In marine anaerobic environments, methane is oxidized where sulfate-rich seawater meets biogenic or thermogenic methane. In those niches, a few phylogenetically distinct microbial types, i.e., anaerobic methanotrophs (ANME), are able to grow through anaerobic oxidation of methane (AOM). Due to the relevance of methane in the global carbon cycle, ANME have drawn the attention of a broad scientific community for 4 decades.
KEYWORDS: anaerobic oxidation of methane, anerobic methanotrophs
SUMMARY
In marine anaerobic environments, methane is oxidized where sulfate-rich seawater meets biogenic or thermogenic methane. In those niches, a few phylogenetically distinct microbial types, i.e., anaerobic methanotrophs (ANME), are able to grow through anaerobic oxidation of methane (AOM). Due to the relevance of methane in the global carbon cycle, ANME have drawn the attention of a broad scientific community for 4 decades. This review presents and discusses the microbiology and physiology of ANME up to the recent discoveries, revealing novel physiological types of anaerobic methane oxidizers which challenge the view of obligate syntrophy for AOM. An overview of the drivers shaping the distribution of ANME in different marine habitats, from cold seep sediments to hydrothermal vents, is given. Multivariate analyses of the abundance of ANME in various habitats identify a distribution of distinct ANME types driven by the mode of methane transport. Intriguingly, ANME have not yet been cultivated in pure culture, despite intense attempts. Further advances in understanding this microbial process are hampered by insufficient amounts of enriched cultures. This review discusses the advantages, limitations, and potential improvements for ANME laboratory-based cultivation systems.
INTRODUCTION
Methane (CH4) is the most abundant and completely reduced form of hydrocarbon. It is the most stable hydrocarbon, which demands +439 kJ mol−1 energy to dissociate the hydrocarbon bond (1). Methane is a widely used energy source, but it is also the second largest contributor to human-induced global warming, after carbon dioxide. Methane concentrations in the atmosphere have increased by 150% (i.e., from about 0.7 to 1.8 ppmv) in the last 200 years (2–4), and experts estimate that this increase is responsible for approximately 20% of the Earth’s warming since preindustrial times (5). On a per mole basis and over a 100-year horizon, the global warming potential of methane is about 28 times more than that of carbon dioxide (6). Therefore, major scientific efforts are being made to resolve detailed maps of methane sources and sinks and how these are affected by the increased levels of this gas in the atmosphere (5).
The global methane cycle is driven largely by microbial processes of methane production (i.e., methanogenesis) and methane oxidation (i.e., methanotrophy). Methane is produced with substrates from the anaerobic degradation of organic matter via microbial food chains and through carbon dioxide reduction (7). These methane production processes occur in diverse anoxic subsurface environments, such as rice paddies, wetlands, landfills, and contaminated aquifers, as well as freshwater and ocean sediments (8). Methane can also be formed physicochemically at specific temperatures of about 150°C to 220°C (thermogenesis). It is estimated that more than half of the methane produced globally is oxidized microbially to CO2 before it reaches the atmosphere (8). Both aerobic and anaerobic methanotrophy are the responsible processes. The first involves the oxidation of methane to methanol and ultimately to carbon dioxide in the presence of molecular oxygen by methanotrophic bacteria (9, 10), whereas the second includes the oxidation of methane to carbon dioxide in the absence of oxygen by a clade of archaea, called anaerobic methanotrophs (ANME), and the process is known as the anaerobic oxidation of methane (AOM).
Large methane reservoirs on Earth (450 to 10,000 gigatonnes of carbon [Gt C]) (11, 12) are found as methane hydrates beneath marine sediments, mostly formed by biogenic processes (13). Methane hydrates, or methane clathrates, are crystalline solids, consisting of large amounts of methane trapped by interlocking water molecules (ice). They are stable at high pressure (>6 MPa) and low temperature (<4°C) (14, 15) and are typically found along continental margins at depths of 600 to 3,000 m below sea level (8, 11, 15). By gravitational and tectonic forces, methane stored in hydrates seeps into the ocean sediment under the form of mud volcanoes, gas chimneys, hydrate mounds, and pock marks (15). These methane seepage manifestations are environments where AOM has been documented, e.g., Black Sea carbonate chimney (16), Gulf of Cadiz mud volcanoes (17), and Gulf of Mexico gas hydrates (18). Besides, AOM also occurs in the sulfate-methane transition zones (SMTZ) of sediments. The SMTZ are quiescent sediment environments, where the upward diffusing (thermogenic and biogenic) methane is oxidized when it meets sulfate, which is transported downward from the overlayer of seawater (Fig. 1). Considering that sulfate is abundant in seawater and that oxygen in seabed sediments is almost absent, AOM coupled with the reduction of sulfate is likely the dominant biological sink of methane in these environments.
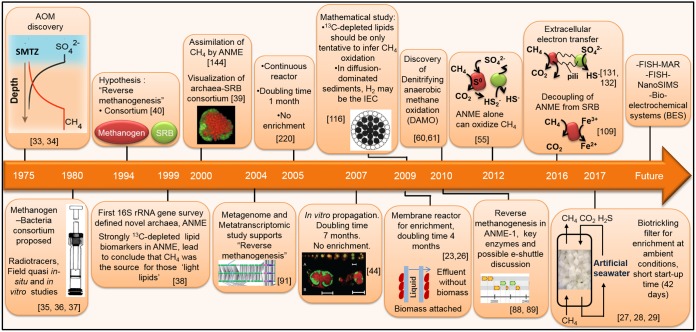
FIG 1.
Time line of relevant research on and discoveries about anaerobic methane oxidation with sulfate as an electron acceptor. The major milestones achieved are depicted in their respective years along with some future possibilities in AOM studies. IEC, interspecies electron carrier; MAR, microautoradiography; NanoSIMS, nanometer-scale secondary ion mass spectrometry.
It is estimated that methane seeps, which generally lay above methane hydrates (19), annually emit 0.01 to 0.05 Gt C, contributing 1% to 5% of the global methane emissions to the atmosphere (15). These emissions would be higher if methane was not scavenged by aerobic or anaerobic oxidation of methane. While aerobic methane oxidation is dominant in shallow oxic seawaters (20), AOM is found in the anoxic zones of the seafloor (8, 21, 22). Due to limited data, it has not been possible to determine the exact global values of methane consumption by AOM. But the rates of AOM in the SMTZ and methane seep environments have been tentatively estimated at 0.05 and 0.01 Gt C per year, respectively (15).
Besides the biogeochemical implications of AOM, this microbial process can have biotechnological applications for the treatment of waste streams rich in sulfate or nitrate/nitrite but low in electron donor. A few studies have highlighted the future of AOM and ANME in environmental biotechnology, where methane is used as the sole electron donor to achieve sulfate reduction in bioreactors (23–29). Biological sulfate reduction is a well-known technique to remove sulfur and metals from wastewaters, and metals can be recovered by metal sulfide precipitation (30). Many industrial wastewaters are deficient in dissolved organic carbon. Hence, supplementation of external carbon sources and electron donors is essential for microbial sulfate reduction. Frequently used electron donors for sulfate-reducing treatment plants are hydrogen/carbon dioxide and ethanol (31), which are costly and are preferably replaced by low-priced electron donors (25). It is estimated that the overall treatment costs would be reduced by a factor of 2 to 4 if methane from natural gas or biogas was used in sulfate-reducing bioreactors as an electron donor instead of hydrogen or ethanol (24). The major limitation for the biotechnological application of AOM is the extremely low growth rates of the ANME, currently with doubling times as long as 2 to 7 months (24).
(This review was adapted from chapter 2 of C.C.’s thesis [32].)
MICROBIOLOGY OF ANAEROBIC METHANE OXIDATION
Discovery of AOM
The anaerobic oxidation of methane coupled to sulfate reduction (AOM-SR) takes place where sulfate (SO42–) meets either biogenic or thermogenic methane. This unique microbiological phenomenon, AOM, has been recognized for 4 decades as a key to close the balance of oceanic carbon (33, 34). Since then, various key discoveries have elucidated the AOM process to some extent, but its exact biochemical mechanism is still unclear (Fig. 1). AOM was first deduced from CH4 and SO42– profile measurements in marine sediments (35–37). The occurrence of AOM yields typical concave-up methane profiles in sediment columns with high methane concentrations in the deep sediment layers and very low CH4 concentrations at the sediment water interface (Fig. 1).
Quasi in situ and in vitro studies using radiotracers have confirmed AOM as a biological process (35–37). Additional in vitro studies have suggested that the AOM process is performed by a unique microbial community (38–40), i.e., the ANME, mostly in association with sulfate-reducing bacteria (SRB) (Fig. 2). The identification of the microorganisms involved in AOM is crucial to explain how methane can be efficiently oxidized with such a low energy yield. By microscopic visualization after hybridization with specifically designed probes, the in situ occurrence of such archaeal-bacterial associations was recorded, showing that the ANME groups are widely distributed throughout marine sediments (39, 41–43). The physicochemical drivers shaping the global distribution of ANME consortia are not fully resolved to date (see Ecology of ANME in Marine Habitats, below). Instead, AOM activity tests and in vitro studies allowed the estimation of their doubling time on the order of 2 to 7 months, revealing the extremely slow growth of ANME on CH4 (44).
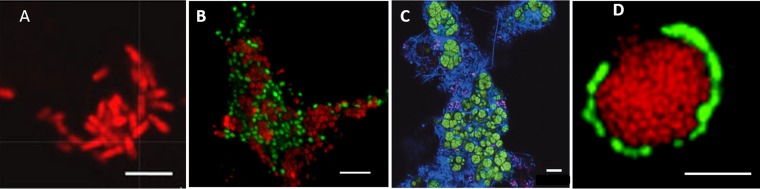
FIG 2.
Fluorescence in situ hybridization images from different ANME types. (A) ANME-1 cells in elongated rectangular shape (red) that inhabit the Guaymas Basin hydrothermal vent as a monospecific clade. Republished from reference 165 with permission from Springer Nature. (B) Aggregate of coccus-shaped ANME-2 (red) and Desulfosarcina/Desulfococcus (green) bacteria, an enrichment sample after 8 years from the Isis Mud Volcano in the Mediterranean Sea. The image was taken from http://www.mpg.de/6619070/marine-methane-oxidation and is republished with permission from Jana Milucka and the Max Planck Institute for Marine Microbiology. (C) Aggregate of large, densely clustered ANME-2d (green) and other (blue) bacteria obtained from a bioreactor enrichment. Republished from reference 61 with permission from Springer Nature. (D) Aggregate of coccus-shaped ANME-3 (red) and Desulfobulbaceae (green) bacteria inhabiting the Haakon Mosby Mud Volcano. Republished from reference 54 with permission. Scale bars, 10 μm.
AOM Coupled to Sulfate Reduction
The ocean is one of the main reservoirs of sulfur, where it occurs mainly as dissolved sulfate in seawater or as a mineral in the form of pyrite (FeS2) and gypsum (CaSO4) in sediments (45). Sulfur exists in different oxidation states, with sulfide (S2–), elemental sulfur (S0), and sulfate (SO42–) as the most abundant and stable species in nature. With a concentration of 29 mM, sulfate is the most dominant anion in ocean water, next to chloride. The reductive part of the sulfur cycle occurring in sediments involves two main microbial processes: (i) bacterial dissimilatory reduction of sulfate to hydrogen sulfide, which can subsequently precipitate with metal ions (mainly iron), and (ii) assimilatory reduction of sulfate to form organic sulfur compounds incorporated in microbial biomass (46). Dissimilatory sulfate reduction by SRB occurs in anoxic marine sediments or in freshwater environments, where SRB use several electron donors, such as hydrogen, and various organic compounds (e.g., ethanol, formate, lactate, pyruvate, fatty acids, methanol, and methanethiol). Methane cannot be used directly as an electron donor by SRB, unless associated with ANME, as in AOM-SR (47). However, the occurrence of both methanotrophy- and dissimilatory sulfate reduction-related genes in the draft genome of Korarchaeota WYZ-LM09 showed the potential utilization of methane and sulfite reduction by the same microorganism (48, 49).
AOM was considered impossible in the past, due to the nonpolar C-H bond of CH4 (1). From a thermodynamic point of view, AOM-SR yields minimal energy: only 17 kJ mol−1 of energy is released during AOM-SR at standard state (Fig. 3, equation 1). In comparison, more energy is released by the hydrolysis of one ATP (31.8 kJ mol−1). Other electron acceptors in the anaerobic environment, such as nitrate, iron, and manganese, provide higher energy yields than sulfate, as deducted by the ΔG° of the different redox reactions (Fig. 3). However, their combined concentration at the marine sediment-water interface is far lower than the sulfate concentration (50). Therefore, AOM-SR usually dominates in marine sediments.
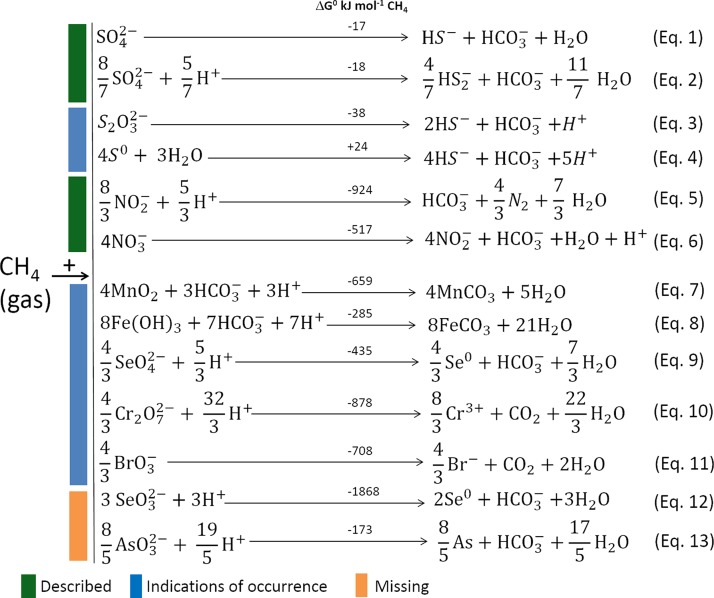
FIG 3.
Described and possible AOM processes with different terminal electron acceptors. AOM with sulfate, nitrate, or nitrite as the electron acceptor is well described, along with the microbes involved (indicated by green bars), whereas AOM with manganese and iron has been shown, but the microbes involved need to be characterized (indicated by blue bars). Other possible electrons are indicated in the bottom part of the figure according to the thermodynamic calculation of the chemical reaction (indicated by an orange bar).
AOM-SR has been suggested to be a cooperative metabolic process of AOM and dissimilatory sulfate reduction, thereby gaining energy by a syntrophic consortium of ANME and SRB (Fig. 3, equation 1) (39, 40). The Desulfosarcina/Desulfococcus and Desulfobulbaceae clades of SRB especially are common associates of ANME for SR. However, the three ANME phylotypes have been visualized without any attached SRB in different marine environments as well (51–54), suggesting that AOM-SR can potentially be performed independently by the ANME themselves (Fig. 3, equation 1, performed solely by ANME). Theoretically, slightly more energy can be released (18 kJ mol−1) if sulfate is reduced to disulfide instead of sulfide (Fig. 3, equation 2) (55).
AOM Coupled to Different Sulfur Compounds as the Electron Acceptor
Microorganisms that mediate AOM-SR could also use S0 or thiosulfate (S2O32–) as the terminal electron acceptor for AOM (Fig. 3, equations 3 and 4). The reduction of 1 mol of S2O32– to 1 mol sulfide would require fewer electrons (4 electrons) than the reduction of SO42– to sulfide (8 electrons). The reduction of S2O32– coupled to CH4 oxidation is also more energetically favorable (Fig. 3, equation 3) than AOM-SR (Fig. 3, equation 1). However, research investigating AOM coupled to S2O32– reduction (28, 29, 56, 57) showed that disproportionation of S2O32– prevailed over its reduction, even if it is theoretically less thermodynamically favorable (ΔG° = −22 kJ mol−1). The presence of known SRB able to metabolize inorganic sulfur compounds by disproportionation, such as Desulfocapsa and Desulfovibrio (58), in the studied sediment (57) might favor S2O32– disproportionation over its reduction with CH4 as the sole electron donor. However, the Desulfosarcina/Desulfococcus and Desulfobulbaceae microorganisms commonly associated with ANME were never shown to metabolize S2O32– via disproportion, even though Desulfosarcina/Desulfococcus microorganisms were once described as putative disulfide-disproportionating bacteria (55).
Differently, the theoretical Gibbs free energy for AOM coupled to S0 is positive (+24 kJ mol−1) (Fig. 3, equation 4). However, in vitro tests showed that this reaction may well proceed and the calculated free energy of reaction under in situ conditions is negative (−84.1 kJ mol−1) (29, 55). In contrast to S2O32– disproportionation, S0 disproportionation requires energy (+41 kJ mol−1) unless an oxidant, such as Fe(III), renders the reaction more energetically favorable (58). Furthermore, S0 may disproportionate in alkaline environments by alkaline halophilic bacteria (59). Therefore, other reactions and mechanisms might be taken into consideration when investigating AOM coupled to the reduction of other sulfur compounds such as S2O32– and S0.
AOM Coupled to Nitrite and Nitrate Reduction
Methanotrophs that utilize nitrite (60) or nitrate (61) have been identified in anaerobic freshwater sediments. Thermodynamically, methane oxidation coupled to nitrite and nitrate yields more energy than AOM-SR, with ΔG° values of −990 kJ mol−1 and −785 kJ mol−1, respectively (Fig. 3, equations 5 and 6). Two specific groups of microbes are involved in the process of AOM coupled with nitrate and nitrite reduction: “Candidatus Methanoperedens nitroreducens” (archaea) and “Candidatus Methylomirabilis oxyfera” (bacteria), respectively.
AOM coupled to denitrification was first hypothesized to occur in a syntrophic manner similar to that of AOM coupled to SR (62). However, Ettwig et al. (60) showed that methane oxidation coupled to nitrite reduction occurs in the absence of archaea. The bacterium “Candidatus Methylomirabilis oxyfera” couples AOM to denitrification, with nitrite being reduced to nitric oxide, which is then converted to nitrogen (N2) and oxygen (O2). The thus-generated intracellular by-product, oxygen, is subsequently used to oxidize CH4 to CO2 (60). Moreover, recent studies revealed that a distinct ANME, affiliated with the ANME-2d subgroup and named “Candidatus Methanoperedens nitroreducens” (Fig. 2C and 3), can carry out AOM using nitrate as the terminal electron acceptor through reversed methanogenesis (61). ANME-2d can perform denitrifying anaerobic methane oxidation (DAMO) without any syntrophic partners (see Fig. 5B). ANME-2d releases nitrite, which can be reduced to N2 by “Candidatus Methylomirabilis oxyfera” or by the anaerobic ammonium-oxidizing (anammox) bacterium in the presence of ammonium. Therefore, different microbial communities dominate in a biological system, depending on the availability of the nitrogen species (nitrate, nitrite, or ammonium) (61).
DAMO, similar to AOM coupled to sulfate reduction, can be applied as sustainable technology to reduce methane emissions and nitrogen levels in aquatic environments. It has been demonstrated that DAMO microorganisms can be enriched in reactors on wastewater (63–67). Moreover, recent studies showed that DAMO and annamox bacteria can be cocultured and enriched in bioreactors, reducing nitrite completely with methane and ammonium as electron donors (63, 68).
AOM Coupled to Iron and Manganese Reduction
Besides sulfate and nitrate, iron and manganese have also been studied as potential electron acceptors for anaerobic methane oxidation. In marine sediments, AOM was found to be coupled to the reduction of manganese or iron in marine sediments (69), coastal sediments (70), and lake sediments (71, 72). An in vitro study by Beal et al. (69) showed that oxide minerals of manganese, i.e., birnessite (simplified as MnO2 in equation 7 of Fig. 3), and iron, i.e., ferrihydrite [simplified as Fe(OH)3 in equation 8 of Fig. 3], can be used as electron acceptors for AOM. Similarly, Sivan et al. (72) showed that 13CH4 was consumed upon Fe(II)-oxide addition to the Lake Kinneret sediment core, and in marine Lake Grevelingen, the iron-dependent AOM rate was 1.32 ± 0.09 μmol cm−3 year−1 (70). The rates of AOM coupled to MnO2 or Fe(OH)3 reduction are lower than AOM-SR rates, but the energy yields (ΔG° of −659 kJ mol−1 and −285 kJ mol−1, respectively) (Fig. 3, equations 7 and 8) are higher. Thus, the potential energy gains of Mn- and Fe-dependent AOM are, respectively, 10 and 2 times higher than those of AOM-SR, inspiring researchers to further investigate these potential processes (69).
Several researchers have investigated the identity of the bacteria present in putative Fe- and Mn-dependent AOM sites and hypothesized their involvement along with ANME (51, 69). In parallel with AOM-SR, Fe- and Mn-dependent AOM are also assumed to be mediated by two cooperative groups of microorganisms. The bacterial 16S rRNA phylotypes found in Fe- and Mn-dependent AOM sites are putative metal reducers, belonging to the Verrucomicrobia phylotypes (51) and the phyla Bacteriodetes, Proteobacteria, and Acidobacteria and are mostly present in heavy-metal-polluted sites and hydrothermal vent systems (69). The ANME-1 clade was identified as the most abundant in metalliferous hydrothermal sediments and in Eel River Basin methane seep sediment. However, the sole identification of specific bacteria and archaea in these marine sediments does not provide evidence for their metal-reducing capacity.
Some studies assumed the direct coupling of AOM to iron reduction. Lake Kinneret sediment incubations with 13CH4 showed an increase in AOM activity after sulfate reduction inhibition by sodium molybdate and addition of amorphous iron (71), showing the occurrence of iron-coupled AOM. Wankel et al. (51) investigated AOM in hydrothermal sediments from the Middle Valley vent field, where AOM dominated by the ANME-1 phylotype occurred in the absence of SR and SRB. Fe-dependent AOM was hypothesized as the process in these sediments, due to the abundance of Fe(III)-bearing minerals, specifically green rust and a mixed ferrous-ferric hydroxide. A higher AOM rate than with sulfate reduction was observed in in vitro incubations with Mn- and Fe-based electron acceptors like birnessite and ferrihydrite (73).
There is also a hypothesis on the possible indirect coupling of AOM with metal reduction (69). Bar-Or et al. (71) suggested the possible role of different microbial consortia in iron-dependent AOM along with archaea (ANME and/or methanogens). Namely, metal oxides catalyze the oxidation of sulfide, present in the sediment, to elemental sulfur and disulfide. The produced sulfur compounds can be disproportionated by bacteria producing transient sulfate, which can be used to oxidize CH4. These sulfur transformations are referred to as cryptic sulfur cycling (74, 75), and its extent can increase if the sediment is rich in microorganisms able to metabolize elemental sulfur and disulfide (76, 77). A recent study of the Bothnian Sea sediment speculated two separate anaerobic regions where AOM occurs: AOM-SR (in the upper anaerobic layer) and Fe-dependent AOM (in the lower anaerobic layer). It was hypothesized that the majority of AOM was coupled directly to iron reduction in the iron-reducing region and that only about 0.1% of AOM-SR was due to cryptic sulfur cycling (70). However, in marine and brackish sediments, probably only a small percentage of CH4 is oxidized by an Fe-dependent process, as sulfate is the most dominant anion in marine water (46).
Other Electron Acceptors for AOM
Theoretically, based on thermodynamics, anaerobic methane-oxidizing microorganisms can also utilize other electron acceptors, including arsenic, selenium, bromate, and chromium (Fig. 3, equations 9, 10, 11, 12, and 13). It should be noted that the chemistry of selenium oxyanions (selenate [SeO42–] and selenite [SeO32–]) is similar to that of sulfur oxyanions (sulfate [SO42–] and sulfite [SO32–]), since both belong to the same group in the periodic table, the so-called chalcogens. Oxidized selenium species, i.e., selenate or selenite, might thus also be used as an electron acceptor for AOM (Fig. 3, equations 9 and 12).
Metal-dependent AOM was also recently reported to be catalyzed by the DAMO archaea, namely “Candidatus Methanoperedens nitroreducens,” which was able to oxidize methane with Fe(III) and Mn(IV) instead of nitrate (78). Moreover, several recent articles on AOM in anaerobic freshwater sediments showed that ANME can oxidize methane using Mn and Fe minerals as electron acceptors (79–82). Recent studies showed that the DAMO archaea can also mediate AOM using other different electron acceptors, such as selenate (83), bromate (84), and chromate (85). Lu et al. (85) proposed that chromate is reduced either by DAMO archaea or by unknown chromate-reducing bacteria coupled to DAMO archaea. However, acetate or other volatile fatty acids could be formed as intermediates and used as the electron acceptor for selenite reduction (83). Indeed, Luo et al. (84) showed that the formation of acetate by methane oxidation was mediated by DAMO microbes under oxygen-limiting conditions and that the acetate was probably used as an electron donor by the bromate-reducing bacteria.
PHYLOGENY OF ANME
ANME Phylogeny
Based on the phylogenetic analysis of 16S rRNA genes previously illustrated by Knittel and Boetius (22), ANME have been grouped into three distinct clades, i.e., ANME-1, ANME-2, and ANME-3 (38, 39, 41, 86). All ANME are phylogenetically related to various groups of methanogenic archaea (22). ANME-2 and ANME-3 are clustered within the order Methanosarcinales, while ANME-1 belongs to a new order that is distantly related to the orders Methanosarcinales and Methanomicrobiales (22). Specifically, ANME-3 is closely related to the genus Methanococcoides. Fluorescence in situ hybridization (FISH) analysis showed that microorganisms belonging to the ANME-2 and ANME-3 groups are coccus shaped, similar to Methanosarcina and Methanococcus methanogens (Fig. 2B and D), while ANME-1 microorganisms exhibit mostly a rod-shape morphology (Fig. 2A).
Samples from marine habitats around the globe have been retrieved over the years, and extensive molecular analysis of such samples has yielded a great number of 16S rRNA and mcrA gene sequences of the archaeal microorganisms inhabiting those sites. The mcrA gene encodes the alpha-subunit of the methyl coenzyme M (methyl-CoM) reductase (MCR), which catalyzes the last step in methanogenesis (87, 88). It is also thought to catalyze the oxidation of methane, since MCR can function in reverse methanogenesis (87, 88). The mcrA gene phylogeny of the various archaeal orders closely parallels that of the 16S rRNA genes (22). Upon phylogenetic analysis based on 16S rRNA and mcrA (22) genes, the three major groups of ANME were identified. ANME-1 is further subgrouped into ANME-1a and ANME-1b. ANME-2 is divided into four subgroups, i.e., ANME-2a, ANME-2b, ANME-2c, and ANME-2d, whereas no subgroups of ANME-3 have been defined so far (22).
Metabolic Pathways: ANME versus Methanogens
MCR is the key carbon-metabolizing enzyme in methanogens responsible for methane formation. MCR constitutes up to 10% of the total protein in ANME (87) and operates in the reverse direction relative to methanogens (89–91). The MCR forward reaction occurring in methanogens releases energy (ΔG° = −30 kJ mol−1) (88); thus, the reverse reaction (occurring in ANME) is endergonic under standard conditions and will not proceed unless an electron acceptor (e.g., sulfate) is added. Besides the addition of an electron acceptor, the larger amount of MCR in ANME relative to that in methanogens (87) might compensate for the slow reaction (92). However, it is still unclear if the thermodynamic conditions and the abundance of MCR alone can ensure AOM; other MCR modifications might play a role in the reverse methanogenesis (92).
The crystal structure of the MCR protein isolated from a Black Sea mat, naturally enriched in ANME-1, reveals that ANME-1 MCR and methanogenic MCR have distinct features in and around the active site of the enzyme (93). In the MCR from ANME-1, the prosthetic group is not the conventional F430 cofactor, but a methylthio-F430 variant. To accommodate the variant of the cofactor, the geometry of the active site of the ANME-1 MCR enzyme is modified so that the amino acid glutamine, which posttranslationally is two-carbon methylated and thus bulky, is replaced by a small valine molecule (93, 94). These distinct features might reflect that ANME-1 are more distantly affiliated to the other ANME groups, for which no such modifications of their MCR seem to exist (93).
Besides their close phylogenetic relationships and usage of MCR, ANME exhibit other similarities with methanogenic archaea. For example, sequenced genomes of ANME-1 and ANME-2 from environmental samples indicate that except for the N5,N10-methylene-tetrahydromethanopterin (H4MPT) reductase in the ANME-1 metagenome (89), these ANME contain homologous genes for the enzymes involved in all seven steps of reverse methanogenesis (61, 89, 90). All the enzymes related to AOM are yet poorly characterized, and the majority of them are known to catalyze reversible reactions (88, 95), except CoM-S-S-CoB. Thus, it is hypothesized that ANME oxidize methane via the methanogenic enzymatic machinery functioning in reverse, i.e., reversal of CO2 reduction to CH4 (Fig. 4) (89, 91).
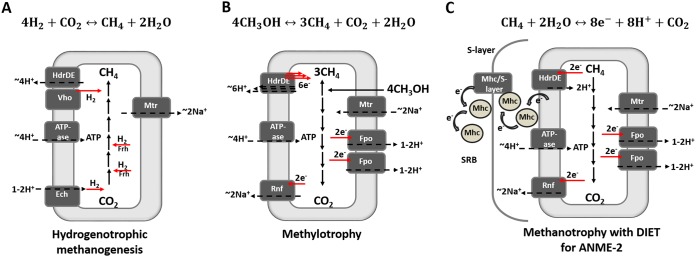
FIG 4.
Diverse physiology in methane-producing and methane-consuming archaea. (A) Hydrogenotrophic methanogenesis by Methanosarcina barkeri; (B) methylotrophy by M. acetivorans; (C) methanotrophy by ANME-2 with direct interspecies electron exchange (DIET). Red arrows indicate electron transfer, and dashed black arrows represent ion translocation steps for energy conservation. Enzyme abbreviations: Frh, F420-reducing hydrogenase; Ech, ferredoxin-dependent hydrogenase; Vho, methanophenazine-reducing hydrogenase; Fpo/Fqo, F420H2:phenazine/quinone oxidoreductase; HdrDE, heterodisulfide reductase; Mhc, multiheme cytochrome; Rnf, Na+-translocating ferredoxin:NAD oxidoreductase; Mtr, Na+-translocating methyl-H4MPT:coenzyme M methyltransferase. The figure is based on reports from McGlynn (108), Timmers et al. (92), Thauer et al. (204), and references therein.
Theoretically, the key AOM enzymes, such as MCR, can also be used to transform methane to alcohols, such as butanol (96, 97): the enzymes responsible for AOM can activate methane and assist in C-C bond formation (97). Nevertheless, there is no formation of a C-C bond during the AOM process. A few recent studies have focused on the conversion of methane to liquid fuels, such as butanol (96, 97). However, only “liquid biofuel precursors,” such as acetate, have been produced from methane (96). This concept is still of interest, because the logistics and infrastructure for handling liquid fuels are more cost-effective than those for utilizing compressed natural gas. A more detailed elucidation of the ANME metabolism is a prerequisite to the development of such biotechnological applications of AOM. Moreover, recent investigations showed that MCR may also use different substrates than methane, such as butane (98).
PHYSIOLOGY OF ANME
Carbon and Nitrogen Metabolism
The difficulty in obtaining enrichment cultures of ANME hampers getting insights into the physiological traits of these microorganisms. Nonetheless, in situ and in vitro activity tests using 13C- or 14C-labeled methane unequivocally revealed that ANME oxidize methane (44). But the physiology of these microorganisms seems to be more intriguing. Recently, it was found that the carbon in ANME biomass is not totally derived from methane, i.e., ANME are not obligate heterotrophs. ANME-2 seem to assimilate carbon from methane and from CO2 at similar amounts (99), whereas carbon within the biomass of ANME-1 is derived from CO2 fixation (16, 100) and ANME-1 also contain genes encoding the CO2 fixation pathway characteristic of methanogens (89). Thus, these ANME are regarded as methane-oxidizing chemoorganoautotrophs (100). In contrast to other typical ANME, ANME-2d microorganisms assimilate mainly methane into their cellular biomass (101).
There is evidence that some ANME-1 and/or ANME-2 present in methane seeps from the Black Sea and the Gulf of Mexico can produce methane (16, 102) from CO2 or from methanol (103). This methanogenic capacity exhibited by these ANME mirrors in turn the methane oxidation capacity displayed by pure cultures of methanogens (104, 105) and by mixed cultures of methanogens present in anaerobic sludge (106), which can oxidize about 1% to 10% of the methane they produce. However, the reported methane oxidation capacity of cultured methanogens is so low that they are not considered to contribute to methane oxidation in marine settings. The detection of important numbers of active ANME-1 cells in both the methane oxidation and the methane production zones of estuary sediments has led to the proposition that this ANME type is not an obligate methane oxidizer but rather a flexible type that can switch and function as a methanogen as well (107).
McGlynn (108) and Timmers et al. (92) recently compared the energy metabolism of different archaea performing AOM: sulfate-reducing ANME, nitrate-reducing ANME, metal-reducing ANME, and methanogens. They showed that the metabolism of the different archaea can be completely reversed by only a few changes in the enzymes present, which are responsible for the electron transfer (Fig. 4, red arrows) and ion movements (Fig. 4, black arrows). In general, in hydrogenotrophic methanogens, e.g., Methanosarcina barkeri (Fig. 4A), CO2 is reduced to CH4 with H2, whereas in methanotrophs (e.g., ANME-2), the reverse process, in which CH4 oxidizes to CO2, occurs (Fig. 4C). Methanogenesis is already partly reversed during methylotrophic methanogenesis (Fig. 4B), in which a carbon compound, such as 4 mol of methanol, is disproportionated to 3 mol of CH4 and 1 mol of CO2 (Fig. 4B). During methylotrophy, in the absence of H2, the methyl group of methanol is partly oxidized to regenerate the reducing equivalents needed in the methanogenic respiratory chain.
The energy-conserving steps in ANME are very similar to the ones shown in the methylotrophic methanogens, as modelled by Methanosarcina acetivorans (Fig. 4C and B, respectively). Among the enzymes embedded in the membrane, some pump ions out of the cell, such as F420H2-phenazine oxidoreductase (Fpo) and sodium-translocating Fd/NAD+ (Rnf), and some let ions into the cell, such as coenzyme B-coenzyme M heterodisulfide reductase (Hdr) and N5-methyl-H4MPT:coenzyme M methyltransferase (Mtr), to promote less-favorable reactions (92, 108).
Methylotrophic methanogens, such as M. acetivorans, contain key elements of carbon metabolism and energy conservation similar to those in ANME, except for the multiheme c-type cytochromes, Mhc (96). Several studies have proposed that the Mhc are in the surface layer (S-layer) of archaea and that electrons are transferred from the archeal membrane to the S-layer cytochromes (Fig. 4C) and from them to the SRB partner (109) or to other electron acceptors such as Fe(III) oxides (70, 109). Thus, the introduction of Mhc would allow Methanosarcina acetivorans to become a methanotroph.
Another intriguing physiological trait is the N2-fixing capacity (i.e., diazotrophy) of ANME-2d. Using 15N2 as the nitrogen source, it was found that ANME-2d cells assimilate 15N in batch incubations of marine mud volcano or methane seep sediments (110, 111). While fixing N2, ANME maintained their methane oxidation rate, but their growth rate was severely reduced. The energetic cost to fix nitrogen is one of the highest among all anabolic processes and requires about 16 ATP molecules, corresponding to 800 kJ mol−1 of nitrogen reduced. Therefore, considering the meager energy gain of AOM (about 30 or 18 kJ mol−1 of CH4 oxidized), it is consistent that the growth rate of ANME is 20 times lower when N2 rather than ammonium (NH4+) is used as the nitrogen source (110). Yet, it is not resolved under which in situ conditions these microorganisms are diazotrophic. Also, whether other ANME types are diazotrophs has not yet been shown. Although the metagenome of ANME-1 reveals the presence of various candidate proteins having similarity to proteins known to be involved in N2 fixation (89), this trait has not yet been tested experimentally.
Electron Transfer between ANME and SRB
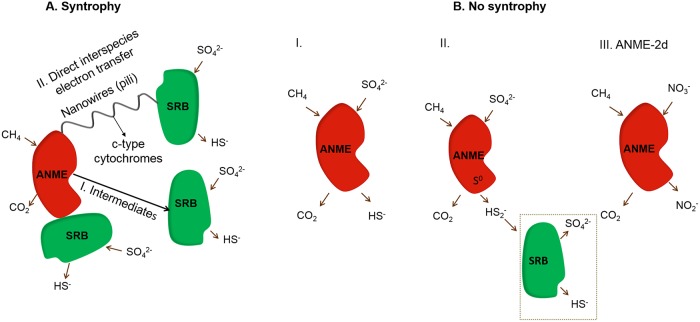
While several theories have been proposed to understand the interaction between ANME archaea and SRB, the most common hypothesis is syntrophy between these two groups (Fig. 5A). Obligate syntrophs share the substrate degradation process, resulting in one partner converting the substrate into an intermediate, which is consumed by the syntrophic partner (112). Unlike other known forms of syntrophy, the intermediate shared by ANME and SRB has not yet been identified. The syntrophy between ANME and SRB is hypothesized on the basis of the tight cooccurrence of ANME and SRB in AOM active sites, as revealed by FISH images (Fig. 2B and D) (39, 41, 113). Isotopic signatures in archaeal and bacterial lipid biomarkers strengthen this hypothesis of syntrophy, assuming transfer of an intermediate carbon compound between the ANME and SRB (39, 114–116). Also, phylogenetic analysis showed the cooccurrence of SRB and ANME in samples from AOM sites, suggesting that their coexistence may play a role in AOM (54, 117, 118).
FIG 5.
Syntrophic and nonsyntrophic anaerobic methanotrophs (ANME) using sulfate as an electron acceptor. (A) In a syntrophic association, ANME can transfer electrons to a sulfate-reducing bacterium (SRB) via different mechanisms: electron transfer via possible intermediate compounds such as formate, acetate, or hydrogen (I) and direct interspecies electron transfer between ANME and SRB mediated by c-type cytochromes, e.g., as in ANME-2 (131), or via nanowires and cytochromes acting, e.g., as in thermophilic ANME-1 (132) (II). (B) In a nonsyntrophic association, ANME can possibly perform the complete AOM process alone without a sulfate-reducing partner by using insoluble iron oxides as external electron acceptors (109) (I), by producing carbon dioxide and disulfide (HS2–) with S0 as an intermediate (the HS2– can be disproportionated by SRB) (55) (II), or by ANME-2d performing DAMO (III).
Hydrogen and other methanogenic substrates, such as acetate, formate, methanol, and methanethiol, were hypothesized as the intermediates between ANME and SRB (Fig. 5A) (40, 119, 120). Formate is the only possible intermediate which would result in free-energy gain, so thermodynamic models support formate as an electron shuttle (e-shuttle) of AOM (120). However, acetate was assumed to be the favorable e-shuttle in high-methane partial-pressure environments (121). Genomic studies have suggested that the putative intermediates for AOM could be acetate, formate, or hydrogen (89, 91). The formate dehydrogenase gene is highly expressed in the ANME-1 genome, and thus formate can be formed by ANME-1 and function as an intermediate (89). Likewise, the ADP-forming acetyl coenzyme A (acetyl-CoA) synthetase (ACS), which converts acetyl-CoA to acetate, was retrieved from an ANME-2a genome (90) and recently also found in ANME-2d, which was linked with its fatty acid metabolism (122). The ADP-forming ACS in ANME-2a has yet to be tested; however, acetate could be formed by ANME-2a and be a possible intermediate.
The first step of AOM is the conversion of methane to methyl-CoM; however, the production of either acetate or hydrogen could be via other pathways and not necessarily by reverse methanogenesis (90, 91). Nevertheless, the addition of hydrogen in an AOM experiment did not lead to any change in AOM rate, in contrast to the typical methanogenesis process (123). Similarly, methane-based SR rates were the same even if these potential intermediates (acetate/formate) were supplied, whereas the reaction should be shifted to lower AOM rates upon the addition of intermediates (123, 124). Moreover, the addition of these potential intermediates induced the growth of SRB other than the Desulfosarcina/Desulfococcus and Desulfobulbaceae groups, which are the assumed syntrophic partner of ANME (125). Therefore, the hypothesis of these compounds being possible AOM e-shuttles remains unconfirmed. Instead, methyl sulfide was proposed to be an intermediate for both methanogenesis and methanotrophy (123). Methyl sulfide was then assumed to be produced by the ANME that can be utilized by the SRB partner (123).
Alternatively, electrons are directly transferred from the ANME to the SRB cells. A few species, such as Geobacter and Methanosarcina species, are known to cooperate by direct electron transfer through conductive structures on the cell surfaces (126, 127). Several mechanisms have been proposed for electron transfer: via microbial nanowires (128), direct electron transfer via Mhc on the cell surfaces (127), or via conductive minerals (129) (Fig. 5A). Mhc (Fig. 4C) were identified in the ANME-1 archaeal genome (89), and the Mhc-specific gene was also well expressed in ANME-2a cells, according to a metatranscriptomic study (90). The importance of Mhc has been extensively discussed in Geobacter species, where the cytochrome can act as electron storage in the cell membrane and, subsequently, extracellular electron transfer occurs (130). These microorganisms use cell membrane cytochromes and pili as biological nanowires to connect with minerals (128). Recent studies have given some other evidence of the direct interspecies electron transfer (DIET) between ANME and SRB, showing a mechanism similar to that for Geobacter (131–133). Thermophilic ANME-1 and bacterial partners showed pilus-like structures and highly expressed genes for outer membrane Mhc, which are putatively responsible for transferring electrons, as shown in Fig. 4C (92, 132). The ANME-2 genome encodes large amounts of Mhc (131). However, whether the aggregates are electrically conductive and whether a conductive matrix can be formed throughout the ANME-SRB aggregates still need to be validated.
Skennerton et al. (134) showed that DIET was methane driven for the SRB partner of ANME. By metagenomics, it was shown that genomes of the SEEP-SRB1 clade, a known bacterial partner of ANME, contain Mhc but lack genes encoding the periplasmic hydrogenases and formate dehydrogenases, typical of sulfate-reducing bacteria that live independently from ANME and use hydrogen or formate as an electron donor. Syntrophy may be obligate not only for some ANME but also for the bacterial partners lacking these genes.
Nonsyntrophic Growth of ANME
Despite the recent discoveries about the cooperation between ANME and SRB, the topic is still under debate. Visualization of ANME and its bacterial partners by FISH showed that for all three clades of ANME, the association with SRB is not obligatory. In some cases, the AOM process may occur by only the ANME without any sulfate-reducing partner. Decoupling of AOM-SR was observed in ANME-1-dominated thermophilic environments (51), and ANME-2 was predicted to be involved in SR as well (55) (Fig. 5B). Moreover, the AOM-SR potential was recently found in the single genome of a group of Korarchaeota, suggesting possible involvement of only one type of phylum in AOM-SR (48, 49).
Occurrence of genes for both SR and AOM in Korarchaeota WYZ-LMO9 further supports the hypothesis of involvement of only one microbial type in AOM-SR (48, 49). Milucka et al. (55) proposed a new AOM mechanism, in which ANME might be responsible for both CH4 oxidation and SR (Fig. 5B). CH4 would be oxidized to bicarbonate, and then the SO42– would be reduced to zero-valent sulfur as an intracellular intermediate in ANME-2 cells. The resulting sulfur would then be released outside the cell as disulfide, which would be converted to sulfide by the SRB. Figure 5B shows that some ANME can sustain the overall AOM reaction without a bacterial partner, even though the Desulfosarcina/Desulfococcum type deltaproteobacteria render the AOM-SR more thermodynamically favorable by scavenging the disulfide by disproportionation or dissimilatory reduction. The disulfide produced by ANME is disproportionated into SO42– and sulfide. The thus-produced SO42– can be used again by the ANME, while sulfide can undergo several conversions, for instance, precipitation as FeS2 or partial (to S0) or complete (to SO42–) oxidation aerobically or anaerobically (in the presence of light by, e.g., purple sulfur bacteria) (135). As described earlier, in the presence of iron oxides, sulfide reacts abiotically, forming more substrates (disulfide and elemental sulfur) for the Deltaproteobacteria. The reaction of sulfide with iron oxides can thus strongly enhance the sulfur cycle, as previously observed in studies with Sulfurospirillum deleyianum (76).
The cooperative/synergistic interaction between ANME and SRB is still unclear, as Milucka et al. (55) stated that a syntrophic partner might not be required for ANME-2, while recent studies have shown that the interactions between these two partners occur via direct electron transfer (131, 132, 136). However, ANME-1 can live without the bacterial partner by using insoluble iron oxides as external electron acceptors (109). Thus, growing ANME separately may be possible in the future, which will contribute to our understanding of the AOM mechanism.
ECOLOGY OF ANME IN MARINE HABITATS
Major Habitats of ANME
ANME are widely distributed in marine habitats, including cold seep systems (gas leakage from methane hydrates), hydrothermal vents (fissures releasing hot liquid and gas in the seafloor), and organic matter-rich sediments with diffusive methane formed by methanogenesis (Fig. 6). The cold seep systems include mud volcanoes, hydrate mounds, carbonate deposits, and gaseous carbonate chimneys (15), which are all abundantly studied ANME habitats. The major controlling factors for ANME distribution are the availability of methane and sulfate or other terminal electron acceptors which can possibly support the AOM, while other environmental parameters, such as temperature, salinity, and alkalinity, also play a decisive role in ANME occurrence. Among the three clades, ANME-2 and ANME-3 apparently inhabit cold seeps, whereas ANME-1 reside in a wide temperature and salinity range. Recently, AOM has been reported in nonsaline and terrestrial environments as well, for instance, in the Apennine terrestrial mud volcanoes (118, 137), in the boreal peat soils of Alaska (138), and even in freshwater sediments (139, 140), tropical wetlands (141), and Italian paddy field soil (142) (Table 1).
FIG 6.
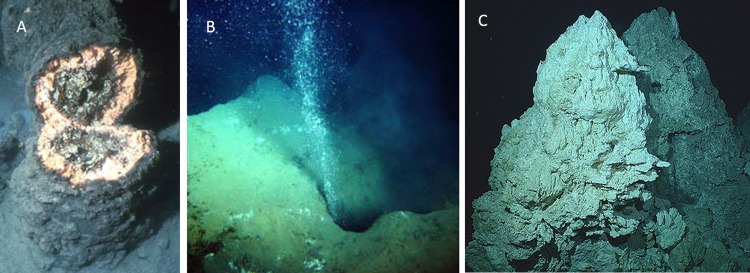
In situ pictures of some of the well-studied ANME habitats. (A) Black and pink microbial mats covering a carbonate chimney in the Black Sea. Republished from reference 143 with permission from AAAS. (B) Methane seeping from the Haakon Mosby Mud Volcano. Republished from reference 205 (image copyright IFREMER, Brest, and Alfred Wegener Institute [AWI] for Polar and Marine Research, Bremerhaven, Germany). (C) Carbonate chimney from the Lost City hydrothermal vent. Republished from reference 164 with permission.
TABLE 1.
Rates of AOM and SR in different natural terrestrial habitatsa
| Location | Soil depth (cm) | Methane (mM) | Sulfate (mM) | AOM rate | SR rate | Reference |
|---|---|---|---|---|---|---|
| Wetland and peat soil | 0–40 | 0.5–1 | 0.1–1 | 265 ± 9 nmol cm−3 day−1 | 300 nmol cm−3 day−1 | 206c |
| Tropical and boreal soils | 10–15 | <1 | 3-21 nmol gdw−1 day−1 | 138b | ||
| Paclele Mici Mud Volcano in Carpathian Mountains | 1.5–2 | 2-4 nmol gdw−1 day−1 | 118b | |||
| Peat land soil from diverse places | 30–50 | 0.03–3 nmol gdw−1 day−1 | 207b |
The different methods of anaerobic methane oxidation (AOM) and sulfate reduction (SR) measurements are indicated in footnotes to the references. gdw, gram (dry weight).
In vitro measurement.
Ex situ radiotracer measurement.
The Black Sea water, a distinct ANME habitat, consists of thick microbial mats of ANME-1 and ANME-2 (2 to 10 cm thick) adhered with carbonate deposits (chimney-like structure) in various water depths of 35 to 2,000 m (53, 113, 143–146). Methane is distributed by vein-like capillaries throughout these carbonate chimneys and finally emanates to the water column (53, 143, 147). These microbial habitats are of different size and nature, such as small preliminary microbial nodules (53), floating microbial mats (147), and large chimneys (143). An immense carbonate chimney from the Black Sea (Fig. 6A), with a height of up to 4 m and a width of up to 1 m, was found to harbor an ANME-1-dominant pink-colored microbial mat with the highest known AOM rates in natural systems (113, 143). Deep-sea carbonate deposits from cold seeps and hydrates are active and massive sites for AOM and ANME habitats (148). Likewise, methane-based authigenic carbonate nodules and methane hydrates which host ANME-1 and ANME-2 (42, 148–151) prevail in the Eel River Basin (offshore, California), a cold seep with an average temperature of 6°C that is known for its gas hydrates (38, 152). Both ANME-1 and ANME-2 are commonly associated with Desulfosarcina/Desulfococcus in the sediments of the Eel River; however, ANME-1 appeared to exist as single filaments or monospecific aggregates in some sites as well (38, 42, 151).
Other cold seep sediments were also extensively studied as ANME habitats. The Gulf of Mexico, a cold seep with a bottom water temperature of 6°C to 8°C, is known for its gas seepage and associated hydrates. These methane hydrates, located at a seawater depth of around 500 m in the Gulf of Mexico, are inhabited by diverse microbial communities. Beggiatoa mats oxidize sulfide and are commonly found in the upper layers of the oxic sediments, while ANME are often found in the bottom part (anaerobic) of the sulfidic sediments in the Gulf of Mexico (18, 102, 153, 154). ANME-1 dominates the sediment of the Gulf of Mexico, particularly in the hypersaline part as a monospecific clade, whereas ANME-2a and -2b are present together with the Desulfosarcina/Desulfococcus groups in the less-saline hydrates (102, 154). Similarly, different mud volcanoes in the Gulf of Cadiz cold seep harbor ANME-2, with the majority being ANME-2a (17), whereas the hypersaline Mercator Mud Volcano of the Gulf of Cadiz hosts ANME-1 (52). Retrieval of ANME-1 in the hypersaline environment suggests that ANME-1 has adaptability to wider salinity ranges than other ANME phylotypes. Mud volcanoes from the Eastern Mediterranean (Kazan and Anaximander Mountains) are inhabited by all three ANME phylotypes, whereas the Kazan Mud Volcano hosts the distinct ANME-2c clade (155–158). Likewise, the Haakon Mosby Mud Volcano (HMMV) in the Barents Sea is the first described habitat for ANME-3, with almost 80% of the microbial cells being ANME-3 and Desulfosarcina/Desulfococcus (Fig. 6B and Fig. 2D) (54, 86).
Some of the hydrothermal vents are well-studied ANME marine habitats for distinct ANME clades and thermophilic AOM. The Guaymas Basin in the California Bay, an active hydrothermal vent with a wide temperature range, is known for the occurrence of different ANME-1 phylotypes, along with unique thermophilic ANME-1 (159–161). ANME-1 is predominant throughout the Guaymas Basin, yet the colder methane seeps of the Sonora Margin host all three ANME phylotypes (ANME-1, ANME-2, and ANME-3) with atypical ANME-2 (ANME-2c Sonora) (161). Likewise, mesophilic to thermophilic AOM carried out by the ANME-1 clade was detected in the Middle Valley vent field on the Juan de Fuca Ridge (51, 162). Another vent site, the Lost City hydrothermal vent, with massive fluid circulation and ejecting hydrothermal fluid of >80°C, predominantly hosts ANME-1 within the calcium carbonate chimneys (Fig. 6C), which are very likely deposited due to bicarbonate formation from AOM (163, 164).
ANME Type Distribution by Temperature
The ANME clades exhibit a distinct pattern of distribution according to temperature (165, 166). Other geochemical parameters, such as salinity, methane concentration, and pressure, can act together with temperature as selection parameters for the distribution of ANME in marine environments (167). ANME-1 was extensively retrieved across the temperature gradient of 2°C to 100°C in the Guaymas Basin from the surface to deep sediments (168). A phylogenetically distinct and deeply branched group of the ANME-1 (ANME-1GBa) was found in the high-temperature Guaymas Basin hydrothermal vent (160) and other geologically diverse marine hydrothermal vents, such as the diffuse hydrothermal vents in the Juan de Fuca Ridge in the Pacific Ocean (10 to 25°C) (169). The thermophilic trait of ANME-1GBa is supported by the GC (guanine and cytosine) content in its 16S rRNA genes, as it has a higher GC percentage (>60 mol%) than other ANME types. As the GC content is positively correlated with the optimum temperature of microbial growth, the elevated GC content of ANME-1GBa suggests that ANME-1GBa is a thermophilic microbial cluster, with an optimum growth temperature of 70°C or above (169). Moreover, when the Guaymas ANME community was enriched in vitro, the highest AOM rate was obtained in the range of 45°C to 60°C, indicating that the community consists predominantly of thermophilic ANME-1 (165).
Other ANME-1 phylotypes (ANME-1a and ANME-1b) were observed in a wide temperature range (3°C to >60°C) (160). ANME-1a and ANME-1b were retrieved from different hydrothermal vent areas and cold seeps, for example, the Guaymas Basin hydrothermal vent at >60°C (160), the Lost City hydrothermal vent (164), the Sonora Margin cold seep of the Guaymas Basin (3°C) (161), mud volcanoes in the Eastern Mediterranean cold seep (14 to 20°C) (170), the Gulf of Mexico (6°C) (154, 171), a Black Sea microbial mat and water column (8°C) (41, 172), and the Eel River Basin (6°C) (38, 151). The occurrence of ANME-1a and ANME-1b in cold seep environments suggests that ANME-1a and ANME-1b are putative mesophiles to psychrophiles. The GC percentage of 16S rRNA genes of ANME-1a and ANME-1b is around 55 mol%, which is common for mesophiles (169).
In contrast, ANME-2 and ANME-3 have a narrow temperature range. ANME-2 clades (2a, 2b, and 2c) appear predominantly in marine cold seeps and in some sulfate-methane transition zones, where the temperature is about 4 to 20°C. The major cold seep environments inhabited by ANME-2 are described above (see “Major Habitats of ANME”). The adaptability of ANME-2 in the cold temperature range is also substantiated by bioreactor enrichments with Eckernförde Bay sediment, where the maximum AOM rate was obtained when the bioreactor was operated at 15°C rather than at 30°C, for ANME-2a (23). Similarly, Eckernförde Bay in vitro measurements showed a steady increment in AOM rates at temperatures from 4°C to 20°C and subsequent decreases at temperatures above 20°C (173). Conversely, the recently described clade of ANME-2d-affiliated “Candidatus Methanoperedens nitroreducens" (Fig. 2C), which was enriched from a mixture of freshwater sediment and wastewater sludge (61), grows optimally at mesophilic (22°C to 35°C) temperatures (174).
ANME-3 is also known to thrive in cold temperature environments, including cold seeps and mud volcanoes. The ANME-3 clade was first retrieved from the Haakon Mosby Mud Volcano at a temperature of about −1°C (86). Later, ANME-3 was found in other cold seep areas as well, such as the Eastern Mediterranean seepages at about 14°C (155, 157) and the Skagerrak seep (Denmark, North Sea) at around 6 to 10°C (175). Recently, ANME-3 was found in a shallow coastal sediment from marine Lake Grevelingen that was rich in methane and with a seasonal temperature variation of ∼1.5°C to 17°C and high sedimentation (176, 177).
Methane Supply Mode as Driver for ANME Distribution
In some seafloor ecosystems, methane is transported by diffusion due to concentration gradients. Diffusion-dominated ecosystems are typically quiescent sediments. In contrast, in seafloor ecosystems with methane seeps, methane is transported by advection of methane-rich fluids. Due to the complex dynamics of methane transport in advection-dominated environments, estimations of in situ methane oxidation rates by geochemical mass balances is rather difficult (178). Based on ex situ tests, the AOM rates are higher in ecosystems where high methane fluxes are sustained by advective transport than in diffusion-dominated ecosystems (15). The velocity of the methane-rich fluid may result in an order of magnitude difference in AOM rates. Higher AOM rates were observed at sites with higher flow velocity (179), and high flows of methane-rich fluid probably support dense ANME populations.
The extent of methane flux and the mode of methane transport (advection versus diffusion) are likely important drivers for ANME population dynamics. Mathematical simulations illustrate that the transport regime can control the activity and abundance of AOM communities (180). Multivariate and cluster analyses show that the mode of methane transport can possibly control AOM communities (Fig. 7). Methane-rich upward fluid flow at active seep systems restricts AOM to a narrow subsurface reaction zone and sustains high methane oxidation rates. In contrast, pore water methane transport dominated by molecular diffusion leads to deeper and broader AOM zones, which are characterized by much lower methane oxidation rates and biomass concentrations (180). In this context, Roalkvam et al. (181) found that the methane flux largely influenced the specific density of ANME populations. However, whether distinct ANME types preferentially inhabit environments dominated by advective or diffusive methane transport is not yet clear. At sites with high seepage activity like the Hydrate Ridge in Oregon, ANME-2 was dominant, whereas ANME-1 apparently was more abundant in the low-seepage locations (182).
FIG 7.
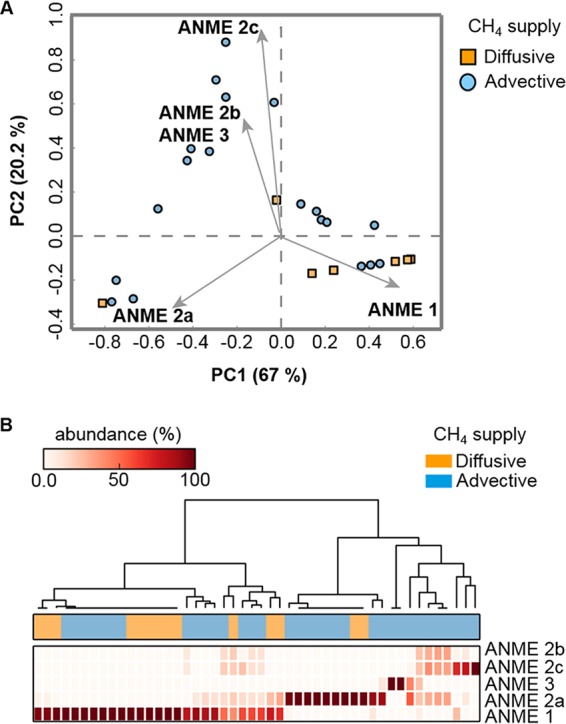
Multivariate (A) and cluster (B) analyses of the mode of CH4 transport, illustrating that it is one of the drivers for the distribution of ANME types in the environment and showing that ANME-2 is dominant mostly in CH4-advective sites. The heat map was generated using the heatmap.2 function in R. Hierarchical clustering was performed using complete linkage.
A rough estimate of the abundance of the ANME type populations, reported in various marine environments, shows that ANME-2 dominates sites where methane is transported by advection, while ANME-1 dominates sites where methane is transported by diffusion or advection (Fig. 7). Recent studies showed that gas eruptions and mud mixing influence the abundance of different ANME phylotypes in the Haakon Mosby Mud Volcano (183) and marine Lake Grevlingen; ANME-3 (176, 177) populations also proliferate in disturbed sediment, unlike ANME-2a. It is advisable that future studies regarding the distribution of the ANME types explicitly indicate the dominant mode of in situ methane transport.
EX SITU ENRICHMENT OF ANME MEDIATING AOM-SR
Need for Enrichment of ANME
Molecular methods allow the recognition of the phylogenetic diversity of ANME microorganisms in a wide range of marine sediments and natural environments. Determination of their detailed physiological and kinetic capabilities has required, until now, the cultivation and isolation of the microorganisms. The culturability of microorganisms inhabiting seawater (0.001 to 0.1%), seafloor (0.00001 to 0.6%), and deep subsea (0.1%) sediments is among the lowest compared to that of other ecosystems (184–186).
Specifically for the enrichment of ANME mediating AOM-SR, the following aspects limit their cultivation: (i) from all known microbial processes, the AOM reaction with sulfate is among those which yield the lowest energy; (ii) the growth rate of these microorganisms is thus very low, with a yield of 0.6 g cell (dry weight) per mol of CH4 oxidized (44); (iii) the dissolved concentrations of their substrate, methane (1.4 mM), at atmospheric pressures is limited to values far lower than the estimated apparent half affinity constant for methane (37 mM) during the AOM-SR process; and (iv) sulfide, which is a product of the bioconversion, can be inhibitory. All of these aspects create a great challenge for the cultivation and isolation of ANME.
It is recognized that culturability can be enhanced when the conditions used for cultivation mimic well those of the natural environment. Cultivation efforts have focused mainly on increasing dissolved methane concentrations. To enrich ANME mediating AOM-SR ex situ, batch and continuous reactors operated at moderate and high pressures have been tested for 24 months. To avoid potential sulfide toxicity, attention has been paid to exchanging the medium so that the sulfide concentrations do not exceed 10 to 14 mM (44, 187).
Conventional In Vitro Enrichment Techniques of ANME Mediating AOM-SR
The conventional in vitro incubation in gas-tight serum bottles provides an opportunity to test the microbial activities, kinetics of the metabolic reactions, and enrichment of the microbes, more specifically for the large number of uncultured anaerobes like ANME. Conventional serum bottles are widely used when the incubation pressures do not exceed 0.25 MPa (69, 112, 188, 189).
A batch bottle experiment provides the flexibility to operate many different experiments in parallel (large numbers of experimental bottles can be handled at the same time) by controlling different environmental conditions, such as temperature, salinity, and alkalinity. The batch incubation-based experiments are relatively easy to control and manipulate, especially with very slowly growing microbes like ANME, which require strictly anaerobic conditions. AOM activity is negligible in the presence of oxygen (173). The commonly used batch serum bottles or culture tubes with thick butyl rubber septa facilitate the sampling while maintaining the redox inside, although there are several other factors which can be key for ANME enrichment, such as the low solubility of methane and the possible accumulation of toxic sulfide levels in the stationary batches.
As shown in Tables 2 and 3, several studies have estimated the AOM rate by in vitro batch incubations (99, 165, 190). Krüger et al. (147) determined AOM rates of 4,000 to 20,000 nmol g (dry weight)−1 day−1 by incubating microbial mats from the Black Sea. Holler et al. (188) estimated AOM at a rate of 250 nmol g (dry weight)−1 day−1 by ANME-1 from the Black Sea (Table 2). ANME-2-dominated communities from the Hydrate Ridge of the northeast Pacific exhibited 20-times-higher specific AOM rates [20 mmol g (dry weight)−1 day−1] than ANME-1 from the Black Sea pink microbial mat (125).
TABLE 2.
Rates of AOM and SR in different natural marine habitats and dominant ANME typesa
| Marine habitat and location | Water depth (m) | Methane (mM) | Sulfate (mM) | Major ANME type(s) | AOM rate | SR rate | Reference(s) |
|---|---|---|---|---|---|---|---|
| Cold seeps (temp, 1.5–20°C) | |||||||
| Eastern Mediterranean Sea | 500–3,200 | 2–8 | 10 | ANME-1, -2, -3 | 100,000–3,600,000 nmol m−2 day−1 | 100,000–66,000,000 nmol m−2 day−1 | 206–219b |
| Black Sea (giant carbonate chimney) | 230 | 2.8 | 17 | ANME-1 | 7,800–21,000 nmol−1 gdw−1 day−1 | 4,300–19,000 nmol gdw−1day−1 | 16, 143b |
| Black Sea (other microbial mats) | 180 | 3.7 | 9–15 | ANME-1, -2 | 2,000–15,000 nmol gdw−1 day−1 | 4,000–20,000 nmol gdw−1 day−1 | 147b |
| Haakon Mosby Mud Volcano, Barents Sea | 1,250 | 0.0003–0.0057 | ANME-2, -3 | 1,233–2,000 nmol cm−2 day−1 | 2,250 nmol cm−2 day−1 | 86c | |
| Gulf of Mexico, hydrate | 550–650 | 2–6 | 20 | ANME-1, -2 | 280 ± 460 nmol cm−2 day−1 | 5,400 ± 9,400 nmol cm−2 day−1 | 18, 102 |
| Eel River Basin, carbonate mounds and hydrates | 500–850 | 3 | 20 | ANME-1, -2 | 200 nmol cm−3 day−1 | 148, 166c | |
| Gulf of Cadiz, mud volcanoes | 810–3,090 | 0.001–1.3 | 10–40 | ANME-2, -1 | 10–104 nmol cm−2 day−1 | 158–189 nmol cm−2 day−1 | 17c |
| Black Sea water | 100–1,500 | 0.011 | ANME-1, -2 | 0.03–3.1 nmol day−1 | 172, 212c | ||
| Tommeliten seepage area, North Sea sediment | 75 | 1.4–2.5 | 30–20 | ANME-1, -2 | 1.4–3 nmol cm−3 day−1 | 3–4.6 nmol cm−3 day−1 | 213c |
| Methane-rich sediments (temp, 4–20°C) | |||||||
| Bothnian Sea sediment | 200 | 2 | 5.5 | 40–90 nmol cm−2 day−1 | 214d | ||
| Baltic Sea/Eckernförde Bay sediment | 25 | 0.001–0.8 | 16–21 | ANME-2 | 1–14 nmol cm−3 day−1 | 20–465 nmol cm−3 day−1 | 173c |
| Skagerrak sediment | 308 | 1.3 | 25 | ANME-2, -3 | 3 nmol cm−3 day−1 | 175e | |
| West African margin sediment | 400–2,200 | <1–19 | 26 | 0.0027 nmol cm−3 day−1 | 215d | ||
| Lake Grevelingen | 45 | 6 | 25 | ANME-3 | 0.05–0.17 μmol gdw−1 day−1 | 5 μmol gdw−1 day−1 | 176, 216b,e |
| Hydrothermal vents (temp, 10 to 100°C) | |||||||
| Guaymas Basin hydrothermal vent | ANME-1 | 1,200 nmol gdw−1 day−1 | 250 nmol gdw−1 day−1 | 165b | |||
| Juan de Fuca Ridge hydrothermal vent | 2,400 | 3 | ANME-1 | 11.1–51.2 nmol cm−3 day−1 | 51d |
The different methods of anaerobic oxidation of methane (AOM) and sulfate reduction (SR) measurements are indicated in footnotes to the references. gdw, gram (dry weight).
In vitro measurement.
Ex situ radiotracer measurement.
Model calculation.
Pore water chemistry measurement.
TABLE 3.
Enrichment conditions and AOM rates for in vitro studies of AOMa
| Enrichment mode | Inoculum and incubation period | Temp (°C) | Pressure (105 Pa) | Enriched ANME type(s) | AOM rate | ANME doubling time (mos) | Growth rate, μ (day−1) | Apparent affinity (Km) | Reference(s) |
|---|---|---|---|---|---|---|---|---|---|
| Culture batches | Isis enrichment, Eastern Mediterranean, 8 yrs | 24 | 3 | ANME-2 | 4 mmol liter−1 | 55b | |||
| Membrane bioreactor, continuous well mixed | Baltic Sea/Eckernförde Bay, 884 days | 15 | 1 | ANME-2a | 286 μmol gdw−1 day−1 | 3.8 | 0.006 | <0.5 mM (for SO42–), 0.075 MPa for CH4 | 23, 112b |
| External ultrafiltration membrane bioreactor | Ginsburg Mud Volcano, Gulf of Cadiz, 726 days | 15 | 1 | ANME-2 | 1.2 μmol gdw−1 day−1 | 26b | |||
| Fed-batch | Hydrate Ridge, North-east Pacific, 700 days | 15 | 14 | ANME-2 | 230 μmol gdw−1 day−1 | 7 | 0.003 | >10 mM (CH4) | 44b |
| Batch | Gulf of Mexico, 150 days | 12 | 15 | ANME-1 | 13.5 μmol gdw−1 day−1 | 2 | 190b | ||
| Batch | Lake Grevelingen, 77 days | 15 | 45 | ANME-3 | 324 μmol gdw−1 day−1 | 1.7 mM (CH4) | 177b | ||
| Fed-batch | Gulf of Cadiz, 286 days | 15 | 80 | ANME-2 | 9.22 μmol gdw−1 day−1 (SR) | 2.5 | 37 mM (CH4) | 196, 197b | |
| Batch | Captain Aryutinov Mud Volcano, Gulf of Cadiz, 80 days | 15 | 100 | ANME-2a | 24 μmol gdw−1 day−1 | 191b | |||
| Batch | Guaymas Basin sediment, 250 days | 42–65 | 2.5 | ANME-1 | 1.2 μmol gdw−1 day−1 | 2.3 | 165b | ||
| Downflow hanging sponge (DHS) bioreactor | Nankai Trough, 2,013 days | 10 | 1 | ANME-1, -2, -3 | 0.375 μmol gdw−1 day−1 | 218b | |||
| Biotrickling filter | Gulf of Cadiz, 248 days | 15 | 1 | ANME-1, -2 | 11.5 μM day−1 | 27, 29b | |||
| Biotrickling filter | Gulf of Cadiz, 238 days | 15 | 1 | ANME-2 | 300 μM day−1 | 221 | |||
| Anaerobic methane incubator system (continuous) | Monterey Bay, 400 days | 5 | 1 | ANME-1, -2 | 9 × 10−3 μmol gdw−1 day−1 (ANME-1), 0.138 μmol gdw−1 day−1 (ANME-2) | 1.1 (ANME-2), 1.4 (ANME-1) | 0.03 (ANME-1), 0.024 (ANME-2) | 220c | |
| Fed-batch | AOM and annamox, 230–290 days | 22–35 | 0.5–1 | ANME-2d | 1,100 μM day−1 | 61d |
The different mineral media used for incubation are indicated in footnotes to the references. gdw, gram (dry weight); SR, sulfate reduction.
Incubation in artificial saltwater mineral medium prepared as described by Widdel and Bak (217).
Incubation with filter-sterilized seawater.
Incubation in freshwater medium with nitrate and ammonium.
During the in vitro incubations under different environmental conditions, unlike the sulfate concentration, pH, and salinity variations, temperature was found to be a major influential parameter for AOM rates in ANME-1 and ANME-2 communities (125). Both ANME communities showed an increment in the AOM rate with elevated methane partial pressure. However, when the microbial mat from the Black Sea with both ANME-1 and ANME-2 was incubated in batch at low methane concentrations, ANME-1 growth was favored over the growth of ANME-2 (189). In vitro incubations of samples from marine Lake Grevelingen at different CH4 partial pressures showed that ANME-3 cells were more pressure sensitive than ANME-2 cells (177). ANME-3 activity was higher (324 μmol g [volatile suspended solids]−1 day−1) at lower (0.1 and 0.45 MPa) than at higher (10, 20, and 40 MPa) CH4 partial pressures (177). Furthermore, Bhattarai et al. (191) showed that both temperature and pressure influence the AOM rate of an enriched ANME-2 (3 years in a continuous-high-pressure reactor at 15°C and 8 MPa) inoculum from Captain Aryutinov Mud Volcano (Gulf of Cadiz) sediment.
Optimum pH, temperature, salinity, and sulfide toxicity were determined to be 7.5, 20°C, 30‰, and 2.5 mM, respectively, for the ANME-2 enrichment from Eckernförde Bay when sediments were incubated in 35-ml serum bottles (112, 173). Likewise, possible electron donors and acceptors involved in the AOM process were studied in batch incubations. The sediment from Eckernförde Bay was also incubated with different methanogenic substrates, which have been speculated to be the interspecies electron carriers between the ANME and SRB (124). The AOM activity with electron acceptors other than sulfate, i.e., Fe(III) and Mn(IV), by Eel River sediment was estimated by batch incubations for the detection of iron/manganese-dependent AOM (69). Moreover, when thermophilic AOM was studied in batch assays within different temperature ranges (up to 100°C) with Guaymas Basin hydrothermal vent sediment, AOM was observed up to 75°C, with the highest AOM rate at 50°C (165).
Modified In Vitro ANME Enrichment Approaches
The growth of ANME-2 was documented (44) in batch incubations using a glass tube connected via a needle to a syringe and placed inside a pressure-proof steel cylinder (187). The syringe, which was filled with medium, transmits the pressure of the cylinder to the medium inside the tube. Using this design, methane hydrate sediment was incubated at 1.4 MPa for 2 years with intermittent replenishment of the supernatant by fresh medium and CH4 (21 mM at 12°C). During the incubation period, the volume of the ANME-2 and SRB consortia, which was tracked using FISH, increased exponentially (44).
A batch incubation, with intermittent replacement of supernatant by fresh medium (i.e., fed-batch system) once a month, was used to successfully achieve enrichment of ANME-2d with a relative abundance of about 78% (61). The inoculum was a mixture of sediment from a local freshwater lake, anaerobic digester sludge, and activated sludge from a wastewater treatment plant in Brisbane, Australia (Table 3) (174). The retention of biomass in the fed-batch system was achieved via a 20-min settling period, prior to the replacement of the supernatant by fresh medium. The cultivation of this freshwater ANME-2d can have the advantage of higher solubility of methane in freshwater than in seawater (192); however, this microorganism was enriched at 35°C, and methane solubility decreases at increased temperatures (174). As previously specified (see “AOM Coupled to Nitrite and Nitrate Reduction”), this ANME-2d microorganism, named “Candidatus Methanoperedens nitroreducens,” utilizes nitrate instead of sulfate as an electron acceptor for AOM. This physiological trait likely contributed to the successful enrichment of this novel ANME clade at high abundance in a relatively short time period (about 2 years), because AOM coupled to nitrate yields about 45-fold more energy than its sulfate-dependent counterpart (Fig. 3).
Continuous Bioreactor-Based Enrichment of ANME Mediating AOM-SR
The design rationale of continuous-flow incubation columns is to provide nutrients and to remove end products at environmentally relevant rates (Table 3) (193). In such systems, 0.2-μm-filtered seawater, reduced with hydrogen sulfide (510 μM) and saturated with methane (1.5 mM) in a conditioning column (4 h at 0.5 MPa), was used to feed cold seep and nonseep sediment cores maintained in polyvinylchloride (PVC) tubes at 0.2 MPa and 5°C (193). The methane oxidation rates before and after incubations of the seep sediments were the same, probably because the incubation time was only 2 weeks. However, some increases in AOM rate and ANME-2c population size were detected in the nonseep sediment incubations. In a second experimental run, the same continuous-flow reactor was used, but the incubations were conducted at 1 MPa. The incubation time was 7.5 months (30 weeks), and a preferential proliferation of ANME-1 over ANME-2 was observed in the nonseep sediments at the highest pore water velocity tested (90 m year−1) (220). In addition, an increase in the AOM activity was reported as measured using batch incubations in serum bottles inoculated with the sediment (seep and nonseep sediments used in the continuous-enrichment experiments) without headspace, using 0.2-μm-filter-sterilized anoxic seawater containing 2.0 mM methane and 1 mM hydrogen sulfide (220).
In efforts to attain methane concentrations close to in situ values, continuous reactors that can handle hydrostatic pressures up to 45 MPa with methane-enriched medium and without free gas in the incubation chamber have been used (194). This reactor configuration is flexible to operate in batch, fed-batch, or continuous mode. Incubation of sediments from the Black Sea showed a 6-fold increase in the volumetric AOM rate when the methane partial pressure was increased from 0.2 to 6 MPa. In all operation modes, AOM rates were estimated based on sulfide production. However, when under otherwise similar operation conditions the CH4-saturated medium was replaced by CH4-free medium, sulfide levels decreased rapidly and stabilized at input levels. This indicated that the sulfide production was indeed coupled to methane oxidation. During continuous operation of such high-pressure reactors, a CH4 concentration of 60 to 65 mM can be readily attained. Noticeably, during continuous operation, the influent sulfate concentration used was 8 mM, which is lower than concentrations in seawater (194). The hydraulic retention time was set at 14 h, which corresponded to a dilution rate of 1.7 day−1. Assuming a reactor containing homogeneously mixed sediment, this means that microorganisms growing at rates of <1.7 day−1 would be washed out from the reactor, which is the case with ANME with much lower growth rates, i.e., 0.006 to 0.03 day−1 (23, 112, 220). Additionally, these tests of continuous operation with CH4 addition lasted only 16 days, and whether and how biomass was retained in the system was not reported (194).
Similar high-pressure systems have been operated at up to 60 MPa hydrostatic pressure and 120°C (195). The flexibility of this system allows the subsampling of medium without loss of pressure, and it can be operated in batch or continuous mode (195). The system was tested by incubating sediments from the Isis Mud Volcano from the Egyptian continental margin (∼991 m below sea level), using artificial seawater preconditioned with 4 MPa of CH4, resulting in dissolved concentrations of ∼96 mM CH4. Following methane saturation, the hydrostatic pressure was increased to 10 MPa using artificial seawater and incubations were conducted for 9 days at 23°C. No measurements of biomass concentration and yield were conducted, but an increase in the sulfide concentration was detected upon addition of methane to the reactor (195).
A continuous high-pressure reactor capable of withstanding up to 8 MPa was used in fed-batch and continuous modes at pressures from 1 to 8 MPa and with a hydraulic retention time of 100 h during a 286-day incubation of sediments from a mud volcano located in the Gulf of Cadiz (196). Under such conditions, the ANME-2 biovolume (count of cells and aggregates) increased about 12-fold (197). There was no description of the biomass retention time and AOM rate in the system.
ANME can also be enriched at moderate pressures or even ambient pressure, provided that biomass retention is applied. Biomass retention can be achieved by introducing a submerged membrane (pore size of 0.2 μm, and effective surface of 0.03 m2) within a reactor with a 2-liter volume (23). Methane was sparged continuously at 190 mmol liter−1 day−1, thus providing mixing, stripping off of the sulfide, and restricting fouling of the membrane. This bioreactor was operated at 15°C and at a slight overpressure (0.0025 MPa) to avoid O2 intrusion. The sulfate loading rate was 3 mmol liter−1 day−1, and the hydraulic retention time was 7 days. Sediment retrieved from the Eckernförde Bay in the Baltic Sea was used as the inoculum, and the reactor was operated for about 3 years. Growth of ANME was inferred by the increase in sulfide production in the membrane reactor, and the increase in AOM rates was monitored by carrying out batch experiments with reactor biomass amended with 13C-labeled CH4 at regular time intervals (23). The ANME in the reactor could be affiliated with ANME-2a, and the doubling time of these microorganisms was estimated at 3.8 months (i.e., growth rate, 0.006 day−1).
Although high-pressure reactors operate at high dissolved CH4 concentrations, their maintenance and operation are cumbersome and various safety criteria must be met for their implementation. When successful enrichment was reported at moderate pressures in fed-batch reactors, a key feature was a good biomass retention via settling (ANME-2d) (61) or membranes (ANME-2c) (23).
Future Development in Ex Situ Enrichment Approaches
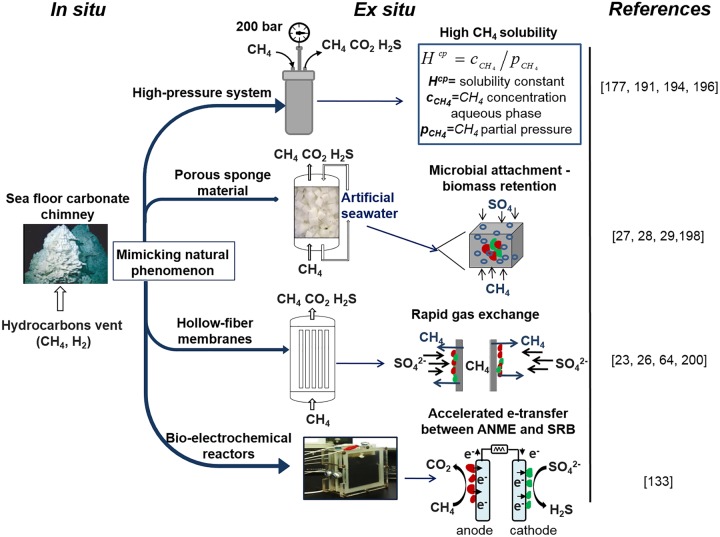
Mimicking the natural conditions in bioreactors can be a fruitful strategy for enrichment of ANME mediating AOM-SR. Reproducing in situ conditions in the laboratory is quite challenging, but artificial material and equipment can be used to mimic the natural environment (Fig. 8). Mimicking natural conditions is possible by using suitable reactors capable of achieving extreme environmental conditions such as high pressure or temperature and with suitable or similar natural packing material.
FIG 8.
Different bioreactor configurations to enhance ex situ growth of ANME mediating AOM-SR mimicking different characteristics of the growth modes of ANME in natural habitats.
The carbonate minerals, where ANME mediating AOM-SR have been found to form microbial reefs, are very porous. This porous natural matrix can harbor aggregates of AOM-performing consortia (148). Similarly, polyurethane sponges are a porous material and can be used as packing material in a packed-bed bioreactor configuration to promote the adhesion, aggregation, and retention of biomass. The collected marine sediment can be entrapped in the porous sponges so that methane can effectively diffuse through them, while the medium containing necessary nutrients and electron acceptor flows through the material (198). In a recent study, fresh bituminous coal and sandstone collected from a coal mine were used in a flowthrough type reactor system at high pressure to simulate and study geological CO2 sequestration and transformation (199). Similarly, naturally occurring materials which assist in biomass retention in the ANME enrichment can be used as packing materials in bioreactors. Recently, biotrickling filters with polyurethane foam as the packing material have been used to enrich ANME-1 with a relative abundance of 40% (27), ANME-2 with a relative abundance of 37%, and SRB consortia with a relative abundance of 55% (28, 29) at ambient pressure and temperature using, respectively, sulfate, thiosulfate, and elemental sulfur as the electron acceptor.
Considering the importance of substrate availability, especially for ANME, which oxidize a poorly soluble compound like methane, membrane reactors can be used to facilitate the contact between substrate and biomass (26). Bhattarai et al. (26) showed that ANME-2 from a cold seep environment could be enriched at ambient temperature and pressure in an external ultrafiltration membrane bioreactor. Moreover, a hollow-fiber membrane reactor was successfully applied for methane-dependent denitrification (64). Methane passes internally through the hollow-fiber membranes and diffuses to the outside layer, where a biofilm of ANME can be retained and grown (Fig. 8). A silicone membrane can also be used as a hollow-fiber membrane, which allows the bubbleless addition of gas to the bioreactor compartment. These gas-diffusive membranes are also applicable for AOM coupled to sulfate reduction, where the diffused methane can be immediately taken up by the ANME consortia that are suspended in the sulfate-containing medium. This mode of methane supply produces minimal bubbles, and the gas supply can be controlled by maintaining the gas pressure inside the membrane. As the microbial metabolism of AOM is slow, the slow diffusion of methane can reduce the large amounts of unused methane released from a bioreactor system, thus reducing the operational costs. Another benefit of the membrane is biomass retention, as the biomass usually develops as a biofilm or flocs (200). Moreover, the sulfide and pH can be continuously monitored by using pH and pS (sulfide sensor) electrodes and the sulfide can be removed before reaching the toxic threshold. Process control algorithms have been developed for the sulfate reduction process (201), which are also applicable for AOM bioreactors.
Several studies have hypothesized an electron transfer between ANME and SRB (see “Electron Transfer between ANME and SRB,” above). Based on this assumption, bioelectrochemical systems (BES) could also be used to study the electron transfer mechanisms (133). The methane oxidation process by the ANME takes place at the anode, and sulfate reduction takes place at the cathode (Fig. 8). Using BES, compounds which can act as e-shuttles (e.g., electron mediators or conductive nanominerals such as iron oxides) between the electrodes and the ANME can be added to facilitate electron transfer from the ANME to the electrode and study of the mechanism of this transfer (202). The electron exchange between the electrodes and the microbes can be determined by measuring the electrode potentials (203). Another advantage of AOM studies using BES is that they might allow isolation of the ANME, as the conductive membrane or electrode can replace the bacterial partner to act as an electron sink, assuming that the bacterial partner is required. In addition, the electrode can be poised at a desired potential to serve as an electron acceptor, and then ANME growth can possibly be maximized by fine-tuning the electrode potential. Thus, the electrodes in BES facilitate experiments with electron transfer of CH4 to the conducting surface and also serve as an electron acceptor by which ANME growth is possibly accelerated.
CONCLUSIONS
The marine habitats hosting ANME mediating AMO-SR have been widely explored in the past, and much was learned about ANME in the last 4 decades. However, details on niche differentiation among the various ANME clades need to be further assessed. Although a few investigations on AOM in freshwater environments have been conducted, unambiguous links between the presence and activity of AOM in these environments are still required. The most challenging drawback is not being able to readily obtain enrichments of sulfate-dependent ANME. With the exception of a few studies in which ANME-2a enrichments were obtained after 8 (55) and 3 (23) years in bioreactors, most biochemical studies have been conducted using naturally ANME-enriched sediments, of which the retrieved small quantities often limit experimental testing. In such a context, proper handling of the ANME biomass from the seafloor to the laboratory as well as enrichment in bioreactor configurations mimicking in situ conditions is a priority.
ACKNOWLEDGMENTS
We thank our past and present coworkers of UNESCO-IHE and Wageningen University, as well as our collaborators and granting agencies, in particular the Erasmus Mundus Joint Doctorate Program ETeCoS3 (Environmental Technologies for Contaminated Solids, Soils and Sediments, grant agreement FPA no. 2010-0009), the Marie Curie Intra European Fellowship (SUREANMetOX-300078), and Science Foundation Ireland Research Professorship programme no. 15/RP/2763.
We thank Graciela Gonzalez-Gil for providing suggestions and help in the preparation of the manuscript, as well as Jiefu Li and Yu Zhang from the Shanghai Jiao Tong University (Shanghai, China) for providing the photograph of the bioelectrochemical system.
Biographies

S. Bhattarai is a postdoc researcher at the University of Minnesota, Minneapolis (USA). She obtained her Ph.D. in Environmental Technology in December 2016 from UNESCO-IHE (the Netherlands), University of Paris-Est (France), and University of Cassino (Italy) as part of the Environmental Technology for Soil, Solid and Sediment (ETeCoS3) Erasmus Joint Doctorate project. She obtained her M.Sc. degree in Environmental Science and Technology from UNESCO-IHE (Delft, the Netherlands). During her Ph.D., she conducted research on “Performance assessment and enrichment of anaerobic methane-oxidizing microbial communities from marine sediments in bioreactors,” focusing on the enrichment of the anaerobic methane oxidizers (ANME) in different bioreactor configurations. She followed the dynamics of enrichments using various chemical and molecular biology approaches. Moreover, she has conducted the genomic characterization of various enriched sediments at the State Key Laboratory of Microbial Metabolism, Shanghai Jiao Tong University (Shanghai, China). Prior to her Ph.D. studies, she worked at the molecular biology lab of EAWAG (Switzerland), where she studied the microbial community in sediment and water of Lake Kivu (East Africa), which is an ecosystem containing large amounts of methane. By analyzing the microbial community and chemical profile from the lake sediment, she found some putative ANME of novel archaeal phylogeny.

C. Cassarini is a postdoc researcher at the National University of Ireland (Galway, Ireland). She obtained her Ph.D. in Environmental Technology in June 2017 from UNESCO-IHE (the Netherlands), University of Paris-Est (France). and University of Cassino (Italy) as part of the Environmental Technology for Soil, Solid and Sediment (ETeCoS3) Erasmus Joint Doctorate project. She obtained her M.Sc. degree in Geochemistry at Utrecht University (the Netherlands). She collaborated closely with Deltares (the Netherlands) and with the Department of Isotope Biogeochemistry of the UFZ (Leipzig, Germany), where she used compound-specific stable isotope analysis (CSIA) to investigate the degradation of chlorinated compounds in an industrial contaminated site. During her M.Sc. studies, she participated in a research cruise on the North Sea to investigate ocean acidification and determine factors controlling the pH dynamics in relation to pelagic processes. She is investigating how various sulfur compounds as potential electron acceptors for AOM impact the anaerobic oxidation of methane in packed-bed bioreactor systems. She uses various approaches, including CSIA, FISH, and FISH-NanoSIMS.

P. N. L. Lens is an established professor of New Energy Technologies at National University Ireland Galway, a professor of Environmental Biotechnology at UNESCO-IHE (Delft, the Netherlands), and an adjunct professor at Tampere University of Technology (Finland) and was previously on the faculty of the Sub-Department of Environmental Technology at Wageningen University, where he still has a zero nomination. Prof. Lens trained in Environmental Sanitation and then obtained his Ph.D. in Environmental Engineering at University Ghent (Belgium). He is founding editor-in-chief of the review journal Reviews in Environmental Science and Bio/Technology and founding editor of the IWA Publishing book series Integrated Environmental Technology. He has (co-)authored over 450 scientific publications and edited 11 book volumes. His awards include the IWA Publishing Award (2002), a Marie Curie Excellence Grant (2004), and nominations as an inaugural IWA fellow (2010) and an IWA distinguished fellow (2015). His research focuses on biofilms, sulfur biotechnology, metal speciation, bioavailability and removal, natural treatment systems, and anaerobic wastewater and waste gas treatment for resource recovery and reuse.
REFERENCES
- 1.Thauer RK, Shima S. 2008. Methane as fuel for anaerobic microorganisms. Ann N Y Acad Sci 1125:158–170. doi: 10.1196/annals.1419.000. [DOI] [PubMed] [Google Scholar]
- 2.Ciais P, Sabine C, Bala G, Bopp L, Brovkin V, Canadell J, Chhabra A, DeFries R, Galloway J, Heimann M. 2014. Carbon and other biogeochemical cycles, p 465–570. In IPCC 2013: climate change 2013. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge University Press, Cambridge, England. [Google Scholar]
- 3.Degelmann DM, Borken W, Drake HL, Kolb S. 2010. Different atmospheric methane-oxidizing communities in European beech and Norway spruce soils. Appl Environ Microbiol 76:3228–3235. doi: 10.1128/AEM.02730-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maxfield P, Hornibrook E, Evershed R. 2006. Estimating high-affinity methanotrophic bacterial biomass, growth, and turnover in soil by phospholipid fatty acid 13C labeling. Appl Environ Microbiol 72:3901–3907. doi: 10.1128/AEM.02779-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kirschke S, Bousquet P, Ciais P, Saunois M, Canadell JG, Dlugokencky EJ, Bergamaschi P, Bergmann D, Blake DR, Bruhwiler L, Cameron-Smith P, Castaldi S, Chevallier F, Feng L, Fraser A, Heimann M, Hodson EL, Houweling S, Josse B, Fraser PJ, Krummel PB, Lamarque J-F, Langenfelds RL, Le Quere C, Naik V, O'Doherty S, Palmer PI, Pison I, Plummer D, Poulter B, Prinn RG, Rigby M, Ringeval B, Santini M, Schmidt M, Shindell DT, Simpson IJ, Spahni R, Steele LP, Strode SA, Sudo K, Szopa S, van der Werf GR, Voulgarakis A, van Weele M, Weiss RF, Williams JE, Zeng G. 2013. Three decades of global methane sources and sinks. Nat Geosci 6:813–823. doi: 10.1038/ngeo1955. [DOI] [Google Scholar]
- 6.Pachauri RK, Allen MR, Barros VR, Broome J, Cramer W, Christ R, Church JA, Clarke L, Dahe Q, Dasgupta P. 2014. Synthesis report In IPCC 2014: climate change 2014. Contribution of Working Groups I, II and III to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge University Press, Cambridge, England. [Google Scholar]
- 7.Nazaries L, Murrell JC, Millard P, Baggs L, Singh BK. 2013. Methane, microbes and models: fundamental understanding of the soil methane cycle for future predictions. Environ Microbiol 15:2395–2417. doi: 10.1111/1462-2920.12149. [DOI] [PubMed] [Google Scholar]
- 8.Reeburgh WS. 2007. Oceanic methane biogeochemistry. Chem Rev 107:486–513. doi: 10.1021/cr050362v. [DOI] [PubMed] [Google Scholar]
- 9.Hanson RS, Hanson TE. 1996. Methanotrophic bacteria. Microbiol Rev 60:439–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chistoserdova L, Vorholt JA, Lidstrom ME. 2005. A genomic view of methane oxidation by aerobic bacteria and anaerobic archaea. Genome Biol 6:208. doi: 10.1186/gb-2005-6-2-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Archer D, Buffett B, Brovkin V. 2009. Ocean methane hydrates as a slow tipping point in the global carbon cycle. Proc Natl Acad Sci U S A 106:20596–20601. doi: 10.1073/pnas.0800885105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wallmann K, Pinero E, Burwicz E, Haeckel M, Hensen C, Dale A, Ruepke L. 2012. The global inventory of methane hydrate in marine sediments: a theoretical approach. Energies 5:2449–2498. doi: 10.3390/en5072449. [DOI] [Google Scholar]
- 13.Pinero E, Marquardt M, Hensen C, Haeckel M, Wallmann K. 2013. Estimation of the global inventory of methane hydrates in marine sediments using transfer functions. Biogeosciences 10:959–975. doi: 10.5194/bg-10-959-2013. [DOI] [Google Scholar]
- 14.Buffett B, Archer D. 2004. Global inventory of methane clathrate: sensitivity to changes in the deep ocean. Earth Planet Sci Lett 227:185–199. doi: 10.1016/j.epsl.2004.09.005. [DOI] [Google Scholar]
- 15.Boetius A, Wenzhöfer F. 2013. Seafloor oxygen consumption fuelled by methane from cold seeps. Nat Geosci 6:725–734. doi: 10.1038/ngeo1926. [DOI] [Google Scholar]
- 16.Treude T, Orphan VJ, Knittel K, Gieseke A, House CH, Boetius A. 2007. Consumption of methane and CO2 by methanotrophic microbial mats from gas seeps of the anoxic Black Sea. Appl Environ Microbiol 73:2271–2283. doi: 10.1128/AEM.02685-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niemann H, Duarte J, Hensen C, Omoregie E, Magalhaes VH, Elvert M, Pinheiro LM, Kopf A, Boetius A. 2006. Microbial methane turnover at mud volcanoes of the Gulf of Cadiz. Geochim Cosmochim Acta 70:5336–5355. doi: 10.1016/j.gca.2006.08.010. [DOI] [Google Scholar]
- 18.Joye SB, Boetius A, Orcutt BN, Montoya JP, Schulz HN, Erickson MJ, Lugo SK. 2004. The anaerobic oxidation of methane and sulfate reduction in sediments from Gulf of Mexico cold seeps. Chem Geol 205:219–238. doi: 10.1016/j.chemgeo.2003.12.019. [DOI] [Google Scholar]
- 19.Suess E. 2014. Marine cold seeps and their manifestations: geological control, biogeochemical criteria and environmental conditions. Int J Earth Sci 103:1889–1916. doi: 10.1007/s00531-014-1010-0. [DOI] [Google Scholar]
- 20.Tavormina PL, Ussler W, Joye SB, Harrison BK, Orphan VJ. 2010. Distributions of putative aerobic methanotrophs in diverse pelagic marine environments. ISME J 4:700–710. doi: 10.1038/ismej.2009.155. [DOI] [PubMed] [Google Scholar]
- 21.Wankel SD, Joye SB, Samarkin VA, Shah SR, Friederich G, Melas-Kyriazi J, Girguis PR. 2010. New constraints on methane fluxes and rates of anaerobic methane oxidation in a Gulf of Mexico brine pool via in situ mass spectrometry. Deep Sea Res Part 2 Top Stud Oceanogr 57:2022–2029. doi: 10.1016/j.dsr2.2010.05.009. [DOI] [Google Scholar]
- 22.Knittel K, Boetius A. 2009. Anaerobic oxidation of methane: progress with an unknown process. Annu Rev Microbiol 63:311–334. doi: 10.1146/annurev.micro.61.080706.093130. [DOI] [PubMed] [Google Scholar]
- 23.Meulepas RJW, Jagersma CG, Gieteling J, Buisman CJN, Stams AJM, Lens P. 2009. Enrichment of anaerobic methanotrophs in sulfate-reducing membrane bioreactors. Biotechnol Bioeng 104:458–470. doi: 10.1002/bit.22412. [DOI] [PubMed] [Google Scholar]
- 24.Meulepas RJW, Stams AJM, Lens P. 2010. Biotechnological aspects of sulfate reduction with methane as electron donor. Rev Environ Sci Biotechnol 9:59–78. doi: 10.1007/s11157-010-9193-8. [DOI] [Google Scholar]
- 25.Gonzalez-Gil G, Meulepas RJW, Lens PNL. 2011. Biotechnological aspects of the use of methane as electron donor for sulfate reduction, p 419–434. In Murray M-Y. (ed), Comprehensive biotechnology, 2nd ed, vol 6 Elsevier B.V, Amsterdam, The Netherlands. [Google Scholar]
- 26.Bhattarai S, Cassarini C, Rene ER, Kümmel S, Esposito G, Lens PN. 2018. Enrichment of ANME‐2 dominated anaerobic methanotrophy from cold seep sediment in an external ultrafiltration membrane bioreactor. Eng Life Sci 18:368–378. doi: 10.1002/elsc.201700148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhattarai S, Cassarini C, Rene ER, Zhang Y, Esposito G, Lens PN. 2018. Enrichment of sulfate reducing anaerobic methane oxidizing community dominated by ANME-1 from Ginsburg Mud Volcano (Gulf of Cadiz) sediment in a biotrickling filter. Bioresour Technol 259:433–441. doi: 10.1016/j.biortech.2018.03.018. [DOI] [PubMed] [Google Scholar]
- 28.Cassarini C, Rene ER, Bhattarai S, Esposito G, Lens PN. 2017. Anaerobic oxidation of methane coupled to thiosulfate reduction in a biotrickling filter. Bioresour Technol 240:214–222. doi: 10.1016/j.biortech.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 29.Cassarini C, Bhattarai S, Rene ER, Vogt C, Musat N, Esposito G, Lens PNL. April 2019. Enrichment of anaerobic methanotrophs in biotrickling filters using different sulfur compounds as electron acceptor. Environ Eng Sci doi: 10.1089/ees.2018.0283. [DOI] [Google Scholar]
- 30.Lens PNL, Vallero M, Esposito G, Zandvoort MH. 2002. Perspectives of sulfate reducing bioreactors in environmental biotechnology. Rev Environ Sci Biotechnol 1:311–325. doi: 10.1023/A:1023207921156. [DOI] [Google Scholar]
- 31.Widdel F, Hansen T. 1992. The dissimilatory sulfate-and sulfur-reducing bacteria, p 582–624. In Balows A, Truper H, Dworkin M, Harder W, Schleifer KH (ed), The prokaryotes: a handbook on the biology of bacteria: ecophysiology, isolation, identification, applications, 2nd ed, vol I Springer-Verlag, Heidelberg, Germany. [Google Scholar]
- 32.Cassarini C. 2017. Anaerobic oxidation of methane coupled to the reduction of different sulfur compounds as electron acceptors in bioreactors. Ph.D. thesis. CRC Press/Balkema, Leiden, Netherlands. [Google Scholar]
- 33.Reeburgh WS. 1976. Methane consumption in Cariaco Trench waters and sediments. Earth Planet Sci Lett 28:337–344. doi: 10.1016/0012-821X(76)90195-3. [DOI] [Google Scholar]
- 34.Martens CS, Berner RA. 1974. Methane production in the interstitial waters of sulfate-depleted marine sediments. Science 185:1167–1169. doi: 10.1126/science.185.4157.1167. [DOI] [PubMed] [Google Scholar]
- 35.Zehnder AJ, Brock TD. 1980. Anaerobic methane oxidation: occurrence and ecology. Appl Environ Microbiol 39:194–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iversen N, Jørgensen BB. 1985. Anaerobic methane oxidation rates at the sulfate‐methane transition in marine sediments from Kattegat and Skagerrak (Denmark). Limnol Oceanogr 30:944–955. doi: 10.4319/lo.1985.30.5.0944. [DOI] [Google Scholar]
- 37.Reeburgh WS. 1980. Anaerobic methane oxidation: rate depth distributions in Skan Bay sediments. Earth Planet Sci Lett 47:345–352. doi: 10.1016/0012-821X(80)90021-7. [DOI] [Google Scholar]
- 38.Hinrichs K-U, Hayes JM, Sylva SP, Brewer PG, DeLong EF. 1999. Methane-consuming archaebacteria in marine sediments. Nature 398:802–805. doi: 10.1038/19751. [DOI] [PubMed] [Google Scholar]
- 39.Boetius A, Ravenschlag K, Schubert CJ, Rickert D, Widdel F, Gieseke A, Amann R, Jørgensen BB, Witte U, Pfannkuche O. 2000. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407:623–626. doi: 10.1038/35036572. [DOI] [PubMed] [Google Scholar]
- 40.Hoehler TM, Alperin MJ, Albert DB, Martens S. 1994. Field and laboratory studies of methane oxidation in an anoxic marine sediment: evidence for a methanogen-sulfate reducer consortium. Global Biogeochem Cycles 8:451–463. doi: 10.1029/94GB01800. [DOI] [Google Scholar]
- 41.Knittel K, Lösekann T, Boetius A, Kort R, Amann R. 2005. Diversity and distribution of methanotrophic archaea at cold seeps. Appl Environ Microbiol 71:467–479. doi: 10.1128/AEM.71.1.467-479.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Orphan VJ, House CH, Hinrichs K-U, McKeegan KD, DeLong EF. 2002. Multiple archaeal groups mediate methane oxidation in anoxic cold seep sediments. Proc Natl Acad Sci U S A 99:7663–7668. doi: 10.1073/pnas.072210299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schreiber L, Holler T, Knittel K, Meyerdierks A, Amann R. 2010. Identification of the dominant sulfate-reducing bacterial partner of anaerobic methanotrophs of the ANME-2 clade. Environ Microbiol 12:2327–2340. doi: 10.1111/j.1462-2920.2010.02275.x. [DOI] [PubMed] [Google Scholar]
- 44.Nauhaus K, Albrecht M, Elvert M, Boetius A, Widdel F. 2007. In vitro cell growth of marine archaeal-bacterial consortia during anaerobic oxidation of methane with sulfate. Environ Microbiol 9:187–196. doi: 10.1111/j.1462-2920.2006.01127.x. [DOI] [PubMed] [Google Scholar]
- 45.Sievert S, Kiene R, Schulz-Vogt H. 2007. The sulfur cycle. Oceanography 20:117–123. doi: 10.5670/oceanog.2007.55. [DOI] [Google Scholar]
- 46.Jørgensen BB, Kasten S. 2006. Sulfur cycling and methane oxidation, p 271–309. In Schulz H, Zabel M (ed), Marine geochemistry, 2nd ed Springer-Verlag, Heidelberg, Germany. [Google Scholar]
- 47.Muyzer G, Stams AJ. 2008. The ecology and biotechnology of sulphate-reducing bacteria. Nat Rev Microbiol 6:441–454. doi: 10.1038/nrmicro1892. [DOI] [PubMed] [Google Scholar]
- 48.Wang Y, Wegener G, Hou J, Wang F, Xiao X. 2019. Expanding anaerobic alkane metabolism in the domain of Archaea. Nat Microbiol 4:595–602. doi: 10.1038/s41564-019-0364-2. [DOI] [PubMed] [Google Scholar]
- 49.McKay L, Dlakic M, Fields M, Jay Z, Eren M, Delmont T, Klingelsmith K, Rusch D, Inskeep W. 2019. Co-occurring genomic capacity for anaerobic methane and dissimilatory sulfur metabolisms discovered in the Korarchaeota. Nat Microbiol 4:614–622. doi: 10.1038/s41564-019-0362-4. [DOI] [PubMed] [Google Scholar]
- 50.D'Hondt S, Rutherford S, Spivack AJ. 2002. Metabolic activity of subsurface life in deep-sea sediments. Science 295:2067–2070. doi: 10.1126/science.1064878. [DOI] [PubMed] [Google Scholar]
- 51.Wankel SD, Adams MM, Johnston DT, Hansel CM, Joye SB, Girguis PR. 2012. Anaerobic methane oxidation in metalliferous hydrothermal sediments: influence on carbon flux and decoupling from sulfate reduction. Environ Microbiol 14:2726–2740. doi: 10.1111/j.1462-2920.2012.02825.x. [DOI] [PubMed] [Google Scholar]
- 52.Maignien L, Parkes RJ, Cragg B, Niemann H, Knittel K, Coulon S, Akhmetzhanov A, Boon N. 2013. Anaerobic oxidation of methane in hypersaline cold seep sediments. FEMS Microbiol Ecol 83:214–231. doi: 10.1111/j.1574-6941.2012.01466.x. [DOI] [PubMed] [Google Scholar]
- 53.Treude T, Knittel K, Blumenberg M, Seifert R, Boetius A. 2005. Subsurface microbial methanotrophic mats in the Black Sea. Appl Environ Microbiol 71:6375–6378. doi: 10.1128/AEM.71.10.6375-6378.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Losekann T, Knittel K, Nadalig T, Fuchs B, Niemann H, Boetius A, Amann R. 2007. Diversity and abundance of aerobic and anaerobic methane oxidizers at the Haakon Mosby mud volcano, Barents Sea. Appl Environ Microbiol 73:3348–3362. doi: 10.1128/AEM.00016-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Milucka J, Ferdelman TG, Polerecky L, Franzke D, Wegener G, Schmid M, Lieberwirth I, Wagner M, Widdel F, Kuypers MM. 2012. Zero-valent sulphur is a key intermediate in marine methane oxidation. Nature 491:541–546. doi: 10.1038/nature11656. [DOI] [PubMed] [Google Scholar]
- 56.Suarez-Zuluaga DA, Timmers PHA, Plugge CM, Stams AJM, Buisman CJN, Weijma J. 2016. Thiosulphate conversion in a methane and acetate fed membrane bioreactor. Environ Sci Pollut Res 23:2467–2478. doi: 10.1007/s11356-015-5344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suarez-Zuluaga DA, Weijma J, Timmers PH, Buisman CJ. 2015. High rates of anaerobic oxidation of methane, ethane and propane coupled to thiosulphate reduction. Environ Sci Pollut Res Int 22:3697–3704. doi: 10.1007/s11356-014-3606-0. [DOI] [PubMed] [Google Scholar]
- 58.Finster K. 2008. Microbiological disproportionation of inorganic sulfur compounds. J Sulfur Chem 29:281–292. doi: 10.1080/17415990802105770. [DOI] [Google Scholar]
- 59.Poser A, Lohmayer R, Vogt C, Knoeller K, Planer-Friedrich B, Sorokin D, Richnow H-H, Finster K. 2013. Disproportionation of elemental sulfur by haloalkaliphilic bacteria from soda lakes. Extremophiles 17:1003–1012. doi: 10.1007/s00792-013-0582-0. [DOI] [PubMed] [Google Scholar]
- 60.Ettwig KF, Butler MK, Le Paslier D, Pelletier E, Mangenot S, Kuypers MMM, Schreiber F, Dutilh BE, Zedelius J, de Beer D, Gloerich J, Wessels HJCT, van Alen T, Luesken F, Wu ML, van de Pas-Schoonen KT, Op den Camp HJM, Janssen-Megens EM, Francoijs K-J, Stunnenberg H, Weissenbach J, Jetten MSM, Strous M. 2010. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464:543–548. doi: 10.1038/nature08883. [DOI] [PubMed] [Google Scholar]
- 61.Haroon MF, Hu S, Shi Y, Imelfort M, Keller J, Hugenholtz P, Yuan Z, Tyson GW. 2013. Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature 500:567–570. doi: 10.1038/nature12375. [DOI] [PubMed] [Google Scholar]
- 62.Raghoebarsing AA, Pol A, van de Pas-Schoonen KT, Smolders AJP, Ettwig KF, Rijpstra WIC, Schouten S, Damsté JSS, Op den Camp HJM, Jetten MSM, Strous M. 2006. A microbial consortium couples anaerobic methane oxidation to denitrification. Nature 440:918–921. doi: 10.1038/nature04617. [DOI] [PubMed] [Google Scholar]
- 63.Kampman C, Hendrickx TLG, Luesken FA, van Alen TA, Op den Camp HJM, Jetten MSM, Zeeman G, Buisman CJN, Temmink H. 2012. Enrichment of denitrifying methanotrophic bacteria for application after direct low-temperature anaerobic sewage treatment. J Hazard Mater 227-228:164–171. doi: 10.1016/j.jhazmat.2012.05.032. [DOI] [PubMed] [Google Scholar]
- 64.Shi Y, Hu S, Lou J, Lu P, Keller J, Yuan Z. 2013. Nitrogen removal from wastewater by coupling anammox and methane-dependent denitrification in a membrane biofilm reactor. Environ Sci Technol 47:11577–11583. doi: 10.1021/es402775z. [DOI] [PubMed] [Google Scholar]
- 65.Wang D, Wang Y, Liu Y, Ngo HH, Lian Y, Zhao J, Chen F, Yang Q, Zeng G, Li X. 2017. Is denitrifying anaerobic methane oxidation-centered technologies a solution for the sustainable operation of wastewater treatment plants? Bioresour Technol 234:456–465. doi: 10.1016/j.biortech.2017.02.059. [DOI] [PubMed] [Google Scholar]
- 66.Wang Y, Wang D, Yang Q, Zeng G, Li X. 2017. Wastewater opportunities for denitrifying anaerobic methane oxidation. Trends Biotechnol 35:799–802. doi: 10.1016/j.tibtech.2017.02.010. [DOI] [PubMed] [Google Scholar]
- 67.Zhu J, Wang Q, Yuan M, Tan G-Y, Sun F, Wang C, Wu W, Lee P-H. 2016. Microbiology and potential applications of aerobic methane oxidation coupled to denitrification (AME-D) process: a review. Water Res 90:203–215. doi: 10.1016/j.watres.2015.12.020. [DOI] [PubMed] [Google Scholar]
- 68.Hu S, Zeng RJ, Haroon MF, Keller J, Lant PA, Tyson GW, Yuan Z. 2015. A laboratory investigation of interactions between denitrifying anaerobic methane oxidation (DAMO) and anammox processes in anoxic environments. Sci Rep 5:8706. doi: 10.1038/srep08706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Beal EJ, House CH, Orphan VJ. 2009. Manganese- and iron-dependent marine methane oxidation. Science 325:184–187. doi: 10.1126/science.1169984. [DOI] [PubMed] [Google Scholar]
- 70.Egger M, Rasigraf O, Sapart CJ, Jilbert T, Jetten MSM, Röckmann T, van der Veen C, Bândă N, Kartal B, Ettwig KF, Slomp CP. 2015. Iron-mediated anaerobic oxidation of methane in brackish coastal sediments. Environ Sci Technol 49:277–283. doi: 10.1021/es503663z. [DOI] [PubMed] [Google Scholar]
- 71.Bar-Or I, Elvert M, Eckert W, Kushmaro A, Vigderovich H, Zhu Q, Ben-Dov E, Sivan O. 2017. Iron-coupled anaerobic oxidation of methane performed by a mixed bacterial-archaeal community based on poorly reactive minerals. Environ Sci Technol 51:12293–12301. doi: 10.1021/acs.est.7b03126. [DOI] [PubMed] [Google Scholar]
- 72.Sivan O, Adler M, Pearson A, Gelman F, Bar-Or I, John SG, Eckert W. 2011. Geochemical evidence for iron‐mediated anaerobic oxidation of methane. Limnol Oceanogr 56:1536–1544. doi: 10.4319/lo.2011.56.4.1536. [DOI] [Google Scholar]
- 73.Segarra KEA, Comerford C, Slaughter J, Joye SB. 2013. Impact of electron acceptor availability on the anaerobic oxidation of methane in coastal freshwater and brackish wetland sediments. Geochim Cosmochim Acta 115:15–30. doi: 10.1016/j.gca.2013.03.029. [DOI] [Google Scholar]
- 74.Aller RC, Rude P. 1988. Complete oxidation of solid phase sulfides by manganese and bacteria in anoxic marine sediments. Geochim Cosmochim Acta 52:751–765. doi: 10.1016/0016-7037(88)90335-3. [DOI] [Google Scholar]
- 75.Canfield DE, Thamdrup B, Hansen JW. 1993. The anaerobic degradation of organic matter in Danish coastal sediments: iron reduction, manganese reduction, and sulfate reduction. Geochim Cosmochim Acta 57:3867–3883. doi: 10.1016/0016-7037(93)90340-3. [DOI] [PubMed] [Google Scholar]
- 76.Straub KL, Schink B. 2004. Ferrihydrite-dependent growth of Sulfurospirillum deleyianum through electron transfer via sulfur cycling. Appl Environ Microbiol 70:5744–5749. doi: 10.1128/AEM.70.10.5744-5749.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wan M, Shchukarev A, Lohmayer R, Planer-Friedrich B, Peiffer S. 2014. Occurrence of surface polysulfides during the interaction between ferric (hydr)oxides and aqueous sulfide. Environ Sci Technol 48:5076–5084. doi: 10.1021/es405612f. [DOI] [PubMed] [Google Scholar]
- 78.Ettwig KF, Zhu B, Speth D, Keltjens JT, Jetten MS, Kartal B. 2016. Archaea catalyze iron-dependent anaerobic oxidation of methane. Proc Natl Acad Sci U S A 113:12792–12796. doi: 10.1073/pnas.1609534113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Oni OE, Friedrich MW. 2017. Metal oxide reduction linked to anaerobic methane oxidation. Trends Microbiol 25:88–90. doi: 10.1016/j.tim.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 80.Fu L, Li S-W, Ding Z-W, Ding J, Lu Y-Z, Zeng RJ. 2016. Iron reduction in the DAMO/Shewanella oneidensis MR-1 coculture system and the fate of Fe(II). Water Res 88:808–815. doi: 10.1016/j.watres.2015.11.011. [DOI] [PubMed] [Google Scholar]
- 81.He Z, Zhang Q, Feng Y, Luo H, Pan X, Gadd GM. 2018. Microbiological and environmental significance of metal-dependent anaerobic oxidation of methane. Sci Total Environ 610-611:759–768. doi: 10.1016/j.scitotenv.2017.08.140. [DOI] [PubMed] [Google Scholar]
- 82.Cai C, Leu AO, Xie GJ, Guo J, Feng Y, Zhao JX, Tyson GW, Yuan Z, Hu S. 2018. A methanotrophic archaeon couples anaerobic oxidation of methane to Fe(III) reduction. ISME J 12:1929–1939. doi: 10.1038/s41396-018-0109-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Luo J-H, Chen H, Hu S, Cai C, Yuan Z, Guo J. 2018. Microbial selenate reduction driven by a denitrifying anaerobic methane oxidation biofilm. Environ Sci Technol 52:4006–4012. doi: 10.1021/acs.est.7b05046. [DOI] [PubMed] [Google Scholar]
- 84.Luo J-H, Wu M, Yuan Z, Guo J. 2017. Biological bromate reduction driven by methane in a membrane biofilm reactor. Environ Sci Technol Lett 4:562–566. doi: 10.1021/acs.estlett.7b00488. [DOI] [Google Scholar]
- 85.Lu Y-Z, Fu L, Ding J, Ding Z-W, Li N, Zeng RJ. 2016. Cr(VI) reduction coupled with anaerobic oxidation of methane in a laboratory reactor. Water Res 102:445–452. doi: 10.1016/j.watres.2016.06.065. [DOI] [PubMed] [Google Scholar]
- 86.Niemann H, Losekann T, de Beer D, Elvert M, Nadalig T, Knittel K, Amann R, Sauter EJ, Schluter M, Klages M, Foucher JP, Boetius A. 2006. Novel microbial communities of the Haakon Mosby mud volcano and their role as a methane sink. Nature 443:854–858. doi: 10.1038/nature05227. [DOI] [PubMed] [Google Scholar]
- 87.Krüger M, Meyerdierks A, Glockner FO, Amann R, Widdel F, Kube M, Reinhardt R, Kahnt R, Bocher R, Thauer RK, Shima S. 2003. A conspicuous nickel protein in microbial mats that oxidize methane anaerobically. Nature 426:878–881. doi: 10.1038/nature02207. [DOI] [PubMed] [Google Scholar]
- 88.Scheller S, Goenrich M, Boecher R, Thauer RK, Jaun B. 2010. The key nickel enzyme of methanogenesis catalyses the anaerobic oxidation of methane. Nature 465:606–608. doi: 10.1038/nature09015. [DOI] [PubMed] [Google Scholar]
- 89.Meyerdierks A, Kube M, Kostadinov I, Teeling H, Glöckner FO, Reinhardt R, Amann R. 2010. Metagenome and mRNA expression analyses of anaerobic methanotrophic archaea of the ANME-1 group. Environ Microbiol 12:422–439. doi: 10.1111/j.1462-2920.2009.02083.x. [DOI] [PubMed] [Google Scholar]
- 90.Wang F-P, Zhang Y, Chen Y, He Y, Qi J, Hinrichs K-U, Zhang X-X, Xiao X, Boon N. 2014. Methanotrophic archaea possessing diverging methane-oxidizing and electron-transporting pathways. ISME J 8:1069–1078. doi: 10.1038/ismej.2013.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hallam SJ, Putnam N, Preston CM, Detter JC, Rokhsar D, Richardson PM, DeLong EF. 2004. Reverse methanogenesis: testing the hypothesis with environmental genomics. Science 305:1457–1462. doi: 10.1126/science.1100025. [DOI] [PubMed] [Google Scholar]
- 92.Timmers PHA, Welte CU, Koehorst JJ, Plugge CM, Jetten MSM, Stams AJM. 2017. Reverse methanogenesis and respiration in methanotrophic archaea. Archaea 2017:1654237. doi: 10.1155/2017/1654237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shima S, Krueger M, Weinert T, Demmer U, Kahnt J, Thauer RK, Ermler U. 2011. Structure of a methyl-coenzyme M reductase from Black Sea mats that oxidize methane anaerobically. Nature 481:98–101. doi: 10.1038/nature10663. [DOI] [PubMed] [Google Scholar]
- 94.Mayr S, Latkoczy C, Krüger M, Günther D, Shima S, Thauer RK, Widdel F, Jaun B. 2008. Structure of an F430 variant from archaea associated with anaerobic oxidation of methane. J Am Chem Soc 130:10758–10767. doi: 10.1021/ja802929z. [DOI] [PubMed] [Google Scholar]
- 95.Rudolf KT. 2011. Anaerobic oxidation of methane with sulfate: on the reversibility of the reactions that are catalyzed by enzymes also involved in methanogenesis from CO2. Curr Opin Microbiol 14:292–299. [DOI] [PubMed] [Google Scholar]
- 96.Soo VWC, McAnulty MJ, Tripathi A, Zhu F, Zhang L, Hatzakis E, Smith PB, Agrawal S, Nazem-Bokaee H, Gopalakrishnan S, Salis HM, Ferry JG, Maranas CD, Patterson AD, Wood TK. 2016. Reversing methanogenesis to capture methane for liquid biofuel precursors. Microb Cell Fact 15:11. doi: 10.1186/s12934-015-0397-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Haynes CA, Gonzalez R. 2014. Rethinking biological activation of methane and conversion to liquid fuels. Nat Chem Biol 10:331–339. doi: 10.1038/nchembio.1509. [DOI] [PubMed] [Google Scholar]
- 98.Chen S-C, Musat N, Lechtenfeld OJ, Paschke H, Schmidt M, Said N, Popp D, Calabrese F, Stryhanyuk H, Jaekel U, Zhu Y-G, Joye SB, Richnow H-H, Widdel F, Musat F. 2019. Anaerobic oxidation of ethane by archaea from a marine hydrocarbon seep. Nature 568:108–111. doi: 10.1038/s41586-019-1063-0. [DOI] [PubMed] [Google Scholar]
- 99.Wegener G, Niemann H, Elvert M, Hinrichs K-U, Boetius A. 2008. Assimilation of methane and inorganic carbon by microbial communities mediating the anaerobic oxidation of methane. Environ Microbiol 10:2287–2298. doi: 10.1111/j.1462-2920.2008.01653.x. [DOI] [PubMed] [Google Scholar]
- 100.Kellermann MY, Wegener G, Elvert M, Yoshinaga MY, Lin Y-S, Holler T, Mollar XP, Knittel K, Hinrichs K-U. 2012. Autotrophy as a predominant mode of carbon fixation in anaerobic methane-oxidizing microbial communities. Proc Natl Acad Sci U S A 109:19321–19326. doi: 10.1073/pnas.1208795109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kurth JM, Smit NT, Berger S, Schouten S, Jetten MMS, Welte CU. 2019. ANME-2d anaerobic methanotrophic archaea differ from other ANME archaea in lipid composition and carbon source. bioRxiv 10.1101/558007. [DOI] [PMC free article] [PubMed]
- 102.Orcutt B, Boetius A, Elvert M, Samarkin V, Joye SB. 2005. Molecular biogeochemistry of sulfate reduction, methanogenesis and the anaerobic oxidation of methane at Gulf of Mexico cold seeps. Geochim Cosmochim Acta 69:4267–4281. doi: 10.1016/j.gca.2005.04.012. [DOI] [Google Scholar]
- 103.Bertram S, Blumenberg M, Michaelis W, Siegert M, Krüger M, Seifert R. 2013. Methanogenic capabilities of ANME-archaea deduced from 13C-labelling approaches. Environ Microbiol 15:2384–2393. doi: 10.1111/1462-2920.12112. [DOI] [PubMed] [Google Scholar]
- 104.Zehnder AJB, Brock TD. 1979. Methane formation and methane oxidation by methanogenic bacteria. J Bacteriol 137:420–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Harder J. 1997. Anaerobic methane oxidation by bacteria employing 14C-methane uncontaminated with 14C-carbon monoxide. Mar Geol 137:13–23. doi: 10.1016/S0025-3227(96)00075-8. [DOI] [Google Scholar]
- 106.Meulepas RJW, Jagersma CG, Zhang Y, Petrillo M, Cai H, Buisman CJN, Stams AJW, Lens P. 2010. Trace methane oxidation and the methane dependency of sulfate reduction in anaerobic granular sludge. FEMS Microbiol Ecol 72:261–271. doi: 10.1111/j.1574-6941.2010.00849.x. [DOI] [PubMed] [Google Scholar]
- 107.Lloyd KG, Alperin MJ, Teske A. 2011. Environmental evidence for net methane production and oxidation in putative ANaerobic MEthanotrophic (ANME) archaea. Environ Microbiol 13:2548–2564. doi: 10.1111/j.1462-2920.2011.02526.x. [DOI] [PubMed] [Google Scholar]
- 108.McGlynn SE. 2017. Energy metabolism during anaerobic methane oxidation in ANME Archaea. Microbes Environ 32:5–13. doi: 10.1264/jsme2.ME16166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Scheller S, Yu H, Chadwick GL, McGlynn SE, Orphan VJ. 2016. Artificial electron acceptors decouple archaeal methane oxidation from sulfate reduction. Science 351:703–707. doi: 10.1126/science.aad7154. [DOI] [PubMed] [Google Scholar]
- 110.Dekas AE, Poretsky RS, Orphan VJ. 2009. Deep-sea archaea fix and share nitrogen in methane-consuming microbial consortia. Science 326:422–426. doi: 10.1126/science.1178223. [DOI] [PubMed] [Google Scholar]
- 111.Dekas AE, Chadwick GL, Bowles MW, Joye SB, Orphan VJ. 2014. Spatial distribution of nitrogen fixation in methane seep sediment and the role of the ANME archaea. Environ Microbiol 16:3012–3029. doi: 10.1111/1462-2920.12247. [DOI] [PubMed] [Google Scholar]
- 112.Meulepas RJW, Jagersma CG, Khadem AF, Buisman CJN, Stams AJM, Lens P. 2009. Effect of environmental conditions on sulfate reduction with methane as electron donor by an Eckemförde Bay enrichment. Environ Sci Technol 43:6553–6559. doi: 10.1021/es900633c. [DOI] [PubMed] [Google Scholar]
- 113.Blumenberg M, Seifert R, Reitner J, Pape T, Michaelis W. 2004. Membrane lipid patterns typify distinct anaerobic methanotrophic consortia. Proc Natl Acad Sci U S A 101:11111–11116. doi: 10.1073/pnas.0401188101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hinrichs K-U, Boetius A. 2003. The anaerobic oxidation of methane: new insights in microbial ecology and biogeochemistry, p 457–477. In Wefer G, Billett D, Hebbeln D, Jørgensen B, Schlüter M, van Weering TE (ed), Ocean margin systems. Springer-Verlag, Heidelberg, Germany. [Google Scholar]
- 115.Hinrichs K-U, Summons RE, Orphan VJ, Sylva SP, Hayes JM. 2000. Molecular and isotopic analysis of anaerobic methane-oxidizing communities in marine sediments. Org Geochem 31:1685–1701. doi: 10.1016/S0146-6380(00)00106-6. [DOI] [Google Scholar]
- 116.Alperin MJ, Hoehler TM. 2009. Anaerobic methane oxidation by archaea/sulfate-reducing bacteria aggregates: 2. Isotopic constraints. Am J Sci 309:958–984. doi: 10.2475/10.2009.02. [DOI] [Google Scholar]
- 117.Stadnitskaia A, Muyzer G, Abbas B, Coolen MJL, Hopmans EC, Baas M, van Weering TCE, Ivanov MK, Poludetkina E, Sinninghe Damsté JS. 2005. Biomarker and 16S rDNA evidence for anaerobic oxidation of methane and related carbonate precipitation in deep-sea mud volcanoes of the Sorokin Trough, Black Sea. Mar Geol 217:67–96. doi: 10.1016/j.margeo.2005.02.023. [DOI] [Google Scholar]
- 118.Alain K, Holler T, Musat F, Elvert M, Treude T, Krüger M. 2006. Microbiological investigation of methane- and hydrocarbon-discharging mud volcanoes in the Carpathian Mountains, Romania. Environ Microbiol 8:574–590. doi: 10.1111/j.1462-2920.2005.00922.x. [DOI] [PubMed] [Google Scholar]
- 119.Valentine DL, Reeburgh WS, Hall R. 2000. New perspectives on anaerobic methane oxidation. Environ Microbiol 2:477–484. doi: 10.1046/j.1462-2920.2000.00135.x. [DOI] [PubMed] [Google Scholar]
- 120.Sørensen K, Finster K, Ramsing N. 2001. Thermodynamic and kinetic requirements in anaerobic methane oxidizing consortia exclude hydrogen, acetate, and methanol as possible electron shuttles. Microb Ecol 42:1–10. doi: 10.1007/s002480000083. [DOI] [PubMed] [Google Scholar]
- 121.Valentine DL. 2002. Biogeochemistry and microbial ecology of methane oxidation in anoxic environments: a review. Antonie Van Leeuwenhoek 81:271–282. doi: 10.1023/A:1020587206351. [DOI] [PubMed] [Google Scholar]
- 122.Arshad A, Speth DR, de Graaf RM, Op den Camp HJ, Jetten MS, Welte CU. 2015. A metagenomics-based metabolic model of nitrate-dependent anaerobic oxidation of methane by Methanoperedens-like archaea. Front Microbiol 18:1423. doi: 10.3389/fmicb.2015.01423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Moran JJ, Beal EJ, Vrentas JM, Orphan VJ, Freeman KH, House CH. 2008. Methyl sulfides as intermediates in the anaerobic oxidation of methane. Environ Microbiol 10:162–173. doi: 10.1111/j.1462-2920.2007.01441.x. [DOI] [PubMed] [Google Scholar]
- 124.Meulepas RJW, Jagersma CG, Khadem A, Stams AJW, Lens P. 2010. Effect of methanogenic substrates on anaerobic oxidation of methane and sulfate reduction by an anaerobic methanotrophic enrichment. Appl Microbiol Biotechnol 87:1499–1506. doi: 10.1007/s00253-010-2597-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nauhaus K, Treude T, Boetius A, Kruger M. 2005. Environmental regulation of the anaerobic oxidation of methane: a comparison of ANME-I and ANME-II communities. Environ Microbiol 7:98–106. doi: 10.1111/j.1462-2920.2004.00669.x. [DOI] [PubMed] [Google Scholar]
- 126.Rotaru A-E, Shrestha PM, Liu F, Markovaite B, Chen S, Nevin K, Lovley D. 2014. Direct interspecies electron transfer between Geobacter metallireducens and Methanosarcina barkeri. Appl Environ Microbiol 80:4599–4605. doi: 10.1128/AEM.00895-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Summers ZM, Fogarty HE, Leang C, Franks AE, Malvankar NS, Lovley DR. 2010. Direct exchange of electrons within aggregates of an evolved syntrophic coculture of anaerobic bacteria. Science 330:1413–1415. doi: 10.1126/science.1196526. [DOI] [PubMed] [Google Scholar]
- 128.Reguera G, McCarthy KD, Mehta T, Nicoll JS, Tuominen MT, Lovley DR. 2005. Extracellular electron transfer via microbial nanowires. Nature 435:1098–1101. doi: 10.1038/nature03661. [DOI] [PubMed] [Google Scholar]
- 129.Kato S, Hashimoto K, Watanabe K. 2012. Microbial interspecies electron transfer via electric currents through conductive minerals. Proc Natl Acad Sci U S A 109:10042–10046. doi: 10.1073/pnas.1117592109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lovley DR. 2008. Extracellular electron transfer: wires, capacitors, iron lungs, and more. Geobiology 6:225–231. doi: 10.1111/j.1472-4669.2008.00148.x. [DOI] [PubMed] [Google Scholar]
- 131.McGlynn SE, Chadwick GL, Kempes CP, Orphan VJ. 2015. Single cell activity reveals direct electron transfer in methanotrophic consortia. Nature 526:531–535. doi: 10.1038/nature15512. [DOI] [PubMed] [Google Scholar]
- 132.Wegener G, Krukenberg V, Riedel D, Tegetmeyer HE, Boetius A. 2015. Intercellular wiring enables electron transfer between methanotrophic archaea and bacteria. Nature 526:587–603. doi: 10.1038/nature15733. [DOI] [PubMed] [Google Scholar]
- 133.Gao Y, Lee J, Neufeld JD, Park J, Rittmann BE, Lee H-S. 2017. Anaerobic oxidation of methane coupled with extracellular electron transfer to electrodes. Sci Rep 7:5099. doi: 10.1038/s41598-017-05180-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Skennerton CT, Chourey K, Iyer R, Hettich RL, Tyson GW, Orphan VJ. 2017. Methane-fueled syntrophy through extracellular electron transfer: uncovering the genomic traits conserved within diverse bacterial partners of anaerobic methanotrophic archaea. mBio 8:e00530-17. doi: 10.1128/mBio.00530-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Dahl C, Prange A. 2006. Bacterial sulfur globules: occurrence, structure and metabolism, p 21–51. In Shively J. (ed), Inclusions in prokaryotes, vol 1 Springer-Verlag, Heidelberg, Germany. [Google Scholar]
- 136.Wegener G, Krukenberg V, Ruff SE, Kellermann MY, Knittel K. 2016. Metabolic capabilities of microorganisms involved in and associated with the anaerobic oxidation of methane. Front Microbiol 7:46. doi: 10.3389/fmicb.2016.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wrede C, Brady S, Rockstroh S, Dreier A, Kokoschka S, Heinzelmann SM, Heller C, Reitner J, Taviani M, Daniel R, Hoppert M. 2012. Aerobic and anaerobic methane oxidation in terrestrial mud volcanoes in the Northern Apennines. Sediment Geol 263-264:210–219. doi: 10.1016/j.sedgeo.2011.06.004. [DOI] [Google Scholar]
- 138.Blazewicz SJ, Petersen DG, Waldrop MP, Firestone MK. 2012. Anaerobic oxidation of methane in tropical and boreal soils: ecological significance in terrestrial methane cycling. J Geophys Res Biogeosci 117:1–9. [Google Scholar]
- 139.Weber HS, Habicht KS, Thamdrup B. 2017. Anaerobic methanotrophic archaea of the ANME-2d cluster are active in a low-sulfate, iron-rich freshwater sediment. Front Microbiol 8:619. doi: 10.3389/fmicb.2017.00619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Martinez-Cruz K, Leewis M-C, Herriott IC, Sepulveda-Jauregui A, Anthony KW, Thalasso F, Leigh MB. 2017. Anaerobic oxidation of methane by aerobic methanotrophs in sub-Arctic lake sediments. Sci Total Environ 607-608:23–31. doi: 10.1016/j.scitotenv.2017.06.187. [DOI] [PubMed] [Google Scholar]
- 141.Valenzuela EI, Prieto-Davó A, López-Lozano NE, Hernández-Eligio A, Vega-Alvarado L, Juárez K, García-González AS, López MG, Cervantes FJ. 2017. Anaerobic methane oxidation driven by microbial reduction of natural organic matter in a tropical wetland. Appl Environ Microbiol 83:e00645-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Vaksmaa A, Guerrero-Cruz S, van Alen TA, Cremers G, Ettwig KF, Lüke C, Jetten MS. 2017. Enrichment of anaerobic nitrate-dependent methanotrophic ‘Candidatus Methanoperedens nitroreducens’ archaea from an Italian paddy field soil. Appl Microbiol Biotechnol 101:7075–7084. doi: 10.1007/s00253-017-8416-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Michaelis W, Seifert R, Nauhaus K, Treude T, Thiel V, Blumenberg M, Knittel K, Gieseke A, Peterknecht K, Pape T, Boetius A, Amann R, Jørgensen BB, Widdel F, Peckmann J, Pimenov NV, Gulin MB. 2002. Microbial reefs in the Black Sea fueled by anaerobic oxidation of methane. Science 297:1013–1015. doi: 10.1126/science.1072502. [DOI] [PubMed] [Google Scholar]
- 144.Thiel V, Peckmann J, Richnow HH, Luth U, Reitner J, Michaelis W. 2001. Molecular signals for anaerobic methane oxidation in Black Sea seep carbonates and a microbial mat. Mar Chem 73:97–112. doi: 10.1016/S0304-4203(00)00099-2. [DOI] [Google Scholar]
- 145.Reitner J, Peckmann J, Reimer A, Schumann G, Thiel V. 2005. Methane-derived carbonate build-ups and associated microbial communities at cold seeps on the lower Crimean shelf (Black Sea). Facies 51:66–79. doi: 10.1007/s10347-005-0059-4. [DOI] [Google Scholar]
- 146.Novikova SA, Shnyukov YF, Sokol EV, Kozmenko OA, Semenova DV, Kutny VA. 2015. A methane-derived carbonate build-up at a cold seep on the Crimean slope, north-western Black Sea. Mar Geol 363:160–173. doi: 10.1016/j.margeo.2015.02.008. [DOI] [Google Scholar]
- 147.Krüger M, Blumenberg M, Kasten S, Wieland A, Känel L, Klock J-H, Michaelis W, Seifert R. 2008. A novel, multi-layered methanotrophic microbial mat system growing on the sediment of the Black Sea. Environ Microbiol 10:1934–1947. doi: 10.1111/j.1462-2920.2008.01607.x. [DOI] [PubMed] [Google Scholar]
- 148.Marlow JJ, Steele JA, Ziebis W, Thurber AR, Levin LA, Orphan VJ. 2014. Carbonate-hosted methanotrophy represents an unrecognized methane sink in the deep sea. Nat Commun 5:5094. doi: 10.1038/ncomms6094. [DOI] [PubMed] [Google Scholar]
- 149.Mason OU, Case DH, Naehr TH, Lee RW, Thomas RB, Bailey JV, Orphan VJ. 2015. Comparison of archaeal and bacterial diversity in methane seep carbonate nodules and host sediments, Eel River Basin and Hydrate Ridge, USA. Microb Ecol 70:766–784. doi: 10.1007/s00248-015-0615-6. [DOI] [PubMed] [Google Scholar]
- 150.Orphan VJ, Hinrichs K-U, Ussler W, Paull CK, Taylor LT, Sylva SP, Hayes JM, Delong EF. 2001. Comparative analysis of methane-oxidizing archaea and sulfate-reducing bacteria in anoxic marine sediments. Appl Environ Microbiol 67:1922–1934. doi: 10.1128/AEM.67.4.1922-1934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Orphan VJ, House CH, Hinrichs K-U, McKeegan KD, DeLong EF. 2001. Methane-consuming archaea revealed by directly coupled isotopic and phylogenetic analysis. Science 293:484–487. doi: 10.1126/science.1061338. [DOI] [PubMed] [Google Scholar]
- 152.Brooks JM, Field ME, Kennicutt MC. 1991. Observations of gas hydrates in marine sediments, offshore northern California. Mar Geol 96:103–109. doi: 10.1016/0025-3227(91)90204-H. [DOI] [Google Scholar]
- 153.Orcutt B, Samarkin V, Boetius A, Joye S. 2008. On the relationship between methane production and oxidation by anaerobic methanotrophic communities from cold seeps of the Gulf of Mexico. Environ Microbiol 10:1108–1117. doi: 10.1111/j.1462-2920.2007.01526.x. [DOI] [PubMed] [Google Scholar]
- 154.Lloyd KG, Lapham L, Teske A. 2006. An anaerobic methane-oxidizing community of ANME-1b archaea in hypersaline Gulf of Mexico sediments. Appl Environ Microbiol 72:7218–7230. doi: 10.1128/AEM.00886-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Heijs SK, Haese RR, van der Wielen PW, Forney LJ, van Elsas JD. 2007. Use of 16S rRNA gene based clone libraries to assess microbial communities potentially involved in anaerobic methane oxidation in a Mediterranean cold seep. Microb Ecol 53:384–398. doi: 10.1007/s00248-006-9172-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kormas K, Meziti A, Dählmann A, De Lange G, Lykousis V. 2008. Characterization of methanogenic and prokaryotic assemblages based on mcrA and 16S rRNA gene diversity in sediments of the Kazan mud volcano (Mediterranean Sea). Geobiology 6:450–460. doi: 10.1111/j.1472-4669.2008.00172.x. [DOI] [PubMed] [Google Scholar]
- 157.Pachiadaki MG, Lykousis V, Stefanou EG, Kormas KA. 2010. Prokaryotic community structure and diversity in the sediments of an active submarine mud volcano (Kazan mud volcano, East Mediterranean Sea). FEMS Microbiol Ecol 72:429–444. doi: 10.1111/j.1574-6941.2010.00857.x. [DOI] [PubMed] [Google Scholar]
- 158.Pachiadaki MG, Kallionaki A, Dählmann A, De Lange GJ, Kormas KA. 2011. Diversity and spatial distribution of prokaryotic communities along a sediment vertical profile of a deep-sea mud volcano. Microb Ecol 62:655–668. doi: 10.1007/s00248-011-9855-2. [DOI] [PubMed] [Google Scholar]
- 159.Larowe DE, Dale AW, Regnier P. 2008. A thermodynamic analysis of the anaerobic oxidation of methane in marine sediments. Geobiology 6:436–449. doi: 10.1111/j.1472-4669.2008.00170.x. [DOI] [PubMed] [Google Scholar]
- 160.Biddle JF, Cardman Z, Mendlovitz H, Albert DB, Lloyd KG, Boetius A, Teske A. 2012. Anaerobic oxidation of methane at different temperature regimes in Guaymas Basin hydrothermal sediments. ISME J 6:1018–1031. doi: 10.1038/ismej.2011.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Vigneron A, Cruaud P, Pignet P, Caprais J-C, Cambon-Bonavita M-A, Godfroy A, Toffin L. 2013. Archaeal and anaerobic methane oxidizer communities in the Sonora Margin cold seeps, Guaymas Basin (Gulf of California). ISME J 7:1595–1608. doi: 10.1038/ismej.2013.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lever MA, Rouxel O, Alt JC, Shimizu N, Ono S, Coggon RM, Shanks WC, Lapham L, Elvert M, Prieto-Mollar X, Hinrichs K-U, Inagaki F, Teske A. 2013. Evidence for microbial carbon and sulfur cycling in deeply buried ridge flank basalt. Science 339:1305–1308. doi: 10.1126/science.1229240. [DOI] [PubMed] [Google Scholar]
- 163.Bradley AS, Fredricks H, Hinrichs K-U, Summons RE. 2009. Structural diversity of diether lipids in carbonate chimneys at the Lost City Hydrothermal Field. Org Geochem 40:1169–1178. doi: 10.1016/j.orggeochem.2009.09.004. [DOI] [Google Scholar]
- 164.Brazelton WJ, Schrenk MO, Kelley DS, Baross JA. 2006. Methane- and sulfur-metabolizing microbial communities dominate the Lost City hydrothermal field ecosystem. Appl Environ Microbiol 72:6257–6270. doi: 10.1128/AEM.00574-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Holler T, Widdel F, Knittel K, Amann R, Kellermann MY, Hinrichs K-U, Teske A, Boetius A, Wegener G. 2011. Thermophilic anaerobic oxidation of methane by marine microbial consortia. ISME J 5:1946–1956. doi: 10.1038/ismej.2011.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Orphan VJ, Ussler W, Naehr TH, House CH, Hinrichs K-U, Paull CK. 2004. Geological, geochemical, and microbiological heterogeneity of the seafloor around methane vents in the Eel River Basin, offshore California. Chem Geol 205:265–289. doi: 10.1016/j.chemgeo.2003.12.035. [DOI] [Google Scholar]
- 167.Timmers PH, Gieteling J, Widjaja-Greefkes HA, Plugge CM, Stams AJ, Lens PN, Meulepas RJ. 2015. Growth of anaerobic methane-oxidizing archaea and sulfate-reducing bacteria in a high-pressure membrane capsule bioreactor. Appl Environ Microbiol 81:1286–1296. doi: 10.1128/AEM.03255-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Teske A, Hinrichs K-U, Edgcomb V, Gomez AD, Kysela D, Sylva SP, Sogin ML, Jannasch HW. 2002. Microbial diversity of hydrothermal sediments in the Guaymas Basin: evidence for anaerobic methanotrophic communities. Appl Environ Microbiol 68:1994–2007. doi: 10.1128/AEM.68.4.1994-2007.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Merkel AY, Huber JA, Chernyh NA, Bonch-Osmolovskaya EA, Lebedinsky AV. 2013. Detection of putatively thermophilic anaerobic methanotrophs in diffuse hydrothermal vent fluids. Appl Environ Microbiol 79:915–923. doi: 10.1128/AEM.03034-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Lazar CS, John Parkes R, Cragg BA, L'Haridon S, Toffin L. 2012. Methanogenic activity and diversity in the centre of the Amsterdam Mud Volcano, Eastern Mediterranean Sea. FEMS Microbiol Ecol 81:243–254. doi: 10.1111/j.1574-6941.2012.01375.x. [DOI] [PubMed] [Google Scholar]
- 171.Lanoil BD, Sassen R, La Duc MT, Sweet ST, Nealson KH. 2001. Bacteria and archaea physically associated with Gulf of Mexico gas hydrates. Appl Environ Microbiol 67:5143–5153. doi: 10.1128/AEM.67.11.5143-5153.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Schubert CJ, Coolen MJL, Neretin LN, Schippers A, Abbas B, Durisch-Kaiser E, Wehrli B, Hopmans EC, Damste JSS, Wakeham S, Kuypers MM. 2006. Aerobic and anaerobic methanotrophs in the Black Sea water column. Environ Microbiol 8:1844–1856. doi: 10.1111/j.1462-2920.2006.01079.x. [DOI] [PubMed] [Google Scholar]
- 173.Treude T, Krüger M, Boetius A, Jørgensen BB. 2005. Environmental control on anaerobic oxidation of methane in the gassy sediments of Eckernförde Bay (German Baltic). Limnol Oceanogr 50:1771–1786. doi: 10.4319/lo.2005.50.6.1771. [DOI] [Google Scholar]
- 174.Hu S, Zeng RJ, Burow LC, Lant P, Keller J, Yuan Z. 2009. Enrichment of denitrifying anaerobic methane oxidizing microorganisms. Environ Microbiol Rep 1:377–384. doi: 10.1111/j.1758-2229.2009.00083.x. [DOI] [PubMed] [Google Scholar]
- 175.Parkes RJ, Cragg BA, Banning N, Brock F, Webster G, Fry JC, Hornibrook E, Pancost RD, Kelly S, Knab N, Jørgensen BB, Rinna J, Weightman AJ. 2007. Biogeochemistry and biodiversity of methane cycling in subsurface marine sediments (Skagerrak, Denmark). Environ Microbiol 9:1146–1161. doi: 10.1111/j.1462-2920.2006.01237.x. [DOI] [PubMed] [Google Scholar]
- 176.Bhattarai S, Cassarini C, Gonzalez-Gil G, Egger M, Slomp CP, Zhang Y, Esposito G, Lens P. 2017. Anaerobic methane-oxidizing microbial community in a coastal marine sediment: anaerobic methanotrophy dominated by ANME-3. Microb Ecol 74:608–622. doi: 10.1007/s00248-017-0978-y. [DOI] [PubMed] [Google Scholar]
- 177.Cassarini C, Zhang Y, Lens PN. 14 January 2019. Pressure selects dominant anaerobic methanotrophic phylotype and sulfate reducing bacteria in coastal marine Lake Grevelingen sediment. Front Environ Sci doi: 10.3389/fenvs.2018.00162. [DOI] [Google Scholar]
- 178.Alperin M, Hoehler T. 2010. The ongoing mystery of sea-floor methane. Science 329:288–289. doi: 10.1126/science.1189966. [DOI] [PubMed] [Google Scholar]
- 179.Krause S, Steeb P, Hensen C, Liebetrau V, Dale AW, Nuzzo M, Treude T. 2014. Microbial activity and carbonate isotope signatures as a tool for identification of spatial differences in methane advection: a case study at the Pacific Costa Rican margin. Biogeosciences 11:507–523. doi: 10.5194/bg-11-507-2014. [DOI] [Google Scholar]
- 180.Dale AW, Van Cappellen P, Aguilera DR, Regnier P. 2008. Methane efflux from marine sediments in passive and active margins: estimations from bioenergetic reaction-transport simulations. Earth Planet Sci Lett 265:329–344. doi: 10.1016/j.epsl.2007.09.026. [DOI] [Google Scholar]
- 181.Roalkvam I, Dahle H, Chen Y, Jørgensen SL, Haflidason H, Steen IH. 2012. Fine-scale community structure analysis of ANME in Nyegga sediments with high and low methane flux. Front Microbiol 3:216. doi: 10.3389/fmicb.2012.00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Marlow JJ, Steele JA, Case DH, Connon SA, Levin LA, Orphan VJ. 2014. Microbial abundance and diversity patterns associated with sediments and carbonates from the methane seep environments of Hydrate Ridge, OR. Front Mar Sci 1:44. doi: 10.3389/fmars.2014.00044. [DOI] [Google Scholar]
- 183.Ruff S, Felden J, Gruber-Vodicka H, Marcon Y, Knittel K, Ramette A, Boetius A. 2019. In situ development of a methanotrophic microbiome in deep-sea sediments. ISME J 13:197–213. doi: 10.1038/s41396-018-0263-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Amann RI, Ludwig W, Schleifer KH. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59:143–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.D'Hondt S, Jørgensen BB, Miller DJ, Batzke A, Blake R, Cragg BA, Cypionka H, Dickens GR, Ferdelman T, Hinrichs K-U, Holm NG, Mitterer R, Spivack A, Wang G, Bekins B, Engelen B, Ford K, Gettemy G, Rutherford SD, Sass H, Skilbeck CG, Aiello IW, Guèrin G, House CH, Inagaki F, Meister P, Naehr T, Niitsuma S, Parkes RJ, Schippers A, Smith DC, Teske A, Wiegel J, Padilla CN, Acosta J. 2004. Distributions of microbial activities in deep subseafloor sediments. Science 306:2216–2221. doi: 10.1126/science.1101155. [DOI] [PubMed] [Google Scholar]
- 186.Parkes RJ, Cragg BA, Wellsbury P. 2000. Recent studies on bacterial populations and processes in subseafloor sediments: a review. Hydrogeol J 8:11–28. doi: 10.1007/PL00010971. [DOI] [Google Scholar]
- 187.Nauhaus K, Boetius A, Kruger M, Widdel F. 2002. In vitro demonstration of anaerobic oxidation of methane coupled to sulphate reduction in sediment from a marine gas hydrate area. Environ Microbiol 4:296–305. doi: 10.1046/j.1462-2920.2002.00299.x. [DOI] [PubMed] [Google Scholar]
- 188.Holler T, Wegener G, Niemann H, Deusner C, Ferdelman TG, Boetius A, Brunner B, Widdel F. 2011. Carbon and sulfur back flux during anaerobic microbial oxidation of methane and coupled sulfate reduction. Proc Natl Acad Sci U S A 108:E1484–E1490. doi: 10.1073/pnas.1106032108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Blumenberg M, Seifert R, Nauhaus K, Pape T, Michaelis W. 2005. In vitro study of lipid biosynthesis in an anaerobically methane-oxidizing microbial mat. Appl Environ Microbiol 71:4345–4351. doi: 10.1128/AEM.71.8.4345-4351.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Kruger M, Wolters H, Gehre M, Joye SB, Richnow H-H. 2008. Tracing the slow growth of anaerobic methane-oxidizing communities by 15N-labelling techniques. FEMS Microbiol Ecol 63:401–411. doi: 10.1111/j.1574-6941.2007.00431.x. [DOI] [PubMed] [Google Scholar]
- 191.Bhattarai S, Zhang Y, Lens P. 2018. Effect of pressure and temperature on anaerobic methanotrophic activities of a highly enriched ANME-2a community. Environ Sci Pollut Res Int 25:30031–30043. doi: 10.1007/s11356-018-2573-2. [DOI] [PubMed] [Google Scholar]
- 192.Yamamoto S, Alcauskas JB, Crozier TE. 1976. Solubility of methane in distilled water and seawater. J Chem Eng Data 21:78–80. doi: 10.1021/je60068a029. [DOI] [Google Scholar]
- 193.Girguis PR, Orphan VJ, Hallam SJ, DeLong EF. 2003. Growth and methane oxidation rates of anaerobic methanotrophic archaea in a continuous-flow bioreactor. Appl Environ Microbiol 69:5472–5482. doi: 10.1128/AEM.69.9.5472-5482.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Deusner C, Meyer V, Ferdelman T. 2010. High-pressure systems for gas-phase free continuous incubation of enriched marine microbial communities performing anaerobic oxidation of methane. Biotechnol Bioeng 105:524–533. doi: 10.1002/bit.22553. [DOI] [PubMed] [Google Scholar]
- 195.Sauer P, Glombitza C, Kallmeyer J. 2012. A system for incubations at high gas partial pressure. Front Microbiol 3:25. doi: 10.3389/fmicb.2012.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Zhang Y, Henriet J-P, Bursens J, Boon N. 2010. Stimulation of in vitro anaerobic oxidation of methane rate in a continuous high-pressure bioreactor. Bioresour Technol 101:3132–3138. doi: 10.1016/j.biortech.2009.11.103. [DOI] [PubMed] [Google Scholar]
- 197.Zhang Y, Maignien L, Zhao X, Wang F, Boon N. 2011. Enrichment of a microbial community performing anaerobic oxidation of methane in a continuous high-pressure bioreactor. BMC Microbiol 11:137. doi: 10.1186/1471-2180-11-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Imachi H, Aoi K, Tasumi E, Saito Y, Yamanaka Y, Saito Y, Yamaguchi T, Tomaru H, Takeuchi R, Morono Y, Inagaki F, Takai K. 2011. Cultivation of methanogenic community from subseafloor sediments using a continuous-flow bioreactor. ISME J 5:1913–1925. doi: 10.1038/ismej.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Ohtomo Y, Ijiri A, Ikegawa Y, Tsutsumi M, Imachi H, Uramoto G-I, Hoshino T, Morono Y, Sakai S, Saito Y, Tanikawa W, Hirose T, Inagaki F. 2013. Biological CO2 conversion to acetate in subsurface coal-sand formation using a high-pressure reactor system. Front Microbiol 4:361. doi: 10.3389/fmicb.2013.00361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Jagersma GC, Meulepas RJW, Heikamp-de Jong I, Gieteling J, Klimiuk A, Schouten S, Sinninghe Damsté JS, Lens PN, Stams AJ. 2009. Microbial diversity and community structure of a highly active anaerobic methane‐oxidizing sulfate‐reducing enrichment. Environ Microbiol 11:3223–3232. doi: 10.1111/j.1462-2920.2009.02036.x. [DOI] [PubMed] [Google Scholar]
- 201.Cassidy J, Lubberding HJ, Esposito G, Keesman KJ, Lens PN. 2015. Automated biological sulphate reduction: a review on mathematical models, monitoring and bioprocess control. FEMS Microbiol Rev 39:823–853. doi: 10.1093/femsre/fuv033. [DOI] [PubMed] [Google Scholar]
- 202.Rabaey K, Rozendal RA. 2010. Microbial electrosynthesis—revisiting the electrical route for microbial production. Nat Rev Microbiol 8:706–716. doi: 10.1038/nrmicro2422. [DOI] [PubMed] [Google Scholar]
- 203.Lovley DR. 2012. Electromicrobiology. Annu Rev Microbiol 66:391–409. doi: 10.1146/annurev-micro-092611-150104. [DOI] [PubMed] [Google Scholar]
- 204.Thauer RK, Kaster A-K, Seedorf H, Buckel W, Hedderich R. 2010. Methanogenic archaea: ecologically relevant differences in energy conservation. Nat Rev Microbiol 6:579–591. doi: 10.1038/nrmicro1931. [DOI] [PubMed] [Google Scholar]
- 205.Niemann H. 2005. Rates and signatures of methane turnover in sediments of continental margins. PhD thesis. University of Bremen, Bremen, Germany. [Google Scholar]
- 206.Segarra KEA, Schubotz F, Samarkin V, Yoshinaga MY, Hinrichs K-U, Joye SB. 2015. High rates of anaerobic methane oxidation in freshwater wetlands reduce potential atmospheric methane emissions. Nat Commun 6:7477. doi: 10.1038/ncomms8477. [DOI] [PubMed] [Google Scholar]
- 207.Gauthier M, Bradley RL, Šimek M. 2015. More evidence that anaerobic oxidation of methane is prevalent in soils: is it time to upgrade our biogeochemical models? Soil Biol Biochem 80:167–174. doi: 10.1016/j.soilbio.2014.10.009. [DOI] [Google Scholar]
- 208.Dupré S, Woodside J, Klaucke I, Mascle J, Foucher J-P. 2010. Widespread active seepage activity on the Nile Deep Sea Fan (offshore Egypt) revealed by high-definition geophysical imagery. Mar Geol 275:1–19. doi: 10.1016/j.margeo.2010.04.003. [DOI] [Google Scholar]
- 209.Girnth AC, Grünke S, Lichtschlag A, Felden J, Knittel K, Wenzhöfer F, de Beer D, Boetius A. 2011. A novel, mat‐forming Thiomargarita population associated with a sulfidic fluid flow from a deep‐sea mud volcano. Environ Microbiol 13:495–505. doi: 10.1111/j.1462-2920.2010.02353.x. [DOI] [PubMed] [Google Scholar]
- 210.Omoregie EO, Niemann H, Mastalerz V, De Lange GJ, Stadnitskaia A, Mascle J, Foucher J-P, Boetius A. 2009. Microbial methane oxidation and sulfate reduction at cold seeps of the deep Eastern Mediterranean Sea. Mar Geol 261:114–127. doi: 10.1016/j.margeo.2009.02.001. [DOI] [Google Scholar]
- 211.Grünke S, Felden J, Lichtschlag A, Girnth AC, De Beer D, Wenzhöfer F, Boetius A. 2011. Niche differentiation among mat‐forming, sulfide‐oxidizing bacteria at cold seeps of the Nile Deep Sea Fan (Eastern Mediterranean Sea). Geobiology 9:330–348. doi: 10.1111/j.1472-4669.2011.00281.x. [DOI] [PubMed] [Google Scholar]
- 212.Durisch-Kaiser E, Klauser L, Wehrli B, Schubert C. 2005. Evidence of intense archaeal and bacterial methanotrophic activity in the Black Sea water column. Appl Environ Microbiol 71:8099–8106. doi: 10.1128/AEM.71.12.8099-8106.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Niemann H, Elvert M, Hovland M, Orcutt B, Judd A, Suck I, Gutt J, Joye S, Damm E, Finster K, Boetius A. 2005. Methane emission and consumption at a North Sea gas seep (Tommeliten area). Biogeosciences 2:335–351. doi: 10.5194/bg-2-335-2005. [DOI] [Google Scholar]
- 214.Slomp CP, Mort HP, Jilbert T, Reed DC, Gustafsson BG, Wolthers M. 2013. Coupled dynamics of iron and phosphorus in sediments of an oligotrophic coastal basin and the impact of anaerobic oxidation of methane. PLoS One 8:e62386. doi: 10.1371/journal.pone.0062386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Sivan O, Schrag DP, Murray RW. 2007. Rates of methanogenesis and methanotrophy in deep-sea sediments. Geobiology 5:141–151. doi: 10.1111/j.1472-4669.2007.00098.x. [DOI] [Google Scholar]
- 216.Egger M, Lenstra W, Jong D, Meysman FJ, Sapart CJ, van der Veen C, Röckmann T, Gonzalez S, Slomp CP. 2016. Rapid sediment accumulation results in high methane effluxes from coastal sediments. PLoS One 11:e0161609. doi: 10.1371/journal.pone.0161609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Widdel F, Bak F. 1992. Gram negative mesophilic sulfate reducing bacteria, p 3352–3378. In Balows A, Truper H, Dworkin M, Harder W, Schleifer KH (ed), The prokaryotes: a handbook on the biology of bacteria: ecophysiology, isolation, identification, applications. Springer-Verlag, New York, NY. [Google Scholar]
- 218.Aoki M, Ehara M, Saito Y, Yoshioka H, Miyazaki M, Saito Y, Miyashita A, Kawakami S, Yamaguchi T, Ohashi A, Nunoura T, Takai K, Imachi H. 2014. A long-term cultivation of an anaerobic methane-oxidizing microbial community from deep-sea methane-seep sediment using a continuous-flow bioreactor. PLoS One 9:e105356. doi: 10.1371/journal.pone.0105356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Pruesse E, Peplies J, Glöckner FO. 2012. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28:1823–1829. doi: 10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Girguis PR, Cozen AE, DeLong EF. 2005. Growth and population dynamics of anaerobic methane-oxidizing archaea and sulfate-reducing bacteria in a continuous-flow bioreactor. Appl Environ Microbiol 71:3725–3733. doi: 10.1128/AEM.71.7.3725-3733.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Cassarini C, Rene ER, Bhattarai S, Vogt C, Musat N, Lens PNL. 3 July 2019. Anaerobic methane oxidation coupled to sulfate reduction in a biotrickling filter: reactor performance and microbial community analysis. Chemosphere doi: 10.1016/j.chemosphere.2019.07.021. [DOI] [PubMed] [Google Scholar]