Abstract
Background
Epithelial-mesenchymal transition (EMT) is critical for cancer cell metastasis. Recently, EMT was reported to be associated with the inflammatory tumour microenvironment and, therefore, might be a predictive biomarker for immune checkpoint blockade agents. However, the underlying mechanism is still unclear.
Methods
Patient survival data for our HCC cohort, TCGA and GEO datasets were determined by Kaplan-Meier analysis. The functional roles of ELMO1 in HCC were demonstrated by a series of in vitro and in vivo experiments. Gene microarray analysis was used to demonstrate potential mechanisms of ELMO1. Data retrieved from the TCGA datasets were used to determine the relationships of ELMO1, EMT and TMB.
Findings
Here, we report an indispensable role for ELMO1 in linking EMT with tumour mutation burden (TMB), which is a promising biomarker for the immune checkpoint blockade agent response. Upregulated ELMO1 expression is associated with a poor prognosis in hepatocellular carcinoma (HCC), as well as increased cell growth, invasion, migration, angiogenesis and EMT in vitro and in vivo. Mechanistically, we provide evidence that ELMO1 regulates SOX10 expression and induces EMT through PI3K/Akt signalling. Moreover, ELMO1 is negatively associated with TMB, indicating a negative relationship between EMT and TMB.
Interpretation
ELMO1 serves as a link between EMT and TMB, providing a mechanistic basis for the further development of ELMO1 as a therapeutic target against HCC and potentially a promising biomarker of the immune checkpoint blockade agent response.
Fund
National Natural Science Foundation of China; Natural Science Foundation of Guangdong Province; Young Teacher Training Program of Sun Yat-sen University; Science and Technology Plan of Guangdong Province; Special Support Program of Guangdong Province, Science and Technology Innovation Youth Talent Support Program; the Pearl River Science and Technology New Talent of Guangzhou City; Medical Scientific Research Foundation of Guangdong Province.
Keywords: ELMO1, Tumour mutation burden, Epithelial-mesenchymal transition, Hepatocellular carcinoma
Abbreviations: HCC, Hepatocellular carcinoma; TMB, Tumour mutation burden; EMT, Epithelial-mesenchymal transition; TCGA, The Cancer Genome Atlas; GEO, Gene Expression Omnibus; ACC, Adrenocortical carcinoma; DLBCL, diffuse large B cell lymphoma; ESCA, Esophageal carcinoma; HNSC, Head and neck squamous; KIRC, Kidney renal clear cell carcinoma; LIHC, Liver hepatocellular carcinoma; PAAD, Pancreatic adenocarcinoma; PCPG, Pheochromocytoma and paraganglioma; SARC, Sarcoma; STAD, Stomach adenocarcinoma; TGCT, Testicular germ cell tumours
Research in context.
Evidence before this study
Recent research as reported that EMT regulates metastasis and affects susceptibility to treatment. EMT is highly associated with the tumour environment and has the potential to predict PD-1/PDL-1 blockade benefit. Furthermore, the tumour mutation burden (TMB) can predict the response to anti–PD-1 or anti–PD-L1 treatments. A lack of a response to anti-PD-L1 treatment with low TMB is associated with the transforming growth factor β(TGF-β) signalling signature, which is a classical regulatory mechanism of EMT. However, whether TMB is associated with EMT and the underlying molecular mechanism are largely unknown.
Added value of this study
In the present study, we report an indispensable role for ELMO1 in linking EMT with the tumour mutation burden (TMB), which is a promising biomarker for immune checkpoint blockade agent response. Upregulated ELMO1 expression is associated with a poor prognosis in hepatocellular carcinoma (HCC), as well as increased cell growth, invasion, migration, angiogenesis and EMT in vitro and in vivo. Mechanistically, we provide evidence that ELMO1 regulates SOX10 expression and induces EMT through PI3K/Akt signalling. Moreover, ELMO1 is negatively associated with TMB, indicating a negative relationship between EMT and TMB.
Implications of all the available evidence
Our study demonstrates that ELMO1 can serve as a link between EMT and TMB, providing a mechanistic basis for further development of ELMO1 as a therapeutic target against HCC as well as a promising biomarker of the immune checkpoint blockade agent response.
Alt-text: Unlabelled Box
1. Introduction
Metastasis accounts for major cancer-related mortality, including HCC, which remains a leading cause of cancer death worldwide [1,2]. Metastasis is a complex process that requires tumour cells with the ability to transit from the epithelial to mesenchymal state, which is considered the primary attribute driving tumour progression from initiation to metastasis [3]. Although epithelial to mesenchymal transition (EMT) has been reported to endow tumour cells to become highly malignant, with considerable invasiveness and an ability to form distant metastases in various cancers [4,5], the underlying regulatory mechanism is still unclear. In particular, it has also been recently reported that EMT is significantly associated with an inflammatory tumour microenvironment in lung adenocarcinoma [6]. A further study has revealed a link between the EMT signature and PD-L1 blockade non-response [7]. However, how EMT impacts the efficiency of immune checkpoint blockade remains unknown.
The majority of studies exploring the role of EMT in cancer focus on metastasis and alteration of susceptibilities to treatment [[8], [9], [10]]. Recent studies indicate that EMT is highly associated with the tumour environment [11] and has the potential to predict PD-1/PDL-1 blockade benefit [12]. Furthermore, tumour mutation burden (TMB) can predict responses to anti–PD-1 or anti–PD-L1 treatments [[13], [14], [15]]. The absence of a response to anti-PD-L1 treatment with low TMB is associated with the transforming growth factor β(TGF-β) signalling signature [7], which is a classical regulatory mechanism of EMT [[16], [17], [18]]. However, whether TMB is associated with EMT and the underlying molecular mechanism are largely unknown.
Engulfment and cell motility protein 1 (ELMO1) is a critical regulator of Rac, a small GTPase that regulates cell motility through cytoskeletal interactions [19,20]. ELMO1 can also interact with dedicator of cytokinesis 180 (Dock180) to control cellular processes, including cell migration and phagocytosis [21]. Increased expression of ELMO1 promotes cell invasion and is an independent predictor of OS for patients with acute myeloid leukaemia (AML) [22]. In addition, ELMO1 is elevated in HCC and promotes a poor prognosis, but the regulatory mechanism remains unclear. Moreover, ELMO1 has been linked to the KRAS mutation [23], resulting in a higher mutation burden via the alteration of genes involved in cell-cycle regulation, DNA replication and damage repair [24], suggesting a potential role for ELMO1 in chromosome instability.
In the present study, we sought to determine the role of ELMO1 in HCC metastasis and how ELMO1 links TMB to EMT, which may supports its use as a predictive biomarker for immune checkpoint blockade treatment.
2. Material and methods
2.1. Patients and follow-up
Two hundred sixty-three patients diagnosed with HCC were enrolled from The First Affiliated Hospital, Sun Yat-Sen University, Guangdong, China between 2006 and 2009. All patients underwent hepatectomy, and surgical specimens were identified histologically as HCC. Patients were regularly followed up at the outpatient clinics every 3 months for the first 2 years, every 6 months for the next 3 years, and once annually thereafter. Patients under aged 18 years, lacking clinical or laboratory data, or lacking follow-up were excluded. The last follow-up ended in December 2012. The disease-free survival (DFS) and overall survival (OS) were calculated from date of surgery to the date of recurrence or HCC-associated death, respectively. Informed consent was obtained for each enrolled patient, and the research was carried out with approval from the Ethics Committee of the First Affiliated Hospital of Sun Yat-sen University (Guangdong, China).
2.2. Haematoxylin and eosin (H&E) and immunohistochemistry (IHC)
For routine histological study, intrahepatic tumours were fixed in 4% paraformaldehyde and embedded in paraffin. Haematoxylin and eosin (H&E) staining was performed according to the manufacturer's instructions.
Tissue microarrays containing tumour samples, as well as the matched adjacent noncancerous tissue, were constructed as previously described [25]. A Dako Real Envision Kit (K5007, Dako Denmark A/S, Denmark) was used for IHC staining. Primary antibodies against ELMO1 (1:100 dilution, Santa Cruz), E-cadherin (1:200 dilution, Cell Signal), N-cadherin (1:200 dilution, Cell Signal), Snail (1:200 dilution, Abcam) and SOX10 (1:100 dilution, Santa Cruz) were used for this study. The degree of immunostaining was reviewed and scored independently by two observers based on the proportion of positively stained tumour cells and intensity of staining as previously reported [26].
2.3. Cell lines, cell culture, signal inhibitor
The liver cancer cell lines HepG2, BEL7402, MHCC97L, MHCC97H, and SMMC7721 and the human normal liver cell line LO2 were purchased from the Cell Bank of the Typical Culture Preservation Committee of Chinese Academy of Science (Shanghai, China). MHCC97L and MHCC97H cell lines have different metastatic potentials but share the same genetic background, and they were used for stable cell line establishment. Cell lines were cultured in DMEM (Invitrogen, Carlsbad, CA, USA) supplemented with 10% FBS (HyClone, Logan, UT, USA) in a humidified atmosphere of 5% CO2 at 37 °C. LY294002, a classic inhibitor of PI3K/Akt signalling, was purchased from Cell Signal (9901; 50 μM).
2.4. Establishment of stable overexpression and knockdown cell lines
ELMO1 lentiviral activation particles and control lentiviral activation particles and ELMO1 shRNA pGFP-C-shLenti vector and scrambled shRNA pGFP-C-shLenti vector were purchased from GeneChem (Shanghai, China). Transfection was performed according to the manufacturer's instructions. Briefly, full-length ELMO1 lentiviral activation particles were transfected into MHCC97L cells to generate MHCC97L-ELMO1 cells, and the ELMO1 shRNA pGFP-C-shLenti vector was transfected into MHCC97H to yield MHCC97H-shELMO1. Additionally, control lentiviral activation particles and scrambled shRNA pGFP-C-shLenti vector were transfected into MHCC97L or MHCC97H as controls, respectively. We then employed puromycin at 3 μg/mL to select the stable clones. For analysis of SOX10 as a substrate of ELMO1, we further transfected SOX10 lentiviral activation particles into MHCC97H-shELMO1 (MHCC97H-shELMO1-SOX10) and SOX10 shRNA pGFP-C-shLenti vector into MHCC97L-ELMO1 (MHCC97L-ELMO1-shSOX10). Additionally, control lentiviral activation particles and scrambled shRNA pGFP-C-shLenti vector were transfected into MHCC97L-ELMO1 or MHCC97H-shELMO1 as controls, respectively. The sequences of the cDNA clone and shRNA are listed in Supplementary Table 1.
2.5. MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide] assay
For cell growth measurements, cells (1 × 103) were seeded in 96-well plates in complete DMEM. Cell density was determined with the Cell Proliferation Kit I (MTT) (#11465007001, Roche) according to the supplier's protocol starting 24 h post-seeding of the cells. The optical density was measured at 490 nm. All experimental points comprised three replicates, and all experiments were repeated at least three times. The data were graphically displayed using GraphPad Prism 5. Each point (mean ± standard deviation) represents the growth of treated compared with untreated cells.
2.6. Western blot analysis
The indicated tissues and cells were suspended in protein lysis buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 0.5% NP-40) with a cocktail of protease and phosphatase inhibitors (cOmplete Mini, Roche; Phosphatase Inhibitor Cocktail 2 and 3, Sigma). The Bradford (Bio-Rad) method was used to quantify the total amount of protein. Equal volumes of total lysate were processed for protein analysis, which were separated by SDS–PAGE and transferred to PVDF (Roche Life Sciences, Switzerland) membrane. Then, the membranes were blocked with 5% milk in TBST (TBS + 0.1% Tween-20) for at least 1 h at room temperature and incubated with primary antibodies diluted in 5% milk in TBST overnight at 4 °C. The membranes were then washed three times with TBST and incubated with horseradish peroxidase-conjugated secondary antibodies for 1 h at room temperature. Primary antibodies were detected with anti–mouse IgG HRP-linked (7076S) or anti–rabbit IgG HRP-linked (7074S) antibody, which were purchased from Cell Signalling and used at a 1:5000 dilution. The antigen-antibody complex was detected with enhanced chemiluminescence reagents (Merck Millipore, Massachusetts, USA) and visualized with the Bio-Rad imaging system. The antibody information is listed in Supplementary Table 2.
2.7. Co-immunoprecipitation (Co-IP)
The co-immunoprecipitation assay was conducted using a co-immunoprecipitation kit (#26149; Pierce; Rockford, IL, USA) according to the kit's protocols. In brief, 500 μg of MHCC97L or MHCC97H cell lysates for each sample was pre-cleared using a control agarose resin and then incubated with the column containing 5 μg antibody against ELMO1 or IgG at 4 °C overnight. After washing to remove non-specific binding, the co-immunoprecipitated proteins were then eluted and analysed by western blotting with the antibody for ELMO1 or SOX10. The input was used as positive control, while IgG was used as negative control.
2.8. Quantitative real-time polymerase chain reaction (qRT-PCR)
Briefly, total RNA was isolated with TRIzol Reagent (Life Technologies, Carlsbad, CA) from frozen tissue samples or HCC cell lines according to the manufacturer's protocol. cDNA was obtained from RNA using a universal cDNA synthesis kit (TAKARA, Shiga, Japan). The resultant products were amplified using a SYBR Green PCR kit (Toyobo, Tokyo, Japan) for qRT-PCR analysis. Amplification plots were analysed using Bio-Rad IQ5 software (Bio-Rad Laboratories Inc., California, USA). All quantifications were normalized to the level of endogenous GAPDH as a control. All primers were purchased from GeneCopia (Rockville, MD, USA), and a representative experiment of at least three independent experiments is shown.
2.9. Gene expression profiling using RNA microarrays
For mRNA expression profiling analysis, tumour tissues in HCC patients and their counterpart non-cancerous tissues (five pairs) were constructed as described above, and total RNA was extracted with TRIzol® Reagent (Thermo Fisher Scientific), respectively. The Agilent Whole Human Genome Oligo Microarray was performed by Kangchen Biotech Inc. (Shanghai, China). Agilent Feature Extraction software (version 11.0.1.1) was performed to analyse the acquired array images. Quantile normalization and subsequent data processing were performed using the GeneSpring GX v12.1 software package (Agilent Technologies). After quantile normalization of the raw data, genes with at least 1 out of all samples containing flags (“All Targets Value”) were chosen for further data analysis. Differentially expressed genes between the two samples were identified through fold change filtering.
2.10. Human epithelial to mesenchymal transition (EMT) RT2 profiler™ PCR array
Changes in expression of EMT-related genes in MHCC97L-ELMO1 versus MHCC97L-control, MHCC97H-shELMO1 versus MHCC97H-control cells were investigated using the EMT specific RT2 Profiler PCR Array (Kangchen Biotech Inc. Shanghai, China) according to the manufacturer's protocol.
2.11. Mutation data analysis
For the discovery set, the somatic mutation data (level 2) of liver hepatocellular carcinoma (LIHC), stomach adenocarcinoma (STAD), lung squamous cell carcinoma (LUSC), diffuse large B cell lymphoma (DLBC), prostate adenocarcinoma (PRAD), colon adenocarcinoma (COAD), and oesophageal carcinoma (ESCA) were retrieved from the TCGA data portal (https://tcga-data.nci.nih.gov/tcga/findArchives.htm). The total number of somatic mutations was adopted to assess the mutation burden, which is convenient and significantly correlated with the number of non-synonymous mutations. Missense, nonsense, nonstop, silent, and frameshift/in-frame insertions and deletions were counted and summed, and germline mutations without somatic mutations were excluded [27]. The frequencies of 6 classes of mutations were calculated by dividing the total number of mutations in each category by the total number of mutations in the 6 categories combined.
2.12. Cell migration and invasion assays
Briefly, 1 × 105 cells in 300 μL of DMEM (with 1% FBS) were seeded in the upper chambers of transwell membranes (Corning, New York, USA). The bottom wells of the chambers were filled with 500 μL DMEM (with 10% FBS) and cultured under hypoxic conditions. After 20 h of incubation, the transwell membranes were fixed with 95% ethanol and then stained with 1% crystal violet. Images of five different fields were captured from each membrane, and the number of migrated cells was counted. A similar procedure was adopted for the cell invasion assay, except that the applied inserts were precoated with Matrigel.
2.13. Immunofluorescence
Cells were seeded on coverslips, then fixed with 4% paraformaldehyde in PBS for 15 min and washed twice with PBS. Primary antibodies against E-cadherin (Proteintech Group) and N-cadherin (Cell Signalling Technology), F-actin (Genecopia, Guangzhou, China) were incubated overnight at 4 °C. E-cadherin and N-cadherin, F-actin were visualized using FITC-conjugated goat anti-mouse or anti-rabbit IgG (Genecopia, Guangzhou, China). DAPI was used as a nuclear counterstain (Genecopia, Guangzhou, China). A Leica DMRA fluorescence microscope (Leica, Wetzlar, German) was used to obtain the images.
2.14. Xenograft studies
All experimental procedures involving animals were in accordance with the Guide for the Care and Use of Laboratory Animals (NIH publication nos. 80–23, revised 1996) and were performed according to the institutional ethical guidelines for mouse experiments. All studies involving mice were approved by the Institutional Animal Care and Use Committee of Sun Yat-sen University. Male BALB/C nude mice (5 weeks old) were raised in an SPF environment in the animal facility during the experimental procedures with free access to diet or water. Briefly, 4 × 106 cells suspended in 100 μl PBS were injected subcutaneously into the flanks of nude mice. Tumour volumes and mouse weights were monitored once per week. Tumour size was measured using a calliper twice weekly, and weights were determined at the same time. Tumour volume was calculated as follows: length × width 2 × 0.51. After 6 weeks, tumours established in this step were collected, and subcutaneous tumours were resected, cut into 1 mm3 pieces, and then implanted into the left liver to mimic primary HCC. Twelve mice were randomly divided into each study group. Eight weeks after implantation, in vivo imaging system (IVIS, PerkinElmer) was used to detect GFP fluorescence in tumour-bearing mice. The mice were then sacrificed, and the liver was analysed by standard histological examination.
2.15. Data availability
Clinical data, mutational profiles and normalized gene expression data from TCGA samples and different types of cancer cell lines were obtained from TCGA datasets (https://portal.gdc.cancer.gov/), GEPIA (http://gepia.cancer-pku.cn/) and Cancer Cell Line Encyclopedia (CCLE, http://www.broadinstitute.org/ccle;ref.25), mutation types of ELMO1 were analysed by cbioportal (http://www.cbioportal.org/), OS and DFS of TCGA patients were analysed by survExpress (http://bioinformatica.mty.itesm.mx:8080/Biomatec/SurvivaX.jsp), and mapping of the ELMOl-involved protein interaction network was performed using STRING (https://string-db.org/cgi/input.pl). The network was then exported and visualized in Cytoscape v.3.4.0 (http://www.cytoscape.org/), and the node sizes were adjusted to be proportional to the average. GSE84598, GSE76427, GSE57957 and GSE62232 were obtained from the NCBI Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/).
2.16. Gene set enrichment analysis (GSEA)
For gene set enrichment analysis (GSEA), the javaGSEA Desktop Application was downloaded from http://software.broadinstitute.org/gsea/index.jsp. GSEA was used to determine statistically significant molecular signatures with the indicated data. The input data for GSEA were a complete table of genes ranked by the test statistics from the cuffdiff results and a catalogue of functional gene sets from the HALLMARK gene sets, GO gene sets (Molecular Signatures Database v6.0, http://software.broadinstitute.org/gsea/msigdb/index.jsp). Default parameters were used. The normalized enrichment score (NES) is the primary statistic for examining gene set enrichment results. The nominal P value estimates the statistical significance of the enrichment score. A gene set with a nominal P ≤ 0.05 was considered to be significantly enriched in genes.
2.17. Statistical analysis
Statistical analyses were performed using SPSS 17.0. Data are expressed as the mean ± standard error of the mean (SEM) from at least three independent experiments. Quantitative data were compared between groups using the Student's t-test. Categorical data were analysed by the c2 or Fisher's exact test. Spearman's rank analysis was used to evaluate correlations between protein expression levels. The Kaplan-Meier and log-rank tests were used to analyse differences in OS and DFS. Regression analysis of survival and recurrence was determined using a Cox proportional hazards model. P values <0.05 were considered significant. Correlations between gene expression levels were conducted using a linear regression test.
3. Results
3.1. ELMO1 is upregulated and can confer risk of a poor prognosis in HCC
To identify gene targets in HCC, we performed a gene microarray assay between tumour and adjacent non-tumour tissues. Of five tumour-non-tumour tissues pairs, ELMO1 was upregulated in three of five tumour tissues (patient No. 3, No. 4 and No. 5) (Supplementary Fig. 1a). The Gene Expression Profiling Interactive Analysis (GEPIA) platform revealed that ELMO1 mRNA was particularly overexpressed in ACC, DLBCL, ESCA, HNSC, KIRC, LIHC, PAAD, PCPG, SARC, STAD, and TGCT (Supplementary Fig. 1b). Indeed, the data showed that TCGA, ELMO1 mRNA expression was significantly upregulated in LIHC (Supplementary Fig. 1c). Moreover, similar results were found in the Gene Expression Omnibus (GEO) databases of HCC (GSE84598, GSE76427, GSE57957 and GSE62232) (Supplementary Fig. 1d). Consistently, western blot analysis revealed a higher level of ELMO1 protein in tumour tissues in 5/6 tumour-non-tumour patient tissue pairs. (Fig. 1a). Together, these results suggest that ELMO1 is upregulated in HCC.
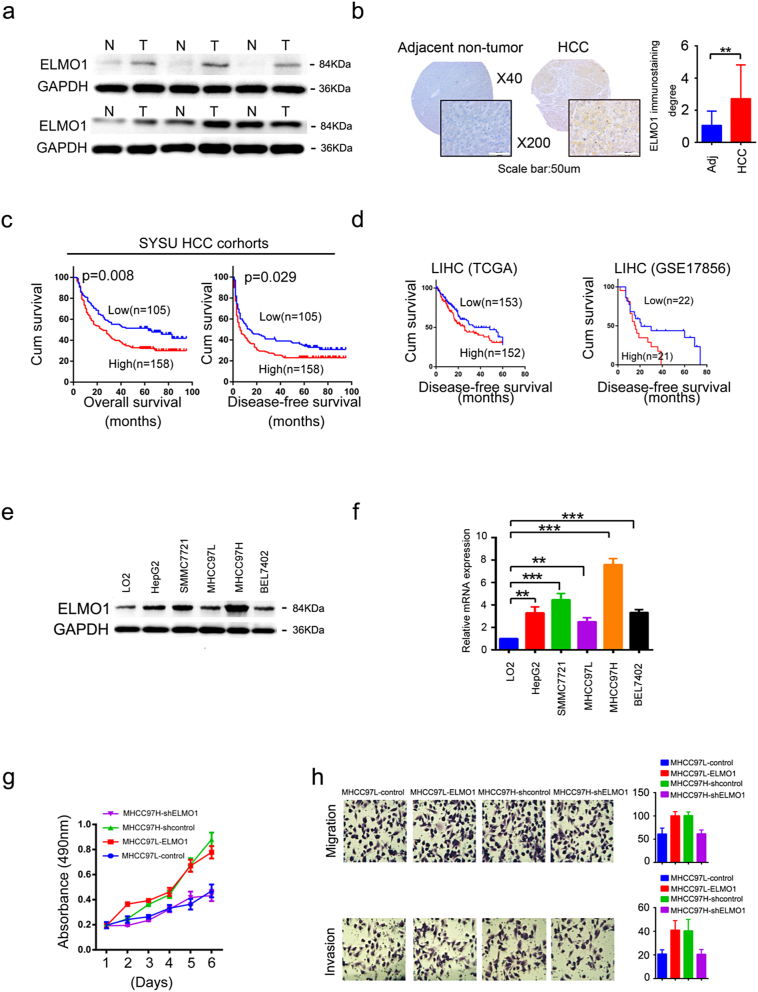
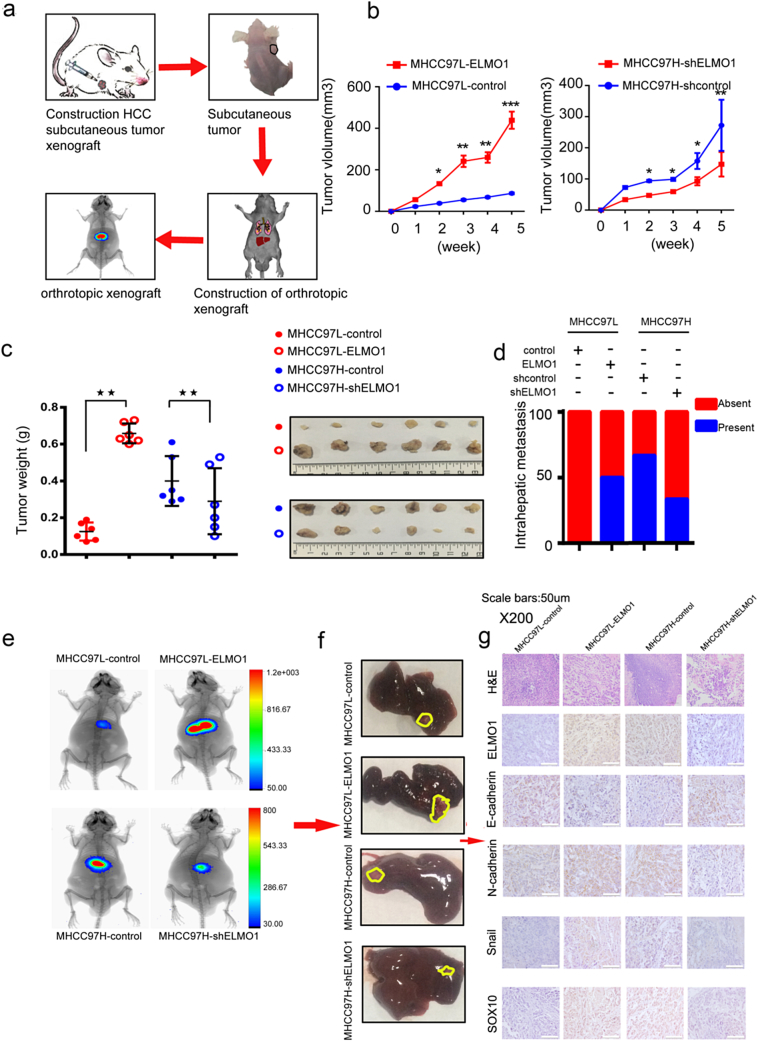
Fig. 1.
Overexpression of ELMO1 is correlated with a poor prognosis and promotes HCC cell metastasis.
(a) Representative western blots of ELMO1 protein expression in paired HCC tissues and adjacent non-tumour tissues. N: non-tumour tissues; T: tumour tissues. Three independently repeated experiments were performed with similar results.
(b) Representative IHC images of ELMO1 protein expression in paired HCC tissues and adjacent non-tumour tissues from TMA. Left panel, top: magnification 40×; bottom: magnification 200×, scale bar: 50um. Right panel: quantification of ELMO1 staining degree, student's t-test: *p < 0.05. **p < 0.01, ***p < 0.001, ns, not significant. Adj: Adjacent non-tumour tissues.
(c, d) OS and DFS association with the expression levels of ELMO1 in the 263 patients in the HCC patient. Left: OS, log-rank test: p = 0.008; Right: DFS, log-rank test: p = 0.029 (c). OS and DFS association with expression levels of ELMO1 in the TCGA and GEO patients cohorts LUAD, log-rank test: p = 0.049; PAAD, log-rank test: p = 0.003; LIHC, log-rank test: p = 0.047; GSE17856, log-rank test: p = 0.043 (d).
(e) Protein expression of ELMO1 in HCC cell lines. Western blot analysis of ELMO1 protein expression levels. Three independently repeated experiments were performed with similar results.
(f) mRNA expression of ELMO1 in HCC cell lines. qRT-PCR analysis of ELMO1 mRNA expression levels, Data are represented as the mean ± SD; Student's t-test: *p < 0.05. **p < 0.01, ***p < 0.001, ns, not significant. Three independently repeated experiments were performed with similar results.
(g, h) MTT (g), transwell migration and transwell invasion (h) assays in HCC cells upon ELMO1 overexpression or knockdown compared with their matched control cells.
To characterize clinical ELMO1, we collected 263 patient-matched tumour-non-tumour liver tissues. Interestingly, IHC staining demonstrated that ELMO1 expression was dramatically enhanced in 41.4% HCC samples (109/263), compared with adjacent nontumourous tissues (Fig. 1b). ELMO1 overexpression was correlated with vascular invasion (Cox regression analysis, p = 0.017), whereas it was associated with other clinical factors such as gender, age, hepatitis B surface antigen level, alpha-fetoprotein level, capsulation, Edmondson grading, tumour size, liver cirrhosis, or number of nodules (Supplementary Table 3). Notably, high expression of ELMO1 was associated with a poor prognosis in our cohort, which demonstrated a shorter DFS and OS (Fig. 1c). Consistently, data from TCGA also revealed a correlation of high ELMO1 expression with shorter DFS in LIHC (Fig. 1d). Furthermore, both univariate and multivariate analyses revealed that the expression of ELMO1, tumour size, capsulation, number of tumours and vascular invasion were independent risk factors of lower overall survival and early tumour recurrence (Supplementary Table 4). These data together show that high ELMO1 expression is negatively associated with OS and DFS in HCC patients, suggesting that ELMO1 may play a predictive role in HCC prognosis.
3.2. ELMO1 is required for the growth, migration and invasion of HCC cells
Little is known regarding the regulation or function of ELMO1 in human cancer cells. We first analysed ELMO1 expression in cancer cell lines. To date, the Cancer Cell Line Encyclopedia (CCLE) project has demonstrated ELMO1 expression across a diversity of cancer types, including liver cancer (Supplementary Fig. 1e). Furthermore, ELMO1 mRNA and protein expression were higher in HepG2, SMMC7721 and MHCC97H but lower in BEL7402, with no significance in MHCC97L, as compared to the human normal liver cell line LO2 (Fig. 1e, f). Based on the previously reported increased metastatic potential of MHCC97H compared with MHCC97L, we identified MHCC97L and MHCC97H as the most appropriate model for our screen. We then overexpressed ELMO1 in MHCC97L and knocked down ELMO1 in MHCC97H as an initial step to investigate the effect of ELMO1 in HCC.
To understand how ELMO1 affects HCC cells, we investigated ELMO1 in the gene expression profiling using mRNA microarray. Gene ontology (GO) analysis showed that ELMO1 could affects cell cycle, cell proliferation, cell migration and vascular angiogenesis (Supplementary Fig. 1f). Then we further explored ELMO1 effects in the TCGA database. Interestingly, ELMO1 was positively correlated with MKI67, MMP3, VEGFA and VEGFB in the TCGA LIHC data (Supplementary Fig. 1g, h, i, j), which are known markers of cell proliferation (MKI67), cell metastasis (MMP3) and angiogenesis (VEGFA and VEGFB). In addition, we monitored cell growth, migration and invasion abilities by generating ELMO1 overexpressed or knocked down HCC cells. Indeed, upregulated ELMO1 expression promoted cell growth (Fig. 1g), migration and invasion (Fig. 1h) capabilities, whereas knocked down ELMO1 resulted in the opposite. Together, these results strongly support a critical role of ELMO1, at least partially, in promoting cell growth, migration and invasion.
3.3. ELMO1 is negatively associated with tumour mutation burden
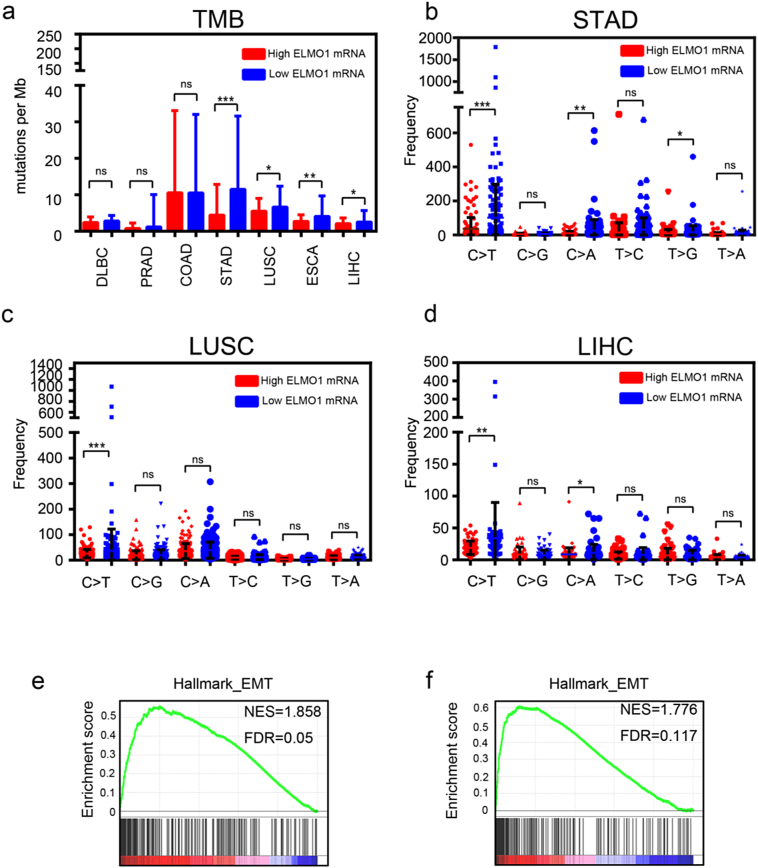
Cell cycle, DNA replication, DNA damage response (DDR) and mismatch repair (MMR) mainly accounted for TMB [28,29], a promising positive predictive biomarker for immune checkpoint blockade. Mutation rates of ELMO1 ranged from 0 to 18.69% in provisional TCGA datasets across various cancer types (Supplementary Fig. 2a), and rates of amplification and deletion ranged from 0 to 5.21%. The majority of the mutations were related to ELMO1, which was overexpressed at the mRNA level across cancer types (Supplementary Fig. 2a), which confirmed ELMO1 upregulated in HCC. Moreover, TMB was significantly decreased in high ELMO1 mRNA expression tumours compared with low ELMO1 mRNA expression tumours in the TCGA datasets (Fig. 2a). To further elucidate the molecular mechanism by which ELMO1 regulates TMB, we examined the mutator phenotype in ELMO1 high or low tumours. Notably, the C > T mutation frequency, which is most commonly seen in MMR-deficient tumours [30], was markedly higher in low ELMO1 tumours (Fig. 2b, c, d), and it was confirmed to be the major type of mutator phenotype in cancer, suggesting that MMR serves as a key node between ELMO1 and TMB. In addition, in the TCGA datasets, the DDR, MMR signature was enriched in ELMO1-high tumours (Supplementary Fig. 2b), which further demonstrated a positive association between ELMO1, DDR and MMR regulators, including ERCC4, PALB2, RPA3 and MSH3, in CCLE (Supplementary Fig. 2c, d, e, f). These findings suggested that ELMO1 might be regulated TMB through DDR or MMR.
Fig. 2.
ELMO1 is associated with TMB.
(a) Tumour mutation burden in ELMO1-high or ELMO1-low tumours of different cancer types. Student's t-test: *p < 0.05. **p < 0.01, ***p < 0.001, ns, not significant.
(b, c, d) Distribution of six types of mutations in ELMO1-high and ELMO1-low tumours of STAD (b), LUSC (c), LIHC (d). Student's t-test: *p < 0.05. **p < 0.01, ***p < 0.001, ns, not significant.
(e) Enrichment plots for EMT signatures from the “Hallmark” gene set collection in low TMB TCGA LIHC patients using GSEA.
(f) Enrichment plots for EMT signatures from the “Hallmark” gene set collection in low TMB with high ELMO1 expression TCGA LIHC patients using GSEA.
3.4. ELMO1 links TMB with EMT in HCC
Recent studies have shown that TMB renders tumours more susceptible to PD-L1 blockade [15], whereas TGFβ exhibit the opposite trend, and further research has revealed that cancer cell lines with high TGFβ-EMT signature tend to carry a lower mutation rate [7], suggesting a negative relationship between TMB and EMT. However, whether TMB is correlated with EMT and the underlying mechanism are unclear. First, we applied GSEA to drive enrichment scores from high or low TMB tumours in the TCGA LIHC datasets. Notably, positive enrichment of EMT was observed in low TMB tumours but was irrelevant in high TMB tumours (Fig. 2e). Moreover, EMT was enriched in high ELMO1 tumours with low TMB (Fig. 2f). Thus, our data support a negative correlation of TMB with EMT, and ELMO1 may participate in the crosstalk.
3.5. ELMO1 induces EMT
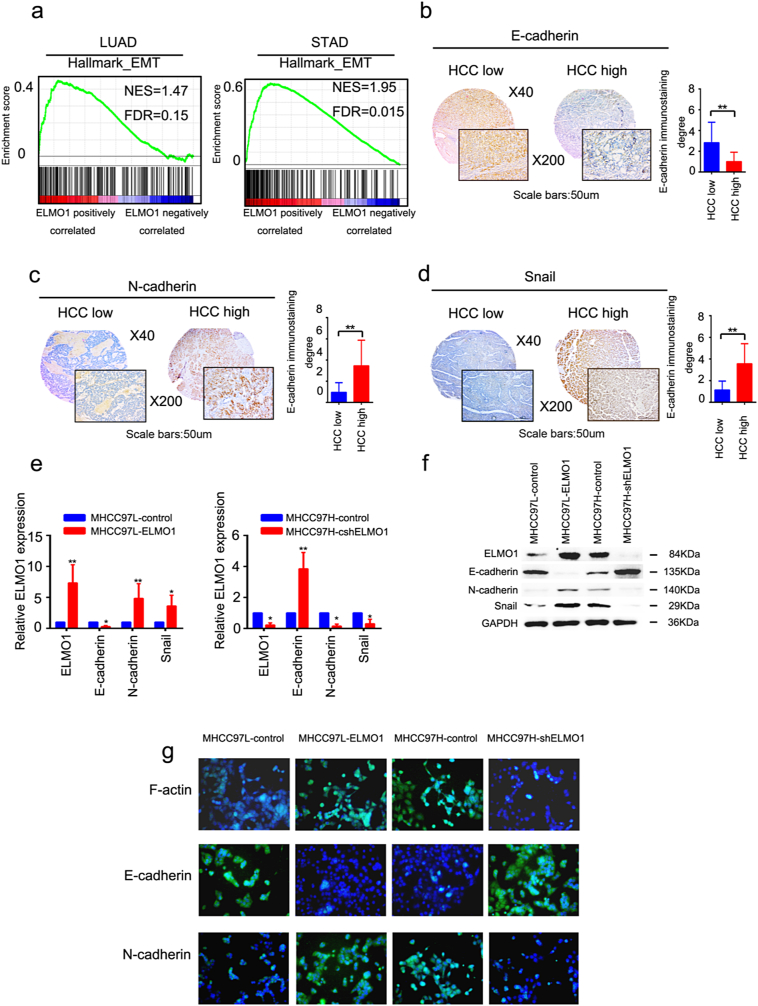
Given that ELMO1 promoted HCC cell invasion and migration, we wondered whether ELMO1 could directly induce EMT. Strikingly, in both the TCGA LUAD and TCGA STAD datasets, a positive correlation between overexpressed ELMO1 and enriched EMT genes was observed (Fig. 3a), suggesting a regulator role of ELMO1 on the EMT signature of cancer. To genetically dissect the contribution of ELMO1 in HCC, we analysed the relationship between ELMO1 protein expression and EMT-related genes in our HCC cohorts. Of note, ELMO1 protein expression level was positively correlated with N-cadherin and Snail, both of which are established essential players in promoting EMT, whereas it showed a negative association with E-cadherin, a canonical marker of suppressed EMT (Fig. 3b, c, d). Furthermore, upregulated N-cadherin and snail mRNA expression in HCC cell lines was observed with overexpressed ELMO1 and downregulated E-cadherin with knocked down ELMO1; in contrast, cell lines with knocked down ELMO1 exhibited the opposite trend (Fig. 3e). Interestingly, comparable observations were observed western blot analyses of HCC cell lines expressing different levels of ELMO1, which demonstrated a significant increase in N-cadherin, snail and decrease in E-cadherin in ELMO1 overexpressed cells. Conversely, decreased ELMO1 expression displayed the opposite trend (Fig. 3f). Moreover, Immunofluorescence assay showed consistent results (Fig. 3g), suggesting that ELMO1 could regulate EMT.
Fig. 3.
ELMO1 induces EMT in HCC.
(a) GSEA plot of the association between gene sets positively correlated with ELMO1 in the TCGA LUAD (left panel) and STAD (right panel) databases and the EMT signatures from “Hallmark” gene set collection.
(b, c, d) Representative IHC images of E-cadherin (b), N-cadherin (c) and Snail (d) in the SYSU cohort. Left panel, top: magnification 40×; bottom: magnification 200×, scale bar: 50um. Right panel: quantification of E-cadherin (b), N-cadherin (c) or Snail (d) immunostaining degree, student's t-test: *p < 0.05. **p < 0.01, ***p < 0.001, ns, not significant.
(e, f) mRNA and protein expression levels of ELMO1, E-cadherin, N-cadherin and Snail in the MHCC97L-control, MHCC-97 L-ELMO1, MHCC97H-shcontrol and MHCC97H-shELMO1 by qRT-PCR (e) and western blot (f) analyses, respectively. GAPDH was used as the internal control. Three independently repeated experiments were performed with similar results.
(g) Representative immunofluorescence (IF) images of F-actin, E-cadherin and N-cadherin in MHCC97L-control, MHCC97L-ELMO1, MHCC97H-shcontrol and MHCC97H- shELMO1.
3.6. ELMO1 activates PI3K/Akt signalling
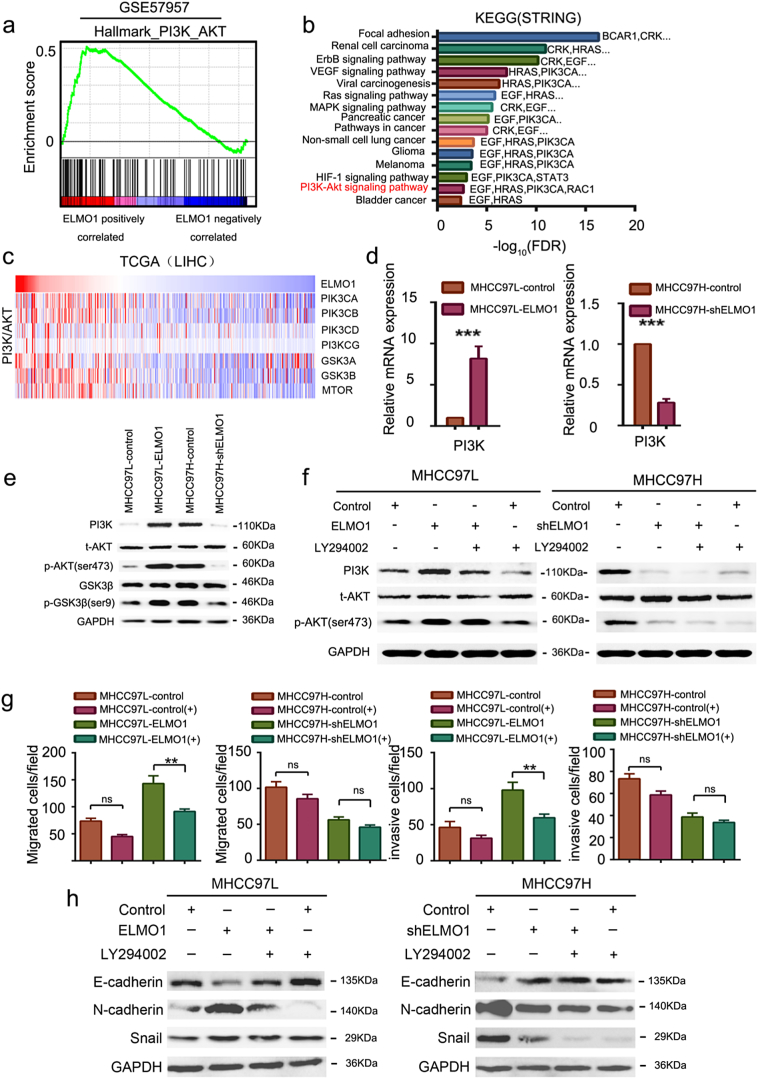
The molecular mechanism linking ELMO1 with EMT is unclear. Notably, gene set enrichment analysis identified PI3K/Akt signalling as the feature associated with ELMO1 from the HCC GEO datasets (Fig. 4a), and genes involved in the PI3K/Akt signalling pathway were significantly overexpressed in ELMO1 overexpressed tumours. Consistently, using Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotations by STRING, the PI3K/Akt signalling pathway was activated, as well as focal adhesion, VEGF signalling, and the MAPK signalling pathway, among others (Fig. 4b), which have been previously reported to be associated with EMT initiation. Of Note, although PI3K/Akt signalling was not the most activated pathway regulated by ELMO1, upregulated PIK3CA, a key member of PI3K/Akt [31], was involved in various signals, such as VEGF signalling, viral carcinogenesis, pancreatic cancer, and non-small cell lung cancer (Fig. 4b). Notably, the PI3K/Akt gene signature was significantly enriched in ELMO1 overexpressed tumours in TCGA LIHC patients (Fig. 4c). We further revealed a positive relationship between ELMO1 and PIK3CA, PIK3CB, PIK3CD, and PIK3CG in TCGA (Supplementary Fig. 3a). Moreover, PIK3CA and PIK3CB were further validated as positively correlated with ELMO1 in CCLE datasets across 760 cancer cell lines (Supplementary Fig. 3b). Importantly, ELMO1 could upregulate PI3K (Fig. 4d, e), and Akt and GSK3β were phosphorylated by overexpressed ELMO1, whereas phosphorylated Akt and pGSK3β were decreased with downregulated ELMO1 in HCC (Fig. 4e). Together, these findings suggest that ELMO1 can activate the PI3K/AKT signalling pathway.
Fig. 4.
ELMO1 activates PI3K/Akt signalling.
(a) Gene set enrichment analysis of the PI3K/Akt gene set signature (Hallmark gene set collection) performed on GSE57957.
(b) KEGG (Kyoto Encyclopedia of Genes and Genomes) analysis of ELMO1-regulated signalling pathways by STRING.
(c) Rows of the heat map display gene expression grouped by PI3K/Akt signalling pathway in TCGA LIHC patients with high or low ELMO1 expression. Data are represented as Z scores.
(d) mRNA expression levels of PI3K in MHCC97L-control, MHCC-97 L-ELMO1, MHCC97H-shcontrol and MHCC97H-shELMO1 cells by qRT-PCR. GAPDH was used as the internal control. Three independently repeated experiments were performed with similar results. Student's t-test: *p < 0.05. **p < 0.01, ***p < 0.001.
(e) Protein expression levels of PI3K, t-Akt, p-Akt, GSK3β, p-GSK3βin MHCC97L-control, MHCC-97 L-ELMO1, MHCC97H-shcontrol and MHCC97H-shELMO1 by western blot analysis. GAPDH was used as the internal control. Three independently repeated experiments were performed with similar results.
(f) Western blot analysis showing PI3K and p-Akt levels in HCC cells in the presence or absence of PI3K/Akt inhibitor (LY294002). Left panel: MHCC97L-control and MHCC97L-ELMO1; Right panel: MHCC97H-shcontrol and MHCC97H-shELMO1. GAPDH was used as the internal control. The results show one representative of three similar experiments.
(g) Transwell assays showing the invasion and migration of MHCC97L-ELMO1, MHCC97H-shELMO1, and their respective parental cells in the presence or absence of LY294002. Student's t-test: *p < 0.05. **p < 0.01, ***p < 0.001, ns, not significant.
(h) Representative blots showing the levels of E-cadherin, N-cadherin, and snail in parental MHCC97L-control and MHCC97L-ELMO1 cells, and parental MHCC97H-shcontrol and MHCC97H-shELMO1 cells, in the presence or absence of LY294002. GAPDH was used as the internal control. The results show one representative of three similar experiments.
To further validate that the PI3K/Akt pathway is downstream of ELMO1, we applied a PI3K/Akt signalling pathway inhibitor, LY294002, which is widely used to attenuate PI3K/Akt pathway expression. Indeed, ELMO1 enhanced PI3K/Akt signalling, as demonstrated by a significant increase in the levels of p-Akt in MHCC97L-ELMO1 cells, and LY294002 could antagonize this effect (Fig. 4f). Furthermore, ELMO1 knocked down the inhibitory Akt signal with a marked decrease in p-Akt levels in HCC cells, and the inhibitory effect was enhanced in the presence of LY294002, as evidenced by an attenuation of the level of p-Akt (Fig. 4f). Altogether, these findings provide evidence that ELMO1 acts as positive regulator of the PI3K/Akt signalling pathway, potentially providing an explanation for the pro-metastatic effect of ELMO1 in HCC.
3.7. ELMO1 induces EMT through the PI3K/Akt signalling pathway
Recent studies have revealed a regulator role of the PI3K/Akt signalling pathway in EMT [32]. Therefore, we considered the possibility that ELMO1 could regulate EMT through the PI3K/Akt signalling pathway. Indeed, ELMO1 protein interacts with the protein of PIK3CA, CDH1 (E-cadherin) and VEGFA derived from STRING, a functional protein association networks database (Supplementary Fig. 3c), suggesting a potential link between ELMO1, PI3K/Akt signalling and EMT. Moreover, overexpression of ELMO1 induced a dramatic rearrangement of F-actin in MHCC97L cells, and inhibition of PI3K/Akt signalling by addition of LY294002 could abolish the inducing effect of ELMO1 on F-actin (Supplementary Fig. 3d). Furthermore, LY294002 could reduce the positive effect of ELMO1 on MHCC97L cell invasion and migration (Fig. 4g). In addition, overexpressed ELMO1 induced a dramatic enhancement of N-cadherin and Snail in MHCC97L cells, whereas inhibition of PI3K/Akt signalling by the addition of LY294002 could abolish the inducing effect of ELMO1 (Fig. 4h). Together, these results suggest that ELMO1 is critically involved in mediating EMT through the Akt signalling pathway.
3.8. ELMO1 promotes cell growth, metastasis and EMT in vivo
We next evaluated ELMO1 overexpression or knocked down cell lines injected subcutaneously in mice (Fig. 5a). Notably, the tumour volume and weight of MHCC97L-ELMO1-derived tumours was significantly increased compared with MHCC97L-control. In contrast, ELMO1 knockdown achieved the opposite effect (Fig. 5b, c). Moreover, we transplanted the subcutaneous tumour into liver (Fig. 5e), although no obvious metastasis was observed in the lung by the end of the experiments (Fig. 5e). ELMO1 enhanced the rate of intrahepatic metastasis in tumours derived from MHCC97L-ELMO1 cells compared with control tumours from parental MHCC97L-control cells (Fig. 5d). Conversely, there was a lower incidence of intrahepatic metastasis in tumours derived from MHCC97H-shELMO1 cells compared with control tumours from parental MHCC97H-shcontrol cells (Fig. 5d). In addition, ELMO1 increased the intrahepatic tumour volume, whereas ELMO1 knockdown resulted in a decreased intrahepatic tumour volume (Fig. 5e, f). Together, these results suggest that ELMO1 promotes cell growth and cell metastasis in vivo.
Fig. 5.
ELMO1 promotes cell growth and EMT in vivo.
(a) Schematic of the experimental workflow with the establishment of subcutaneous ELMO1 nude mice and the ELMO1 luciferase orthotropic mouse model in the liver of nude mice and monitoring of tumour growth and metastasis in vivo.
(b) Tumour growth in mice subcutaneously injected with the indicated stable cell lines at the indicated time points. n = 6 mice per group. Representative images are shown below.
(c) Left panel: Tumours derived from the indicated HCC cells were weighed. n = 6 for each group. Right panel: representative images of subcutaneous tumours derived from the indicated HCC cells. Student's t-test: *p < 0.05. **p < 0.01, ***p < 0.001.
(d) The intrahepatic metastatic proportion of the liver was compared in the orthotropic xenograft. c2 test: *p < 0.05. **p < 0.01, ***p < 0.001.
(e) Representative bioluminescence images of each indicated group.
(f) Representative images of orthotopic tumours derived from the indicated HCC cells.
(g) Representative H&E and IHC images of E-cadherin, N-cadherin, Snail and SOX10 in orthotopic tumours derived from the indicated HCC cells. Magnification: X200, scale bar:50um.
We further examined the relationship between ELMO1 and EMT-associated markers in intrahepatic tumours by IHC. Similar to the in vitro results, increases in N-cadherin, Snail and SOX10 protein expression were detected in tumours from ELMO1 overexpression cells, and decreased E-cadherin protein levels were observed in tumours from ELMO1 knockdown cells. In contrast, tumours derived from MHCC97H-shELMO1 HCC cells exhibited the opposite effects (Fig. 5g). These findings suggest that, in vivo, the availability of EMT-associated factors can be affected by ELMO1, thus revealing a critical role of ELMO1 in the induction of EMT of HCC cells.
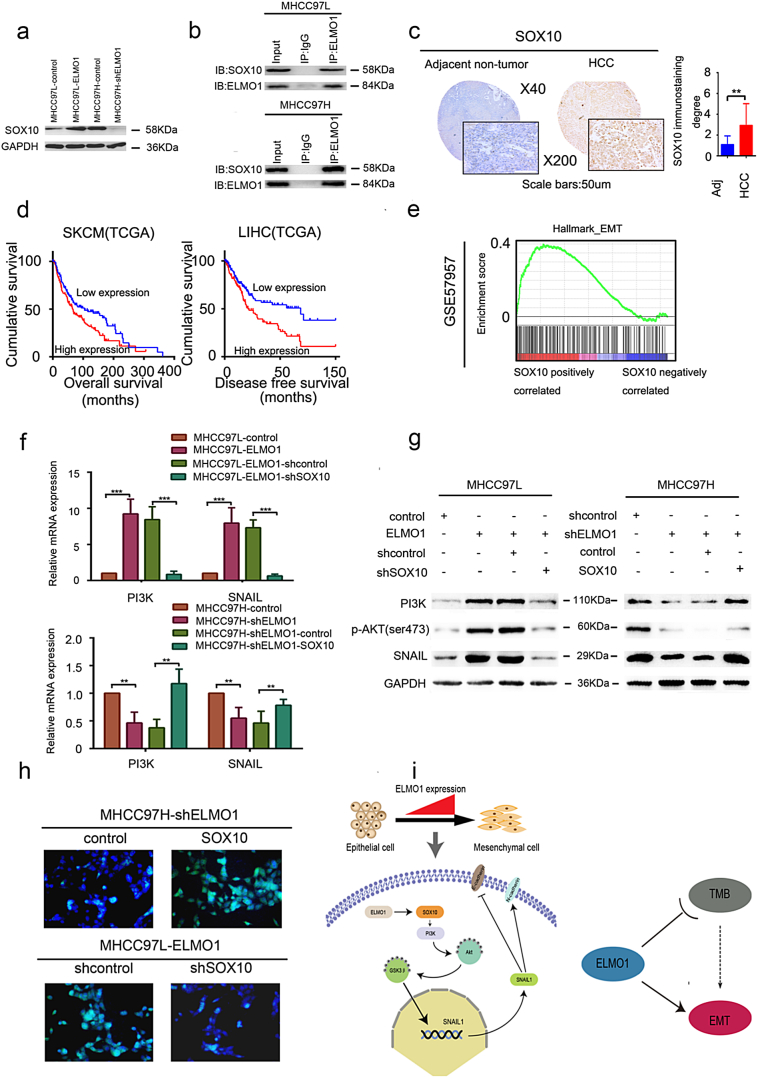
3.9. SOX10 is critical for ELMO1-enhanced cell invasion, metastasis and PI3K/Akt signalling
Our data support ELMO1-activated Akt signalling as a cause of HCC metastasis, but the full mechanism was still unclear. We performed EMT-associated regulator analyses using human the Epithelial to Mesenchymal Transition (EMT) RT2 Profiler™ PCR Array, and we identified various EMT-associated regulators that could be affected by ELMO1 (Supplementary Fig. 4a, b). Notably, sex determining region Y (SRY) and the related HMG box-containing factor 10 (SOX10), a member of the SOX family transcription factors, were upregulated in the ELMO1 overexpression HCC cells and suppressed in ELMO1 knockdown HCC cells, suggesting that SOX10 expression is likely responsible for the increased or decreased ELMO1 expression levels. This result was further confirmed by western blot analysis in ELMO1 overexpression or knockdown HCC cells (Fig. 6a). Interestingly, we found that ELMO1 could directly bind to SOX10 (Fig. 6b). Additionally, SOX10 protein expression was higher than in adjacent non-tumour tissues by IHC (Fig. 6c). Furthermore, a positive relationship between ELMO1 and SOX10 was observed in orthotopic HCC (Fig. 5g). Therefore, it seemed likely that ELMO1 mediated SOX10 expression in HCC. These data support the conclusion that ELMO1 can increase SOX10 expression in HCC.
Fig. 6.
ELMO1 regulates SOX10.
(a) SOX10 expression levels were determined by western blot analysis. GAPDH was used as the internal control. Three independent experiments were performed with similar results.
(b) Co-IP between ELMO1 and SOX10 on MHCC97L or MHCC97H cell contexts.
(c) Representative IHC images of SOX10 expression in the SYSU cohorts. Left panel, top: magnification 40×; bottom: magnification 200×, scale bar: 50um. Right panel: quantification of SOX10 immunostaining degree, student's t-test: *p < 0.05. **p < 0.01, ***p < 0.001, ns, not significant. Adj: Adjacent non-tumour tissues.
(d) Kaplan-Meier analysis of OS of SOX10 in TCGA SKCM patients and DFS of SOX10 in TCGA LIHC patients.
(e) Enrichment plot for the EMT signature from the “Hallmark” gene set collection in GSE57957.
(f) mRNA expression level of PI3K, Snail in the indicated HCC cells by qRT-PCR. GAPDH was used as the internal control. Three independent experiments were performed with similar results. Student's t-test: *p < 0.05. **p < 0.01, ***p < 0.001.
(g) Protein expression levels of PI3K, p-Akt, and Snail in the indicated HCC cells by western blot analysis. GAPDH was used as the internal control. Three independent experiments were performed with similar results.
(h) Representative IF images of F-actin staining of the indicated HCC cells.
(i) A proposed model for all results. Left panel: ELMO1 regulates SOX10 and subsequently activates PI3K/Akt signalling and dramatically upregulates Snail expression, thus inducing EMT. Right panel: ELMO1 can link TMB with EMT.
To evaluate the clinical relevance of SOX10 in cancer, we explored the TCGA datasets. Notably, although SOX10 had no significant effect on OS in LIHC patients, LIHC patients with high SOX10 expression showed a shorter DFS (Fig. 6d). Furthermore, in human SKCM patient cohorts of TCGA, patients with high SOX10 displayed a poorer OS when compared with patients with low SOX10 (Fig. 6d). Thus, we concluded that SOX10 could predict the prognosis of HCC patients, at least partially, indicating DFS in HCC.
Recent studies have indicated that SOX10 regulates EMT in cancer [33,34]. This finding was further supported in our GSEA of GEO databases using HALLMARK gene collection (Fig. 6e), which revealed an enhancement of the EMT signature in ELMO1 overexpression tumours, suggesting that SOX10 is involved in initializing EMT in HCC.
Triggered by our initial finding of ELMO1 mediating PI3K/Akt signalling and subsequently inducing EMT, we wondered whether SOX10 was involved in this axis. However, to the best of our knowledge, the relationship between SOX10 and PI3K/Akt signalling has not yet been explored, and recent studies have reported that SOX9, another member of SOX family, can activate PI3K/Akt signalling [35]. Therefore, we hypothesized that SOX10 might mediate PI3K/Akt activation by ELMO1. As expected, KEGG analysis from platform STRING revealed an enrichment of PI3K/Akt signalling in the presence of SOX10, confirming the involvement of SOX10 in PI3K/Akt signalling activation, although PI3K/Akt was not the most enriched signalling pathway (Supplementary Fig. 4c). Furthermore, we evaluated whether SOX10 was essential for ELMO1 mediation of the PI3K/Akt/Snail axis. SOX10 silencing resulted in reduced mRNA and protein levels of PI3K and snail, as well as Akt phosphorylation. However, overexpression of SOX10 showed the opposite effect (Fig. 6f, g), suggesting that SOX10 functions as a downstream target of ELMO1 that leads to PI3K/Akt signalling activation. Additionally, upregulated SOX10 in the ELMO1-low HCC cell line led to enhanced cytoskeletal staining, whereas downregulated SOX10 in the ELMO1-high HCC cell line demonstrated an opposite effect (Fig. 6h), supporting a role for ELMO1 in promoting cell metastasis through regulating SOX10. Together, these results suggest that ELMO1 regulates cell metastasis and PI3K/Akt signalling in a manner dependent on ELMO1 regulatory activity towards SOX10.
4. Discussion
In the present study, we demonstrated that increased ELMO1 expression is associated with HCC metastasis and a poor prognosis. ELMO1 induces EMT via the SOX10/PI3K/Akt axis. Moreover, increased ELMO1 is correlated with a lower TMB and positively with DDR (ERCC4, PALB2), MMR (RPA3), MSI (MSH3). In addition, ELMO1 can serve as a link between EMT and TMB. These results provide insight into the regulatory mechanism underlying EMT and how EMT is linked to TMB in HCC, which may improve our understanding of predictive biomarkers in immune checkpoint blockade.
This study indicated the importance of ELMO1 in HCC prognosis, which is in agreement with previous research implicated in AML [22]. Furthermore, it should be emphasized that the current findings are consistent with prior studies [36], suggesting that ELMO1 expression is upregulated in HCC and associated with microscopic vein invasion in HCC. Together, the results imply the potential of ELMO1 as a predictive prognostic biomarker in HCC and its association with the HCC vascular invasion phenotype. Furthermore, we extended these findings in a larger LIHC patient cohort, in TCGA, combined with the notion that plasma markers of methylated ELMO1 detect 86% of gastric adenocarcinoma with 95% specificity [37], supporting ELMO1 as a promising non-invasive predictive biomarker of HCC screening, surveillance and prognostic prediction. However, further studies corroborated in a larger cohort in HCC are still warranted, and whether mRNA- or protein-based quantification of ELMO1 for HCC prognosis is a better predictor is unclear.
Notably, we further revealed a link between ELMO1 and EMT. Although it has been previously reported that ELMO1 can stabilize snail via NF-kB signalling, this phenomenon only occurred in the context of IL-8 binding to CXCR1 [38]. Therefore, whether ELMO1 upregulated in the absence of the indicated background can induce EMT remains unclear. Furthermore, a recently published article has challenged the classic function of EMT in cancer metastasis [39,40]. In our study, we found that ELMO1 could upregulate SOX10 and induce EMT via PI3K/Akt signalling in HCC. SOX10 is a member of the SOX transcription factor family. In this study, SOX10 was found to be overexpressed in HCC and was a predictor of DFS, although it failed to predict OS in the TCGA LIHC cohort. Although the importance of SOX10 in cancer initiation and progression is well-recognized in several cancer types [41], the regulation of SOX10 remains poorly characterized. Our data suggested ELMO1 could directly regulate SOX10, which means that ELMO could act upstream of SOX10. SOX10 has been reported to play a pivotal role in driving EMT and to link Stem- and EMT-like characteristics in breast cancer [42]. Consistently, we demonstrated SOX10 associated with the EMT phenotype with PI3K/Akt signalling as a key downstream target of ELMO1/SOX10. However, whether the combination of ELMO1 and SOX10 inhibition could increase the antitumour effect in cancer requires further investigation.
TMB is an important biomarker of the immune checkpoint blockade response in tumour therapy [[13], [14], [15]], based on increased expression of tumour neoantigens that are recognized by immune cells [43]. Interestingly, higher ELMO1 expression resulted in lower TMB. Furthermore, ELMO was associated with the DDR, MMR phenotype, a pivotal regulatory mechanism in tumour mutability. These results implied that ELMO1 might decrease TMB through DDR and MMR. In brief, ELMO1 has the potential to predict the response to immune checkpoint blockade therapy, lending insight into personalized cancer therapy strategies. However, the mechanism by which ELMO1 regulates tumour mutability through DDR or MMR remains to be explored.
Although most studies have focused on the effect of EMT in cancer metastasis and drug resistance, recent studies have explored the role of EMT in reprogramming the tumour immunity microenvironment. A strong association between EMT status and an inflammatory tumour microenvironment with elevation of PD-L1, PD-L2, PD-1, TIM-3, B7-H3, BTLA and CTLA-4 was identified in NSCLC [44]. Despite a failure to identify an association between EMT and tumour mutation burden in NSCLC, Harbhajanka et al. revealed a higher frequency of STK11 or KEAP1 mutations in “epithelial” than “mesenchymal” lung adenocarcinomas [45], suggesting that mesenchymal tumours have fewer neoantigen mutations. Moreover, one recent report has demonstrated that low TMB is correlated with TGFβ signalling, which is known as a main driver of EMT [12]. Notably, this study provides evidence that EMT is inversely correlated with TMB in HCC and that ELMO1 links the two phenotypes. However, it is noteworthy that EMT was correlated with elevated PD-L1 expression, which is a potential biomarker for anti-PD-1/PD-L1 therapy [7]. Therefore, this finding seems contradictory to our results showing that EMT was negatively correlated with TMB, which was recently reported as a biomarker of better clinical outcomes for immune checkpoint blockade therapy. Notably, some patients with high PD-L1 expression benefit little from immune checkpoint blockade therapy, while some patients with low PD-L1 expression show a greater benefit [46], suggesting that the PD-L1 expression level is not an exclusive biomarker for the response to immune checkpoint blockade therapy. Therefore, our study provides insights into ELMO1, which links EMT and TMB. The use of TMB detection is restricted due to its high cost [47]. Therefore, examination of ELMO1 expression levels could represent a substitute solution with a much lower cost.
In conclusion, ELMO1 is an independent prognostic factor of HCC and some other cancer types. ELMO1 overexpression promotes HCC cell proliferation, invasion, and migration via EMT in vitro and in vivo by regulating SOX10/PI3K/Akt signalling, suggesting a tumour-activator role of ELMO1 in HCC. Furthermore, ELMO1 is inversely correlated with TMB by regulating MMR or DDR. In addition, ELMO1 links EMT with TMB. Close monitoring of ELMO1 levels in patients has the potential to predict the response to immune checkpoint blockade therapy in HCC either alone or combined with TMB. This study highlights the role of ELMO1 as a biomarker for HCC prognosis and the immune checkpoint blockade therapeutic response and as a potential therapeutic target for HCC therapy.
Financial support information
This work was supported by National Natural Science Foundation of China (81302142, 81172039, 81671805, 81602723, 81760112), Natural Science Foundation of Guangdong Province (S2011010005864; 2014A030313108, 2018A030313529), Young Teacher Training Program of Sun Yat-sen University (15ykpy15), Science and Technology Plan of Guangdong Province (2013B021800134, 2017A020215125, 2017b020247057), Special Support Program of Guangdong Province, Science and technology innovation youth talent support program (No. 201627015), the Pearl River Science and Technology New Talent of Guangzhou City (No. 201806010076), and Medical Scientific Research Foundation of Guangdong Province (A2015313).
Author contributions
Study design: Shunli Shen and Baogang Peng. Conducting the experiments: Hong Peng, Yi Zhang, Zhiwei Zhou, Yu Guo. Data analysis: Yi Zhang, Yu Guo. Manuscript writing: Hong Peng, Yi Zhang, Xiaohui Huang, Zhaohui Zhang, Yupeng Hua, Shaoqiang Li, Kenneth D. Westover, Bin Chen, Ruiyun Xu, Nan Lin. All authors read and approved the final manuscript. Hong Peng, Yi Zhang, Zhiwei Zhou, and Yu Guo contributed equally to this work. All authors agreed with the results and conclusions.
Declaration of Competing Interest
The authors declare that they have no conflicts of interest.
Acknowledgements
We thank our colleagues who supported this work at our institution, with special thanks to Professor Sarah Comerford from the Molecular Genetics, Green Center for Systems Biology, University of Texas Southwestern Medical Center at Dallas, and Jiawei Zhao and Yuemeng Jia from the University of Texas Southwestern Medical Center at Dallas for review and correction of the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.07.002.
Contributor Information
Baogang Peng, Email: pengbg@mail.sysu.edu.cn.
Shunli Shen, Email: shenshli@sysu.edu.cn.
Appendix A. Supplementary data
Supplementary material
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.El-Serag H.B. Hepatocellular carcinoma. N Engl J Med. 2011;365:1118–1127. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- 3.Pastushenko I., Brisebarre A., Sifrim A., Fioramonti M., Revenco T., Boumahdi S. Identification of the tumour transition states occurring during EMT. Nature. 2018;556:463–468. doi: 10.1038/s41586-018-0040-3. [DOI] [PubMed] [Google Scholar]
- 4.Nieto M.A., Huang R.Y., Jackson R.A., Thiery J.P. EMT: 2016. Cell. 2016;166:21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 5.Lamouille S., Xu J., Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lou Y., Diao L., Cuentas E.R., Denning W.L., Chen L., Fan Y.H. Epithelial- mesenchymal transition is associated with a distinct tumor microenvironment including elevation of inflammatory signals and multiple immune checkpoints in lung adenocarcinoma. Clin Cancer Res. 2016;22:3630–3642. doi: 10.1158/1078-0432.CCR-15-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mariathasan S., Turley S.J., Nickles D., Castiglioni A., Yuen K., Wang Y. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature. 2018;554:544–548. doi: 10.1038/nature25501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu L.Y., Zhang W.M., Yang X.M., Cui L., Li J., Zhang Y.L. Silencing of MICAL- L2 suppresses malignancy of ovarian cancer by inducing mesenchymal-epithelial transition. Cancer Lett. 2015;363:71–82. doi: 10.1016/j.canlet.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 9.Aiello N.M., Brabletz T., Kang Y., Nieto M.A., Weinberg R.A., Stanger B.Z. Upholding a role for EMT in pancreatic cancer metastasis. Nature. 2017;547:E7–E8. doi: 10.1038/nature22963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thiery J.P., Lim C.T. Tumor dissemination: an EMT affair. Cancer Cell. 2013;23:272–273. doi: 10.1016/j.ccr.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 11.Kolijn K., Verhoef E.I., Smid M., Böttcher R., Jenster G.W., Debets R. Epithelial-mesenchymal transition in human prostate cancer demonstrates enhanced immune evasion marked by IDO1 expression. Cancer Res. 2018;78:4671–4679. doi: 10.1158/0008-5472.CAN-17-3752. [DOI] [PubMed] [Google Scholar]
- 12.Wang L., Saci A., Szabo P.M., Chasalow S.D., Castillo-Martin M., Domingo-Domenech J. EMT-and stroma-related gene expression and resistance to PD- 1 blockade in urothelial cancer. Nat Commun. 2018;9:3503. doi: 10.1038/s41467-018-05992-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Migden M.R., Rischin D., Schmults C.D., Guminski A., Hauschild A., Lewis K.D. PD-1 blockade with Cemiplimab in advanced cutaneous squamous-cell carcinoma. N Engl J Med. 2018;379:341–351. doi: 10.1056/NEJMoa1805131. [DOI] [PubMed] [Google Scholar]
- 14.Forde P.M., Chaft J.E., Smith K.N., Anagnostou V., Cottrell T.R., Hellmann M.D. Neoadjuvant PD-1 blockade in resectable lung cancer. N Engl J Med. 2018;378:1976–1986. doi: 10.1056/NEJMoa1716078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yarchoan M., Hopkins A., Jaffee E.M. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med. 2017;7:2500–2501. doi: 10.1056/NEJMc1713444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giannelli G., Villa E., Lahn M. Transforming growth factor-β as a therapeutic target in hepatocellular carcinoma. Cancer Res. 2014;74:1890–1894. doi: 10.1158/0008-5472.CAN-14-0243. [DOI] [PubMed] [Google Scholar]
- 17.Bertran E., Crosas-Molist E., Sancho P., Caja L., Lopez-Luque J., Navarro E. Overactivation of the TGF-β pathway confers a mesenchymal-like phenotype and CXCR4-dependent migratory properties to liver tumor cells. Hepatology. 2013;58:2032–2044. doi: 10.1002/hep.26597. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y.P., Yu G.R., Lee M.J., Lee S.Y., Chu I.S., Leem S.H. Lipocalin-2 negatively modulates the epithelial-to-mesenchymal transition in hepatocellular carcinoma through the epidermal growth factor (TGF-beta1)/Lcn2/Twist1 pathway. Hepatology. 2013;58:1349–1361. doi: 10.1002/hep.26467. [DOI] [PubMed] [Google Scholar]
- 19.Sanui T., Inayoshi A., Noda M., Iwata E., Stein J.V., Sasazuki T. DOCK2 regulates Rac activation and cytoskeletal reorganization through interaction with ELMO1. Blood. 2003;102:2948–2950. doi: 10.1182/blood-2003-01-0173. [DOI] [PubMed] [Google Scholar]
- 20.Li H., Yang L., Fu H., Yan J., Wang Y., Guo H. Association between Gαi2 and ELMO1/Dock180 connects chemokine signalling with Rac activation and metastasis. Nat Commun. 2013;4:1706. doi: 10.1038/ncomms2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grimsley C.M., Kinchen J.M., Tosello-Trampont A.C., Brugnera E., Haney L.B., Lu M. Dock180 and ELMO1 proteins cooperate to promote evolutionarily conserved Rac-dependent cell migration. J Biol Chem. 2004;279:6087–6097. doi: 10.1074/jbc.M307087200. [DOI] [PubMed] [Google Scholar]
- 22.Capala M.E., Vellenga E., Schuringa J.J. ELMO1 is upregulated in AML CD34+ stem/progenitor cells, mediates chemotaxis and predicts poor prognosis in normal karyotype AML. PLoS One. 2014;9 doi: 10.1371/journal.pone.0111568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yagi K., Akagi K., Hayashi H., Nagae G., Tsuji S., Isagawa T. Three DNA methylation epigenotypes in human colorectal cancer. Clin Cancer Res. 2011;16:21–33. doi: 10.1158/1078-0432.CCR-09-2006. [DOI] [PubMed] [Google Scholar]
- 24.Dong Z.Y., Zhong W.Z., Zhang X.C., Su J., Xie Z., Liu S.Y. Potential predictive value of TP53 and KRAS mutation status for response to PD-1 blockade immunotherapy in lung adenocarcinoma. Clin Cancer Res. 2017;23:3012–3024. doi: 10.1158/1078-0432.CCR-16-2554. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y., Wang W., Wang Y., Huang X., Zhang Z., Chen B. NEK2 promotes hepatocellular carcinoma migration and invasion through modulation of the epithelial- mesenchymal transition. Oncol Rep. 2018;39:1023–1033. doi: 10.3892/or.2018.6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo Y., Wang J., Zhang L., Shen S., Guo R., Yang Y. Theranostical nanosystem- mediated identification of an oncogene and highly effective therapy in hepatocellular carcinoma. Hepatology. 2016;63:1240–1255. doi: 10.1002/hep.28409. [DOI] [PubMed] [Google Scholar]
- 27.Shen J., Ju Z., Zhao W., Wang L., Peng Y., Ge Z. ARID1A deficiency promotes mutability and potentiates therapeutic antitumor immunity unleashed by immune checkpoint blockade. Nat Med. 2018;24:556–562. doi: 10.1038/s41591-018-0012-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Devarakonda S., Rotolo F., Tsao M.S., Lanc I., Brambilla E., Masood A. Tumor mutation burden as a biomarker in resected non-small-cell lung cancer. J Clin Oncol. 2018;36:2995–3006. doi: 10.1200/JCO.2018.78.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vanderwalde A., Spetzler D., Xiao N., Gatalica Z., Marshall J. Microsatellite instability status determined by next-generation sequencing and compared with PD-L1 and tumor mutational burden in 11,348 patients. Cancer Med. 2018;7:746–756. doi: 10.1002/cam4.1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kandoth C., McLellan M.D., Vandin F., Ye K., Niu B., Lu C. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lv D., Jia F., Hou Y., Sang Y., Alvarez A.A., Zhang W. Histone acetyltransferase KAT6A upregulates PI3K/AKT signaling through TRIM24 binding. Cancer Res. 2017;77:6190–6201. doi: 10.1158/0008-5472.CAN-17-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yin T., Wang G., He S., Shen G., Su C., Zhang Y. Malignant pleural effusion and ascites induce epithelial-mesenchymal transition and cancer stem-like cell properties via the vascular endothelial growth factor (VEGF)/phosphatidylinositol 3-kinase (PI3K)/Akt/mechanistic target of rapamycin (mTOR) pathway. J Biol Chem. 2016;291:26750–26761. doi: 10.1074/jbc.M116.753236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y., Xu Y., Li Z., Zhu Y., Wen S., Wang M. Identification of the key transcription factors in esophageal squamous cell carcinoma. J Thorac Dis. 2018;10:148–161. doi: 10.21037/jtd.2017.12.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nelson E.R., Sharma R., Argani P., Cimino-Mathews A. Utility of Sox10 labeling in metastatic breast carcinomas. Hum Pathol. 2017;67:205–210. doi: 10.1016/j.humpath.2017.08.011. [DOI] [PubMed] [Google Scholar]
- 35.Hong Y., Chen H., Rao Z., Peng B., Hu H., Lin S. In vitro study on the role of SOX9 in trastuzumab resistance of adenocarcinoma of the esophagogastric junction. Exp Ther Med. 2018;5:3103–3107. doi: 10.3892/etm.2018.5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.J1 Jiang, Liu G., Miao X., Hua S., Zhong D. Overexpression of engulfment and cell motility 1 promotes cell invasion and migration of hepatocellular carcinoma. Exp Ther Med. 2011;2:505–511. doi: 10.3892/etm.2011.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anderson B.W., Suh Y.S., Choi B., Lee H.J., Yab T.C., Taylor W.R. Detection of gastric cancer with novel methylated DNA markers: discovery, tissue validation, and pilot testing in plasma. Clin Cancer Res. 2018;24:5724–5734. doi: 10.1158/1078-0432.CCR-17-3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang B., Shi L., Lu S., Sun X., Liu Y., Li H. Autocrine IL-8 promotes F-actin polymerization and mediate mesenchymal transition via ELMO1-NF-κB-snail signaling in glioma. Cancer Biol Ther. 2015;16:898–911. doi: 10.1080/15384047.2015.1028702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fischer K.R., Durrans A., Lee S., Sheng J., Li F., Wong S.T. Epithelial- to- mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015;527:472–476. doi: 10.1038/nature15748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng X., Carstens J.L., Kim J., Scheible M., Kaye J., Sugimoto H. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527:525–530. doi: 10.1038/nature16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han S., Ren Y., He W., Liu H., Zhi Z., Zhu X. ERK-mediated phosphorylation regulates SOX10 sumoylation and targets expression in mutant BRAF melanoma. Nat Commun. 2018;9:28. doi: 10.1038/s41467-017-02354-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meng Z., Chen G., Chen J., Yang B., Yu M., Feng L. Tumorigenicity analysis of heterogeneous dental stem cells and its self-modification forchromosome instability. Cell Cycle. 2015;14:3396–3407. doi: 10.1080/15384101.2015.1036204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hellmann M.D., Callahan M.K., Awad M.M., Calvo E., Ascierto P.A., Atmaca A. Tumor mutational burden and efficacy of nivolumab monotherapy and in combination with ipilimumab in small-cell lung cancer. Cancer Cell. 2018;33:853–861. doi: 10.1016/j.ccell.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hellmann M.D., Nathanson T., Rizvi H., Creelan B.C., Sanchez-Vega F., Ahuja A. Genomic features of response to combination immunotherapy in patients with advanced non-small-cell lung Cancer. Cancer Cell. 2018;33:843–852. doi: 10.1016/j.ccell.2018.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hodges T.R., Ott M., Xiu J., Gatalica Z., Swensen J., Zhou S. Mutational burden, immune checkpoint expression, and mismatch repair in glioma: implications for immune checkpoint immunotherapy. Neuro Oncol. 2017;19:1047–1057. doi: 10.1093/neuonc/nox026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meng X., Huang Z., Teng F., Xing L., Yu J. Predictive biomarkers in PD-1/PD-L1 checkpoint blockade immunotherapy. Cancer Treat Rev. 2015;41:868–876. doi: 10.1016/j.ctrv.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 47.Steuer C.E., Ramalingam S.S. Tumor mutation burden: leading immunotherapy to the era of precision medicine? J Clin Oncol. 2018;36:631–632. doi: 10.1200/JCO.2017.76.8770. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
Clinical data, mutational profiles and normalized gene expression data from TCGA samples and different types of cancer cell lines were obtained from TCGA datasets (https://portal.gdc.cancer.gov/), GEPIA (http://gepia.cancer-pku.cn/) and Cancer Cell Line Encyclopedia (CCLE, http://www.broadinstitute.org/ccle;ref.25), mutation types of ELMO1 were analysed by cbioportal (http://www.cbioportal.org/), OS and DFS of TCGA patients were analysed by survExpress (http://bioinformatica.mty.itesm.mx:8080/Biomatec/SurvivaX.jsp), and mapping of the ELMOl-involved protein interaction network was performed using STRING (https://string-db.org/cgi/input.pl). The network was then exported and visualized in Cytoscape v.3.4.0 (http://www.cytoscape.org/), and the node sizes were adjusted to be proportional to the average. GSE84598, GSE76427, GSE57957 and GSE62232 were obtained from the NCBI Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/).