Summary
The generation of humanized ectopic ossicles (hOss) in mice has been proposed as an advanced translational and fundamental model to study the human hematopoietic system. The approach relies on the presence of human bone marrow-derived mesenchymal stromal cells (hMSCs) supporting the engraftment of transplanted human hematopoietic stem and progenitor cells (HSPCs). However, the functional distribution of hMSCs within the humanized microenvironment remains to be investigated. Here, we combined genetic tools and quantitative confocal microscopy to engineer and subsequently analyze hMSCs′ fate and distribution in hOss. Implanted hMSCs reconstituted a humanized environment including osteocytes, osteoblasts, adipocytes, and stromal cells associated with vessels. By imaging full hOss, we identified rare physical interactions between hMSCs and human CD45+/CD34+/CD90+ cells, supporting a functional contact-triggered regulatory role of hMSCs. Our study highlights the importance of compiling quantitative information from humanized organs, to decode the interactions between the hematopoietic and the stromal compartments.
Subject Areas: Biological Sciences, Stem Cells Research, Tissue Engineering
Graphical Abstract
Highlights
-
•
Mesenchymal cells can generate human bone organs with tailored molecular signature
-
•
Mesenchymal cells reconstitute a human niche environment capable of regulating HSPCs
Biological Sciences; Stem Cells Research; Tissue Engineering
Introduction
The lifelong production of all human blood cell lineages is ensured by hematopoietic stem cells (HSCs) (Méndez-Ferrer et al., 2010, Morrison and Scadden, 2014). In adults, HSCs′ functions are maintained and tightly regulated in the specialized bone marrow (BM) microenvironment, referred to as the BM niche (Méndez-Ferrer et al., 2010, Morrison and Scadden, 2014). This environment is defined by unique physical properties (Engler et al., 2006, Guilak et al., 2009, Keung et al., 2010) and includes differentiated cells, extracellular matrix, and signaling factors (Rieger et al., 2009, Zhang and Lodish, 2008) essential for cell differentiation, survival (Knapp et al., 2016), and self-renewal (Kunisaki and Frenette, 2012, Méndez-Ferrer et al., 2010). However, the precise cellular and molecular composition of the human hematopoietic niche remains elusive (Bourgine et al., 2018, van Pel et al., 2015). Our understanding of human hematopoiesis largely relies on the analogy made with the mouse system (Schepers et al., 2015). In reality, despite commonly inherited genetic traits, HSC basic biology differs across species and the corresponding interactions with their niches are not fully conserved (Doulatov et al., 2012, van Pel et al., 2015). In consequence, information derived from murine studies does not systematically correlate with the human system, raising concerns about their direct relevance toward therapeutic developments (Doulatov et al., 2012).
Advanced xenotransplantation models offer robust engraftment and development of human hematopoiesis in mouse bones (Rongvaux et al., 2014). This has significantly contributed to the progressive understanding of human HSC functions in healthy and pathological setups (Antonelli et al., 2016, Reinisch et al., 2016). However, such humanized mouse models are incompatible with the organizational and functional study of the HSC niche, because the BM microenvironment remains entirely murine.
As an alternative, the possibility to engineer ectopic humanized ossicles (hOss) using human BM-derived mesenchymal stromal cells (hMSCs) is receiving increasing attention (Bourgine et al., 2014, Reinisch et al., 2016, Scotti et al., 2013), with demonstration of robust human blood engraftment in both healthy and malignant scenarios (Abarrategi et al., 2017, Martine et al., 2017, Reinisch et al., 2016). Although this is attributed to the presence of hMSCs, their contribution in the functional organization of the niche remains to be investigated.
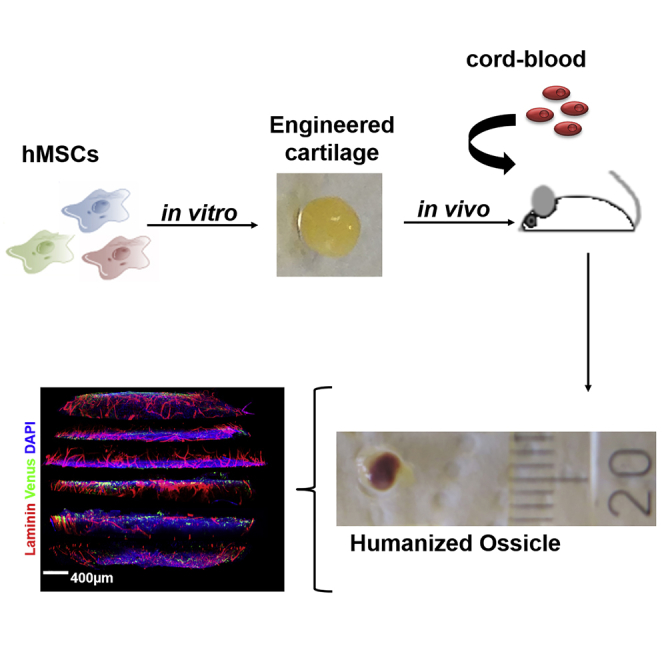
Here, we propose the genetic engineering of hMSCs together with the use of a recently developed deep multicolor imaging confocal analysis (Coutu et al., 2017) to achieve a first quantitative assessment of the distribution and role of hMSCs in hOss. We previously reported a developmental approach to bone organ formation, by in vitro chondrogenic priming of hMSCs (Bourgine et al., 2014, Scotti et al., 2013). Following hypertrophic cartilage (hyC) formation, the generated tissues remodel into hOss upon subcutaneous implantation in humanized mice (Fritsch et al., 2018) by recapitulating the endochondral ossification process (Kronenberg and Kronenberg, 2003). We target the further exploitation of this approach to engineer and characterize customized hematopoietic bone organs, here exemplified by the generation of niches overexpressing stromal-derived factor 1 alpha (SDF1α), through compositional and distributional assessment of their human cellular compartments.
The validation of the methodology bears relevance toward deciphering the human hematopoietic and skeletal systems using advanced and modular or tunable models of higher translational relevance.
Results
Primary hMSCs Can Be Genetically Engineered without Altering Their Capacity to Form Hypertrophic Cartilage
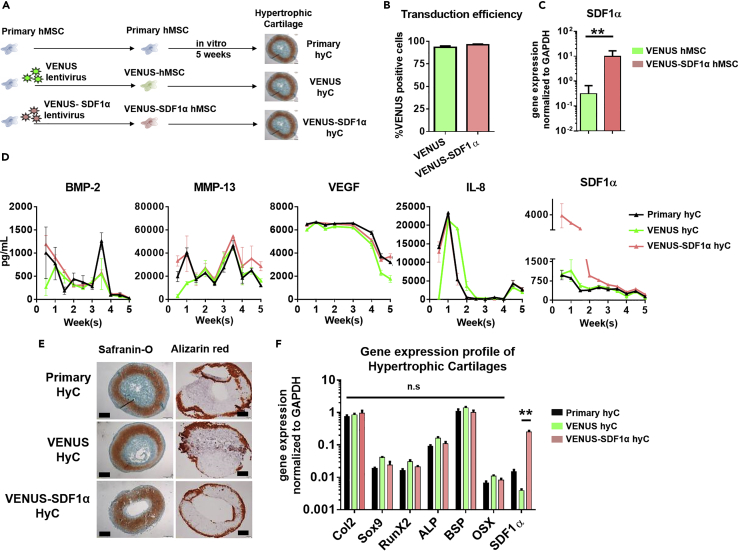
Before cartilage formation, hMSCs were transduced (Figure 1A) using a VENUS (mock control) or VENUS-SDF1α lentivirus (Figure S1). The transduction allowed the generation of homogeneous VENUS and VENUS-SDF1α hMSCs populations (>93% and 96% using VENUS and VENUS-SDF1α viruses, respectively, Figure 1B). The VENUS-SDF1α transduction led to a significant SDF1α overexpression (31-fold increase in RNA levels when compared with VENUS hMSCs, Figure 1C), with transduced and untransduced MSCs displaying unchanged phenotypes (Figure S2).
Figure 1.
Primary hMSCs Can Be Genetically Engineered without Altering Their Capacity to Form Hypertrophic Cartilage
(A) Experimental design for generation of hypertrophic cartilage (hyC). SDF1α, stromal-derived factor 1 alpha.
(B) Primary hMSCs were successfully transduced with the VENUS and VENUS-SDF1α lentiviruses, as assessed by flow cytometry. n ≥ 4 biological replicates.
(C) The VENUS-SDF1α transduction led to a significantly higher expression of SDF1α levels in corresponding cells before hyC formation. ∗∗p < 0.01, using non-parametric Mann-Whitney t test. n ≥ 5 biological replicates.
(D) All hyC display similar protein secretion patterns during in vitro culture time, but VENUS-SDF1α hyC releases higher amounts of SDF1α. n ≥ 3 biological replicates. BMP-2, bone morphogenetic protein-2; MMP-13, matrix metalloproteinase-13; VEGF, vascular endothelial growth factor; IL-8, interleukin-8.
(E) VENUS and VENUS-SDF1α successfully displayed features of mature hypertrophic cartilage tissue following 5 weeks of in vitro culture, as assessed by histological analysis. Safranin O staining reveals the presence of glycosaminoglycans (red), whereas alizarin red reveals the presence of mineralized tissue (red). Scale bars, 500 μm.
(F) After 5 weeks of in vitro culture, VENUS-SDF1α hyC successfully displayed a typical hypertrophic molecular profile while exhibiting a significant SDF1α increase.
∗∗p < 0.01, using one-way ANOVA. n ≥ 4 biological replicates. Col2, collagen type 2; RunX2, Runt-related transcription factor 2; ALP, alkaline phosphatase; BSP, bone sialoprotein; OSX, osterix. Data are represented as mean ± SEM.
Cells were subsequently seeded on collagen meshes and primed to form hyC. Over the 5-week course of in vitro culture, the total number of cells in the hyC remained stable (1.3 × 106 and 1.2 × 106 at weeks 1 and 5 respectively, Figure S3). During this period, the monitoring of proteins in hyC supernatant revealed comparable release profiles of angiogenic (vascular endothelial growth factor), osteoinductive (bone morphogenetic protein-2), bone remodeling (matrix metalloproteinase-13), and inflammatory (interleukin-8) factors, suggesting similar development of the templates by untransduced (primary hyC) or transduced hMSCs (VENUS and VENUS-SDF1α hyC) (Figure 1D). VENUS-SDF1α hyC secreted significantly higher amounts of SDF1α (4-fold increase at day 3, Figure 1D), although a progressive decrease was observed over time.
At the end of the in vitro culture, histological analysis indicated the successful formation of mature hyC in all groups, characterized by the large presence of glycosaminoglycans (safranin O, Figure 1E) and a mineralized ring at the periphery of the tissue (alizarin red, Figure 1E). Differentiation was confirmed by RT-PCR, revealing activation of chondrogenic (Collagen 2, Sox 9, Figure 1F) and hypertrophic genes (RUNX2, ALP, BSP, OSX, Figure 1F) in all hyCs. Importantly, VENUS-SDF1α hyCs were shown to maintain a marked SDF1α overexpression (Figure 1F) when compared with primary and VENUS hyCs.
Hypertrophic Cartilage with a Targeted SDF1α Enrichment Can Be Generated
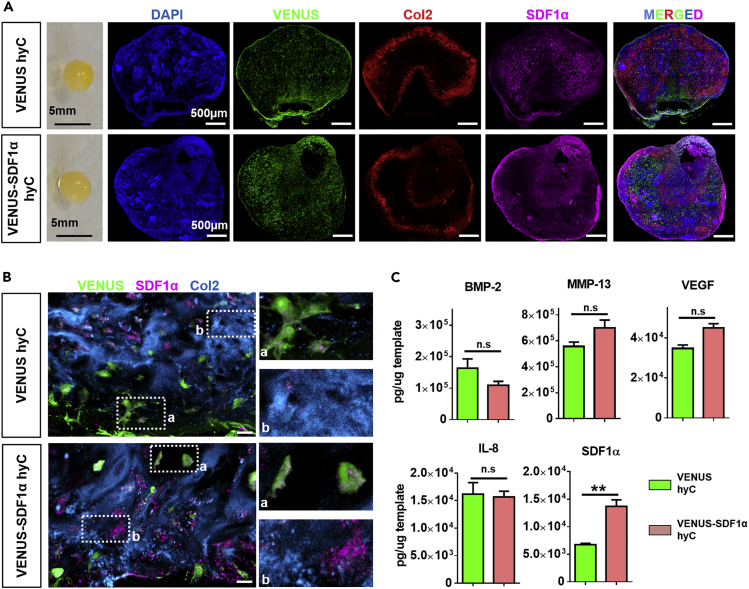
After demonstrating the similar quality of hyCs and hOss derived from primary or VENUS hMSCs (Figure S4), VENUS hyCs were further used as the control group allowing for the tracing of hMSCs via the VENUS signal.
Multicolor confocal analysis of thick hyC sections was performed to investigate the presence and distribution of cells and SDF1α in the templates. VENUS cells were homogenously distributed within the tissue, largely embedded into a collagen type 2-rich matrix with detectable SDF1α proteins (Figure 2A). High-resolution imaging revealed presence of the chemokine intracellularly in both VENUS and VENUS-SDF1α cells in their corresponding hyC (Figure 2B). The SDF1α protein was also found associated with the extracellular matrix (ECM), as shown by colocalization with collagen type 2, in a more abundant fashion in VENUS-SDF1α samples (Figure 2B). To confirm microscopic observations, hyCs were lysed and assessed for their content in a panel of growth factors, including SDF1α. VENUS and VENUS-SDF1α hyCs displayed comparable protein contents (Figure 2C) except a 2-fold SDF1α enrichment in the VENUS-SDF1α templates (Figure 2C). Consistent with previous observations (Dalonneau et al., 2014, Pelletier et al., 2000), the reported decrease in secreted SDF1α over culture time (Figure 1D) can be explained by SDF1α′s capacity to bind to the ECM, leading to its progressive embedding. We thus report the successful tuning of cartilage tissue′s composition, through a targeted enrichment of SDF1α content.
Figure 2.
Hypertrophic Cartilage with a Targeted SDF1α Enrichment Can Be Generated
(A) VENUS and VENUS-SDF1α hyC consist in cartilage pellets (macroscopic view, left) in which hMSCs (VENUS positive) and the SDF1α protein are found abundantly in the collagen-rich matrix, as assessed by immunofluorescence analysis of thick hyC sections. Col2, collagen type 2. Scale bars, 500 μm.
(B) SDF1α is more abundant in the ECM of VENUS-SDF1α hyC, as revealed by high-resolution immunofluorescent imaging. Scale bars, 20 μm. Col2, collagen type 2.
(C) A significant and specific SDF1α enrichment is obtained in the VENUS-SDF1α hyC, as assessed by protein quantification. n = 3 biological repeats. ∗∗p < 0.01, using one-way ANOVA test. Data are represented as mean ± SEM.
Molecularly Engineered hyC Can Remodel into Humanized Bone Organs of Distinct Blood Compositions
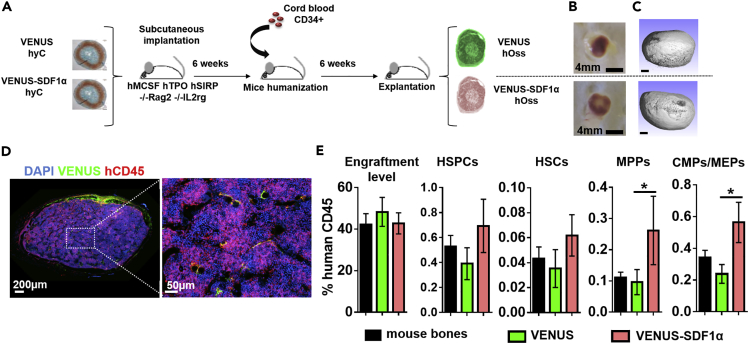
In vitro-engineered VENUS and VENUS-SDF1α hyCs were subcutaneously implanted in mice (Figure 3A). After 6 weeks, when hyCs are expected to be remodeled into bone tissue, animals were intravenously transplanted with CD34+ cord-blood-derived hematopoietic cells to reconstitute human hematopoiesis (Figure 3A). After a total in vivo period of 12 weeks, VENUS and VENUS-SDF1α hyCs remodeled into ectopic ossicles exhibiting macroscopic evidence of vascularization (Figure 3B). Microtomography scans (Figure 3C) revealed the formation of mature bone tissue with no quality differences between the two hOss types, consisting in a spheroid organ of 18 ± 2.1 mm3 (Figure S5). Confocal microscopy allowed to identify human blood cells in the hOss, forming heterogeneous “islets” of human hematopoiesis, indicating successful engraftment (Figure 3D).
Figure 3.
Molecularly Engineered hyC Can Remodel into Humanized Bone Organs of Distinct Blood Compositions
(A) Experimental design for the generation of humanized ossicles (hOss). Engineered hyCs are implanted into immunocompromised animals. Mice are humanized 6 weeks later by intravenous transplantation of human CD34+ isolated from cord blood. Constructs are retrieved 6 weeks later for analysis.
(B and C) (B) VENUS and VENUS-SDF1α hyCs successfully remodeled into ossicles, as shown macroscopically by blood colonization and (C) by microtomographic scans revealing the formation of mature bone tissue. Ossicles were retrieved after 12 weeks in vivo. Scale bars, 0.5 mm.
(D) Humanized ossicles display a chimeric blood composition organized in “islets” of human hematopoieisis.
(E) SDF1α-overexpressing ossicles displayed higher frequencies of human blood populations, as assessed by flow cytometry. n ≥ 18 biological replicates from four independent experiments. ∗p < 0.5, using one-way ANOVA test. CMPs, common myeloid progenitors; HSCs, hematopoietic stem cells; HSPCs, hematopoietic stem and progenitor cells; MEPs, megakaryocyte-erythroid progenitors; MPPs, multipotent progenitors. Data are represented as mean ± SEM.
Human blood populations were quantified by flow cytometry in retrieved hOss and corresponding mouse bones (Figures S6A and S6B). Engraftment of human blood cells was similar with an average hCD45 chimerism level of 40% (Figure 3E). VENUS hOss and mouse bones displayed comparable frequencies of naive and more committed blood populations (Figures 3E and S6C), whereas hOss overexpressing SDF1α showed significantly higher frequencies of multipotent progenitors (MPPs) and common myeloid progenitors (CMPs)/megakaryocyte-erythroid progenitor (MEPs) (2.7- and 2.4-fold increase, respectively) and superior HSPC and HSC content (1.8- and 1.9-fold increase, respectively), although not reaching significance.
The functionality of hCD45/CD34+ cells retrieved from mouse bones or hOss was evaluated by in vitro colony formation unit (CFU) assays. Cells were capable of efficiently giving rise to all myeloid colonies, but the hCD45/CD34+ fraction derived from hOss displayed a significantly higher potential to form hematopoietic colonies, including GEmM, than the corresponding population retrieved from mouse bones (Figure S6D). No differences in CFU activity were observed between cells retrieved from VENUS or VENUS-SDF1α hOss, suggesting that the SDF1α overexpression did not affect stem and progenitor functionalities. We thus validate the generation of SDF1α-customized hOss, composed of an increased frequency of HSPCs without alterations of their functionality.
Quantitative Microscopy of Bone Organs Reveals the Reconstitution of a Human Mesenchymal Niche including Rare Mesenchymal-Hematopoietic Physical Interactions
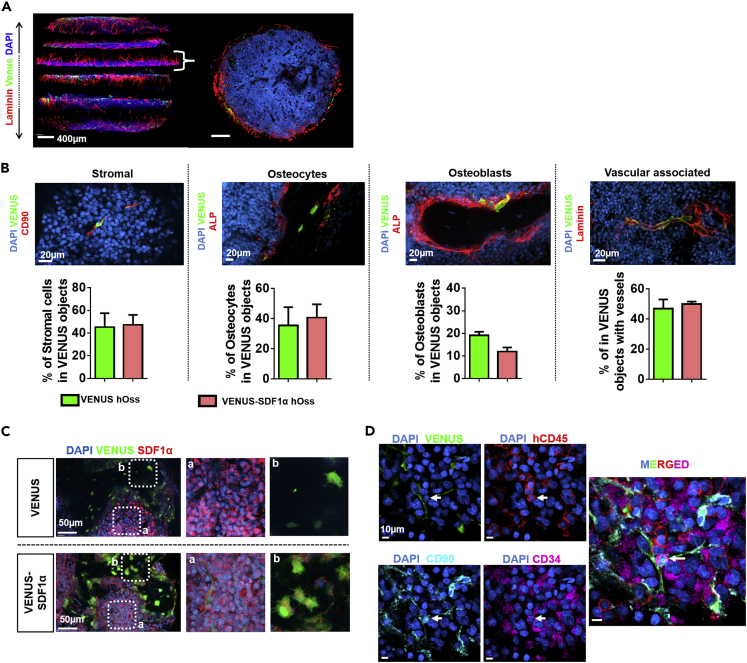
Deep confocal analysis on both types of hOss offered a quantitative understanding of the reconstituted BM environment. Serial sectioning of hOss allowed gathering comprehensive 3D information (Figures 4A and S7A) and indicated an intense vascularization surrounding the hOss with its cavity largely filled by BM cells. The hOss were also connected to the host nervous system as shown by evidences of innervation (peripherin, Figure S7B).
Figure 4.
Quantitative Microscopy of Bone Organs Reveals the Reconstitution of a Human Mesenchymal Niche Including Rare Mesenchymal-Hematopoietic Physical Interactions
(A) Multidimensional confocal immunofluorescence imaging allows for the reconstitution of hOss for 3D quantitative information retrieval (left). Top view of a transverse hOss section (right) illustrating the internal bone marrow cavity (DAPI) and intense peripheral vascularization (lamimin). Scale bars, 400 μm.
(B) Top: implanted hMSCs (VENUS positive) demonstrate fate plasticity by acquiring multiple niche cell phenotypes. Scale bars, 20 μm. Bottom: fate quantification of VENUS hMSCs based on the segmentation of immunofluorescence data. Stromal cells, venus+/CD90+/stroma localization; osteocytes, venus+/ALP-/localization in bone nodules; osteoblasts, venus+/ALP+/localization at the bone and bone marrow interface; vascular associated, venus+/distance to lamimin+ vessels < 1 μm; ALP, alkaline phosphatase. n ≥ 3 biological replicates.
(C) The SDF1α protein was expressed by blood cells and also found more abundantly in VENUS cells from the VENUS-SDF1α ossicles. Right and b are magnification panels.
(D) Deep confocal analysis of hOss allows for the identification and localization of a rare HSPC subset (arrow, hCD45/CD34/CD90), found directly in contact with hMSC-derived niche cells. n = 10 events detected in 5.9 mm3 of tissue scanned. Data are represented as mean ± SEM.
The presence of hMSCs in both VENUS and VENUS-SDF1α organs was identified by VENUS expression (Figure S8A). The segmentation of VENUS cells (Figure S8A) in corresponding sections (Figure S8B) allowed determining the average number of hMSCs per hOss. From the 1.2 × 106 hMSCs present in the hyC at the time of implantation, only 0.1 × 106 were still populating the hOss, corresponding to a 90% decrease (Figure S8C). To corroborate this finding, flow cytometry quantification of VENUS cells after hOss digestion was also performed, giving a number of hMSCs per hOss of 0.06 × 106 (Figure S8C). The difference is likely due to difficulty to retrieve bone-embedded hMSCs and adipocytes. The observed hMSC death was not due to the animal irradiation, because non-irradiated hOss gave similar numbers (0.07 × 106, Figure S8C).
Remarkably, in the engineered hOss, hMSCs were associated with an important fate diversity following the remodeling of hyC. HMSCs′ fates in bone organs were quantified by specific segmentation strategies (c.f. Methods section and Figures S8 and S9). First, no significant differences could be observed between VENUS and VENUS-SDF1α hOss, indicating that the hMSCs′ genetic modification did not impact their subsequent fate decisions upon hOss formation. The capacity to manipulate hMSCs without impairing their endochondral program allows the stringent assessment of SDF1α effects. As such, the previously observed distinct human blood composition in VENUS-SDF1α hOss can be strictly attributed to the factor overexpression.
In all hOss, hMSCs were abundantly found within the BM stroma (stromal, 45% and 47% in VENUS and VENUS-SDF1α hOss respectively, Figure 4B), exhibiting a fibroblast-like shape and positivity for CD90 (Figure S9A). A large fraction of them (47% and 50%, respectively; Figure 4B) was directly associated with vasculature (0–1 μm distance to vessels). The hMSCs′ enrichment at vessel sites was confirmed by comparison to a random dots distribution (Figure S9C). HMSCs also differentiated into the osteogenic lineage in the form of osteocytes embedded in the bone matrix (osteocytes, Figures 4B and S9A), accounting for 36% (VENUS hOss) and 41% (VENUS-SDF1α hOss) of total VENUS cells. Lining osteoblasts were less abundant (19% in VENUS hOss versus 12% in VENUS-SDF1α hOss, Figures 4B and S9A). To a lower extent, we also identified hMSCs differentiated into the adipogenic lineage, as shown by the presence of VENUS-positive adipocytes (Figure S7D).
To obtain a better understanding of the observed in vivo fate diversity, we further investigated the proportion of hMSCs committed to certain lineages before implantation, at the hyC stage (5 weeks in vitro culture). The Sox-9 and RUNX2 transcription factors are typically associated with chondrogenic (Akiyama et al., 2002) and osteoblastic (Ducy et al., 1997) differentiation, respectively; 41.3% (±5.6) of the cells were positive for Sry-box 9 (Sox-9), 6.3% (±5) expressed the Runt-related transcription factor 2 (RUNX2), and 10.8% (±5.6) were both RUNX2 and Sox-9 positive (Figure S10). No cells showed positivity for the adipocytic marker peroxisome proliferator-activated receptor gamma (Lefterova et al., 2014). As such, the majority of our cells were chondrocytes (Sox-9 positivity), hypertrophic chondrocytes (Sox-9 and/or RUNX2 positivity), or osteoblasts (RUNX2 positivity). Interestingly, despite the 5-week course of differentiation, 29.9% ± 1.6% maintained the expression of Stro-1, a progenitor marker associated with multipotency (Lin et al., 2011). This indicates that some hMSCs are not fully committed at the end of the in vitro differentiation stage and acquire their definitive function after implantation.
Further immunofluorescence analysis of the ossicles was performed. SDF1α staining revealed the presence of the protein in the marrow (Figure 4C), expressed by blood cells (Dar et al., 2006). The SDF1α protein could also be detected in hMSCs-derived cells from VENUS-SDF1α hOss (Figure 4C). Acquisition of deep multicolor staining was further performed for the identification of hMSC-derived niche cells (positive for VENUS) and particular HSPC populations within engineered hOss. This allowed the localization of a very rare subset of HSPCs (hCD45+/CD34+/CD90+) described to be enriched for functional HSCs, which was consistently in physical contact (less than 1 μm distance) with VENUS cells, in both VENUS and VENUS-SDF1α niches (n = 10 events detected in 5.9 mm3 of tissue scanned, Figure 4D). To ensure that those interactions did not result from a random distribution, we investigated the probability for HSPCs touching an hMSC (Figure S11). This probability was found to be 36% (±11), although 100% of the HSPCs we found were in contact with hMSCs.
This recurrent physical interaction between the human stromal and naive hematopoietic compartments, combined with the finding that SDF1α customization leads to changes in frequencies of hematopoietic populations, supports a functional contact-triggered regulation of HSPCs by the human mesenchymal compartment, to date only reported in mouse studies (Gomariz et al., 2018, Rongvaux et al., 2011).
Discussion
We report the engineering and characterization of customized human hematopoietic bone organs. The method relies on the genetic modification of primary hMSCs, their priming to recapitulate the developmental program of endochondral ossification (Kronenberg and Kronenberg, 2003), and quantitative multidimensional imaging of the reconstituted human BM environment.
The study of hematopoiesis in a humanized context is a primary challenge. The generation of transgenic animals supporting human engraftment is associated with some limitations (Devoy et al., 2012), including time-consuming single-gene targeting, the unpredictable biological outcome (e.g., embryonic lethality, low efficiency, absence of recognizable phenotypes), and the often non-tissue-specific nature if at all conditional. Instead, our strategy relies on the exploitation and characterization of the hOss model, using hMSCs as cellular vectors for the targeted delivery of factors influencing the composition of the human blood compartment. The introduced modification is thus confined within the BM tissue as ensured by hMSC-derived niche cells.
The biological validation of the method was performed using SDF1α as a known factor influencing stem cell behavior. SDF1α has been reported both as stem cell chemoattractant (Aiuti et al., 1997, Lapidot and Kollet, 2002, Mohle et al., 1998) and pro-quiescent molecule (Itkin and Lapidot, 2011, Tzeng et al., 2011), thus offering multiple readouts to validate the effects of its overexpression in hOss. Pre-existing molecular engineering approach (Carretta et al., 2017, Chen et al., 2012) models did not investigate the effect of the genetic modification on the reconstituted human niche environment. As a direct consequence, impact on the blood compartment could not be strictly attributed to the factor′s secretion. Here, the SDF1α overexpression was shown to specifically affect the human blood composition while not affecting the fate and distribution of implanted hMSCs. The capacity to manipulate hMSCs without impairing their endochondral program is a pre-requisite for the direct assessment of SDF1α effects. We observed a specific enrichment in CMPs/MEPs, MPPs, and HSC populations in SDF1α-overexpressing hOss. All these populations express the CXCR4 receptor (Toni et al., 2011), thus being sensitive to the SDF1α chemoattractant effect (Peled et al., 1999). The previously reported SDF1α-driven mobilization of CD34+ cells by stromal cells included the recruitment of more committed erythroid, lymphoid, and myeloid lineages (Bleul et al., 2002). Our observations are thus in line with the existing literature, although our study is the first exploiting a humanized approach to evidence an SDF1α-triggered effect. Although our model was previously validated for the engraftment of fully functional HSCs (Fritsch et al., 2018), secondary transplantation would be required to assess putative SDF1α effects on the self-renewal of long-term repopulating stem cells.
Interestingly, the association of SDF1α with proteoglycans—the main constituents of cartilaginous ECM (Roughley and Lee, 1994)—was reported to strongly promote the migration of HSPCs (Netelenbos et al., 2002). This might suggest that the observed effects result from preferential homing at the time of engraftment (Lapidot and Kollet, 2002), although a different cycling rate of HSPCs cannot be excluded.
In our study, we used advanced microscopy tools to monitor hMSCs within the engineered tissues, from the in vitro hyC stage to the fully remodeled hOss. As easily accessible organs tunable in size, quantitative 3D information on the hOss cellular composition could be retrieved and offered a comprehensive understanding of the human niche in this in vivo setting. This revealed a remarkable degree of hMSCs′ plasticity in the model, giving rise to several niche phenotypes, including lining osteoblasts, osteocytes, stromal cells, and adipocytes. Distance analyses indicated not only a strong association of hMSCs with vasculature but also direct physical interactions with HSPCs. The observed fate diversity may derive from a pool of hMSCs with no signs of chondrogenic or osteoblastic commitment at the hyC stage. However, lineage-committed hMSCs have also been described as capable of transdifferentiation (Song and Tuan, 2004).
Although previous studies reported the presence of hMSCs in ossicles (Abarrategi et al., 2017, Martine et al., 2017, Reinisch et al., 2016), their functional status has not been rigorously demonstrated beyond their support for human blood engraftment. Our work evidences for the first time a functional regulatory role of hMSCs in the model, validated by the SDF1α customization leading to distinct changes in frequencies of hematopoietic populations. These were achieved despite the relatively low number of hMSCs composing the niche and supporting human blood cell engraftment. Collectively, these findings reinforce the notion that hMSCs are essential niche players (Méndez-Ferrer et al., 2010, Morrison and Scadden, 2014) in the engineered ossicles and support a contact-triggered regulation of HSPCs by the mesenchymal compartment, only previously reported in mouse studies (Rongvaux et al., 2011). The detection of a high number of interactions of a more specific HSC phenotype (CD45+/CD34+/CD38-/CD45RA-/CD90+/CD49f+) (Fares et al., 2014), which requires challenging immunofluorescence multiplexing, would, however, be required to provide direct evidence of a physical interaction between hMSCs and functional HSCs.
The functional role of hMSCs in the hOss model supports their use for the molecular engineering of human niches. The importance of the proximity between hMSCs and HSPCs could be further evaluated by overexpression of signaling molecules requiring direct contact of adjacent cells for influencing their fate decisions, e.g., Notch ligands (Artavanis-Tsakonas et al., 1999). The paradigm of customization can also be further explored with additional factors putatively affecting stem cell homing/localization/function in pathological scenarios (e.g., by engraftment of leukemic primary material) and include the impact assessment on the human niche compartment. In fact, the hOss could be valuable for the identification of specific human niche cell populations, derived from the implanted hMSCs. This is of particular importance in pathologic scenarios, in which the role of the stroma and associated factors in disease evolution remains elusive (Schepers et al., 2015, Schepers et al., 2013). Our model could thus help deciphering the complex phenotypes and functions associated with hMSCs (Mo et al., 2016, Nombela-Arrieta et al., 2011) in myeloid or lymphoid malignancies. Along the same line, our model could also be exploited to study the engraftment of metastatic solid tumors (e.g., breast, prostate) that naturally migrate to bones, including those for which SDF1α has been shown to be highly expressed in BM sites of tumor metastasis (Roccaro et al., 2014).
Toward these objectives, the exploitation of dedicated cell lines may not only facilitate molecular engineering of hMSCs but also potentially lead to their higher survival in the BM. This would maximize the therapeutic delivery of factors impacting engrafted healthy or malignant blood populations. However, so far no hMSC line was proved capable of recapitulating the endochondral process.
Finally, using similar knockout or knockin strategies in hMSCs, we also envision the possibility of identifying key molecular players of the endochondral program, by studying their impact on hMSC fate decision. The ossicle would thus be exploited as a developmental model of human bone formation.
Limitations of the Study
For immunofluorescence analysis, the hCD45+/CD34+/CD90 + phenotype was used to identify HSPCs. Of this population, 24.6% are CD34+/CD38-/CD45RA-CD90+, of which 5% were reported to be functional HSCs in freshly isolated cord blood (Notta et al., 2011). The hCD45+/CD34+/CD90 + population thus contains ca. 1.2% functional HSCs. Detection of functional HSCs will require improved HSC markers. In addition, owing to the large required effort, only a small number of these cells were imaged in ossicles. A higher number will allow robust statistical analysis of human MSC-HSPC interactions.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
This study was supported in part by the SystemsX (StemSysMed grant #2014/266 to M.G.M., T.S., and I.M.) and Division III Programs (grant # 31003A_156430 to I.M.) of the Swiss National Science Foundation.
Author Contributions
P.E.B., I.M., T.S., and M.G.M. designed the research. P.E.B., K.F., and S.P. performed experiments. P.E.B., K.F., S.P., H.T., L.K., K.D.K., and D.L.C. analyzed data. P.E.B., T.S., I.M., and M.G.M. wrote the manuscript. P.E.B., T.S., I.M., and M.G.M. financed the project.
Declaration of Interests
The authors declare no competing interests.
Published: September 27, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2019.08.006.
Contributor Information
Markus G. Manz, Email: markus.manz@usz.ch.
Ivan Martin, Email: ivan.martin@usb.ch.
Timm Schroeder, Email: timm.schroeder@bsse.ethz.ch.
Supplemental Information
References
- Abarrategi A., Foster K., Hamilton A., Mian S.A., Passaro D., Gribben J., Mufti G., Bonnet D. Versatile humanized niche model enables study of normal and malignant human hematopoiesis. J. Clin. Invest. 2017;127:543–548. doi: 10.1172/JCI89364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiuti A., Webb I.J., Bleul C., Springer T., Gutierrez-Ramos J.C. The chemokine SDF-1 is a chemoattractant for human CD34+ hematopoietic progenitor cells and provides a new mechanism to explain the mobilization of CD34+ progenitors to peripheral blood. J. Exp. Med. 1997;185:111–120. doi: 10.1084/jem.185.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama H., Chaboissier M.C., Martin J.F., Schedl A., De Crombrugghe B. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 2002;16:2813–2828. doi: 10.1101/gad.1017802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonelli A., Noort W.A., Jaques J., de Boer B., de Jong-Korlaar R., Brouwers-Vos A.Z., Lubbers-Aalders L., van Velzen J.F., Bloem A.C., Yuan H. Establishing human leukemia xenograft mouse models by implanting human bone marrow-like scaffold-based niches. Blood. 2016;128:2949–2959. doi: 10.1182/blood-2016-05-719021. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S., Rand M.D., Lake R.J. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Bleul C., Webb I.J., Springer T., Gutierrez-Ramos J.C., Aiuti A. The chemokine SDF-1 is a chemoattractant for human CD34 + hematopoietic progenitor cells and provides a new mechanism to explain the mobilization of CD34 + progenitors to peripheral blood. J. Exp. Med. 2002 doi: 10.1084/jem.185.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgine P.E., Martin I., Schroeder T. Engineering human bone marrow proxies. Cell Stem Cell. 2018;22:298–301. doi: 10.1016/j.stem.2018.01.002. [DOI] [PubMed] [Google Scholar]
- Bourgine P.E., Scotti C., Pigeot S., Tchang L.A., Todorov A., Martin I. Osteoinductivity of engineered cartilaginous templates devitalized by inducible apoptosis. Proc. Natl. Acad. Sci. U S A. 2014;111:17426–17431. doi: 10.1073/pnas.1411975111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carretta M., de Boer B., Jaques J., Antonelli A., Horton S.J., Yuan H., de Bruijn J.D., Groen R.W.J., Vellenga E., Schuringa J.J. Genetically engineered mesenchymal stromal cells produce IL-3 and TPO to further improve human scaffold-based xenograft models. Exp. Hematol. 2017;51:36–46. doi: 10.1016/j.exphem.2017.04.008. [DOI] [PubMed] [Google Scholar]
- Chen Y., Jacamo R., Shi Y.X., Wang R.Y., Battula V.L., Konoplev S., Strunk D., Hofmann N.A., Reinisch A., Konopleva M., Andreeff M. Human extramedullary bone marrow in mice: a novel in vivo model of genetically controlled hematopoietic microenvironment. Blood. 2012;119:4971–4980. doi: 10.1182/blood-2011-11-389957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutu D.L., Kokkaliaris K.D., Kunz L., Schroeder T. Three-dimensional map of non-hematopoetic bone and bone marrow cells and molecules. Nat. Biotechnol. 2017;35:1202–1210. doi: 10.1038/nbt.4006. [DOI] [PubMed] [Google Scholar]
- Dalonneau F., Liu X.Q., Sadir R., Almodovar J., Mertani H.C., Bruckert F., Albiges-Rizo C., Weidenhaupt M., Lortat-Jacob H., Picart C. The effect of delivering the chemokine SDF-1α in a matrix-bound manner on myogenesis. Biomaterials. 2014;35:4525–4535. doi: 10.1016/j.biomaterials.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dar A., Kollet O., Lapidot T. Mutual, reciprocal SDF-1/CXCR4 interactions between hematopoietic and bone marrow stromal cells regulate human stem cell migration and development in NOD/SCID chimeric mice. Exp. Hematol. 2006;34:967–975. doi: 10.1016/j.exphem.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Devoy A., Bunton-Stasyshyn R.K.A., Tybulewicz V.L.J., Smith A.J.H., Fisher E.M.C. Genomically humanized mice: technologies and promises. Nat. Rev. Genet. 2012;13:14–20. doi: 10.1038/nrg3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doulatov S., Notta F., Laurenti E., Dick J.E. Hematopoiesis: a human perspective. Cell Stem Cell. 2012;10:120–136. doi: 10.1016/j.stem.2012.01.006. [DOI] [PubMed] [Google Scholar]
- Ducy P., Zhang R., Geoffroy V., Ridall A.L., Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- Engler A.J., Sen S., Sweeney H.L., Discher D.E. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- Fares I., Chagraoui J., Gareau Y., Gingras S., Ruel R., Mayotte N., Csaszar E., Knapp D.J.H.F., Miller P., Ngom M. Pyrimidoindole derivatives are agonists of human hematopoietic stem cell self-renewal. Science. 2014;345:1509–1512. doi: 10.1126/science.1256337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch K., Pigeot S., Feng X., Bourgine P.E., Schroeder T., Martin I., Manz M.G., Takizawa H. Engineered humanized bone organs maintain human hematopoiesis in vivo. Exp. Hematol. 2018;61:45–51.e5. doi: 10.1016/j.exphem.2018.01.004. [DOI] [PubMed] [Google Scholar]
- Gomariz A., Helbling P.M., Isringhausen S., Suessbier U., Becker A., Boss A., Nagasawa T., Paul G., Goksel O., Székely G. Quantitative spatial analysis of haematopoiesis- regulating stromal cells in the bone marrow microenvironment by 3D microscopy. Nat. Commun. 2018;9:2532. doi: 10.1038/s41467-018-04770-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilak F., Cohen D.M., Estes B.T., Gimble J.M., Liedtke W., Chen C.S. Control of stem cell fate by physical interactions with the extracellular matrix. Cell Stem Cell. 2009;5:17–26. doi: 10.1016/j.stem.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itkin T., Lapidot T. SDF-1 keeps HSC quiescent at home. Blood. 2011;117:373–374. doi: 10.1182/blood-2010-09-307843. [DOI] [PubMed] [Google Scholar]
- Keung A.J., Healy K.E., Kumar S., Schaffer D.V. Biophysics and dynamics of natural and engineered stem cell microenvironments. Wiley Interdiscip. Rev. Syst. Biol. Med. 2010;2:49–64. doi: 10.1002/wsbm.46. [DOI] [PubMed] [Google Scholar]
- Knapp D.J.H.F., Hammond C.A., Aghaeepour N., Miller P.H., Pellacani D., Beer P.A., Sachs K., Qiao W., Wang W., Humphries R.K. Distinct signaling programs control human hematopoietic stem cell survival and proliferation. Blood. 2016;129:307–319. doi: 10.1182/blood-2016-09-740654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronenberg H.M., Kronenberg H.M. Developmental regulation of the growth plate. Nature. 2003;423:332–336. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- Kunisaki Y., Frenette P.S. The secrets of the bone marrow niche: enigmatic niche brings challenge for HSC expansion. Nat. Med. 2012;18:864–865. doi: 10.1038/nm.2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapidot T., Kollet O. The essential roles of the chemokine SDF-1 and its receptor CXCR4 in human stem cell homing and repopulation of transplanted immune-deficient NOD/SCID and NOD/SCID/B2mnull mice. Leukemia. 2002;16:1992–2003. doi: 10.1038/sj.leu.2402684. [DOI] [PubMed] [Google Scholar]
- Lefterova M.I., Haakonsson A.K., Lazar M.A., Mandrup S. PPARγ and the global map of adipogenesis and beyond. Trends Endocrinol. Metab. 2014;25:293–302. doi: 10.1016/j.tem.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin G., Liu G., Banie L., Wang G., Ning H., Lue T.F., Lin C.-S. Tissue distribution of mesenchymal stem cell marker stro-1. Stem Cells Dev. 2011;20:1747–1752. doi: 10.1089/scd.2010.0564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martine L.C., Holzapfel B.M., McGovern J.A., Wagner F., Quent V.M., Hesami P., Wunner F.M., Vaquette C., De-Juan-Pardo E.M., Brown T.D. Engineering a humanized bone organ model in mice to study bone metastases. Nat. Protoc. 2017;12:639–663. doi: 10.1038/nprot.2017.002. [DOI] [PubMed] [Google Scholar]
- Méndez-Ferrer S., Michurina T.V., Ferraro F., Mazloom A.R., Macarthur B.D., Lira S.A., Scadden D.T., Ma’ayan A., Enikolopov G.N., Frenette P.S. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010;466:829–834. doi: 10.1038/nature09262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo M., Wang S., Zhou Y., Li H., Wu Y. Mesenchymal stem cell subpopulations: phenotype, property and therapeutic potential. Cell. Mol. Life Sci. 2016;73:3311–3321. doi: 10.1007/s00018-016-2229-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohle R., Bautz F., Rafii S., Moore M.A., Brugger W., Kanz L. The chemokine receptor CXCR-4 is expressed on CD34+ hematopoietic progenitors and leukemic cells and mediates transendothelial migration induced by stromal cell-derived factor-1. Blood. 1998;91:4523–4530. [PubMed] [Google Scholar]
- Morrison S.J., Scadden D.T. The bone marrow niche for haematopoietic stem cells. Nature. 2014;505:327–334. doi: 10.1038/nature12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netelenbos T., Zuijderduijn S., Van Den Born J., Kessler F.L., Zweegman S., Huijgens P.C., Dräger A.M. Proteoglycans guide SDF-1-induced migration of hematopoietic progenitor cells. J. Leukoc. Biol. 2002;72:353–362. [PubMed] [Google Scholar]
- Nombela-Arrieta C., Ritz J., Silberstein L.E. The elusive nature and function of mesenchymal stem cells. Nat. Rev. Mol. Cell Biol. 2011;12:126–131. doi: 10.1038/nrm3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notta F., Doulatov S., Laurenti E., Poeppl A., Jurisica I., Dick J.E. Isolation of single human hematopoietic stem cells capable of long-term multilineage engraftment. Science. 2011;333:218–221. doi: 10.1126/science.1201219. [DOI] [PubMed] [Google Scholar]
- Peled A., Petit I., Kollet O., Magid M., Ponomaryov T., Byk T., Nagler A., Ben-Hur H., Many A., Shultz L. Dependence of human stem cell engraftment and repopulation of NOD/SCID mice on CXCR4. Science. 1999;283:845–848. doi: 10.1126/science.283.5403.845. [DOI] [PubMed] [Google Scholar]
- Pelletier A.J., van der Laan L.J.W., Hildbrand P., Siani M.A., Thompson D.A., Dawson P.E., Torbett B.E., Salomon D.R. Presentation of chemokine SDF-1α by fibronectin mediates directed migration of T cells. Blood. 2000;96:2682–2690. [PubMed] [Google Scholar]
- Reinisch A., Thomas D., Corces M.R., Zhang X., Gratzinger D., Hong W.-J., Schallmoser K., Strunk D., Majeti R. A humanized bone marrow ossicle xenotransplantation model enables improved engraftment of healthy and leukemic human hematopoietic cells. Nat. Med. 2016;22:812–821. doi: 10.1038/nm.4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieger M.A., Hoppe P.S., Smejkal B.M., Eitelhuber A.C., Schroeder T. Hematopoietic cytokines can instruct lineage choice. Science. 2009;325:217–218. doi: 10.1126/science.1171461. [DOI] [PubMed] [Google Scholar]
- Roccaro A.M., Sacco A., Purschke W.G., Moschetta M., Buchner K., Maasch C., Zboralski D., Zöllner S., Vonhoff S., Mishima Y. SDF-1 inhibition targets the bone marrow niche for cancer therapy. Cell Rep. 2014;9:118–128. doi: 10.1016/j.celrep.2014.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rongvaux A., Willinger T., Martinek J., Strowig T., Gearty S.V., Teichmann L.L., Saito Y., Marches F., Halene S., Palucka A.K. Development and function of human innate immune cells in a humanized mouse model. Nat. Biotechnol. 2014;32:364–372. doi: 10.1038/nbt.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rongvaux A., Willinger T., Takizawa H., Rathinam C., Auerbach W., Murphy A.J., Valenzuela D.M., Yancopoulos G.D., Eynon E.E., Stevens S. Human thrombopoietin knockin mice efficiently support human hematopoiesis in vivo. Proc. Natl. Acad. Sci. U S A. 2011;108:2378–2383. doi: 10.1073/pnas.1019524108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roughley P.J., Lee E.R. Cartilage proteoglycans: structure and potential functions. Microsc. Res. Tech. 1994;28:385–397. doi: 10.1002/jemt.1070280505. [DOI] [PubMed] [Google Scholar]
- Schepers K., Campbell T.B., Passegué E. Normal and leukemic stem cell niches: insights and therapeutic opportunities. Cell Stem Cell. 2015;16:254–267. doi: 10.1016/j.stem.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepers K., Pietras E.M., Reynaud D., Flach J., Binnewies M., Garg T., Wagers A.J., Hsiao E.C., Passegué E. Myeloproliferative neoplasia remodels the endosteal bone marrow niche into a self-reinforcing leukemic niche. Cell Stem Cell. 2013;13:285–299. doi: 10.1016/j.stem.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotti C., Piccinini E., Takizawa H., Todorov A., Bourgine P., Papadimitropoulos A., Barbero A., Manz M.G., Martin I. Engineering of a functional bone organ through endochondral ossification. Proc. Natl. Acad. Sci. U S A. 2013;110:3997–4002. doi: 10.1073/pnas.1220108110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L., Tuan R.S. Transdifferentiation potential of human mesenchymal stem cells derived from bone marrow. FASEB J. 2004;18:980–982. doi: 10.1096/fj.03-1100fje. [DOI] [PubMed] [Google Scholar]
- Toni R., Tampieri A., Zini N., Strusi V., Sandri M., Dallatana D., Spaletta G., Bassoli E., Gatto A., Ferrari A., Martin I. Ex situ bioengineering of bioartificial endocrine glands: a new frontier in regenerative medicine of soft tissue organs. Ann. Anat. 2011;193:381–394. doi: 10.1016/j.aanat.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Tzeng Y.S., Li H., Kang Y.L., Chen W.G., Cheng W., Lai D.M. Loss of Cxcl12/Sdf-1 in adult mice decreases the quiescent state of hematopoietic stem/progenitor cells and alters the pattern of hematopoietic regeneration after myelosuppression. Blood. 2011;117:429–439. doi: 10.1182/blood-2010-01-266833. [DOI] [PubMed] [Google Scholar]
- van Pel M., Fibbe W.E., Schepers K. The human and murine hematopoietic stem cell niches: are they comparable? Ann. N. Y. Acad. Sci. 2015;1370:55–64. doi: 10.1111/nyas.12994. [DOI] [PubMed] [Google Scholar]
- Zhang C.C., Lodish H.F. Cytokines regulating hematopoietic stem cell function. Curr. Opin. Hematol. 2008;15:307–311. doi: 10.1097/MOH.0b013e3283007db5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.