A wide variety of mechanisms that control gene expression in bacteria are based on conditional transcription termination. Generally, in these mechanisms, a transcription terminator is located between a promoter and a downstream gene(s), and the efficiency of the terminator is controlled by a regulatory effector that can be a metabolite, protein, or RNA.
KEYWORDS: RNA-binding proteins, Rho-dependent termination, transcription attenuation, bacterial gene regulation, intrinsic termination, reiterative transcription, riboswitches
SUMMARY
A wide variety of mechanisms that control gene expression in bacteria are based on conditional transcription termination. Generally, in these mechanisms, a transcription terminator is located between a promoter and a downstream gene(s), and the efficiency of the terminator is controlled by a regulatory effector that can be a metabolite, protein, or RNA. The most common type of regulation involving conditional termination is transcription attenuation, in which the primary regulatory target is an essential element of a single terminator. The terminator can be either intrinsic or Rho dependent, with each presenting unique regulatory targets. Transcription attenuation mechanisms can be divided into five classes based primarily on the manner in which transcription termination is rendered conditional. This review summarizes each class of control mechanisms from a historical perspective, describes important examples in a physiological context and the current state of knowledge, highlights major advances, and discusses expectations of future discoveries.
INTRODUCTION
With the establishment of the operon model in bacteria in the 1960s, influential scientists pressed the idea that all gene expression would be negatively regulated by a repressor protein that bound to the promoter and prevented transcription initiation by RNA polymerase (RNAP) (1). Strict adherence to this idea hindered the acceptance of the next major advancement in the study of bacterial gene regulation, namely, positive control, in which an activator protein bound near the promoter to stimulate transcription initiation. The lack of acceptance of positive control occurred even though it shared many features of negative regulation. It is not surprising, therefore, that the discovery of the next new general mechanism of bacterial gene regulation took nearly two more decades, because it did not resemble in any way previously discovered examples of negative or positive control. The new mechanism was based on conditional transcription termination at a site (the terminator) located between a promoter and a downstream gene(s), with the efficiency of termination controlled by a specific physiological effector that interacted with the nascent RNA transcript (2). As a consequence, premature termination occurs when downstream gene activities are not needed by the cell. Even more surprising was the discovery, over time, that the examples of this new type of gene regulation employed many different ways to make transcription termination conditional in response to a multitude of physiological effectors (3). This diversity allows the regulation of expression of genes involved in many aspects of cellular metabolism in a wide variety of bacteria.
The discovery of gene regulation by conditional transcription termination was closely linked to seminal studies in the 1970s on the mechanisms of transcription termination in bacteria (4, 5). These studies revealed two distinct mechanisms, now called intrinsic and Rho-dependent termination (6), both of which can participate in control of gene expression. The role of each termination mechanism is to disrupt a highly stable transcription elongation complex (EC) that includes RNAP, DNA that is partially unwound to form a roughly 12-residue transcription bubble, and approximately 14 nucleotides (nt) of newly transcribed RNA (6). Within the active cleft of RNAP, the 3′-most region of the nascent RNA transcript forms a 9- to 10-bp RNA-DNA hybrid with the template strand of the transcription bubble, while ∼5 residues of adjacent transcript fill the RNA exit channel of RNAP (7). The stability of the EC is due in large part to contacts between RNAP and both the RNA-DNA hybrid and single-stranded RNA in the exit channel (8). Disruption of these interactions and the extraction of the RNA from the EC are required for both intrinsic and Rho-dependent transcription termination, although these objectives are accomplished differently in the two mechanisms. Understanding the molecular details of these mechanisms is critical to understanding how termination is rendered conditional in gene regulation.
In intrinsic termination, a canonical DNA sequence specifies two RNA elements that are sufficient to cause RNAP to immediately terminate elongation and release the nascent transcript (6). The upstream region of the conserved DNA sequence is an approximately 20-bp hyphenated dyad symmetry, which specifies a stem-loop structure called the terminator hairpin. The stem of the terminator hairpin is G+C rich and usually contains at least 8 bp. The hyphenated dyad symmetry is immediately followed by a tract of typically 8 T residues, or mostly T residues, in the nontemplate strand, which specifies U or mostly U residues in the RNA. The first step in intrinsic termination is transcription pausing at the end of the T/U tract. This pause provides time for the terminator hairpin to form in the RNAP exit channel, which disrupts RNAP-nucleic acid interactions that stabilize the EC and causes 3 to 4 bp of the upstream RNA-DNA hybrid to melt. With these (and perhaps other) structural perturbations and with the remainder of the RNA-DNA hybrid consisting of only exceptionally weak rU-dA base pairs (9), the nascent transcript is free to dissociate from the EC.
In contrast to intrinsic termination, Rho-dependent termination requires a factor called Rho, a homohexameric protein with both RNA translocase and helicase activities. In addition to Rho, there are two other elements needed for termination: an approximately 80- to 90-nucleotide-long Rho-binding site called the rut (Rho utilization) site in the nascent RNA transcript and a transcription pause site specified by downstream DNA (6). The first step in termination is the binding of Rho to the rut site, which is followed by translocation in the 5′ to 3′ direction on the nascent transcript via ATP hydrolysis. During translocation, Rho remains bound to the rut site and threads RNA through its central pore, a process called tethered tracking (10). When Rho encounters a paused RNAP, it apparently causes disruptive conformational changes in the EC and uses its helicase activity to shear the RNA-DNA hybrid, thereby extracting the nascent RNA transcript (6).
Most mechanisms of gene regulation based on conditional transcription termination fall into two categories called transcription attenuation (or simply attenuation) and antitermination (4, 11). The major differences between these two types of control mechanisms are the number of terminators regulated and the nature of the regulatory target. In attenuation, typically one transcription terminator is involved, and it is directly targeted by the control mechanism. In antitermination, a large number of terminators are affected, and the target is RNAP. In this review, I focus only on attenuation control and refer the reader to two excellent reviews to learn about antitermination (12, 13). The goals of this review are to summarize attenuation control mechanisms from a historical perspective; to define general mechanistic classes; and to describe representative examples in a physiological context, highlight major advances, and discuss expectations of future discovery.
TRANSCRIPTION ATTENUATION
The term transcription attenuation arose from studies by Takashi Kasai on histidine (his) operon expression in Salmonella enterica serovar Typhimurium (referred to here as Salmonella) in which he proposed that an “attenuator” region in the DNA downstream of the promoter blocked transcription into the genes encoding the histidine-biosynthetic enzymes (14). It was subsequently shown that this attenuator was in fact a transcription terminator, and the term attenuator is now generally used to define a terminator that functions conditionally in gene regulation. Today, there are many examples of gene regulation by transcription attenuation, but they can be divided into five classes based on common themes in the regulatory mechanisms.
This division is somewhat arbitrary, however, due to overlap in mechanistic elements. The first four classes involve regulation of an intrinsic transcription terminator.
CLASS I: RIBOSOME-MEDIATED TRANSCRIPTION ATTENUATION
Tryptophan Operon of E. coli
Pioneering studies by Charles Yanofsky and colleagues in the 1970s led to the discovery of transcription attenuation control of the trpEDCBA (or simply trp) operon of Escherichia coli, which encodes the enzymes required for synthesis of l-tryptophan (Trp) (15, 16). The key elements required for attenuation control of trp operon expression are found in a 162-bp leader region, which is defined as the region between the transcription start site and the first gene of the operon (Fig. 1A). These elements include four segments in the leader transcript (i.e., segments 1, 2, 3, and 4) capable of forming two alternative transcript conformations. The first conformation includes an upstream 1:2 hairpin, which can induce transcription pausing, and a downstream 3:4 hairpin, which is the G+C-rich hairpin of an intrinsic transcription terminator (the trp attenuator). The alternative conformation contains only a 2:3 hairpin, the formation of which precludes formation of the 3:4 terminator hairpin, thereby permitting readthrough transcription into the genes of the trp operon. The final regulatory element is a 14-codon open reading frame (ORF) that extends through the end of leader transcript segment 1. The leader ORF also contains two Trp “control” codons at positions 10 and 11.
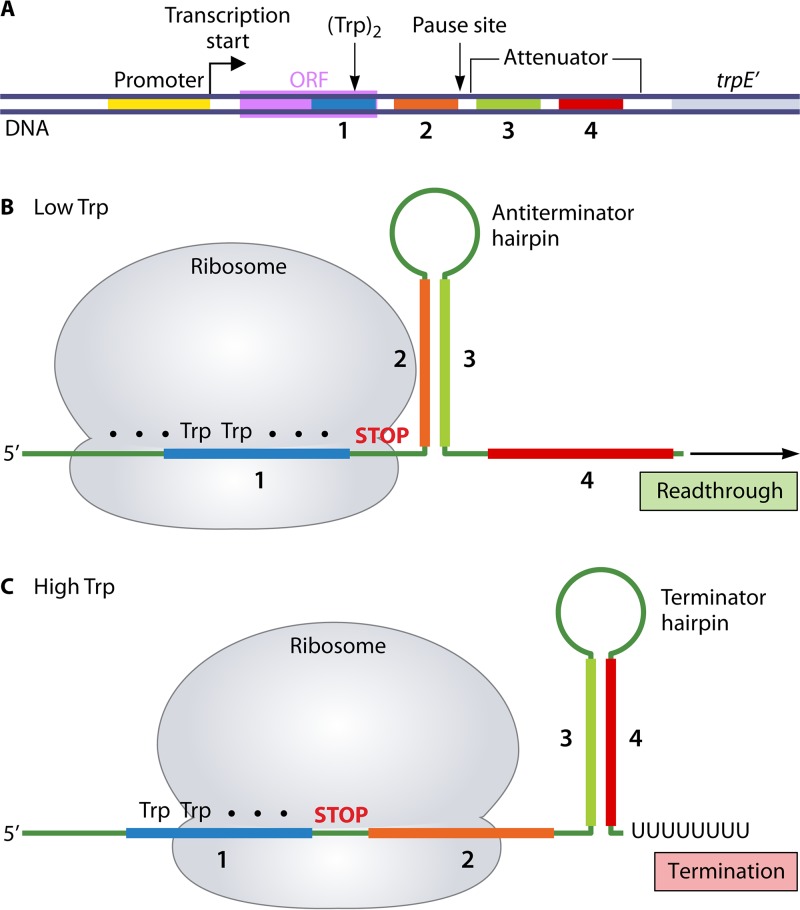
FIG 1.
Transcription attenuation control of trp operon expression in E. coli. (A) Key regulatory elements in the trp leader region. The leader region contains four segments (1, 2, 3, and 4) capable of specifying hairpins 1:2, 2:3, and 3:4 in the leader transcript. The leader region also contains a 14-codon ORF with Trp codons at positions 10 and 11 that extends through segment 1, and the trp attenuator in which segments 3 and 4 specify the terminator hairpin. Formation of the 1:2 hairpin induces transcription pausing, which is released by disruption of the hairpin by a ribosome translating the leader ORF. (B) When Trp (and Trp-tRNATrp) levels are low, the ribosome translating the leader ORF pauses at the tandem Trp codons in segment 1, while transcription continues downstream. With segment 1 sequestered, the 2:3 antiterminator hairpin forms immediately, which precludes formation of the 3:4 terminator hairpin and allows readthrough transcription. (C) When Trp (and Trp-tRNATrp) levels are high, translation proceeds through the entire leader ORF, which allows the ribosome to physically cover both segments 1 and 2 in the transcript. With segment 2 sequestered, further downstream, transcription allows the 3:4 terminator hairpin to form, which leads to transcription termination at the end of the adjacent U8 tract.
According to the regulatory model (17), RNAP initiates transcription at the trp promoter and moves rapidly through the leader region that specifies transcript segments 1 and 2, which then form the 1:2 pause hairpin. RNAP stalls at this site to permit a ribosome to begin translation of the leader ORF. Early in translation, the ribosome physically disrupts the 1:2 hairpin to release the stalled RNAP, and the ribosome then proceeds to the Trp control codons. When the intracellular level of Trp is limiting, causing a low level of Trp-tRNATrp, the ribosome pauses at the tandem Trp codons (Fig. 1B). During this time, the reengaged RNAP synthesizes transcript segment 3, permitting formation of the 2:3 antiterminator hairpin. As transcription continues, the nascent transcript is extended through leader segment 4 (without formation of the 3:4 terminator hairpin) and eventually through the entire operon. Translation of the full-length trp mRNA produces the biosynthetic enzymes needed to overcome Trp limitation. Conversely, when there is ample intracellular Trp and Trp-tRNATrp, the ribosome translating the leader transcript does not pause at the tandem Trp codons but proceeds to, and effectively pauses at, the stop codon at the end of the leader ORF (Fig. 1C) (18). At this position, the ribosome occupies leader transcript segments 1 and 2 (19, 20). Continuing transcription synthesizes leader transcript segments 3 and 4 and an adjacent 8-residue poly(U) tract, which allows formation of the 3:4 terminator hairpin and transcription termination at the end of the U tract. Consequently, additional trp-biosynthetic enzymes are not synthesized when there is sufficient Trp to support optimal cell growth. Attenuation control regulates trp operon expression over an approximately 8-fold range. In E. coli, inhibition of transcription initiation by the Trp repressor independently controls trp expression over a 70- to 80-fold range. Together, attenuation and repression control mechanisms regulate the expression of the trp operon over an approximately 600-fold range in response to intracellular levels of Trp (21).
Although attenuation control of trp operon expression in E. coli is generally considered the first example of this type of regulation, concurrent studies by Bruce Ames, John Roth, and their colleagues led to the discovery of an analogous mechanism responsible for attenuation control of his operon expression in Salmonella (22, 23). The major difference between the trp and his mechanisms is that, in the case of the his operon, the control codons are 7 histidine codons in a row (24). Within the decade after the elucidation of the trp and his attenuation control mechanisms, similar mechanisms were discovered for several other amino acid-biosynthetic operons and an aminoacyl-tRNA synthetase operon of enteric bacteria (17). In each case, ribosome stalling at cognate control codons regulated the formation of alternative secondary structures in the leader transcript, one permitting and the other precluding intrinsic transcription termination.
The discovery of these first examples of attenuation control was extremely exciting; however, the similarities between the control mechanisms raised the possibility that this new type of gene regulation was limited to amino acid-related operons and to a single mechanism for regulating transcription termination. This idea was soon dispelled by the discovery of a mechanistically different type of attenuation control involved in regulating pyrBI expression in E. coli.
pyrBI Operon of E. coli
The pyrBI operon encodes the catalytic (pyrB) and regulatory (pyrI) subunits of the pyrimidine nucleotide-biosynthetic enzyme aspartate transcarbamoylase. This enzyme catalyzes the first committed step in pyrimidine nucleotide biosynthesis, and its activity is allosterically regulated by pyrimidine nucleotide inhibitors and purine nucleotide activators (25). Expression of the pyrBI operon is negatively regulated over an approximately 350-fold range by the intracellular level of UTP, with most of this regulation (i.e., 50-fold) occurring through a transcription attenuation control mechanism (26). This mechanism relies on three regulatory elements located in a 156-bp leader region preceding the pyrB gene: a 44-codon ORF, an intrinsic transcription terminator (the pyrB attenuator) located within and near the end of the ORF, and UTP-sensitive transcription pause sites (i.e., sites for U addition, often as U tracts, in the nascent transcript) within the ORF and preceding the attenuator (Fig. 2A). Transcription pausing at U residues, but not at non-U residues, is due to the unique ability of the intracellular UTP pool to fall (e.g., during pyrimidine starvation) to levels that significantly slow transcription elongation (27).
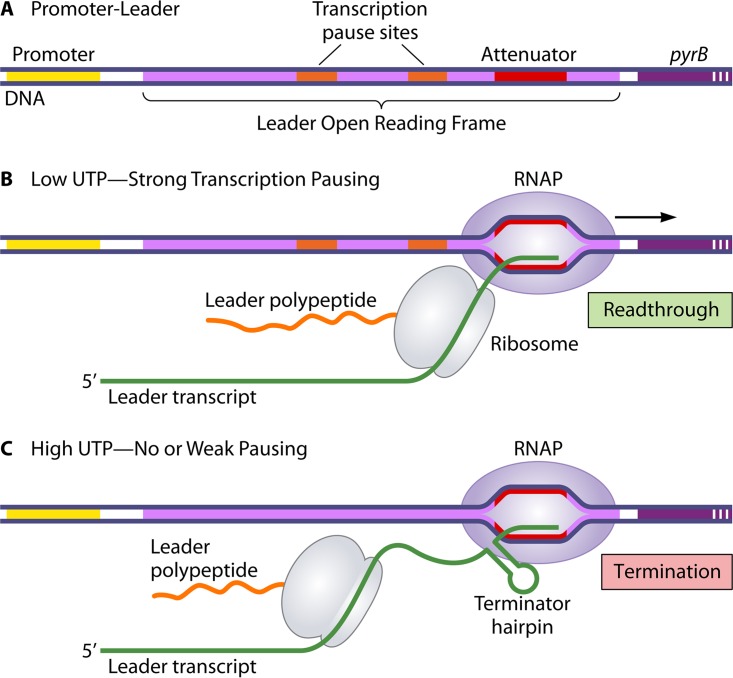
FIG 2.
Ribosome-mediated attenuation control of pyrBI expression in E. coli. The diagram shows the key regulatory elements in the pyrBI leader region (A) and the relative positions of RNAP and the translating ribosome on the leader transcript when intracellular UTP levels are either low (B) or high (C). In the model for regulation, low-UTP-induced transcription pausing in the leader region promotes tightly coupled transcription and translation. This coupling permits the ribosome to physically prevent the formation of the attenuator-encoded terminator hairpin, resulting in readthrough transcription. At high UTP levels, transcription and translation are not tightly coupled. In fact, according to the model, there is no requirement for ribosome binding to or translation of the leader transcript at high UTP levels. See the text for additional details.
According to the model of attenuation control, transcription initiates constitutively at the pyrBI promoter and continues into the leader region (28, 29). When the level of UTP in the cell is low, RNAP stalls at the UTP-sensitive pause sites, which provides time for a ribosome to begin translation of the leader ORF and proceed up to the stalled RNAP, establishing tightly coupled transcription and translation. When RNAP eventually escapes the pause sites and transcribes the attenuator, formation of the attenuator-specified terminator RNA hairpin is physically blocked by the adjacent translating ribosome (Fig. 2B). In this case, transcription termination at the attenuator is precluded, and RNAP continues transcription into the pyrBI genes. The result is an increase in the capacity of the cell to produce more UTP under conditions of UTP limitation. In contrast, when the level of UTP in the cell is high, RNAP transcribes the leader region without stalling at the UTP-sensitive pause sites. Consequently, there is insufficient time for a ribosome to translate the leader ORF and establish tight coupling with RNAP before the formation of the terminator hairpin and transcription termination at the attenuator (Fig. 2C). The result is that additional aspartate transcarbamoylase is not produced when there is no need for an increase in intracellular UTP production.
Shortly after the mechanism for attenuation control of pyrBI expression was elucidated, an analogous UTP-sensitive attenuation mechanism that controls expression of the pyrE gene of E. coli was discovered (30). The pyrE gene encodes the pyrimidine nucleotide-biosynthetic enzyme orotate phosphoribosyltransferase, and its expression is regulated over an approximately 30-fold range by transcription attenuation. The major difference between the pyrBI and pyrE control mechanisms is that the pyrE “leader ORF” is actually the 238-codon rph gene, which encodes the tRNA-processing exoribonuclease RNase PH (31). Thus, the pyrE gene is the second gene of an rph-pyrE operon, and UTP-sensitive transcription pausing and tight coupling of transcription and translation within the rph ORF are used to prevent intrinsic transcription termination at an attenuator preceding the pyrE gene. It is noteworthy that the rph and pyrE genes appear to be metabolically unrelated and that the pyrE attenuator can double as a terminator for a monocistronic rph transcript. Another interesting difference between the pyrBI and pyrE mechanisms is that the pyrE attenuator is not in the rph ORF but is located 10 bp downstream. Nevertheless, based on the size of the ribosome footprint (∼12 to 15 nt on either side of the ribosomal P site) (19, 32), translation to the end of the rph cistron would disrupt formation of the terminator hairpin and allow readthrough transcription.
Historically, with the elucidation of the pyrBI and pyrE attenuation control mechanisms, there were now two distinct mechanisms for rendering transcription termination conditional, namely, formation of an alternative leader transcript secondary structure that excludes the terminator hairpin and tightly coupled transcription and translation that allow a translating ribosome to physically interfere with terminator hairpin formation. However, in both cases, attenuation control was mediated by a translating ribosome, again suggesting a limitation in the scope of attenuation control mechanisms. However, the reality was that a treasure chest of attenuation control mechanisms was just beginning to open. The next examples of attenuation control did not rely on a translating ribosome but employed RNA-binding proteins to control the formation of the terminator hairpin.
CLASS II: RNA-BINDING-PROTEIN-MEDIATED TRANSCRIPTION ATTENUATION
bgl Operon of E. coli
The first example of transcription attenuation control mediated by an RNA-binding protein was provided by the mechanism for β-glucoside-sensitive regulation of bgl operon expression in E. coli (33–35). The bgl operon contains four genes, bglG, bglF, bglB, and bglH, which encode proteins that enable cells to use certain aromatic β-glucosides as carbon sources (Fig. 3A). Two of the proteins, BglG and BglF, are involved in regulation. As a dimer, BglG is an RNA-binding protein that positively regulates operon expression. BglF is a membrane-bound, β-glucoside-specific enzyme II of the phosphoenolpyruvate-dependent phosphotransferase system (PTS) that catalyzes transport and concomitant phosphorylation of β-glucosidic sugars (Fig. 3B) (36). BglF also catalyzes reversible phosphorylation of BglG (BglG∼P), and when phosphorylated, BglG∼P is monomeric and unable to bind RNA (Fig. 3C). In most wild-type strains of E. coli, the bgl operon is cryptic but can be activated by a variety of cis- and trans-acting mutations (37, 38). Once activated, full bgl expression requires a β-glucoside inducer and also cyclic AMP (cAMP) and the cAMP receptor protein (CRP) (39).
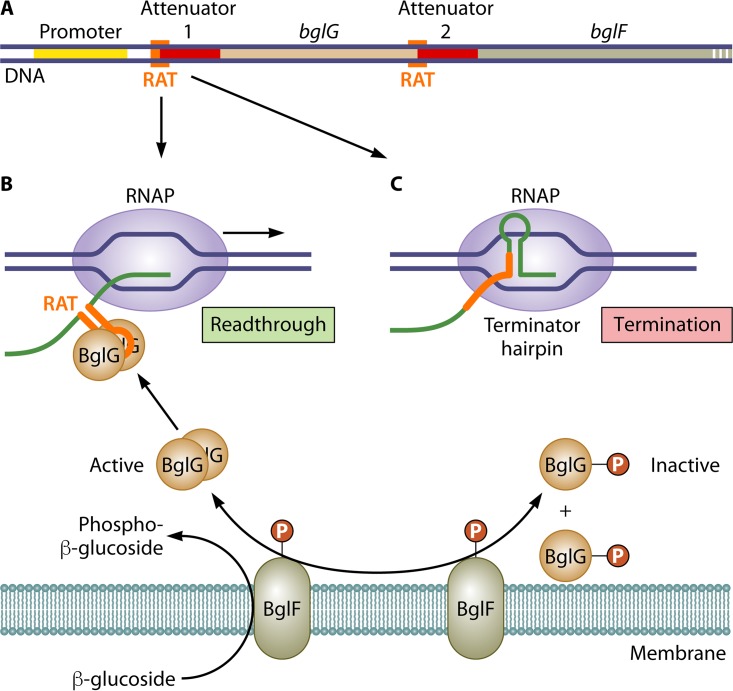
FIG 3.
Basic mechanism of BglG-mediated attenuation control of bgl operon expression in E. coli. (A) Regulatory elements of the bgl operon including the attenuators and partially overlapping RAT sites preceding the bglG and bglF genes. (B) In the presence of a β-glucoside substrate, BglF-catalyzed dephosphorylation of monomeric BglG∼P allows formation of active (i.e., RNA-binding-proficient) BglG dimers. These dimers bind to and stabilize RAT secondary structures in the nascent leader transcript that prevent terminator hairpin formation at attenuators 1 and 2. The result is readthrough transcription at each site. (C) In the absence of β-glucosides, BglF-catalyzed phosphorylation of dimeric BglG produces monomeric and membrane-bound BglG∼P that is unable to bind the nascent bgl transcript and disrupt terminator hairpin formation. Consequently, most transcripts initiated at the bgl promoter are terminated at either attenuator 1 or 2, and only minimal levels of BglG, BglF, BglB, and BglH are synthesized.
According to the basic model for β-glucoside-inducible attenuation control, in the absence of a β-glucoside substrate, BglF recruits BglG to the cell membrane, where its ability to bind RNA is inactivated by phosphorylation (40). With BglG sequestered, most transcripts initiated at the activated bgl promoter are terminated at an intrinsic transcription terminator (bgl attenuator 1) located just upstream of the bglG gene (41–43) (Fig. 3C). Additional premature termination of bgl transcripts occurs at a second intrinsic transcription terminator (bgl attenuator 2) located between the bglG and adjacent bglF genes (Fig. 3A). Consequently, only minimal levels of BglG and BglF (as well as BglB and BglH) are synthesized. On the other hand, in the presence of β-glucosides, BglF acquires phosphate residues for sugar phosphorylation from BglG∼P, which releases unphosphorylated BglG into the cytoplasm, where it dimerizes (40). Dimeric BglG then binds independently to specific transcript sites called RAT (ribonucleic antiterminator) sites that precede and partially overlap each attenuator-specified terminator hairpin (Fig. 3A), thereby preventing termination at attenuators 1 and 2 and allowing expression of the operon and β-glucoside catabolism (Fig. 3B). The basic model became a bit more complicated with the discovery that BglG is phosphorylated at a second site and that this phosphorylation is required for dimeric BglG binding to RNA. Phosphorylation at the second site is catalyzed by the PTS enzyme HPr, which is not specific for a particular sugar, and its paralog, FruB. Accordingly, when other PTS sugars become available in addition to β-glucosides, second-site phosphorylation of BglG is restricted and BglG activity is suppressed (43).
Overall, the critical feature in attenuation control of bgl expression is the binding of dimeric BglG to RAT sites in the nascent transcript, which physically blocks formation of attenuator-encoded terminator hairpins. RAT site sequences form an alternative RNA hairpin that is stabilized by the binding of BglG (Fig. 3B) (41, 44, 45). The RNA-binding activity of BglG is regulated through antagonistic phosphorylation events: one ensures that high levels of the enzymes required for β-glucoside catabolism are produced only when β-glucoside substrates are available, and the other couples bgl expression to the general carbohydrate supply. Subsequent to the discovery of attenuation control of bgl expression in E. coli, several analogous regulatory mechanisms for operons involved in PTS-transported sugar catabolism and employing a BglG-like antiterminator protein were found in other bacteria, especially Gram-positive bacteria (46). Additionally, a steadily growing number of other attenuation control mechanisms that rely on unique RNA-binding proteins to control transcription termination have been discovered. Two of the best studied are TRAP and PyrR of Bacillus subtilis.
TRAP and PyrR of B. subtilis
Expression of the B. subtilis trpEDCFBA (or trp) operon, which contains six of the seven genes needed for Trp biosynthesis, is regulated over an approximately 100-fold range by Trp-sensitive transcription attenuation (47). A 203-bp leader region can specify two alternative RNA hairpins, an antiterminator (A:B) hairpin and a downstream (C:D) terminator hairpin specified by an intrinsic transcription terminator (the trp attenuator) (Fig. 4). The two hairpins are mutually exclusive due to a 4-base overlap of leader transcript segments B and C (48). An RNA-binding protein called TRAP (trp RNA-binding attenuation protein) controls formation of the alternative hairpins. TRAP is composed of 11 identical 75-amino-acid subunits, each encoded by the mtrB gene (49), which assemble into a β-wheel with a large central hole (50). TRAP is activated to bind RNA by positive cooperative binding of up to 11 molecules of Trp (51). The TRAP binding site in the leader transcript consists of 11 repeats of GAG or UAG separated by 2- or 3-nt nonconserved spacers (52). The triplet repeats are located between leader residues 36 and 91, which overlap segment A and the 5′ portion of segment B of the antiterminator hairpin (Fig. 4A). When bound to TRAP. the leader transcript wraps around the outside of the protein ring (50, 52).
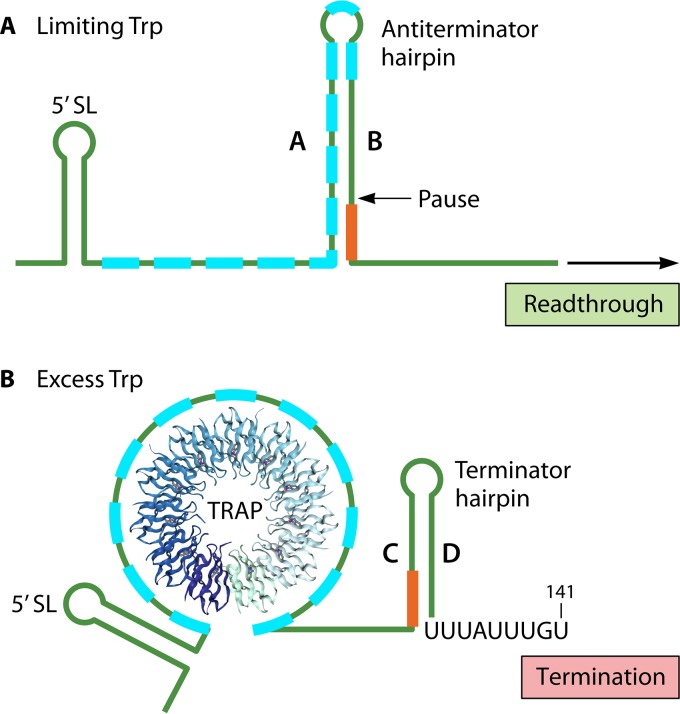
FIG 4.
TRAP-mediated attenuation control of trp expression in B. subtilis. The leader transcripts shown include the 11 GAG or UAG sequences (blue boxes) to which TRAP subunits can bind, the 5′ stem-loop (5′ SL) and the transcription pause site (Pause) that enhance TRAP binding, the A:B antiterminator hairpin, the C:D terminator hairpin, and the 4-base overlap of transcript segments B and C (red line). (A) Under conditions of limiting Trp, TRAP is not activated to bind the leader transcript and the A and B segments form the antiterminator hairpin as soon as they are synthesized. Formation of the antiterminator hairpin precludes terminator hairpin formation, thereby allowing readthrough transcription of the trp structural genes. (B) Under conditions of excess Trp, TRAP is activated to bind the (G/U)AG repeat region, thereby blocking formation of the antiterminator hairpin. Transcript segments C and D then form the terminator hairpin, and transcription is terminated before the structural genes.
According to the standard model for attenuation control (44), transcription initiation at the B. subtilis trp promoter occurs constitutively, and transcript elongation proceeds into the leader region. Under conditions of limiting Trp, TRAP is not activated and does not bind the nascent leader transcript, which permits the antiterminator hairpin to form as soon as segments A and B are synthesized. As transcription proceeds downstream, formation of the terminator hairpin is precluded, allowing transcription into and expression of the structural genes of the operon (Fig. 4A). Alternatively, when cells are growing in the presence of excess Trp, TRAP is activated and binds the (G/U)AG repeat region of the nascent transcript, inhibiting formation of the A:B antiterminator hairpin. This inhibition permits formation of the alternative C:D terminator hairpin, which leads to premature leader transcript termination at the trp attenuator and suppression of trp operon expression (Fig. 4B).
Over the years, the standard model has been expanded to include regulatory roles for leader features, such as a 5′ stem-loop (53) and NusA- and NusG-stimulated transcription pausing (54, 55), which enhance TRAP binding, and a role for an anti-TRAP protein, the production of which is induced by uncharged tRNATrp (56, 57). More recent studies even suggest a second role for TRAP in attenuation control. The trp leader terminator hairpin is only 40% G+C rich, which should make the trp attenuator a weak transcription terminator. However, transcription termination at the trp attenuator is highly efficient in vivo. To account for this apparent contradiction, it has been proposed that TRAP, in addition to its role in controlling leader hairpin formation, interacts with RNAP and perhaps other cellular factors to directly stimulate transcription termination at the trp attenuator (58, 59). Although not directly related to attenuation control, TRAP also represses translation initiation of trpE and several other genes involved in tryptophan metabolism (60).
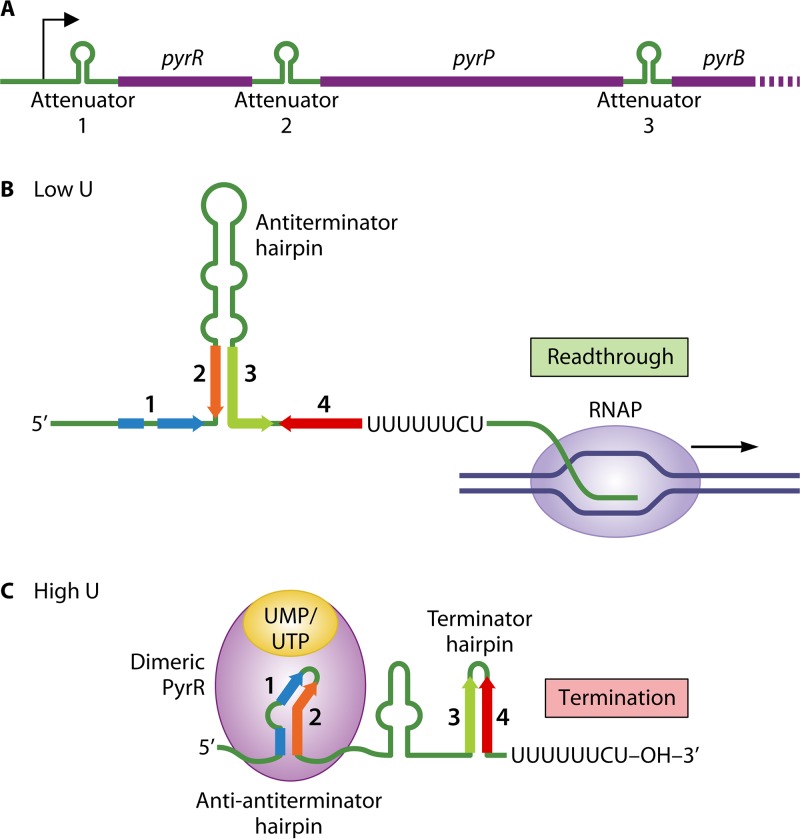
Like TRAP, the PyrR protein of B. subtilis is an RNA-binding protein that regulates transcription termination, but their mechanisms of action are quite distinct. PyrR regulates expression of the pyrRPBC(AA)(AB)KDFE (or pyr) operon, the last eight genes of which encode the six enzymes required for de novo synthesis of UMP (29, 61). The first and second genes of the operon, pyrR and pyrP, encode PyrR and uracil permease, respectively. The operon also contains three similar-strength intrinsic transcription terminators that function as attenuators (numbered 1, 2, and 3), the first located in the leader region and the second and third in untranslated regions between pyrR and pyrP and pyrP and pyrB, respectively (Fig. 5A). The RNA specified by all three of these untranslated regions is also capable of folding into a large antiterminator hairpin that sequesters residues in the 5′ segment of each downstream terminator hairpin, thereby preventing termination at each attenuator (Fig. 5B). Furthermore, at each location, the RNA embedded in the 5′ half of each antiterminator can form a third hairpin called the antiantiterminator, which precludes antiterminator formation and thus allows termination at the attenuator (62). Each antiantiterminator hairpin includes structures and conserved sequences that constitute a PyrR binding site (Fig. 5C) (63, 64). PyrR binding to these sites is stimulated by uridine nucleotides and phosphoribosyl pyrophosphate (PRPP) and antagonized by guanosine nucleotides, all at physiological concentrations (63, 65, 66). Binding of these effectors also affects the aggregation state of PyrR, with activator and inhibitor binding resulting in dimer and tetramer formation, respectively (67).
FIG 5.
PyrR-mediated attenuation control of pyr operon expression in B. subtilis. (A) Regulatory elements of the pyr operon, including the attenuators preceding the pyrR, pyrP, and pyrB genes. For simplicity, transcription at only one of the attenuator regions is shown. (B) Under conditions of low levels of uridine nucleotides and inactive PyrR, formation of the antiterminator hairpin (including segments 2 and 3) is favored in the nascent transcript, which prevents transcription termination and allows readthrough transcription into the downstream gene(s). (C) At high levels of uridine nucleotides, UMP/UTP-bound dimeric PyrR binds to and stabilizes the antiantiterminator hairpin (including segments 1 and 2), which promotes formation of the attenuator-encoded terminator hairpin (including segments 3 and 4) and immediate transcription termination. See the text for additional details.
In the model for regulation, transcription of the pyr operon is initiated at a constitutive promoter, and expression of the pyrimidine nucleotide-biosynthetic genes is negatively regulated over a ≥200-fold range by uridine nucleotide-sensitive attenuation (68). When intracellular levels of uridine nucleotides (e.g., UMP and UTP) are low, PyrR forms tetramers (or hexamers) that do not bind RNA (69). In this case, antiterminator hairpin formation is favored in the nascent transcript, preventing premature termination of pyr transcripts and allowing production of enzymes needed to overcome pyrimidine nucleotide deficiency (Fig. 5B). At high levels, uridine nucleotides bind PyrR and promote the formation of dimers that rapidly bind to and stabilize each antiantiterminator hairpin (Fig. 5C). This event precludes formation of the antiterminator hairpins, thereby permitting terminator hairpin formation and transcription termination at each attenuator. Consequently, there is no unnecessary increase in pyrimidine nucleotide-biosynthetic capacity.
It was also observed that guanosine (e.g., GMP and GTP) and uridine nucleotides compete for the same PyrR binding site (65, 67). Thus, a high ratio of guanine to uridine nucleotides results in guanine nucleotide-bound PyrR, which forms non-RNA-binding tetramers. In this case, expression of the pyr operon is increased. This effect provides a means of coordinating the rate of pyrimidine nucleotide biosynthesis with the size of intracellular guanine nucleotide pools. Additionally, PRPP stimulates PyrR dimerization and binding to antiantiterminator hairpins, which leads to reduced pyr operon expression. This response makes physiological sense, because an increase in the intracellular PRPP level is a consequence of guanosine nucleotide shortage and hence a diminished demand for pyrimidine nucleotide synthesis. One cautionary note about the PyrR regulatory models described above is that correlation between the aggregation state of PyrR and RNA binding is just the current hypothesis; it has not been rigorously demonstrated.
Another interesting aspect of PyrR is that it possesses uracil phosphoribosyltransferase activity; however, the activity is weak and apparently plays a negligible role in uracil salvage in vivo (70). A close structural resemblance of GMP-bound PyrR to an evolutionarily distant hypoxanthine-guanine phosphoribosyltransferase suggests that the two enzymes share ancestry (65). Such a relationship might explain the ability of PyrR to bind guanine nucleotides. It seems clear, though, that PyrR was derived from a phosphoribosyltransferase that later acquired the ability to bind RNA. Finally, homologs of PyrR have been identified in several hundred distinct bacterial species. In many cases, these homologs appear to be involved in transcription attenuation (29).
Although the discovery of attenuation control based on RNA-binding proteins was a major advance in the field, the next class of control mechanisms revolutionized the study of gene regulation. These mechanisms relied on a new type of RNA called a riboswitch.
CLASS III: RIBOSWITCH-MEDIATED TRANSCRIPTION ATTENUATION
Definition of a Riboswitch
A riboswitch is broadly defined as a cis-acting regulatory segment of an mRNA that directly senses a physiological signal, which causes a change in RNA structure that impacts downstream gene expression (71). In many instances, the change in RNA structure controls the formation of an attenuator-specified terminator hairpin (72). When the term riboswitch was first coined by Ron Breaker in 2002, it was defined more narrowly to mean a regulatory segment of mRNA that specifically binds a small-molecule cellular metabolite (73). However, the first example of a riboswitch that meets the broad definition of the term was discovered a decade earlier, and in this case, the mRNA-binding ligand was a tRNA (74–76).
tRNA-Binding Riboswitches
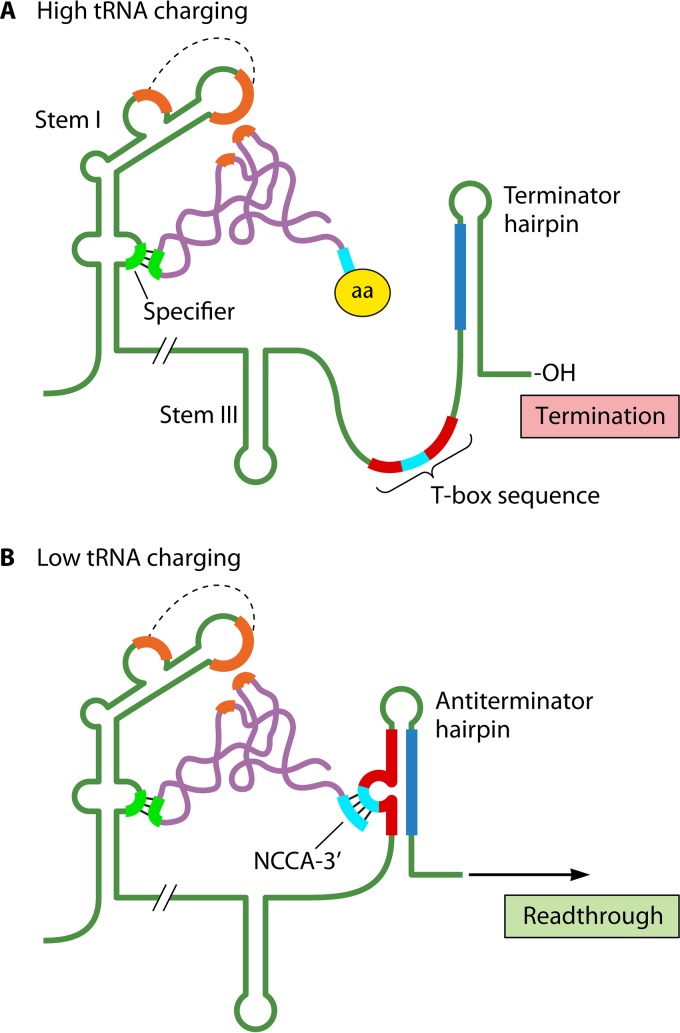
In 1992, analysis of the B. subtilis tyrS operon, which encodes tyrosyl-tRNA synthetase, indicated that tyrS expression was regulated by tyrosine-sensitive transcription attenuation, with conditional termination occurring at an intrinsic terminator located near the end of an ∼300-bp leader region. Transcription past the attenuator was dependent on a 14-base transcript sequence, designated the T box, which was located immediately upstream of the terminator hairpin (Fig. 6A) (75). It was quickly realized that T-box sequences and downstream intrinsic terminators were present in long leader regions of numerous aminoacyl-tRNA synthetase and amino acid-biosynthetic operons in different Bacillus species, suggesting a common attenuation control mechanism (74). Additional analysis revealed that the leader transcripts of each of these operons were capable of forming three conserved stem-loop structures (I, II, and III) that preceded the T box. Each stem I contains a bulge region that includes a triplet sequence called the specifier sequence, which is actually a codon that specifies the amino acid associated with the operon being regulated (Fig. 6A). This result suggested that the specifier sequence is part of a binding site for a cognate tRNA, which was confirmed by further experimentation (77). Inspection of sequences downstream of stem III showed that the region could form an antiterminator hairpin by base pairing a segment of the T-box sequence with residues in the upstream portion of the terminator hairpin (Fig. 6B). In each antiterminator hairpin, the central 7 bases of the T-box sequence formed a bulge that included the sequence UGGN, which is complementary to the NCCA sequence present at the 3′ acceptor ends of cognate tRNAs (Fig. 6B). Interaction of these 4-base sequences was subsequently shown to provide a second leader binding site for an uncharged—but not a charged—cognate tRNA (77).
FIG 6.
T-box mechanism. (A) High-tRNA charging. The anticodon (chartreuse) of a charged cognate tRNA (purple) base pairs with the specifier sequence (chartreuse) in stem I of the leader RNA. The amino acid (aa) at the 3′ end of the tRNA prevents its interaction with the T-box sequence (red with central 7 bases in cyan), resulting in terminator hairpin formation and transcription termination within the leader region. (B) Low-tRNA charging. The anticodon of an uncharged cognate tRNA base pairs with the specifier sequence as described above, and the sequence NCCA at the 3′ end of the tRNA (cyan) hybridizes to a complementary sequence (cyan) within the central 7 bases of the T-box sequence. The latter interaction stabilizes formation of the antiterminator hairpin, which precludes terminator hairpin formation and allows transcription into the structural gene(s). Also shown in both panels is a recently discovered stacking interaction (orange) between the elbow of the cognate tRNA and the stem I platform. The dashed lines indicate interacting sequence motifs. For clarity, stem II in the leader transcript was omitted in both panels.
Based on the features described above, a novel mechanism to regulate expression of amino acid-related operons in Bacillus and closely related species was proposed (74, 77). According to the model, when the cell has ample levels of a particular amino acid, its cognate tRNA is efficiently aminoacylated. This charged tRNA can bind to the specifier sequence but not to the T-box bulge, which prevents stabilization of the antiterminator hairpin. Consequently, the terminator hairpin forms, and the leader transcript is terminated at the attenuator (Fig. 6A). Conversely, when the cell is starved for the amino acid, cognate uncharged tRNA accumulates and interacts with both tRNA-binding sites of the leader region. This binding stabilizes formation of the antiterminator hairpin (78), which allows transcription past the attenuator and into the structural genes of the operon (Fig. 6B). In this model, the regulatory mechanism monitors the ratio of charged to uncharged tRNA and, in response, adjusts the level of associated operon expression to meet the physiological needs of the cell (71). This mechanism has now been shown to be widely used by members of the phyla Firmicutes and Actinobacteria, with the number of examples per genome ranging from 1 to 40 (79, 80). More recent studies have focused on identifying additional tRNA-leader transcript contacts and contacts that are tRNA specific (Fig. 6) (81–84).
Small-Molecule-Binding Riboswitches
Most examples of riboswitches bind molecules much smaller than a tRNA, with the first of these identified about 20 years ago (85–87). To date, more than 40 different classes of riboswitches that bind a variety of metabolites, coenzymes, signaling molecules, and inorganic ions have been discovered (88–90). These riboswitches control the formation of alternative leader transcript secondary structures that often affect intrinsic transcription termination, although they can also control translation initiation and RNA stability and splicing (91). When riboswitches control transcription termination, they dictate the formation of an RNA secondary structure that either includes or excludes the attenuator-encoded terminator hairpin. Leader regions that include riboswitches are atypically long (usually >200 nt) and contain aptamers that bind a target molecule with high selectivity and specificity.
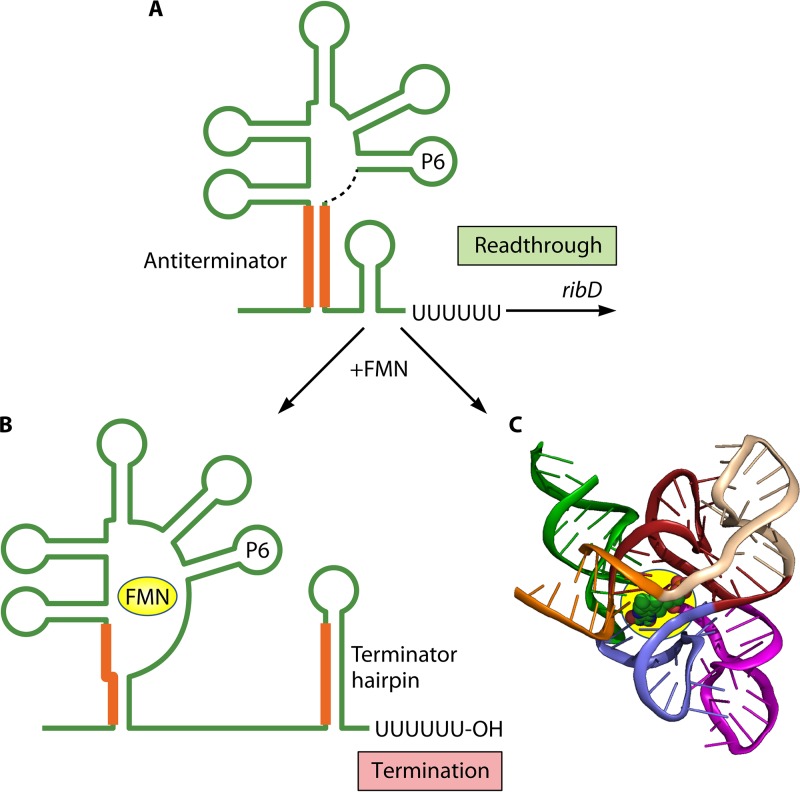
One of the earliest examples of a small-molecule-binding riboswitch was the mechanism controlling expression of the riboflavin-biosynthetic operon ribDEAHT in B. subtilis (92). Riboflavin is a precursor of flavin mononucleotide (FMN), and ribDEAHT expression is controlled by the binding of FMN to an aptamer (called an rfn box) formed by the ribD leader transcript. When FMN levels are low, it fails to bind its aptamer, and the ribD leader transcript adopts a secondary structure that includes an antiterminator stem (Fig. 7A). The downstream segment of this stem is shared with a potential attenuator-encoded terminator hairpin located further downstream. Therefore, formation of the antiterminator stem precludes formation of the terminator hairpin, allowing readthrough transcription into the structural genes of the operon. At high levels, FMN binds its aptamer, which promotes the formation of an alternative leader transcript secondary structure that includes the terminator hairpin (Fig. 7B). As a result, transcription is terminated upstream of the structural genes of the operon under conditions of ample riboflavin and FMN. FMN riboswitches like the one described above are now known to control expression of genes responsible for the biosynthesis and transport of riboflavin and FMN in many bacteria (88). In some cases, the structure of FMN bound by its aptamer has been determined (Fig. 7C).
FIG 7.
Control of riboflavin-biosynthetic operon ribDEAHT expression by an FMN-binding riboswitch in B. subtilis. (A) In the absence of FMN binding to the ribD leader transcript, the RNA forms a secondary structure that includes an antiterminator stem (orange lines). Because the downstream segment of this stem is shared with a potential attenuator-encoded terminator hairpin located further downstream, formation of the antiterminator stem permits readthrough transcription into the structural genes. The dashed black line immediately downstream of the stem-loop labeled P6 represents a long segment of unstructured leader RNA. (B) When FMN binds its aptamer, the leader transcript assumes a secondary structure that includes the terminator hairpin, resulting in transcription termination at the attenuator. (C) Crystal structure of the aptamer domain of an FMN riboswitch bound to its ligand (140).
Pioneered by the Breaker laboratory, a productive strategy for discovering small-molecule-binding riboswitches has been the use of computer algorithms to compare sequences of noncoding regions of bacterial genomes and the RNA structures that they encode (88, 93–98). This method uncovers variants of known riboswitches that can be subsequently verified by genetic, biochemical, and biophysical studies. The limitation of this method is that it fails to detect rare classes of riboswitches, and it is possible that many undiscovered riboswitches fall into this category (99).
CLASS IV: TRANSCRIPTION ATTENUATION WITHOUT AN RNA-BINDING ELEMENT
A Class of One, So Far, and Reiterative Transcription
All the examples of attenuation control described above employ an RNA-binding element; however, there is one known example where this is not the case. In this example, attenuation control relies on conditional (but permanent) modification of the leader transcript primary sequence by a noncanonical form of transcription called reiterative transcription (100). This reaction is defined by repetitive addition of nucleotides to the 3′ end of a nascent transcript due to one or more rounds of a 1-base upstream shift of the transcript without movement of the DNA template. Accordingly, the same nucleotide in the template specifies multiple (or extra) residues in the transcript (101). Reiterative transcription occurs primarily within a ≥3-residue homopolymeric tract in the DNA template and can involve the repetitive addition of any nucleotide substrate of RNAP.
pyrG Operon of B. subtilis
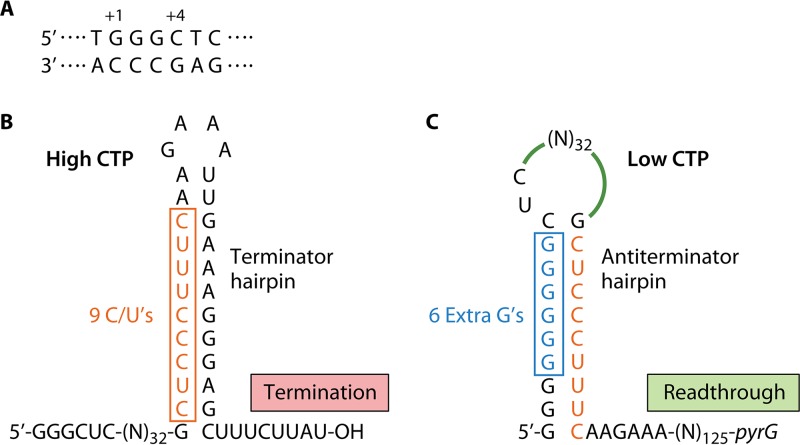
Expression of the pyrG operon of B. subtilis, which encodes the pyrimidine-biosynthetic enzyme CTP synthetase, is regulated over a ≥20-fold range by a CTP-sensitive attenuation control mechanism that targets an intrinsic transcription terminator near the downstream end of a 189-bp leader region. The mechanism that renders the pyrG attenuator conditional relies on reiterative transcription that involves repetitive addition of G residues during transcription initiation (102). This reaction occurs during transcription of the pyrG initially transcribed region (Fig. 8A), which specifies the sequence 5′-GGGCU at the start of the pyrG transcript. Another key control element is an attenuator-encoded terminator hairpin in which most of the upstream segment of the stem consists of the 9-residue pyrimidine tract 5′-CUCCCUUUC (Fig. 8B), all of which can base pair with a poly(G) tract of RNA (Fig. 8C) (103).
FIG 8.
Reiterative transcription-mediated attenuation control of pyrG expression in B. subtilis. (A) The pyrG initially transcribed region with positions +1 and +4 indicated. (B) When CTP levels are high, the nascent pyrG leader transcript is a faithful copy of the DNA template, including an attenuator-specified terminator hairpin and an adjacent U-rich tract that cause immediate transcription termination. Atypically, the upstream segment of the terminator hairpin contains a track of 9 pyrimidine residues. (C) When CTP levels are low, transcription pausing at position +4 C promotes reiterative transcription and the addition of extra (6 are shown) G residues prior to the resumption of normal transcription. When the upstream segment of the terminator hairpin is synthesized, it immediately base pairs with the poly(G) tract at the 5′ end of the transcript to form an antiterminator hairpin. This structure precludes terminator hairpin formation and transcription termination at the attenuator, allowing transcription of the pyrG gene.
According to the model for regulation (102, 104), when the intracellular level of CTP is high (reflecting ample pyrimidine availability), transcription of the pyrG leader region is rapid and produces transcripts that are copied base for base from the DNA template. However, these transcripts are efficiently terminated at the pyrG attenuator (Fig. 8B). Thus, when CTP is abundant, transcription of the pyrG gene is repressed. On the other hand, when the intracellular level of CTP is low (reflecting pyrimidine limitation), transcription of the pyrG leader region pauses at position +4 C (i.e., after the synthesis of the 5′-GGG transcript) because of insufficient substrate. The time provided by this pause allows the nascent transcript to slip upstream relative to the DNA template (i.e., into the 3′-CCC tract), which directs the addition of an extra G residue to the transcript. This process can be repeated up to at least 10 times, until eventually a C residue is added to the 3′ end of the transcript by canonical transcription. Transcript elongation then continues normally until RNAP transcribes the attenuator sequence specifying the upstream segment of the terminator hairpin, which includes the long pyrimidine tract. This tract immediately base pairs with the poly(G) tract at the 5′ end of the transcript to form an antiterminator hairpin (Fig. 8C). Formation of a highly effective antiterminator hairpin requires a minimum of three extra G residues in the poly(G) tract (103, 105). As RNAP continues to elongate the pyrG transcript, the antiterminator hairpin precludes terminator hairpin formation, which allows the synthesis of full-length transcripts. Translation of these transcripts produces more CTP synthetase, which is needed to overcome the CTP deficiency. Although the model is described in binary terms of high and low intracellular CTP concentrations, regulation can occur continuously over a range of CTP concentrations that affect the extent of pausing at position +4 (104).
Although attenuation control of pyrG expression in B. subtilis is the only example in its class to date, it is reasonable to expect that analogous mechanisms will be discovered. One difficulty in this process is that there are no computer algorithms that identify sites of reiterative transcription. Reiterative transcription that might be involved in attenuation control could be detected by the presence of extra nucleotides in the leader regions of sequenced RNA transcripts. However, the discovery of extra nucleotides is typically considered a sequencing error and dismissed as unimportant. There is also the possibility that attenuation control without an RNA-binding element could rely on something other than reiterative transcription, although that “something” remains to be discovered.
CLASS V: TRANSCRIPTION ATTENUATION AT RHO-DEPENDENT TERMINATORS
Finding Rho-Dependent Terminators, Their Expanding Role in Gene Regulation, and Familiar Regulators
Unlike intrinsic terminators, Rho-dependent terminators have been difficult to find because they lack a consensus sequence (6). For example, the only requirements for a rut site are that it is long enough to accommodate all six Rho subunits; that it is rich in cytidines, which preferentially bind Rho; and that it is free of ribosomes and extensive secondary structure that can preclude Rho binding (6, 106). Consequently, the discovery of a role for Rho in gene regulation was initially a slow process. However, with the advent in the last decade of high-throughput sequencing techniques capable of identifying the 3′ ends of RNA transcripts, the pace of discovery has quickened dramatically (107). Today, we recognize a variety of regulatory roles for Rho, including control of individual gene expression (106), as a housekeeper of genome-wide transcription (108, 109) and genome integrity (110), and even in the determination of cell fate (111).
Perhaps not surprisingly, the regulatory factors that mediate transcription attenuation at Rho-dependent terminators are often the same as those involved in making intrinsic terminators function conditionally. The difference is the target: the terminator hairpin of intrinsic attenuators and primarily the rut site for Rho-dependent attenuation. As with the first examples of attenuation involving an intrinsic terminator, the first example of Rho-dependent transcription attenuation employed ribosome positioning on the leader transcript to control termination, but with interesting twists.
Ribosome-Mediated Rho-Dependent Attenuation
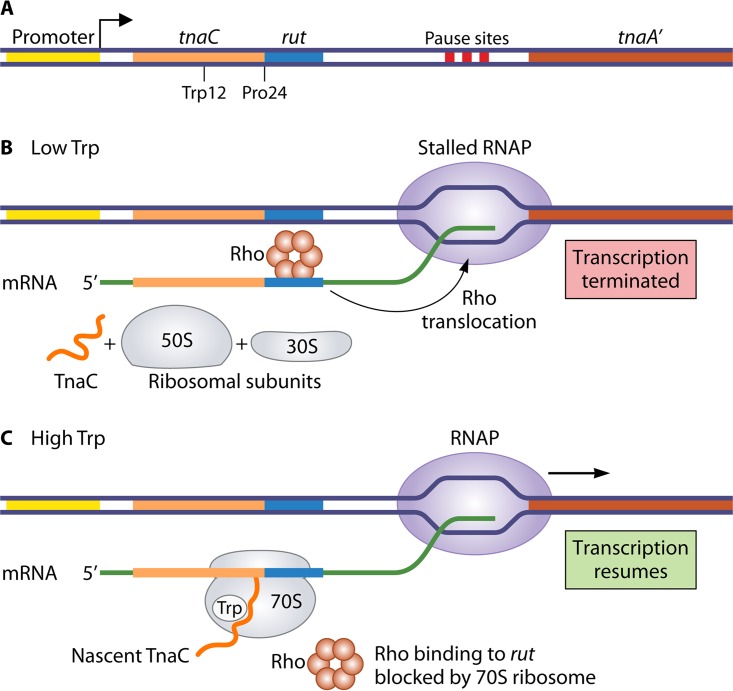
The first well-studied example of Rho-dependent attenuation was the mechanism controlling Trp-induced (and also cAMP/CRP-dependent) expression of the Trp-degradative tnaCAB (or tna) operon of E. coli (112). The 320-bp leader region of the operon includes three features required for regulation: a 24-codon ORF designated tnaC, a rut site, and transcription pause sites, all of which precede the structural genes encoding tryptophanase (tnaA) and tryptophan permease (tnaB) (Fig. 9A). According to an early regulatory model, after synthesizing the tnaC segment of the nascent tna transcript, RNAP pauses in the tnaC-tnaA intergenic space to allow a ribosome to begin translating the leader ORF (113). When intracellular levels of Trp are low, translation of the leader region is completed, and the translating ribosome is released from the leader transcript, thereby exposing the rut site. Rho then binds to this site and translocates downstream until it reaches RNAP stalled at a pause site, where it terminates transcription (Fig. 9B). On the other hand, when Trp levels are high, the ribosome stalls at a proline codon at position 24 of tnaC and blocks the rut site. Consequently, Rho cannot bind to the leader transcript, and the paused RNAP can resume transcription and proceed through the structural genes (Fig. 9C). The ribosome stalled at tnaC codon 24 is eventually released and recycled.
FIG 9.
Mechanism of ribosome-mediated Rho-dependent attenuation control of tna expression in E. coli. (A) Organization of the tna promoter-leader region, including tnaC, with residues Trp12 and Pro24 marked, and the following spacer region that contains a rut site and downstream transcription pause sites. Following transcription through most of the leader region and RNAP stalling at a pause site, a ribosome begins translating the tnaC cistron. (B) When the level of Trp is low, the translating ribosome completes synthesis of the TnaC polypeptide and dissociates from the transcript. Rho factor then binds the exposed rut site, translocates downstream until it contacts the stalled RNAP, and terminates transcription. (C) When the level of Trp is high, it binds to the nascent TnaC polypeptide while it is within the exit channel of the translating ribosome. This interaction causes the ribosome to stall at codon Pro24, which masks the rut site and prevents Rho-dependent transcription termination. Eventually, the stalled RNAP resumes transcription, allowing expression of the proteins capable of catabolizing excess Trp.
This model describes how tna expression occurs only when intracellular levels of Trp are high, but it does not explain how Trp levels control the extent of translation in the leader ORF. The answer was a huge surprise. Further studies revealed that when translation of the leader region reaches codon Pro24, particular residues of the nascent TnaC peptide (e.g., Trp12 and Ile19) and the 23S rRNA form a binding pocket for free Trp within the exit channel of the ribosome (114–116). Binding of Trp to this pocket causes the ribosome to stall, thereby masking the rut site and preventing premature Rho-dependent termination within the leader region (Fig. 9C). The TnaC peptide was an early example of regulatory peptides called ribosome arrest peptides, which induce translational arrest to control either transcription or translation of downstream genes in the same operon. Today, there are numerous examples of ribosome arrest peptides that are involved in a variety of regulatory mechanisms (117, 118).
A recent and equally interesting example of ribosome-mediated Rho-dependent attenuation is the mechanism controlling expression of the mgtA operon of Salmonella, which encodes the Mg2+ transport protein MgtA and is regulated over a 100-fold range by the intracellular level of Mg2+ (119–121). The leader region of the operon includes a short, proline-rich ORF, mgtL, and mutually exclusive secondary structures, only one of which has an exposed rut site. Rapid and complete translation of mgtL permits formation of the secondary structure with the exposed rut site, resulting in Rho-dependent termination at a transcription pause site preceding the mgtA gene (122). Efficient translation of mgtL, specifically the included proline codons, is thought to require high concentrations of Mg2+. This putative requirement apparently reflects the involvement of Mg2+-dependent components of the translation machinery that are uniquely involved in translation of proline codons (123, 124). Thus, when Mg2+ levels are high, mgtA expression is repressed. On the other hand, when Mg2+ levels are low, the proline codons in mgtL presumably present an impediment to translation. The resulting slow translation directs formation of the leader secondary structure that occludes the rut site, thereby preventing Rho-dependent termination and allowing mgtA expression. This mechanism shares many key features with the archetypical trp attenuation control mechanism of E. coli (Fig. 1). In both cases, the efficiency of translating control codons in a leader ORF dictates transcript secondary structures that control the availability of an RNA element that is essential for transcription termination (i.e., a rut site or terminator hairpin).
It should be noted that a somewhat different model for regulation of mgtA expression proposes a different role for Mg2+ in controlling the formation of alternative transcript secondary structures. Instead of the indirect role proposed above, this alternative model proposes that it is the extent of direct Mg2+ binding to the leader transcript that dictates secondary-structure formation. In other words, the leader transcript is a multi-ion Mg2+-sensitive riboswitch (125). Consistent with this model, changes in Mg2+ concentrations within the physiological range alter the secondary structure of the mgtA leader transcript in ways that affect transcription elongation and termination, and the regulation by Mg2+ was recapitulated in vitro using only DNA, nucleotides, and RNAP (i.e., no ribosomes) (120). In addition, replacing the proline codons in mgtL with other sense codons did not eliminate the response to Mg2+ (126). Furthermore, ribosome profiling failed to show slow translation at proline codons at low Mg2+ concentrations (127). The differences in the two proposed mechanisms for attenuation control of mgtA expression remain to be resolved, although it is possible that both mechanisms are correct.
Protein- and Riboswitch-Mediated Rho-Dependent Attenuation
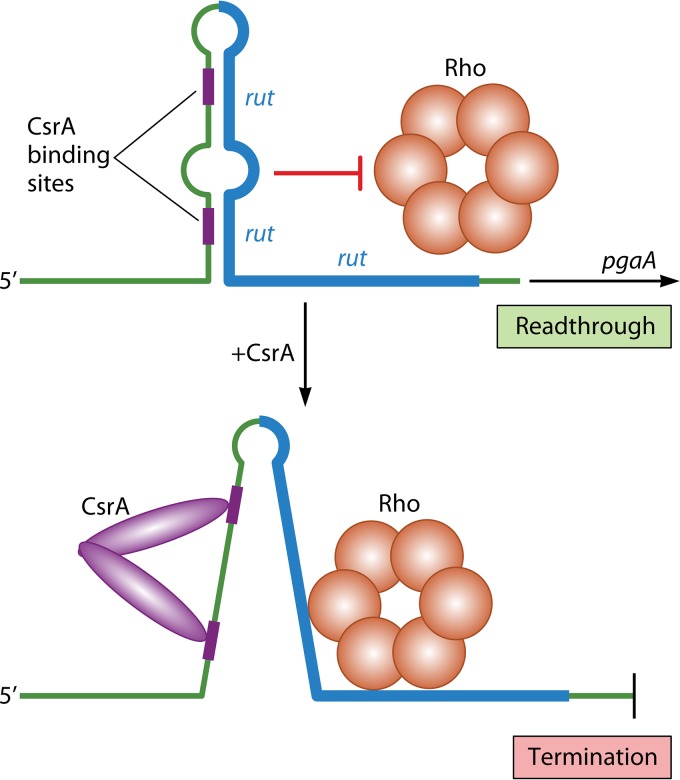
In addition to the ribosome, RNA-binding proteins and riboswitches can target rut site accessibility as a means of controlling Rho-dependent attenuation; however, there are presently few examples of these mechanisms. The archetype regulatory protein is CsrA (carbon storage regulator A) of E. coli, a key regulator of carbon and stationary-phase metabolism, biofilm formation, quorum sensing, and motility (128). One of the targets of CsrA binding is the leader transcript of the pgaABCD operon, which encodes proteins responsible for the production, modification, and export of the biofilm adhesin poly-β(1-6)-N-acetyl-d-glucosamine. CsrA binding negatively regulates pgaABCD expression and associated biofilm formation (in part) by remodeling leader transcript secondary structure to unmask a rut site, which permits Rho-dependent termination before transcription of the structural genes (129) (Fig. 10). This regulation allows biofilm formation to be linked to the carbon metabolism-related signals that control the RNA-binding activity of CsrA (128).
FIG 10.
Model for CsrA-induced Rho-dependent attenuation control of pgaABCD expression in E. coli. The upstream region of the pgaA leader transcript folds into an imperfect hairpin that includes two potential CsrA binding sites in the upstream segment of the stem. The downstream segment of the stem includes a large part of a rut site, and its inclusion in the secondary structure prevents Rho binding and allows transcription to read through into the structural genes of the pgaABCD operon. In the presence of active CsrA, a dimeric form of the protein binds both CsrA binding sites simultaneously and unfolds the leader hairpin to expose the entire rut site. This configuration allows Rho to bind to the rut site and cause transcription termination within the leader region.
The clearest example of riboswitch-mediated Rho-dependent attenuation appears to be the mechanism controlling expression of the ribB operon of E. coli, which encodes an enzyme required for riboflavin synthesis (125). The ribB leader transcript contains a highly conserved aptamer for FMN, which closely resembles the previously described rfn box formed by the ribD leader transcript of B. subtilis (Fig. 7). Binding of FMN to the ribB leader transcript results in a secondary structure that enhances Rho-dependent transcription termination within the leader region. Presumably, the FMN-induced secondary structure unmasks a rut site, but that important detail remains to be determined. The only other reported examples of riboswitch-mediated Rho-dependent attenuation are Mg2+-sensitive mechanisms that regulate expression of the mgtA, mgtCRB, and corA operons of Salmonella. Each operon encodes a separate Mg2+ uptake system. The riboswitch-mediated mechanism that controls mgtA expression was discussed above, and apparently, analogous mechanisms control mgtCBR and corA expression (120, 121, 130, 131).
RNA-Mediated Mechanisms of Rho-Dependent Attenuation and Regulatory Targets Other than the rut Site
Recent evidence suggests that small RNAs (sRNAs) capable of binding leader transcripts are important regulators of Rho-dependent attenuation (132). For example, such a mechanism is proposed to regulate expression of the E. coli rpoS gene, which encodes the stationary-phase sigma factor, σs (132). The small RNAs DsrA, ArcZ, and RprA apparently bind to the 5′ untranslated region of nascent mRNA and inhibit Rho activity, thereby stimulating rpoS expression. The mechanism of action of the sRNAs has not been defined for this or any other mechanism of this type, and it could be something other than controlling rut site availability. Additionally, it has been reported that cis-acting RNAP-binding aptamers (RAPs) are present in the leader regions of many E. coli transcripts and that these RAPs promote Rho-dependent transcription termination. The model is that RAPs promote termination either by uncoupling transcription and translation to expose rut sites or by enhancing transcription pausing (133).
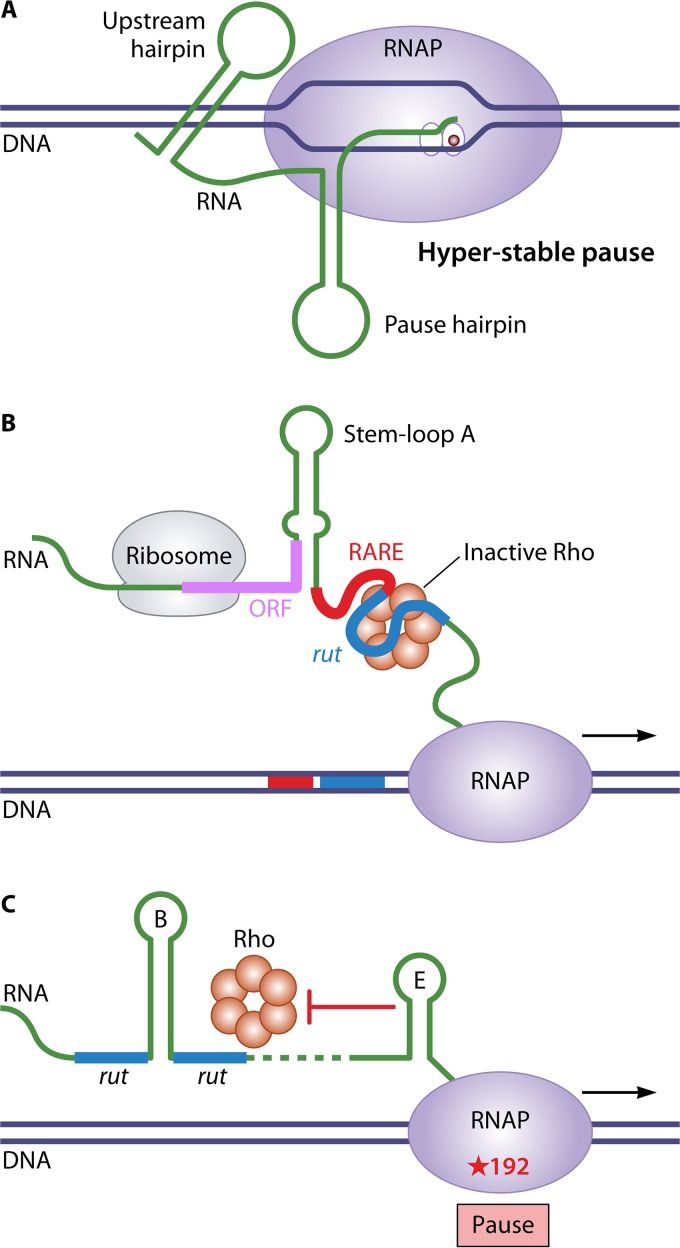
Direct evidence for regulatory targets other than the rut site comes from studies of Rho-dependent attenuation control of mgtA, mgtCBR, and corA expression in Salmonella. In each case, the leader transcript can adopt two mutually exclusive conformations that control rut site accessibility. In the case of mgtA, the secondary structure that exposes the rut site under conditions of high Mg2+ levels also contains elements that create a long-lived pause site that corresponds to the only Rho-dependent termination site in the leader region (Fig. 11A). Furthermore, this strong pause site is required for Mg2+-sensitive regulation of mgtA expression (122). In the case of mgtBCR, the leader transcript contains a Rho-antagonizing RNA element (RARE) that precedes the rut site. The RARE does not block Rho binding to the rut site but, when single stranded, traps Rho in an inactive complex. Under conditions of low Mg2+, inefficient translation of a short leader ORF promotes formation of a transcript secondary structure that exposes a single-stranded RARE, which can inactivate Rho and permit transcription through the mgtBCR structural genes (Fig. 11B) (134). In the case of corA, the leader transcript conformation that exposes the rut site also promotes pausing at a site >100 bases downstream at which Rho-dependent termination can occur. In addition, a stem-loop structure that forms downstream of the alternative secondary structures controls the location at which Rho-dependent termination occurs (135). The stem-loop blocks Rho’s access to RNAP when paused at position 192 but not at a subsequent pause site at position 240 (Fig. 11C). These results indicate that multiple steps in the process of Rho-dependent termination can be targets for gene regulation.
FIG 11.
Examples of non-rut site targets for regulating Rho-dependent attenuation. (A) Prolonged transcription pausing in the mgtA leader region. Under conditions of high Mg2+ levels, an upstream secondary structure in the leader transcript interacts with RNAP stalled at a hairpin-stabilized pause site to promote unique hyperstable transcription pausing. (B) RARE-mediated inactivation of Rho in the mgtCBR leader region. Under conditions of low Mg2+ levels, translation of a short leader ORF is inefficient, which promotes formation of a transcript secondary structure that exposes a single-stranded RARE immediately upstream of a rut site. When Rho binds the rut site, a single-stranded RARE can trap Rho in an inactive state, thereby permitting transcription through the mgtBCR structural genes. (C) Controlling the location of transcription termination in the corA leader region. Rho can bind the leader transcript at a discontinuous rut site interrupted by stem-loop B and translocate toward RNAP paused at position 192. However, stem-loop E prevents Rho from accessing RNAP at this site, thus promoting downstream Rho-mediated termination, typically at a pause site at position 240.
Finally, another type of Rho-dependent attenuation broadly referred to as Rho-dependent polarity is worth mentioning. In this mechanism, uncoupled transcription and translation within an early gene of an operon exposes latent ribosome-free rut sites that permit Rho-dependent termination before the transcription of downstream genes. Any regulatory scheme that prevents translation of an early cistron can be employed, although typically translation initiation is the target, using regulators such as riboswitches and sRNAs (136, 137). Rho-dependent polarity differs fundamentally from the other attenuation control mechanisms described above in that translation is the primary regulatory target and transcription termination occurs within the structural genes of an operon.
CONCLUSIONS
This review describes nearly a half century of seminal research on the regulation of bacterial gene expression based on conditional transcription termination. Perhaps the three things that best characterize these studies are as follows: there are a lot of these mechanisms, and they are everywhere; the mechanisms that render terminators conditional are incredibly diverse; and the discovery of each class of regulatory mechanisms was a huge surprise. For example, the discovery of the trp mechanism in E. coli certainly made it easier to discover the pyrBI mechanism in the same bacterium, because both mechanisms employ the position of a translating ribosome as the central regulatory feature. However, these studies did little to aid the discovery of attenuation mechanisms that rely on RNA-binding proteins. The same can be said about attenuation mechanisms based on riboswitches and reiterative transcription. Finally, could anyone have guessed that the central element of Rho-dependent attenuation control of the E. coli tna operon was binding of the amino acid Trp to a nascent peptide while it was still in the exit channel of the ribosome?
Although the mechanisms discussed in this review represent only a small sample of many interesting examples of attenuation control, they were chosen because they highlight both major and subtle elements of this type of gene regulation. Certainly, they represent each class of known attenuation control mechanisms. They also demonstrate the economy, versatility, dynamic range, and physiological importance of attenuation control mechanisms. Many of the mechanisms rely only on the basic machinery of transcription or transcription and translation, and none rely on DNA binding regulatory proteins, suggesting that attenuation control has an ancient origin (138). The mechanisms also take advantage of subtle effects. For example, many rely on the fact that RNA secondary structures form quickly, so that when there are competing structures, formation of the more upstream one is strongly favored. Additionally, the mechanism controlling tna expression in E. coli and the mechanisms controlling expression of the Mg2+ transporters in Salmonella appear to use uniquely hard-to-translate Pro codons as speed bumps to appropriately position a translating ribosome. Finally, how much more ingenious can riboswitches and reiterative transcription be?
The discovery of new transcription attenuation mechanisms will be easier in the future. Computational analyses can find virtually every intrinsic transcription terminator, common riboswitch, short ORF, and long leader region in any sequenced bacterial genome. Powerful new sequencing techniques and perhaps a reliable algorithm will make it much easier to find Rho-dependent terminators (107, 139). Even sites of reiterative transcription might be recognizable in genomic sequences in the near future. However, the history of attenuation control mechanisms strongly suggests that many of the most important new examples will not be so easy to define, because the regulatory elements will be unlike anything seen before. The elucidation of these new mechanisms, as with so many of the old ones, will require thorough examination of individual operons and researchers who are challenged by unexpected and puzzling results.
ACKNOWLEDGMENTS
I thank Paul Babitzke, Ron Breaker, Keith Corbino, Mike Gray, Eduardo Groisman, Tina Henkin, Patrick Lane, Bob Landick, Tony Romeo, and Bob Switzer for their help with the manuscript.
Work on bacterial gene regulation in my laboratory was supported by National Institutes of Health grants GM29466 and GM94466.
I declare no conflict of interest.
Biography

Charles L. Turnbough, Jr., received a Ph.D. in biochemistry from the University of Illinois, Champaign-Urbana, and studied bacterial gene expression as a postdoctoral fellow at the Berkeley and Davis campuses of the University of California. In 1980, he joined the faculty of the Department of Microbiology at the University of Alabama at Birmingham, where he has spent his entire academic career. His primary research interest during this time has been the regulation of gene expression in bacteria, focusing on the genes involved in pyrimidine nucleotide biosynthesis and salvage. These studies have resulted in the discovery of a number of new regulatory mechanisms, most of which employ only the basic machinery of transcription and translation. Several of these are described in this review. He is now a Professor Emeritus and actively continues his research.
REFERENCES
- 1.Beckwith J. 1996. The operon: an historical account, p. 1227−1231. In Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE (ed), Escherichia coli and Salmonella: cellular and molecular biology, vol 1 American Society for Microbiology, Washington, DC. [Google Scholar]
- 2.Yanofsky C. 1981. Attenuation in the control of expression of bacterial operons. Nature 289:751–758. doi: 10.1038/289751a0. [DOI] [PubMed] [Google Scholar]
- 3.Henkin TM, Yanofsky C. 2002. Regulation by transcription attenuation in bacteria: how RNA provides instructions for transcription termination/antitermination decisions. Bioessays 24:700–707. doi: 10.1002/bies.10125. [DOI] [PubMed] [Google Scholar]
- 4.Adhya S, Gottesman M. 1978. Control of transcription termination. Annu Rev Biochem 47:967–996. doi: 10.1146/annurev.bi.47.070178.004535. [DOI] [PubMed] [Google Scholar]
- 5.Rosenberg M, Court D. 1979. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet 13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- 6.Ray-Soni A, Bellecourt MJ, Landick R. 2016. Mechanisms of bacterial transcription termination: all good things must end. Annu Rev Biochem 85:319–347. doi: 10.1146/annurev-biochem-060815-014844. [DOI] [PubMed] [Google Scholar]
- 7.Malinen AM, Turtola M, Parthiban M, Vainonen L, Johnson MS, Belogurov GA. 2012. Active site opening and closure control translocation of multisubunit RNA polymerase. Nucleic Acids Res 40:7442–7451. doi: 10.1093/nar/gks383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. 2007. Structural basis for transcription elongation by bacterial RNA polymerase. Nature 448:157–162. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- 9.Martin FH, Tinoco I Jr. 1980. DNA-RNA hybrid duplexes containing oligo(dA:rU) sequences are exceptionally unstable and may facilitate termination of transcription. Nucleic Acids Res 8:2295–2299. doi: 10.1093/nar/8.10.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gocheva V, Le Gall A, Boudvillain M, Margeat E, Nollmann M. 2015. Direct observation of the translocation mechanism of transcription termination factor Rho. Nucleic Acids Res 43:2367–2377. doi: 10.1093/nar/gkv085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yanofsky C. 1988. Transcription attenuation. J Biol Chem 263:609–612. [PubMed] [Google Scholar]
- 12.Santangelo TJ, Artsimovitch I. 2011. Termination and antitermination: RNA polymerase runs a stop sign. Nat Rev Microbiol 9:319–329. doi: 10.1038/nrmicro2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goodson JR, Winkler WC. 2018. Processive antitermination. Microbiol Spectrum 6:RWR-0031-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kasai T. 1974. Regulation of the expression of the histidine operon in Salmonella typhimurium. Nature 249:523–527. doi: 10.1038/249523a0. [DOI] [PubMed] [Google Scholar]
- 15.Lee F, Yanofsky C. 1977. Transcription termination at the trp operon attenuators of Escherichia coli and Salmonella typhimurium: RNA secondary structure and regulation of termination. Proc Natl Acad Sci U S A 74:4365–4369. doi: 10.1073/pnas.74.10.4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oxender DL, Zurawski G, Yanofsky C. 1979. Attenuation in the Escherichia coli tryptophan operon: role of RNA secondary structure involving the tryptophan codon region. Proc Natl Acad Sci U S A 76:5524–5528. doi: 10.1073/pnas.76.11.5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landick R, Turnbough CL Jr, Yanofsky C. 1996. Transcription attenuation, p 1263–1286. In Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE (ed), Escherichia coli and Salmonella: cellular and molecular biology, vol 1 American Society for Microbiology, Washington, DC. [Google Scholar]
- 18.Roesser JR, Yanofsky CL Jr. 1988. Ribosome release modulates basal level expression of the trp operon of Escherichia coli. J Biol Chem 263:14251–14255. [PubMed] [Google Scholar]
- 19.Roland KL, Liu C, Turnbough CL Jr. 1988. Role of the ribosome in suppressing transcriptional termination at the pyrBI attenuator of Escherichia coli K-12. Proc Natl Acad Sci U S A 85:7149–7153. doi: 10.1073/pnas.85.19.7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zurawski G, Elseviers D, Stauffer GV, Yanofsky C. 1978. Translational control of transcription termination at the attenuator of the Escherichia coli tryptophan operon. Proc Natl Acad Sci U S A 75:5988–5992. doi: 10.1073/pnas.75.12.5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yanofsky C, Kelley RL, Horn V. 1984. Repression is relieved before attenuation in the trp operon of Escherichia coli as tryptophan starvation becomes increasingly severe. J Bacteriol 158:1018–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Artz SW, Broach JR. 1975. Histidine regulation in Salmonella typhimurium: an activator attenuator model of gene regulation. Proc Natl Acad Sci U S A 72:3453–3457. doi: 10.1073/pnas.72.9.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnston HM, Barnes WM, Chumley FG, Bossi L, Roth JR. 1980. Model for regulation of the histidine operon of Salmonella. Proc Natl Acad Sci U S A 77:508–512. doi: 10.1073/pnas.77.1.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barnes WM. 1978. DNA sequence from the histidine operon control region: seven histidine codons in a row. Proc Natl Acad Sci U S A 75:4281–4285. doi: 10.1073/pnas.75.9.4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cockrell GM, Zheng Y, Guo W, Peterson AW, Truong JK, Kantrowitz ER. 2013. New paradigm for allosteric regulation of Escherichia coli aspartate transcarbamoylase. Biochemistry 52:8036–8047. doi: 10.1021/bi401205n. [DOI] [PubMed] [Google Scholar]
- 26.Liu C, Turnbough CL Jr. 1989. Multiple control mechanisms for pyrimidine-mediated regulation of pyrBI operon expression in Escherichia coli K-12. J Bacteriol 171:3337–3342. doi: 10.1128/jb.171.6.3337-3342.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Donahue JP, Turnbough CL Jr. 1994. Nucleotide-specific transcriptional pausing in the pyrBI leader region of Escherichia coli K-12. J Biol Chem 269:18185–18191. [PubMed] [Google Scholar]
- 28.Turnbough CL Jr, Hicks KL, Donahue JP. 1983. Attenuation control of pyrBI operon expression in Escherichia coli K-12. Proc Natl Acad Sci U S A 80:368–372. doi: 10.1073/pnas.80.2.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turnbough CL Jr, Switzer RL. 2008. Regulation of pyrimidine biosynthetic gene expression in bacteria: repression without repressors. Microbiol Mol Biol Rev 72:266–300. doi: 10.1128/MMBR.00001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bonekamp F, Clemmesen K, Karlström O, Jensen KF. 1984. Mechanism of UTP-modulated attenuation at the pyrE gene of Escherichia coli: an example of operon polarity control through the coupling of translation to transcription. EMBO J 3:2857–2861. doi: 10.1002/j.1460-2075.1984.tb02220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ost KA, Deutscher MP. 1991. Escherichia coli orfE (upstream of pyrE) encodes RNase PH. J Bacteriol 173:5589–5591. doi: 10.1128/jb.173.17.5589-5591.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steitz JA. 1969. Polypeptide chain initiation: nucleotide sequences of the three ribosomal binding sites in bacteriophage R17 RNA. Nature 224:957–964. doi: 10.1038/224957a0. [DOI] [PubMed] [Google Scholar]
- 33.Amster-Choder O, Houman F, Wright A. 1989. Protein phosphorylation regulates transcription of the β-glucoside utilization operon in E. coli. Cell 58:847–855. doi: 10.1016/0092-8674(89)90937-9. [DOI] [PubMed] [Google Scholar]
- 34.Mahadevan S, Wright A. 1987. A bacterial gene involved in transcription antitermination: regulation at a rho-independent terminator in the bgl operon of E. coli. Cell 50:485–494. doi: 10.1016/0092-8674(87)90502-2. [DOI] [PubMed] [Google Scholar]
- 35.Schnetz K, Rak B. 1988. Regulation of the bgl operon of Escherichia coli by transcriptional antitermination. EMBO J 7:3271–3277. doi: 10.1002/j.1460-2075.1988.tb03194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amster-Choder O. 2005. The bgl sensory system: a transmembrane signaling pathway controlling transcriptional antitermination. Curr Opin Microbiol 8:127–134. doi: 10.1016/j.mib.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 37.Harwani D. 2014. Regulation of gene expression: cryptic β-glucoside (bgl) operon of Escherichia coli as a paradigm. Braz J Microbiol 45:1139–1144. doi: 10.1590/s1517-83822014000400003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lopilato J, Wright A. 1990. Mechanisms of activation of the cryptic bgl operon of Escherichia coli K-12, p 435–444. In Drlica K, Riley M (ed), The bacterial chromosome. American Society for Microbiology, Washington, DC. [Google Scholar]
- 39.Reynolds AE, Mahadevan S, LeGrice SF, Wright A. 1986. Enhancement of bacterial gene expression by insertion elements or by mutation in a CAP-cAMP binding site. J Mol Biol 191:85–95. doi: 10.1016/0022-2836(86)90424-9. [DOI] [PubMed] [Google Scholar]
- 40.Lopian L, Nussbaum-Shochat A, O'Day-Kerstein K, Wright A, Amster-Choder O. 2003. The BglF sensor recruits the BglG transcription regulator to the membrane and releases it on stimulation. Proc Natl Acad Sci U S A 100:7099–7104. doi: 10.1073/pnas.1037608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Amster-Choder O, Wright A. 1993. Transcriptional regulation of the bgl operon of Escherichia coli involves phosphotransferase system-mediated phosphorylation of a transcriptional antiterminator. J Cell Biochem 51:83–90. doi: 10.1002/jcb.240510115. [DOI] [PubMed] [Google Scholar]
- 42.Raveh H, Lopian L, Nussbaum-Shochat A, Wright A, Amster-Choder O. 2009. Modulation of transcription antitermination in the bgl operon of Escherichia coli by the PTS. Proc Natl Acad Sci U S A 106:13523–13528. doi: 10.1073/pnas.0902559106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rothe FM, Bahr T, Stülke J, Rak B, Görke B. 2012. Activation of Escherichia coli antiterminator BglG requires its phosphorylation. Proc Natl Acad Sci U S A 109:15906–15911. doi: 10.1073/pnas.1210443109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gollnick P, Babitzke P. 2002. Transcription attenuation. Biochim Biophys Acta 1577:240–250. doi: 10.1016/S0167-4781(02)00455-4. [DOI] [PubMed] [Google Scholar]
- 45.Houman F, Diaz-Torres MR, Wright A. 1990. Transcriptional antitermination in the bgl operon of E. coli is modulated by a specific RNA binding protein. Cell 62:1153–1163. doi: 10.1016/0092-8674(90)90392-R. [DOI] [PubMed] [Google Scholar]
- 46.Stülke J. 2002. Control of transcription termination in bacteria by RNA-binding proteins that modulate RNA structures. Arch Microbiol 177:433–440. doi: 10.1007/s00203-002-0407-5. [DOI] [PubMed] [Google Scholar]
- 47.Shimotsu H, Kuroda MI, Yanofsky C, Henner DJ. 1986. Novel form of transcription attenuation regulates expression of the Bacillus subtilis tryptophan operon. J Bacteriol 166:461–471. doi: 10.1128/jb.166.2.461-471.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuroda MI, Henner D, Yanofsky C. 1988. cis-acting sites in the transcript of the Bacillus subtilis trp operon regulate expression of the operon. J Bacteriol 170:3080–3088. doi: 10.1128/jb.170.7.3080-3088.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gollnick P, Ishino S, Kuroda MI, Henner DJ, Yanofsky C. 1990. The mtr locus is a two-gene operon required for transcription attenuation in the trp operon of Bacillus subtilis. Proc Natl Acad Sci U S A 87:8726–8730. doi: 10.1073/pnas.87.22.8726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Antson AA, Otridge J, Brzozowski AM, Dodson EJ, Dodson GG, Wilson KS, Smith TM, Yang M, Kurecki T, Gollnick P. 1995. The structure of trp RNA-binding attenuation protein. Nature 374:693–700. doi: 10.1038/374693a0. [DOI] [PubMed] [Google Scholar]
- 51.Yang M, Chen X, Militello K, Hoffman R, Fernandez B, Baumann C, Gollnick P. 1997. Alanine-scanning mutagenesis of Bacillus subtilis trp RNA-binding attenuation protein (TRAP) reveals residues involved in tryptophan binding and RNA binding. J Mol Biol 270:696–710. doi: 10.1006/jmbi.1997.1149. [DOI] [PubMed] [Google Scholar]
- 52.Babitzke P, Bear DG, Yanofsky C. 1995. TRAP, the trp RNA-binding attenuation protein of Bacillus subtilis, is a toroid-shaped molecule that binds transcripts containing GAG or UAG repeats separated by two nucleotides. Proc Natl Acad Sci U S A 92:7916–7920. doi: 10.1073/pnas.92.17.7916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McGraw AP, Bevilacqua PC, Babitzke P. 2007. TRAP-5' stem loop interaction increases the efficiency of transcription termination in the Bacillus subtilis trpEDCFBA operon leader region. RNA 13:2020–2033. doi: 10.1261/rna.719507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mondal S, Yakhnin AV, Babitzke P. 2017. Modular organization of the NusA- and NusG-stimulated RNA polymerase pause signal that participates in the Bacillus subtilis trp operon attenuation mechanism. J Bacteriol 199:e00223-17. doi: 10.1128/JB.00223-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yakhnin AV, Babitzke P. 2002. NusA-stimulated RNA polymerase pausing and termination participates in the Bacillus subtilis trp operon attenuation mechanism in vitro. Proc Natl Acad Sci U S A 99:11067–11072. doi: 10.1073/pnas.162373299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ihms EC, Zhou M, Zhang Y, Kleckner IR, McElroy CA, Wysocki VH, Gollnick P, Foster MP. 2014. Gene regulation by substoichiometric heterocomplex formation of undecameric TRAP and trimeric anti-TRAP. Proc Natl Acad Sci U S A 111:3442–3447. doi: 10.1073/pnas.1315281111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Valbuzzi A, Yanofsky C. 2001. Inhibition of the B. subtilis regulatory protein TRAP by the TRAP-inhibitory protein, AT. Science 293:2057–2059. doi: 10.1126/science.1062187. [DOI] [PubMed] [Google Scholar]
- 58.McAdams NM, Patterson A, Gollnick P. 2017. Identification of a residue (Glu60) in TRAP required for inducing efficient transcription termination at the trp attenuator independent of binding tryptophan and RNA. J Bacteriol 199:e00710-16. doi: 10.1128/JB.00710-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McAdams NM, Gollnick P. 2014. The Bacillus subtilis TRAP protein can induce transcription termination in the leader region of the tryptophan biosynthetic (trp) operon independent of the trp attenuator RNA. PLoS One 9:e88097. doi: 10.1371/journal.pone.0088097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Babitzke P, Baker CS, Romeo T. 2009. Regulation of translation initiation by RNA binding proteins. Annu Rev Microbiol 63:27–44. doi: 10.1146/annurev.micro.091208.073514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Turner RJ, Lu Y, Switzer RL. 1994. Regulation of the Bacillus subtilis pyrimidine biosynthetic (pyr) gene cluster by an autogenous transcriptional attenuation mechanism. J Bacteriol 176:3708–3722. doi: 10.1128/jb.176.12.3708-3722.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lu Y, Turner RJ, Switzer RL. 1996. Function of RNA secondary structures in transcriptional attenuation of the Bacillus subtilis pyr operon. Proc Natl Acad Sci U S A 93:14462–14467. doi: 10.1073/pnas.93.25.14462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bonner ER, D’Elia JN, Billips BK, Switzer RL. 2001. Molecular recognition of pyr mRNA by the Bacillus subtilis attenuation regulatory protein PyrR. Nucleic Acids Res 29:4851–4865. doi: 10.1093/nar/29.23.4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hobl B, Mack M. 2007. The regulator protein PyrR of Bacillus subtilis specifically interacts in vivo with three untranslated regions within pyr mRNA of pyrimidine biosynthesis. Microbiology 153:693–700. doi: 10.1099/mic.0.2006/003772-0. [DOI] [PubMed] [Google Scholar]
- 65.Chander P, Halbig KM, Miller JK, Fields CJ, Bonner HK, Grabner GK, Switzer RL, Smith JL. 2005. Structure of the nucleotide complex of PyrR, the pyr attenuation protein from Bacillus caldolyticus, suggests dual regulation by pyrimidine and purine nucleotides. J Bacteriol 187:1773–1782. doi: 10.1128/JB.187.5.1773-1782.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jørgensen CM, Fields CJ, Chander P, Watt D, Burgner JW II, Smith JL, Switzer RL. 2008. pyr RNA binding to the Bacillus caldolyticus PyrR attenuation protein. Characterization and regulation by uridine and guanosine nucleotides. FEBS J 275:655–670. doi: 10.1111/j.1742-4658.2007.06227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hove-Jensen B, Andersen KR, Kilstrup M, Martinussen J, Switzer RL, Willemoës M. 2017. Phosphoribosyl diphosphate (PRPP): biosynthesis, enzymology, utilization, and metabolic significance. Microbiol Mol Biol Rev 81:e00040-16. doi: 10.1128/MMBR.00040-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lu Y, Turner RJ, Switzer RL. 1995. Roles of the three transcriptional attenuators of the Bacillus subtilis pyrimidine biosynthetic operon in the regulation of its expression. J Bacteriol 177:1315–1325. doi: 10.1128/jb.177.5.1315-1325.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Turner RJ, Bonner ER, Grabner GK, Switzer RL. 1998. Purification and characterization of Bacillus subtilis PyrR, a bifunctional pyr mRNA-binding attenuation protein/uracil phosphoribosyltransferase. J Biol Chem 273:5932–5938. doi: 10.1074/jbc.273.10.5932. [DOI] [PubMed] [Google Scholar]
- 70.Martinussen J, Glaser P, Andersen PS, Saxild HH. 1995. Two genes encoding uracil phosphoribosyltransferase are present in Bacillus subtilis. J Bacteriol 177:271–274. doi: 10.1128/jb.177.1.271-274.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sherwood AV, Henkin TM. 2016. Riboswitch-mediated gene regulation: novel RNA architectures dictate gene expression responses. Annu Rev Microbiol 70:361–374. doi: 10.1146/annurev-micro-091014-104306. [DOI] [PubMed] [Google Scholar]
- 72.Winkler WC, Breaker RR. 2003. Genetic control by metabolite-binding riboswitches. Chembiochem 4:1024–1032. doi: 10.1002/cbic.200300685. [DOI] [PubMed] [Google Scholar]
- 73.Winkler W, Nahvi A, Breaker RR. 2002. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 74.Grundy FJ, Henkin TM. 1993. tRNA as a positive regulator of transcription antitermination in B. subtilis. Cell 74:475–482. doi: 10.1016/0092-8674(93)80049-K. [DOI] [PubMed] [Google Scholar]
- 75.Henkin TM, Glass BL, Grundy FJ. 1992. Analysis of the Bacillus subtilis tyrS gene: conservation of a regulatory sequence in multiple tRNA synthetase genes. J Bacteriol 174:1299–1306. doi: 10.1128/jb.174.4.1299-1306.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Henkin TM. 2014. The T box riboswitch: a novel regulatory RNA that utilizes tRNA as its ligand. Biochim Biophys Acta 1839:959–963. doi: 10.1016/j.bbagrm.2014.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Henkin TM. 1994. tRNA-directed transcription antitermination. Mol Microbiol 13:381–387. doi: 10.1111/j.1365-2958.1994.tb00432.x. [DOI] [PubMed] [Google Scholar]
- 78.Zhang J, Ferré-D'Amaré AR. 2014. Direct evaluation of tRNA aminoacylation status by the T-box riboswitch using tRNA-mRNA stacking and steric readout. Mol Cell 55:148–155. doi: 10.1016/j.molcel.2014.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gutiérrez-Preciado A, Henkin TM, Grundy FJ, Yanofsky C, Merino E. 2009. Biochemical features and functional implications of the RNA-based T-box regulatory mechanism. Microbiol Mol Biol Rev 73:36–61. doi: 10.1128/MMBR.00026-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kreuzer KD, Henkin TM. 2018. The T-box riboswitch: tRNA as an effector to modulate gene regulation. Microbiol Spectr 6:RWR-0028-2018. doi: 10.1128/microbiolspec.RWR-0028-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Grigg JC, Chen Y, Grundy FJ, Henkin TM, Pollack L, Ke A. 2013. T box RNA decodes both the information content and geometry of tRNA to affect gene expression. Proc Natl Acad Sci U S A 110:7240–7245. doi: 10.1073/pnas.1222214110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sherwood AV, Frandsen JK, Grundy FJ, Henkin TM. 2018. New contacts facilitate ligand binding in a Mycobacterium smegmatis T box riboswitch. Proc Natl Acad Sci U S A 115:3894–3899. doi: 10.1073/pnas.1721254115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang J, Ferré-D'Amaré AR. 2013. Co-crystal structure of a T-box riboswitch stem I domain in complex with its cognate tRNA. Nature 500:363–366. doi: 10.1038/nature12440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang J, Ferré-D’Amaré AR. 2016. The tRNA elbow in structure, recognition and evolution. Life 6:E3. doi: 10.3390/life6010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gelfand MS, Mironov AA, Jomantas J, Kozlov YI, Perumov DA. 1999. A conserved RNA structure element involved in the regulation of bacterial riboflavin synthesis genes. Trends Genet 15:439–442. doi: 10.1016/S0168-9525(99)01856-9. [DOI] [PubMed] [Google Scholar]
- 86.Miranda-Ríos J, Navarro M, Soberón M. 2001. A conserved RNA structure (thi box) is involved in regulation of thiamin biosynthetic gene expression in bacteria. Proc Natl Acad Sci U S A 98:9736–9741. doi: 10.1073/pnas.161168098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stormo GD, Ji Y. 2001. Do mRNAs act as direct sensors of small molecules to control their expression? Proc Natl Acad Sci U S A 98:9465–9467. doi: 10.1073/pnas.181334498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.McCown PJ, Corbino KA, Stav S, Sherlock ME, Breaker RR. 2017. Riboswitch diversity and distribution. RNA 23:995–1011. doi: 10.1261/rna.061234.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Serganov A, Nudler E. 2013. A decade of riboswitches. Cell 152:17–24. doi: 10.1016/j.cell.2012.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sherlock ME, Sudarsan N, Breaker RR. 2018. Riboswitches for the alarmone ppGpp expand the collection of RNA-based signaling systems. Proc Natl Acad Sci U S A 115:6052–6057. doi: 10.1073/pnas.1720406115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Breaker RR. 2018. Riboswitches and translation control. Cold Spring Harb Perspect Biol 10:a032797. doi: 10.1101/cshperspect.a032797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Winkler WC, Cohen-Chalamish S, Breaker RR. 2002. An mRNA structure that controls gene expression by binding FMN. Proc Natl Acad Sci U S A 99:15908–15913. doi: 10.1073/pnas.212628899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Barrick JE, Corbino KA, Winkler WC, Nahvi A, Mandal M, Collins J, Lee M, Roth A, Sudarsan N, Jona I, Wickiser JK, Breaker RR. 2004. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc Natl Acad Sci U S A 101:6421–6426. doi: 10.1073/pnas.0308014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kim JN, Roth A, Breaker RR. 2007. Guanine riboswitch variants from Mesoplasma florum selectively recognize 2'-deoxyguanosine. Proc Natl Acad Sci U S A 104:16092–16097. doi: 10.1073/pnas.0705884104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mandal M, Breaker RR. 2004. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat Struct Mol Biol 11:29–35. doi: 10.1038/nsmb710. [DOI] [PubMed] [Google Scholar]
- 96.McCown PJ, Liang JJ, Weinberg Z, Breaker RR. 2014. Structural, functional, and taxonomic diversity of three preQ1 riboswitch classes. Chem Biol 21:880–889. doi: 10.1016/j.chembiol.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Weinberg Z, Barrick JE, Yao Z, Roth A, Kim JN, Gore J, Wang JX, Lee ER, Block KF, Sudarsan N, Neph S, Tompa M, Ruzzo WL, Breaker RR. 2007. Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline. Nucleic Acids Res 35:4809–4819. doi: 10.1093/nar/gkm487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Weinberg Z, Wang JX, Bogue J, Yang J, Corbino K, Moy RH, Breaker RR. 2010. Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes. Genome Biol 11:R31. doi: 10.1186/gb-2010-11-3-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Atilho RM, Perkins KR, Breaker RR. 2019. Rare variants of the FMN riboswitch class in Clostridium difficile and other bacteria exhibit altered ligand specificity. RNA 25:23–34. doi: 10.1261/rna.067975.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Turnbough CL., Jr 2011. Regulation of gene expression by reiterative transcription. Curr Opin Microbiol 14:142–147. doi: 10.1016/j.mib.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Anikin M, Molodtsov V, Temiakov D, McAllister WT. 2010. Transcript slippage and recoding, p 409–432. In Atkins JF, Gesteland RF (ed), Recoding: expansion of decoding rules enriches gene expression, vol 24 Springer, New York, NY. [Google Scholar]
- 102.Meng Q, Turnbough CL Jr., Switzer RL. 2004. Attenuation control of pyrG expression in Bacillus subtilis is mediated by CTP-sensitive reiterative transcription. Proc Natl Acad Sci U S A 101:10943–10948. doi: 10.1073/pnas.0403755101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Varani G, McClain WH. 2000. The G·U wobble base pair. A fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Rep 1:18–23. doi: 10.1093/embo-reports/kvd001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jensen-MacAllister IE, Meng Q, Switzer RL. 2007. Regulation of pyrG expression in Bacillus subtilis: CTP-regulated antitermination and reiterative transcription with pyrG templates in vitro. Mol Microbiol 63:1440–1452. doi: 10.1111/j.1365-2958.2007.05595.x. [DOI] [PubMed] [Google Scholar]
- 105.Elsholz AKW, Jørgensen CM, Switzer RL. 2007. The number of G residues in the Bacillus subtilis pyrG initially transcribed region governs reiterative transcription-mediated regulation. J Bacteriol 189:2176–2180. doi: 10.1128/JB.01611-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kriner MA, Sevostyanova A, Groisman EA. 2016. Learning from the leaders: gene regulation by the transcription termination factor Rho. Trends Biochem Sci 41:690–699. doi: 10.1016/j.tibs.2016.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Peters JM, Mooney RA, Kuan PF, Rowland JL, Keles S, Landick R. 2009. Rho directs widespread termination of intragenic and stable RNA transcription. Proc Natl Acad Sci U S A 106:15406–15411. doi: 10.1073/pnas.0903846106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Grylak-Mielnicka A, Bidnenko V, Bardowski J, Bidnenko E. 2016. Transcription termination factor Rho: a hub linking diverse physiological processes in bacteria. Microbiology 162:433–447. doi: 10.1099/mic.0.000244. [DOI] [PubMed] [Google Scholar]
- 109.Peters JM, Mooney RA, Grass JA, Jessen ED, Tran F, Landick R. 2012. Rho and NusG suppress pervasive antisense transcription in Escherichia coli. Genes Dev 26:2621–2633. doi: 10.1101/gad.196741.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Washburn RS, Gottesman ME. 2011. Transcription termination maintains chromosome integrity. Proc Natl Acad Sci U S A 108:792–797. doi: 10.1073/pnas.1009564108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bidnenko V, Nicolas P, Grylak-Mielnicka A, Delumeau O, Auger S, Aucouturier A, Guerin C, Repoil F, Bardowski J, Aymerich S, Bidnenko E. 2017. Termination factor Rho: from the control of pervasive transcription to cell fate determination in Bacillus subtilis. PLoS Genet 13:e1006909. doi: 10.1371/journal.pgen.1006909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Stewart V, Landick R, Yanofsky C. 1986. Rho-dependent transcription termination in the tryptophanase operon leader region of Escherichia coli K-12. J Bacteriol 166:217–223. doi: 10.1128/jb.166.1.217-223.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gong F, Ito K, Nakamura Y, Yanofsky C. 2001. The mechanism of tryptophan induction of tryptophanase operon expression: tryptophan inhibits release factor-mediated cleavage of TnaC-peptidyl-tRNA(Pro). Proc Natl Acad Sci U S A 98:8997–9001. doi: 10.1073/pnas.171299298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cruz-Vera LR, Yanofsky C. 2008. Conserved residues Asp16 and Pro24 of TnaC-tRNAPro participate in tryptophan induction of tna operon expression. J Bacteriol 190:4791–4797. doi: 10.1128/JB.00290-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gong F, Yanofsky C. 2002. Instruction of translating ribosome by nascent peptide. Science 297:1864–1867. doi: 10.1126/science.1073997. [DOI] [PubMed] [Google Scholar]
- 116.Martínez AK, Gordon E, Sengupta A, Shirole N, Klepacki D, Martinez-Garriga B, Brown LM, Benedik MJ, Yanofsky C, Mankin AS, Vazquez-Laslop N, Sachs MS, Cruz-Vera LR. 2014. Interactions of the TnaC nascent peptide with rRNA in the exit tunnel enable the ribosome to respond to free tryptophan. Nucleic Acids Res 42:1245–1256. doi: 10.1093/nar/gkt923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cruz-Vera LR, Sachs MS, Squires CL, Yanofsky C. 2011. Nascent polypeptide sequences that influence ribosome function. Curr Opin Microbiol 14:160–166. doi: 10.1016/j.mib.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 118.Seip B, Innis CA. 2016. How widespread is metabolite sensing by ribosome-arresting nascent peptides? J Mol Biol 428:2217–2227. doi: 10.1016/j.jmb.2016.04.019. [DOI] [PubMed] [Google Scholar]
- 119.Cromie MJ, Groisman EA. 2010. Promoter and riboswitch control of the Mg2+ transporter MgtA from Salmonella enterica. J Bacteriol 192:604–607. doi: 10.1128/JB.01239-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cromie MJ, Shi Y, Latifi T, Groisman EA. 2006. An RNA sensor for intracellular Mg2+. Cell 125:71–84. doi: 10.1016/j.cell.2006.01.043. [DOI] [PubMed] [Google Scholar]
- 121.Gall AR, Datsenko KA, Figueroa-Bossi N, Bossi L, Masuda I, Hou YM, Csonka LN. 2016. Mg2+ regulates transcription of mgtA in Salmonella Typhimurium via translation of proline codons during synthesis of the MgtL peptide. Proc Natl Acad Sci U S A 113:15096–15101. doi: 10.1073/pnas.1612268113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hollands K, Sevostiyanova A, Groisman EA. 2014. Unusually long-lived pause required for regulation of a Rho-dependent transcription terminator. Proc Natl Acad Sci U S A 111:E1999–E2007. doi: 10.1073/pnas.1319193111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Doerfel LK, Wohlgemuth I, Kothe C, Peske F, Urlaub H, Rodnina MV. 2013. EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science 339:85–88. doi: 10.1126/science.1229017. [DOI] [PubMed] [Google Scholar]
- 124.Pavlov MY, Watts RE, Tan Z, Cornish VW, Ehrenberg M, Forster AC. 2009. Slow peptide bond formation by proline and other N-alkylamino acids in translation. Proc Natl Acad Sci U S A 106:50–54. doi: 10.1073/pnas.0809211106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hollands K, Proshkin S, Sklyarova S, Epshtein V, Mironov A, Nudler E, Groisman EA. 2012. Riboswitch control of Rho-dependent transcription termination. Proc Natl Acad Sci U S A 109:5376–5381. doi: 10.1073/pnas.1112211109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Park SY, Cromie MJ, Lee EJ, Groisman EA. 2010. A bacterial mRNA leader that employs different mechanisms to sense disparate intracellular signals. Cell 142:737–748. doi: 10.1016/j.cell.2010.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Baek J, Lee J, Yoon K, Lee H. 2017. Identification of unannotated small genes in Salmonella. G3 (Bethesda) 7:983–989. doi: 10.1534/g3.116.036939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Romeo T, Babitzke P. 2018. Global regulation by CsrA and its RNA antagonists. Microbiol Spectr 6:RWR-0009-2017. doi: 10.1128/microbiolspec.RWR-0009-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Figueroa-Bossi N, Schwartz A, Guillemardet B, D'Heygère F, Bossi L, Boudvillain M. 2014. RNA remodeling by bacterial global regulator CsrA promotes Rho-dependent transcription termination. Genes Dev 28:1239–1251. doi: 10.1101/gad.240192.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kriner MA, Groisman EA. 2015. The bacterial transcription termination factor Rho coordinates Mg2+ homeostasis with translational signals. J Mol Biol 427:3834–3849. doi: 10.1016/j.jmb.2015.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lee EJ, Choi J, Groisman EA. 2014. Control of a Salmonella virulence operon by proline-charged tRNAPro. Proc Natl Acad Sci U S A 111:3140–3145. doi: 10.1073/pnas.1316209111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sedlyarova N, Shamovsky I, Bharati BK, Epshtein V, Chen J, Gottesman S, Schroeder R, Nudler E. 2016. sRNA-mediated control of transcription termination in E. coli. Cell 167:111–121. doi: 10.1016/j.cell.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sedlyarova N, Rescheneder P, Magán A, Popitsch N, Rziha N, Bilusic I, Epshtein V, Zimmermann B, Lybecker M, Sedlyarov V, Schroeder R, Nudler E. 2017. Natural RNA polymerase aptamers regulate transcription in E. coli. Mol Cell 67:30–43. doi: 10.1016/j.molcel.2017.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sevostyanova A, Groisman EA. 2015. An RNA motif advances transcription by preventing Rho-dependent termination. Proc Natl Acad Sci U S A 112:E6835–E6843. doi: 10.1073/pnas.1515383112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kriner MA, Groisman EA. 2017. RNA secondary structures regulate three steps of Rho-dependent transcription termination within a bacterial mRNA leader. Nucleic Acids Res 45:631–642. doi: 10.1093/nar/gkw889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bastet L, Chauvier A, Singh N, Lussier A, Lamontagne AM, Prévost K, Massé E, Wade JT, Lafontaine DA. 2017. Translational control and Rho-dependent transcription termination are intimately linked in riboswitch regulation. Nucleic Acids Res 45:7474–7486. doi: 10.1093/nar/gkx434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bossi L, Schwartz A, Guillemardet B, Boudvillain M, Figueroa-Bossi N. 2012. A role for Rho-dependent polarity in gene regulation by a noncoding small RNA. Genes Dev 26:1864–1873. doi: 10.1101/gad.195412.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Nelson JW, Breaker RR. 2017. The lost language of the RNA world. Sci Signal 10:eaam8812. doi: 10.1126/scisignal.aam8812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Nadiras C, Eveno E, Schwartz A, Figueroa-Bossi N, Boudvillain M. 2018. A multivariate prediction model for Rho-dependent termination of transcription. Nucleic Acids Res 46:8245–8260. doi: 10.1093/nar/gky563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Vicens Q, Mondragón E, Batey RT. 2011. Molecular sensing by the aptamer domain of the FMN riboswitch: a general model for ligand binding by conformational selection. Nucleic Acids Res 39:8586–8598. doi: 10.1093/nar/gkr565. [DOI] [PMC free article] [PubMed] [Google Scholar]