3.7 | 79.7 | 4.5 | 0.81 |
| ZFPM2 | 43.3 | 11.2 | 44.6 | 11.9 | 48.9 | 13.5 | 0.01 |
| ZNF483 | 36.1 | 3.6 | 35.5 | 3.1 | 35.9 | 4.0 | 0.23 |
p value of Kruskal-Wallis test
IQR: The interquartile range
Figure 1. Methylation differences by p,p’-DDT in IGF1, and ZFPM2.
The boxplot displays the distribution of CpG methylation values (percent) by different p,p’-DDT exposure group (low exposure, < 8.09 μg/L; median exposure, 8.09–13.90 μg/L; high exposure, > 13.90 μg/L) in 2 gene regions (IGF1 and ZFPM2) examined. The middle bold line represents the median methylation. Each dot represents the methylation value of each samples. The distribution of percentage of IGF1 methylation (left) by each p,p’-DDT exposure group: low exposure (median: 81.1%, Q1-Q3=(75.8%, 84.3%), Min-Max (68.2%, 93.8%)); median exposure (median:83.8%, Q1-Q3=(81.0%, 85.9%), Min-Max=(70.6%, 92.6%)); high exposure (median: 84.2%, Q1-Q3=(80.9%, 86.7%), Min-Max=(56.0%, 91.1%)). The distribution of percentage of ZFPM2 methylation (right) by each p,p’-DDT exposure group: low exposure (median: 44.1%, Q1-Q3=(38.4%, 49.6%), Min-Max= (17.4%, 82.9%); median exposure (median: 42.5%, Q1-Q3=(36.1%, 48.6%), Min-Max=(16.9%, 69.6%)); high exposure (median:46.6%, Q1-Q3=(41.1%, 52.1%), Min-Max=(26.9%, 75.3%)).
Table 3.
Distribution of methylation levels in 30 genes by p,p’-DDE exposure of daughters in the Child Health and Development Studies Pregnancy Cohort
| p,p’-DDE ≤35.23, μg/L | p,p’-DDE :35.23–58.49, μg/L | p,p’-DDE >58.49, μg/L | |||||
|---|---|---|---|---|---|---|---|
| Gene | Median, % | IQR | Median, % | IQR | Median, % | IQR | P.value |
| ADRB1 | 23.5 | 9.3 | 24.3 | 8.6 | 26.5 | 9.8 | 0.14 |
| ARHGEF7 | 44.0 | 9.5 | 45.9 | 8.9 | 44.5 | 9.1 | 0.29 |
| BRCA1 | 80.7 | 14.0 | 82.4 | 10.2 | 82.9 | 11.3 | 0.30 |
| CCDC85A | 34.7 | 7.7 | 34.2 | 8.2 | 36.4 | 8.4 | 0.02 |
| CCNL1 | 0.3 | 0.8 | 0.3 | 0.8 | 0.2 | 0.6 | 0.49 |
| CDH1 | 7.3 | 2.0 | 7.3 | 2.6 | 7.7 | 3.0 | 0.19 |
| CELF4 | 55.7 | 11.1 | 57.3 | 11.3 | 55.7 | 13.1 | 0.54 |
| chr11 | 7.7 | 3.9 | 8.5 | 5.2 | 8.2 | 5.2 | 0.32 |
| chr12 | 78.7 | 4.6 | 78.1 | 5.1 | 78.2 | 5.2 | 0.32 |
| chr16 | 81.6 | 5.4 | 82.0 | 6.4 | 82.1 | 6.3 | 0.21 |
| chr4 | 82.3 | 8.0 | 81.4 | 8.4 | 80.9 | 9.8 | 0.36 |
| CYP1A1 | 50.0 | 20.1 | 51.8 | 17.3 | 55.0 | 19.2 | 0.03 |
| DLGAP2 | 97.2 | 1.1 | 96.9 | 1.2 | 97.1 | 1.0 | 0.17 |
| ESR1 | 55.1 | 19.6 | 55.0 | 20.6 | 56.0 | 23.7 | 0.88 |
| GAB2 | 96.9 | 2.4 | 97.2 | 2.7 | 97.0 | 3.4 | 0.70 |
| GNA12 | 37.9 | 21.1 | 35.7 | 21.4 | 39.7 | 32.4 | 0.63 |
| IGF1 | 82.2 | 4.3 | 83.2 | 7.4 | 84.5 | 6.6 | 0.23 |
| MCHR2 | 13.4 | 3.5 | 12.2 | 3.9 | 13.8 | 3.9 | 0.07 |
| OBSCN | 92.5 | 1.7 | 92.5 | 2.1 | 92.2 | 1.7 | 0.25 |
| PCDHGB1 | 40.6 | 7.8 | 40.3 | 8.0 | 39.9 | 9.1 | 0.73 |
| PEX14 | 67.0 | 7.1 | 65.8 | 7.5 | 67.5 | 8.9 | 0.25 |
| RAD51L1 | 98.1 | 1.5 | 98.1 | 1.7 | 98.0 | 1.9 | 0.97 |
| RECQL4 | 86.7 | 4.6 | 86.8 | 4.5 | 87.5 | 3.6 | 0.16 |
| SLC39A14 | 87.3 | 9.7 | 87.3 | 10.0 | 87.2 | 10.9 | 0.25 |
| SLC6A3 | 73.6 | 4.9 | 74.5 | 6.0 | 74.1 | 5.5 | 0.72 |
| TCF7L2 | 52.3 | 13.5 | 50.3 | 13.9 | 54.5 | 16.6 | 0.23 |
| TERT | 96.8 | 2.7 | 96.6 | 2.9 | 97.1 | 2.5 | 0.50 |
| XRCC3 | 79.6 | 3.6 | 79.9 | 3.2 | 79.7 | 3.3 | 0.94 |
| ZFPM2 | 44.2 | 11.4 | 44.5 | 11.9 | 44.6 | 10.9 | 0.59 |
| ZNF483 | 35.9 | 3.4 | 36.0 | 3.4 | 35.4 | 4.0 | 0.86 |
p value of Kruskal-Wallis test
IQR: The interquartile range
Figure 2. Methylation differences by p,p’-DDE in CCDC85A, and CYP1A1.
The boxplot displays the distribution of CpG methylation values (percent) by different p,p’-DDE exposure group (low exposure, ≤ 35.23 μg/L; median exposure, > 35.23–58.49 μg/L; high exposure, > 58.49 μg/L).) in 2 gene regions (CCDC85A and CYP1A1) examined. The middle bold line represents the median methylation. Each dot represents the methylation value of each samples. The distribution of percentage of CCDC85A methylation (left) by each p,p’-DDE exposure group: low exposure (median: 34.7%, Q1-Q3=(30.7%, 38.5%), Min-Max(22.5%, 48.3%)); median exposure (median:34.2%, Q1-Q3=(30.7.0%, 38.9%), Min-Max=(21.0%, 49.9%)); high exposure (median: 36.4%, Q1-Q3=(32.3%, 40.7%), Min-Max=(19.8%, 50.9%)). The distribution of percentage of CYP1A1 methylation (right) by each p,p’-DDE exposure group: low exposure (median: 50.0%, Q1-Q3=(38.1%, 58.2%), Min-Max= (16.8%, 76.1%); median exposure (median: 51.8%, Q1-Q3=(41.0%, 58.3%), Min-Max=(9.5%, 82.5%)); high exposure (median:55.0%, Q1-Q3=(45.4%, 64.6%), Min-Max=(20.2%, 79.6%)).
Figure 3. Methylation differences by o,p’-DDT in CCDC85A, RECQL4 and ZFPM2.
The boxplot displays the distribution of CpG methylation values (percent) by different o,p’-DDT exposure group (low exposure, ≤ 0.42 μg/L; median exposure, 0.43–0.72 μg/L; high exposure, > 0.72 μg/L) in 3 gene regions (CCDC85A, RECQL4 and ZFPM2) examined. The middle bold line represents the median methylation. Each dot represents the methylation value of each samples. The distribution of percentage of CCDC85A methylation (top left) by each o,p’-DDT exposure group: low exposure (median: 34.3%, Q1-Q3=(30.5%, 38.5%), Min-Max(21.0%, 48.3%)); median exposure (median:34.8%, Q1-Q3=(31.0%, 39.1%), Min-Max=(19.8%, 50.9%)); high exposure (median: 36.4%, Q1-Q3=(33.0%, 41.5%), Min-Max=(21.0%, 49.3%)). The distribution of percentage of RECQL4 methylation (top right) by each o,p’-DDT exposure group: low exposure (median: 86.7%, Q1-Q3=(84.4%, 89.0%), Min-Max= (79.4%, 95.3%); median exposure (median: 87.8%, Q1-Q3=(85.8%, 89.8%), Min-Max=(80.1%, 95.4%)); high exposure (median:87.5%, Q1-Q3=(85.5%, 89.4%), Min-Max=(80.9%, 94.3%)). The distribution of percentage of ZFPM2 methylation (down left) by each o,p’-DDT exposure group: low exposure (median: 43.3%, Q1-Q3=(37.8%, 49.0%), Min-Max(16.9%, 82.9%)); median exposure (median:44.6%, Q1-Q3=(37.8%, 49.7%), Min-Max=(17.4%, 67.8%)); high exposure (median: 48.9%, Q1-Q3=(41.0%, 54.5%), Min-Max=(25.7%, 75.3%)).
We present analyses adjusted for age and race/ethnicity in Table 5; the associations with CCDC85A, CYP1A1 and ZFPM2 and DDT remained. Higher p,p’-DDT, and p,p’-DDE were associated with 2.05% and 1.89 % higher CCDC85A methylation. Methylation in CYP1A1 was 3.68% (95%CI=−0.19, 7.56) higher in daughter participants with highest p,p’-DDT, 4.30% (95%CI=0.27,8.32) higher in daughter participants with the highest p,p’-DDE and 4.14% (95%CI=0.33, 7.94) higher daughter participants with the highest o,p’-DDT. Daughter participants with the highest p,p’-DDT had 3.79% (95%CI=0.76, 6.81) higher methylation in ZFPM2, and daughter participants with the highest o,p’-DDT had 4.31% (95%CI=1.33, 7.29) higher methylation. We did not see any significant interaction with a family history of breast cancer.
Table 5.
Multivariable Association between DNA Methylation and DDT exposure of daughters in the Child Health and Development Studies Pregnancy Cohort
| Median Exposure* | High Exposure* | ||||
|---|---|---|---|---|---|
| Gene | DDT Markers | β Estimate | 95% CL | β Estimate | 95% CL |
| CCDC85A | p-p’-DDT | 0.50 | (−1.21, 2.21) | 0.35 | (−1.50, 2.20) |
| p-p’-DDE | −0.46 | (−2.00, 1.08) | 2.05 | (0.17, 3,94) | |
| o-p’-DDT | 0.82 | (−0.83, 2.46) | 1.89 | (0.08, 3.70) | |
| CYP1A1 | p-p’-DDT | 0.12 | (−3.48, 3.71) | 3.68 | (−0.19, 7.56) |
| p-p’-DDE | 0.78 | (−2.48, 4.04) | 4.30 | (0.27, 8.32) | |
| o-p’-DDT | 0.40 | (−3.09, 3.89) | 4.14 | (0.33, 7.94) | |
| IGF1 | p-p’-DDT | 2.80 | (0.33, 5.26) | 2.37 | (−0.31, 5.05) |
| p-p’-DDE | 1.14 | (−1.14, 3.42) | 1.50 | (−1.21, 4.22) | |
| o-p’-DDT | −0.65 | (−3.14, 1.85) | 0.04 | (−2.55, 2.63) | |
| RECQL4 | p-p’-DDT | −0.22 | (−1.09, 0.66) | 0.53 | (−0.41, 1.48) |
| p-p’-DDE | −0.14 | (−0.94, 0.66) | 0.40 | (−0.59, 1.38) | |
| o-p’-DDT | 0.86 | (0.01, 1.71) | 0.72 | (−0.21, 1.65) | |
| ZFPM2 | p-p’-DDT | −1.52 | (−4.33, 1.29) | 3.79 | (0.76, 6.81) |
| p-p’-DDE | 1.21 | (−1.40, 3.82) | 0.62 | (−2.64, 3.88) | |
| o-p’-DDT | −0.55 | (−3.33, 2.23) | 4.31 | (1.33, 7.29) | |
Adjust for age and race/ethnicity
Cl: confidence interval
The cutpoint value of median exposure for p,p´-DDT (low exposure, < 8.09 μg/L; median exposure, 8.09–13.90 μg/L; high exposure, > 13.90 μg/L), o,p´-DDT (low exposure, ≤ 0.42 μg/L; median exposure, 0.43–0.72 μg/L; high exposure, > 0.72 μg/L) and p,p´-DDE (low exposure, ≤ 35.23 μg/L; median exposure, > 35.23–58.49 μg/L; high exposure, > 58.49 μg/L).
Discussion
Due to the estrogenic properties of DDT compounds,(3–8) a number of epidemiological studies have investigated DDT exposure in relation to breast cancer (Reviewed in(14)). Most evidence is based on DDT measured in blood specimens collected at midlife or later during time periods after DDT was banned. Evidence from these studies is weak and inconsistent.(16, 17, 71–80) The strongest evidence for DDT associations with breast cancer in human populations is based on the CHDS where DDT could be measured in blood specimens obtained from young women during active DDT use.(16) DDT levels measured at the time of cancer diagnosis were associated with stage/ aggressiveness of breast cancer.(81, 82) We observed the largest DDT associations with breast cancer for women exposed to DDT in utero or before puberty.(16, 17, 80) Here we investigated the possible contribution of DDT-related changes in DNA methylation of breast-cancer associated genes to breast cancer risk in the CHDS.
Increasing evidence suggests epigenetic effects of EDCs on human health.(45–48, 83, 84) We compared the methylation status of breast-cancer associated genes for daughter participants with different levels of prenatal DDT exposure to identify specific DMRs associated with breast cancer susceptibility. We observed three DDT DMRs located in genes that are involved in growth and development and breast cancer susceptibility, CCDC85A, CYP1A1 and ZFPM2, respectively.(85–89) All three DDT DMRs consistently showed higher methylation in daughter participants with higher prenatal DDT exposure measured by the primary constituents of commercial DDT; o,p´-DDT and p,p´-DDT, and p,p’-DDE, the primary metabolite of p,p´-DDT. Higher prenatal exposure to p,p´-DDT or o,p´-DDT was associated with higher methylation in the promoter regions of CCDC85A and ZFPM2, genes related to puberty development, and CYP1A1 which is a breast cancer susceptibility gene. Although the biological function of the DMRs in these regions is unclear, DNA methylation in promoter regions usually correlates inversely with gene expression.(90, 91) Methylation of the DDT associated-DMRs in ZFPM2, CCDC85A and CYP1A1 showed high inter-individual variability (Figures 1–3), suggesting the presence of methylation quantitative trait loci (mQTLs; a.k.a. meQTLs).
Animal and epidemiological studies implicate EDCs as a significant concern to public health.(92) EDCs can interfere with the endocrine system, resulting in adverse health outcomes.(93) Many animal studies have linked EDCs such as diethylstilbestrol (DES), polychlorinated biphenyl (PCBs) and DDT exposure to epigenetic modifications including DNA methylation changes, resulting in alteration in gene expression and chromosomal stability.(94–99) In particular, the mouse model had clearly established the ability of environmental factors to influence epigenetics thus promoting phenotypic changes later in development.(100, 101) Compared with controls, young rats exposed to DDT had lower methylation overall.(98) Rats treated in utero and postnatally with organochlorine pesticides and PCBs also showed decreased methylation in the tumor suppressor gene p16 (INK4a) compared to controls.(99) An in vitro study of rat ovarian cells observed that o,p’-DDT can suppress the expression of selected genes (such as cytochrome P450 side chain cleavage enzyme (P450scc), progesterone receptor (PR), and epidermal growth factor epiregulin (EREG)) in very low doses through an estrogen receptor-independent pathway.(102)
DNA methylation may play an important role in causing disease by silencing genes through hypermethylation or activating genes through hypomethylation.(103–106) Moreover, global decrease in 5-methylcytosine content might contribute to the reactivation of transposable elements and the generation of chromosomal instability.(45, 46, 107, 108) Many epidemiological studies of DNA methylation have focused on global DNA methylation measured as methylation value in repetitive elements such as LINE-1 and Alu or in CCGG sequences which is quantified by the LUMA assay as an indication of overall DNA methylation value in the sample. Studies focusing on identification of DMRs can measure epigenetic wide DNA methylation profile using sequence- and array-based technologies.(109) Advantages of the bis-seq method are the ability to examine methylation across multiple CpGs. Evidence related to EDCs and DNA methylation in humans is more limited. Using information from the Sister Study of 200 women, half of whose mothers retrospectively reported taking DES during pregnancy, Harlid et al.(110) examined the association of genome-wide DNA methylation with intrauterine DES exposure. Although they found 22 CpGs had nominal p values < 10−4, none achieved genome-wide significance after considering multiple comparisons (q<0.05).(110) The association of prenatal exposure to DDT and other organic pollutants measured in blood from mothers during pregnancy or at delivery with global DNA methylation markers including LINE1 and Alu in cord blood DNA was examined in a birth cohort of Mexican-American children. The cohort was established in 1999–2000 when DDT use was continuing in Mexico for Malaria control.(107) Higher prenatal DDT exposure was associated with lower Alu methylation at birth; with an β value (95%CI) of −0.37 (−0.69,−0.05) for o,p′ -DDT, and an β value (95%CI) of −0.33 (−0.64,−0.01) for p,p′-DDE.(107) They also examined prenatal DDT exposure and DNA methylation in 9-year old children, and found the same but nonsignificant trend of lower Alu methylation with prenatal polybrominated diphenyl ethers (PBDE) exposure.(107) In contrast, other studies examined global methylation in adults and found lower methylation in Alu was associated with high level of serum DDT.(45, 46) Our study found prenatal DDT exposure is associated with persistent change in DNA methylation of breast-cancer associated genes, in particular genes that are important to puberty development.
CYP1A1, the main cytochrome P450 enzymes, play an important role in the detoxification of environmental carcinogens.(111) Increasing evidence suggests that exposures including smoking can modify methylation in CYP1A1. Multiple CpG sites in the promoter regions of CYP1A1 have been associated with maternal smoking during prenatal exposure and some of these associations have extended to midlife.(43, 112, 113) Microarray analyses in human endometrial endothelial cells revealed that DDT affected biological processes such as the cell cycle, cell division, and lipid metabolism.(114) Wójtowicz et al. (115) studied the effects of DDT on gene expression in placental cells and found DDT/DDE inhibited the expression of CYP1A1 and AhR within 48 h after treatment. This study suggests that DDT exposure might affect the AhR/CYP1A1 signaling pathway. Studying the association between genetic polymorphisms and gene expression of CYP1A1 in breast tissue, Goth-Goldstein et al. (116) found there is a large variation in CYP1A1 expression in breast tissue and the variations could not explained by the variant genotype. Alteration in DNA methylation might be the underlying mechanism by which DDT regulates gene expression. Higher methylation in the enhancer of CYP1A1 was associated with lower mRNA levels.(117)
The breast undergoes many changes in early life and environmental factors may have a stronger effect on breast cancer risk during development and maturation of breast tissue.(78) The effect of DDT on breast cancer observed in the developmental stage of exposure such as prenatal and early life;(16, 17) contrasts with the lack of a clear association when DDT is measured in midlife may be due to this being outside of specific windows of susceptibility.(75, 76) Exposures during a crucial time of development can alter genome activity associated with the differentiation programming of cells or organ systems through epigenetic mechanism. Modification of the epigenome may continue throughout development, subsequently affecting the adult transcriptome and making tissues, such as the breast, susceptible to developing disease.(118)
A key strength of our study is that we prospectively examined the association between in utero exposure to DDT measured in maternal early postpartum serum with adult DNA methylation changes. However, DNA methylation was only estimated one time using blood collected at midlife in daughters. To better understand the long-term effect of prenatal DDT exposure on DNA methylation changes, repeated measurements of DNA methylation profiles across the lifecourse including infancy, childhood, adolescence, and adulthood are needed. Although we selected the list of candidate genes based on a priori considerations, if we were to conservatively divide the p value by the number of tests we conducted, our results may be due to chance. In this study, the DNA methylation profile was measured in DNA that was derived from whole blood which contains a mixture of different cells. As DNA methylation profile is cell-type specific,(119) we recognize that the abundance of specific cell subtypes which may have different levels of methylation in the genes of interest may impact our results. However, comparing methylation in these DDT associated-DMRs in ZFPM2, CCDC85A and CYP1A1 across different cell types using data from Illumina HumanMethylation450 BeadChips (GEO accession: GSE35069), we did not find any differences in methylation by cell type including WBC, peripheral blood mononuclear cells, monocytes and neutrophils,(120) suggesting the methylation in these three DMRs are not different by blood cell types.
The ability of prenatal DDT exposure to alter methylation in breast cancer genes has been suggested. A recent study using Illumina Infinium HumanMethylation 450 K BeadChips measured cord blood from 24 subjects and examined the association of prenatal DDT exposure with fetal genome-wide DNA methylation.(121) Comparing DNA methylation profiles between subjects with no detectable DDT and subjects with detectable DDT, the authors identified 1,131 CpG sites with differences≥5% associated with intrauterine DDT exposure. Most of these CpG sites were located in the open sea regions and only 22% DMRs were in CpG islands. These CpG sites included 690 hypermethylation sites and 441 hypomethylation sites. The authors further validated the association between methylation of BRCA1 and intrauterine DDT exposure in another group of 126 subjects and found both DNA methylation and gene expression were statistically significantly different between the different exposure groups of p,p′-DDE, o,p′-DDD, o,p′-DDT, and p,p′-DDT (P < 0.05).(121) Unfortunately, our BRCA1 amplicon is about 700 bp away from their amplicon so we were unable to replicate this result in our study.
Our prospective study examining the effect of prenatal DDT exposure on adult DNA methylation changes suggests the persistent effect of environmental exposure may be through epigenetic alteration. The observation of alteration in DNA methylation profiles in genes important to breast cancer such as genes associated with menarche suggests a potential molecular mechanism involved in prenatal DDT exposure in breast cancer development. If verified, DNA methylation patterns associated with intrauterine DDT exposure may become clinically useful biomarkers of risk in current populations of women in midlife who were heavily exposed to DDT in utero in the 1960’s. Verification should include investigation of biomarker relevance to future risk in animal, in vitro, and clinical studies and eventually investigation of the potential for mitigating risk via restoring methylation marks to lower risk patterns.
Supplementary Material
Highlights.
Prenatal DDTs exposure is associated with DNA methylation in key genes that are potentially important to breast cancer.
Differentially methylated regions (DMRs) in CCDC85A, CYP1A1 and ZFPM2 are associated with markers of DDTs
Higher methylation in genes that are involved in growth and development and breast cancer susceptibility are associated with higher in utero DDT exposure
Prenatal DDT exposure may have life-long consequence through alteration in genes relevant to breast cancer.
Acknowledgments:
The authors thank the Epigenetics Shared Resource and the Molecular Pathology Share Resource of the Herbert Irving Comprehensive Cancer Center for performing the targeted bisulfite sequencing. We thank the CHDS families for their participation in this study. We acknowledge the late Jacob Yerushalmy who had the foresight to design and implement the CHDS; the late Barbara van den Berg, the second Director of the CHDS, whose steadfast efforts were responsible for preserving the data and serum archive, thus granting the CHDS longevity.
Funding: This work was supported by the Breast Cancer and the Environment Research Program (BCERP) grant U01 ES019457 and P30 ES009089 from the National Institute of Environmental Health Sciences (NIEHS) and the National Cancer Institute (NCI), NIH, DHHS; and, by funding from the California Breast Cancer Research Program through the Special Research Initiative under Grant 15ZB-0186 and the Breast Cancer Research Foundation. This research was funded in part through the NIH/NCI Cancer Center Support Grant P30CA013696.
Abbreviations:
- CHDS
Child Health and Development Studies
- bis-seq
bisulfite sequencing
- BMI
body mass index
- DDT
dichlorodiphenyltrichloroethane
- DMR
differentially methylated regions
- EDCs
endocrine disrupting chemicals
- o,p´-DDT
[1,1,1-trichloro-2-(p-chlorophenyl)-2-(o-chlorophenyl) ethane]
- p,p´-DDE
[1,1´-dichloro-2,2´-bis(p-chlorophenyl) ethylene]
- p,p´-DDT
[1,1,1-trichloro-2,2-bis(pchlorophenyl) ethane]
- WBC
white blood cells
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests
The authors declare that they have no competing interests
References
- 1.Toxicological Profile for DDT, DDE, and DDD. Atlanta, GA: U.S. Department of Health and Human Services, Public Health Service, 2002. [Google Scholar]
- 2.EPA US. DDT, a Review of Scientific and Economic Aspects of the Decision to Ban Its Use as a Pesticide: EPA-540/1-75-022. Washington, DC:U.S. Environmental Protection Agency, 1975. [Google Scholar]
- 3.Welch RM, Levin W, Conney AH. Estrogenic action of DDT and its analogs. Toxicology and Applied Pharmacology 1969;14:358–367. [DOI] [PubMed] [Google Scholar]
- 4.Gellert RJ, Leroy Heinrichs W, Swerdloff RS. DDT Homologues: Estrogen-Like Effects on the Vagina, Uterus and Pituitary of the Rat. Endocrinology 1972;91:1095–1100. [DOI] [PubMed] [Google Scholar]
- 5.Kuiper GGJM, Lemmen JG, Carlsson B, Corton JC, Safe SH, van der Saag PT, van der Burg B, et al. Interaction of Estrogenic Chemicals and Phytoestrogens with Estrogen Receptor β. Endocrinology 1998;139:4252–4263. [DOI] [PubMed] [Google Scholar]
- 6.Shanle EK, Xu W. Endocrine disrupting chemicals targeting estrogen receptor signaling: Identification and mechanisms of action. Chemical research in toxicology 2011;24:6–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klotz DM, Beckman BS, Hill SM, McLachlan JA, Walters MR, Arnold SF. Identification of environmental chemicals with estrogenic activity using a combination of in vitro assays. Environmental Health Perspectives 1996;104:1084–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qin X-Y, Zaha H, Nagano R, Yoshinaga J, Yonemoto J, Sone H. Xenoestrogens down-regulate aryl-hydrocarbon receptor nuclear translocator 2 mRNA expression in human breast cancer cells via an estrogen receptor alpha-dependent mechanism. Toxicology Letters 2011;206:152–157. [DOI] [PubMed] [Google Scholar]
- 9.Robison AK, Sirbasku DA, Stancel GM. DDT supports the growth of an estrogen-responsive tumor: Mammary tumor growth; pesticides. Toxicology Letters 1985;27:109–113. [DOI] [PubMed] [Google Scholar]
- 10.Johnson NA, Ho A, Cline JM, Hughes CL, Foster WG, Davis VL. Accelerated Mammary Tumor Onset in a HER2/Neu Mouse Model Exposed to DDT Metabolites Locally Delivered to the Mammary Gland. Environmental Health Perspectives 2012;120:1170–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han EH, Kim HG, Hwang YP, Choi JH, Im JH, Park B, Yang JH, et al. The role of cyclooxygenase-2-dependent signaling via cyclic AMP response element activation on aromatase up-regulation by o,p′-DDT in human breast cancer cells. Toxicology Letters 2010;198:331–341. [DOI] [PubMed] [Google Scholar]
- 12.White AJ, D’Aloisio AA, Nichols HB, DeRoo LA, Sandler DP. Breast cancer and exposure to tobacco smoke during potential windows of susceptibility. Cancer Causes & Control 2017;28:667–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Biro FM, Deardorff J. Identifying Opportunities for Cancer Prevention During Pre-Adolescence and Adolescence: Puberty as a Window of Susceptibility. The Journal of adolescent health: official publication of the Society for Adolescent Medicine 2013;52:S15–S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodgers KM, Udesky JO, Rudel RA, Brody JG. Environmental chemicals and breast cancer: An updated review of epidemiological literature informed by biological mechanisms. Environmental Research 2018;160:152–182. [DOI] [PubMed] [Google Scholar]
- 15.Fenton SE, Reed C, Newbold RR. Perinatal Environmental Exposures Affect Mammary Development, Function, and Cancer Risk in Adulthood. Annual Review of Pharmacology and Toxicology 2012;52:455–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohn BA, Wolff MS, Cirillo PM, Sholtz RI. DDT and Breast Cancer in Young Women: New Data on the Significance of Age at Exposure. Environmental Health Perspectives 2007;115:1406–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cohn BA, La Merrill M, Krigbaum NY, Yeh G, Park J-S, Zimmermann L, Cirillo PM. DDT Exposure in Utero and Breast Cancer. The Journal of Clinical Endocrinology and Metabolism 2015;100:2865–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forman MR, Winn DM, Collman GW, Rizzo J, Birnbaum LS. Environmental exposures, breast development and cancer risk: Through the looking glass of breast cancer prevention. Reproductive Toxicology 2015;54:6–10. [DOI] [PubMed] [Google Scholar]
- 19.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 1992;69:915–926. [DOI] [PubMed] [Google Scholar]
- 20.Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature 1993;366:362. [DOI] [PubMed] [Google Scholar]
- 21.Stability Reik W. and flexibility of epigenetic gene regulation in mammalian development. Nature 2007;447:425. [DOI] [PubMed] [Google Scholar]
- 22.Martin EM, Fry RC. Environmental Influences on the Epigenome: Exposure- Associated DNA Methylation in Human Populations. Annual Review of Public Health 2018;39:309–333. [DOI] [PubMed] [Google Scholar]
- 23.Terry MB, Delgado-Cruzata L, Vin-Raviv N, Wu HC, Santella RM. DNA methylation in white blood cells: association with risk factors in epidemiologic studies. Epigenetics 2011;6:828–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joubert BR, Haberg SE, Nilsen RM, Wang X, Vollset SE, Murphy SK, Huang Z, et al. 450K epigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy. Environ Health Perspect 2012;120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li L, Choi J-Y, Lee K-M, Sung H, Park SK, Oze I, Pan K-F, et al. DNA Methylation in Peripheral Blood: A Potential Biomarker for Cancer Molecular Epidemiology. Journal of Epidemiology 2012;22:384–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu H, Delgado-Cruzata L, Flom JD, Perrin M, Liao Y, Ferris JS, Santella RM, et al. Repetitive element DNA methylation levels in white blood cell DNA from sisters discordant for breast cancer from the New York site of the Breast Cancer Family Registry. Carcinogenesis 2012;33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delgado-Cruzata L, Wu HC, May P, Liao Y, Kappil AM, Ferris J, Flom J, et al. Global DNA methylation levels in white blood cell DNA from sisters discordant for breast cancer from the New York site of the Breast Cancer Family Registry. Epigenetics 2012;7:868–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Esteller M, Silva J, Dominguez G, Bonilla F, Matias-Guiu X, Lerma E, Bussaglia E, et al. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J Natl Cancer Inst 2000;92:564–569. [DOI] [PubMed] [Google Scholar]
- 29.Esteller M, Fraga MF, Guo M, Garcia-Foncillas J, Hedenfalk I, Godwin AK, Trojan J, et al. DNA methylation patterns in hereditary human cancers mimic sporadic tumorigenesis. Human Molecular Genetics 2001;10:3001–3007. [DOI] [PubMed] [Google Scholar]
- 30.Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res 2001;61:3225–3229. [PubMed] [Google Scholar]
- 31.Zhang L, Long X. Association of BRCA1 promoter methylation with sporadic breast cancers: Evidence from 40 studies. 2015;5:17869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wong EM, Southey MC, Fox SB, Brown MA, Dowty JG, Jenkins MA, Giles GG, et al. Constitutional Methylation of the BRCA1 Promoter Is Specifically Associated with BRCA1 Mutation-Associated Pathology in Early-Onset Breast Cancer. Cancer Prevention Research 2011;4:23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cho YH, McCullough LE, Gammon MD, Wu H-C, Zhang Y-J, Wang Q, Xu X, et al. Promoter Hypermethylation in White Blood Cell DNA and Breast Cancer Risk. Journal of Cancer 2015;6:819–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang L, Long X. Association of BRCA1 promoter methylation with sporadic breast cancers: Evidence from 40 studies. Scientific Reports 2015;5:17869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knower KC, To SQ, Leung Y-K, Ho S-M, Clyne CD. Endocrine disruption of the epigenome: a breast cancer link. Endocrine-related cancer 2014;21:T33–T55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goodrich JM, Dolinoy DC, Sánchez BN, Zhang Z, Meeker JD, Mercado-Garcia A, Solano-González M, et al. Adolescent epigenetic profiles and environmental exposures from early life through peri-adolescence. Environmental Epigenetics 2016;2:dvw018–dvw018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tobi EW, Goeman JJ, Monajemi R, Gu H, Putter H, Zhang Y, Slieker RC, et al. DNA methylation signatures link prenatal famine exposure to growth and metabolism. Nat Commun 2014;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richmond RC, Simpkin AJ, Woodward G, Gaunt TR, Lyttleton O, McArdle WL, Ring SM, et al. Prenatal exposure to maternal smoking and offspring DNA methylation across the lifecourse: findings from the Avon Longitudinal Study of Parents and Children (ALSPAC). Human Molecular Genetics 2015;24:2201–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chatterton Z, Hartley BJ, Seok M-H, Mendelev N, Chen S, Milekic M, Rosoklija G, et al. In utero exposure to maternal smoking is associated with DNA methylation alterations and reduced neuronal content in the developing fetal brain. Epigenetics & Chromatin 2017;10:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Joubert Bonnie R, Felix Janine F, Yousefi P, Bakulski Kelly M, Just Allan C, Breton C, Reese SE, et al. DNA Methylation in Newborns and Maternal Smoking in Pregnancy: Genome-wide Consortium Meta-analysis. The American Journal of Human Genetics 2016;98:680–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rzehak P, Saffery R, Reischl E, Covic M, Wahl S, Grote V, Xhonneux A, et al. Maternal Smoking during Pregnancy and DNA-Methylation in Children at Age 5.5 Years: Epigenome-Wide-Analysis in the European Childhood Obesity Project (CHOP)-Study. PLOS ONE 2016;11:e0155554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prince C, Hammerton G, Taylor AE, Anderson EL, Timpson NJ, Davey Smith G, Munafò MR, et al. Investigating the impact of cigarette smoking behaviours on DNA methylation patterns in adolescence. Human Molecular Genetics 2019;28:155–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tehranifar P, Wu H-C, McDonald JA, Jasmine F, Santella RM, Gurvich I, Flom JD, et al. Maternal cigarette smoking during pregnancy and offspring DNA methylation in midlife. Epigenetics 2017:00–00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vaiserman A Early-life Exposure to Endocrine Disrupting Chemicals and Later-life Health Outcomes: An Epigenetic Bridge? Aging and Disease 2014;5:419–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim K-Y, Kim D-S, Lee S-K, Lee I-K, Kang J-H, Chang Y-S, Jacobs DR, et al. Association of Low-Dose Exposure to Persistent Organic Pollutants with Global DNA Hypomethylation in Healthy Koreans. Environmental Health Perspectives 2010;118:370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rusiecki JA, Baccarelli A, Bollati V, Tarantini L, Moore LE, Bonefeld-Jorgensen EC. Global DNA Hypomethylation Is Associated with High Serum-Persistent Organic Pollutants in Greenlandic Inuit. Environmental Health Perspectives 2008;116:1547–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Itoh H, Iwasaki M, Kasuga Y, Yokoyama S, Onuma H, Nishimura H, Kusama R, et al. Association between serum organochlorines and global methylation level of leukocyte DNA among Japanese women: a cross-sectional study. Science of The Total Environment 2014;490:603–609. [DOI] [PubMed] [Google Scholar]
- 48.Lind L, Penell J, Luttropp K, Nordfors L, Syvänen A-C, Axelsson T, Salihovic S, et al. Global DNA hypermethylation is associated with high serum levels of persistent organic pollutants in an elderly population. Environment International 2013;59:456–461. [DOI] [PubMed] [Google Scholar]
- 49.La Merrill M, Cirillo PM, Terry MB, Krigbaum NY, Flom JD, Cohn BA. Prenatal Exposure to the Pesticide DDT and Hypertension Diagnosed in Women before Age 50: A Longitudinal Birth Cohort Study. Environmental Health Perspectives 2013;121:594–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Link BG, Susser ES, Factor-Litvak P, March D, Kezios KL, Lovasi GS, Rundle AG, et al. Disparities in self-rated health across generations and through the life course. Social science & medicine (1982) 2017;174:17–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gammon MD, Wolff MS, Neugut AI, Eng SM, Teitelbaum SL, Britton JA, Terry MB, et al. Environmental Toxins and Breast Cancer on Long Island. II. Organochlorine Compound Levels in Blood. Cancer Epidemiology Biomarkers & Prevention 2002;11:686–697. [PubMed] [Google Scholar]
- 52.Purrington KS, Slager S, Eccles D, Yannoukakos D, Fasching PA, Miron P, Carpenter J, et al. Genome-wide association study identifies 25 known breast cancer susceptibility loci as risk factors for triple-negative breast cancer. Carcinogenesis 2014;35:1012–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, Klemm A, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Research 2014;42:D1001–D1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dvornyk V, Waqar-ul-Haq. Genetics of age at menarche: a systematic review. Human Reproduction Update 2012;18:198–210. [DOI] [PubMed] [Google Scholar]
- 55.Horikoshi M, Yaghootkar H, Mook-Kanamori DO, Sovio U, Taal HR, Hennig BJ, Bradfield JP, et al. New loci associated with birth weight identify genetic links between intrauterine growth and adult height and metabolism. Nature genetics 2013;45:76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Engel SM, Joubert BR, Wu MC, Olshan AF, Håberg SE, Ueland PM, Nystad W, et al. Neonatal Genome-Wide Methylation Patterns in Relation to Birth Weight in the Norwegian Mother and Child Cohort. American Journal of Epidemiology 2014;179:834–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu H-C, Do C, Andrulis IL, John EM, Daly MB, Buys SS, Chung WK, et al. Breast Cancer Family History and Allele-Specific DNA Methylation in the Legacy Girls Study. Epigenetics 2018:01–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, et al. Systematic analysis of chromatin state dynamics in nine human cell types. Nature 2011;473:43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Karolchik D, Hinrichs AS, Kent WJ. The UCSC Genome Browser Current protocols in bioinformatics / editoral board, Baxevanis Andreas D. … [et al. ] 2009;CHAPTER:Unit1.4-Unit1.4. [DOI] [PubMed] [Google Scholar]
- 60.Sur I, Taipale J. The role of enhancers in cancer. Nat Rev Cancer 2016;16:483–493. [DOI] [PubMed] [Google Scholar]
- 61.Yang J, Corces VG. Chromatin Insulators: A Role in Nuclear Organization and Gene Expression. Advances in cancer research 2011;110:43–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lay FD, Liu Y, Kelly TK, Witt H, Farnham PJ, Jones PA, Berman BP. The role of DNA methylation in directing the functional organization of the cancer epigenome. Genome Research 2015;25:467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Robertson KD. DNA methylation and human disease. Nat Rev Genet 2005;6:597–610. [DOI] [PubMed] [Google Scholar]
- 64.Yotova I, Hsu E, Do C, Gaba A, Sczabolcs M, Dekan S, Kenner L, et al. Epigenetic Alterations Affecting Transcription Factors and Signaling Pathways in Stromal Cells of Endometriosis. PLoS ONE 2017;12:e0170859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Do C, Lang CF, Lin J, Darbary H, Krupska I, Gaba A, Petukhova L, et al. Mechanisms and Disease Associations of Haplotype-Dependent Allele-Specific DNA Methylation. American Journal of Human Genetics 2016;98:934–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Do C, Lang Charles F, Lin J, Darbary H, Krupska I, Gaba A, Petukhova L, et al. Mechanisms and Disease Associations of Haplotype-Dependent Allele-Specific DNA Methylation. The American Journal of Human Genetics 2016;98:934–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics 2002;18:1427–1431. [DOI] [PubMed] [Google Scholar]
- 68.Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 2011;27:1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Terry MB, Ferris JS, Pilsner R, Flom JD, Tehranifar P, Santella RM, Gamble MV, et al. Genomic DNA methylation among Women in a Multi-ethnic New York City Birth Cohort. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology 2008;17: 10.1158/1055-9965.EPI-1108-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ciccarone F, Tagliatesta S, Caiafa P, Zampieri M. DNA methylation dynamics in aging: how far are we from understanding the mechanisms? Mechanisms of Ageing and Development 2017. [DOI] [PubMed] [Google Scholar]
- 71.DeBruin LS, Josephy PD. Perspectives on the chemical etiology of breast cancer. Environmental Health Perspectives 2002;110:119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Snedeker SM. Pesticides and breast cancer risk: a review of DDT, DDE, and dieldrin. Environmental Health Perspectives 2001;109:35–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.White AJ, Teitelbaum SL, Wolff MS, Stellman SD, Neugut AI, Gammon MD. Exposure to fogger trucks and breast cancer incidence in the Long Island Breast Cancer Study Project: a case–control study. Environmental Health 2013;12:24–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Louis LM, Lerro CC, Friesen MC, Andreotti G, Koutros S, Sandler DP, Blair A, et al. A prospective study of cancer risk among Agricultural Health Study farm spouses associated with personal use of organochlorine insecticides. Environmental Health 2017;16:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.López-Cervantes M, Torres-Sánchez L, Tobías A, López-Carrillo L. Dichlorodiphenyldichloroethane burden and breast cancer risk: a meta-analysis of the epidemiologic evidence. Environmental Health Perspectives 2004;112:207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ingber SZ, Buser MC, Pohl HR, Abadin HG, Edward Murray H, Scinicariello F. DDT/DDE and breast cancer: A meta-analysis. Regulatory Toxicology and Pharmacology 2013;67:421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Park J-H, Cha ES, Ko Y, Hwang M-S, Hong J-H, Lee WJ. Exposure to Dichlorodiphenyltrichloroethane and the Risk of Breast Cancer: A Systematic Review and Meta-analysis. Osong Public Health and Research Perspectives 2014;5:77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rodgers KM, Udesky JO, Rudel RA, Brody JG. Environmental chemicals and breast cancer: An updated review of epidemiological literature informed by biological mechanisms. Environmental Research 2018;160:152–182. [DOI] [PubMed] [Google Scholar]
- 79.Cohn BA. Developmental and Environmental Origins of Breast Cancer DDT as a Case Study. Reproductive Toxicology (Elmsford, N.y.) 2011;31:302–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Soto AM, Sonnenschein C. DDT, endocrine disruption and breast cancer. Nature reviews. Endocrinology 2015;11:507–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Arrebola JP, Fernández-Rodríguez M, Artacho-Cordón F, Garde C, Perez-Carrascosa F, Linares I, Tovar I, et al. Associations of persistent organic pollutants in serum and adipose tissue with breast cancer prognostic markers. Science of The Total Environment 2016;566–567:41–49. [DOI] [PubMed] [Google Scholar]
- 82.Parada H, Wolff MS, Engel LS, White AJ, Eng SM, Cleveland RJ, Khankari NK, et al. Organochlorine insecticides DDT and chlordane in relation to survival following breast cancer. International journal of cancer. Journal international du cancer 2016;138:565–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ladd-Acosta C, Fallin MD. The role of epigenetics in genetic and environmental epidemiology. Epigenomics 2015;8:271–283. [DOI] [PubMed] [Google Scholar]
- 84.Bakulski KM, Fallin MD. Epigenetic epidemiology: Promises for public health research. Environmental and Molecular Mutagenesis 2014;55:171–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Demerath EW, Liu C-T, Franceschini N, Chen G, Palmer JR, Smith EN, Chen CTL, et al. Genome-wide association study of age at menarche in African-American women. Human Molecular Genetics 2013;22:3329–3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tanikawa C, Okada Y, Takahashi A, Oda K, Kamatani N, Kubo M, Nakamura Y, et al. Genome Wide Association Study of Age at Menarche in the Japanese Population. PLOS ONE 2013;8:e63821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Au Yeung SL, Jiang C, Cheng KK, Xu L, Zhang W, Lam TH, Leung GM, et al. Age at menarche and cardiovascular risk factors using Mendelian randomization in the Guangzhou Biobank Cohort Study. Preventive Medicine 2017;101:142–148. [DOI] [PubMed] [Google Scholar]
- 88.Dvornyk V, Waqar ul H. Genetics of age at menarche: a systematic review. Human Reproduction Update 2012;18:198–210. [DOI] [PubMed] [Google Scholar]
- 89.Chen C, Huang Y, Li Y, Mao Y, Xie Y. Cytochrome P450 1A1 (CYP1A1) T3801C and A2455G polymorphisms in breast cancer risk: a meta-analysis. Journal Of Human Genetics 2007;52:423. [DOI] [PubMed] [Google Scholar]
- 90.Breast Cancer Risk Assessment Tool. http://www.cancer.gov/bcrisktool/.
- 91.Feng S, Cokus SJ, Zhang X, Chen P-Y, Bostick M, Goll MG, Hetzel J, et al. Conservation and divergence of methylation patterning in plants and animals. Proceedings of the National Academy of Sciences 2010;107:8689–8694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Acerini CL, Hughes IA. Endocrine disrupting chemicals: a new and emerging public health problem? Archives of Disease in Childhood 2006;91:633–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schug TT, Janesick A, Blumberg B, Heindel JJ. Endocrine Disrupting Chemicals and Disease Susceptibility. The Journal of steroid biochemistry and molecular biology 2011;127:204–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li Y, Hamilton KJ, Lai AY, Burns KA, Li L, Wade PA, Korach KS. Diethylstilbestrol (DES)-Stimulated Hormonal Toxicity is Mediated by ERα Alteration of Target Gene Methylation Patterns and Epigenetic Modifiers (DNMT3A, MBD2, and HDAC2) in the Mouse Seminal Vesicle. Environmental Health Perspectives 2014;122:262–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bromer JG, Wu J, Zhou Y, Taylor HS. Hypermethylation of Homeobox A10 by in Utero Diethylstilbestrol Exposure: An Epigenetic Mechanism for Altered Developmental Programming. Endocrinology 2009;150:3376–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Haddad R, Kasneci A, Mepham K, Sebag IA, Chalifour LE. Gestational exposure to diethylstilbestrol alters cardiac structure/function, protein expression and DNA methylation in adult male mice progeny. Toxicology and Applied Pharmacology 2013;266:27–37. [DOI] [PubMed] [Google Scholar]
- 97.Morgan HD, Sutherland HGE, Martin DIK, Whitelaw E. Epigenetic inheritance at the agouti locus in the mouse. Nature Genetics 1999;23:314. [DOI] [PubMed] [Google Scholar]
- 98.Shutoh Y, Takeda M, Ohtsuka R, Haishima A, Yamaguchi S, Fujie H, Komatsu Y, et al. Low dose effects of dichlorodiphenyltrichloroethane (DDT) on gene transcription and DNA methylation in the hypothalamus of young male rats: implication of hormesis-like effects. The Journal of Toxicological Sciences 2009;34:469–482. [DOI] [PubMed] [Google Scholar]
- 99.Desaulniers D, Xiao G-h, Lian H, Feng Y-L, Zhu J, Nakai J, Bowers WJ. Effects of Mixtures of Polychlorinated Biphenyls, Methylmercury, and Organochlorine Pesticides on Hepatic DNA Methylation in Prepubertal Female Sprague-Dawley Rats. International Journal of Toxicology 2009;28:294–307. [DOI] [PubMed] [Google Scholar]
- 100.Li S, Washburn KA, Moore R, Uno T, Teng C, Newbold RR, McLachlan JA, et al. Developmental Exposure to Diethylstilbestrol Elicits Demethylation of Estrogen-responsive Lactoferrin Gene in Mouse Uterus. Cancer Research 1997;57:4356–4359. [PubMed] [Google Scholar]
- 101.Dolinoy DC, Huang D, Jirtle RL. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proceedings of the National Academy of Sciences of the United States of America 2007;104:13056–13061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liu J, Zhao M, Zhuang S, Yang Y, Yang Y, Liu W. Low Concentrations of o,p’-DDT Inhibit Gene Expression and Prostaglandin Synthesis by Estrogen Receptor-Independent Mechanism in Rat Ovarian Cells. PLoS ONE 2012;7:e49916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet 2002;3:415–428. [DOI] [PubMed] [Google Scholar]
- 104.Feinberg AP, Tycko B. The history of cancer epigenetics. Nature Reviews Cancer 2004;4. [DOI] [PubMed] [Google Scholar]
- 105.Feinberg A, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature 1983;301. [DOI] [PubMed] [Google Scholar]
- 106.Genetic Tycko B. and Epigenetic Mosaicism in Cancer Precursor Tissues. Annals of the New York Academy of Sciences 2003;983:43–54. [DOI] [PubMed] [Google Scholar]
- 107.Huen K, Yousefi P, Bradman A, Yan L, Harley KG, Kogut K, Eskenazi B, et al. Effects of age, sex, and persistent organic pollutants on DNA methylation in children. Environmental and molecular mutagenesis 2014;55:209–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ehrlich M DNA methylation in cancer: too much, but also too little. Oncogene 2002;21:5400–5413. [DOI] [PubMed] [Google Scholar]
- 109.Dao-Peng C, Ying-Chao L, Fann CSJ. Methods for identifying differentially methylated regions for sequence- and array-based data. Briefings in Functional Genomics 2016;15:485–490. [DOI] [PubMed] [Google Scholar]
- 110.Harlid S, Xu Z, Panduri V, D’Aloisio AA, DeRoo LA, Sandler DP, Taylor JA. In Utero Exposure to Diethylstilbestrol and Blood DNA Methylation in Women Ages 40–59 Years from the Sister Study. PLOS ONE 2015;10:e0118757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Androutsopoulos VP, Tsatsakis AM, Spandidos DA. Cytochrome P450 CYP1A1: wider roles in cancer progression and prevention. BMC Cancer 2009;9:187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Küpers LK, Xu X, Jankipersadsing SA, Vaez A, la Bastide-van Gemert S, Scholtens S, Nolte IM, et al. DNA methylation mediates the effect of maternal smoking during pregnancy on birthweight of the offspring. International Journal of Epidemiology 2015;44:1224–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Richmond RC, Joubert BR. Contrasting the effects of intra-uterine smoking and one-carbon micronutrient exposures on offspring DNA methylation. Epigenomics 2017;9:351–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bredhult C, Sahlin L, Olovsson M. Gene expression analysis of human endometrial endothelial cells exposed to op′-DDT. MHR: Basic science of reproductive medicine 2008;14:97–106. [DOI] [PubMed] [Google Scholar]
- 115.Wójtowicz AK, Honkisz E, Zięba-Przybylska D, Milewicz T, Kajta M. Effects of two isomers of DDT and their metabolite DDE on CYP1A1 and AhR function in human placental cells. Pharmacological Reports 2011;63:1460–1468. [DOI] [PubMed] [Google Scholar]
- 116.Goth-Goldstein R, Stampfer MR, Erdmann CA, Russell M. Interindividual variation in CYP1A1 expression in breast tissue and the role of genetic polymorphism. Carcinogenesis 2000;21:2119–2122. [DOI] [PubMed] [Google Scholar]
- 117.Tekpli X, Zienolddiny S, Skaug V, Stangeland L, Haugen A, Mollerup S. DNA methylation of the CYP1A1 enhancer is associated with smoking-induced genetic alterations in human lung. International Journal of Cancer 2012;131:1509–1516. [DOI] [PubMed] [Google Scholar]
- 118.Bowers EC, McCullough SD. Linking the Epigenome with Exposure Effects and Susceptibility: The Epigenetic Seed and Soil Model. Toxicological Sciences 2017;155:302–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wu HC, Delgado-Cruzata L, Flom JD, Kappil M, Ferris JS, Liao Y, Santella RM, et al. Global methylation profiles in DNA from different blood cell types. Epigenetics 2011;6:76–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Reinius LE, Acevedo N, Joerink M, Pershagen G, Dahlén S-E, Greco D, Söderhäll C, et al. Differential DNA Methylation in Purified Human Blood Cells: Implications for Cell Lineage and Studies on Disease Susceptibility. PLOS ONE 2012;7:e41361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yu X, Zhao B, Su Y, Zhang Y, Chen J, Wu W, Cheng Q, et al. Association of prenatal organochlorine pesticide-dichlorodiphenyltrichloroethane exposure with fetal genome-wide DNA methylation. Life Sciences 2018;200:81–86. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.