Abstract
Curcumin is a major component of the spice, turmeric (Curcuma longa) often used in food or as a dietary supplement. Many preclinical studies on curcumin suggest health benefit in many diseases due to its antioxidant/anti-inflammatory and epigenetic effects. The few human studies and curcumin’s unfavorable pharmacokinetics (PK) have limited its potential, leading researchers to study and develop formulations to improve its PK. The purpose of this clinical study is to describe the acute pharmacokinetics and pharmacodynamics (PK/PD) of commercially marketed curcumin in normal, healthy human volunteers. Twelve volunteers received 4 g dose of curcumin capsules with standard breakfast. Plasma samples were collected at specified time points and analyzed for curcumin and its glucuronide levels. RNA was extracted from leukocytes and analyzed for expression of select antioxidant and epigenetic histone deacetylase (HDAC) genes. Plasma levels of parent curcumin were below the detection limit by HPLC-ITMS/MS/MS. However, curcumin-O-glucuronide (COG), a major metabolite of curcumin, was detected as soon as 30 min. These observations of little to no curcumin and some levels of metabolite are in line with previous studies. PD markers antioxidant genes NRF2, HO-1, NQO1, and epigenetic genes HDAC1, HDAC2, HDAC3, and HDAC4 were quantified by qPCR. COG PK are well-described by a one-compartment model and the PK/PD of COG and its effect on antioxidant and epigenetic gene expression are captured by indirect response model (IDR). A structural population PK model was sequentially established using the nonlinear mixed-effect model program (Monolix Lixoft, Orsay, France). Physiologically based pharmacokinetic modeling (PBPK) and simulation using Simcyp correlated well with the observed data. Taken together these results show that the bioavailability of the parent curcumin compound is low, and oral administration of curcumin can still deliver detectable levels of curcumin glucuronide metabolite. But most importantly, it elicits antioxidant and epigenetic effects which could contribute to the overall health beneficial effects of curcumin.
Keywords: Curcumin, pharmacokinetics/pharmacodynamics, NRF2, oxidative stress, inflammation
Graphical Abstract:
Introduction
Curcumin, a natural polyphenol, is the major component of rhizomes from the spice, turmeric (Curcuma longa) often used in food and in traditional Ayurvedic medicine. Many studies have investigated the therapeutic potential of curcumin across many diseases such as cancer, diabetes, osteoarthritis, and antianxiety 1. Curcumin’s reported benefits and its safety at even high doses of up to 12g/day 2 has made curcumin a popular health supplement 1. In 2016, curcumin remained the top-selling herbal supplement in natural retail stores in United States with sales reaching $47,654,008 and analysis of Google search queries shows spike in interest, classifying curcumin as a “breakout star” 3. Therefore, its growing popularity has drawn researchers to investigate its mechanisms and potential.
The wide range of curcumin’s pharmacological activity is due to its observed pleotropic activities 4. Although there appear to be countless benefits of curcumin supplementation, the majority of these benefits can be attributed to its antioxidant and anti-inflammatory effects 5. Curcumin’s antioxidant activity is due in part to its modulation of nuclear factor (erythroid-derived 2)-like 2 (NRF2), an important regulator of cellular response to oxidative stress 6 that activates downstream antioxidant genes such as heme oxygenase-1 (HO-1) 7, 8, NAD(P)H dehydrogenase [quinone] 1 (NQO1) 9 and glutathione S-transferase P1 (GSTP1) 10. Knockout of NRF2 attenuates curcumin’s antioxidant activity in mouse macrophages 11 and curcumin’s antioxidant effects have also been reported in neuroprotection 12, cancer prevention 13, 14, and chronic kidney disease 15. Curcumin’s ability to suppress inflammation have also been widely reported 16. In human tenocytes stimulated with IL-1β, curcumin downregulated NF-κB signaling and decreased cyclooxygenase-2 production and matrix metalloproteinase-1, −9, and −13 gene expression 17. Curcumin can also inhibit nitric oxide synthase (NOS) induction and nitric oxide production in lipopolysaccharide (LPS) and interferon-γ stimulated RAW 264.7 macrophages 18.
Recent investigations have revealed that curcumin may exhibit epigenetic modifying effects 19, 20. Curcumin exerts effects on multiple epigenetic regulators such as DNA methyltransferases (DNMTs), histone deacetylases (HDACs), histone acetyltransferases (HATs), and micro RNAs (miRNAs) 13. Curcumin has been shown to inhibit HDAC expression and increase Ac-histone H4 in Raji cells 21. In colorectal cancer cells, curcumin shows anti-cancer activity in part by modulating and demethylating CpG loci of certain genes 22. Additional studies have revealed curcumin can inhibit DNMTs and HDACs to restore gene expression of NRF2 23, Neurog1 24, and DLEC1 25. Recently, in an azoxymethane-dextran sulfate sodium (AOM-DSS) colon cancer mouse model, DNA methylation of Tnf was discovered to be hypermethylated in mice fed with curcumin compared to AOM-DSS alone, providing further evidence of curcumin’s chemopreventive effects, possibly via epigenetic mechanisms 26.
Due to curcumin’s reported antioxidant and anti-inflammatory effects and widespread use as a supplement, several clinical trials have characterized its pharmacokinetic (PK) and pharmacodynamic (PD) profile and curcumin’s potential use in humans across a wide range of disease states 27. However, curcumin possesses poor oral bioavailability both in human and in preclinical models due to its poor absorption, fast metabolism and elimination 28. To address these challenges, various formulations of curcumin have been developed and tested. One method has been to use piperine, a major component of black pepper, and combining it with curcumin to inhibit metabolism by UDG-glucoronosyl transferase (UGTs) and thus enhance curcumin’s bioavailability 29, 30. While many studies on curcumin have focused on improving bioavailability and on investigating its effects in disease, to date, few studies have matched PK and PD of commercially marketed curcumin and no studies have been done to study the PK/PD of curcumin on antioxidant and epigenetic gene activity. Our current study aims to describe the effects of oral curcumin on the antioxidant and epigenetic modifiers in healthy human volunteers using a PK/PD modeling approach.
Materials and Methods
Subjects
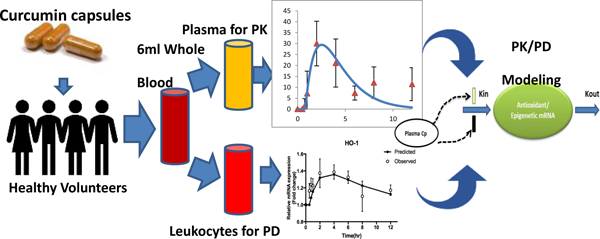
Twelve healthy volunteers aged 18–27 years (seven men and five women) were recruited into the study following approval from the Rutgers University institutional review board (protocol ID 20160000736). Subjects aged 18 to 65 years of age with a body mass index between 18.0 – 29.9 kg/m2 were eligible for inclusion. The subjects agreed to avoid curcumin 14 days prior to arriving at study site. A standardized breakfast with 750 calories and 35 g of fat was provided to delay gastric emptying 31 and help enhance curcumin absorption. All volunteer samples were collected at Robert Wood Johnson University Hospital-Somerset. Exclusion criteria included allergy to curcumin or black pepper, history of malignancy, autoimmune disease, pancreatic or biliary tract disease, renal or hepatic disease, or diagnosis of anemia (hemoglobin < 10mg/dL); any active gastrointestinal condition; history of large bowel resection; women of childbearing age not on oral contraceptives; women who are pregnant or nursing; bleeding disorders or on anticoagulation. All twelve volunteers completed the study.
Study Design
Upon arrival, subjects reviewed consent and expectations for the study. Once consent was obtained, inclusion and exclusion criteria were reviewed to ensure eligibility. Hemoglobin levels were measured, and allergy history of subjects were collected. Vitals including temperature, blood pressure, heart rate, and pulse oximetry were obtained. Thirty minutes prior to curcumin administration, subjects consumed a standardized breakfast and a saline lock device was established to facilitate blood draws. Curcumin was provided as Turmeric Extract Curcumin C3® With BioPerine® capsules obtained from Nature’s Lab (Nature’s Lab, Las Vegas, NV). Each capsule contained 500 mg of curcuminoids and 2.5 mg of bioperine. Each subject was given 8 capsules for a total dose of 4 g of curcuminiods and 20 mg of bioperine. Using LC/MS, the weight ratio of curcumin in the capsule was 77.85±1.47 (%, weight ratio).
Analytical methods
Blood samples (6 mL) at 0, 0.5, 0.75, 1, 2, 4, 6, 8, and 12 h were collected in EDTA vacutainers. Two aliquots of 500 µL of whole blood were collected and mixed with 1.3 mL of RNAlater solution and stored in −80°C for further analysis. Remaining whole blood was centrifuged at 3,000 rpm for 10 minutes and resulting plasma layer was aliquoted into two tubes and stored in −80°C until assayed. Our analytical method was performed according to previously published and validated triple stage ion trap mass spectrometry coupled with high performance liquid chromatography (HPLC-ITMS/MS/MS) method, in which the validation procedure was done according to FDA standard and the recovery rate was 96.69% - 109.26% and matrix effect of 95.40% - 110.98%32. Briefly, Isotope labeled curcumin-d6 was used as the internal standard. The sample preparation was acetonitrile precipitation coupled with molecular weight cut-off size exclusion method. The lower limit of detection (LLOD) and lower limit of quantification (LLOQ) for curcumin and curcumin-O-glucuronide (COG) in rat or human plasma were 0.1 ng/mL and 1 ng/mL, respectively. Figure 1 shows the chemical structure of curcumin and its major metabolite (COG).
Figure 1.
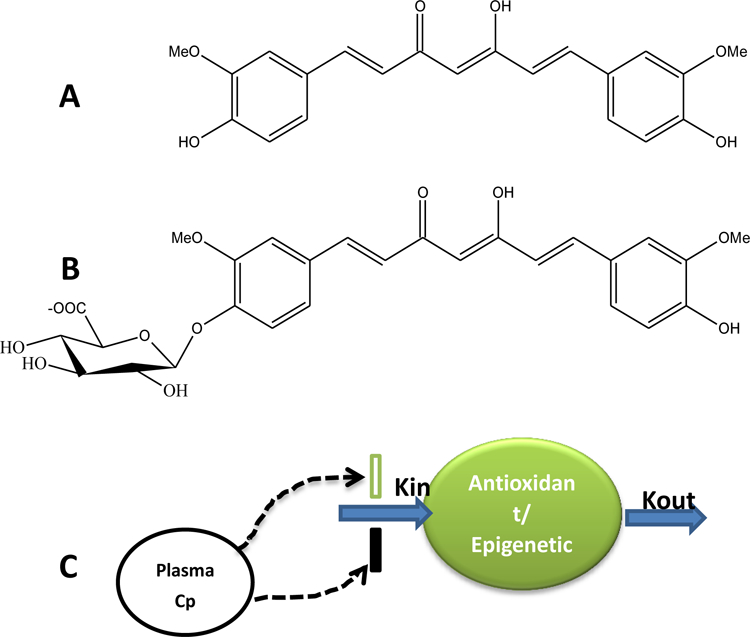
(A) Chemical structures of curcumin and (B) its metabolite curcumin-O-glucuronide (COG). (C) Plasma concentration (Cp) of curcumin/curcumin-O-glucuronide mediated pharmacodynamic response of antioxidant and epigenetic gene expression.
RNA extraction and quantitative real-time PCR
RNA samples preserved in RNAlater stabilization solution (Thermo Fisher Scientific, Rockford, IL) were thawed and RNA was extracted using RiboPure RNA Purification Kit, Blood per protocol. RNA concentrations were measured on an Infinite M200 PRO and cDNA was synthesized using Taqman Reverse Transcription reagents (Thermo Fisher Scientific, Rockford, IL). Quantitative real-time PCR was performed with Power SYBR Green PCR Master mix. Gene expression was measured on a Quantstudio 5 Real-Time PCR System (Thermo Fisher Scientific, Rockford, IL) and quantified by comparative ΔΔCt method.
Pharmacokinetic (PK) and pharmacodynamic (PD) modeling
We next investigated the pharmacokinetics (PK) of curcumin/curcumin-O-glucuronide (COG) metabolite in healthy human volunteers 0–12 hour after a single oral dose. Curcumin was administered with a total 4 g dose to each subject (N=12). Plasma samples were quantified for free curcumin and its glucuronide metabolite COG, since curcumin predominantly undergoes glucuronidation in the gut wall. Non-compartmental analysis (NCA) was first conducted on the time course of curcumin metabolites concentration in plasma. Plasma area under curve (AUC) was determined by the linear trapezoidal rule. The COG concentration (Cm) of 12 human subjects at time t was analyzed by one compartment PK model (1CM) or two compartment (2CM) PK model (Eq. 1, 2, 3, 4), with first order kmixed (a mixed of absorption/UGT metabolism/transport) and ke elimination in Phoenix WinNonlin (version 7.0,Pharsight, USA). The differential equations are shown in the following:
| (1) |
1CM
| (2) |
2CM
| (3) |
| (4) |
where kmixed is the mixed absorption/UGT metabolism/transport rate constant, F is the oral curcumin bioavailability and tlag is the lag time for absorption/UGT metabolism/transport of UGT glucuronide metabolite. Eqs. 1, 2, 3 and 4 show the differential equations for 1CM and 2CM oral dosing, where Vc and Vp represent the volume of distribution in the central and peripheral compartment, respectively. CL and CLd represent the total clearance from the central compartment and inter-compartment distribution clearance, respectively. Ac and Ap represent the amount of COG in the central and peripheral compartments; thus, the plasma concentration C equals Ac/Vc. A population PK model (popPK) was developed using MONOLIX (Lixoft, Orsay, France). To describe the inter-individual variability, the popPK model used to simultaneously described total COG concentration in plasma with aim of a good characterization of the absorption and the elimination phase. The model evaluation was performed using goodness-of-fit plots (GOF) and visual predictive check (VPC), which is a common diagnostic tool that makes a comparison between statistics obtained from the simulated data using the estimated population parameters and the true observed data.
To quantitatively evaluate the PD response of the induction of anti-oxidant (NRF2, HO-1, NQO1) and suppression of epigenetic histone deacetylase (HDAC) effects of COG, these two groups of gene biomarkers were measured as the PD response (R) and modeled using an linear Indirect Response Model (IDR) 33–35The intensity of the biomarkers induction are related to hypothetic biophase curcumin concentration by S, which is a single parameter (linear model). The parameter S relates to the observed intensity of effect (E) to COG biophase concentrations (Cm), according to the equation: E = E0+SC, where mRNA0 is the initial condition defines as mRNA0 = 1 represents the gene expression before Curcumin administration. The differential equations shown in Eq (5–7), respectively:
| (5) |
| (6) |
| (7) |
kin is the zero-order rate constant for the production of antioxidant and epigenetic genes’ mRNA, kout is the first-order rate constant for the degradation of the mRNA,and mRNA0 is fixed to 1 as the initial condition before curcumin administration. The PK parameters obtained from the final 1CM PK model were used as input variables to predict drug concentrations in plasma for PD modeling using the Adapt program 36 (Biomedical Simulations Resource, University of Southern California, Los Angeles, CA).
Simcyp Simulation of PK and drug metabolism of curcumin and its metabolite COG
The simulation of the PK and drug metabolism of the parent compound curcumin was conducted using the developed model available in Simcyp (V17). The “Sim-Healthy” volunteer population with randomly selected individual subjects aged 18–27 years with a male and female ratio of 7:5 was used for the PK and intestinal UGT glucuronidation metabolism simulation. For the initial PK model development, one trial of 12 human subjects receiving a single oral capsule of 4 g dose of curcumin was simulated. For this human PK simulation, the phytochemical properties and in vitro parameters used for the simulation building obtained from ADMET® Predictor (Simulations Plus) and literatures (Table 1) 37, 38
Table1:
Physicochemical and metabolism related in vitro characteristics of curcumin used in SimCYP PK profile simulation
| Parameters | Value | source |
|---|---|---|
| Molecular Weight (g/mol) | 368.4 | ADMET® Predictor |
| pKa | 9.5, 8.96, 8.32 | ADMET® Predictor |
| logPo:w | 2.9 | ADMET® Predictor |
| Caco-2 permeability(10−6 cm/s) | 1.13 | Julia S. Dempe, etc. (37) |
| %Fu in human plasma | 0.1637 | ADMET® Predictor |
| CLint in human liver microsomes (mL/min/kg) | 5.613 | ADMET® Predictor |
| Dose (g, Oral) | 4 | Experiment design |
| UGT involved metabolism | UGT1A1, 1A8, 1A9, 1A10 | ADMET® Predictor& (38) |
| rCYP1A2 in vitro Clint | 11.068 | ADMET® Predictor |
| CYP1A2 Vmax and Km | 24.123, 113.336 | ADMET® Predictor |
In vitro studies of curcumin glucuronide
HepG2-C8-ARE-luciferase cells were treated with curcumin metabolite COG (Santa Cruz Biotechnology, Dallas, TX) for 24 hours at varying concentrations. In vitro study was performed as previously reported 39. COG cytotoxicity was measured using CellTiter 96® AQueous One Solution Cell Proliferation Assay kit (Promega, Madison, WI). ARE-Luciferase activity was measured on a Lucetta Luminometer (Lonza, Basel, Switzerland) and gene expression was measured by qPCR as described above.
Results
Curcumin pharmacokinetics
Several PK studies on curcumin have been conducted on various formulations and high purity curcumin. Our current study investigates the PK/PD of commercially marketed curcumin in healthy human volunteers. Similar to other studies, plasma concentrations of curcumin were below the level of detection due to its poor bioavailability, high metabolism, among others. 1CM and popPK Concentration-time profile of COG is displayed in Figure 2 and 3.
Figure 2. Plasma concentration-time profile of curcumin-O-glucuronide.
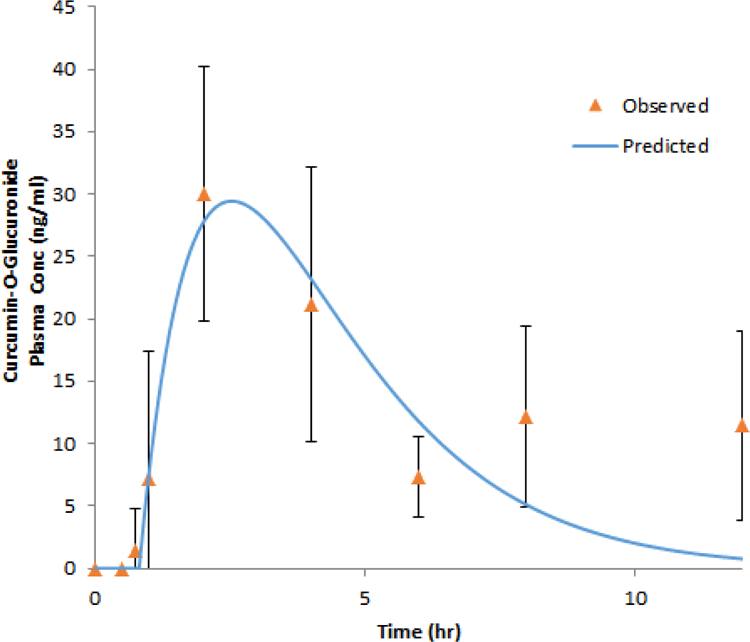
Concentration-time profile of curcumin-O-glucuronide (COG) as described by a one-compartment model. Experimental observation data are shown as the mean +/− SD and the solid line represents the Phoenix WinNonlin model predicted curves after a 4 g oral dose of curcumin to 12 healthy subjects.
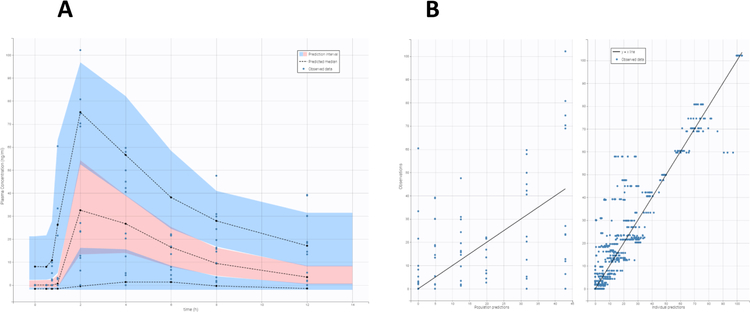
Figure 3. Population Pharmacokinetic study.
VPC of the final popPK model (Figure 3A) described using the total COG plasma concetration based on 1000 simulated replicates of the original data. The dashed lines show the 10th, 50th and 90th percentiles of simulated data; the blue shaded areas represents 90% confidence interval for the corresponding popPK model predicted percentile. Goodness-of-fit plots for the COG PK using total COG plasma concentrations (3B). In the observed versus model predicted plots, the solid lines indicate the linear regression fit.
Plasma levels of the COG were fitted with both one-compartment (1CM) and two-compartment (2CM) PK models. The performance of 1CM (Figure 2) and 2CM (Supplementary Figure 1) were evaluated by goodness-of-fit criteria including visual inspection of the fitted curves, Akaike Information Criteria (AIC), and Coefficients of Variation (CV%) of the estimated parameters. For the 1CM, the fitted PK parameters, maximum concentration of metabolite, Cmax is 29.44 ng/ml, time to maximum concentration, tmax, is 2.53 hours, absorption lag time tlag is 0.99 h, and the kmixed for absorption, UGT metabolism and transport rat rate constant is 0.16 h−1. The AIC of 1CM is 43.886. For the 2CM predicted parameters, the predicted maximum concentration is 30.44 ng/ml, tmax is 2.088 h, kmixed is 9.6h−1 which would bounce many times with high CV%. The AIC of 2CM model is 53.778. The predicted PK parameters for the 1CM and 2CM are shown in Table 2 and Supplementary table 1. Inter-individual variability was reported by the popPK modeling. GOF plots presented in Figure 3B illustrate that the proposal PK model adequately characterized COG plasma concentration in human body. The individual and population predicted values are in good agreement with the observed total plasma as PK input in the model (Figure 3B). VPC (Figure 3A) indicate that the final model effectively explained the observed COG plasma concentration. The final population pharmacokinetics parameters are listed in Table 3.
TABLE 2.
Pharmacokinetic (PK) parameters of curcumin-O-glucuronide in human plasma in 1 CM
| PK Parameters | Description | Estimation (CV%) |
|---|---|---|
| kmixed (h−1) | First order Intestinal absorption, UGT metabolism and transport rate constant | 0.16 (26.87) |
| tlag (h) | Absorption delay | 0.99 (7.21) |
| Cmax (ng/ml) | Maximum concentration predicted | 29.44(15.80) |
| tmax (h) | Time to reach maximum concentration | 2.53 (10.41) |
| AUC0–12h (ng/ml*h) | Area under the curve 0 to 12h | 160.87 (15.72) |
| AUC0−∞ (ng/ml*h) | Area under the curve 0 to infinity | 250.10 (5.18) |
Table 3:
Final population pharmacokinetics parameters
| Parameters | Values | S.E | R.S.E (%) |
|---|---|---|---|
| tlag (h) | 1.01 | 0.216 | 21.4 |
| ka (h−1) | 0.119 | 0.029 | 23.9 |
| Standard Deviation of the Random Effects | |||
| omega_tlag | 0.581 | 0.149 | 25.6 |
| omega_ka | 0.418 | 0.206 | 49.2 |
| Residual variability | |||
| b | 0.513 | 0.059 | 11.5 |
| AIC | 741.15 | -- | -- |
Note: Relative standard error = RSE % = (estimate/standard error) × 100; AIC: Akaike information criterion
Antioxidant and epigenetic gene expression
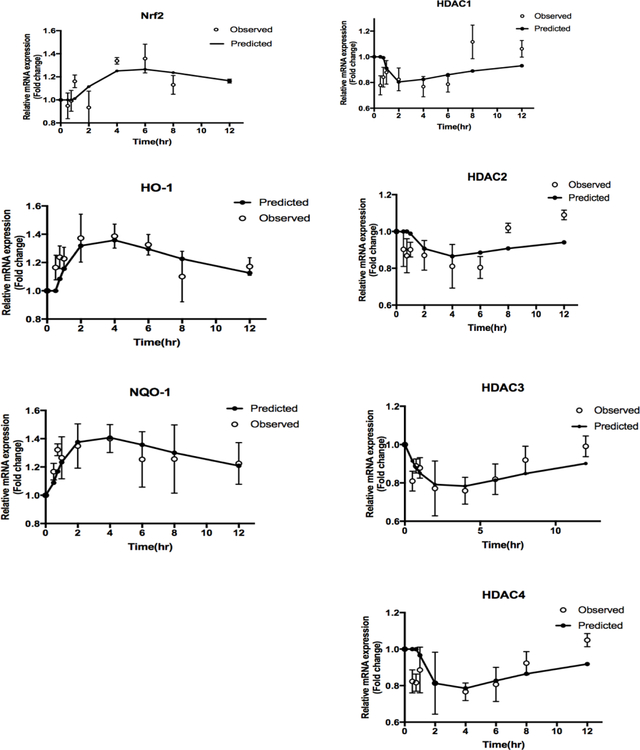
The expression of antioxidant genes such as NRF2, HO-1, and NQO1 increased over time in the subjects, reaching a maximum peak value at approximately 4 to 6 h (Figure 4). The levels of epigenetic marker genes including HDAC1, HDAC2, HDAC3, and HDAC4 decreased, reaching a nadir around 2 to 4 h, and then recovered over time. The estimated PD parameters are listed in Table 4. Epigenetic gene expression reached its lowest point before 5 h after oral curcumin administration. A PK/PD modeling approach will be utilized to describe the induced antioxidant and suppressed epigenetic gene expression below.
Figure 4. Antioxidant and epigenetic gene expression changes as described by indirect response model.
Phase II anti-oxidant and Epigenetic gene expression, including NRF2, HO-1, NQO1, and HDACs (1, 2, 3, 4) as described by the designated IDR model. The open circles represent the mean of the observed data in rat leukocytes, and the black lines represent the model prediction.
TABLE 4.
Pharmacodynamic parameters of mRNA expression estimated from indirect response model
| Gene | kin/kout (h-1) (CV%) | S (CV%) | mRNA0 |
|---|---|---|---|
| Nrf2 | 0.566 (90.63) | 0.016 (30.67) | 1.00 (Fixed) |
| HO-1 | 0.769 (37.91) | 0.021 (33.89) | 1.00 (Fixed) |
| NQO-1 | 0.804 (25.13) | 0.030 (32.14) | 1.00 (Fixed) |
| HDAC1 | 1.296 (72.49) | 0.022 (85.28) | 1.00 (Fixed) |
| HDAC2 | 1.532 (24.13) | 0.0064 (49.08) | 1.00 (Fixed) |
| HDAC3 | 1.276 (37.19) | 0.015 (48.52) | 1.00 (Fixed) |
| HDAC4 | 1.026 (49.73) | 0.010 (30.01) | 1.00 (Fixed) |
Simcyp PBPK modeling and simulation of curcumin and its metabolite COG
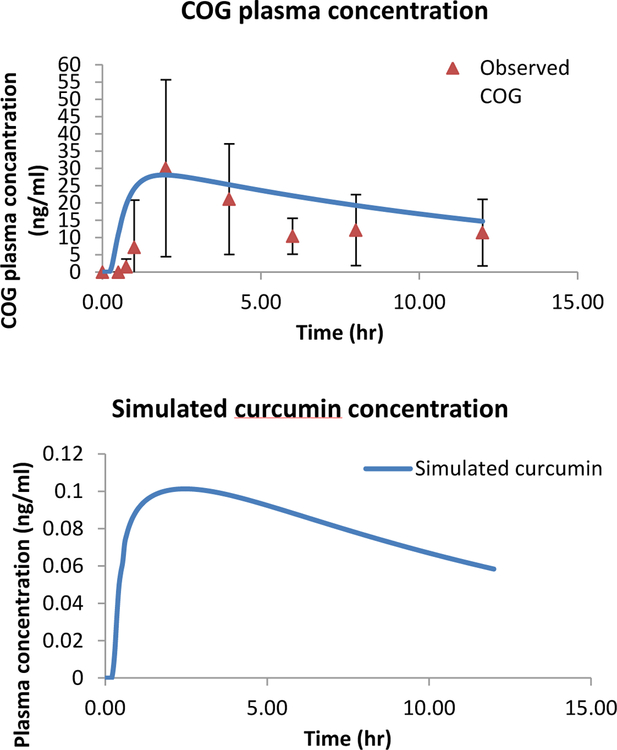
The Simcyp built-in PBPK model for curcumin was able to simulate the parent curcumin compound and the metabolite COG PK profile following a 4 g oral dose of curcumin (Figure 5). The simulated curcumin plasma concentrations versus time profile were below the level of detection of 0.1 ng/ml (Figure 5B), which would be similar to other published human studies confirming poor bioavailability, as well as what we observed in the rats (unpublished observations). Figure 5A shows the simulated curcumin metabolite COG concentrations, which appear to capture reasonably well the observed human data. The simulated metabolite COG’s Cmax (28.1 ng/ml) and AUC (258 ng/ml*h) by Simcyp are similar to the observed Cmax (29.44 ng.ml) and AUC (250.1ng/ml*h). Previous reports show that curcumin is mostly metabolized in the liver and small intestine and that little to no curcumin or its metabolites are present in the organ tissues 40–43.
Figure 5. Simulation of curcumin and COG using Simcyp.
Simulated and observed plasma concentration-time profiles of COG (A) and simulated curcumin plasma concentration-time profile (B) following a single 4 g oral administration. The blue line represents the mean concentration for the simulated population (1 trial X 12 human subjects). The quadrangles denote the mean values (n=12) from the clinical trial.
In vitro studies of curcumin glucuronide
HepG2C8 cells were treated with curcumin metabolite COG for 24 hours and measured for cytotoxicity, antioxidant, and epigenetic gene expression activity. The IC50 of COG was around 100 μM (Supplementary Figure 2A ). Nrf2-ARE luciferase activity increased starting at 100 μM coupled with increase of Nrf2 target gene HO1 antioxidant gene expression (Supplementary Figure 2B). HDAC1 mRNA gene expression decreased at higher dose of curcumin glucuronide while HDAC2 and HDAC3 were not observed to change (Supplementary Figure 2C). Curcumin metabolite COG decreased DNMT1 and DNMT3A, but not DNMT3B gene expression (Supplementary Figure 2D).
Discussion
Curcumin is a dietary phytochemical and health supplement widely consumed for its purported antioxidant and anti-inflammatory properties44. Despite the plethora of research supporting curcumin’s benefits, the low oral bioavailability presents a challenge for curcumin to potentially reach sufficient in vivo concentration levels in order to achieve adequate response 45. Because of this, many have developed and tested different formulations to achieve more favorable PKPD parameters. Some have micronized curcumin 46 or administered curcumin intravenously through liposomes 47. But one common approach is to formulate with piperine, which can enhance bioavailability of curcumin due to its potential to inhibit intestinal and hepatic metabolism 29, 48. In this study, we investigated the antioxidant and epigenetic effects of a commercially marketed formulation of curcumin and characterize its acute effects using a PK/PD modeling approach.
Similar to previous studies 43, the plasma levels of curcumin were below the detection limit of 0.1 ng/mL and only the metabolite, curcumin glucuronide COG was observed. COG was detected at the earliest time point of 30 minutes after curcumin administration and achieved maximal concentrations around 2.7 hours (Table 2), in line with previous work 43. Thus, the metabolism of curcumin appears to occur quite rapidly forming the glucuronide conjugate. The presence of curcumin glucuronide can be explained by the important role UGTs play in the metabolism of curcumin. Glucuronidation is a major pathway in curcumin metabolism in humans, particularly in the intestine. Several isoforms such as UGT1A1, UGT1A7, UGT1A9, and UGT1A10 in human liver and human intestinal microsomes have been reported to be responsible although majority of UGT isoforms were able to catalyze glucuronidation 38. Because only curcumin glucuronide metabolite was detected, curcumin glucuronide was used to model and predict the characteristics of the PK/PD effects of the parent curcumin. The PK of COG were better described by a 1CM (Figure 2) compared to a 2CM (Supplementary Figure 1), in agreement with previous work 43. COG was then used for further characterization of the PD response of antioxidant and epigenetic gene expression. Interestingly, these PK/PD effects of curcumin in human recapitulated our recently reported PK/PD studies in rats 49. The developed PBPK model with intestinal UGT enzymes (UGT1A1, 7, 8, 10) used for curcumin and COG simulation using Simcyp shows that the metabolite COG PK profile correlates well with the observed human data.
The beneficial effects of curcumin can be explained in part via its actions on NRF2 50 and on epigenetic regulators 13. Numerous studies have observed upregulation of NRF2 signaling by curcumin in preventing disease. To better understand the biological mechanism and potential of curcumin’s antioxidant effects, antioxidant gene expression was measured. NRF2, HO-1, and NQO1 increased over time and these changes were predicted by the IDR model (Figure 4A). Pure curcumin glucuronide COG was also tested in vitro in HepG2-C8 cells. COG treatment increased antioxidant NRF2-ARE activity, although in much higher concentrations than the in vivo scenario (Supplementary Figure 2). The reasons for these differences between in vitro cell culture versus in vivo animal models including human, are discussed in our recent review 51. Altogether, the curcumin metabolite COG, exerts antioxidant effects and its presence in human plasma may potential contribute curcumin’s overall antioxidant activity.
Curcumin has been described to possess multiple epigenetic mechanisms 13 and for HDACs has pan-HDAC inhibitory activity in cancer 52, 53. Curcumin, a derivative of caffeic acid, was screened in molecular docking assay in HeLa nuclear extracts and demonstrated more potent HDAC inhibition than other screened compounds 54. In in vitro and in vivo models of medulloblastoma, curcumin inhibited HDAC activity, induced apoptosis, and inhibited tumor growth 55. In this study, oral curcumin decreased HDAC1, HDAC2, HDAC3, and HDAC4 expression which may be mediated by curcumin glucuronide and predicted by the IDR model (Figure 4B, Table 4). Similar to antioxidant genes, increasing AUC of curcumin metabolite COG resulted in linear decrease in HDAC responses. These observations in human are in line with a previous study in rats administered lipopolysaccharide (LPS) with or without curcumin. Curcumin attenuated LPS induction of HDACs and reduced HDAC2, HDAC3, and HDAC4 gene expression in rat lymphocytes 49.
Our results demonstrate that oral curcumin administration induced antioxidant gene expression and reduced epigenetic HDAC expression in normal, healthy human volunteers. The dose used in this study is within the range of previous studies and was well-tolerated. To date, there are limited clinical studies that have investigated the PK/PD of commercially marketed curcumin, an herbal medicinal product often taken as an over-the-counter botanical supplement. Similar to previous studies and reports, the overall bioavailability of curcumin was low and only curcumin metabolite, curcumin glucuronide, was detectable in blood plasma. The presence of curcumin glucuronide is due to intestinal metabolism by UGTs. Despite the absence of parent curcumin in the blood/plasma, antioxidant and epigenetic modulatory effects was nevertheless observed in plasma, presumably due to the presence of COG and may partially explain some of the potential overall health beneficial effects of curcumin.
In conclusion, the results of our current study are highly relevant to the over-the-counter use of botanical curcumin in a real-world scenario. Our current study of oral administration of curcumin resulted in detectable levels of its metabolite, curcumin glucuronide, but more importantly increases the gene expression of antioxidant genes NRF2, HO-1, and NQO1 suppresses epigenetic genes HDAC1, HDAC2, HDAC3, and HDAC4. This suggests there may be potential health benefits of taking curcumin and challenges negative perceptions on the lack of health benefit or efficacy of taking oral curcumin.
Supplementary Material
Acknowledgment
We thank all the members of Kong lab for their suggestions and helpful discussion in the preparation of this manuscript. This work was supported in part by institutional funds and R01AT007065 from the National Center for Complementary and Alternative Medicines (NCCAM) and the Office of Dietary Supplements (ODS).
References:
- 1.Soleimani V; Sahebkar A; Hosseinzadeh H, Turmeric (Curcuma longa) and its major constituent (curcumin) as nontoxic and safe substances: Review. Phytother Res 2018, 32 (6), 985–995. [DOI] [PubMed] [Google Scholar]
- 2.Lao CD; Ruffin M. T. t.; Normolle D; Heath DD; Murray SI; Bailey JM; Boggs ME; Crowell J; Rock CL; Brenner DE, Dose escalation of a curcuminoid formulation. BMC Complement Altern Med 2006, 6, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith T; Kawa K; Eckl V; Morton C; Stredney R, Herbal Supplement Sales in US Increase 7.7% in 2016 Consumer preferences shifting toward ingredients with general wellness benefits, driving growth of adaptogens and digestive health products www.herbalgram.org 2017, (115), 56–65. [Google Scholar]
- 4.Shehzad A; Shahzad R; Lee YS, Curcumin: A Potent Modulator of Multiple Enzymes in Multiple Cancers. Enzymes 2014, 36, 149–74. [DOI] [PubMed] [Google Scholar]
- 5.Hewlings SJ; Kalman DS, Curcumin: A Review of Its’ Effects on Human Health. Foods 2017, 6 (10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma Q, Role of nrf2 in oxidative stress and toxicity. Annu Rev Pharmacol Toxicol 2013, 53, 401–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balogun E; Hoque M; Gong P; Killeen E; Green CJ; Foresti R; Alam J; Motterlini R, Curcumin activates the haem oxygenase-1 gene via regulation of Nrf2 and the antioxidant-responsive element. Biochem J 2003, 371 (Pt 3), 887–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Motterlini R; Foresti R; Bassi R; Green CJ, Curcumin, an antioxidant and anti-inflammatory agent, induces heme oxygenase-1 and protects endothelial cells against oxidative stress. Free Radic Biol Med 2000, 28 (8), 1303–12. [DOI] [PubMed] [Google Scholar]
- 9.Xie Y; Zhao QY; Li HY; Zhou X; Liu Y; Zhang H, Curcumin ameliorates cognitive deficits heavy ion irradiation-induced learning and memory deficits through enhancing of Nrf2 antioxidant signaling pathways. Pharmacol Biochem Behav 2014, 126, 181–6. [DOI] [PubMed] [Google Scholar]
- 10.Nishinaka T; Ichijo Y; Ito M; Kimura M; Katsuyama M; Iwata K; Miura T; Terada T; Yabe-Nishimura C, Curcumin activates human glutathione S-transferase P1 expression through antioxidant response element. Toxicol Lett 2007, 170 (3), 238–47. [DOI] [PubMed] [Google Scholar]
- 11.Boyanapalli SS; Paredes-Gonzalez X; Fuentes F; Zhang C; Guo Y; Pung D; Saw CL; Kong AN, Nrf2 knockout attenuates the anti-inflammatory effects of phenethyl isothiocyanate and curcumin. Chem Res Toxicol 2014, 27 (12), 2036–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong W; Yang B; Wang L; Li B; Guo X; Zhang M; Jiang Z; Fu J; Pi J; Guan D; Zhao R, Curcumin plays neuroprotective roles against traumatic brain injury partly via Nrf2 signaling. Toxicol Appl Pharmacol 2018, 346, 28–36. [DOI] [PubMed] [Google Scholar]
- 13.Boyanapalli SS; Kong AT, “Curcumin, the King of Spices”: Epigenetic Regulatory Mechanisms in the Prevention of Cancer, Neurological, and Inflammatory Diseases. Curr Pharmacol Rep 2015, 1 (2), 129–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen G; Xu C; Hu R; Jain MR; Gopalkrishnan A; Nair S; Huang MT; Chan JY; Kong AN, Modulation of nuclear factor E2-related factor 2-mediated gene expression in mice liver and small intestine by cancer chemopreventive agent curcumin. Mol Cancer Ther 2006, 5 (1), 39–51. [DOI] [PubMed] [Google Scholar]
- 15.Ali BH; Al-Salam S; Al Suleimani Y; Al Kalbani J; Al Bahlani S; Ashique M; Manoj P; Al Dhahli B; Al Abri N; Naser HT; Yasin J; Nemmar A; Al Za’abi M; Hartmann C; Schupp N, Curcumin Ameliorates Kidney Function and Oxidative Stress in Experimental Chronic Kidney Disease. Basic Clin Pharmacol Toxicol 2018, 122 (1), 65–73. [DOI] [PubMed] [Google Scholar]
- 16.Shehzad A; Rehman G; Lee YS, Curcumin in inflammatory diseases. Biofactors 2013, 39 (1), 69–77. [DOI] [PubMed] [Google Scholar]
- 17.Buhrmann C; Mobasheri A; Busch F; Aldinger C; Stahlmann R; Montaseri A; Shakibaei M, Curcumin modulates nuclear factor kappaB (NF-kappaB)-mediated inflammation in human tenocytes in vitro: role of the phosphatidylinositol 3-kinase/Akt pathway. J Biol Chem 2011, 286 (32), 28556–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brouet I; Ohshima H, Curcumin, an anti-tumour promoter and anti-inflammatory agent, inhibits induction of nitric oxide synthase in activated macrophages. Biochem Biophys Res Commun 1995, 206 (2), 533–40. [DOI] [PubMed] [Google Scholar]
- 19.Fu S; Kurzrock R, Development of curcumin as an epigenetic agent. Cancer 2010, 116 (20), 4670–6. [DOI] [PubMed] [Google Scholar]
- 20.Rahman I, Dietary polyphenols mediated regulation of oxidative stress and chromatin remodeling in inflammation. Nutr Rev 2008, 66 Suppl 1, S42–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu HL; Chen Y; Cui GH; Zhou JF, Curcumin, a potent anti-tumor reagent, is a novel histone deacetylase inhibitor regulating B-NHL cell line Raji proliferation. Acta Pharmacol Sin 2005, 26 (5), 603–9. [DOI] [PubMed] [Google Scholar]
- 22.Link A; Balaguer F; Shen Y; Lozano JJ; Leung HC; Boland CR; Goel A, Curcumin modulates DNA methylation in colorectal cancer cells. PLoS One 2013, 8 (2), e57709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khor TO; Huang Y; Wu TY; Shu L; Lee J; Kong AN, Pharmacodynamics of curcumin as DNA hypomethylation agent in restoring the expression of Nrf2 via promoter CpGs demethylation. Biochem Pharmacol 2011, 82 (9), 1073–8. [DOI] [PubMed] [Google Scholar]
- 24.Shu L; Khor TO; Lee JH; Boyanapalli SS; Huang Y; Wu TY; Saw CL; Cheung KL; Kong AN, Epigenetic CpG demethylation of the promoter and reactivation of the expression of Neurog1 by curcumin in prostate LNCaP cells. AAPS J 2011, 13 (4), 606–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo Y; Shu L; Zhang C; Su ZY; Kong AN, Curcumin inhibits anchorage-independent growth of HT29 human colon cancer cells by targeting epigenetic restoration of the tumor suppressor gene DLEC1. Biochem Pharmacol 2015, 94 (2), 69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo Y; Wu R; Gaspar JM; Sargsyan D; Su ZY; Zhang C; Gao L; Cheng D; Li W; Wang C; Yin R; Fang M; Verzi MP; Hart RP; Kong AN, DNA methylome and transcriptome alterations and cancer prevention by curcumin in colitis-accelerated colon cancer in mice. Carcinogenesis 2018, 39 (5), 669–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsu CH; Cheng AL, Clinical studies with curcumin. Adv Exp Med Biol 2007, 595, 471–80. [DOI] [PubMed] [Google Scholar]
- 28.Sharma RA; Steward WP; Gescher AJ, Pharmacokinetics and pharmacodynamics of curcumin. Adv Exp Med Biol 2007, 595, 453–70. [DOI] [PubMed] [Google Scholar]
- 29.Shoba G; Joy D; Joseph T; Majeed M; Rajendran R; Srinivas PS, Influence of piperine on the pharmacokinetics of curcumin in animals and human volunteers. Planta Med 1998, 64 (4), 353–6. [DOI] [PubMed] [Google Scholar]
- 30.Lestari ML; Indrayanto G, Curcumin. Profiles Drug Subst Excip Relat Methodol 2014, 39, 113–204. [DOI] [PubMed] [Google Scholar]
- 31.Stacher G; Granser GV; Bergmann H; Kugi A; Stacher-Janotta G; Hobart J, Slow gastric emptying induced by high fat content of meal accelerated by cisapride administered rectally. Dig Dis Sci 1991, 36 (9), 1259–65. [DOI] [PubMed] [Google Scholar]
- 32.Li W; Yang H; Buckley B; Wang L; Kong AN, A Novel Triple Stage Ion Trap MS method validated for curcumin pharmacokinetics application: A comparison summary of the latest validated curcumin LC/MS methods. J Pharm Biomed Anal 2018, 156, 116–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun YN; Jusko WJ, Transit compartments versus gamma distribution function to model signal transduction processes in pharmacodynamics. J Pharm Sci 1998, 87 (6), 732–7. [DOI] [PubMed] [Google Scholar]
- 34.Dayneka NL; Garg V; Jusko WJ, Comparison of four basic models of indirect pharmacodynamic responses. J Pharmacokinet Biopharm 1993, 21 (4), 457–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piergies AA; Ruo TI; Jansyn EM; Belknap SM; Atkinson AJ Jr., Effect kinetics of N-acetylprocainamide-induced QT interval prolongation. Clin Pharmacol Ther 1987, 42 (1), 107–12. [DOI] [PubMed] [Google Scholar]
- 36.D’Argenio DZ, S. A., Wang X, ADAPT 5 User’s Guide: Pharmacokinetic/Pharmacodynamic Systems Analysis Software 2009. [Google Scholar]
- 37.Dempe JS; Scheerle RK; Pfeiffer E; Metzler M, Metabolism and permeability of curcumin in cultured Caco-2 cells. Mol Nutr Food Res 2013, 57 (9), 1543–9. [DOI] [PubMed] [Google Scholar]
- 38.Hoehle SI; Pfeiffer E; Metzler M, Glucuronidation of curcuminoids by human microsomal and recombinant UDP-glucuronosyltransferases. Mol Nutr Food Res 2007, 51 (8), 932–8. [DOI] [PubMed] [Google Scholar]
- 39.Saw CL; Yang AY; Cheng DC; Boyanapalli SS; Su ZY; Khor TO; Gao S; Wang J; Jiang ZH; Kong AN, Pharmacodynamics of ginsenosides: antioxidant activities, activation of Nrf2, and potential synergistic effects of combinations. Chem Res Toxicol 2012, 25 (8), 1574–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharma RA; McLelland HR; Hill KA; Ireson CR; Euden SA; Manson MM; Pirmohamed M; Marnett LJ; Gescher AJ; Steward WP, Pharmacodynamic and pharmacokinetic study of oral Curcuma extract in patients with colorectal cancer. Clin Cancer Res 2001, 7 (7), 1894–900. [PubMed] [Google Scholar]
- 41.Ireson C; Orr S; Jones DJ; Verschoyle R; Lim CK; Luo JL; Howells L; Plummer S; Jukes R; Williams M; Steward WP; Gescher A, Characterization of metabolites of the chemopreventive agent curcumin in human and rat hepatocytes and in the rat in vivo, and evaluation of their ability to inhibit phorbol ester-induced prostaglandin E2 production. Cancer Res 2001, 61 (3), 1058–64. [PubMed] [Google Scholar]
- 42.Garcea G; Jones DJ; Singh R; Dennison AR; Farmer PB; Sharma RA; Steward WP; Gescher AJ; Berry DP, Detection of curcumin and its metabolites in hepatic tissue and portal blood of patients following oral administration. Br J Cancer 2004, 90 (5), 1011–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vareed SK; Kakarala M; Ruffin MT; Crowell JA; Normolle DP; Djuric Z; Brenner DE, Pharmacokinetics of curcumin conjugate metabolites in healthy human subjects. Cancer Epidemiol Biomarkers Prev 2008, 17 (6), 1411–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jamwal R, Bioavailable curcumin formulations: A review of pharmacokinetic studies in healthy volunteers. J Integr Med 2018. [DOI] [PubMed] [Google Scholar]
- 45.Storka A; Vcelar B; Klickovic U; Gouya G; Weisshaar S; Aschauer S; Bolger G; Helson L; Wolzt M, Safety, tolerability and pharmacokinetics of liposomal curcumin in healthy humans. Int J Clin Pharmacol Ther 2015, 53 (1), 54–65. [DOI] [PubMed] [Google Scholar]
- 46.Kesarwani K; Gupta R; Mukerjee A, Bioavailability enhancers of herbal origin: an overview. Asian Pac J Trop Biomed 2013, 3 (4), 253–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boyanapalli SSS; Huang Y; Su Z; Cheng D; Zhang C; Guo Y; Rao R; Androulakis IP; Kong AN, Pharmacokinetics and Pharmacodynamics of Curcumin in regulating anti-inflammatory and epigenetic gene expression. Biopharm Drug Dispos 2018, 39 (6), 289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trujillo J; Chirino YI; Molina-Jijon E; Anderica-Romero AC; Tapia E; Pedraza-Chaverri J, Renoprotective effect of the antioxidant curcumin: Recent findings. Redox Biol 2013, 1, 448–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramirez CN; Li W; Zhang C; Wu R; Su S; Wang C; Gao L; Yin R; Kong AN, In Vitro-In Vivo Dose Response of Ursolic Acid, Sulforaphane, PEITC, and Curcumin in Cancer Prevention. AAPS J 2017, 20 (1), 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soflaei SS; Momtazi-Borojeni AA; Majeed M; Derosa G; Maffioli P; Sahebkar A, Curcumin: A Natural Pan-HDAC Inhibitor in Cancer. Curr Pharm Des 2018, 24 (2), 123–129. [DOI] [PubMed] [Google Scholar]
- 51.Teiten MH; Dicato M; Diederich M, Curcumin as a regulator of epigenetic events. Mol Nutr Food Res 2013, 57 (9), 1619–29. [DOI] [PubMed] [Google Scholar]
- 52.Bora-Tatar G; Dayangac-Erden D; Demir AS; Dalkara S; Yelekci K; Erdem-Yurter H, Molecular modifications on carboxylic acid derivatives as potent histone deacetylase inhibitors: Activity and docking studies. Bioorg Med Chem 2009, 17 (14), 5219–28. [DOI] [PubMed] [Google Scholar]
- 53.Lee SJ; Krauthauser C; Maduskuie V; Fawcett PT; Olson JM; Rajasekaran SA, Curcumin-induced HDAC inhibition and attenuation of medulloblastoma growth in vitro and in vivo. BMC Cancer 2011, 11, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.