Abstract
RNA analysis at the cellular resolution in the human brain is challenging. Here, we describe an optimised approach for detecting single RNA transcripts in a cell-type specific manner in frozen human brain tissue using multiplexed fluorescent RNAscope probes. We developed a new robust analytical approach for RNAscope quantification. Our method shows that low RNA integrity does not significantly affect RNAscope signal, recapitulates bulk RNA analysis and provides spatial context to transcriptomic analysis of human post-mortem brain at single-cell resolution. In summary, our optimised method allows the usage of frozen human samples from brain banks to perform quantitative RNAscope analysis.
Subject terms: Transcription, Molecular neuroscience
Introduction
Molecular characterisation of different cell-types in complex human tissues such as the brain is of crucial importance for both fundamental and applied research. The human brain is composed of potentially hundreds of different cell types displaying complex morphologies and often long, ramified and entwined processes, making its dissociation into single cells a difficult process. RNA-seq of immunopanned human brain cell types has been performed from fresh biopsies obtained from neurosurgical site1, but these samples are non-existent for neurodegenerative diseases; instead, neuroscientists normally access frozen or fixed post-mortem human brain samples from brain banks. However, single-cell preparation from snap frozen tissue is extremely difficult, thus making single-cell sorting virtually impossible. To overcome these limitations, several laboratories have performed single-nucleus RNA-seq. This method can be readily applied to frozen tissue samples and it has been shown to well recapitulate the cellular transcriptome2,3. Nonetheless, this method has its own limitations, notably the loss of any compartmentalised cytoplasmic RNA3. Single-cell or single-nucleus-based transcriptomics fail to capture spatial context, cell-to-cell relationships and circuitry structure. In situ hybridisation (ISH), which visualises specific mRNA molecules at a cellular level, is ideal to study gene expression in different cell types because it uses intact tissue sections, therefore maintaining tissue integrity, cell structure and morphology4,5. Additionally, ISH has been successfully used for systematic analyses of RNA expression in the rodent brain (Allen Brain Atlas). Over the last decade, new and more sensitive ISH methods have been developed4, including single molecule RNA fluorescent in situ hybridization (smFISH)6,7, enabling quantification of RNA expression levels in specific cell types at single-cell resolution. Crucially, post-mortem human brain tissue presents higher rates of RNA degradation8, making bulk and single-cell or single-nuclei RNA analyses more complex.
Alzheimer’s disease (AD) is a devastating neurodegenerative disorder that accounts for two thirds of total dementia cases, and is characterised by progressive cognitive impairment and memory loss leading to difficulties in daily tasks9. The AD brain displays extracellular amyloid beta (Aβ) deposition and intracellular tau aggregates (neurofibrillary tangles), resulting in synapse loss and neuronal death10. In addition, AD has been associated with changes in the molecular profile of microglia and astrocytes11,12. Thus, studying molecular changes in human AD samples at single-cell resolution by a combination of smFISH and image analysis methods is critical for elucidating the mechanisms underlying neurodegenerative processes, including the investigation of cell-cell relationships and the impact of secondary features (e.g. Aβ plaques) in spatial context.
In this study, we show that multiplexed RNAscope, a commercially available smFISH assay13, can be successfully employed in frozen human hippocampal samples obtained from brain banks. In addition, we developed an analysis pipeline to quantify levels of single RNA molecules in major cell-type populations of the human brain with single-cell resolution. Importantly, we show that low RNA integrity score (RNA Quality Indicator; RQI = 2.9) does not significantly affect smFISH signal, thus frozen samples with partially degraded RNA could be used for smFISH. By analysing the expression of three cell-type specific genes (SLC1A2 for astrocytes, MAP2 for neurons and P2RY12 for microglia), we demonstrate that our adapted smFISH protocol is suitable to discriminate RNA expression in a cell-type specific manner. Finally, we use our optimised smFISH protocol and pipeline analysis to study gene expression in the context of neurodegeneration. We performed multiplexed smFISH to detect three cell-type specific genes known to be up- or downregulated in AD: TREM2 in microglia, SNAP25 in neurons and DKK1 in neurons and astrocytes. Our results for TREM2, SNAP25 and DKK1 compare favourably to bulk qPCR and recapitulate previously reported changes in expression between control and AD cases.
Results
RNAscope can be used in frozen human brain samples
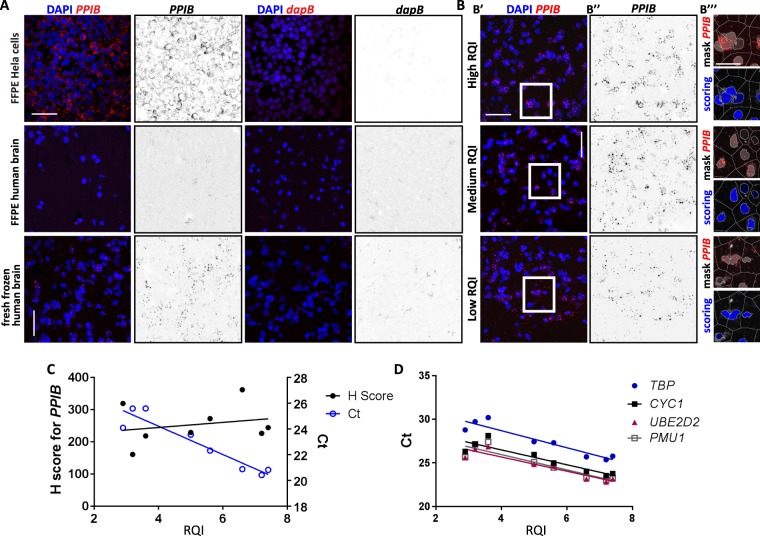
The aims of this study were to establish a protocol for visualising specific mRNA species in different human brain cell types, and to develop a method to quantify the data. We used this technique on tissue samples from brain banks, which are preserved as formalin-fixed paraffin-embedded (FFPE) tissue blocks or fresh-frozen material. First, we performed smFISH in FFPE HeLa cells (Fig. 1A) and FFPE human hippocampal sections (Fig. 1A). Probes against the widely expressed Peptidylprolyl Isomerase B (PPIB) and against the bacterial gene dapB were used as positive and negative controls, respectively. Although a study successfully used chromogenic-RNAscope in human hippocampal sections14, our fluorescence read-out was not conclusive, with only a few cells showing expression of PPIB. Another study reported the use of chromogenic-BaseScope (an RNAscope approach using shorter probes) in human brain tissue15. We therefore performed BaseScope in FFPE human hippocampal sections but we could not detect any consistent signal (data not shown), and the fact that BaseScope cannot be multiplexed limits the applicability of this method. Based on these results, we hypothesised that long formalin fixation (2–3 weeks) and/or deparaffinisation might degrade RNA or block accessibility of the probes to the target RNA, thus impairing smFISH staining.
Figure 1.
Use of RNAscope to visualise gene expression in human hippocampal biopsies from healthy individuals. (A) Confocal images of Hela cell FFPE pellets (top panel), FFPE (middle panel) and fresh frozen (bottom panel) hippocampal human sections showing no signal for the negative control (dapB) and variable signal levels of the positive control probe (PPIB; red). DAPI indicates nuclear staining (blue). Scale bars 50 µm. (B) Confocal fluorescent RNAscope shows expression of PPIB in fresh frozen hippocampal sections with high (7.2–7.4, top panel), medium (5–6.6, middle panel) and low (2.9–3.6, right panel) RNA Quality Indicator (RQI). B’ shows double staining for DAPI (blue) and PPIB (red). B” shows PPIB expression (black). (B’’’) Mark-up overlay illustrating the cell by cell segmentation and scoring of the ISH signals (nuclear colour coding indicates the expression level 0+ = black, increasingly brighter shades of blue indicating 1+, 2+, 3+ and 4+ cells). Scale bars 50 µm, zoom imaged 20 µm. (C) Black filled circles: quantification of the number of puncta/cell (H-score) in PPIB-positive cells showing no differences in sections with different RQIs (R = 0.227; p = 0.5889). Blue hollow circles: qPCR values (Ct) for PPIB gene showing a correlation between RQI and Ct (R = −0.942; p = 0.0004). (D) Quantitative qPCR showing a correlation between Ct and RQI of four housekeeping genes, including PMU1 (R = −0.918; 0.0013), TBP (R = −0.941; p = 0.0004), CYC1 (R = −0.916; p = 0.0013) and UBE2D2 (R = −0.936; p = 0.0006).
To overcome the limitations of long-term fixation of FFPE protocols used by brain banks, we hypothesised that smFISH could be successfully implemented by using frozen tissue with high RNA integrity. We extracted RNA from different hippocampal samples and analysed RNA integrity by determining the RNA Quality Indicator (RQI) score (from 1 to 10, with 10 being the best quality). RQI of our samples ranged from 2.9 to 7.4. Samples with high RQI (above 7) were selected to test whether human hippocampal sections from frozen samples can be used for smFISH assays. A clear signal for the positive probe (PPIB) was detected in comparison with the negative control probe (dapB; Fig. 1A). These experiments demonstrate that RNAscope can be successfully used in frozen human brain samples with high RNA integrity.
RNA integrity does not affect RNAscope signal
The brain is characterised by a higher RNA degradation rate than other tissues8. It has been shown that low RNA quality (RQI < 3.9) negatively impacts on qPCR amplification8 and is not optimal for RNA-seq16. Samples with low RQIs must therefore be excluded from the experimental datasets. To determine if low RQI influences RNAscope signal, we performed smFISH on frozen brain tissues from three different RQI groups: high (7.2–7.4), medium (5–6.6) and low (2.9–3.6; Fig. 1B’,B”). We did not observe qualitative differences in smFISH puncta per cell between the different RQI groups. However, to conclude if low RQI impacts on smFISH signal, proper quantification of fluorescent puncta and their correct attribution to the cognate cell is paramount.
To quantify smFISH puncta, we have implemented a new approach using the HALO v2.3.2089.18 (Indica Labs, Corrales, NM, USA) software platform. This approach allows reproducible quantification of mRNA signals in multiplexed smFISH at single-cell resolution and provides spatial coding and morphometric characterization of the cell populations of interest (see Methods for details). Briefly, we performed colour deconvolution to optimise the detection of probes and nuclear counterstain. We optimised nuclear segmentation and contour detection using the colour and intensity attributes of the nuclear counterstain in combination with cell classification features. Absolute cell counts were derived for each region analysed. Probe detection was performed based on channel deconvolution and probe signals were assigned to each nucleus by proximity. Cell-by-cell smFISH signal counts were derived for each probe on a continuous scale. Graphical mark-ups were generated to visualize cell-by-cell probe quantification within the microarchitecture of the hippocampus (Fig. 1B’’’ and Supp. Fig. 1, see Methods for further detail). By using this new quantitative approach, we analysed PPIB levels (Fig. 1C) and observed that low RQI is not associated with a reduced number of smFISH puncta per cell (H-score; Fig. 1C black filled dots, R = 0.227, p = 0.5889). In contrast, RQI affected qPCR amplification of PPIB and other reference genes in the same samples (Fig. 1C,D blue hollow dots, R = −0.942, p = 0.0004), as previously reported8. To further demonstrate that low RQI does not significantly impact on the count of smFISH signals, we quantified the levels of another common housekeeping gene (TATA-Box Binding Protein; TBP) in samples covering a range of RQI, from low to high. As shown for PPIB, low RNA integrity does not result in different levels of smFISH signal for TBP (Supp. Fig. 2). Altogether, these data indicate that RNAscope can be successfully employed in frozen human brain sections even with low RNA integrity (RQI ≥ 2.9).
Cell-type discrimination of differentially expressed genes by RNAscope
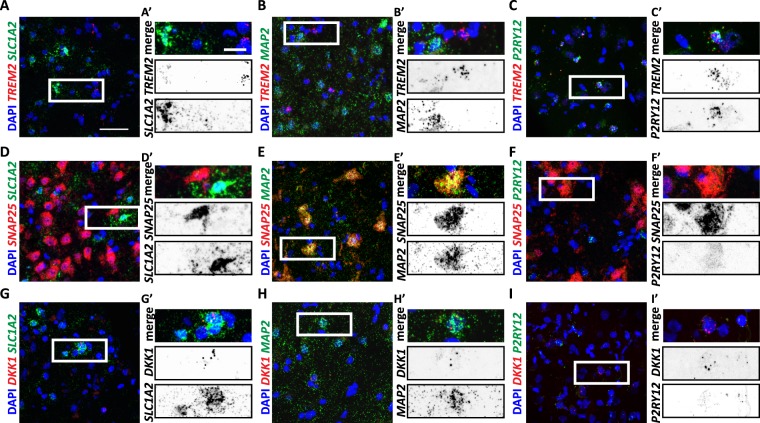
The secondary aim of this study was to establish a multiplexed smFISH protocol to study changes in RNA levels in a cell-type specific manner in the human brain. Therefore, we next demonstrated smFISH signal in three major cell types in the brain. Astrocytes, neurons and microglia were detected using cell-type specific probes against SLC1A2, MAP2 and P2RY12, respectively (Fig. 2)1. We have also tested alternative probes for astrocytes (SLC1A3), neurons (RBFOX3) and microglia (ITGAM and CST7; data not shown). The evaluation of probe suitability was based on puncta distribution and levels of expression. The astrocytic probe against SLC1A3 and the neuronal probe against RBFOX3 are manly localised in the cytoplasmic compartment, making its attribution to a particular nucleus more difficult. The cytoplasmic localization of SLC1A3 smFISH puncta is supported by the work of Bayraktar and collegues17. In addition, SLC1A3 is also detected in some microglia1. Our results also show that ITGAM and CST7 are less expressed than our primary microglial probe candidate, P2RY12. Indeed, single microglia RNA-seq show that ITGAM and CST7 are less expressed than P2RY1218. Therefore, we suggest the use of SLC1A2, MAP2 and P2RY12 probes for labelling the above cell types in human frozen brain sections.
Figure 2.
Single-cell resolution of TREM2, SNAP25 and DKK1 mRNAs present differential cell-type expression. Confocal images of control hippocampal slices showing TREM2, SNAP25 and DKK1 co-localising with different cell-type-specific markers including SLC1A2 (astrocytic), MAP2 (neuronal) and P2RY12 (microglial). (A–C) RNAscope of TREM2 (red) showing no colocalization with SLC1A2 (A-A’; green) nor MAP2 (B-B’; green), but P2RY12 (C-C’; green). (D–F) In situ hybridisation of SNAP25 (red) showing no colocalization with SLC1A2 (D-D’; green) or P2RY12 (F-F’; green), but MAP2 (E-E’, green) (G–I) RNAscope of DKK1 (red) showing colocalization with SLC1A2 (G-G’; green) and MAP2 (H-H’, green) but not with P2RY12 (I-I’; green). DAPI indicates nuclear staining (blue) in all cases. Scale bars 50 µm, zoom imaged 20 µm.
Subsequently, we used smFISH to determine the cell-type distribution of mRNA transcripts of three genes that are relevant to neurodegeneration: Triggering Receptor Expressed on Myeloid cells 2 (TREM2), Synaptosome Associated Protein 25 (SNAP25) and Dickkopf 1 (DKK1). TREM2 is mainly expressed in microglia cells1 and polymorphisms in this gene are associated with AD19 and other neurodegenerative diseases. SNAP25, which is expressed in neurons and is essential at synapses, is decreased in AD20. DKK1, a secreted Wnt signalling antagonist, is expressed in neurons and astrocytes1, is upregulated in AD21, and mediates Aβ-induced synapse loss22. These genes are expressed at different levels, with SNAP25 being highly expressed and DKK1 expressed at low levels in healthy individuals1. Therefore, these three genes allow us to further analyse our smFISH staining at a wide range of gene expression levels. Consistent with previous reports, in control hippocampal samples, TREM2 expression is restricted to microglia (Fig. 2A–C)1, SNAP25 to neurons (Fig. 2D–F)1, and DKK1 to neurons and astrocytes (Fig. 2G–I)1, as shown by colocalization with the cell-type specific probes. As expected, we observed differential expression levels of these three genes: SNAP25 is abundantly expressed, whereas DKK1 is sparsely expressed. Based on these data, we conclude that the use of frozen brain tissue for RNAscope is a sensitive and robust method for the visualisation of mRNA transcripts.
Differential cell-type RNA expression in Alzheimer’s disease
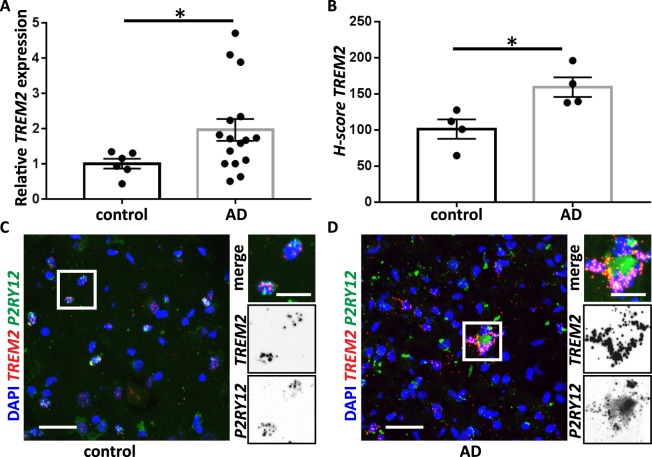
Finally, to further test our modified RNAscope protocol and analysis pipeline, we compared the expression of TREM2, SNAP25 and DKK1 in hippocampal samples from 4 control and 4 AD tissue samples (RQI 3.9–7.4). This analysis was performed using the same pipeline described above (Supp. Fig. 1): signal counts for the cell-lineage specific genes, MAP2, SLC1A2 and P2RY12, were used for cell-type identification and test probes (TREM2, SNAP25 or DKK1) were analysed in particular cell types. qPCR analyses showed that TREM2 expression is elevated in AD brains (Fig. 3A)23. Consistently, H-score quantification of smFISH signal showed upregulation of TREM2 expression in AD samples (Fig. 3B–D). Note that in AD samples, the green channel shows a high background signal, even when using the autofluorescence quencher TrueBlack (Supp. Fig. 3). This high background level might be due to the presence of large protein aggregates in AD brain tissue. Therefore, this channel is not ideal for analyses of samples from patients with diseases presenting autofluorescent aggregates24,25. Bulk qPCR analyses revealed that SNAP25 expression is decreased in AD20 (Supp. Fig. 4A). smFISH also showed a decrease in SNAP25 (Supp. Fig. 4B–D), accompanied by reduced MAP2 expression. However, SNAP25 could not be quantified using the same method used for TREM2, as single RNA puncta could not be detected. Thus, for highly expressed genes we propose quantification of total smFISH intensity per positive cell. Finally, we analysed DKK1 expression. Bulk qPCR confirmed an increased expression of DKK1 in the hippocampus of AD patients21 (Supp. Fig. 5). Although the DKK1 smFISH signal was above background, we could not quantify it because very few cells express DKK1. Altogether, these data indicate that our optimised smFISH protocol and analysis pipeline can be used to analyse cell-type specific changes in RNA levels in human frozen tissues from control and neurodegenerative cases at the single-cell resolution.
Figure 3.
Microglia-specific upregulation of TREM2 in fresh frozen human AD biopsies. (A) Bulk TREM2 mRNA expression in human hippocampal samples analysed by qPCR showed a 1.96 fold increase of TREM2 mRNA in AD (n = 16) vs. control (n = 6) samples. (*Indicates p = 0.045 by Mann-Whitney U Test). (B) Quantification of the number of dots/cell (H-score) in TREM2-positive cells for control (C; n = 4) and AD (D; n = 4) samples (*Indicates p = 0.0286 by Mann-Whitney U Test). (C,D) Representative confocal images of TREM2 mRNA expression in control (C) and AD (D) samples. TREM2 is shown in red, P2RY12 in green. DAPI indicates nuclear staining (blue) in all confocal images. Scale bars 50 µm, zoom images 20 µm. Graphs showing mean value ± SEM.
Discussion
Our study describes a multiplexed smFISH methodology optimised to enable detection of RNA molecules at single-cell resolution in frozen human brain tissue. In addition, we describe an analysis pipeline that enables the quantification of multiplexed smFISH data at the single-cell level, allowing visualisation of single RNA transcripts and cell-type discrimination. Importantly, we demonstrate that RNA integrity is not a critical factor for using smFISH in frozen human samples. Together, this approach has widespread utility for the quantification of gene expression in complex human tissues at single-cell resolution, thus complementing large scale unbiased genomic approaches such as single-cell and single-nucleus RNA-seq.
A recent study showed that fresh human brain biopsies could be utilised for multiplexed smFISH17. These samples were taken from the neurosurgical site and processed for smFISH following standard PFA fixation, sucrose placement and OCT mounting. However, research laboratories outside clinical environments have limited access to samples of this nature. In addition, fresh biopsies from neurodegenerative cases are virtually non-existent. Instead, laboratories obtain human samples from brain banks, which generally provide FFPE or frozen samples. Although positive results can be achieved by chromogenic-RNAscope and BaseScope in human FFPE brain sections14, or by smFISH in human FFPE preserved gut26,27 or lymphoma28 tissues, there are no studies reporting multiplexed smFISH in human FFPE brain samples. Indeed, our results show that FFPE human brain sections are not suitable for fluorescent RNAscope (Fig. 1). In contrast, we show that smFISH can be successfully used to analyse sections of frozen human brain tissue. A previous report also showed that smFISH can be deployed in human cortex29. However, the smFISH staining seems weaker than the ones presented in this study. These differences may arise from section thickness, the fixation method or other steps of RNAscope staining and imaging, which are not clearly explained in the text. Therefore, our optimised RNAscope approach could potentially be used by any laboratory as frozen human brain samples are accessible through brain banks and we provide a detailed and replicable methodological section.
RNA integrity is a critical issue when analysing gene expression by qPCR or sequencing as fragmented RNA impairs qPCR amplification8 and also interferes with RNA-seq analyses16. However, how RNA integrity impacts on RNAscope staining had not been previously investigated. Our results show that low-quality RNA (RQI = 2.9) does not significantly affect smFISH detection of two housekeeping genes (PPIB and TBP), the microglial gene TREM2 or the neuronal gene SNAP25 (Fig. 1 and Supp. Fig. 2). A possible explanation for this difference between smFISH and qPCR sensitivity to low RQI RNA is that qPCR requires the intact cDNA sequence for amplification (from the 5′ of the forward primer to the 3′ end of the reverse primer), whereas RNAscope requires the annealing of only 3 pairs of the 20 possible double Z probes to produce a detectable signal. Therefore, shorter, partially degraded RNA molecules could still be detected using RNAscope probes, positioning smFISH for the analysis of human frozen samples with variable RNA integrity. Note that our human sample set presented a large range of RQI scores (from 2.9 to 7.4), of which 51.8% present an RQI score lower than 3.9. Therefore, more than half of our sample set would need to be excluded for qPCR experiments. However, studies on RNA expression could be performed on the entire sample set across a wide range of RQI values with our optimised protocol for RNAscope in frozen human samples. This approach could be used in any given collection of frozen human samples, strongly increasing the number of samples available for in situ transcriptomics analysis.
To prove that RNA integrity has no impact on smFISH, and to analyse genes related to neurodegeneration in a cell-type manner, we have established a digital pathology analysis pipeline using the HALO software. This approach uses single-cell segmentation to properly attribute smFISH puncta to cognate cells. Correct ascription of RNA molecules is paramount for obtaining a reliable H-score (mRNA expression levels per cell), in particular for multiplexed smFISH aiming to analyse RNA levels in a cell-type specific manner. A scarcity of methods has previously limited the quantification of RNA expression by smFISH. Most studies in the rodent brain used smFISH to determine if a particular cell-type expresses a given gene30–32, to quantify changes in smFISH as dots per area of the brain33 or used semi-quantitative scoring34. In the human brain only one other pipeline analysis using Harmony in the Perkin-Elmer workflow has been previously reported17. We have used HALO because analysis of smFISH puncta is possible regardless of the input imaging platform, making our method broadly applicable to different research settings. An important requirement for determining the H-score of an individual probe is the capacity to detect and quantify single smFISH puncta, as successfully demonstrated in this study for a combination of six genes relevant for lineage determination, AD pathogenesis and housekeeping in the human brain. The H-score of highly expressed genes, like SNAP25, cannot be calculated because single RNA molecules are indistinguishable. Therefore, when studying highly expressed genes we suggest analysing overall RNAscope intensity per cell. In addition, super-resolution microscopy might allow detection of a discrete smFISH signal, facilitating H-score quantification with the analytical pipeline described here. Quantification of genes expressed at very low levels also presents limitations. In this report we have shown that DKK1 is expressed by very sparsely distributed cells, indicated by the absence of puncta in most fields of view. This pattern of expression is very likely to introduce a selection bias by the operator when acquiring the images. To overcome this potential issue, and to increase the analysis throughput of RNAscope, we suggest performing whole slide imaging with scanning fluorescence microscopes, coupled with our image analysis pipeline for quantification. When applied to multiplexed staining, this approach would allow systematic screening for the expression of different genes across large tissue samples.
Our analysis pipeline assigns all probe signals to the nearest nucleus by proximity within a maximum radius of 25 µm, approximately corresponding to the maximum diameter of the soma of the largest cortical neuron35. As shown in Supp. Fig. 1, cell borders are extended outwards from the geometric center of each nucleus to the nearest adjacent cellular border. smFISH puncta that are not associated with cell nuclei on the image plane are excluded for cell classification and scoring. In the absence of reference markers to determine cellular boundaries, these settings represent a bona fide approximation of the cellular arrangement in coherent tissue where one cell physiologically borders the adjacent. In areas presenting high cellular density, this approach models the increasing spatial compression, whereas in areas with low cellular density a maximum radius of 25 µm from the centre of the nucleus is accepted, which approximates the largest physiological extension of a cell soma of a neuron in situ35. Limitations in the use of this analysis include sectioning artefacts that can lead to inclusions/exclusion of smFISH puncta. On one hand, smFISH puncta located in cellular processes coming from cells outside the image plane could be included in the quantification. On the other hand, smFISH puncta located in cellular processes further than 25 µm away from the centre of the nucleus would be excluded from the quantification. Stochastically, these are rare events, as we observed the highest signal density in direct proximity to the corresponding nuclei (Fig. 2). Cell isolation and culture techniques are likely to provide the most accurate results when analysing smFISH signals located in dendrites, axonal and other cellular processes. To optimally capture cell morphology in culture, we expect that adaptation of the present protocol to include cytoplasmic or membranous staining for cell segmentation will be required.
Last, we used our optimised smFISH protocol and pipeline analysis to study differential RNA expression in AD. Our RNAscope results compare favourably to bulk qPCR experiments and recapitulate previously reported changes in genes associated with neurodegeneration. In this type of study, it is important to have age- and gender-matched control and AD groups, but this is often limited by the available samples. In our case, younger samples are overrepresented in the control group (Supp. Table 1). To test if age has an impact on the results presented in this study, we analysed the correlation between gene expression and age at death. Our results show that the expression of DKK1, SNAP25 and TREM2 is not correlated to the age of death in our control sample set (Supp. Fig. 6). For the AD group, our results show no correlation between age at death and gene expression for DKK1, SNAP25 or TREM2 when analysed by qPCR, nor for TREM2 when analysed by smFISH. However, the age of the AD patient is correlated with lower expression of SNAP25 (Supp. Fig. 6). This result is in line with previous reports showing that older AD patients exhibit greater neuronal loss in the hippocampus36. All together, these results show that our optimised approach is a sensitive and robust method to analyse changes in RNA levels by multiplexed RNAscope, opening the possibility to validate transcriptomics data at the single cell-type level in the intact tissue. This validation step will be important to support conclusions from single-cell and/or single-nuclei RNA-seq, such as the hypothesised subtypes of the major brain cell types37 or that perturbed molecular profiles of different brain cell types could be involved in distinct processes in neurodegenerative diseases, such as AD18,37.
In summary, we show that the multiplexed fluorescent RNAscope assay can be successfully used in frozen human brain tissue to analyse single RNA molecules in a cell-type specific manner at the single-cell resolution. In addition, we provide a new pipeline analysis for smFISH quantification with broad applicability to human tissue samples and increased methodological robustness permitting the use of samples with a wide range of RNA integrity values. These results broaden the opportunity to use human frozen tissue samples from brain banks and provide new opportunities for research on cell-type specific RNA expression in neurodegenerative diseases.
Methods
Human tissue
Anonymised human samples from control and AD subjects were obtained from Cambridge Brain Bank (CBB), Division of the Human Research Tissue Bank, Addenbrooke’s Hospital, Cambridge UK. All samples were obtained with informed consent under CBB license (NRES 10/HO308/56) approved by the NHS Research Ethic Services. Tissue was either stored at −80 °C or fixed in 10% neutral buffered formalin for at least 2 weeks before being embedded in paraffin. All experiments were performed in accordance with relevant guidelines and regulations. Demographic data, Braak stages, RQI and post-mortem intervals for each sample set are shown in Supp. Table 1.
RNAscope in-situ hybridization
To detect single mRNA molecules, RNAscope was performed on frozen vs FFPE-fixed control and AD hippocampal slices. 15μm sections were cut from frozen biopsies, dried for 10 min at 40 °C and kept at −80 °C. 9 µm sections were cut from FFPE blocks. In this study, one positive (Homo sapiens PPIB), one negative (Escherichia coli DapB) control probe and 6 different probes against genes of interest were used (Supp. Table 2). In situ hybridization (ISH) was performed according to the protocol of the RNAscope Multiplex Fluorescent Reagent Kit v2 (Cat. No. 320293).
Briefly, slides from frozen brains were dried in the oven at 40 °C for 3–4 min prior to incubation in cold 4% PFA for 25 min. Slides were then dehydrated in 50%, 70% and 100% ethanol for 5 min each at room temperature (RT). After drying the slides for 5 min at RT, H2O2 was added for 10 min at RT. For antigen accessibility, slides were treated with Protease IV for 20 min at RT. For FFPE-fixed sections and HeLa cells, slides were baked at 60 °C for 1 hour before being deparaffinised in xylene and 100% ethanol. After drying the slides 5 min at RT, H2O2 was added for 10 min at RT. For antigen accessibility, slides were incubated in boiling antigen retrieval solution (<98 °C) for 15 min, washed in water twice, dehydrated in 100% ethanol and finally treated with Protease Plus for 40 min at 40 °C.
Frozen and FFPE-fixed sections were washed twice in PBS. C2 probes were diluted in C1 probes at a 1:50 ratio and incubated on the slides for 2hrs at 40 °C. C1 probes were detected with TSA-Cy3 (Perkin Elmer, REF FP1170) and C2 probes with TSA-Fluorescein or TSA-Cy5 (Perkin Elmer, REF FP1168). To quench the autofluorescence due to the accumulation of lipofuscin or other protein aggregates, slides were incubated with TrueBlack (Biotium, 23007) for 30 sec at RT. Before mounting the slices, DAPI (Perkin Elmer, REF 323108) was added to label the nuclei. A one-day protocol has been used in all experiments to preserve the quality of the slices.
Image acquisition and pre-processing
Images were acquired using a Zeiss 880 confocal microscope (x40 objective, 1.3 NA). Settings were established during the first acquisition and not modified afterwards. Stack images of 10 steps with a 0.5 µm interval were used. Two representative sections per samples were selected and 3–4 regions of interest were imaged at 1024 × 1024 pixels for image analysis. All images were pre-processed using ImageJ (maximum z-projection).
Image analysis
Quantification of MAP2, P2RY12, PPIB, SLC1A2, TBP and TREM2
All images were reviewed by a board-certified pathologist and expert in image analysis (VHK). Regions with technical artefacts or unspecific background staining were annotated in.xml overlays and excluded from analyses. We then developed an image analysis approach for scoring multiplex mRNA smFISH of the lineage specific probes SLC1A2 (astrocytes), MAP2 (neurons) and P2RY12 (microglia) and the genes of interest PPIB, TBP and TREM2 in brain tissue using the HALO smFISH quantification for multiple probes v1.1 module. Briefly, colour deconvolution was performed and excitation channels were assigned for detection of nuclear counterstains and mRNA probes (DAPI, nuclear, 405 nm blue channel; C1 probe, 568 nm, red channel; C2 probe, 488 nm or 647 nm, green channel). Nuclear segmentation and contour detection were optimised using the DAPI counterstain and pathologist-supervised cell classification features (nuclear size, roundness, nuclear intensity, nuclear contrast threshold and segmentation aggressiveness) in real-time tuning mode using graphical overlays. Cell nuclei and X-Y location coordinates were recorded and nuclear morphometric parameters were quantified for each cell. Probe detection was optimised based on signal size (minimum signal size 0.45 µm2), intensity of positive probe pixels and contrast threshold parameter settings. Probe signals were recorded both as number of individual fluorescent spots as well as total signal area in µm2 per cell on a continuous scale with conservative settings for spot segmentation and cluster counting (probe copy intensity threshold = 0.15, spot segmentation aggressiveness = 0.5). For absolute quantification of probe signals, physically separated puncta were always counted as one probe copy. A spot was classified as a cluster when its intensity surpassed the set probe copy intensity threshold. The probe count of each cluster is derived from absolute intensity divided by the copy intensity parameter. Thresholding for the copy intensity parameter was performed using individual probe signals in lineage-positive cells as internal positive controls taking into account any background. Probe signals were assigned to each nucleus by proximity within a maximum cell radius of 25 µm. Total counts for the lineage specific probes, P2RY12, SLC1A2 and MAP2, were used to determine cell identity (neurons, astrocytes and microglia). We considered a cell positive for a particular linage when presenting 2 or more counts. Signal counts for the test probes PPIB, TBP and TREM2 were quantified for each cell and were used for the classification of each cell as negative (0 copies), 1 + (Minimum Copies/Cell: 1), 2 + (Minimum Copies/Cell: 4); 3 + (Minimum Copies/Cell: 10); or 4 + (Minimum Copies/Cell: 16), as shown in Fig. 1B and Supp. Fig. 1. The total number of probe counts associated with a cell give it the number used when generating the H-score for the entire area analysed. H-scores were calculated as follows: H-Score = (1 × % Probe 1 + Cells) + (2 × % Probe 2 + Cells) + (3 × % Probe 3 + Cells) + (4 × % Probe 4 + Cells), following recommendations by Cell Diagnostics USA (Newark, CA, USA) for the evaluation of RNAscope assays. Final scores derived by this metric will have a range between 0 and 400. A file containing the pipeline analysis ready to use in HALO can be sent upon request.
Quantification of SNAP25
Due to high levels of expression, SNAP25 gene expression was further measured as total intensity per cell SNAP25 staining was quantified using ImageJ: overall intensity of the SNAP25 signal was measured in arbitrary units and was normalised to the number of neurons counted per image (Supp. Fig. 4).
RNA extraction and RNA integrity analysis
Total RNA from frozen hippocampal tissue was extracted with TRIzol Reagent (Thermo Fisher Scientific ref: 15596018) and DirectZol Miniprep RNA Kit (Zymo Research ref: R2052). Briefly, 20–50 mg of tissue was homogenised in 800 µL of TRIzol using pellet pestles (Sigma-Aldrich ref: Z359947) coupled to pellet pestles cordless motor (Sigma-Aldrich ref: Z359971) in 1.5 ml tubes. Once homogenised, samples were centrifuged for 5 min at 16,000 g at 4 °C to remove any remaining tissue pieces and the supernatant was transferred to clean 1.5 mL tubes. 200 µL of chloroform were added, tubes were vortexed for 5 sec and left at RT for 10 min. To obtain the phase separation, samples were centrifuged for 15 min at 10,000 g at 4 °C. The aqueous upper face containing the RNA was transferred into a clean tube and RNA was extracted with DirectZol Miniprep RNA Kit following manufacturer’s instructions and including DNase in column treatment. RNA was quantified by measuring absorbance at 260 nm using a Nanodrop ND-100 (Thermo Fisher Scientific). RNA integrity (RNA Quality Indicator; RQI) was analysed using RNA HighSense Assay chips (Bio-Rad ref: 7007155) and run in Experion™ Automated Electrophoresis Station (Bio-Rad). Briefly, samples were diluted to 1–5 ng/µL, loaded into RNA HighSense Assay chips and run. 18S and 28S peaks were detected and RQI was determined. Only samples with a RQI 3.9 or above were used for relative qPCR comparing control and AD cases38.
Retrotranscription and qPCR
Retrotranscription to first strand cDNA was performed using RevertAid H Minus First Strand cDNA Synthesis kit (Themo Fisher Scientific ref: K1632). Briefly, 2000 ng of total RNA was used for cDNA synthesis following manufacturer’s instructions. 5 ng of original RNA was used to perform fast qPCR using GoTaq qPCR Master Mix (Promega ref: A6002) in a LigherCycler 480 (Roche) with the manufacturer’s protocol (Supp. Table 3). Primers were designed using OligoPerfect design (Thermo Fisher Scientific) and validated using in silico PCR (UCSC genome Browser) and Ensembl BLAST (Ensemble.org). Primers were purchased from Sigma-Aldrich (Supp. Table 4) and used at 0.5 μM final concentration. Finally, primer efficiency was tested using serial dilutions from a mixed pool of cDNAs from all the samples. All primers used showed an efficiency of 1.9 or above (Supp. Table 4).
Reference genes selection for qPCR
21 possible reference genes were tested (Supp. Table 4). GeNorm from GenEx package was used to select the best combination of reference genes39. Briefly, intra- and inter-group (Control and AD) variability was calculated. A cut-off of 0.5 was applied for intra-group and a cut-off of 0.2 was applied for inter-group variances (Supp. Table 5). The leftover genes were utilised to calculate SD for each gene regardless of groups and associated SD when pooling reference genes (Supp. Fig. 7). Based on these results, 4 reference genes were selected to perform relative qPCR (PMU1, TBP, CYC1 and UBE2D2).
Statistics
All statistical analyses were performed using SPSS 25 (IBM). Outliers were assessed by box plot and extreme outliers (1.5 x interquartile ranges (IQR) above the third or below the first quartile) were removed from the dataset. Pearson correlation (R) was tested with a bivariate correlation tool. Exact p-values are indicated. qPCR and RNAscope comparisons between control and AD cases were analysed by Mann-Whitney U test. In the figures, asterisks indicate p-values (*p < 0.05).
Supplementary information
Acknowledgements
This work was funded by the MRC (PCS: MR/M024083/1), Alzheimer's Society (PCS: AS-PG-18-008), Alzheimer’s Research UK (PCS and EP: ARUK-PG2018A-002; PW: ARUK-2018DDI-UCL), European Commission Horizon 2020 (PCS and EP: H2020 MSCA-IF 749209) and Promedica Foundation (VHK: F-87701-41-01). We tahnk The Cambridge Brain Bank for provind human samples. The Human Research Tissue Bank is supported by the NIHR Cambridge Biomedical Research CentreWe thank Anne Hellebust, Sean Griffiths and Billy Heseltine from Indica for their input on the H-Score generation and support in the installs used to run the analysis.
Author Contributions
S.J., V.L., C.S.F., P.C.S., P.W. and E.P. conceived this study. S.J., V.L. and L.M. performed RNAscope experiments. E.P. performed qPCR experiments. V.H.K. developed the digital image analysis pipeline and analysed RNAscope images.
Data Availability
Data generated for this study and a file containing the pipeline analysis ready to use in HALO re available upon request.
Competing Interests
V.H.K. is participant of a patent application co-owned by the Netherlands Cancer Institute (NKI-AVL) and the University of Basel on the assessment of cancer immunotherapy biomarkers by digital pathology and has served as an invited speaker on behalf of Indica Labs.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Sarah Jolly, Verena Lang and Viktor Hendrik Koelzer contributed equally.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-48787-w.
References
- 1.Zhang Y, et al. Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse. Neuron. 2016;89:37–53. doi: 10.1016/j.neuron.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Habib N, et al. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat. Methods. 2017;14:955–958. doi: 10.1038/nmeth.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bakken TE, et al. Single-nucleus and single-cell transcriptomes compared in matched cortical cell types. PLoS One. 2018;13:e0209648. doi: 10.1371/journal.pone.0209648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lein E, Borm LE, Linnarsson S. The promise of spatial transcriptomics for neuroscience in the era of molecular cell typing. Science. 2017;358:64–69. doi: 10.1126/science.aan6827. [DOI] [PubMed] [Google Scholar]
- 5.Crosetto N, Bienko M, van Oudenaarden A. Spatially resolved transcriptomics and beyond. Nat. Rev. Genet. 2015;16:57–66. doi: 10.1038/nrg3832. [DOI] [PubMed] [Google Scholar]
- 6.Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of Single RNA Transcripts in Situ. Science (80-.). 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- 7.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koppelkamm A, Vennemann B, Lutz-Bonengel S, Fracasso T, Vennemann M. RNA integrity in post-mortem samples: influencing parameters and implications on RT-qPCR assays. Int. J. Legal Med. 2011;125:573–80. doi: 10.1007/s00414-011-0578-1. [DOI] [PubMed] [Google Scholar]
- 9.Alzheimer’s Association, 2014 Alzheimer’s disease facts and figures. Alzheimer’s & Dementia10, e47–e92 (2014). [DOI] [PubMed]
- 10.Spires-Jones TL, Hyman BT. The Intersection of Amyloid Beta and Tau at Synapses in Alzheimer’s Disease. Neuron. 2014;82:756–771. doi: 10.1016/j.neuron.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liddelow SA, Barres BA. Reactive Astrocytes: Production, Function, and Therapeutic Potential. Immunity. 2017;46:957–967. doi: 10.1016/j.immuni.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 12.Li Q, Barres BA. Microglia and macrophages in brain homeostasis and disease. Nat. Rev. Immunol. 2017;18:225–242. doi: 10.1038/nri.2017.125. [DOI] [PubMed] [Google Scholar]
- 13.Wang F, et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J. Mol. Diagn. 2012;14:22–9. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bialas AR, et al. Microglia-dependent synapse loss in type I interferon-mediated lupus. Nature. 2017;546:539. doi: 10.1038/nature22821. [DOI] [PubMed] [Google Scholar]
- 15.Selvaraj BT, et al. C9ORF72 repeat expansion causes vulnerability of motor neurons to Ca2+-permeable AMPA receptor-mediated excitotoxicity. Nat. Commun. 2018;9:347. doi: 10.1038/s41467-017-02729-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gallego Romero I, Pai AA, Tung J, Gilad Y. RNA-seq: impact of RNA degradation on transcript quantification. BMC Biol. 2014;12:42. doi: 10.1186/1741-7007-12-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bayraktar, O. A. et al. Single-cell in situ transcriptomic map of astrocyte cortical layer diversity. bioRxiv432104, 10.1101/432104 (2018).
- 18.Sala Frigerio C, et al. The Major Risk Factors for Alzheimer’s Disease: Age, Sex, and Genes Modulate the Microglia Response to Aβ Plaques. Cell Rep. 2019;27:1293–1306.e6. doi: 10.1016/j.celrep.2019.03.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carmona S, et al. The role of TREM2 in Alzheimer’s disease and other neurodegenerative disorders. Lancet. Neurol. 2018;17:721–730. doi: 10.1016/S1474-4422(18)30232-1. [DOI] [PubMed] [Google Scholar]
- 20.Furuya TK, et al. Analysis of SNAP25 mRNA expression and promoter DNA methylation in brain areas of Alzheimer’s Disease patients. Neuroscience. 2012;220:41–46. doi: 10.1016/j.neuroscience.2012.06.035. [DOI] [PubMed] [Google Scholar]
- 21.Buechler J, Salinas PC. Deficient Wnt Signaling and Synaptic Vulnerability in Alzheimer’s Disease: Emerging Roles for the LRP6 Receptor. Front. Synaptic Neurosci. 2018;10:38. doi: 10.3389/fnsyn.2018.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Purro SA, Dickins EM, Salinas PC. The secreted Wnt antagonist Dickkopf-1 is required for amyloid β-mediated synaptic loss. J. Neurosci. 2012;32:3492–8. doi: 10.1523/JNEUROSCI.4562-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Celarain N, et al. TREM2 upregulation correlates with 5-hydroxymethycytosine enrichment in Alzheimer’s disease hippocampus. Clin. Epigenetics. 2016;8:37. doi: 10.1186/s13148-016-0202-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moreno-García A, Kun A, Calero O, Medina M, Calero M. An Overview of the Role of Lipofuscin in Age-Related Neurodegeneration. Front. Neurosci. 2018;12:464. doi: 10.3389/fnins.2018.00464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grune T, Jung T, Merker K, Davies KJA. Decreased proteolysis caused by protein aggregates, inclusion bodies, plaques, lipofuscin, ceroid, and ‘aggresomes’ during oxidative stress, aging, and disease. Int. J. Biochem. Cell Biol. 2004;36:2519–2530. doi: 10.1016/j.biocel.2004.04.020. [DOI] [PubMed] [Google Scholar]
- 26.Fernández‐Barral, A. et al. Vitamin D differentially regulates colon stem cells in patient‐derived normal and tumor organoids. FEBS J. febs. 14998, 10.1111/febs.14998 (2019). [DOI] [PMC free article] [PubMed]
- 27.Ziskin JL, et al. In situ validation of an intestinal stem cell signature in colorectal cancer. Gut. 2013;62:1012–23. doi: 10.1136/gutjnl-2011-301195. [DOI] [PubMed] [Google Scholar]
- 28.Tamma R, et al. STAT-3 RNAscope Determination in Human Diffuse Large B-Cell Lymphoma. Transl. Oncol. 2019;12:545–549. doi: 10.1016/j.tranon.2018.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lake BB, et al. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science (80-.). 2016;352:1586–1590. doi: 10.1126/science.aaf1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Q-R, et al. Cannabinoid type 2 receptors in dopamine neurons inhibits psychomotor behaviors, alters anxiety, depression and alcohol preference. Sci. Rep. 2017;7:17410. doi: 10.1038/s41598-017-17796-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang L, et al. GPR139 and Dopamine D2 Receptor Co-express in the Same Cells of the Brain and May Functionally Interact. Front. Neurosci. 2019;13:281. doi: 10.3389/fnins.2019.00281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baho E, et al. p75 Neurotrophin Receptor Activation Regulates the Timing of the Maturation of Cortical Parvalbumin Interneuron Connectivity and Promotes Juvenile-like Plasticity in Adult Visual Cortex. J. Neurosci. 2019;39:4489–4510. doi: 10.1523/JNEUROSCI.2881-18.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tora D, et al. Cellular Functions of the Autism Risk Factor PTCHD1 in Mice. J. Neurosci. 2017;37:11993–12005. doi: 10.1523/JNEUROSCI.1393-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, H., Illenberger, J. M., McLaurin, K. A., Mactutus, C. F. & Booze, R. M. Identification of Dopamine D1-Alpha Receptor Within Rodent Nucleus Accumbens by an Innovative RNA In situ Detection Technology. J. Vis. Exp. e57444, 10.3791/57444 (2018). [DOI] [PMC free article] [PubMed]
- 35.Fiala, J. C. & Harris, K. M. Dendrite structure, Dendrite, Oxford University Press (1999).
- 36.West MJ, Coleman PD, Flood DG, Troncoso JC. Differences in the pattern of hippocampal neuronal loss in normal ageing and Alzheimer’s disease. Lancet (London, England) 1994;344:769–72. doi: 10.1016/s0140-6736(94)92338-8. [DOI] [PubMed] [Google Scholar]
- 37.Mathys H, et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature. 2019;570:332–337. doi: 10.1038/s41586-019-1195-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rydbirk R, et al. Assessment of brain reference genes for RT-qPCR studies in neurodegenerative diseases. Sci. Rep. 2016;6:37116. doi: 10.1038/srep37116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vandesompele, J. et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, RESEARCH0034 (2002). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data generated for this study and a file containing the pipeline analysis ready to use in HALO re available upon request.