Abstract
Background
Metastatic colorectal cancer (CRC) remains a deadly disease. Identifying locally advanced CRC patients with high risk of developing metastasis and improving outcome of metastatic CRC patients require discovering master regulators of metastasis. In this context, the non-coding part of the human genome is still largely unexplored.
Methods
To interrogate the non-coding part of the human genome and disclose regulators of CRC metastasis, we combined a transposon-based forward genetic screen with a novel in vitro assay, which forces cells to grow deprived of cell-substrate and cell-cell contacts (i.e. forced single cell suspension assay - fSCS).
Findings
We proved that fSCS selects CRC cells with mesenchymal and pro-metastatic traits. Moreover, we found that the transposon insertions conferred CRC cells resistance to fSCS and thus metastatic advantage. Among the retrieved transposon insertions, we demonstrated that the one located in the 3′UTR of BTBD7 disrupts miR-23b::BTBD7 interaction and contributes to pro-metastatic traits. In addition, miR-23b and BTBD7 correlate with CRC metastasis both in preclinical experiments and in clinical samples.
Interpretation
fSCS is a simple and scalable in vitro assay to investigate pro-metastatic traits and transposon-based genetic screens can interrogate the non-coding part of the human genome (e.g. miRNA::target interactions). Finally, both Btbd7 and miR-23b represent promising prognostic biomarkers and therapeutic targets in CRC.
Fund
This work was supported by Marie Curie Actions (CIG n. 303877) and Friuli Venezia Giulia region (Grant Agreement n°245574), Italian Association for Cancer Research (AIRC, MFAG n°13589), Italian Ministry of Health (GR-2010-2319387 and PE-2016-02361040) and 5x1000 to CRO Aviano.
Keywords: spleeping beauty, DNA transposons, microRNA target, Colorectal cancer metastasis
Research in context.
Evidence before this study
The biological contribution of the non-coding part of the human genome to colorectal cancer (CRC) metastasis is difficult to tackle. In vitro resistance to anoikis has been used to identify genes involved in CRC metastasis. Using a microRNA-library screen, miR-23b was found as a negative regulator of CRC metastasis. Transposon based forward genetic screens are generally harnessed to target protein coding genes. Epithelial mesenchymal transition (EMT) is required for metastasis occurrence. Btbd7 is an EMT regulator with prognostic implications in hepatocellular carcinoma and lung cancer.
Added value of this study
We showed that transposon genetic screens, when devoid of protein trapping cassettes, can find in an unbiased manner non-coding elements relevant for CRC metastasis. We developed a modified in vitro anoikis assay, named forced single cell suspension assay (fSCS), which is easy and scalable, able to select cancer cells with EMT traits and metastatic advantages. Finally, we found that Btbd7 is a target of miR-23b, and together they participate to EMT and CRC metastasis formation.
Implications of all the available evidence
miR-23b, rather than an anti-metastatic miRNA, should be considered a miRNA with different roles in the different steps of metastasis. miR-23b in primary tumors of metastatic patients favors cancer cell extravasation, whereas at distant sites it inhibits overt metastasis. Instead Btbd7 expression has to be high to allow distant metastasis formation. Thus, miR-23b and Btbd7 represent novel prognostic markers to asses the probability of metastasis development in patients with CRC. Further prospective studies are warranted to further confirm miR-23b and Btbd7 clinical relevance, both as prognostic biomarkers and as therapeutic targets.
Alt-text: Unlabelled Box
1. Introduction
Metastasis (i.e. dissemination of cancer cells from the site of tumor origin to other tissues) is the primary cause of cancer-related deaths worldwide [1,2]. In such sense, colorectal cancer (CRC) is a paradigm: CRC is the third most common tumor in industrialized countries [3] and almost 50% of CRC patients develop metastasis with a consequent 5-years overall survival rate of 10% [[4], [5], [6], [7]]. Metastasis is a multi-step and plastic process: at the site of tumor origin, tumor cells gain increased invasion capability at expenses of proliferation; vice versa, when tumor cells colonize distant sites, they decrease their invasion capability favoring proliferation [[8], [9], [10]]. The plasticity of metastatic cells relies in part on their ability to undergo epithelial-mesenchymal transition (EMT), during which cells loose contact with neighboring cells and the extracellular matrix, and acquire an invasive phenotype and resistance to anoikis (death induced by loss of adhesion), all traits necessary to intravasate and survive into the circulation. When metastatic cells reach distant sites, to colonize these, they must undergo the inverse process, mesenchymal to epithelial transition (MET), and gain back epithelial features with high proliferation index [10]. Identifying locally advanced CRC patients with high risk of developing metastasis and improving outcome of metastatic CRC patients requires the identification of master regulators of metastasis. In this context, the non-coding part of the human genome is still largely unexplored.
The human genome counts 3 × 109 base pairs, of which <2% codes for proteins (coding part) and >98% is transcribed but does not code for proteins (non-coding part). The non-coding part of the human genome expanded during evolution far more than the protein-coding part [11]; very likely, evolution and biologic complexity relies on protein regulation rather than on protein number. Consistently, complex biological processes (e.g. cancer metastasis) are finely tuned by non-coding genomic elements, including transcribed ones (e.g. long non-coding RNAs and microRNAs) [12,13]. In fact, microRNAs regulate protein expression in a reversible manner and shape cellular phenotype accordingly, and thus they are ideal molecular on-off switches of cancer cell invasive and proliferative capabilities during metastasis.
Genetic screens permit to link predefined phenotypes (e.g. metastasis) with genes or genomic positions. Reverse genetic screens (based on cDNA, siRNA, and microRNA libraries, or more recently CRISPR) target mainly known genes to be overexpressed, silenced or mutated; whereas, forward genetic screens use a mutagen (e.g. chemicals, viruses or transposons) that randomly disrupts genetic elements in the entire genome. Compared with other genetic screens (e.g. retroviral-based screens that prefer the 5′ portion of active transcribed units) [14,15] transposon based screens interrogate the entire genome randomly and have already been used to retrieve non-coding regions promoting tumorigenesis [16]. In particular, sleeping beauty transposon (SB) is a mobile DNA element delimited by two inverted repeats (IR) able to insert in the host genome through a “cut and paste” mechanism; by doing so, SB can simultaneously cause insertional mutations and it can be used as tag to retrieve the genomic locations of its own insertions. SB integration occurs at any TA dinucleotides within the host genome without gross biases for any chromosome or chromosomal regions [17], which makes it an ideal tool to explore the entire human genome in search of new regulators of CRC metastasis.
To identify novel master regulators of CRC cancer metastasis, we combined a novel in vitro assay, which selects for cells able to survive in the concomitant absence of cell-cell contacts and cell-ECM adhesion, with the SB transposon-based forward genetic screen. For the functional part of the screen, we developed a modified in vitro anoikis assay. In fact in vitro anoikis, i.e. cell growth in the absence of cell-matrix contacts, has been used to select pro-metastatic cells [18,19]; however, this functional assay is not always as stringent as expected and resistance to anoikis is a common feature of most cancer cells.
By this approach, we found that the transposon insertions conferred CRC cells metastatic advantage. Among the retrieved transposon insertions, we demonstrated that the one located in the 3′UTR of BTBD7 disrupts miR-23b::BTBD7 interaction and contributes to pro-metastatic traits. In addition, both miR-23b and BTBD7 participate in CRC metastasis with possible clinical implications.
2. Materials and methods
2.1. Cell lines
HCT116, HT29, SW480, SW620, COLO205 CRC-lines and MCF7 and MDAMB231 breast cancer cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA) and were maintained in RPMI 1640 or DMEM high glucose, supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine (Life Technologies, Grand Island, NY, USA). The Rat intestine cell line RIE was provided by Dr. Peeper's laboratory and grown in DMEM high glucose 10% FBS. 293-FT cells were obtained from Invitrogen (#R70007, Thermo Fisher Scientific, Eugene, OR, USA), and were maintained in DMEM high glucose, supplemented with 10% FBS, 0.1 mM MEM Non-Essential Amino Acids (NEAA), 6 mM L-glutamine, 1 mM MEM Sodium Pyruvate, and 500 μg/ml G418.
Cell lines were grown at 37 °C, under 5% CO2, in humidified incubators and routinely tested using MycoAlert detection Kit (Lonza, Cologne, Germany) for mycoplasma contamination. Only mycoplasma negative cells were used for experiments.
2.2. Cell transfections and lentiviral transductions
Lipofectamine 2000 reagent (#1668019, Life Technologies) was used for plasmids or miRNA precursors' transfection (negative control and miR-23b-3p, #AM17110 and #AM17100, Life Technologies) as per manufacturer instruction. miRNA precursors were transfected at 100 nM final concentration. The calcium phosphate precipitation method was used for 293-FT transfection.
pEGFP-N1 empty vector was purchased by Clontech and pEGFP-Btbd7 was gently provided by Dr. Kenneth M. Yamada (National Institute of Health, Bethesda, Maryland, USA). HCT116 pEGFP empty or pEGFP-Btbd7 stable clones were obtained by selection of cells with 500 μg/ml G418 followed by FACS-sorting of pEGFP positive cells.
TWIST-ER, SNAIL-ER or BTBD7-sh CRC-cell lines were generated by lentiviral transduction. The lentiviral expression vectors used were: pWZL-TWIST-ER (#18799, Addgene), pWZL-Snail-ER (#18798, Addgene), and pLKO.1-puro carrying either human BTBD7-shRNAs (sh1_TRCN0000136584, sh2_TRCN0000137476, sh3_TRCN0000365745, sh4_ TRCN0000365743, sh5_ TRCN0000365741, MISSION shRNAs from Sigma-Aldrich (St. Louis, MO, USA)) or non-target shRNA control (SHC016; Sigma-Aldrich). Briefly, 293-FT cells were transfected with the above lentiviral expression vectors together with ViraPower™ Packaging Mix (pLP1, pLP2, and VSV-G) (#K497500, Invitrogen, Thermo Fisher Scientific, Eugene, OR, USA). Viral supernatants were collected 72 h later and transducing units per ml were determined by limiting dilution titration in HCT116 cells. A MOI of approximately 5 was used for transducing cells. After five days of selection with either 10 μg/ml blasticydin or 1 μg/ml puromycin (for pWZL or pLKO constructs, respectively) cells were immediately used for in vitro experiments, protein and RNA extraction. Polybrene (#TR1003G, Sigma-Aldrich) at a final concentration of 8 μg/ml was used to increase transduction efficiency.
pT2-vector, carrying the sleeping beauty transposon, pCMV-SB100x-vector, carrying the sleeping beauty transposase with high enzymatic activity or pCMV-SBD3-vector carrying the mutated sleeping beauty transposase with low enzymatic activity [50] were provided by Dr. Ivics (Max Delbrück Center for Molecular Medicine, Berlin, Germany). As mutagenic tool of the forward genetic screen used herein, we modified the pT2 transposon vector in order to carry the coding sequence of EGFP under the control of CMV promoter, but in the absence of any polyA signal, splicing acceptor or donor sites. By co-trasfecting the pT2-CMV-EGFP vector with the pCMV-SB100x wt trasposase, the SB trasposon was stably integrated in the DNA of the transfected cells to randomly mutagenize their genome. Co-trasfection with pCMV-SBD3 was used as negative control.
2.3. Cell treatments
For 4-Hydroxytamoxifen treatment (4OHT)(#SML1666, Sigma) cells were plated 24 h earlier at low density and then exposed to 0.1 μM 4OHT in complete medium for 3–6 days, with daily treatment medium change, and were passaged by trypsinization before confluence was reached. At the end of the treatment period, cells were immediately used for in vitro anoikis and fSCS assays and for protein and RNA extraction.
Cell response to chemotherapy was done by treating exponentially growing cells in 24 multiwell plates with 100 or 500 μg/ml of Fluorouracil (5-FU) for 24 h in complete medium. Cell viability was assed 24 h later by MTT (#M2003, Sigma) staining. MTT stock solution was added to cells at a final concentration of 0.28 mg/ml and incubated for 2 h at 37 °C. Supernatant was then discarded and cells were air-dried. Reduced MTT, measure of cellular metabolic activity and index of cell viability, was dissolved by adding 200 μl of DMSO per 24-well and measured by reading the absorbance at 580 nm with Infinite M1000 microplate reader (Tecan, Männedorf, Switzerland). MTT experiments were performed in technical triplicates at least two times (independent biological replicates).
2.4. In vitro anoikis and forced single cell suspension (fSCS) assays
For the in vitro anoikis assay 1 × 105 cells detached by trypsin were resuspended in serum free medium containing 0.1% BSA (SFM) and kept on ultra-low attachment (ULA) six-well plates (#3471, Corning) for 24 h. Next, cells were collected and washed with PBS without calcium phosphate, treated for 5 min with trypsin for the disruption of cell aggregates, plated on standard 10 cm dishes in complete medium and let grow until surviving cells formed visible colonies (approximately 3–6 days). For the fSCS assay, 1 × 105 cells were suspended in SFM and kept on ULA six-well plates for 24 h as above, but either with the addition of 1 mM EDTA (Ethylenediaminetetraacetic Acid, #405463, Carlo Erba) or of 1.33 mg/ml ES tested Hyaluronidase (#37326–33-3, Sigma-Aldrich), to maintain cells separated from each other. Next, cells were collected, washed with PBS and either tested by FACS for propidium iodide staining of death cells or plated on standard 10 cm dishes for 7–14 days. For both anoikis and fSCS assays, colonies formed by surviving cells on 10 cm dishes were stained with 20% methanol-crystal violet (#C3886, Sigma) solution. Crystal violet stained dishes were acquired using UVItec transilluminator (UVItec, Cambridge). Cell colonies or surviving cells were counted either manually our automatically using Fiji “cell counter” plug-in or the “analyze particle” tool [51]. For automatic analyses, images of stained cell dishes were converted as 8-bit binary. Shape descriptors from the Fiji “set measurement” tool were annotated to calculate colony circularity index.
To obtain fSCS-resistant pools of cells from parental HCT116 cells (T0 cells that never underwent fSCS), sequential rounds of fSCS were performed on ULA T75 flasks (#3814, Corning). Cells surviving from the first round of fSCS were pooled together and expanded to generate T1 progeny. By subjecting T1 cells to an additional round of fSCS assay T2 progeny was generated and so on.
For physical mean-based fSCS, single cells were confined into individual water-in-oil droplets. To this aim, cell suspension (at 1 × 106 cells/ml in SFM) was divided into a monodisperse water-in-oil microdroplet population, using the Droplet generator microfluidic chip (microfluidic ChipShop). In particular, the cell suspension was mixed at a flow-focusing junction with a continuous phase consisting of a fluorinated oil (3 m HFE-7500) and 2% (w/w) of surfactant (RAN biotechnologies). Surfactant was added to avoid droplet fusion after generation. Microfluidics was operated by syringe pumps (Cetoni), at a continuous:dispersed flow rate of 3:1 (continuous phase 900 μl/h, dispersed phase 300 μl/h). The average volume of generated droplets was ~110 pl, leading to a ~89% probability to generate an empty droplet, and a ~99.4% probability to generate a droplet containing at most a single cell, according to Poisson distribution. The emulsion was incubated for 24 h at 37 °C, under 5% CO2, in a humidified incubator. Next, droplets were broken as described in [52], by adding an equal volume of cell culture medium to the emulsion, then by adding a volume of perfluorooctanol (Sigma) equal to the volume of the continuous phase. After a gentle hand-mix and a couple of minutes of incubation the top phase containing the cell suspension was recovered with a micropipette, plated on standard 10 cm dishes and let grow for 7–14 days. Colonies formed by surviving cells were fixed, stained and counted as described above.
Cell morphology during or following growth in suspension (either anoikis or fSCS) or 4OHT treatment was detected by using an inverted microscope (Eclipse TS/100, Nikon) coupled with photomicrographic systems (DS camera control unit DS L2, Nikon).
2.5. Migration and invasion assays
Migration and invasion assays were performed using fluoroblock transwells with 8 μm pore size (24 well format) (#351152, BD falcon). For migration experiments the bottom-side of transwell membranes was coated with fibronectin (10 μg/ml in PBS for 1 h at 37 °C, subsequently blocked with 1% BSA in PBS 1 h at 37 °C). For invasion experiments the top-side of transwell porous membranes was coated with Matrigel (8 μg/100 μl overnight at 4 °C and let solidify two hours at 37 °C prior to use). Cells were starved overnight, detached with trypsin, counted and stained in SFM with DiI (#V-22885, Molecular Probes) staining solution diluted 1:200 for 20 min at 37 °C. Either 150′000 or 250′000 cells in 150 μl of SFM were transferred to the top of transwell for migration or invasion experiments, respectively. Complete medium was used to cover the bottom of the transwells. Cell migration or invasion was detected analyzing bottom and top fluorescence using the Infinite M1000 microplate reader (Tecan, Männedorf, Switzerland) (excitation wavelength 535 nm, emission 575 nm). Percentage of migrating or invading cells was calculated for each time point as the bottom emission minus the background measured at time zero, over the top fluorescence at time zero.
2.6. Matrigel cell evasion assay
Matrigel was diluted with SFM at a final concentration of 6 μg/ul. 2,500 cells were included in 10 μl matrigels drops, plating 4 drops in 6 well plates. Drops were let solidify by rotating the plate upside down at 37 °C, under 5% CO2, in a humidified incubator. The plate was then rotated again and 2 ml of complete medium was added per well. At time 0 pictures were taken to check cell density. After 7 days the experiment was stopped by fixing and staining cells with 20% methanol-crystal violet solution.
2.7. Luciferase assay
To confirm miR-23b direct interaction with BTBD7–3′UTR predicted target site, we performed luciferase assay as described in [53]. We cloned BTBD7–3′UTR fragments (657 nt) carrying the predicted miR-23b target site inside the pGL3-control vector at the XbaI site. The mutant pGL3-BTBD7–3′UTR construct was deleted for the miR-23b 8 seed sequence using the QuickChange Site-Directed mutagenesis kit (#200519, Agilent) as per kit instructions. HCT116 cells were seeded in 24-well plates (150 × 103 cells per well) the day before transfection. In each well, 0.4 μg of pGL3 (luciferase) construct together with 80 ng of pRLTK (renilla) vector (both from Promega) were cotransfected with 100 nM of precursor miR-23b or negative control. Twenty-four hours later, cells were lysed in 100 μl of passive lysis buffer according to the Dual-Luciferase Reporter Assay protocol (#E1910, Promega); 10 μl of the lysate was used for the luciferase activity measurements with Infinite M1000 microplate reader (Tecan). The relative reporter activity was obtained by normalization to the renilla activity. Luciferase assays were done in three technical replicates and at least three times.
2.8. Nucleic acid purification and polymerase chain reactions
RNA was extracted using the Isol-RNA Lysis Reagent (#2302700, 5 Prime, Hamburg, Germany) according to protocol instructions. Extraction was followed by PCA (Phenol: Chloroform: Isoamyl Alcohol 5:24:1, #P2069, Sigma Aldrich), procedure that reduces salt contamination, and by DNase digestion (#2238G Turbo-Dnase, Ambion, Thermo Fisher Scientific). RNA quality was assessed using agarose gel electrophoresis after RNA incubation at 70 °C for 5 min. cDNA was synthesized from 1 μg of RNA using the AMV Reverse Transcriptase (#M5108, Promega) with random primers (#C1181, Promega). cDNA was diluted 10 times and used for qRT-PCR using iQ SYBR Green Supermix (#1708880, Biorad) with the appropriate primers (Supplementary Table 4).
For miR-23b-3p quantification total RNA was first retrotranscribed with TaqMan® MiRNA Reverse Transcription Kit (#4366597 Applied Biosystems, Thermo Fisher Scientific) and then qRT-PCR was carried out using TaqMan™ Universal Master Mix II, no UNG (#4440040, Applied Biosystems) with TaqMan™ MicroRNA assays (ID 000400, for miR-23b-3p and ID 001973 for U6snRNA, # 4427975, Applied Biosystems).
Genomic DNA was extracted using Gentra Puregene Cell Kit (Qiagen Sciences, Maryland, USA), according to manufacturer's instructions. DNA quality was assessed using agarose gel electrophoresis. RNA and DNA concentration (absorbance at 260 nm) and purity (ratio between absorbance at 260 nm and absorbance at 280 nm) were determined by using a spectrophotometer (Nanodrop, Thermo Fisher Scientific).
qRT-PCR reactions were carried out either in 96-well optical reaction plates using Two-Colour Real-Time PCR Detection System MyiQ2 (Biorad) or in 384-well optical reaction plates using Applied Biosystems 7900HT Fast Real-Time PCR System (Thermo Fisher Scientific), according to manufacturer's protocol. The 2 − ΔΔCt method was used to calculate gene relative abundance, using U6snRNA, or GAPDH expression as housekeeping controls.
2.9. Linker mediated PCR
To identify the genomic position of TN insertion in HCT116 transposed cells, we performed the linker mediated PCR (LM-PCR) according to [35]. In brief, genomic DNA from transposed cells was digested at 37 °C with BfaI (for cloning off the left IRDR) or NlaIII (for cloning off the right IRDR). Linkers were ligated to NlaIII- (right-side) or BfaI- (left side) digested genomic DNA using T4 DNA ligase. A secondary digestion (XhoI, right side; BamHI, left side) was performed to destroy concatemer-generated products. Primary and secondary PCRs were performed using primers specific for linker and SB transposon sequences. PCR products were run and separated on a 1% agarose gel. To purify digested DNA and PCR products QIAquick PCR purification kit from Qiagen was used following kit directions. Final PCR products were ligated in pCR.2.1 Topo TA vector (Thermo Scientific) and transformed into DH5α competent cells. Two separate PCRs and nested PCRs were performed to obtain the left and the right boundaries of pT2-CMV-EGFP insertions in the genome.
DNA sequencing of minipreps (Promega) from topo TA clones containing LM-PCR products was done by direct Sanger sequencing, using BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Fostercity, CA, USA). Primers were designed using Primer 3 Plus [54] and the Agilent QuickChange Primer design tool; linker sequences were obtained from [35]. All primers and linkers were purchased from Sigma-Aldrich and are listed in Supplemental Table 3.
2.10. Western blotting
To obtain protein extracts, cells were incubated for 20 min on ice in NP40 lysis buffer (0.5% NP40, 50 mM HEPES, 250 mM NaCl, 5 mM EDTA, 0.5 mM EGTA, 1 mM DTT, containing Protease Inhibitors (#04693116001, Complete, Roche, Mannheim, Germany) and Phosphatase Inhibitor Cocktail 1 (#P5726, Sigma Aldrich), vortexing every 5 min. Cell lysates were clarified by centrifugation at maximum speed for 20 min. Protein concentration was quantified using the Protein Assay Dye Reagent Concentrate (#500–0006, Biorad, Hercules, CA, USA). For immunoblotting analysis, approximately 40 μg of proteins were separated using 4–20% SDS-PAGE Criterion-TGX-stain free Precast Gels (#5678094, Biorad) and transferred to Nitrocellulose membranes (#10600003, Biorad). Membranes were blocked with 5% non-fat dried milk in TBS-0.1% Tween20 (#P7949, Sigma), and incubated overnight at 4 °C with the following primary antibodies: goat-anti-Btbd7 1:500 (EB10630, Everest Biotech); mouse-anti-e-Cadherin (610181, BD Transduction Laboratories) 1:1000; mouse-anti-Twist (sc-81417, Santa Cruz) 1:200; and goat-anti-Vinculin (sc-7649, Santa Cruz) 1:1000. Incubation with secondary antibodies, ECL IgG-HRP-linked anti-rabbit, anti-mouse or anti-goat (GE Healthcare) was performed at room temperature for 1 h at a dilution of 1:3000–1:5000.
2.11. Flow cytometry
Human epithelial adhesion molecule (EpCAM) was used as a marker of cellular differentiation and was measured by FACS using APC Mouse Anti-Human EpCAM (#347200, BD Biosciences). To this aim HCT116 cells subjected to increasing rounds of fSCS were detached from culture dishes using 2 mM EDTA in PBS, pelleted and suspended in PBS containing 10% FBS at a final concentration 10 × 106 cells/ml. Following 30 min blocking on ice, 100 μl of cell suspension was stained on ice with 5 μl of antibody for 30 min and washed with ice cold PBS. For cell death detection immediately following fSCS, dead cells were stained with propidium iodide (0.4 mg/ml). FACS analysis was performed using the 633 nm excitation laser of the BD FACSCantoTM flow cytometer system.
2.12. In vivo experiments
Animal experiments were reviewed and approved by the CRO-Aviano Institutional Animal Care and Use Committee and were conducted according to the Committee's and the Italian Ministry of Health guidelines. For extravasation assay we used 4 to 6-weeks-old athymic nude mice (Charles River, UK Foxn1nu, females), following the protocol described in [30]. In particular, HCT116 cells (T0, T2, TN4_sorted, TN4_20, pEGFP-Empty or pEGFP-Btbd7) were stained with DiI (#D282, Molecular Probes) 1:200 for 30 min at 37 °C in serum free medium. Then, 1.5 × 106 cells were suspended in 200 μl of PBS containing 0.1% BSA, 1 mM EDTA, and 0.1% fluorescein and retro-orbitally inoculated in previously anesthetized nude mice to reach blood circulation. Fluorescein was used to detect the homogeneity of injection in all mice using the in vivo imaging system (Ivis Lumina, Xenogen) after subtraction of the background signal at time 0. Fluorescein signal was not detectable any longer 72 h later, when mice were humanly culled by cervical dislocation for analyses. During mice necroscopy lungs were inflated through the trachea with 1 ml of ice cold 4% PFA. Lungs were immediately observed at the stereomicroscope (Leica M205FA) and pictures of whole lungs from both top and bottom were taken. Cryostat-cut 10 μm sections were then obtained from OTC lung inclusions. To detect the presence of DiI stained HCT116 cells infiltrated into the lungs of mice following in vivo extravasation assay, lung sections from OCT inclusions were fixed with PFA 4% and nuclei of mice lungs were counterstained with TOPRO (#T3605, Molecular Probes). Stained tissue specimens were observed using a confocal laser-scanning module (TSP2 Leica) linked to a Leica DMIRE 2 fluorescence microscope. Quantification of cell infiltrates was calculated, using Image J or Volocity Software (Perkin Elmer), as the ratio between the integrated density of red signal (DiI stained cells) over the integrated density of blue signal (total nuclei of mice lungs) in 8-bit thresholded images analyzing at least two different lung sections per mice. For the analyses of HCT116 cell lung infiltrates at the stereomicroscope, 8 ROI (region of interest) per mice were analyzed to calculate the integrated density of the red signal (2 from the top and 2 from the bottom of left and right lung).
Orthotopic mice injections: 6 to 8-weeks-old severe combined immunodeficiency (SCID) mice (Charles River, UK) were anesthetized prior to midline laparotomy and exteriorization of the caecum. A 1:1 suspension of cells and Matrigel was injected submucosally into the caecal wall under magnified vision, raising a bleb on the caecum. For each animal, 0.5 × 106 cells (TN4-sorted or TN4_20) were implanted orthotopically, with the entire experiment conducted in duplicate. Primary tumors grew in all animals. When showing signs of disease or > 10% weight loss, mice were humanely culled, and colon, liver and lungs were harvested.
2.13. Histopathology and Immunohistochemistry
Formalin-fixed organs from orthotopic injection experiments were embedded in paraffin; tissue sections were stained by haematoxylin-eosin and were evaluated by an expert pathologist from CRO-Aviano's Pathology Division. Samples from normal colon mucosa (n = 10), colorectal cancer primary tumors (n = 11 without metastasis at diagnosis - M0; n = 10 with metastasis at diagnosis - M1) and CRC liver metastasis were obtained from CRO-Aviano, National Cancer Institute (Italy) according to [55], after approval by CRO Aviano's Internal Research and Ethics Committee. All samples were obtained, with patient's informed consent, at the time of surgery and snap frozen prior to being deposited at CRO-Aviano's Biobank. Patient's clinical information was registered and follow-up data was recorded at checkup.
CRO Aviano's Pathology Division evaluated tumor content in each sample using Haematoxylin and Eosin staining of sample slides. Only CRC samples with a tumor percentage >60% were used for this study. Approximately ten cryostat-cut tissue slices of 10 μm each were used for RNA extraction by Trizol.
2.14. TCGA microRNA and gene expression analysis of CRC samples
The analysis was performed on data from 16 CRC primary samples with low microsatellite instability (L-MSI). Specifically, a group of 8 CRC patients with metastasis at the time of diagnosis (M1) was compared with a group of 8 CRC patients without metastasis at the time of diagnosis (M0). CRC samples from both groups were paired for patients' pT and pN status at diagnosis and for age and gender (Suppl. Fig. S3A). MicroRNA expression profiling was conducted using TCGA row data (level 1) provided by GDC Data Portal (portal.gdc.cancer.gov) upon authorized approval. Downloaded BAM files were converted in FASTQ and mapped using Bowtie on a miRNA library obtained from [56] not allowing any mismatch. Statistical analysis was performed in R using DESeq2 package and differences were considered significant at p < .05.
Gene expression analysis was conducted using TCGA data (level 3) downloaded from GDC Data Portal (portal.gdc.cancer.gov). Data normalization was performed in R calculating the reads per millions (RPM) of all the mapped reads in each sample.
2.15. Statistical analysis
Graphs presented in figures and statistical analyses were obtained using GraphPad Prism v6.0d. Comparisons between two groups of data were examined using unpaired two-tailed Student t-test when assuming equal variances or using unpaired two-tailed Mann-Whitney test for unequal variances. When comparing multiple groups with unequal variances Kruskal-Wallis non-parametric test was applied. Differences were considered significant at p < .05.
3. Results
3.1. Preventing cell-matrix and cell-cell contacts in vitro selects tumor cells with pro-metastatic traits
In our effort to study the molecular mechanisms governing CRC metastasis, we tested the ability of several CRC cell lines to survive during loss of cell-matrix contacts (i.e. anoikis resistance assay - AR), as previously shown in [18,19]. We seeded both CRC and normal intestine cell lines (i.e. HCT116, HT29, SW480, SW620 and RIE, respectively) on ultralow attachment plates in serum-free medium for 24 h. Next, we collected cells and re-plated them on adherence-proficient tissue culture plates in 10% serum medium (standard conditions). We let cells grow until they formed visible colonies; finally, we fixed and stained colonies with crystal violet. As a result, we observed that tumor and normal cells survived similarly to AR assay (Fig. 1a, Suppl. Fig. S1a), and we concluded that AR assay was not stringent enough to investigate pro-metastatic ability of our CRC cell models.
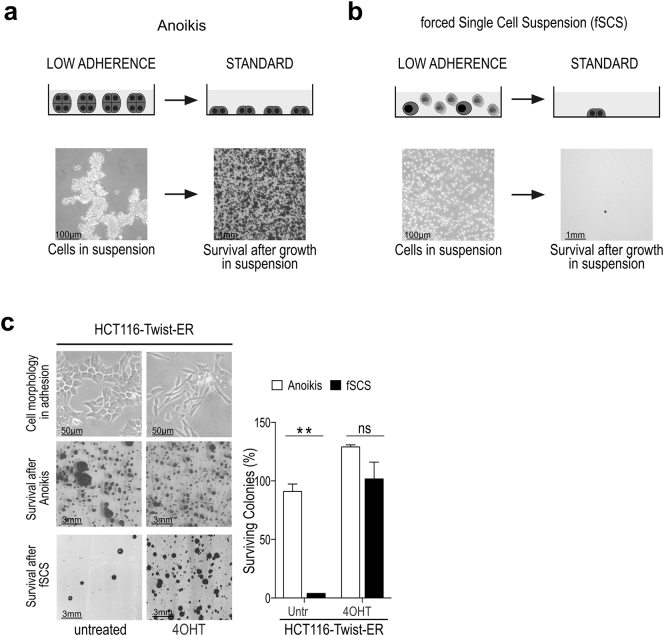
Fig. 1.
Forcing tumor cells to a single cell suspension (fSCS) reduces cell survival compared to anoikis. (a) Left panels: scheme and representative image at 10× magnification of HCT116 cells during culture in low adherence plates with serum free medium (anoikis). Right panels: scheme and representative image of fixed and stained colonies formed in standard plates by surviving HCT116 cells after 24 h in anoikis. (b) Left panels: scheme and representative image at 10× magnification of HCT116 cells kept on low adherence plates in serum free medium with the addition of 1 mM EDTA (forced single cell suspension = fSCS). Right panels: scheme and representative image of fixed and stained colonies formed in standard plates by surviving HCT116 cells after 24 h in fSCS. (c) Upper panels: cell morphology at 10× magnification of HCT116 cells stably overexpressing TWIST-ER construct either untreated (untr) or treated with 4-hydroxytamoxifen (4OHT). Middle and lower panels show representative images of fixed and stained colonies formed by surviving HCT116-TWIST-ER cells either untreated (untr) or treated with 4OHT following anoikis and fSCS, respectively. Bars in right graph represent mean +/− sem, expressed as percentage compared to anoikis, of surviving colonies of one representative experiment performed in triplicate (ns, non-significant; **p < .005 using unpaired t-test assuming equal variances).
In agreement with [20,21], we observed that during AR both cancer and normal intestine cells strengthened cell-cell contacts and formed cell aggregates (Fig. 1a and Suppl. Fig. S1c). Yet, it is acknowledged that to metastasize epithelial tumor cells lose epithelial traits (e.g. cell-cell adhesion molecules and proliferation), gain mesenchymal traits (e.g. reshape cytoskeleton and motility) [22] and ultimately survive to loss of cell-cell and cell-matrix contacts [[23], [24], [25]] by activating the genetic program named epithelial to mesenchymal transition (EMT) [2,22,26,27]. Therefore, we hypothesized that by simultaneously preventing cell-matrix and cell-cell contacts, we could exert a stronger selective pressure than AR assay and ultimately select cells with mesenchymal traits.
To simultaneously prevent cell-matrix and cell-cell contacts, we seeded cells on ultralow attachment plates in serum-free medium with the addition of EDTA, a Ca2+ chelator, thus disrupting calcium-dependent cell-cell contacts (Fig. 1b). We named this assay forced single cell suspension (fSCS). We used 1 mM EDTA, because this was the lowest concentration that significantly reduced cell survival (Suppl. Fig. S1b). After 24 h of fSCS, we collected cells, seeded them in standard conditions and let them grow until they formed visible colonies; eventually, we fixed and stained colonies with crystal violet. As expected, the number of surviving colonies after fSCS assay was much less than after AR assay (Fig. 1a-b and Suppl. Fig. S1a-b). To exclude that impaired survival during fSCS assay was due to Ca2+ deprivation rather than loss of cell-cell contacts, we used both enzymatic (i.e. hyaluronidase treatment) [21] and physical strategies (i.e. cell inclusions into water-in-oil microdroplets) to force cells into a single-cell suspension in the presence of Ca2+. Similarly to fSCS, using these strategies, we observed a decrease in the number of surviving colonies (Suppl. Fig. S1c).
To challenge our hypothesis that tumor cells with mesenchymal traits survive more than epithelial tumor cells during fSCS assay [28], we first tested both CRC (SW480, COLO205) and breast cancer (MCF7, MDAMB231) cell lines with epithelial (SW480, MCF7) or mesenchymal (COLO205, MDAMB231) traits. We observed that all these cell lines survived similarly to AR assay; whereas, mesenchymal cells (COLO205, MDAMB231) survived to fSCS assay more than epithelial cells (SW480, MCF7) (Suppl. Fig. S2a-b; p < .05, using paired t-test with unequal variances). Secondly, we tested an EMT CRC cell model by overexpressing inducible forms of TWIST and SNAIL, two known EMT transcription factors [29], in CRC cell lines (HCT116 and HT29). Upon treatment with 4-hydroxy-tamoxifen (4OHT), which activates Twist-ER and Snail-ER proteins (Suppl. Fig. S2c), cells lost cell-cell contacts, became scattered and acquired a spindled morphology (Fig. 1c, Suppl. Fig. S2e-f), and gene expression and migration ability changed accordingly (Suppl. Fig. S2d). When we tested HCT116 and HT29 overexpressing Twist-ER or Snail-ER in fSCS assay, we observed that 4OHT-treated cells (i.e. mesenchymal cells) survived more than untreated cells (Fig. 1c and Suppl. Fig. S2e-f; p < .005 for HCT116 and p < 0.5 for HT29 models, using unpaired t-test assuming equal variances).
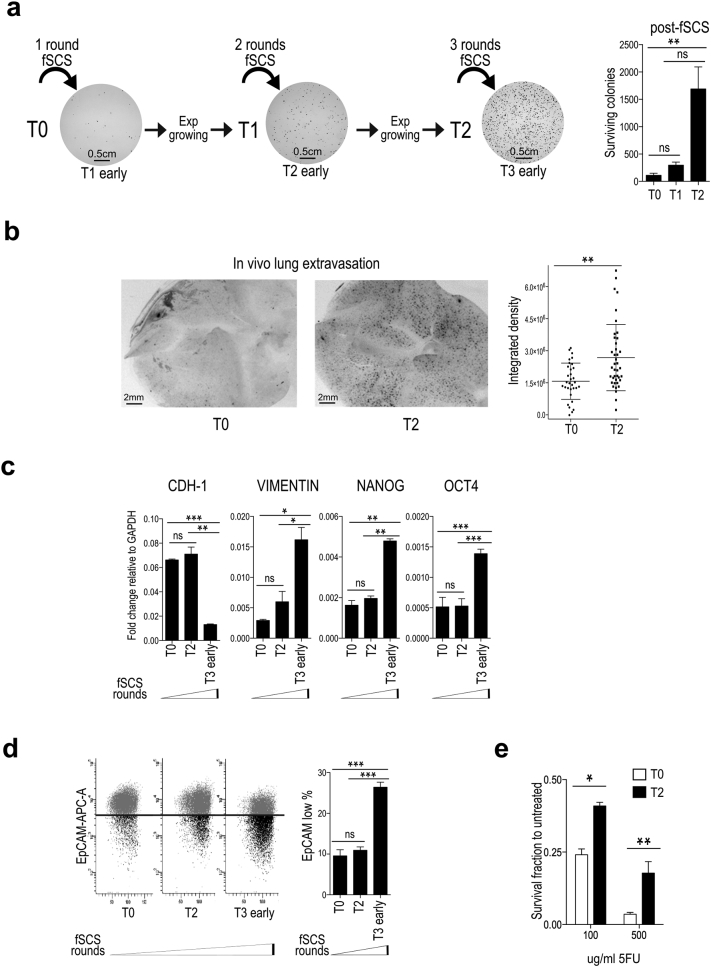
At last, we hypothesized that in the context of cell lines that survive poorly to fSCS, those few surviving colonies are more mesenchymal compared with the original bulk of parental cells. To test this hypothesis, we subjected HCT116 parental cells (T0) to fSCS, we collected and seeded them in standard conditions till the surviving cells replenished the culture plate, and eventually we cultured them in exponentially growing condition (T1 progeny). We repeated this protocol with T1 progeny to obtain T2 progeny and so on for several rounds (see scheme in Fig. 2a). By these means, we observed that T2 and higher progenies survived more to fSCS and that surviving colonies had a more scattered morphology compared to T1 and T0 parental cells (Fig. 2a and Suppl. Fig. S3a-b; p < .005 using paired t-test assuming unequal variances). To assess whether fSCS resistant cells gained survival in blood circulation and ability to extravasate from the bloodstream, we performed an in vivo extravasation assay in mice, as described in [30]. When intravenously injected in mice, T2 cells showed greater ability to colonize lungs compared to T0 cells (Fig. 2b and Suppl. Fig. S3c; p < .005, using non-parametric Mann-Whitney test). Despite T2 progeny showed increased ability to survive during fSCS assay and to extravasate from the blood stream, exponentially growing T2 cells were morphologically indistinguishable from exponentially growing T0 cells (data not shown). Nevertheless, when we measured the expression of mesenchymal markers in exponentially growing T0, T2, and in surviving colonies after T2 was challenged with fSCS (early T3), we observed that early T3 cells showed molecular mesenchymal traits: decreased CDH1, increased VIMENTIN, OCT4 and NANOG mRNA expression levels (Fig. 2c; ns not significant between T0 and T1, all other comparisons p < .05, using paired t-test assuming equal variances) and reduced EpCAM protein levels (Fig. 2d). Because mesenchymal traits are associated with cancer chemoresistance [31], we also tested the sensitivity of T0 and T2 cells to 5-fluorouracil (5FU), which is commonly used in CRC patients, and we observed that T2 progeny was more chemo-resistant than T0 parental cells (Fig. 2e; p < .05 or p < .005, with 100 or 500 μg/ml of 5FU respectively, using paired t-test assuming unequal variances).
Fig. 2.
Multiple rounds of forced single cell suspension assay (fSCS) enrich for cells showing increased resistance to fSCS, in vivo extravasation ability and chemo-resistance. (a) Left: scheme of the experimental procedure to obtain fSCS-resistant cells and representative images of surviving colonies. Right: bars in graph represent mean +/− std. of the number of colonies generated by T0, T1 and T2 cells after fSCS (ns, non-significant; **p < .005, using paired t-test assuming unequal variances). (b) In vivo extravasation assay. Left: representative stereomicroscope images (1.5× magnification) of nude mice whole lungs at 72 h after intravenous injection of DiI labeled T0 and T2 cells, (black spots = DiI positive infiltrated cells). Right: dot Plot indicates mean +/− std. of the signal from DiI stained infiltrated cells per field in lungs of 5 mice per condition, quantified as described in Materials and Methods (**p < .005, using non-parametric Mann-Whitney test). (c) qRT-analysis of CDH1, VIMENTIN, NANOG, OCT4 expression in HCT116 cells subjected to multiple rounds of fSCS. Bars indicate mean +/− sem of two independent biological replicates (ns, non-significant; *p < .05, **p < .005; ***p < .0005, using paired t-test assuming equal variances). (d) Analysis of EpCAM cell surface expression by FACS in HCT116 cells subjected to multiple rounds of fSCS; left: representative scatter plot images; right: graph with bars indicating mean +/− sem of the fraction of EpCAM low population from three independent biological replicates (ns, non-significant; ***p < .0005 using paired t-test assuming equal variances). (e) Cell viability of HCT116 T0 and T2 after 16 h treatment with the indicated doses of 5-Fluorouracil (5FU) assessed by MTT 48 h after treatment stopped. Bars represent the mean +/− std. of two independent experiments (*p < .05; **p < .005, using paired t-test assuming unequal variances).
Altogether, these data show that fSCS is a stringent in vitro assay that selects cells with mesenchymal traits and pro-metastatic potential in vivo.
3.2. A transposon based genetic screen generates fSCS-resistant cells with EMT traits
To disclose novel regulators of CRC metastasis, we hypothesized that by combining the ability of fSCS to select CRC cells with pro-metastatic potential together with the ability of forward genetic screens to generate random mutations, we could be able to retrieve fSCS-resistant cells that carry genetic mutations in regulators of CRC metastasis. In particular, we were keen to retrieve mutagenic insertions located in the non-coding part of the human genome; since the non-coding part of the human genome outnumbers the coding part by a rough factor of 99 to 1, the sleeping beauty DNA transposon elements (SB) seemed the ideal tool. Indeed, SB integrates at any TA dinucleotides within the host genome without gross biases for any chromosome or chromosomal regions [17]. The SB mutagenic system that we used is composed by two plasmids, one carrying the DNA SB transposon vector (pT2BH) and the other carrying the SB100 transposase enzyme (pSB100) necessary for SB-DNA integration [32]. The pT2BH vector is devoid of any protein coding gene-trapping elements (e.g. 5′-gene trap cassettes splicing acceptors, donors, early terminators, poly-A signals) [33], thus has no preferences for coding regions compared to non-coding regions and it acts solely by insertional mutagenesis. We modified pT2BH to carry a fluorescent reporter (pT2-CMV-EGFP), in order to identify transposed cells (Suppl. Fig. S4a). We performed the SB mutagenic screen in HCT116 CRC cell line, because this cell line was already successfully used in genetic screens exploring CRC metastasis [34], and it has a diploid number of chromosomes thus offering greater chances to develop a phenotype following SB insertions. We co-transfected pT2-CMV-EGFP with pSB100 (Suppl. Fig. S4a-b); fourteen days after transfection, we sorted by FACS transposed EGFP-expressing HCT116 cells (TN4_sorted) (Suppl. Fig. S4b-c). Next, we challenged TN4_sorted cells to fSCS assay and seeded them in standard conditions until single cell clones originated (Suppl. Fig. S4b). Approximately, one hundred clones were collected and, to select the fSCS-resistant ones, they were re-challenged to fSCS assay. Only 4 transposed clones displayed overt fSCS resistance, and among these, TN4_20 was the most resistant one (Suppl. Fig. S4d and Fig. 3a).
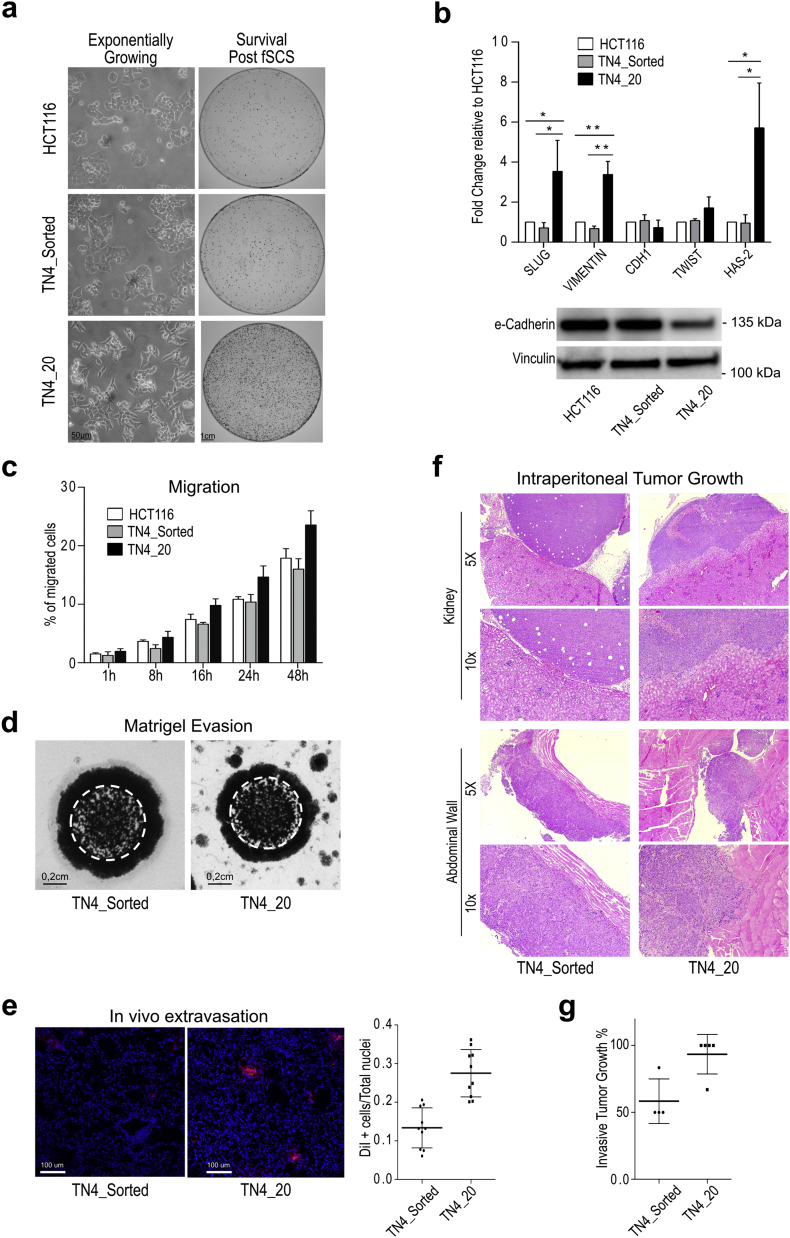
Fig. 3.
The combination of fSCS assay with a TN-based genetic screen generated TN4_20 clone, which is resistant to fSCS and shows EMT and pro-invasive features. (a) Left panels: representative images at 10× magnification showing the morphology of exponentially growing HCT116, TN4_Sorted and TN4_20 cells. Right panels: representative images of fixed and stained colonies of HCT116, TN4_Sorted and TN4_20 post-fSCS (i.e. Survival post fSCS) in 10 cm plates. (b) Upper panel: qRT-analysis in HCT116, TN4_Sorted and TN4_20 cells. Bars indicate mean +/− std. of three independent biological replicates (*p < .05; **p < .005, using paired t-test assuming equal variances). Lower panel: Western Blot analysis of e-Cadherin expression in HCT116, TN4_Sorted and TN4_20 cells. (c) Transwell migration assay: quantification of HCT116, TN4_Sorted and TN4_20 cells motility. Bars indicate mean +/− std. of 2 independent biological replicates. (d) Matrigel evasion assay. Representative images of fixed and stained TN4_Sorted and TN4_20 cells evaded from matrigel drops in which cells were included two weeks earlier. White dashed line delimits the size of the matrigel drop. (e) In vivo extravasation assay. Left: representative confocal images (10× magnification) of nude mice lung sections at 72 h after intravenous injection of DiI labeled TN4_Sorted and TN4_20 cells (Red = DiI stained infiltrated cells; Blue = lung mice nuclei counterstained with Topro). Right: dot plot indicates mean +/− std. of the fraction of infiltrated DiI stained cells in lungs of 5 mice per condition, quantified as described in Material and Methods (***p < .0005, using unpaired t-test with unequal variances). (f) Representative images of H&E staining (5× and 10× magnification) of sections from kidney and abdominal wall lesions found in mice intra-peritoneally injected with TN4_Sorted and TN4_20 cells. (g) Dot plot describes % of intra-peritoneal tumor lesions with invasive behavior over total lesions found in each inoculated mouse. Bars indicate mean +/− std. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Interestingly, we observed that TN4_20 cells displayed elongated shape and grew in a scattered fashion instead of being compact and cuboidal as HCT116 and TN4_sorted cells (Fig. 3a). Consistently, TN4_20 cells expressed higher mRNA levels of mesenchymal markers (SLUG, VIMENTIN and HAS2) and decreased e-Cadherin protein levels compared to TN4_sorted or parental HCT116 cells (Fig. 3b). TN4_20 showed greater in vitro invading ability compared with TN4_sorted cells both in transwell motility (Fig. 3c and Suppl. Fig. S5a) and in matrigel evading (Fig. 3d) assays. In vivo experiments paralleled in vitro evidences: intra-caecally TN4_20 injected mice developed distant lung metastasis, which was not the case for TN4_sorted cells (Suppl. Fig. S5b). Similarly, TN4_20 extravasated more efficiently from the bloodstream (Fig. 3e; p < .0005 using unpaired t-test with unequal variances) and showed greater invasive tumor growth when injected intra-peritoneally compared with TN4_sorted cells (Fig. 3f-g). In fact, almost all tumor nodules formed by TN4_20 cells had infiltrative margins, compared to nodules formed by TN4_sorted cells, which instead showed prevalently expansive growth (Fig. 3f-g).
All together, these results indicate that fSCS selection coupled with the SB mutagenic tool retrieved the TN4_20 cell clone that showed mesenchymal traits and greater in vitro and in vivo pro-metastatic potential.
3.3. miR-23b targets BTBD7 gene and impairs fSCS survival
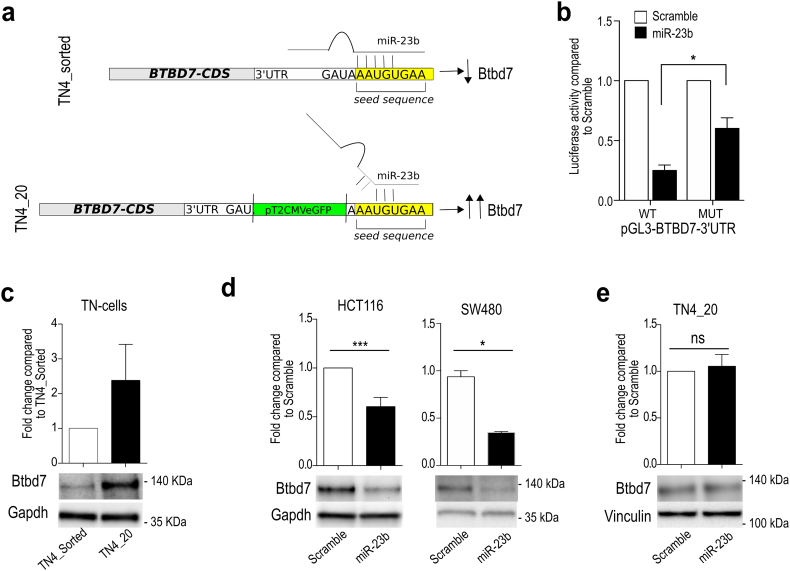
To retrieve the genomic positions of pT2-CMV-EGFP insertions in TN4_20 clone, we used the linker mediated PCR [35] (Suppl. Fig. S6a) and found that TN4_20 cells carried seven pT2-CMV-EGFP insertions, all inside non-protein-coding regions (introns, promoter region and 3′UTR) (Suppl. Table 1). We decided to focus on the insertion located within the 3′UTR of BTB/POZ containing domain protein 7 (BTBD7) for the following reasons: first, BTBD7 regulates EMT during salivary gland branching [36] and during tumor invasiveness and metastasis [37,38]; second, the SB insertion in the 3′UTR of BTBD7 is located within the predicted target site of miR-23b, a known anti-metastatic miRNA [34,39] (Suppl. Fig. S6b-c); ultimately, no published data described miR-23b::BTBD7 interaction.
We hypothesized that TN4_20 clone was able to survive to fSCS because SB insertion within BTBD7–3′UTR disrupted the predicted direct interaction between miR-23b and BTBD7, leading to BTBD7 deregulation (Fig. 4a). In agreement with this hypothesis, luciferase assay confirmed direct interaction between miR-23b and BTBD7 3′UTR (Fig. 4b; p < .05 using paired t-test assuming equal variances) and Btbd7 protein expression levels were higher in TN4_20 cells compared to TN4_sorted cells (Fig. 4c); yet, BTBD7 mRNA levels were not affected (Suppl. Fig. S7a). Consistently, transfection of synthetic miR-23b precursor decreased Btbd7 protein levels (Fig. 4d) without affecting BTBD7 mRNA levels in HCT116 and SW480 cells (Suppl. Fig. S7b-c). On the contrary, overexpression of miR-23b precursor did not affect Btbd7 protein expression levels in TN4_20 cells (Fig. 4e).
Fig. 4.
TN4_20 cell clone contains a SB insertion in the 3′UTR of BTBD7 gene that disrupts the interaction between BTBD7 and miR-23b. (a) Schematic model of BTBD7::miR-23b interaction. Upper panel: in TN4-sorted cells, miR-23b targets the 3′UTR of BTBD7 gene and down-regulates Btbd7 protein expression. In TN4_20 cell clone, the presence of TN insertion in the 3′UTR of BTBD7 gene close to miR-23b seed sequence impairs BTBD7::miR-23b interaction and increases Btbd7 protein levels. Gray rectangle = BTBD7 cds; White rectangle = 3′UTR; Yellow rectangle = miR-23b seed sequence; Green rectangle = pT2 CMV EGFP TN. (b) Luciferase assay in HCT116 transfected with pGL3-BTBD7–3′UTR WT or pGL3-BTBD7–3′UTR MUT (deleted for miR-23b seed sequence) and miR-23b precursor or scramble control. Bars indicate mean +/− std. of 3 independent biological replicates (*p < .05 using paired t-test assuming equal variances). (c) Western Blot analysis of Btbd7 expression in TN4_Sorted and TN4_20 cells and relative quantifications. Bars indicate mean +/− std. of two independent biological replicates. d) Western Blot analysis of Btbd7 expression after overexpression of miR-23b precursor in HCT116 (left) and SW480 (right) and relative quantifications. Bars indicate mean +/− std. of four independent biological replicates for HCT116 and two technical replicates for SW480. (e) Western Blot analysis of Btbd7 expression after overexpression of miR-23b precursor in TN4–20 and relative quantifications. Bars indicate mean +/− std. of two independent biological replicates. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
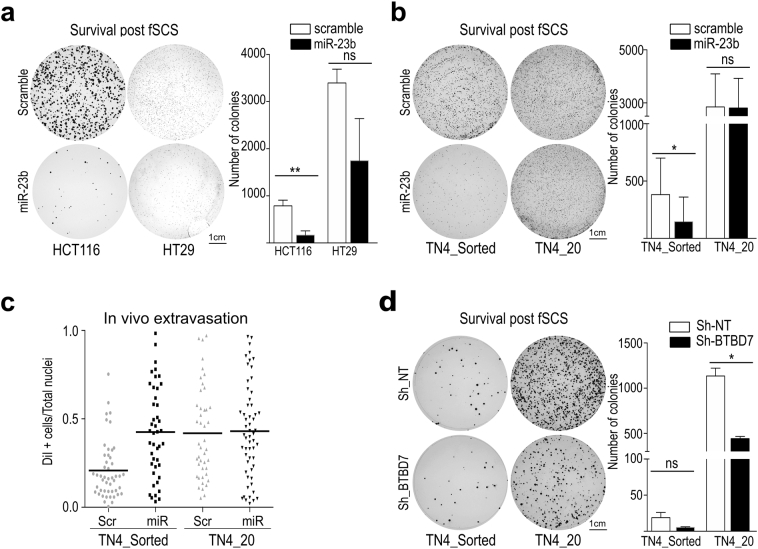
To investigate whether BTBD7::miR-23b interaction affects survival during fSCS, we transfected miR-23b precursor in HCT116 parental, TN4_sorted, TN4_20, and in other CRC cell lines (HT29, SW480 and SW620) and tested them with the fSCS assay. We observed that overexpression of miR-23b prevented fSCS survival in all these cell lines except for TN4_20 cells (Fig. 5a-b and Suppl. Fig. S8a-b; p < .005, using paired t-test with unequal variances). Similarly, miR-23b transfection was unable to affect in vivo extravasation ability of TN4_20 cells compared with TN4_sorted (Fig. 5c).
Fig. 5.
miR-23b overexpression and Btbd7 silencing both impair fSCS resistance in CRC cell lines and regulate in vivo cell invasiveness. (a) Left panel: representative images of fixed and stained post-fSCS colonies formed in 10 cm plates by HCT116 and HT29 after transfection of miR-23b precursor or scramble. Right panel: bars in the graph represent mean +/− std. of the number of colonies generated by scramble and miR23b overexpressing cells (ns, non-significant; **p < .005, using paired t-test with unequal variances) (b) Left panel: representative images of fixed and stained post-fSCS colonies formed in 10 cm plates by TN4_Sorted and TN4_20 cells after transfection of miR-23b precursor or scramble. Right panel: bars in the graph represent mean +/− std. of the number of colonies generated by TN4_Sorted and TN4_20 cells. (ns, non-significant; *p < .05, using paired t-test with unequal variances) (c) In vivo extravasation assay. Mice were intravenously injected with DiI labeled TN4_Sorted and TN4_20 cells overexpressing miR-23b precursor or scramble. After 72 h, infiltrated cells in the lung parenchyma were quantified as described in Material and Methods. Dot plot indicates mean +/− std. of the fraction of infiltrated cells over total nuclei in lungs of 3 mice per condition. (d) Left panel: representative images of fixed and stained post-fSCS colonies formed in 10 cm plates by TN4_Sorted and TN4_20 cells after silencing of BTBD7 with Sh-BTBD7. Right panel: bars in the graph represent mean +/− std. of the number of colonies generated by TN4_Sorted and TN4_20 cells (ns, non-significant; *p < .05, using paired t-test assuming unequal variances).
To investigate whether TN4–20 fSCS resistance was dependent on Btbd7 expression levels, we silenced BTBD7 by short hairpin RNAs (shRNA) (Suppl. Fig. S8c) and observed that BTBD7 silencing impaired fSCS resistance in both TN4_sorted and TN4_20 cells (Fig. 5d; p < .05, using paired t-test assuming unequal variances).
To sum up, these data suggest that miR-23b targets BTBD7, that the interaction between miR-23b and BTBD7 reduces fSCS survival, and that TN4_20 resistance to fSCS relies on the impaired miR23b::BTBD7 interaction due to SB insertion.
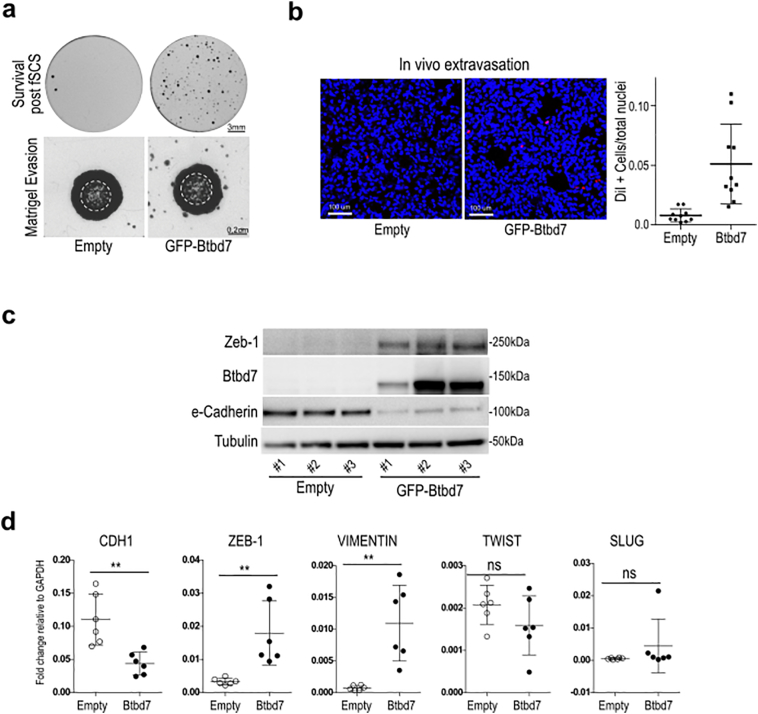
3.4. Btbd7 protein overexpression confers EMT traits and pro-metastatic potential
To further assess the importance of Btbd7 in fSCS resistance and in pro-metastatic phenotype, we generated HCT116 clones stably overexpressing ectopic pEGFP-Btbd7 (Suppl. Fig.S8d). In agreement with [36], we observed that pEGFP-Btbd7 showed a diffuse cytoplasmic localization and was absent from the nucleus (Suppl. Fig.S8d). Btbd7 overexpressing cells displayed greater ability in fSCS, matrigel evasion, and in vivo extravasation assays compared with control cells (Fig. 6a-b; p < .005, using unpaired t-test with unequal variances). To start shedding light on Btbd7 molecular function, we investigated whether EMT genes changed after Btbd7 overexpression and we found e-Cadherin downregulation, Vimentin and Zeb1 upregulation both at the protein and mRNA level (Fig. 6c-d; p < .005, using paired t-test assuming equal variances); whereas, TWIST and SLUG did not change (Fig. 6d).
Fig. 6.
Overexpression of Btbd7 protein confers resistance to fSCS and EMT/pro-invasive traits. (a) Upper panel: representative images of fixed and stained post-fSCS colonies formed in 6-well plates by pEGFP-Btbd7 clone #3 or Empty clone #1 (= Survival post fSCS). Lower panel: matrigel evasion assay. Representative images of fixed and stained pEGFP-Btbd7 clone #3 or pEGFP-Empty clone #1 evaded from matrigel drops in which they were included two weeks earlier. White dashed line delimits size of matrigel drop. (b) In vivo extravasation assay. Left: representative confocal images (10× magnification) of lung sections from nude mice at 72 h after intravenous injection of DiI labeled pEGFP-Btbd7 and pEGFP-Empty HCT116 cells (Red = DiI stained infiltrated cells; Blue = lung mice nuclei counterstained with Topro). Right: dot plot indicates mean +/− std. of the fraction of infiltrated cells over total nuclei in lungs of 5 mice per condition, quantified as described in Material and Methods (***p < .005, using unpaired t-test with unequal variances). (c) Western Blot analysis of Zeb1, Btbd7 and e-Cadherin expression in pEGFP-Btbd7 clones #1, #2, #3 or pEGFP-Empty clones #1, #2, #3. (d). CDH1, ZEB-1, VIMENTIN, TWIST and SLUG qRT-analysis in pEGFP-Btbd7 clones #1, #2, #3 or pEGFP-Empty clones #1, #2, #3. Dot plot indicate mean +/− std. of two independent biological replicates (ns, non-significant; **p < .005, using paired t-test assuming equal variances). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Taken together, these data suggest that ectopic expression of Btbd7 resembles mesenchymal gene expression and pro-metastatic traits in CRC cells, in agreement with [[36], [37], [38]].
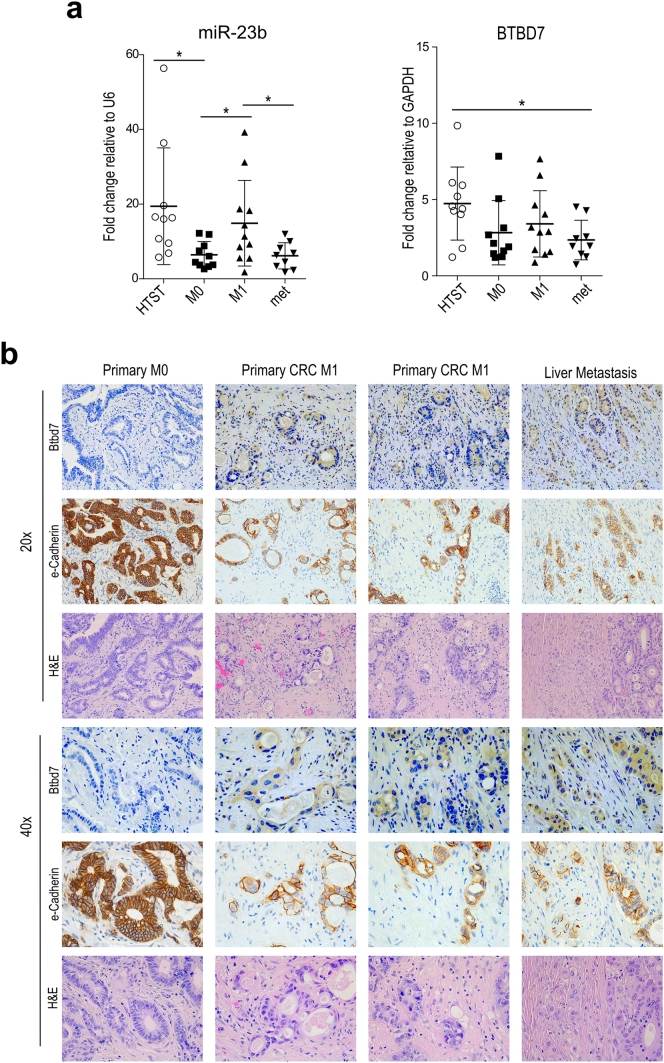
3.5. miR-23b is downregulated and Btbd7 is upregulated in CRC liver metastasis
To better understand the clinical significance of miR-23b in CRC patients, we explored The Cancer Genome Atlas (TCGA) gene expression CRC dataset [40]. We selected two groups of primary CRC samples with or without distant metastasis (M0 and M1; 8 versus 9 samples) and matched for tumors size, lymph node involvement, gender, age, and microsatellite stability (Suppl. Table 2). We found that miR-23b was upregulated in M1 versus M0 primary samples; whereas, changes in BTBD7 expression were non-significant (Suppl. Fig. S9a; p < .05; using unpaired t-test with unequal variances). CDH1 was lower and VIMENTIN was higher in M1 versus M0 CRC tumors although not in a statistically significant manner, suggestive of an EMT switch, instead ZEB1 was found unchanged (Suppl. Fig. S9a).
To evaluate miR-23b expression in CRC liver metastasis, which was not addressed in previous analyses [34], we used a cohort of CRC samples that included healthy tissues surrounding tumor (HTST; n = 10), primary CRC samples of patients without and with metastasis at diagnosis (M0 and M1, n = 10 and n = 11 respectively), and samples from liver metastasis (met; n = 9) (Suppl. Table 3). Similarly to the TCGA dataset, CDH1 and VIMENTIN expression changed between M0 and M1, although not in a statistically significant manner (Suppl. Fig. S9b). Concordantly to the TCGA data analysis (Suppl. Fig. S9a) and to [34], we found that miR-23b was upregulated in M1 compared to M0 primary samples and was down-regulated in liver metastasis compared to primary M1 (Fig. 7a; p < .05, using unpaired t-test assuming unequal variances), underscoring its possible role as metastasis-regulator also in the clinical setting. BTBD7 expression levels showed a trend similar to miR-23b, but differences were not significant across all groups (Fig. 7a; using unpaired t-test assuming unequal variances). Because miR-23b regulates BTBD7 at the protein level, we explored Btbd7 protein expression levels by IHC in the same cohort of CRC samples, and found that Btbd7 was not detectable both in normal mucosa and in primary CRC samples of non-metastatic patients (n = 6 and n = 5). On the other hand, Btbd7 was present in one primary CRC sample of a metastatic patient (n = 8) and in 2 out of the 4 available metastatic samples (Fig. 7b). Although the positive Btbd7 samples were only three, all of them had miR-23b expression levels below the absolute median value. Interestingly, we observed that CRC cancer cells expressing Btbd7 were not inside the bulk of the primary tumor or metastatic lesion, but rather dispersed within the normal surrounding tissue, forming smaller cell nests, and that these Btbd7 positive cells had low expression levels of e-Cadherin consistently to in vitro findings (Fig. 6c-d).
Fig. 7.
miR-23b and Btbd7 are inversely expressed in CRC patient samples (a) qRT-analysis of miR-23b (left panel) and BTBD7 (right panel) in healthy tissues surrounding tumor (HTST; n = 10), in primary CRC samples without (M0; n = 10), or with metastasis at diagnosis (M1; n = 11), and in liver metastatic samples (MET; n = 9). Dot plot indicate mean +/− std. of the selected biological CRC patient samples (*p < .05, using unpaired t-test assuming unequal variances). (b) Representative images of IHC staining for Btbd7, e-Cadherin and H&E (20× and 40× magnification) on tissue sections derived from M0, M1 and MET CRC samples.
4. Discussion
The driving objective of the present paper was to identify regulatory genetic elements that regulate CRC metastasis. We accomplished this by combining an in vitro assay that selects cancer cells with mesenchymal traits (i.e. forced single cell suspension assay - fSCS), with an SB-based forward genetic screen. We discovered the miR-23b::BTBD7–3′UTR interaction, we showed that this interaction is crucial for pro-metastatic cell traits in preclinical models (i.e. cell lines and mice), and we found that miR-23b and Btbd7 are promising biomarkers to identify CRC patients with greater risk of developing metastasis.
Loss of epithelial traits (e.g. adherens junctions) and gain of mesenchymal traits are prerequisites for cells to detach from neighboring cells, to invade surrounding tissues and to proceed along the metastatic cascade [[8], [9], [10]]. Herein, we presented a novel in vitro assay (i.e. fSCS), which exerts a double selective pressure over tumor cells (i.e. it prevents anchorage dependent growth and cell-cell contacts) and thus it selects mesenchymal and pro-metastatic tumor cells. fSCS is easily scalable as shown in this manuscript (from 96 well plate to flask format) and it can be used in several applications (e.g. library of compounds [41] or other genetic screens) to further identify additional genes involved in EMT [42].
<2% of the human genome codes for proteins (coding part) and >98% is transcribed but does not code for proteins (non-coding part). The annotation of the non-coding part is improving thanks to novel tools to discover transcripts [43,44] or regulatory elements [45]; however, the function of most non-coding elements is still largely unexplored. In this manuscript, we suggest that SB-based genetic screens are valuable tools to improve the understanding of the function of the non-coding part of the human genome. In particular, by this approach we identified the novel miR-23b::BTBD7–3′UTR interaction, which proved to be relevant for the phenotype investigated (i.e. pro-metastatic traits). However, it must be recalled that in the TN4_20 fSCS resistant CRC cell clone, SB insertions were also located inside other non-coding regions in addition to BTBD7–3′UTR; at present, we cannot either confirm or rule out their role in fSCS resistance and CRC metastasis, but this offers further opportunities to investigate the non-coding part of the human genome. At the same time, our strategy suggests that SB-based genetic screen can be used to identify non-coding genes relevant to other phenotypes if combined with different functional assays.
In agreement with our data, by interrogating hundreds of human microRNAs, Zhang et al. previously showed that miR-23b impairs pro-metastatic traits in vitro [34]. However, miR-23b function in in vivo models of CRC metastasis and in CRC clinical samples appears conflicting: Zhang et al. showed that miR-23b impaired distant metastasis formation when CRC cell lines are injected in mice [34]; on the contrary, we observed that miR-23b increased CRC cell extravasation in the lung parenchyma (Fig. 5c). Counter-intuitively to former preclinical models, miR-23b gene expression was higher in primary CRC tumors that metastasized (M1) compared with those that did not (M0) in three independent cohorts (both from this manuscript - Fig. 7a and Suppl. Fig. 9a - and [34]). However, when we analyzed miR-23b expression in distant liver metastasis (i.e. the last step of the metastatic cascade), we observed that miR-23b expression is indeed decreased in liver metastatic samples compared to primary CRC lesions (Fig. 7a). To sum up, miR-23b appears to promote the early steps of the metastatic cascade, which is corroborated by miR-23b higher expression in M1 primary CRC samples compared with M0 and its greater pro-extravasation effect in mice; at the same time, miR-23b appears to prevent the later steps as demonstrated by its decreased expression in liver metastasis compared with primary CRC tissues and the impaired ability to generate distant metastasis in miR-23b overexpressing cell lines. All these considerations underscore the complexity and the plasticity of the metastatic cascade and how the same microRNA at different steps along this cascade exerts opposite effects with presumably different targets.
Herein, we discovered that mir-23b targets BTBD7–3′UTR and regulates Btbd7 expression levels. Onodera et al. showed that BTBD7 regulates salivary gland branching in mouse by decreasing cell-cell adhesions and increasing cell invasion with loss of e-Cadherin. Consistently, we observed that Btbd7 expression was mainly present in isolated clumps of CRC cells rather than in the bulk of the liver metastasis, and that Btbd7 expression inversely correlated with e-Cadherin expression levels (Fig. 7b). Altogether these data suggest that Btbd7 is expressed in the invading tumor cells. We also started to shed light on the function of Btbd7: we confirmed that Btbd7 down-regulates the expression of e-Cadherin in agreement with [[36], [37], [38]] and found that it up-regulates the expression of Zeb-1 transcription factor. How exactly BTBD7 controls EMT gene reprogramming and metastasis still remains to be thoroughly addressed. On one hand, Btbd7 has a predicted N-myristilation motif [46] that should redirect it to lipid rafts of the plasma membrane [47]. Indeed, lipid rafts have been described to regulate EMT and tumor plasticity [48]. Furthermore, we showed that Btbd7 localizes in the cytoplasm (Fig. 7b and Suppl. Fig. S8d lower panels), and it affects both mRNA and protein expression levels of CDH1 and ZEB1 genes. Taking all these evidences into account, we speculate that Btbd7 could exert its effect at lipid rafts and controls EMT gene expression by an indirect transcriptional activation.
In conclusion, our data indicate that detection of high miR-23b and Btbd7 levels in the primary tumor of CRC patients can be correlated with the development of cancer metastasis and thus patients with these biological features should be treated more aggressively and have closer follow-ups. Although further prospective studies are required, both miR-23b and Btbd7 could serve as prognostic biomarkers for CRC patients. We can also envision the use of strategies to modulate miR-23b expression at the primary tumor to arrest CRC dissemination to distant sites or systemically when overt metastasis has already occurred to induce metastasis regression [49]. The correct timing and site of miR-23b modulation must be first determined with appropriate pre-clinical models.
Funding sources
This work was supported by grants from Marie Curie Actions (CIG n. 303,877) to MSN, from Talents Marie Curie Actions/Regione Friuli Venezia Giulia (Grant Agreement n°245,574) to RS, from Italian Association for Cancer Research (AIRC, MFAG n°13,589), from Italian Ministry of Health (GR-2010-2319387 and PE-2016-02361040) and, from 5 × 1000 to CRO Aviano. Funding bodies had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript and eventually in the decision to submit the manuscript.
Author contributions
Literature search (EG, MC, RS, MSN); figures (EG, MC, RS, MSN, LC); study design (EG, MC, RS, MSN); experiments (EG, MC, RS, MSN, LZ, LC, AP, MM, FDB, GB, EB, MT, ED, AM, VC); data collection (EG, MC, RS, MSN, LZ, LC, AP, MM, FDB, GB, EB, MT, AM, GRV, AV); data interpretation (EG, MC, RS, MSN, LZ, LC, MM, FDB, GB, EB, MT, ED, AM, VC, GRV, AV); data analysis (EG, MC, RS, MSN, LC, FDB, GB, EB, ED, GRV, AV); writing (EG, MC, RS, MSN). No medical writer was involved in the creation of this manuscript. RS has full access to all the data in the study and had final responsibility for the decision to submit for publication.
Declaration of Competing Interest
Dr. Grisard has nothing to disclose; Dr. Coan has nothing to disclose; Dr. Cesaratto has nothing to disclose; Dr. Rigo has nothing to disclose; Dr. Zandonà has nothing to disclose; Dr. Pauliti has nothing to disclose; Dr. Andreuzzi has nothing to disclose; Dr. Rampioni Vinciguerra has nothing to disclose; Dr. Poletto has nothing to disclose; Dr. Del Ben has nothing to disclose; Dr. Brisotto has nothing to disclose; Dr. Biscontin has nothing to disclose; Dr. Turetta has nothing to disclose; Dr. Dassi has nothing to disclose; Dr. Mirnezami has nothing to disclose; Dr. Canzonieri has nothing to disclose; Dr. Vecchione has nothing to disclose; Dr. Baldassarre has nothing to disclose; Dr. Mongiat has nothing to disclose; Dr. Spizzo has nothing to disclose; Dr. Nicoloso has nothing to disclose.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.06.044.
Appendix A. Supplementary data
Supplementary material
References
- 1.Mehlen P., Puisieux A. Metastasis: a question of life or death. Nat Rev Cancer. 2006;6(6):449–458. doi: 10.1038/nrc1886. [DOI] [PubMed] [Google Scholar]
- 2.Valastyan S., Weinberg R.A. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147(2):275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arvelo F., Sojo F., Cotte C. Biology of colorectal cancer. Ecancermedicalscience. 2015;9:520. doi: 10.3332/ecancer.2015.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Society. AC . American Cancer Society; Atlanta, GA: 2014. Colorectal cancer facts & figures 2014-2016. [Google Scholar]
- 5.Markowitz S.D., Bertagnolli M.M. Molecular origins of cancer: molecular basis of colorectal cancer. N Engl J Med. 2009;361(25):2449–2460. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.House M.G., Ito H., Gonen M. Survival after hepatic resection for metastatic colorectal cancer: trends in outcomes for 1,600 patients during two decades at a single institution. Journal of the American College of Surgeons. 2010;210(5) doi: 10.1016/j.jamcollsurg.2009.12.040. 744-52, 52-5. [DOI] [PubMed] [Google Scholar]
- 7.Weiser M.R., Jarnagin W.R., Saltz L.B. Colorectal cancer patients with oligometastatic liver disease: what is the optimal approach? Oncology. 2013;27(11):1074–1078. [PubMed] [Google Scholar]
- 8.Brabletz T. To differentiate or not--routes towards metastasis. Nat Rev Cancer. 2012;12(6):425–436. doi: 10.1038/nrc3265. [DOI] [PubMed] [Google Scholar]
- 9.Brabletz T., Kalluri R., Nieto M.A., Weinberg R.A. EMT in cancer. Nat Rev Cancer. 2018;18(2):128–134. doi: 10.1038/nrc.2017.118. [DOI] [PubMed] [Google Scholar]
- 10.Gawrzak S., Rinaldi L., Gregorio S. MSK1 regulates luminal cell differentiation and metastatic dormancy in ER(+) breast cancer. Nat Cell Biol. 2018;20(2):211–221. doi: 10.1038/s41556-017-0021-z. [DOI] [PubMed] [Google Scholar]
- 11.Lander E.S., Linton L.M., Birren B. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 12.Grisard E., Nicoloso M.S. Following MicroRNAs through the Cancer metastatic cascade. Int Rev Cell Mol Biol. 2017;333:173–228. doi: 10.1016/bs.ircmb.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 13.Spizzo R., Almeida M.I., Colombatti A., Calin G.A. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012;31(43):4577–4587. doi: 10.1038/onc.2011.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Copeland N.G., Jenkins N.A. Harnessing transposons for cancer gene discovery. Nat Rev Cancer. 2010;10(10):696–706. doi: 10.1038/nrc2916. [DOI] [PubMed] [Google Scholar]
- 15.Burckstummer T., Banning C., Hainzl P. A reversible gene trap collection empowers haploid genetics in human cells. Nat Methods. 2013;10(10):965–971. doi: 10.1038/nmeth.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rad R., Rad L., Wang W. A conditional piggyBac transposition system for genetic screening in mice identifies oncogenic networks in pancreatic cancer. Nat Genet. 2015;47(1):47–56. doi: 10.1038/ng.3164. [DOI] [PubMed] [Google Scholar]
- 17.Ivics Z., Izsvak Z. The expanding universe of transposon technologies for gene and cell engineering. Mobile DNA. 2010;1(1):25. doi: 10.1186/1759-8753-1-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu Z., Sanchez-Sweatman O., Huang X. Anoikis and metastatic potential of cloudman S91 melanoma cells. Cancer Res. 2001;61(4):1707–1716. [PubMed] [Google Scholar]
- 19.Douma S., Van Laar T., Zevenhoven J., Meuwissen R., Van Garderen E., Peeper D.S. Suppression of anoikis and induction of metastasis by the neurotrophic receptor TrkB. Nature. 2004;430(7003):1034–1039. doi: 10.1038/nature02765. [DOI] [PubMed] [Google Scholar]
- 20.Hofmann C., Obermeier F., Artinger M. Cell-cell contacts prevent anoikis in primary human colonic epithelial cells. Gastroenterology. 2007;132(2):587–600. doi: 10.1053/j.gastro.2006.11.017. [DOI] [PubMed] [Google Scholar]
- 21.Mueller S., Cadenas E., Schonthal A.H. p21WAF1 regulates anchorage-independent growth of HCT116 colon carcinoma cells via E-cadherin expression. Cancer Res. 2000;60(1):156–163. [PubMed] [Google Scholar]
- 22.Nieto M.A., Huang R.Y., Jackson R.A., Thiery J.P. Emt: 2016. Cell. 2016;166(1):21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 23.Frisch S.M., Schaller M., Cieply B. Mechanisms that link the oncogenic epithelial-mesenchymal transition to suppression of anoikis. J Cell Sci. 2013;126:21–29. doi: 10.1242/jcs.120907. Pt 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guadamillas M.C., Cerezo A., Del Pozo M.A. Overcoming anoikis--pathways to anchorage-independent growth in cancer. J Cell Sci. 2011;124:3189–3197. doi: 10.1242/jcs.072165. Pt 19. [DOI] [PubMed] [Google Scholar]
- 25.Huang R.Y., Wong M.K., Tan T.Z. An EMT spectrum defines an anoikis-resistant and spheroidogenic intermediate mesenchymal state that is sensitive to e-cadherin restoration by a src-kinase inhibitor, saracatinib (AZD0530) Cell Death Dis. 2013;4:e915. doi: 10.1038/cddis.2013.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bates R.C., Mercurio A.M. The epithelial-mesenchymal transition (EMT) and colorectal cancer progression. Cancer Biol Ther. 2005;4(4):365–370. doi: 10.4161/cbt.4.4.1655. [DOI] [PubMed] [Google Scholar]
- 27.Brabletz T., Hlubek F., Spaderna S. Invasion and metastasis in colorectal cancer: epithelial-mesenchymal transition, mesenchymal-epithelial transition, stem cells and beta-catenin. Cells Tissues Organs. 2005;179(1–2):56–65. doi: 10.1159/000084509. [DOI] [PubMed] [Google Scholar]
- 28.Kalluri R., Weinberg R.A. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119(6):1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mani S.A., Guo W., Liao M.J. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133(4):704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Penna EaT D. In vivo Extravasation Assay. Bio-protocol. 2014;4(4) [Google Scholar]
- 31.Fischer K.R., Durrans A., Lee S. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015;527(7579):472–476. doi: 10.1038/nature15748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ivics Z., Hackett P.B., Plasterk R.H., Izsvak Z. Molecular reconstruction of sleeping beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91(4):501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 33.Ivics Z., Li M.A., Mates L. Transposon-mediated genome manipulation in vertebrates. Nat Methods. 2009;6(6):415–422. doi: 10.1038/nmeth.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang H., Hao Y., Yang J. Genome-wide functional screening of miR-23b as a pleiotropic modulator suppressing cancer metastasis. Nat Commun. 2011;2:554. doi: 10.1038/ncomms1555. [DOI] [PubMed] [Google Scholar]
- 35.Largaespada D.A., Collier L.S. Transposon-mediated mutagenesis in somatic cells: identification of transposon-genomic DNA junctions. Methods Mol Biol. 2008;435:95–108. doi: 10.1007/978-1-59745-232-8_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Onodera T., Sakai T., Hsu J.C., Matsumoto K., Chiorini J.A., Yamada K.M. Btbd7 regulates epithelial cell dynamics and branching morphogenesis. Science. 2010;329(5991):562–565. doi: 10.1126/science.1191880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tao Y.M., Huang J.L., Zeng S. BTB/POZ domain-containing protein 7: epithelial-mesenchymal transition promoter and prognostic biomarker of hepatocellular carcinoma. Hepatology. 2013;57(6):2326–2337. doi: 10.1002/hep.26268. [DOI] [PubMed] [Google Scholar]
- 38.Fan C., Miao Y., Zhang X. Btbd7 contributes to reduced E-cadherin expression and predicts poor prognosis in non-small cell lung cancer. BMC Cancer. 2014;14:704. doi: 10.1186/1471-2407-14-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pellegrino L., Stebbing J., Braga V.M. miR-23b regulates cytoskeletal remodeling, motility and metastasis by directly targeting multiple transcripts. Nucleic Acids Res. 2013;41(10):5400–5412. doi: 10.1093/nar/gkt245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gupta P.B., Onder T.T., Jiang G. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138(4):645–659. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Evers B., Jastrzebski K., Heijmans J.P., Grernrum W., Beijersbergen R.L., Bernards R. CRISPR knockout screening outperforms shRNA and CRISPRi in identifying essential genes. Nat Biotechnol. 2016;34(6):631–633. doi: 10.1038/nbt.3536. [DOI] [PubMed] [Google Scholar]
- 43.Frankish A., Diekhans M., Ferreira A.M. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019;47(D1) doi: 10.1093/nar/gky955. (D766-D73) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Noguchi S., Arakawa T., Fukuda S. FANTOM5 CAGE profiles of human and mouse samples. Sci. Data. 2017;4 doi: 10.1038/sdata.2017.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klann T.S., Black J.B., Chellappan M. CRISPR-Cas9 epigenome editing enables high-throughput screening for functional regulatory elements in the human genome. Nat Biotechnol. 2017;35(6):561–568. doi: 10.1038/nbt.3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.UniProt C. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47(D1) doi: 10.1093/nar/gky1049. (D506-D15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Resh M.D. Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim Biophys Acta. 1999;1451(1):1–16. doi: 10.1016/s0167-4889(99)00075-0. [DOI] [PubMed] [Google Scholar]
- 48.Zuo W., Chen Y.G. Specific activation of mitogen-activated protein kinase by transforming growth factor-beta receptors in lipid rafts is required for epithelial cell plasticity. Mol Biol Cell. 2009;20(3):1020–1029. doi: 10.1091/mbc.E08-09-0898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu S.Y., Lopez-Berestein G., Calin G.A., Sood A.K. RNAi therapies: drugging the undruggable. Sci Transl Med. 2014;6(240):240ps7. doi: 10.1126/scitranslmed.3008362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mates L., Chuah M.K., Belay E. Molecular evolution of a novel hyperactive sleeping beauty transposase enables robust stable gene transfer in vertebrates. Nat Genet. 2009;41(6):753–761. doi: 10.1038/ng.343. [DOI] [PubMed] [Google Scholar]
- 51.Schindelin J., Arganda-Carreras I., Frise E. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mazutis L., Gilbert J., Ung W.L., Weitz D.A., Griffiths A.D., Heyman J.A. Single-cell analysis and sorting using droplet-based microfluidics. Nat Protoc. 2013;8(5):870–891. doi: 10.1038/nprot.2013.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nicoloso M.S., Sun H., Spizzo R. Single-nucleotide polymorphisms inside microRNA target sites influence tumor susceptibility. Cancer Res. 2010;70(7):2789–2798. doi: 10.1158/0008-5472.CAN-09-3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Untergasser A., Nijveen H., Rao X., Bisseling T., Geurts R., Leunissen J.A. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007;35(Web Server):W71–W74. doi: 10.1093/nar/gkm306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cervo S., Rovina J., Talamini R. An effective multisource informed consent procedure for research and clinical practice: an observational study of patient understanding and awareness of their roles as research stakeholders in a cancer biobank. BMC Med Ethics. 2013;14:30. doi: 10.1186/1472-6939-14-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Muller H., Marzi M.J., Nicassio F. IsomiRage: from functional classification to differential expression of miRNA isoforms. Front Bioeng Biotechnol. 2014;2:38. doi: 10.3389/fbioe.2014.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material