Abstract
Phenytoin drug reaction with eosinophilia and systemic symptoms (DRESS) in 3 Aboriginal Australians positive for HLA‐B*56:02 has been previously reported. We report the allele frequency of HLA‐B*56:02 in 2 South Australian populations, 1 Aboriginal (4.8%, 95% confidence interval 2.4–7.8%) and the other European (0%). We compared the frequency with publicly available information on HLA‐B*56:02 status in other Indigenous Australian (n = 4) and European Australian cohorts (n = 1). In the Indigenous Australian cohorts, HLA‐B*56:02 allele frequency ranged from 1.3 to 19%. We also describe an additional case of phenytoin DRESS (RegiSCAR DRESS score 7) in an Aboriginal Australian that was associated with HLA‐B*56:02 and with CYP2C9*1/*3 genotype. In Aboriginal Australians, phenytoin DRESS appears distinctly linked to HLA‐B*56:02 with an allele carriage rate substantially higher than in Europeans, but also with considerable regional variation. Investigations of human leucocyte antigen and other contributing genes and severe adverse drug reactions in understudied non‐European populations are required to optimize safe medication use and inform risk mitigation strategies.
Keywords: drug reaction with eosinophilia and systemic symptoms, drug toxicity, human leucocyte antigen, indigenous Australians, phenytoin
What is already known about this subject
Phenytoin hypersensitivity reactions including Stevens–Johnson syndrome/toxic epidermal necrolysis (SJS/TEN) and drug reaction with eosinophilia and systemic symptoms (DRESS) in Europeans are rare but can be life threatening.
An association has been found between phenytoin SJS/TEN and HLA‐B*15:02 but only in Southeast Asians.
Phenytoin associated DRESS has been reported in Aboriginal Australians who carry HLA‐B*56:02.
An association has been described between slow metabolism of phenytoin (CYP2C9*3) for multiple phenotypes of phenytoin hypersensitivity including maculopapular exanthem >> DRESS >> SJS/TEN.
What this study adds
HLA‐B*56:02 shows allele frequencies of 1.3–19% in different geographic Indigenous Australian cohorts compared with 0% in Europeans.
We report an additional case of phenytoin DRESS in an Aboriginal Australian who carried HLA‐B*56:02.
There is considerable variability in the frequency of HLA‐B*56:02 within Australia's indigenous populations.
HLA genotype associated with severe adverse drug reactions may be different in understudied non‐European populations.
1. INTRODUCTION
Phenytoin has a narrow therapeutic index with common adverse effects including ataxia, nystagmus and agitation that occur predictably with high concentrations. Phenytoin is also associated with uncommon but severe adverse drug reactions (ADR) which appear unrelated to systemic concentrations and may have delayed onset. An example is severe cutaneous adverse reaction (SCAR), including Stevens–Johnson syndrome (SJS), toxic epidermal necrolysis (TEN), drug reaction with eosinophilia and systemic symptoms (DRESS), and drug hypersensitivity syndrome. Phenytoin SJS/TEN has been strongly linked to the HLA‐B*15:02 allele and less commonly to other human leucocyte antigen (HLA)‐B alleles of the same B75 serotype (HLA‐B*15:21, B*15:11, B*15:08)1, 2 originally reported in Han Chinese from Taiwan but subsequently confirmed in other Asian countries including Malaysia, China, Thailand and India, where its frequency can be >10%.1 In Taiwan where screening for HLA‐B*15:02 is reimbursed, the use of carbamazepine and cases of carbamazepine‐related SCAR have declined.3 In Korean, Japanese and European populations, phenytoin‐associated DRESS due to HLA‐B*15:02 is rare due to very low allele frequency (< 0.5%). In a Malay population,4 a different HLA‐B allele in the B75 serotype, HLA‐B*15:13 was associated with phenytoin SJS/TEN and DRESS. HLA‐B*15:13 frequency ranges from 5.5% in Burmese5 to 13.3% in Javanese6 subjects, hence showing a high frequency in Southeast Asian populations but not in Han Chinese or Thai populations where frequency is <2%.6
CYP2C9 is the major enzyme metabolizing phenytoin with a lesser contribution from CYP2C19.1 The decreased function CYP2C9*3 has a higher frequency in South Asian populations (11.3%) compared to Europeans (5.6%) and East Asians (3.4%),7 and has also been linked to phenytoin associated SCAR in South Asian populations.8, 9, 10, 11
HLA‐B*15:02 was not carried in 3 cases of phenytoin‐associated DRESS reported in Aboriginal Australians in 2012.12 Two of these were fatal and all carried HLA‐B*56:02, with 1 being homozygous. CYP2C9 was not assessed.
We report the allele frequency of HLA‐B*56:02 in 2 South Australian populations, 1 Aboriginal and the other European. We compare this with publicly available information on HLA‐B*56:02 status in other Indigenous Australian (n = 4) and European Australian (n = 1) cohorts. We also describe an additional case of phenytoin DRESS (RegiSCAR DRESS score 7) in an Aboriginal Australian associated with HLA‐B*56:02 and with CYP2C9*1/*3 genotype, providing further evidence that phenytoin‐associated DRESS in Aboriginal Australians is linked to HLA‐B*56:02 and not B*15:02.
2. METHODS
2.1. HLA frequency in South Australian aboriginal and European populations
This was conducted to ascertain the HLA‐B locus allele frequency in a cohort of Aboriginal Australians compared to Europeans living in Australia; we have used the group name European as recommended.13 Ethics approval was from the Aboriginal Health Council of SA Inc. [Reference No. 04–14‐592] and the University of Adelaide: Human Research Ethics Committee [Project No. H‐117‐2011]. Following informed and signed consent, both groups of participants provided information on age, sex, Aboriginal or European ancestry (parents/grandparents), presence of disease, smoking and alcohol (current/previous/never), residential location, and a saliva sample was collected (Oragene DNA Saliva Kit‐DNA Genotek Inc. Canada) for genotyping.
2.2. Population allele frequency database
A publicly available allele frequency database (http://www.allelefrequencies.net/) was searched to find data on HLA‐B*56:02, B*15:02 and B*15:13 frequencies in different Indigenous Australian communities and Europeans living in Australia.14
2.3. Case report
A 30 year‐old Australian Aboriginal woman, of Western Aranda and Pintupi heritage (Figure 1), presented to hospital with hypotension, recent onset of fevers and 1 month of rash, after initially developing acute oedema of her face, arms and legs. Phenytoin had been commenced 7 months previously following multiple seizures, and she had recent influenza B 5 weeks prior to hospitalization, treated with benzylpenicillin and doxycycline. Height was 155 cm, weight 65 kg (body mass index 27.1 kg/m2) and temperature up to 38.6°C. There was widespread (>50% extent) rash (pruritic, ichthyotic and scaling), which was very severe on the palms (Figure 2), feet and neck. Investigations revealed early severe eosinophilia of 1.22 × 109/L (reference: 0.04–0.4 × 109/L), acute kidney injury (serum creatinine 231 [reference: 50–100] μmol/L, creatinine clearance [Cockcroft–Gault formula] 32 ml/min) and hypoalbuminaemia of 22 (reference: 34–48) g/L. Gamma‐glutamyl transferase was 478 U/L (reference: <60), alkaline phosphatase was also elevated at 717 U/L (reference: 30–110) with minor elevation of transaminases (alanine 75 U/L [reference: <55], aspartate 53 U/L [reference: <45]); bilirubin was normal. Coeliac antibodies were negative, imaging showed mediastinal and inguinal lymphadenopathy up to 19 mm, and endoscopy demonstrated normal oesophagus, with minor erosive gastropathy and was positive for Helicobacter pylori. Multiple blood cultures were negative as were hepatitis B, C and human immunodeficiency virus, and other viral and infective conditions. Multiple autoimmune serologies were negative.
Figure 1.
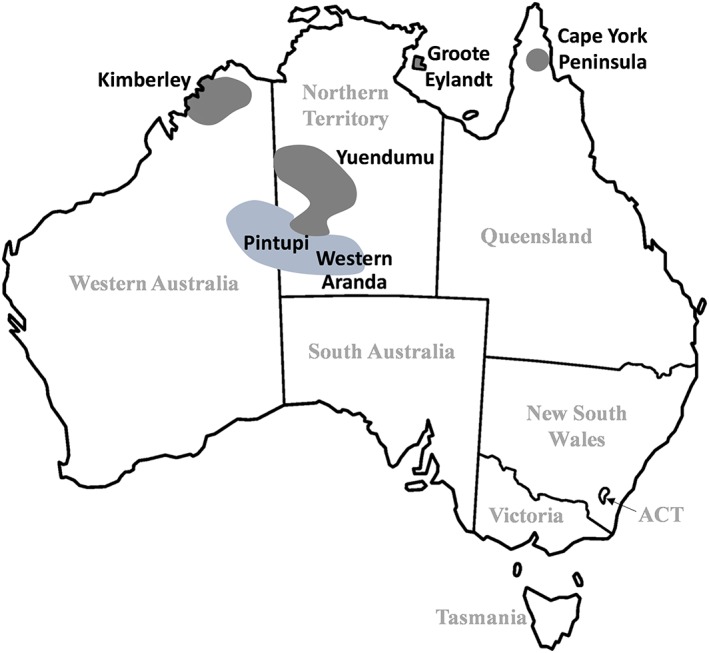
Location of the sites (Cape York Peninsula, Groote Eylandt, Kimberley, Yuendumu) of the 4 indigenous Australian cohorts who had been previously tested for HLA‐B status. Data obtained from http://allelefrequencies.net (HLA & Adverse Drug Reaction Section). West Aranda and Pintupi is the heritage of the subject in the clinical case of DRESS
Figure 2.
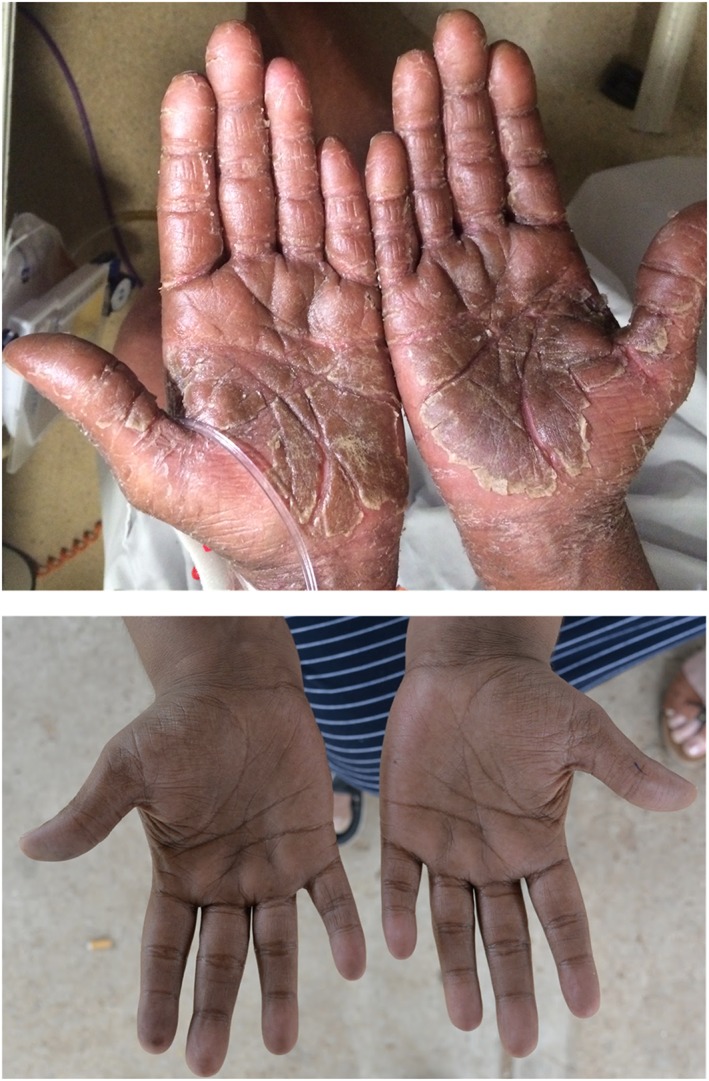
Photo of subject's palms at time of DRESS diagnosis (top) and at 2 years and 4 months after phenytoin was ceased
Medications at admission were phenytoin (300 mg daily) for 7 months, pantoprazole 40 mg orally for 11 weeks and prednisolone, reducing dose commenced at 20 mg daily 3 weeks prior to admission. Plasma phenytoin was 12 (reference: 40–80) μmol/L (albumin corrected phenytoin concentration using the Sheiner–Tozer equation15 was 22 μmol/L). Clinical features were scored for DRESS using RegiSCAR criteria (Score 7),16, 17 and a clinical diagnosis was made of DRESS associated with phenytoin. Phenytoin was ceased and replaced with levetiracetam. Rechallenge was not done due to disease severity.
Following phenytoin cessation there was a slow resolution of rash over weeks to months (Figure 2). Minor flaking of skin persisted at 5 months; however, at 2 years the abnormalities of skin had resolved completely. Eosinophilia resolved over 5 months. The patient gave written informed consent for reporting the case including photographs, and for research blood taken for genotyping.
2.4. Genotyping
For the South Australian Aboriginal and European populations and the Case Report patient, HLA‐B locus was genotyped (Olerup SBT HLA‐B Sequencing Based Typing kit [CareDX Pty Ltd, Fremantle, WA, Australia] with allele calling by Assign SBT v471 [CareDX Pty Ltd] using IMGT database 3.29.0.0 2017‐07‐10) at the South Australian Transplantation and Immunogenetics Service (Australian Red Cross Blood Service, Adelaide, SA, Australia). CYP2C9 *2 (rs1799853) and *3 (rs1057910) were assayed using Sanger sequencing. Amplicons surrounding the rs1799853 and rs1057910 loci were amplified separately by polymerase chain reaction (rs1799853 primers: 5′‐ATGGGGAGGATGGAAAACAGAGACTT‐3′ and 5′‐AGTAGAGAAGATAGTAGTCCAGTAAGGTCAGTGATATG‐3′; or rs1057910 primers: 5’‐CTCTTTAAGTTTGCATATACTTCCAGCAC‐3′ and 5′‐TGAGTTATGCACTTCTCTCACCCGG‐3′; Sigma‐Aldrich Pty Ltd, Castle Hill, NSW, Australia). Bidirectional sequencing (BigDye Terminator v3.1 using primers above) was conducted at the Australian Genome Research Facility (Melbourne, VIC, Australia).
2.5. Statistics
Frequencies (and 95% confidence intervals [for allele n > 3]) of HLA‐B*56:02, B*15:02 and B*15:13 alleles within the Indigenous Australian and Europeans cohorts were calculated.
3. RESULTS
3.1. HLA frequency in a South Australian aboriginal population
Of the 147 Aboriginal Australians, 33 were non‐South Australian being mainly from the Northern Territory (NT) or Western Australia (WA; Figure 1). Median age was 30 (range: 18–67) years, and 69% were female. The European cohort comprised 158 participants residing in South Australia of median age 37 (18–78) years, and 63% were female. In the Aboriginal cohort, 13 individuals (9%) were positive for HLA‐B*56:02, including 1 homozygous, with an overall HLA‐B*56:02 allele frequency of 4.8%. One of the 13 was from the Tanami Desert region containing Yuendumu (NT). For the European cohort, none (0%) were HLA‐B*56:02 positive (Table 1). For B*15:02, only 1 Aboriginal (overall allele frequency 0.3%) was positive compared to none in the European cohort, and no B*15:13 alleles were detected in either population (Table 1).
Table 1.
HLA‐B*56:02, ‐B*15:02 and ‐B*15:13 allele frequencies in indigenous Australian compared to European cohorts living in Australia
| Cohort | N | B*56:02, % [95% CI] (n) | B*15:02, % (n) | B*15:13, % (n) |
|---|---|---|---|---|
| Aboriginal (South Australia) | 147 | 4.8 [2.4–7.2] (14)a | 0.3 (1) | 0 (0) |
| European (South Australia) | 158 | 0 (0) | 0 (0) | 0 (0) |
| European (NSW)b | 134 | 0.4 (1) | 0 (0) | 0 (0) |
| Aboriginal/Torres Strait Islander (Cape York Peninsula)b | 103 | 3 [0.6–5.4] (6) | ND | ND |
| Aboriginal (Groote Eylandt)b | 75 | 2.7 [0.1–5.2] (4) | 0.7 (1) | ND |
| Aboriginal (Kimberly)b | 41 | 1.3 (1) | ND | ND |
| Aboriginal (Yuendumu)b | 191 | 19 [15–23] (73) | 0 (0) | 0 (0) |
N = number of individuals; CI = confidence interval; n = number of alleles.
includes 1 homozygous individual;
data extracted from http://allelefrequencies.net. ND = not determined.
Note: Australia has 2 distinct indigenous populations, Aboriginals and Torres Strait Islanders.
3.2. Population allele frequency database
Data on 4 separate cohorts of indigenous Australians who had been tested for HLA‐B status were found (n = 41 [Kimberly region] to 191 [Yuendumu]; Figure 1). HLA‐B*56:02 allele frequency ranged from 1.3% (Kimberley) to 19% (Yuendumu; Table 1). For B*15:02, none were found in Yuendumu and 0.7% in Groote Eylandt. For B*15:13, none were found in Yuendumu, and it was not determined in other indigenous cohorts (Table 1). The database also included a European population (n = 134) from New South Wales (Australia). The B*56:02 allele frequency was 0.4% and was zero percent for B*15:02 and B*15:13.
3.3. Case report
The patient carried HLA‐B*56:02 and CYP2C9*1/*3.
4. DISCUSSION
We provide further evidence that phenytoin DRESS in Aboriginal Australians is associated with HLA‐B*56:02 and that the carriage of this allele is prevalent, but also variable in different geographic indigenous Australian populations compared to Europeans. This supports previous findings of mitochondrial genome variation in Aboriginal Australians.18
In the case report, the diagnosis of DRESS was based on meeting at least 5 of 9 inclusion criteria for a potential case16 and a validation RegiSCAR score of 7 (definite case) based on fever >38.5°C, lymphadenopathy >1 cm at 2 sites, eosinophilia, extensive rash, involvement of internal organs (liver) and exclusion of other disorders with multiple negative investigations.16 The time course favoured DRESS with delayed onset after commencement of phenytoin and slow resolution after cessation. In 3 cases reported by Harding et al12 the onset of symptoms was 2–3 weeks (2 patients) but about 3 months in the other case. In our patient, the onset of symptoms was somewhat atypical in that it occurred 7 months after phenytoin initiation. The presence of early severe eosinophilia was predictive of a severe reaction and systemic disease, although unfortunately formal diagnosis of phenytoin associated DRESS was delayed for some weeks.19 The occurrence of this ADR was communicated to the patient, the primary care practitioner, other treating clinicians and reported to the hospital drug information service, and the national regulator. The patient was also CYP2C9*1/*3, indicative of slower phenytoin metabolism, which has been implicated in phenytoin SCAR in Southeast Asian populations.8 The mechanism is unclear but Chung et al8 found that those with SJS/TEN had delayed phenytoin clearance but not those with DRESS.
Phenytoin DRESS has been convincingly associated with HLA‐B*15:02 especially in South Asian populations but not in Europeans as the allele frequency is substantially higher with Asian ancestry.1 Unlike the association of HLA‐B*15:02 and carbamazepine SJS/TEN in Southeast Asians where the negative predictive value is 100%, a substantial percentage of phenytoin SCAR would not be explained by HLA‐B*15:02 in Southeast Asians. Although the mechanism by which the variant causes this reaction is unknown, it is of interest that slow phenytoin metabolism (CYP2C9*3 plays a significant role in risk for phenytoin SCAR) and other HLA‐B alleles have been associated with phenytoin DRESS. Tassaneeyaku et al20 identified several other HLA‐B variants that were associated with phenytoin SJS/TEN (13:01, 38:02, 51:01, 56:02) and DRESS (51:01) in Thai patients but when corrected for multiple comparisons, statistical significance was lost. Yampayon et al,11 in another Thai population, identified that 56:02/04 was associated with phenytoin DRESS, which remained statistically significant after multiple comparison correction.
Our case report supports that of Harding et al12 in which 2 of the 3 cases were fatal, of phenytoin DRESS in Aboriginal Australians and who were carriers (one was homozygous) for HLA‐B*56:02. The population frequency of this variant is relatively high, ranging from 1.3 to 4% in Northern Australia, to 4.8% in South Australia to 19% in Central Australia (Yuendumu). The latter is the highest frequency reported for this allele in any location and the reasons for such are unknown, but a founder effect is probable. For most T‐cell mediated drug reactions, it is suspected that the drug noncovalently binds to an immune receptor such as HLA and/or TCR in a dose‐dependent fashion. It is also possible that the combination of carriage of HLA‐B*56:02 and a slower metabolizing CYP2C9 genotype are important for the development of phenytoin SCAR (or DRESS) in this population.
The positive and negative predictive values of HLA‐B*56:02 in Indigenous Australian populations cannot be estimated; however, the clinical implications from this finding in Yuendumu have already been addressed. Alice Springs Hospital is the major acute hospital for Central Australia and has reported several cases of serious phenytoin ADRs, such that, since 2014, phenytoin has only been available in the pharmacy for intensive care unit use in status epilepticus. HLA‐B genotyping is not routinely available in clinical practice, but the link to B*56:02 12 and the high frequency of this allele in their patient catchment area led to this pragmatic decision (Tsai, Personal Communication). This decision had not been systematically relayed to prescribers in other parts of Australia, as the national warning system is limited. Although of central Australian heritage, our patient could not benefit from this local decision as she was residing outside central Australia at the time of initiation of phenytoin. Although B*56:02 allele frequency is near zero in Europeans, it can be up to 5% in other Indigenous Australian populations (excluding those in Central Australia). It is incumbent on both national regulatory bodies and pharmaceutical companies to take action when life‐threatening adverse drug reactions are affected by regional and not necessarily global pharmacogenetic factors.
The study illustrates the large global diversity in pharmacogenetic variation13 and highlights the need to investigate life‐threatening adverse drug events in understudied indigenous populations21 including in Oceanians whose pharmacogenetic profile can be very different from Europeans.22, 23
COMPETING INTERESTS
There are no competing interests to declare.
CONTRIBUTORS
A.A.S.: study design, funding, conduct of experiments, data analyses and interpretation, preparation of manuscript. D.T.B.: study design, conduct of experiments, data analyses, review of manuscript. E.J.P.: data interpretation, review of manuscript. K.M.: case report documentation, review of manuscript. F.I.: case report documentation, patient clinical care, review of manuscript. G.M.B.: patient consent and clinical care, data interpretation, review of manuscript. All authors have approved the final manuscript and have agreed to be accountable for all aspects of the work.
5.
ACKNOWLEDGEMENTS
We would like to sincerely acknowledge the contribution of our patient to this paper. We also thank Assoc. Prof. Peter Bardy for expediting the HLA testing on our patient; Dr Damien Harding for discussions on his 3 patients, Charles Conrad for photography and Danny Tsai (Pharmacy Department, Alice Springs Hospital) for information of phenytoin removal from the Imprest System. The study was supported by NHMRC Project Grant APP108798.
Somogyi AA, Barratt DT, Phillips EJ, Moore K, Ilyas F, Gabb GM. High and variable population prevalence of HLA‐B*56:02 in indigenous Australians and relation to phenytoin‐associated drug reaction with eosinophilia and systemic symptoms. Br J Clin Pharmacol. 2019;85:2163–2169. 10.1111/bcp.14025
The authors confirm that the Principal Investigator for this paper is Andrew A. Somogyi and that Genevieve M. Gabb had direct clinical responsibility for the patient
Data Availability Statement:The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.
REFERENCES
- 1. Caudle KE, Rettie AE, Whirl‐Carrillo M, et al. Clinical pharmacogenetics implementation consortium guidelines for CYP2C9 and HLA‐B genotypes and phenytoin dosing. Clin Pharmacol Ther. 2014;96(5):542‐548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Phillips EJ, Sukasem C, Whirl‐Carrillo M, et al. Clinical pharmacogenetics implementation consortium guideline for HLA genotype and use of carbamazepine and oxcarbazepine: 2017 update. Clin Pharmacol Ther. 2018;103(4):574‐581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lin CW, Hwang WI, Chao PH, Chen WW, Hsiao FY. Temporal trends and patterns in carbamazepine use, related severe cutaneous adverse reactions, and HLA‐B*15:02 screening: a nationwide survey. Epilepsia. 2018;12:2325‐2339. [DOI] [PubMed] [Google Scholar]
- 4. Chang C‐C, Ng C‐C, Loo C‐L, et al. Association of HLA‐B*15:13 and HLA‐B*15:02 with phenytoin‐induced severe cutaneous adverse reactions in a Malay population. Pharmacogenomics J. 2017;17(2):170‐173. [DOI] [PubMed] [Google Scholar]
- 5. Kongmaroeng C, Romphruk A, Ruangwerayut R, et al. HLA‐B*15 subtypes in Burmese population by sequence‐based typing. Tissue Antigens. 2009;74(2):164‐167. [DOI] [PubMed] [Google Scholar]
- 6. Gonzalez‐Galarza FF, Christmas S, Middleton D, Jones AR. Allele frequency net: a database and online repository for immune gene frequencies in worldwide populations. Nucleic Acids Res. 2011;39(Suppl 1):D913‐D919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhou Y, Ingelman‐Sundberg M, Lauschke VM. Worldwide distribution of cytochrome P450 alleles: a meta‐analysis of population‐scale sequencing projects. Clin Pharmacol Ther. 2017;102(4):688‐700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chung W‐H, Chang W‐C, Lee W‐S, et al. Genetics variants associated with phenytoin‐related severe cutaneous adverse reactions. JAMA. 2014;312(5):525‐534. [DOI] [PubMed] [Google Scholar]
- 9. Suvichapanich S, Jittikoon J, Wichukchinda N, et al. Association analysis of CYP2C9*3 and phenytoin‐induced severe cutaneous adverse reactions in Thai epilepsy children. J Hum Genet. 2015;60(8):413‐417. [DOI] [PubMed] [Google Scholar]
- 10. Lee A‐Y, Kim M‐J, Chey W‐Y, Choi J, Kim BG. Genetic polymorphism of cytochrome P 450 2C9 in diphenylhydantoin‐induced cutaneous adverse drug reactions. Eur J Clin Pharmacol. 2004;60(3):155‐159. [DOI] [PubMed] [Google Scholar]
- 11. Yampayon K, Sukasem C, Limwongse C, et al. Influence of genetic and non‐genetic factors on phenytoin‐induced severe cutaneous adverse drug reactions. Eur J Clin Pharmacol. 2017;73(7):855‐865. [DOI] [PubMed] [Google Scholar]
- 12. Harding DJ, Subramaniam K, MacQuillin G, Davis J, Nolan D. Severe drug‐induced hypersensitivity syndrome with a shared HLA allele. Med J Aust. 2012;197(7):411‐413. [DOI] [PubMed] [Google Scholar]
- 13. Huddart R, Fohner AE, Whirl‐Carrillo M, et al. Standardized biogeographic grouping system for annotating populations in pharmacogenetic research. Clin Pharmacol Ther. 2019;105(5):1256‐1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ghattaoraya GS, Dundar Y, González‐Galarza FF, et al. A web resource for mining HLA associations with adverse drug reactions. HLA‐ADR Database (Oxford). 2016;2016:1‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sheiner LB, Tozer TN. Clinical pharmacokinetics: the use of plasma concentrations of drugs In: Melmon KL, Morelli HF, eds. Clinical Pharmacology: Basic Principles in Therapeutics. New York: Macmillan; 1978:71‐109. [Google Scholar]
- 16. Karduan SH, Sidoroff A, Valeyrie‐Allanore L, et al. Variability in the clinical pattern of cutaneous side‐effects of drugs with systemic symptoms: does a DRESS syndrome really exist. Br J Dermatol. 2007;156(3):609‐611. [DOI] [PubMed] [Google Scholar]
- 17. Karduan SH, Sekula P, Valeyrie‐Allanore L, et al. Drug reaction with eosinophilia with systemic symptoms (DRESS): an original multisystem adverse drug reaction. Results from the prospective RegiSCAR study. Br J Dermatol. 2013;169(5):1071‐1080. [DOI] [PubMed] [Google Scholar]
- 18. Tobler R, Rohrlach A, Sobrier J, et al. Aboriginal mitogenomes reveal 50,000 years of regionalism in Australia. Nature. 2017;544(7649):180‐184. [DOI] [PubMed] [Google Scholar]
- 19. Ramírez E, Medrano‐Casique N, Tong HY, et al. Eosinophilic drug reactions detected by a prospective pharmacovigilance programme in a tertiary hospital. Br J Clin Pharmacol. 2017;83(2):400‐415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tassaneeyakul W, Prabmeechai N, Sukasem C, et al. Associations between HLA class I and cytochrome P450 2C9 genetic polymorphisms and phenytoin‐related severe cutaneous adverse reactions in a Thai population. Pharmacogenet Genomics. 2016;26(5):225‐234. [DOI] [PubMed] [Google Scholar]
- 21. Thynne T, Gabb GM. Therapeutic drug safety for indigenous Australians: how do we close the gap. Med J Aust. 2016;204(1):16‐17. [DOI] [PubMed] [Google Scholar]
- 22. Tucci JD, Pumuye PP, Helsby NA, et al. Pharmacogenomics in Papua new Guineans: unique profiles and implications for enhancing drug efficacy whilst improving drug safety. Pharmacogenet Genomics. 2018;28(6):153‐164. [DOI] [PubMed] [Google Scholar]
- 23. Helsby NA. CYP2C19 and CYP2D6 genotypes in Pacific peoples. Br J Clin Pharmacol. 2016;82:1302‐1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.


