Abstract
the bile duct system and pancreas show many similarities due to their anatomical proximity and common embryological origin. Consequently, preneoplastic and neoplastic lesions of the bile duct and pancreas share analogies in terms of molecular, histological and pathophysiological features. Intraepithelial neoplasms are reported in biliary tract, as biliary intraepithelial neoplasm (BilIN), and in pancreas, as pancreatic intraepithelial neoplasm (PanIN). Both can evolve to invasive carcinomas, respectively cholangiocarcinoma (CCA) and pancreatic ductal adenocarcinoma (PDAC). Intraductal papillary neoplasms arise in biliary tract and pancreas. Intraductal papillary neoplasm of the biliary tract (IPNB) share common histologic and phenotypic features such as pancreatobiliary, gastric, intestinal and oncocytic types, and biological behavior with the pancreatic counterpart, the intraductal papillary mucinous neoplasm of the pancreas (IPMN). All these neoplastic lesions exhibit similar immunohistochemical phenotypes, suggesting a common carcinogenic process. Indeed, CCA and PDAC display similar clinic-pathological features as growth pattern, poor response to conventional chemotherapy and radiotherapy and, as a consequence, an unfavorable prognosis. The objective of this review is to discuss similarities and differences between the neoplastic lesions of the pancreas and biliary tract with potential implications on a common origin from similar stem/progenitor cells.
Keywords: Biliary, Pancreatic, Progenitors, Preneoplastic, Common, Tumor
Core tip: The carcinogenesis process of the biliary tract and the pancreas has common histological, biologic and molecular characteristics possibly due to similar stem/progenitors origin. Such findings could set the base for studies aimed at identifying novel strategies for prevention, surveillance and treatment of these deadly diseases.
INTRODUCTION
The biliary tract and the pancreas share a common embryologic origin[1]. In the primitive duodenum, the bilio-pancreatic progenitor differentiates in distinct lineages driven by specific transcription factors such as Hes1 in the case of the hepatobiliary fate, or Pdx1/Ngn3/MafA in the case of the pancreatic fate[2]. Stem/progenitors for biliary tree and pancreas exist at early stages of human development and both derive from the definitive anterior endoderm forming foregut[3]. During human development, the hepatic diverticulum emerges from the foregut[4]. While the cranial part of the diverticulum forms the liver and the intrahepatic bile ducts (IHBDs), the extrahepatic bile ducts (EHBDs) and ventral pancreas arise from its caudal part[5]. The large IHBDs and the EHBDs share the presence of glands in their duct walls, which are named peribiliary glands (PBGs)[6]. PBGs are more commonly located at branching points of the biliary tree and at the hepatopancreatic ampulla. It has been clearly demonstrated that specialized glands of the biliopancreatic tract contain niches of stem/progenitors cells that could have an important role in the carcinogenesis process, when they are activated in response to chronic stimuli[7]. Niches of biliary tree stem cells (BTSCs) are located at the bottom of PBGs, near the fibromuscular layer; these cells express markers of stem/progenitors’ cells of endodermal origin and can proliferate and self-renew[8]. BTSCs are able to differentiate in vitro and in vivo, in experimental models of liver diseases or streptozotocin-induced diabetes, into cholangiocytes and, hepatocytes, or pancreatic islets[9]. Interestingly multipotency property and the capability to respond to hyperglycemia in vivo are suggested by data obtained in animal models of diabetes and in humans affected by type II diabetes mellitus, where PBGs undergo proliferation and expansion in response to hyperglycemia differentiating toward insulin-producing cells[10]. These data open to the exciting hypothesis that the PBG niche commitment to endocrine pancreatic fate may contribute to the homeostatic pancreatic islet regeneration response observed in diabetes[7]. Studies of human anatomy support the hypothesis that BTSCs are organized in a proximal-to-distal axis, in which the hepatopancreatic ampulla is considered the proximal site where the most primitive stem biliary tree and pancreas reside. In fact, pancreatic stem cell traits persist in the adult biliary tree, especially at the ampullary region[11].
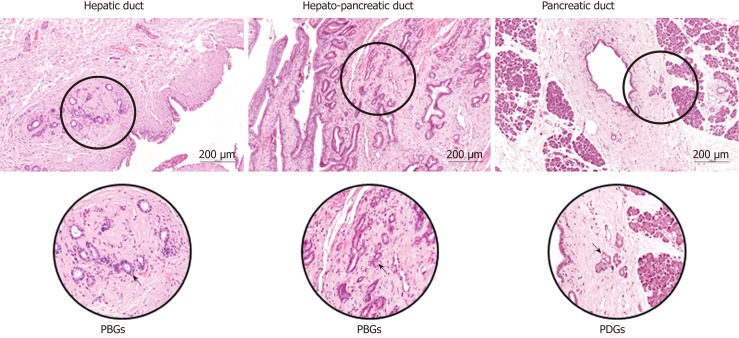
In the pancreas, pancreatic ductal glands (PDGs) containing pancreatic committed progenitors are observed within the walls of larger pancreatic ducts (Figure 1). PDGs are more numerous in the main pancreatic duct at the head of the pancreas than in the tail and in larger interlobular ducts, where PDGs progressively reduce and finally, disappear in interlobular ducts with a diameter < 300 μm[12]. PDGs are therefore considered the pancreatic counterpart of PBGs[13], they are most abundant at the hepatopancreatic ampulla and their distribution along larger pancreatic ducts shows similarities with the distribution of PBGs in the biliary tree[7,12] (Figure 1).
Figure 1.
Common and Distinctive Features of peribiliary and pancreatic ductal glands. The peribiliary glands are tubule-alveolar glands present within the walls of larger biliary ducts, more commonly located at branching points of the biliary tree, in particular at the hepatopancreatic region. The pancreatic ductal glands containing pancreatic committed progenitors are observed within the walls of larger pancreatic ducts. PBGs: Peribiliary glands; PDGs: Pancreatic ductal glands.
To sum up, on the basis of the progenitor segregation, multiple stem cell niches persist in specific anatomical locations within adult human organs: (1) BTSCs in PBGs along EHBD and large IHBDs; and (2) Pancreatic stem cells that appear confined to the biliary tree, particularly the hepatopancreatic common duct, while their descendants, committed progenitors, are observed within the pancreatic ductal system in PDGs[11] (Figure 1). These stem/progenitor cell niches may contribute to repair pervasive, chronic injuries, whereas the replication of mature parenchymal cells ensures physiological turnover and restoration of parenchyma after minor injuries.
The above mentioned similarities in the embryological and histological features between the biliary tree and the pancreas are reflected in the clinic-pathologic similarities of several diseases affecting these two organs, such as their involvement in IgG4 related-disease, preneoplastic lesions and cancers[14-16]. Accordingly, cholangiocarcinoma (CCA) and pancreatic ductal adenocarcinoma (PDAC) seem to share molecular pathways and cells of origin as recently suggested by the results of the wide molecular characterization of determined clinical-pathological classified malignancies conducted in The Cancer Genome Atlas results[17,18]. Interestingly, there is also evidence that common risk factors, such as obesity and diabetes mellitus, are associated with both CCA[19] and PDAC[20].
The present review article will discuss available evidence concerning common features between neoplastic and preneoplastic lesions of the biliary tree and the pancreas.
INTRAEPITHELIAL NEOPLASMS OF THE BILIARY TRACT AND PANCREAS
One of the most relevant malignant precursors of biliary tract cancers is represented by the biliary intraepithelial neoplasm (BilIN), a microscopic not macroscopically identifiable lesion, with a micropapillary, pseudopapillary or flat growth pattern, involved in the process of multistep cholangiocarcinogenesis[21]. BilIN represents the biliary counterpart of pancreatic intraepithelial neoplasia (PanIN)[13,22]. BilIN are usually encountered in the epithelium lining the EHBDs and large IHBDs and may also be found in the gallbladder[23,24]. Furthermore, these preneoplastic lesions have been identified and characterized in chronic hepatobiliary diseases such as hepatolithiasis, primary sclerosing cholangitis (PSC) and choledocal cyst, which are associated with an increased risk of developing CCA, particularly the perihilar CCA[23-27]. BilINs are also observed by histological examination in the biliary epithelium close to invasive biliary tract cancers[28]. Based on the degree of cellular and architectural atypia, BilINs have been classified into three categories: BilIN-1 (low grade dysplasia) showing the mildest changes compared to non-neoplastic epithelium of the bile ducts; BilIN-2 (intermediate grade dysplasia) with increased nuclear atypia and focal anomalies of cellular polarity as compared to BilIN-1; BilIN-3 (high grade dysplasia or carcinoma in situ), which are usually identified in proximity of CCA areas[29] (Figure 2).
Figure 2.
Common and distinctive features of pancreatic intraepithelial neoplasias and biliary intraepithelial neoplasms. A: Pancreatic intraepithelial neoplasia, high grade (PanIN-3, 10 ×) sorrounded by infiltrating adenocarcinoma. B: Biliary intraepithelial neoplasia, high grade (BilIN-3, 10 ×) with nearby infiltrating adenocarcinoma Both lesions are microscopic precancerous lesions of pancreas (A) and biliary tree (B), not identifiable by radiological imaging.
Regarding the expression of cell cycle-related molecules, it has been shown that p21 and cyclin D1 are progressively upregulated with the increase of BilIN grades, while the expression of the tumor suppressor gene SMAD4 gets down-regulated. Interestingly, a significant upregulation of p53 is observed in BilIN-3 and invasive CCA in contrast to BilIN-1 and BilIN-2, suggesting an important role of p53 in the acquisition of invasive growth capacity in the final step of transformation into CCA. Importantly, p21, cyclin D1 and p53 are not expressed in the non-neoplastic epithelium of bile ducts[30].
The expression pattern of cytoplasmic and luminal surface mucins is also altered during the increase in the degree of BilIN, with some evidences showing that the expression of MUC1 increases with the increase of BilIN grades, while the expression of MUC5AC is high in all BilIN grades[31]. Interestingly, BilIN are sporadically associated with intestinal metaplasia that could explained the focal immu-nohistochemical expression of MUC2, whereas this mucin is not identified in PanINs[22]. Moreover, the immunohistochemical expression of S100 proteins has been found to be increased in BilIN compared to the non-neoplastic epithelium of bile ducts. In particular, S100P is specifically expressed in BilIN-3 and invasive CCA rather than in BilIN-1/-2, suggesting an involvement of S100P in the late phases of the multistep cholangiocarcinogenesis process[32].
The demonstration that PBGs are stem cell niches of large IHBDs and EHBDs have increased the attention on these structures in the neoplastic transformation of the bile ducts. BilIN are also reported in PBGs and are graded as BilIN-1/-2/-3 as well[33]. Several studies suggested a role of PBGs in the response to chronic inflammatory stimuli of bile ducts such as those occurring in PSC, through a process leading to the occurrence of dysplastic lesions and consequently CCA development[34]. BilIN in PBGs also showed a strong immunoreactivity to S100P, and carcinoembryonic and carbohydrate antigen 19-9[35].
The pancreatic counterpart of intraepithelial neoplasm of the biliary tract is represented by PanINs, defined as a microscopic flat or micropapillary noninvasive lesions, usually inferior to 5 mm, and considered the most common malignant precursors of PDAC, with a lower proportion of cases originating from intraductal papillary mucinous neoplasms of the pancreas (IPMNs) and mucinous cystic neoplasms (MCNs)[36]. PanINs have been classified, according to the degree of cellular and architectural atypia, into low grade (previously classified as PanIN-1 and PanIN-2) with mild-moderate cytological atypia and basally located nuclei, and high grade (previously classified PanIN-3) with severe cytological atypia, loss of polarity and mitoses[37,38]. The findings of loss of nuclear polarity, increased nucleus-to-cytoplasm ratio, nuclear hyperchromasia, and architectural atypia are basically similar between BilINs and PanINs[39].
As for the expression of cell cycle-related molecules, the immunohistochemical expression of cyclin D1, p21 and p53 which is absent in nonneoplastic pancreatic ductal epithelium, has been reported in high grade PanIN compared to low grade PanINs, as well as in BilINs[22]. The loss of SMAD4 expression which is a typical late event in pancreatic carcinogenesis has been demonstrated in high-grade but not in low-grade PanINs[40]. As far as the expression of mucins is concerned, except for MUC2 expression, that is almost absent in PanINs, a similar expression pattern is found in BilINs and PanINs. MUC1 is progressively over-expressed in the process from PanIN to PDAC, while MUC5AC is also expressed in PanIN, especially when occurring in PDAC compared to normal pancreata[22,41]. The finding of upregulated expression of the above mentioned mucins associated to higher PanIN grades supports their role in the early phases of pancreatic carcinogenesis for which they might be useful biomarkers. The expression of S100P is also increased in high grade PanINs and PDAC compared to the low grade PanINs[42,43]. The most common mutated gene found in in over 90% of PanINs is Kras, especially in the lowest grades[44]. Mutated Kras is considered the initiating genetic event in PDAC development but it cannot alone explain the progression to invasive carcinoma. Hedgehog (HH) signaling pathway, crucial during embryonic pancreatic development and stem cell regulation[45], resulted aberrantly activated both in PanINs and in invasive carcinoma[46,47]. Sonic Hedgehog (SHH) is one of the three protein of the Hh signaling pathway family. Overexpression of SHH is reported in PanINs and PDAC but not in normal pancreas, many studies showed a synergic crosstalk between mutated Kras and activation of SHH in the early development of cancer[48]. In the biliary counterpart, Kras mutations are reported in about 30% of cases of BilINs, thus less frequently compared to PanINs but, similarly to what is reported in the pancreatic counterpart, they occurr from the lowest grades during the progression to CCA[49]. Regarding the role of HH signaling pathway, studies revealed that Gli1, a transcriptional activator target of HH pathway, when overexpressed is associated with tumor invasion and metastasis in iCCA[50].
Similarly, to what has been reported regarding the activation of PBGs in disorders associated with increased risk of carcinogenesis in the biliary tract, within in pancreas, specialized PDGs, containing pancreatic committed progenitors, are activated in response to longstanding stimuli such as in chronic pancreatitis[51]. The upregulation of MUC-6 and a de-novo expression of MUC5AC has been reported in PDGs, supporting the hypothesis that these niches might contribute to mucinous metaplasia with features of PanIN-like lesions, and to the development of dysplastic pancreatic lesions[52].
In conclusion, there is solid evidence that BilINs and PanINs share common histological and immunohistochemical features and similar molecular changes associated with the multistep carcinogenetic process respectively of CCA and PDAC.
INTRADUCTAL PAPILLARY NEOPLASMS OF PANCREATO-BILIARY SYSTEMS
Intraductal papillary neoplasms of the biliary tract (IPNBs) are a rare variant of bile duct tumors that are considered the biliary counterpart of pancreatic IPMNs. Both arise within the ductal system and show a predominantly intraductal papillary growth pattern[53]. IPNBs can arise along all the biliary tree from both EHBDs and IHBDs and represent an alternative carcinogenesis pathway, distinct from BilIN[54,55]. Histologically, IPNBs show papillary proliferation of neoplastic biliary epithelium with delicate fibrovascular cores associated with luminal dilatation of the affected bile ducts, sometimes with mucin hypersecretion and, occasionally with prominent cystic changes[56]. The neoplastic epithelium of IPNBs is characterized by a spectrum of cytologic and architectural atypia. Based on these features, IPNBs are classified into low or intermediate grade intraepithelial neoplasia, high grade intraepithelial neoplasia and IPNB with invasive carcinoma[57,58] (Figure 3). Regarding the immunophenotype and the histology, IPNBs are classified into four epithelial subtypes: (1) Pancreatobiliary; (2) Gastric; (3) Intestinal; and (4) Oncocytic types, according to the immunophenotype classification of IPMNs[59,60]. The pancreatobiliary subtype is the most common subtype of IPNB followed by the intestinal, gastric and oncocytic ones[61]. Immunohistochemically, mucin core proteins are associated with the epithelial subtypes in IPNB, as well as in IPMNs[62]. MUC-1 has been reported to be expressed mainly in the pancreatobiliary subtype but not in the others, while MUC-2 is expressed in the intestinal subtype and MUC5AC in all subtypes including the oncocytic one, which also shows a focal expression of MUC-1 and/or MUC-2[63]. For this reason, the oncocytic form could be considered a variant of the pancreatobiliary subtype. While mucin hypersecretion is a typical and frequent feature of IPMNs, its presence in IPNBs is more frequently reported in the gastric and intestinal subtypes rather than in the pancreatobiliary one[64].
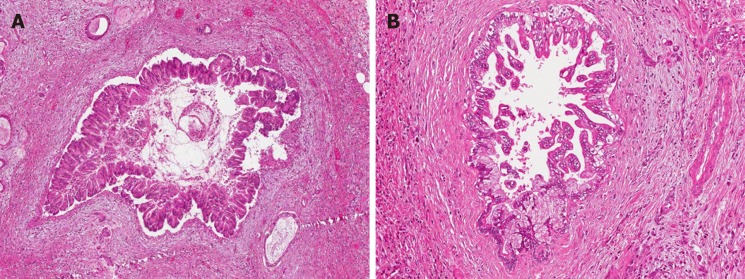
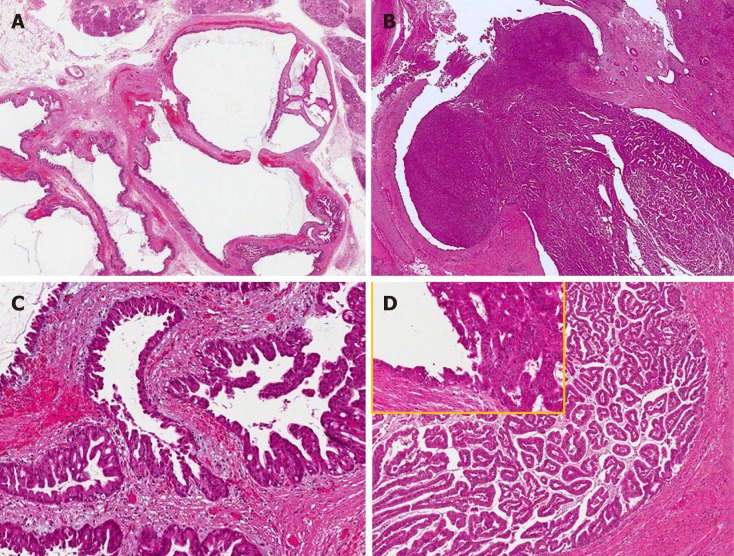
Figure 3.
Common and distinctive features of pancreatic and biliary intraductal papillary mucinous neoplasms. Low power view of intraductal papillary mucinous neoplasm of the pancreas (A) with oncocytic aspects (B), causing evident dilatation of the Wirsung duct which was detected radiologically as a macroscopic lesion. Low power view of intraductal papillary neoplasm of the bile duct (C) characterized by a polypoid growth inside the common hepatic duct, composed of multiple papillary projections with a fibrovascular stroma and covered by oncocytic cells (D). In the upper left part of panel D at higher magnification, the transition between the normal bile duct epithelium and the papillary neoplastic proliferation is evident.
The expression of several cytokeratins has also been related to the different subtypes. Particularly, CK-20 expression has been associated with the intestinal subtype, whereas CK-7 with the gastric subtype[65].
Several studies suggested a role of PBGs in the development of IPNBs[33,66]. A possible hint toward the origin of IPNBs from PBGs is represented by the high level of expression of another gastric mucin MUC6, which is not normally expressed in the biliary epithelium, both in PBGs and IPNBs, supporting the hypothesis of PBGs-derived IPNBs[67].
As far as regards the pancreas, IPMNs are defined as a grossly visible noninvasive epithelial neoplasm constituted of mucin-producing cells which could arise from the main pancreatic duct (IPMN-MD) and/or from the branch ducts (IPMN-BD)[68]. Microscopically, IPMNs show a papillary proliferation with delicate fibrovascular cores[69]. Based on cytoarchitectural atypia, IPMNs are classified according to WHO classification into: (1) Low grade dysplasia characterized by regular tall columnar cells with no or only minor signs of atypia; (2) Intermediate grade dysplasia with moderate cytological atypia, such as hyperchromasia and enlarged nuclei; and (3) High grade dysplasia, which displays marked nuclear atypia with loss of polarity, enlarged and pleomorphic nuclei[57,70].
IPMNs are extremely common incidentally diagnosed lesions with a prevalence of some 7% in the healthy population[71], the majority being represented by IPMN-BD, that are associated with a relatively low risk of neoplastic transformation estimated in 0-7% per year[72]. In contrast, IPNBs are rare lesions, mostly reported in far eastern nations like Taiwan, Japan, and Korea where chronic diseases such as hepatolithiasis and clonorchiasis are endemic[62,73]. The most common epithelial subtype of IPMN-BD is represented by the gastric subtype, which is MUC-1 negative, MUC5AC positive, and CK20 negative, meanwhile the most common epithelial subtypes reported in IPMN-MD are the intestinal and the pancreatobiliary subtypes, which are respectively MUC-2/MUC5AC positive and MUC-1 negative, and MUC-1/MUC5AC positive and MUC-2 negative[74].
Several studies reported that the pattern of expression of mucins is related with two distinct types of tumor progression that might occur in IPMNs: the MUC1positive pathway leading to tubular adenocarcinoma and the MUC2positive pathway which evolves into mucinous adenocarcinoma[75]. There is some evidence suggesting that the pancreatobiliary subtype is strongly associated with invasive tubular carcinoma which holds the poorest prognosis due to the more advanced stage at presentation and high recurrence after curative resection[76], and that MUC-1 expression is associated with a shorter overall survival, similarly to what reported in IPNBs[56,77,78].
Similarly to what has been reported for the possible origin of IPNBs from PBGs, it has been hypothesized that PDGs might have a central role in the development of IPMNs, as suggested by the observed high levels of expression of Trefoil factor family 2 (TFF2) in IPMNs, a protein expressed also by PDGs[79]. As mentioned above, the expression patterns of mucin core proteins and cytokeratins of IPMNs, especially the main duct ones, resembles that observed in IPNBs[80].
Beyond all the similarities, there are also several differences between IPMNs and IPNBs: (1) Mucin production is observed in almost all cases of IPMNs whereas it is reported in only one third of IPNB cases; (2) The gastric subtype of IPNB is rare compared to what observed for IPMNs and; (3) The expression of CK20 is far more common in IPNBs compared with IPMNs[80,81]. In conclusion, while there are evidences suggesting that IPNBs and IPMNs of the MD share common histologic and phenotypic features associated with a relatively high risk of malignant transformation, supporting the case of an almost identical entity, on the contrary, most IPMN-BDs have a different molecular profile and biological and clinical behavior.
CHOLANGIOCARCINOMA AND PANCREATIC DUCTAL ADENOCARCINOMA
As discussed above, both microscopic (PanINs and BilINs) and macroscopic (IPMNs and IPNBs) precursor lesions of pancreatic ductal adenocarcinoma (PDAC) and cholangiocarcinoma (CCA) share many common aspects, which may be explained by the described common embryologic development of the biliary tract and the pancreatic duct system.
CCA is the second most common primary liver tumors[82] and its incidence is increasing worldwide[83]. CCA constitutes a heterogeneous group of malignancies arising within the biliary tree[84,85], and based on its anatomical location, it is currently classified into intrahepatic CCA (iCCA), perihilar CCA (pCCA), and distal CCA (dCCA)[86-88]. While, the pCCA and dCCA are typically conventional mucin-producing adenocarcinomas or papillary tumors, iCCA may present histologically as conventional adenocarcinoma, cholangiolocarcinoma (CLC), or as rare histological variants[88]. Recently it appeared that the conventional iCCA comprise two separate histological subtypes: The small bile duct-type iCCA, described as nodular invasive peripheral tubular or acinar adenocarcinoma with no or minimal mucin production, and the large bile duct-type iCCA characterized by the mucinous production and by the involvement of the large IHBDs[88]. Interestingly the large bile duct-type iCCA, but not small bile duct-type iCCA, share similar characteristics with pCCA and PDAC[89].
Based on embryology, iCCA subtypes reflect the different cells of origin. In fact, several studies reported that the small bile duct-type iCCA could emerge from the hepatic/stem progenitor cells located at the level of canals of Hering or ductules, in opposition to the p/dCCA and large bile duct-type iCCA which could originate from cholangiocytes or PBG-cells[90,91]. A large body of evidences indicates that large bile duct-type iCCAs and p/dCCA originate from PBGs[92] on the basis of: (1) The anatomical location of PBGs is similar to those of large bile duct-type iCCA and p/dCCA origin in the biliary tree; (2) PBGs activation can lead to mucinous metaplasia favoring the occurrence of preneoplastic lesions; and (3) Cells with stem/progenitor markers exclusively located at the bottoms of PBGs, are detected in cancer stem cells identified in large bile duct-type iCCAs and p/dCCA[93].
PDAC is the one of leading causes of cancer mortality in developed countries, and one of the most lethal malignant neoplasms across the world[94,95]. PDAC represents nearly 90% of all pancreatic neoplasms[96] and can be considered as the pancreatic counterpart of p/dCCA. Based on these premises, we will focus on the common aspects between p/dCCA and PDAC. In fact, these malignancies both appear macroscopically as firm grayish infiltrating masses with a nodular-sclerosing pattern of growth; histologically, both can produce mucin, are generally well differentiated tubular adenocarcinomas sometimes with a micropapillary component, commonly present perineural and lymphovascular invasion and are characterized by abundant fibrous stroma[97]. On the basis of histological characteristics and in particular for the large amount of stroma, PDAC and CCA share many cross-sectional imaging features both at computed tomography (CT) scans and magnetic resonance imaging (MRI). CCA appears as a hypodense lesion on non-contrast-enhanced CT scans, and after contrast media injection in the early arterial phase presents an irregular peripheral enhancement with only minor enhancement in the center of the tumor, followed by progressive enhancement in the portal and the delayed phase, with the late phase accentuated by the presence of fibrous stroma[98,99]. On MRI, CCA usually appears as a hypo to isointense lesion on T1-weighted MR images and variably hyperintense in T2-weighted images[100]. On CT-scan PDAC behaves like a hypoattenuating mass that shows less or no enhancement compared to the normal peripheral pancreatic parenchyma. On MRI it appears as a hypointense lesion on T1-weighted with a variable appearance on T2 weighted MRI images[101,102]. New advanced endoscopic techniques such as peroral cholangioscopy and pancreatoscopy are encouraging in order to better identify p/dCCA and PDAC, their diagnostic role is useful to differentiate indeterminate biliary and pancreatic duct strictures, distinguishing benign and malignant lesions through a bioptic mapping[103,104].
From an immunohistochemical point of view, PDAC and p/dCCA are both characterized by mucin overexpression[105]. Notably, while in the healthy pancreatic ducts mucins are rarely expressed, their expression in biliopancreatic cancers is upregulated, particularly that of MUC1, MUC4, and MUC5AC[106]. MUC1 in particular, has been associated with tumor size and high grade of dysplasia. Some 80% of PDAC samples express MUC1[107-109]. Interestingly, an increased expression of MUC1 in p/dCCA is also reported, particularly, patients with high levels of MUC1 expression carry a significantly worse survival when compared to those with low MUC1 expression and high MUC2 expression[110]. A correlation between MUC1 and the presence of metastasis has also been reported in CCA[111].
Regarding MUC4, it is not expressed in normal pancreas, but it has been reported to be expressed in the earliest PanIN lesions[112]. Furthermore, MUC4 expression has been associated with the severity of dysplasia, as during carcinogenesis, a gradual expression of MUC4 has been demonstrated. An association between MUC4 expression and PDAC poor prognosis has also been reported[113,114]. In the normal biliary epithelium, MUC4 is not expressed[115], whereas in CCA, MUC4 expression is significantly related to worse prognosis, similarly to what is observed in PDAC[116,117]. Moreover, in both biliopancreatic tumors MUC5AC is overexpressed[118-120] and in studies in which p/dCCA and iCCA are distinctly evaluated, p/dCCAs more frequently express MUC5AC than iCCA[121,122].
The two most commonly mutated genes found in PDAC are Kras and p53. PDAC is arguably the most Ras-addicted cancer, in fact, Kras mutations are recognized in virtually 100% of PDAC cases, from the earliest stages[123-125]. In CCA, Kras mutations have also been reported, but in a smaller quote of cases compared to the pancreas[126]. Several studies revealed that Kras mutations occur early in BilIN during the cholangiocarcinogenic process[127]. The tumor suppressor gene p53 is also commonly mutated in both CCA and PDAC and seem to be involved in the latest phases of tumorigenesis process[128,129]. Regarding the immunophenotype profile, both PDAC and CCA are characterized by common phenotypic traits such as the expression of S100P[42,130,131]. Furthermore, they share common tumor markers such as cytokeratin-7, cytokeratin-17 and cytokeratin-19[132]. In the past decades, with the improvement of technology many studies aimed to determine the gene-expression profiles of PDAC using microarray techniques and RNA sequencing[133]. PDAC presents a significant inter-tumor and an intra-tumor heterogeneity and based on patterns of gene expression profiling, researchers classified PDAC into sub-groups with different molecular features such as (1) squamous; (2) pancreatic progenitor; (3) immunogenic; and (4) aberrantly differentiated endocrine exocrine, that reflect a different clinical behavior with a potential for specific personalized treatment[134-136]. Genomic profiling studies have highlighted distinct subtypes also regarding CCA[137], some evidences demonstrated two main classes: the inflammation and the proliferation, with the latter being associated with worse outcome compared to the inflammation class[138]. In addition, in the past few years, an increasing attention has been paid on the potential role of noncoding RNAs (ncRNAs), classified according to their length into long (> 200 nucleotides) and small (18 to 200 nucleotides), for their involvement in cell proliferation, growth, tumor progression and drug resistance of several tumors[139,140]. Among small noncoding RNAs, microRNAs (miRNAs), composed by 18 to 24 nucleotides in length, are the most investigated and they act as master gene regulators involved in a variety of cellular pathways crucial to regular cellular functions[141,142]. The earliest evidences demonstrated a role of miRNAs in cholangiocarcinogenesis, in fact, the overexpression of certain miRNAs, for example miR-21, miR-25, miR-26a, miR-191, and miR-221, is involved in proliferation, tumor growth, migration and invasion of CCA[143-147]. Many efforts have been made to isolate and characterize miRNAs on CCA specimens and body fluids (serum, bile and urine) for their possible usefulness in clinic as promising diagnostic and prognostic biomarkers[148-152]. MiRNAs are extensively studied also in pancreatic carcinogenesis; they have been classified in oncogenic and tumor suppressor miRNAs according to their role in carcinogenic process. The upregulation of certain miRNAs such as miR-21, miR-196a, miR-221/222, is involved in mechanisms of cell proliferation, migration, inhibition of apoptosis and chemoresistance of PDAC, and several evidences showed miRNAs’ potential role as prognostic and diagnostic biomarkers[153-156]. In addition, in PDAC samples certain miRNAs acting as tumor suppressor are found to be downregulated, such as miR-126 and let-7 involved in the control of migration and invasion of PDAC[157,158].
These two tumor entities, therefore, share a wide range of common features such as histological aspects and immunophenotype profiles. These molecular characteristics could explain the similar dismal prognosis of both PDAC and CCA, which is substantially due to advanced stage at time of presentation and poor response to conventional chemotherapy and radiotherapy (Table 1).
Table 1.
Main similarities and differences in terms of embryologic origin and molecular characteristics between biliary and pancreatic neoplastic lesions
| Biliary tract | Pancreas | Ref. | |
| Embryologic origin of the ductal system | EHBDs and large IHBD from ventral part of foregut; small IHBD from Mesenchymal cells surrounding the septum transversum | Ventral part of MPD from the ventral part of foregut. Dorsal part of MPD from the dorsal bud that obliterates completely; the ducts of the dorsal bud fuse with the ductal system of the ventral part | [1-5] |
| Niches of Stem/progenitors’ cells | PBGs located in EHBDs and large IHBDs. Canals of Hering | PDGs located in MPD and larger interlobular ducts > 300 μm | [6-12] |
| Microscopic (intraepithelial) precursors of cancer | BilIN-1 low grade, BilIN-2 intermediate grade, BilIN-3 high grade | PanIN low grade, previously -1, -2 PanIN high grade, previously -3 | [21-26,29,37-39] |
| Macroscopic (intraductal) precursors of cancer | IPNB | IPMN | [54-62] |
| Kras role in cancer | Mutated in 15%-30% | Mutated in > 90% | [44,123,124,127,129] |
| p53 role in cancer | Mutated in 35%-60% | Mutated 40%-87% | [49,125-129] |
| SHH role in cancer | Overexpressed ~ 50% | Overexpressed ~ 70% | [159,160] |
| Mucin expression in biliopancreatic cancers | MUC-1+ 45%-70% MUC-4+ 35%-50% (worse prognosis compared to MUC4 negative) MUC5AC+ 30%-60% | MUC-1 + ~ 80% MUC-4+ 30%-80% (worse prognosis compared to MUC4 negative) MUC5AC+ 10%-70% | [106-122] |
| S100P expression | upregulated in ~75% | upregulated in ~ 90% | [42,43,130,131] |
EHBDs: Extrahepatic bile ducts; IHBDs: Intrahepatic bile ducts; MPD: Main pancreatic duct; PBGs: Peribiliary glands; PDGs: Pancreatic ductal glands; BilIN: Biliary intraepithelial neoplasm; PanIN: Pancreatic intraepithelial neoplasm; IPNB: Intraductal papillary neoplasm of the biliary tract; IPMN: Intraductal papillary mucinous neoplasm of the pancreas.
CONCLUSION
In conclusion, preneoplastic and neoplastic lesions of the biliary tract and the pancreas share similar histologic, biologic and molecular characteristics. These features are principally due to their anatomical proximity and common embryological origin. On the basis of these important similarities, it would be relevant to conduct studies aimed at identifying new strategies to prevent and treat biliopancreatic neoplastic lesions, considering them as a common disease entity
Footnotes
Conflict-of-interest statement: No potential conflicts of interest.
Peer-review started: April 1, 2019
First decision: April 30, 2019
Article in press: July 19, 2019
Specialty type: Gastroenterology and hepatology
Country of origin:
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Barreto SG, Kawaida H, Kupeli S, Löhr JM S-Editor: Gong ZM L-Editor: A E-Editor: Ma YJ
Contributor Information
Piera Zaccari, Department of Internal Medicine and Medical Specialties, Gastroenterology Unit, Sapienza University of Rome, Rome 00161, Italy.
Vincenzo Cardinale, Department of Medico-Surgical Sciences and Biotechnologies, Sapienza University of Rome, 00161 Rome, Italy.
Carola Severi, Department of Internal Medicine and Medical Specialties, Gastroenterology Unit, Sapienza University of Rome, Rome 00161, Italy.
Federica Pedica, Pathology Department, Pancreas Translational and Clinical Research Center, San Raffaele Scientific Institute IRCCS, Milan 20132, Italy.
Guido Carpino, Department of Movement, Human and Health Sciences, Division of Health Sciences, University of Rome "Foro Italico", Rome 00161, Italy.
Eugenio Gaudio, Department of Anatomical, Histological, Forensic Medicine and Orthopedics Sciences, Division of Human Anatomy, Sapienza University of Rome, Rome 00161, Italy.
Claudio Doglioni, Pathology Department, Pancreas Translational and Clinical Research Center, San Raffaele Scientific Institute IRCCS, Milan 20132, Italy.
Maria Chiara Petrone, PancreatoBiliary Endoscopy and EUS Division, Pancreas Translational and Clinical Research Center, San Raffaele Scientific Institute IRCCS, Milan 20132, Italy.
Domenico Alvaro, Department of Translational and Precision Medicine, Sapienza University of Rome, Rome 00161, Italy.
Paolo Giorgio Arcidiacono, PancreatoBiliary Endoscopy and EUS Division, Pancreas Translational and Clinical Research Center, San Raffaele Scientific Institute IRCCS, Milan 20132, Italy, arcidiacono.paologiorgio@hsr.it..
Gabriele Capurso, PancreatoBiliary Endoscopy and EUS Division, Pancreas Translational and Clinical Research Center, San Raffaele Scientific Institute IRCCS, Milan 20132, Italy.
References
- 1.Gray H. 2000. Anatomy of the Human Body. Lea Febiger, 1918, Philadelphia [Google Scholar]
- 2.Zhu Y, Liu Q, Zhou Z, Ikeda Y. PDX1, Neurogenin-3, and MAFA: critical transcription regulators for beta cell development and regeneration. Stem Cell Res Ther. 2017;8:240. doi: 10.1186/s13287-017-0694-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ando H. Embryology of the biliary tract. Dig Surg. 2010;27:87–89. doi: 10.1159/000286463. [DOI] [PubMed] [Google Scholar]
- 4.Roskams T, Desmet V. Embryology of extra- and intrahepatic bile ducts, the ductal plate. Anat Rec (Hoboken) 2008;291:628–635. doi: 10.1002/ar.20710. [DOI] [PubMed] [Google Scholar]
- 5.Tremblay KD, Zaret KS. Distinct populations of endoderm cells converge to generate the embryonic liver bud and ventral foregut tissues. Dev Biol. 2005;280:87–99. doi: 10.1016/j.ydbio.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 6.Carpino G, Cardinale V, Onori P, Franchitto A, Berloco PB, Rossi M, Wang Y, Semeraro R, Anceschi M, Brunelli R, Alvaro D, Reid LM, Gaudio E. Biliary tree stem/progenitor cells in glands of extrahepatic and intraheptic bile ducts: an anatomical in situ study yielding evidence of maturational lineages. J Anat. 2012;220:186–199. doi: 10.1111/j.1469-7580.2011.01462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lanzoni G, Cardinale V, Carpino G. The hepatic, biliary, and pancreatic network of stem/progenitor cell niches in humans: A new reference frame for disease and regeneration. Hepatology. 2016;64:277–286. doi: 10.1002/hep.28326. [DOI] [PubMed] [Google Scholar]
- 8.Cardinale V, Wang Y, Carpino G, Mendel G, Alpini G, Gaudio E, Reid LM, Alvaro D. The biliary tree--a reservoir of multipotent stem cells. Nat Rev Gastroenterol Hepatol. 2012;9:231–240. doi: 10.1038/nrgastro.2012.23. [DOI] [PubMed] [Google Scholar]
- 9.Cardinale V, Wang Y, Carpino G, Cui CB, Gatto M, Rossi M, Berloco PB, Cantafora A, Wauthier E, Furth ME, Inverardi L, Dominguez-Bendala J, Ricordi C, Gerber D, Gaudio E, Alvaro D, Reid L. Multipotent stem/progenitor cells in human biliary tree give rise to hepatocytes, cholangiocytes, and pancreatic islets. Hepatology. 2011;54:2159–2172. doi: 10.1002/hep.24590. [DOI] [PubMed] [Google Scholar]
- 10.Carpino G, Puca R, Cardinale V, Renzi A, Scafetta G, Nevi L, Rossi M, Berloco PB, Ginanni Corradini S, Reid LM, Maroder M, Gaudio E, Alvaro D. Peribiliary Glands as a Niche of Extrapancreatic Precursors Yielding Insulin-Producing Cells in Experimental and Human Diabetes. Stem Cells. 2016;34:1332–1342. doi: 10.1002/stem.2311. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, Lanzoni G, Carpino G, Cui CB, Dominguez-Bendala J, Wauthier E, Cardinale V, Oikawa T, Pileggi A, Gerber D, Furth ME, Alvaro D, Gaudio E, Inverardi L, Reid LM. Biliary tree stem cells, precursors to pancreatic committed progenitors: evidence for possible life-long pancreatic organogenesis. Stem Cells. 2013;31:1966–1979. doi: 10.1002/stem.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carpino G, Renzi A, Cardinale V, Franchitto A, Onori P, Overi D, Rossi M, Berloco PB, Alvaro D, Reid LM, Gaudio E. Progenitor cell niches in the human pancreatic duct system and associated pancreatic duct glands: an anatomical and immunophenotyping study. J Anat. 2016;228:474–486. doi: 10.1111/joa.12418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakanuma Y. A novel approach to biliary tract pathology based on similarities to pancreatic counterparts: is the biliary tract an incomplete pancreas? Pathol Int. 2010;60:419–429. doi: 10.1111/j.1440-1827.2010.02543.x. [DOI] [PubMed] [Google Scholar]
- 14.Okazaki K, Yanagawa M, Mitsuyama T, Uchida K. Recent advances in the concept and pathogenesis of IgG4-related disease in the hepato-bilio-pancreatic system. Gut Liver. 2014;8:462–470. doi: 10.5009/gnl14107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katabathina VS, Flaherty EM, Dasyam AK, Menias CO, Riddle ND, Lath N, Kozaka K, Matsui O, Nakanuma Y, Prasad SR. "Biliary Diseases with Pancreatic Counterparts": Cross-sectional Imaging Findings. Radiographics. 2016;36:374–392. doi: 10.1148/rg.2016150071. [DOI] [PubMed] [Google Scholar]
- 16.Schmuck RB, de Carvalho-Fischer CV, Neumann C, Pratschke J, Bahra M. Distal bile duct carcinomas and pancreatic ductal adenocarcinomas: postulating a common tumor entity. Cancer Med. 2016;5:88–99. doi: 10.1002/cam4.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoadley KA, Yau C, Hinoue T, Wolf DM, Lazar AJ, Drill E, Shen R, Taylor AM, Cherniack AD, Thorsson V, Akbani R, Bowlby R, Wong CK, Wiznerowicz M, Sanchez-Vega F, Robertson AG, Schneider BG, Lawrence MS, Noushmehr H, Malta TM Cancer Genome Atlas Network, Stuart JM, Benz CC, Laird PW. Cell-of-Origin Patterns Dominate the Molecular Classification of 10,000 Tumors from 33 Types of Cancer. Cell. 2018;173:291–304.e6. doi: 10.1016/j.cell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farshidfar F, Zheng S, Gingras MC, Newton Y, Shih J, Robertson AG, Hinoue T, Hoadley KA, Gibb EA, Roszik J, Covington KR, Wu CC, Shinbrot E, Stransky N, Hegde A, Yang JD, Reznik E, Sadeghi S, Pedamallu CS, Ojesina AI, Hess JM, Auman JT, Rhie SK, Bowlby R, Borad MJ Cancer Genome Atlas Network, Zhu AX, Stuart JM, Sander C, Akbani R, Cherniack AD, Deshpande V, Mounajjed T, Foo WC, Torbenson MS, Kleiner DE, Laird PW, Wheeler DA, McRee AJ, Bathe OF, Andersen JB, Bardeesy N, Roberts LR, Kwong LN. Integrative Genomic Analysis of Cholangiocarcinoma Identifies Distinct IDH-Mutant Molecular Profiles. Cell Rep. 2017;18:2780–2794. doi: 10.1016/j.celrep.2017.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petrick JL, Yang B, Altekruse SF, Van Dyke AL, Koshiol J, Graubard BI, McGlynn KA. Risk factors for intrahepatic and extrahepatic cholangiocarcinoma in the United States: A population-based study in SEER-Medicare. PLoS One. 2017;12:e0186643. doi: 10.1371/journal.pone.0186643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang H, Dong X, Hassan M, Abbruzzese JL, Li D. Body mass index and obesity- and diabetes-associated genotypes and risk for pancreatic cancer. Cancer Epidemiol Biomarkers Prev. 2011;20:779–792. doi: 10.1158/1055-9965.EPI-10-0845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sato Y, Harada K, Sasaki M, Nakanuma Y. Histological characteristics of biliary intraepithelial neoplasia-3 and intraepithelial spread of cholangiocarcinoma. Virchows Arch. 2013;462:421–427. doi: 10.1007/s00428-013-1384-6. [DOI] [PubMed] [Google Scholar]
- 22.Sato Y, Harada K, Sasaki M, Nakanuma Y. Histological Characterization of Biliary Intraepithelial Neoplasia with respect to Pancreatic Intraepithelial Neoplasia. Int J Hepatol. 2014;2014:678260. doi: 10.1155/2014/678260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katabi N. Neoplasia of gallbladder and biliary epithelium. Arch Pathol Lab Med. 2010;134:1621–1627. doi: 10.5858/2009-0580-RAR.1. [DOI] [PubMed] [Google Scholar]
- 24.Nakanuma Y, Sasaki M, Sato Y, Ren X, Ikeda H, Harada K. Multistep carcinogenesis of perihilar cholangiocarcinoma arising in the intrahepatic large bile ducts. World J Hepatol. 2009;1:35–42. doi: 10.4254/wjh.v1.i1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zen Y, Aishima S, Ajioka Y, Haratake J, Kage M, Kondo F, Nimura Y, Sakamoto M, Sasaki M, Shimamatsu K, Wakasa K, Park YN, Chen MF, Atomi Y, Nakanuma Y. Proposal of histological criteria for intraepithelial atypical/proliferative biliary epithelial lesions of the bile duct in hepatolithiasis with respect to cholangiocarcinoma: preliminary report based on interobserver agreement. Pathol Int. 2005;55:180–188. doi: 10.1111/j.1440-1827.2005.01816.x. [DOI] [PubMed] [Google Scholar]
- 26.Akbarzadeh L, Geramizadeh B, Kazemi K, Nikeghbalian S, Malekhosseini S. Biliary Intraepithelial Neoplasia (BilIN) in Primary Sclerosing Cholangitis: The First Report from Iran. Hepat Mon. 2016;16:e38726. doi: 10.5812/hepatmon.38726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katabi N, Pillarisetty VG, DeMatteo R, Klimstra DS. Choledochal cysts: a clinicopathologic study of 36 cases with emphasis on the morphologic and the immunohistochemical features of premalignant and malignant alterations. Hum Pathol. 2014;45:2107–2114. doi: 10.1016/j.humpath.2014.06.016. [DOI] [PubMed] [Google Scholar]
- 28.Matthaei H, Lingohr P, Strässer A, Dietrich D, Rostamzadeh B, Glees S, Roering M, Möhring P, Scheerbaum M, Stoffels B, Kalff JC, Schäfer N, Kristiansen G. Biliary intraepithelial neoplasia (BilIN) is frequently found in surgical margins of biliary tract cancer resection specimens but has no clinical implications. Virchows Arch. 2015;466:133–141. doi: 10.1007/s00428-014-1689-0. [DOI] [PubMed] [Google Scholar]
- 29.Zen Y, Adsay NV, Bardadin K, Colombari R, Ferrell L, Haga H, Hong SM, Hytiroglou P, Klöppel G, Lauwers GY, van Leeuwen DJ, Notohara K, Oshima K, Quaglia A, Sasaki M, Sessa F, Suriawinata A, Tsui W, Atomi Y, Nakanuma Y. Biliary intraepithelial neoplasia: an international interobserver agreement study and proposal for diagnostic criteria. Mod Pathol. 2007;20:701–709. doi: 10.1038/modpathol.3800788. [DOI] [PubMed] [Google Scholar]
- 30.Nakanishi Y, Zen Y, Kondo S, Itoh T, Itatsu K, Nakanuma Y. Expression of cell cycle-related molecules in biliary premalignant lesions: biliary intraepithelial neoplasia and biliary intraductal papillary neoplasm. Hum Pathol. 2008;39:1153–1161. doi: 10.1016/j.humpath.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 31.Zen Y, Sasaki M, Fujii T, Chen TC, Chen MF, Yeh TS, Jan YY, Huang SF, Nimura Y, Nakanuma Y. Different expression patterns of mucin core proteins and cytokeratins during intrahepatic cholangiocarcinogenesis from biliary intraepithelial neoplasia and intraductal papillary neoplasm of the bile duct--an immunohistochemical study of 110 cases of hepatolithiasis. J Hepatol. 2006;44:350–358. doi: 10.1016/j.jhep.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 32.Sato Y, Harada K, Sasaki M, Nakanuma Y. Clinicopathological significance of S100 protein expression in cholangiocarcinoma. J Gastroenterol Hepatol. 2013;28:1422–1429. doi: 10.1111/jgh.12247. [DOI] [PubMed] [Google Scholar]
- 33.Nakanuma Y, Sato Y. Cystic and papillary neoplasm involving peribiliary glands: a biliary counterpart of branch-type intraductal papillary mucinous [corrected] neoplasm? Hepatology. 2012;55:2040–2041. doi: 10.1002/hep.25590. [DOI] [PubMed] [Google Scholar]
- 34.Carpino G, Cardinale V, Renzi A, Hov JR, Berloco PB, Rossi M, Karlsen TH, Alvaro D, Gaudio E. Activation of biliary tree stem cells within peribiliary glands in primary sclerosing cholangitis. J Hepatol. 2015;63:1220–1228. doi: 10.1016/j.jhep.2015.06.018. [DOI] [PubMed] [Google Scholar]
- 35.Terada T, Nakanuma Y. Pathological observations of intrahepatic peribiliary glands in 1,000 consecutive autopsy livers. II. A possible source of cholangiocarcinoma. Hepatology. 1990;12:92–97. doi: 10.1002/hep.1840120115. [DOI] [PubMed] [Google Scholar]
- 36.Gnoni A, Licchetta A, Scarpa A, Azzariti A, Brunetti AE, Simone G, Nardulli P, Santini D, Aieta M, Delcuratolo S, Silvestris N. Carcinogenesis of pancreatic adenocarcinoma: precursor lesions. Int J Mol Sci. 2013;14:19731–19762. doi: 10.3390/ijms141019731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Basturk O, Hong SM, Wood LD, Adsay NV, Albores-Saavedra J, Biankin AV, Brosens LA, Fukushima N, Goggins M, Hruban RH, Kato Y, Klimstra DS, Klöppel G, Krasinskas A, Longnecker DS, Matthaei H, Offerhaus GJ, Shimizu M, Takaori K, Terris B, Yachida S, Esposito I, Furukawa T Baltimore Consensus Meeting. A Revised Classification System and Recommendations From the Baltimore Consensus Meeting for Neoplastic Precursor Lesions in the Pancreas. Am J Surg Pathol. 2015;39:1730–1741. doi: 10.1097/PAS.0000000000000533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hruban RH, Adsay NV, Albores-Saavedra J, Compton C, Garrett ES, Goodman SN, Kern SE, Klimstra DS, Klöppel G, Longnecker DS, Lüttges J, Offerhaus GJ. Pancreatic intraepithelial neoplasia: a new nomenclature and classification system for pancreatic duct lesions. Am J Surg Pathol. 2001;25:579–586. doi: 10.1097/00000478-200105000-00003. [DOI] [PubMed] [Google Scholar]
- 39.Nakanuma Y, Sudo Y. Biliary tumors with pancreatic counterparts. Semin Diagn Pathol. 2017;34:167–175. doi: 10.1053/j.semdp.2016.12.013. [DOI] [PubMed] [Google Scholar]
- 40.Wilentz RE, Iacobuzio-Donahue CA, Argani P, McCarthy DM, Parsons JL, Yeo CJ, Kern SE, Hruban RH. Loss of expression of Dpc4 in pancreatic intraepithelial neoplasia: evidence that DPC4 inactivation occurs late in neoplastic progression. Cancer Res. 2000;60:2002–2006. [PubMed] [Google Scholar]
- 41.Matsuyama M, Kondo F, Ishihara T, Yamaguchi T, Ito R, Tsuyuguchi T, Tawada K, Yokosuka O. Evaluation of pancreatic intraepithelial neoplasia and mucin expression in normal pancreata. J Hepatobiliary Pancreat Sci. 2012;19:242–248. doi: 10.1007/s00534-011-0401-x. [DOI] [PubMed] [Google Scholar]
- 42.Ohuchida K, Mizumoto K, Egami T, Yamaguchi H, Fujii K, Konomi H, Nagai E, Yamaguchi K, Tsuneyoshi M, Tanaka M. S100P is an early developmental marker of pancreatic carcinogenesis. Clin Cancer Res. 2006;12:5411–5416. doi: 10.1158/1078-0432.CCR-06-0298. [DOI] [PubMed] [Google Scholar]
- 43.Dowen SE, Crnogorac-Jurcevic T, Gangeswaran R, Hansen M, Eloranta JJ, Bhakta V, Brentnall TA, Lüttges J, Klöppel G, Lemoine NR. Expression of S100P and its novel binding partner S100PBPR in early pancreatic cancer. Am J Pathol. 2005;166:81–92. doi: 10.1016/S0002-9440(10)62234-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanda M, Matthaei H, Wu J, Hong SM, Yu J, Borges M, Hruban RH, Maitra A, Kinzler K, Vogelstein B, Goggins M. Presence of somatic mutations in most early-stage pancreatic intraepithelial neoplasia. Gastroenterology. 2012;142:730–733.e9. doi: 10.1053/j.gastro.2011.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang L, Xie G, Fan Q, Xie J. Activation of the hedgehog-signaling pathway in human cancer and the clinical implications. Oncogene. 2010;29:469–481. doi: 10.1038/onc.2009.392. [DOI] [PubMed] [Google Scholar]
- 46.Morris JP. 4th, Wang SC, Hebrok M. KRAS, Hedgehog, Wnt and the twisted developmental biology of pancreatic ductal adenocarcinoma. Nat Rev Cancer. 2010;10:683–695. doi: 10.1038/nrc2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thayer SP, di Magliano MP, Heiser PW, Nielsen CM, Roberts DJ, Lauwers GY, Qi YP, Gysin S, Fernández-del Castillo C, Yajnik V, Antoniu B, McMahon M, Warshaw AL, Hebrok M. Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature. 2003;425:851–856. doi: 10.1038/nature02009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pasca di Magliano M, Sekine S, Ermilov A, Ferris J, Dlugosz AA, Hebrok M. Hedgehog/Ras interactions regulate early stages of pancreatic cancer. Genes Dev. 2006;20:3161–3173. doi: 10.1101/gad.1470806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hsu M, Sasaki M, Igarashi S, Sato Y, Nakanuma Y. KRAS and GNAS mutations and p53 overexpression in biliary intraepithelial neoplasia and intrahepatic cholangiocarcinomas. Cancer. 2013;119:1669–1674. doi: 10.1002/cncr.27955. [DOI] [PubMed] [Google Scholar]
- 50.Tang L, Tan YX, Jiang BG, Pan YF, Li SX, Yang GZ, Wang M, Wang Q, Zhang J, Zhou WP, Dong LW, Wang HY. The prognostic significance and therapeutic potential of hedgehog signaling in intrahepatic cholangiocellular carcinoma. Clin Cancer Res. 2013;19:2014–2024. doi: 10.1158/1078-0432.CCR-12-0349. [DOI] [PubMed] [Google Scholar]
- 51.Yamaguchi J, Liss AS, Sontheimer A, Mino-Kenudson M, Castillo CF, Warshaw AL, Thayer SP. Pancreatic duct glands (PDGs) are a progenitor compartment responsible for pancreatic ductal epithelial repair. Stem Cell Res. 2015;15:190–202. doi: 10.1016/j.scr.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Strobel O, Rosow DE, Rakhlin EY, Lauwers GY, Trainor AG, Alsina J, Fernández-Del Castillo C, Warshaw AL, Thayer SP. Pancreatic duct glands are distinct ductal compartments that react to chronic injury and mediate Shh-induced metaplasia. Gastroenterology. 2010;138:1166–1177. doi: 10.1053/j.gastro.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nakanuma Y, Zen Y, Harada K, Ikeda H, Sato Y, Uehara T, Sasaki M. Tumorigenesis and phenotypic characteristics of mucin-producing bile duct tumors: an immunohistochemical approach. J Hepatobiliary Pancreat Sci. 2010;17:211–222. doi: 10.1007/s00534-009-0158-7. [DOI] [PubMed] [Google Scholar]
- 54.Wan XS, Xu YY, Qian JY, Yang XB, Wang AQ, He L, Zhao HT, Sang XT. Intraductal papillary neoplasm of the bile duct. World J Gastroenterol. 2013;19:8595–8604. doi: 10.3748/wjg.v19.i46.8595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Young TH, Chen TK, Lee HS. Electronic clinical challenges and images in GI. Noninvasive and minimally invasive papillary carcinomas of the common bile duct. Gastroenterology. 2008;134:e1–e3. doi: 10.1053/j.gastro.2008.03.051. [DOI] [PubMed] [Google Scholar]
- 56.Bennett S, Marginean EC, Paquin-Gobeil M, Wasserman J, Weaver J, Mimeault R, Balaa FK, Martel G. Clinical and pathological features of intraductal papillary neoplasm of the biliary tract and gallbladder. HPB (Oxford) 2015;17:811–818. doi: 10.1111/hpb.12460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bosman FT, Carneiro F, Hruban RH, Theise ND. WHO classification of tumours of the digestive system. 4th ed. Lyon: IARC;; 2010. p. 417. [Google Scholar]
- 58.Nakanuma Y, Sasaki M, Ishikawa A, Tsui W, Chen TC, Huang SF. Biliary papillary neoplasm of the liver. Histol Histopathol. 2002;17:851–861. doi: 10.14670/HH-17.851. [DOI] [PubMed] [Google Scholar]
- 59.Adsay NV, Merati K, Basturk O, Iacobuzio-Donahue C, Levi E, Cheng JD, Sarkar FH, Hruban RH, Klimstra DS. Pathologically and biologically distinct types of epithelium in intraductal papillary mucinous neoplasms: delineation of an "intestinal" pathway of carcinogenesis in the pancreas. Am J Surg Pathol. 2004;28:839–848. doi: 10.1097/00000478-200407000-00001. [DOI] [PubMed] [Google Scholar]
- 60.Fukumura Y, Nakanuma Y, Kakuda Y, Takase M, Yao T. Clinicopathological features of intraductal papillary neoplasms of the bile duct: a comparison with intraductal papillary mucinous neoplasm of the pancreas with reference to subtypes. Virchows Arch. 2017;471:65–76. doi: 10.1007/s00428-017-2144-9. [DOI] [PubMed] [Google Scholar]
- 61.Ohtsuka M, Kimura F, Shimizu H, Yoshidome H, Kato A, Yoshitomi H, Furukawa K, Takeuchi D, Takayashiki T, Suda K, Takano S, Kondo Y, Miyazaki M. Similarities and differences between intraductal papillary tumors of the bile duct with and without macroscopically visible mucin secretion. Am J Surg Pathol. 2011;35:512–521. doi: 10.1097/PAS.0b013e3182103f36. [DOI] [PubMed] [Google Scholar]
- 62.Rocha FG, Lee H, Katabi N, DeMatteo RP, Fong Y, D'Angelica MI, Allen PJ, Klimstra DS, Jarnagin WR. Intraductal papillary neoplasm of the bile duct: a biliary equivalent to intraductal papillary mucinous neoplasm of the pancreas? Hepatology. 2012;56:1352–1360. doi: 10.1002/hep.25786. [DOI] [PubMed] [Google Scholar]
- 63.Lüttges J, Zamboni G, Longnecker D, Klöppel G. The immunohistochemical mucin expression pattern distinguishes different types of intraductal papillary mucinous neoplasms of the pancreas and determines their relationship to mucinous noncystic carcinoma and ductal adenocarcinoma. Am J Surg Pathol. 2001;25:942–948. doi: 10.1097/00000478-200107000-00014. [DOI] [PubMed] [Google Scholar]
- 64.Kim KM, Lee JK, Shin JU, Lee KH, Lee KT, Sung JY, Jang KT, Heo JS, Choi SH, Choi DW, Lim JH. Clinicopathologic features of intraductal papillary neoplasm of the bile duct according to histologic subtype. Am J Gastroenterol. 2012;107:118–125. doi: 10.1038/ajg.2011.316. [DOI] [PubMed] [Google Scholar]
- 65.Ohtsuka M, Shimizu H, Kato A, Yoshitomi H, Furukawa K, Tsuyuguchi T, Sakai Y, Yokosuka O, Miyazaki M. Intraductal papillary neoplasms of the bile duct. Int J Hepatol. 2014;2014:459091. doi: 10.1155/2014/459091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nakanishi Y, Zen Y, Hirano S, Tanaka E, Takahashi O, Yonemori A, Doumen H, Kawakami H, Itoh T, Nakanuma Y, Kondo S. Intraductal oncocytic papillary neoplasm of the bile duct: the first case of peribiliary gland origin. J Hepatobiliary Pancreat Surg. 2009;16:869–873. doi: 10.1007/s00534-009-0070-1. [DOI] [PubMed] [Google Scholar]
- 67.Nakanishi Y, Nakanuma Y, Ohara M, Iwao T, Kimura N, Ishidate T, Kijima H. Intraductal papillary neoplasm arising from peribiliary glands connecting with the inferior branch of the bile duct of the anterior segment of the liver. Pathol Int. 2011;61:773–777. doi: 10.1111/j.1440-1827.2011.02738.x. [DOI] [PubMed] [Google Scholar]
- 68.Tanaka M, Chari S, Adsay V, Fernandez-del Castillo C, Falconi M, Shimizu M, Yamaguchi K, Yamao K, Matsuno S International Association of Pancreatology. International consensus guidelines for management of intraductal papillary mucinous neoplasms and mucinous cystic neoplasms of the pancreas. Pancreatology. 2006;6:17–32. doi: 10.1159/000090023. [DOI] [PubMed] [Google Scholar]
- 69.Shi C, Hruban RH. Intraductal papillary mucinous neoplasm. Hum Pathol. 2012;43:1–16. doi: 10.1016/j.humpath.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 70.Castellano-Megías VM, Andrés CI, López-Alonso G, Colina-Ruizdelgado F. Pathological features and diagnosis of intraductal papillary mucinous neoplasm of the pancreas. World J Gastrointest Oncol. 2014;6:311–324. doi: 10.4251/wjgo.v6.i9.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zerboni G, Signoretti M, Crippa S, Falconi M, Arcidiacono PG, Capurso G. Systematic review and meta-analysis: Prevalence of incidentally detected pancreatic cystic lesions in asymptomatic individuals. Pancreatology. 2019;19:2–9. doi: 10.1016/j.pan.2018.11.014. [DOI] [PubMed] [Google Scholar]
- 72.Crippa S, Capurso G, Cammà C, Fave GD, Castillo CF, Falconi M. Risk of pancreatic malignancy and mortality in branch-duct IPMNs undergoing surveillance: A systematic review and meta-analysis. Dig Liver Dis. 2016;48:473–479. doi: 10.1016/j.dld.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 73.Jung G, Park KM, Lee SS, Yu E, Hong SM, Kim J. Long-term clinical outcome of the surgically resected intraductal papillary neoplasm of the bile duct. J Hepatol. 2012;57:787–793. doi: 10.1016/j.jhep.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 74.Nakamura A, Horinouchi M, Goto M, Nagata K, Sakoda K, Takao S, Imai K, Kim YS, Sato E, Yonezawa S. New classification of pancreatic intraductal papillary-mucinous tumour by mucin expression: its relationship with potential for malignancy. J Pathol. 2002;197:201–210. doi: 10.1002/path.1109. [DOI] [PubMed] [Google Scholar]
- 75.Adsay NV, Merati K, Andea A, Sarkar F, Hruban RH, Wilentz RE, Goggins M, Iocobuzio-Donahue C, Longnecker DS, Klimstra DS. The dichotomy in the preinvasive neoplasia to invasive carcinoma sequence in the pancreas: differential expression of MUC1 and MUC2 supports the existence of two separate pathways of carcinogenesis. Mod Pathol. 2002;15:1087–1095. doi: 10.1097/01.MP.0000028647.98725.8B. [DOI] [PubMed] [Google Scholar]
- 76.Distler M, Kersting S, Niedergethmann M, Aust DE, Franz M, Rückert F, Ehehalt F, Pilarsky C, Post S, Saeger HD, Grützmann R. Pathohistological subtype predicts survival in patients with intraductal papillary mucinous neoplasm (IPMN) of the pancreas. Ann Surg. 2013;258:324–330. doi: 10.1097/SLA.0b013e318287ab73. [DOI] [PubMed] [Google Scholar]
- 77.Kang MJ, Lee KB, Jang JY, Han IW, Kim SW. Evaluation of clinical meaning of histological subtypes of intraductal papillary mucinous neoplasm of the pancreas. Pancreas. 2013;42:959–966. doi: 10.1097/MPA.0b013e31827cddbc. [DOI] [PubMed] [Google Scholar]
- 78.Rong Y, Wang D, Xu C, Ji Y, Jin D, Wu W, Xu X, Kuang T, Lou W. Prognostic value of histological subtype in intraductal papillary mucinous neoplasm of the pancreas: A retrospective analysis of outcome from one single center. Medicine (Baltimore) 2017;96:e6599. doi: 10.1097/MD.0000000000006599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yamaguchi J, Mino-Kenudson M, Liss AS, Chowdhury S, Wang TC, Fernández-Del Castillo C, Lillemoe KD, Warshaw AL, Thayer SP. Loss of Trefoil Factor 2 From Pancreatic Duct Glands Promotes Formation of Intraductal Papillary Mucinous Neoplasms in Mice. Gastroenterology. 2016;151:1232–1244.e10. doi: 10.1053/j.gastro.2016.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zen Y, Fujii T, Itatsu K, Nakamura K, Minato H, Kasashima S, Kurumaya H, Katayanagi K, Kawashima A, Masuda S, Niwa H, Mitsui T, Asada Y, Miura S, Ohta T, Nakanuma Y. Biliary papillary tumors share pathological features with intraductal papillary mucinous neoplasm of the pancreas. Hepatology. 2006;44:1333–1343. doi: 10.1002/hep.21387. [DOI] [PubMed] [Google Scholar]
- 81.Nakanuma Y, Kakuda Y, Uesaka K, Miyata T, Yamamoto Y, Fukumura Y, Sato Y, Sasaki M, Harada K, Takase M. Characterization of intraductal papillary neoplasm of bile duct with respect to histopathologic similarities to pancreatic intraductal papillary mucinous neoplasm. Hum Pathol. 2016;51:103–113. doi: 10.1016/j.humpath.2015.12.022. [DOI] [PubMed] [Google Scholar]
- 82.Massarweh NN, El-Serag HB. Epidemiology of Hepatocellular Carcinoma and Intrahepatic Cholangiocarcinoma. Cancer Control. 2017;24:1073274817729245. doi: 10.1177/1073274817729245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cardinale V. Classifications and misclassification in cholangiocarcinoma. Liver Int. 2019;39:260–262. doi: 10.1111/liv.13998. [DOI] [PubMed] [Google Scholar]
- 84.Nakanuma Y, Curabo MP, Franceschi S, Gores G, Paradis V, Sripa B, et al. Intrahepatic cholangiocarcinoma. In: Bosman FT, Carneiro F, Hruban RH, Theise ND, editors. WHO classification of tumours of the digestive system; world health organization of tumours, 4th edn. Lyon: IARC;; 2010. pp. 217–224. [Google Scholar]
- 85.Nakanuma Y, Miyata T, Uchida T. Latest advances in the pathological understanding of cholangiocarcinomas. Expert Rev Gastroenterol Hepatol. 2016;10:113–127. doi: 10.1586/17474124.2016.1104246. [DOI] [PubMed] [Google Scholar]
- 86.Nakeeb A, Pitt HA, Sohn TA, Coleman J, Abrams RA, Piantadosi S, Hruban RH, Lillemoe KD, Yeo CJ, Cameron JL. Cholangiocarcinoma. A spectrum of intrahepatic, perihilar, and distal tumors. Ann Surg. 1996;224:463–73; discussion 473-5. doi: 10.1097/00000658-199610000-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Han Y, Glaser S, Meng F, Francis H, Marzioni M, McDaniel K, Alvaro D, Venter J, Carpino G, Onori P, Gaudio E, Alpini G, Franchitto A. Recent advances in the morphological and functional heterogeneity of the biliary epithelium. Exp Biol Med (Maywood) 2013;238:549–565. doi: 10.1177/1535370213489926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kendall T, Verheij J, Gaudio E, Evert M, Guido M, Goeppert B, Carpino G. Anatomical, histomorphological and molecular classification of cholangiocarcinoma. Liver Int. 2019;39 Suppl 1:7–18. doi: 10.1111/liv.14093. [DOI] [PubMed] [Google Scholar]
- 89.Bragazzi MC, Ridola L, Safarikia S, Matteo SD, Costantini D, Nevi L, Cardinale V. New insights into cholangiocarcinoma: multiple stems and related cell lineages of origin. Ann Gastroenterol. 2018;31:42–55. doi: 10.20524/aog.2017.0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Banales JM, Cardinale V, Carpino G, Marzioni M, Andersen JB, Invernizzi P, Lind GE, Folseraas T, Forbes SJ, Fouassier L, Geier A, Calvisi DF, Mertens JC, Trauner M, Benedetti A, Maroni L, Vaquero J, Macias RI, Raggi C, Perugorria MJ, Gaudio E, Boberg KM, Marin JJ, Alvaro D. Expert consensus document: Cholangiocarcinoma: current knowledge and future perspectives consensus statement from the European Network for the Study of Cholangiocarcinoma (ENS-CCA) Nat Rev Gastroenterol Hepatol. 2016;13:261–280. doi: 10.1038/nrgastro.2016.51. [DOI] [PubMed] [Google Scholar]
- 91.Aishima S, Oda Y. Pathogenesis and classification of intrahepatic cholangiocarcinoma: different characters of perihilar large duct type versus peripheral small duct type. J Hepatobiliary Pancreat Sci. 2015;22:94–100. doi: 10.1002/jhbp.154. [DOI] [PubMed] [Google Scholar]
- 92.Nakagawa H, Hayata Y, Yamada T, Kawamura S, Suzuki N, Koike K. Peribiliary Glands as the Cellular Origin of Biliary Tract Cancer. Int J Mol Sci. 2018;19 doi: 10.3390/ijms19061745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cardinale V, Renzi A, Carpino G, Torrice A, Bragazzi MC, Giuliante F, DeRose AM, Fraveto A, Onori P, Napoletano C, Franchitto A, Cantafora A, Grazi G, Caporaso N, D'Argenio G, Alpini G, Reid LM, Gaudio E, Alvaro D. Profiles of cancer stem cell subpopulations in cholangiocarcinomas. Am J Pathol. 2015;185:1724–1739. doi: 10.1016/j.ajpath.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Howlader N, Noone AM, Krapcho M, Miller D, Bishop K, Altekruse SF, Kosary CL, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis DR, Chen HS, Feuer EJ, Cronin KA. 2016. SEER Cancer Statistics Review, 1975-2013, National Cancer Institute. Bethesda, MD. Available from: http://seer.cancer.gov/csr/1975_2013/ [Google Scholar]
- 95.Raimondi S, Maisonneuve P, Lowenfels AB. Epidemiology of pancreatic cancer: an overview. Nat Rev Gastroenterol Hepatol. 2009;6:699–708. doi: 10.1038/nrgastro.2009.177. [DOI] [PubMed] [Google Scholar]
- 96.Luchini C, Capelli P, Scarpa A. Pancreatic Ductal Adenocarcinoma and Its Variants. Surg Pathol Clin. 2016;9:547–560. doi: 10.1016/j.path.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 97.Nakanuma Y, Sato Y. Hilar cholangiocarcinoma is pathologically similar to pancreatic duct adenocarcinoma: suggestions of similar background and development. J Hepatobiliary Pancreat Sci. 2014;21:441–447. doi: 10.1002/jhbp.70. [DOI] [PubMed] [Google Scholar]
- 98.Valls C, Gumà A, Puig I, Sanchez A, Andía E, Serrano T, Figueras J. Intrahepatic peripheral cholangiocarcinoma: CT evaluation. Abdom Imaging. 2000;25:490–496. doi: 10.1007/s002610000079. [DOI] [PubMed] [Google Scholar]
- 99.Ringe KI, Wacker F. Radiological diagnosis in cholangiocarcinoma: Application of computed tomography, magnetic resonance imaging, and positron emission tomography. Best Pract Res Clin Gastroenterol. 2015;29:253–265. doi: 10.1016/j.bpg.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 100.Jhaveri KS, Hosseini-Nik H. MRI of cholangiocarcinoma. J Magn Reson Imaging. 2015;42:1165–1179. doi: 10.1002/jmri.24810. [DOI] [PubMed] [Google Scholar]
- 101.Park HS, Lee JM, Choi HK, Hong SH, Han JK, Choi BI. Preoperative evaluation of pancreatic cancer: comparison of gadolinium-enhanced dynamic MRI with MR cholangiopancreatography versus MDCT. J Magn Reson Imaging. 2009;30:586–595. doi: 10.1002/jmri.21889. [DOI] [PubMed] [Google Scholar]
- 102.Tamm EP, Bhosale PR, Vikram R, de Almeida Marcal LP, Balachandran A. Imaging of pancreatic ductal adenocarcinoma: State of the art. World J Radiol. 2013;5:98–105. doi: 10.4329/wjr.v5.i3.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.De Luca L, Repici A, Koçollari A, Auriemma F, Bianchetti M, Mangiavillano B. Pancreatoscopy: An update. World J Gastrointest Endosc. 2019;11:22–30. doi: 10.4253/wjge.v11.i1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ogawa T, Ito K, Koshita S, Kanno Y, Masu K, Kusunose H, Sakai T, Murabayashi T, Hasegawa S, Noda Y. Usefulness of cholangioscopic-guided mapping biopsy using SpyGlass DS for preoperative evaluation of extrahepatic cholangiocarcinoma: a pilot study. Endosc Int Open. 2018;6:E199–E204. doi: 10.1055/s-0043-117949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Moschovis D, Bamias G, Delladetsima I. Mucins in neoplasms of pancreas, ampulla of Vater and biliary system. World J Gastrointest Oncol. 2016;8:725–734. doi: 10.4251/wjgo.v8.i10.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jonckheere N, Skrypek N, Van Seuningen I. Mucins and pancreatic cancer. Cancers (Basel) 2010;2:1794–1812. doi: 10.3390/cancers2041794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Suh H, Pillai K, Morris DL. Mucins in pancreatic cancer: biological role, implications in carcinogenesis and applications in diagnosis and therapy. Am J Cancer Res. 2017;7:1372–1383. [PMC free article] [PubMed] [Google Scholar]
- 108.Levi E, Klimstra DS, Andea A, Basturk O, Adsay NV. MUC1 and MUC2 in pancreatic neoplasia. J Clin Pathol. 2004;57:456–462. doi: 10.1136/jcp.2003.013292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chhieng DC, Benson E, Eltoum I, Eloubeidi MA, Jhala N, Jhala D, Siegal GP, Grizzle WE, Manne U. MUC1 and MUC2 expression in pancreatic ductal carcinoma obtained by fine-needle aspiration. Cancer. 2003;99:365–371. doi: 10.1002/cncr.11857. [DOI] [PubMed] [Google Scholar]
- 110.Tamada S, Goto M, Nomoto M, Nagata K, Shimizu T, Tanaka S, Sakoda K, Imai K, Yonezawa S. Expression of MUC1 and MUC2 mucins in extrahepatic bile duct carcinomas: its relationship with tumor progression and prognosis. Pathol Int. 2002;52:713–723. doi: 10.1046/j.1440-1827.2002.01414.x. [DOI] [PubMed] [Google Scholar]
- 111.Mall AS, Tyler MG, Ho SB, Krige JE, Kahn D, Spearman W, Myer L, Govender D. The expression of MUC mucin in cholangiocarcinoma. Pathol Res Pract. 2010;206:805–809. doi: 10.1016/j.prp.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 112.Andrianifahanana M, Moniaux N, Schmied BM, Ringel J, Friess H, Hollingsworth MA, Büchler MW, Aubert JP, Batra SK. Mucin (MUC) gene expression in human pancreatic adenocarcinoma and chronic pancreatitis: a potential role of MUC4 as a tumor marker of diagnostic significance. Clin Cancer Res. 2001;7:4033–4040. [PubMed] [Google Scholar]
- 113.Gautam SK, Kumar S, Cannon A, Hall B, Bhatia R, Nasser MW, Mahapatra S, Batra SK, Jain M. MUC4 mucin- a therapeutic target for pancreatic ductal adenocarcinoma. Expert Opin Ther Targets. 2017;21:657–669. doi: 10.1080/14728222.2017.1323880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Saitou M, Goto M, Horinouchi M, Tamada S, Nagata K, Hamada T, Osako M, Takao S, Batra SK, Aikou T, Imai K, Yonezawa S. MUC4 expression is a novel prognostic factor in patients with invasive ductal carcinoma of the pancreas. J Clin Pathol. 2005;58:845–852. doi: 10.1136/jcp.2004.023572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Vandenhaute B, Buisine MP, Debailleul V, Clément B, Moniaux N, Dieu MC, Degand P, Porchet N, Aubert JP. Mucin gene expression in biliary epithelial cells. J Hepatol. 1997;27:1057–1066. doi: 10.1016/s0168-8278(97)80150-x. [DOI] [PubMed] [Google Scholar]
- 116.Tamada S, Shibahara H, Higashi M, Goto M, Batra SK, Imai K, Yonezawa S. MUC4 is a novel prognostic factor of extrahepatic bile duct carcinoma. Clin Cancer Res. 2006;12:4257–4264. doi: 10.1158/1078-0432.CCR-05-2814. [DOI] [PubMed] [Google Scholar]
- 117.Shibahara H, Tamada S, Higashi M, Goto M, Batra SK, Hollingsworth MA, Imai K, Yonezawa S. MUC4 is a novel prognostic factor of intrahepatic cholangiocarcinoma-mass forming type. Hepatology. 2004;39:220–229. doi: 10.1002/hep.20031. [DOI] [PubMed] [Google Scholar]
- 118.Jinfeng M, Kimura W, Hirai I, Sakurai F, Moriya T, Mizutani M. Expression of MUC5AC and MUC6 in invasive ductal carcinoma of the pancreas and relationship with prognosis. Int J Gastrointest Cancer. 2003;34:9–18. doi: 10.1385/IJGC:34:1:09. [DOI] [PubMed] [Google Scholar]
- 119.Yonezawa S, Horinouchi M, Osako M, Kubo M, Takao S, Arimura Y, Nagata K, Tanaka S, Sakoda K, Aikou T, Sato E. Gene expression of gastric type mucin (MUC5AC) in pancreatic tumors: its relationship with the biological behavior of the tumor. Pathol Int. 1999;49:45–54. doi: 10.1046/j.1440-1827.1999.00823.x. [DOI] [PubMed] [Google Scholar]
- 120.Okano K, Yoshizawa T, Miura T, Ishido K, Kudo D, Kimura N, Wakiya TI, Wu Y, Morohashi S, Hakamada K, Kijima H. Impact of the histological phenotype of extrahepatic bile duct carcinoma. Mol Clin Oncol. 2018;8:54–60. doi: 10.3892/mco.2017.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Park SY, Roh SJ, Kim YN, Kim SZ, Park HS, Jang KY, Chung MJ, Kang MJ, Lee DG, Moon WS. Expression of MUC1, MUC2, MUC5AC and MUC6 in cholangiocarcinoma: prognostic impact. Oncol Rep. 2009;22:649–657. doi: 10.3892/or_00000485. [DOI] [PubMed] [Google Scholar]
- 122.Lee MJ, Lee HS, Kim WH, Choi Y, Yang M. Expression of mucins and cytokeratins in primary carcinomas of the digestive system. Mod Pathol. 2003;16:403–410. doi: 10.1097/01.MP.0000067683.84284.66. [DOI] [PubMed] [Google Scholar]
- 123.Almoguera C, Shibata D, Forrester K, Martin J, Arnheim N, Perucho M. Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell. 1988;53:549–554. doi: 10.1016/0092-8674(88)90571-5. [DOI] [PubMed] [Google Scholar]
- 124.Waters AM, Der CJ. KRAS: The Critical Driver and Therapeutic Target for Pancreatic Cancer. Cold Spring Harb Perspect Med. 2018;8 doi: 10.1101/cshperspect.a031435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Boschman CR, Stryker S, Reddy JK, Rao MS. Expression of p53 protein in precursor lesions and adenocarcinoma of human pancreas. Am J Pathol. 1994;145:1291–1295. [PMC free article] [PubMed] [Google Scholar]
- 126.Liu XF, Zhang H, Zhu SG, Zhou XT, Su HL, Xu Z, Li SJ. Correlation of p53 gene mutation and expression of P53 protein in cholangiocarcinoma. World J Gastroenterol. 2006;12:4706–4709. doi: 10.3748/wjg.v12.i29.4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rashid A, Ueki T, Gao YT, Houlihan PS, Wallace C, Wang BS, Shen MC, Deng J, Hsing AW. K-ras mutation, p53 overexpression, and microsatellite instability in biliary tract cancers: a population-based study in China. Clin Cancer Res. 2002;8:3156–3163. [PubMed] [Google Scholar]
- 128.Khan SA, Thomas HC, Toledano MB, Cox IJ, Taylor-Robinson SD. p53 Mutations in human cholangiocarcinoma: a review. Liver Int. 2005;25:704–716. doi: 10.1111/j.1478-3231.2005.01106.x. [DOI] [PubMed] [Google Scholar]
- 129.Lu L, Zeng J. Evaluation of K-ras and p53 expression in pancreatic adenocarcinoma using the cancer genome atlas. PLoS One. 2017;12:e0181532. doi: 10.1371/journal.pone.0181532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ali A, Brown V, Denley S, Jamieson NB, Morton JP, Nixon C, Graham JS, Sansom OJ, Carter CR, McKay CJ, Duthie FR, Oien KA. Expression of KOC, S100P, mesothelin and MUC1 in pancreatico-biliary adenocarcinomas: development and utility of a potential diagnostic immunohistochemistry panel. BMC Clin Pathol. 2014;14:35. doi: 10.1186/1472-6890-14-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hamada S, Satoh K, Hirota M, Kanno A, Ishida K, Umino J, Ito H, Kikuta K, Kume K, Masamune A, Katayose Y, Unno M, Shimosegawa T. Calcium-binding protein S100P is a novel diagnostic marker of cholangiocarcinoma. Cancer Sci. 2011;102:150–156. doi: 10.1111/j.1349-7006.2010.01757.x. [DOI] [PubMed] [Google Scholar]
- 132.Perysinakis I, Margaris I, Kouraklis G. Ampullary cancer--a separate clinical entity? Histopathology. 2014;64:759–768. doi: 10.1111/his.12324. [DOI] [PubMed] [Google Scholar]
- 133.Mao Y, Shen J, Lu Y, Lin K, Wang H, Li Y, Chang P, Walker MG, Li D. RNA sequencing analyses reveal novel differentially expressed genes and pathways in pancreatic cancer. Oncotarget. 2017;8:42537–42547. doi: 10.18632/oncotarget.16451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, Miller DK, Christ AN, Bruxner TJ, Quinn MC, Nourse C, Murtaugh LC, Harliwong I, Idrisoglu S, Manning S, Nourbakhsh E, Wani S, Fink L, Holmes O, Chin V, Anderson MJ, Kazakoff S, Leonard C, Newell F, Waddell N, Wood S, Xu Q, Wilson PJ, Cloonan N, Kassahn KS, Taylor D, Quek K, Robertson A, Pantano L, Mincarelli L, Sanchez LN, Evers L, Wu J, Pinese M, Cowley MJ, Jones MD, Colvin EK, Nagrial AM, Humphrey ES, Chantrill LA, Mawson A, Humphris J, Chou A, Pajic M, Scarlett CJ, Pinho AV, Giry-Laterriere M, Rooman I, Samra JS, Kench JG, Lovell JA, Merrett ND, Toon CW, Epari K, Nguyen NQ, Barbour A, Zeps N, Moran-Jones K, Jamieson NB, Graham JS, Duthie F, Oien K, Hair J, Grützmann R, Maitra A, Iacobuzio-Donahue CA, Wolfgang CL, Morgan RA, Lawlor RT, Corbo V, Bassi C, Rusev B, Capelli P, Salvia R, Tortora G, Mukhopadhyay D, Petersen GM Australian Pancreatic Cancer Genome Initiative, Munzy DM, Fisher WE, Karim SA, Eshleman JR, Hruban RH, Pilarsky C, Morton JP, Sansom OJ, Scarpa A, Musgrove EA, Bailey UM, Hofmann O, Sutherland RL, Wheeler DA, Gill AJ, Gibbs RA, Pearson JV, Waddell N, Biankin AV, Grimmond SM. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 135.Zhao L, Zhao H, Yan H. Gene expression profiling of 1200 pancreatic ductal adenocarcinoma reveals novel subtypes. BMC Cancer. 2018;18:603. doi: 10.1186/s12885-018-4546-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Collisson EA, Sadanandam A, Olson P, Gibb WJ, Truitt M, Gu S, Cooc J, Weinkle J, Kim GE, Jakkula L, Feiler HS, Ko AH, Olshen AB, Danenberg KL, Tempero MA, Spellman PT, Hanahan D, Gray JW. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med. 2011;17:500–503. doi: 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhong W, Dai L, Liu J, Zhou S. Cholangiocarcinoma‑associated genes identified by integrative analysis of gene expression data. Mol Med Rep. 2018;17:5744–5753. doi: 10.3892/mmr.2018.8594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sia D, Hoshida Y, Villanueva A, Roayaie S, Ferrer J, Tabak B, Peix J, Sole M, Tovar V, Alsinet C, Cornella H, Klotzle B, Fan JB, Cotsoglou C, Thung SN, Fuster J, Waxman S, Garcia-Valdecasas JC, Bruix J, Schwartz ME, Beroukhim R, Mazzaferro V, Llovet JM. Integrative molecular analysis of intrahepatic cholangiocarcinoma reveals 2 classes that have different outcomes. Gastroenterology. 2013;144:829–840. doi: 10.1053/j.gastro.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339:159–166. doi: 10.1016/j.canlet.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 141.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 142.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Selaru FM, Olaru AV, Kan T, David S, Cheng Y, Mori Y, Yang J, Paun B, Jin Z, Agarwal R, Hamilton JP, Abraham J, Georgiades C, Alvarez H, Vivekanandan P, Yu W, Maitra A, Torbenson M, Thuluvath PJ, Gores GJ, LaRusso NF, Hruban R, Meltzer SJ. MicroRNA-21 is overexpressed in human cholangiocarcinoma and regulates programmed cell death 4 and tissue inhibitor of metalloproteinase 3. Hepatology. 2009;49:1595–1601. doi: 10.1002/hep.22838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Razumilava N, Bronk SF, Smoot RL, Fingas CD, Werneburg NW, Roberts LR, Mott JL. miR-25 targets TNF-related apoptosis inducing ligand (TRAIL) death receptor-4 and promotes apoptosis resistance in cholangiocarcinoma. Hepatology. 2012;55:465–475. doi: 10.1002/hep.24698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhang J, Han C, Wu T. MicroRNA-26a promotes cholangiocarcinoma growth by activating β-catenin. Gastroenterology. 2012;143:246–56.e8. doi: 10.1053/j.gastro.2012.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Li H, Zhou ZQ, Yang ZR, Tong DN, Guan J, Shi BJ, Nie J, Ding XT, Li B, Zhou GW, Zhang ZY. MicroRNA-191 acts as a tumor promoter by modulating the TET1-p53 pathway in intrahepatic cholangiocarcinoma. Hepatology. 2017;66:136–151. doi: 10.1002/hep.29116. [DOI] [PubMed] [Google Scholar]
- 147.Li J, Yao L, Li G, Ma D, Sun C, Gao S, Zhang P, Gao F. miR-221 Promotes Epithelial-Mesenchymal Transition through Targeting PTEN and Forms a Positive Feedback Loop with β-catenin/c-Jun Signaling Pathway in Extra-Hepatic Cholangiocarcinoma. PLoS One. 2015;10:e0141168. doi: 10.1371/journal.pone.0141168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wang LJ, He CC, Sui X, Cai MJ, Zhou CY, Ma JL, Wu L, Wang H, Han SX, Zhu Q. MiR-21 promotes intrahepatic cholangiocarcinoma proliferation and growth in vitro and in vivo by targeting PTPN14 and PTEN. Oncotarget. 2015;6:5932–5946. doi: 10.18632/oncotarget.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zheng B, Jeong S, Zhu Y, Chen L, Xia Q. miRNA and lncRNA as biomarkers in cholangiocarcinoma(CCA) Oncotarget. 2017;8:100819–100830. doi: 10.18632/oncotarget.19044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Voigtländer T, Gupta SK, Thum S, Fendrich J, Manns MP, Lankisch TO, Thum T. MicroRNAs in Serum and Bile of Patients with Primary Sclerosing Cholangitis and/or Cholangiocarcinoma. PLoS One. 2015;10:e0139305. doi: 10.1371/journal.pone.0139305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Silakit R, Loilome W, Yongvanit P, Thongchot S, Sithithaworn P, Boonmars T, Koonmee S, Titapun A, Khuntikeo N, Chamadol N, Techasen A, Namwat N. Urinary microRNA-192 and microRNA-21 as potential indicators for liver fluke-associated cholangiocarcinoma risk group. Parasitol Int. 2017;66:479–485. doi: 10.1016/j.parint.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 152.Li L, Masica D, Ishida M, Tomuleasa C, Umegaki S, Kalloo AN, Georgiades C, Singh VK, Khashab M, Amateau S, Li Z, Okolo P, Lennon AM, Saxena P, Geschwind JF, Schlachter T, Hong K, Pawlik TM, Canto M, Law J, Sharaiha R, Weiss CR, Thuluvath P, Goggins M, Shin EJ, Peng H, Kumbhari V, Hutfless S, Zhou L, Mezey E, Meltzer SJ, Karchin R, Selaru FM. Human bile contains microRNA-laden extracellular vesicles that can be used for cholangiocarcinoma diagnosis. Hepatology. 2014;60:896–907. doi: 10.1002/hep.27050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hu GY, Tao F, Wang W, Ji KW. Prognostic value of microRNA-21 in pancreatic ductal adenocarcinoma: a meta-analysis. World J Surg Oncol. 2016;14:82. doi: 10.1186/s12957-016-0842-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Dillhoff M, Liu J, Frankel W, Croce C, Bloomston M. MicroRNA-21 is overexpressed in pancreatic cancer and a potential predictor of survival. J Gastrointest Surg. 2008;12:2171–2176. doi: 10.1007/s11605-008-0584-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Huang F, Tang J, Zhuang X, Zhuang Y, Cheng W, Chen W, Yao H, Zhang S. MiR-196a promotes pancreatic cancer progression by targeting nuclear factor kappa-B-inhibitor alpha. PLoS One. 2014;9:e87897. doi: 10.1371/journal.pone.0087897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Xu Q, Li P, Chen X, Zong L, Jiang Z, Nan L, Lei J, Duan W, Zhang D, Li X, Sha H, Wu Z, Ma Q, Wang Z. miR-221/222 induces pancreatic cancer progression through the regulation of matrix metalloproteinases. Oncotarget. 2015;6:14153–14164. doi: 10.18632/oncotarget.3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Frampton AE, Krell J, Jacob J, Stebbing J, Castellano L, Jiao LR. Loss of miR-126 is crucial to pancreatic cancer progression. Expert Rev Anticancer Ther. 2012;12:881–884. doi: 10.1586/era.12.67. [DOI] [PubMed] [Google Scholar]
- 158.Jiao LR, Frampton AE, Jacob J, Pellegrino L, Krell J, Giamas G, Tsim N, Vlavianos P, Cohen P, Ahmad R, Keller A, Habib NA, Stebbing J, Castellano L. MicroRNAs targeting oncogenes are down-regulated in pancreatic malignant transformation from benign tumors. PLoS One. 2012;7:e32068. doi: 10.1371/journal.pone.0032068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Riedlinger D, Bahra M, Boas-Knoop S, Lippert S, Bradtmöller M, Guse K, Seehofer D, Bova R, Sauer IM, Neuhaus P, Koch A, Kamphues C. Hedgehog pathway as a potential treatment target in human cholangiocarcinoma. J Hepatobiliary Pancreat Sci. 2014;21:607–615. doi: 10.1002/jhbp.107. [DOI] [PubMed] [Google Scholar]
- 160.Gu D, Schlotman KE, Xie J. Deciphering the role of hedgehog signaling in pancreatic cancer. J Biomed Res. 2016;30:353–360. doi: 10.7555/JBR.30.20150107. [DOI] [PMC free article] [PubMed] [Google Scholar]