Abstract
The steep increase in the incidence of type 1 diabetes (T1D), in the Western world after World War II, cannot be explained solely by genetic factors but implies that this rise must be due to crucial interactions between predisposing genes and environmental changes. Three parallel phenomena in early childhood – the dynamic development of the immune system, maturation of the gut microbiome, and the appearance of the first T1D-associated autoantibodies – raise the question whether these phenomena might reflect causative relationships. Plenty of novel data on the role of the microbiome in the development of T1D has been published over recent years and this review summarizes recent findings regarding the associations between islet autoimmunity, T1D, and the intestinal microbiota.
Keywords: Type 1 diabetes, Microbiota, Mycobiota, Virome, Dysbiosis
1. Introduction
Type 1 diabetes (T1D) is a chronic immune-mediated disease in which the insulin-producing beta cells of the pancreatic islets are destroyed. The disease process starts often at a young age, during the first years of life. In T1D-affected patients, the lack of endogenous insulin causes a life-long need of exogenous insulin therapy [1,2]. The rate of T1D has increased conspicuously after World War II in most developed countries. For example, in Finland the incidence among children under the age of 15 years has increased from 12 to 65 new cases/100,000/year in five decades [3]. The increasing disease rate cannot be explained by genetic factors but implies that these changes are an outcome of interactions between the modern westernized environment and predisposing genes.
The human intestinal microbiome maturates during the first years of life, after which its composition resembles the composition observed in adults [4,5]. The maturation of the gut microbiome is closely linked to the development of the immune system [6]. Three contemporary phenomena occurring in early childhood – the dynamic development of the immune system, th the maturation of the gut microbiome, and the appearance of the first T1D-associated autoantibodies – raise the question whether these phenomena are only temporarily associated, or do they reflect causative relationships. Even if the field of microbiome studies has expanded remarkably during recent years, causal relationships have not been established. Since our last review in which we explored the potential role of the gut microbiota in the development of T1D [7], ample new data has been generated. In this review, we set out to summarize recent findings from the microbiome studies.
2. Intestinal bacterial microbiota
2.1. Lessons learned from experimental studies
The idea that intestinal microbiota might play an important role in the development of immune-mediated diseases, such as T1D, was presented already in the 1980's [8]. To test this hypothesis, genetically modified rodents have been raised in germ-free conditions; colonized with various microbial communities; treated with probiotics, prebiotics, and antibiotics; fed with distinct diets, and given intestinal microbiota from other animals (Supplementary Table 1). Considering the interplay between the ntestinal microbiota and the developing immunity, several key findings have been observed (Table 1). Comparisons between these findings and those observed in man (Table 2) will be discussed in the next paragraphs [[9], [10], [11], [12], [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31]]. While interpreting the results, one should bear in mind that autoimmune diabetes observed in NOD mice is not identical with T1D in humans [32]. Considering these two species, two major differences associate with the timing of the disease development: (i) in man, seroconversions peak during the first three years of life, while NOD mice typically seroconvert close to their sexual maturity, and (ii) approximately half of the cases appear before adulthood in humans, whereas in NOD mice, nearly all cases occur in adult animals, the typical age of onset being 12–30 weeks. Importantly, by 30 weeks of age, >80% of the NOD females and >50 of the NOD males are typically overtly diabetic, while in human populations the prevalence of T1D is under 1% [33].
Table 1.
Summary on findings from experimental studies on the interplay between the intestinal microbiota and exogenous and host-related factors.
|
Table 2.
Recent studies investigating the associations between gastrointestinal microbiota and islet autoimmunity leading to type 1 diabetes (T1D) in humans.
| Ref. # | Author | Age (years) | Study groups; Geographical locations | Findings related to T1D and/or islet autoimmunity |
|---|---|---|---|---|
| [9] | Mejía-León ME et al. 2014 | 7–18 | Children with T1D at diagnosis (n = 8) and after 2 years of insulin treatment (n = 13) vs. healthy controls (n = 8); Mexico | At diagnosis, T1D patients had high levels of Bacteroides, whereas controls had Prevotella dominance. After 2 years of insulin treatment, cases resembled the controls. |
| [10] | Kemppainen KM et al. 2015 | 0–2 | Seronegative young children with high risk HLA genotype (n = 15 from each site); Finland, Sweden, Germany, and USA | Bacterial diversity differed by geographical location. Children from Finland and Colorado had lower bacterial diversity, and children from Sweden and Washington state had Bifidobacterium dominance. |
| [11] | Soyucen E et al. 2014 | 6–15 | Children with newly diagnosed T1D (n = 35) and healthy controls (n = 35); Turkey | Compared to controls, Bifidobacterium colonization was lower in patients with T1D. Candida albicans and Enterobacteriaceae (other than Echerichia coli) were increased in controls. |
| [12] | Kostic AD et al. 2015 | 0–3 | Young children with islet autoimmunity (n = 11) and healthy controls (n = 22). All have HLA-conferred risk for T1D; Finland and Estonia | Drop in alpha-diversity between seroconversion and T1D diagnosis, accompanied with an increase in proinflammatory organisms, changes in gene functions, and serum and stool metabolites. Microbial relationships were shared across most subjects. Strain compositions were highly variable between individuals, but stable within individuals throughout infancy. Metabolic composition and metabolic pathway abundances were constant across time. |
| [13] | Endesfelder D et al. 2016 | 0–3 | Young children with persistent islet autoimmunity (n = 22) and healthy controls (n = 22); Germany | Bacteroides-dominated subgroup associated with early introduction of non-milk diet, increased risk for early autoantibody development, and lower abundances of genes for butyrate production. Alterations in the composition of mucin-degrading bacteria associated with early development of islet autoimmunity. Functional associations between diet, gut microbiome and islet autoimmunity existed in microbial co-occurrence networks. |
| [14] | Heintz-Buschart A et al. 2016 | 5–62 | Four families with ≥2 generations of ≥2 cases of T1D per family (n = 20); Netherlands | Family membership has a pronounced effect on the structural and functional composition of the gastrointestinal microbiome. No differences in taxonomic or functional diversity and richness between T1D cases and their family members. Protein expression findings implicated that T1D cases may have slightly dysfunctional exocrine pancreas. |
| [15] | Maffeis C et al. 2016 | 6–16 | Children with islet autoimmunity (10) and healthy controls (n = 10); Italy | Group-specific differentiation between cases and controls was challenging. Intestinal permeability was higher in children with islet autoimmunity. Dialister invisus, Gemella sanguinis and Bifidobacterium longum were associated with prediabetes. |
| [16] | Qi CJ et al. 2016 | 10–15 | Children with T1D (n = 15) and healthy controls (n = 15); China | T1D cases had decreased bacterial richness, decreased Haemophilus, Lachnospira, Dialister, and Acidaminococcus, and increased Blautia levels. Percentage of Blautia correlated with HbA1c, number of T1D-associated autoantibodies, and IA-2A levels. |
| [17] | Vatanen T et al. 2016 | 0–3 | Young children with HLA-associated risk for T1D (n = 74 from each country); Finland, Estonia, and Russia | Bacteroides strains were more abundant in Finnish infants compared to Russian Karelian infants, while E. coli was more frequent on Finnish infants Bacteroides LPS had immunoinhibitory properties that may preclude early immune education and contribute to development of T1D, while E. coli derived LPS was strongly immunostimulatory. Metabolism of human milk oligosaccharides (HMOs) maintained Bifidobacterium-dominant versus Bacteroides-dominant gut microbiota in the first year of life. |
| [18] | Cinek O et al. 2017 | 0–3 | Young children with islet autoimmunity progressing to T1D (n = 18) and healthy controls (18); Finland | Four operational taxonomic units were less abundant in children developing islet autoimmunity, most markedly Bacteroides vulgatus and Bifidobacterium bifidum. Potential relation existed between CrAssphage and Bacteroides dorei. No differences were observed between cases and controls in alpha or beta diversity, or the taxonomic levels of bacterial phyla, classes or genera. |
| [19] | de Groot PF et al. 2017 | 25–45 | Adults with T1D (n = 53) and healthy controls (n = 50); Netherlands | Both oral and gut microbiota differed between cases and controls. Oral microbiota had higher abundance of streptococci in cases, while gut microbiota showed decreased butyrate-producing species and less butyryl-CoA transferase genes. Plasma levels of acetate and propionate were lower in T1D. Christensenella and Subdoligranulum strains correlated with glycaemic control, inflammatory parameters, and level of short-chain fatty acids (SCFAs). |
| [20] | Pellegrini S et al. 2017 | 1–65 | Children and adults with T1D (n = 19) or celiac disease (CD, n = 19) vs. controls (n = 16); Italy | T1D-specific increase in monocyte/macrophage infiltration in the intestinal biopsies. Cases had increase in Firmicutes and Firmicutes/Bacteroidetes ratio, and a reduction in Proteobacteria and Bacteroidetes in the duodenal mucosa. The expression levels of genes specific for T1D inflammation were associated with the abundance of specific bacteria in the duodenum. |
| [21] | Pinto E et al. 2017 | 8–11 | Children with T1D (n = 3) and healthy controls (n = 3); Portugal | T1D cases had a microbial intestinal proteome enriched with proteins of clostridial cluster XVa and cluster IV and Bacteroides, while controls had more often bifidobacterial proteins. T1D children had lower levels of exocrine pancreatic enzymes. |
| [22] | Stewart CJ et al. 2017 | 25–30 | Young adults with T1D (n = 10) and healthy controls (n = 10); UK | Faecalibacterium sp., Roseburia sp. and Bacteroides sp. were the most abundant microbes in both groups. Each bacterial profile was individual and no significant differences in the profiles or in the diversity indices were observed between the groups. |
| [23] | Cinek O et al. 2018 | 7–15 | Children at T1D onset (n = 73; 14–20 per country) and healthy controls (n = 104); Azerbaijan, Jordan, Nigeria, and Sudan | Genus Escherichia spp. correlated with T1D. T1D cases showed an inverse association with Eubacterium and Roseburia (Firmicutes), and clostridial clusters IV and XIVa. No T1D-associations for microbial richness or enterotypes were observed. |
| [24] | Gao X et al. 2018 | 0–3 | Young children with islet autoimmunity (n = 11) and healthy controls (n = 22). All have HLA-conferred risk for T1D; Finland and Estonia | More microbial interactions and the inhibition of Clostridia by Gammaproteobacteria were maintained throughout the first 3 years of life in healthy children. Gammaproteobacteria inhibited Bacteroidia in the cases, but not in controls. The inhibitory effects from Actinobacteria and Bacilli on Bacteroidia, from Bacteroidia on Clostridia, and the beneficial effect from Clostridiao on Bacteroidia were shared between cases and controls. |
| [25] | Gavin PG et al. 2018 | 2–45 | Children and adults with recent-onset T1D (33) or islet autoimmunity (n = 17), and low-risk autoantibody-negative subjects (n = 29) and healthy controls (n = 22); USA | T1D patients had increased intestinal inflammation and decreased barrier function. Microbial taxa capable of promoting the host's mucous barrier, microvilli adhesion, and functions of the exocrine pancreas were depleted in T1D. Pancreatic exocrine dysfunction appeared in high-risk individuals before the disease onset. Both host-derived and microbial-derived proteins were able to differentiate the new-onset and the islet autoantibody-positive subjects from the low-risk subjects. |
| [26] | Higuchi BS et al. 2018 | 15–35 | Teenagers and young adults with T1D (n = 20) and healthy controls (n = 28); Brazil | T1D patients had intestinal dysbiosis with prevalent Gram-negative bacteria like Bacteroides vulgatus, Bacteroides rodentium, Prevotella copri, and Bacteroides xylanisolvens. Cases had increased plasma levels of the proinflammatory interleukin-6. Poor glycaemic control associated with the relative abundance of Bacteroidetes, Lactobacillales, and Bacteroides dorei. |
| [27] | Huang Y et al. 2018 | 18–25 | Young adults with T1D (n = 12) and healthy controls (n = 10); China | Various bacterial taxonomic clades differed between cases and controls. Bacteroidetes and Firmicutes were the dominant phyla in cases and controls, respectively. Abundance of Faecalibacterium correlated inveresly with HbA1c levels. Numbers of islet autoantibodies correlated positively with Bacteriodes and Bilophila abundances and inversely with Streptococcus and Ruminococcaceae abundances. |
| [28] | Leiva-Gea I et al. 2018 | 9–16 | Children with T1D (n = 15) or MODY2 (n = 15), and healthy controls (n = 13); Spain | T1D associated with (i) lower microbial diversity, (II) higher relative abundance of Bacteroides, Ruminococcus, Veillonella, Blautia, and Streptococcus genera, and (III) lower relative abundance of Bifidobacterium, Roseburia, Faecalibacterium, and Lachnospira. Proinflammatory cytokines and lipopolysaccharides were increased in T1D cases, as were the expression of genes related to lipid and amino acid metabolism, ATP-binding cassette transport, lipopolysaccharide biosynthesis, arachidonic acid metabolism, antigen processing and presentation, and chemokine signalling pathways. MODY2 associated with higher Prevotella abundance and a lower Ruminococcus and Bacteroides abundance. Intestinal permeability was increased both in T1D and in MODY2 cases. |
| [29] | Mejía-León ME et al. 2018 | 9–14 | Children with newly diagnosed T1D (n = 10) | The increasing Bacteroides proportion (during the first months after diagnosis) correlated with the consumption of saturated fat and carbohydrates at 3 months. After adjusting for HbA1C, consumption of carbohydrates associated with decreased Bacteroides abundance over time. |
| [30] | Stewart CJ et al. 2018 | 0–4 | Young children (n = 903) with high risk HLA genotype; Finland, Sweden, Germany, and USA | Feeding with breast milk was the most significant factor associated with the microbiome structure. It associated with higher levels of Bifidobacterium sp. (B. breve and B. bifidum). Bacteroides associated with increased microbial diversity and faster maturation of the gut. Only subtle associations were observed between microbial taxonomy and islet autoimmunity: i) higher relative abundance of an unclassified Erysipelotrichaceae in children with autoimmunity; ii) five bacterial genera associated with T1D onset (Parabacteroides the most significant); iii) eleven bacterial genera were lower in T1D cases, including four unclassified Ruminococcaceae, Lactococcus, Streptococcus, and Akkermansia. |
| [31] | Vatanen T et al. 2018 | 0–4 | Young children (n = 783) with high risk HLA genotype developing persistent islet autoimmunity or T1D vs. controls; Finland, Sweden, Germany, and USA | Control children had more genes associated with fermentation and biosynthesis of SCFAs. T1D-associated microbial compositions were taxonomically diffuse but functionally more coherent. Compared to children with autoimmunity, controls had higher levels of Lactobacillus rhamnosus and Bifidobacterium dentium, while children with autoimmunity had higher abundance of Streptococcus group mitis/oralis/pneumoniae species. Compared to children developing T1D, controls had higher levels of Streptococcus thermophiles and Lactococcus lactis species. T1D cases had higher levels of Bifidobacterium pseudocatenulatum, Roseburia hominis, and Alistipes shahii. |
2.2. The interactions between the genetic setup, nutrition, microbial composition, and susceptibility to islet autoimmunity
The genetic setup of the host may affect the host's microbial composition and function, the activation of innate and adaptive immunity and susceptibility to various diseases; both in animal models and in man [[34], [35], [36]]. In mice, the host's inherited disease susceptibility can be modified by colonizing with selected microbes, providing mice with specific diets, or treating them with antibiotics. Results from these interventions range from accelerated autoimmunity to total prevention of the disease (Supplementary Table 1). In humans, dietary modifications, such as weaning infants to highly hydrolysed formula after exclusive breast-feeding [37] or delaying the introduction of gluten-containing foods in infancy [38], have not provided any disease protection. The same holds true with oral antigens [39] and nicotinamide, a form of vitamin B3 suggested to protect beta-cells from destruction [40]. However, one observation from children with HLA-conferred disease susceptibility implies that early oral exposure to probiotics may decrease the risk of islet autoimmunity [41]. The protective effect was restricted to children carrying the high-risk HLA DR3/4 genotype. Recently, two intervention studies aimed at preservation of endogenous insulin secretion have been launched: one with L. rhamnosus GG and B. lactis Bb12 in children with newly diagnosed T1D, and one pilot study with prebiotic oligofructose-enriched inulin in children with a T1D duration of at least one year [42,43].
2.3. Intestinal microbiota matures early in life and affects the incidence of immune-mediated diseases
The human microbiota maturates early in life. After being born from nearly sterile conditions, children encounter a lot of microbes and environmental antigens before reaching their third year of life, by which the intestinal microbiota has reached its adult-like composition and relative stability. From early on, the main modifiers of the microbial composition are the route of delivery, early nutrition (especially breast-feeding and introduction of supplementary feeding), use of antibiotics, and the microbial exposure and the hygiene level of the immediate environment [44,45]. Considering T1D, significant changes occur in the gut microbiota before any systemic signs of islet autoimmunity. These changes occur early in life and have a far-reaching effect on the development of the immune system [12,30,46,47]. In rodents, progression to autoimmune diabetes associates with altered composition and diversity of the intestinal microbial colonies, reduced abundance of Firmicutes in relation to Bacteroidetes, and decrease in butyrate-producing bacteria subgroups. This dysbiosis coincide with increased intestinal permeability, translocation of microbial materials through the epithelium, and increased and aberrant presentation of foreign and self-antigens. These alterations activate proinflammatory pathways in the gut, in the regional lymph nodes, and in the pancreas (Supplementary Table 1). In humans developing T1D, two comparable findings have been reported regardless of geographical location: the proinflammatory environment in the gut and the combination of an increased relative abundance of Bacteroidetes together with decreased levels of Firmicutes (Table 2). More specifically, in a recent study where young children with HLA-conferred disease susceptibility and signs of islet autoimmunity were compared to matched controls, the controls had higher levels of Lactobacillus rhamnosus and Bifidobacterium dentium, while children with autoimmunity had higher abundance of the Streptococcus group mitis/oralis/pneumoniae species. When children progressing to T1D were compared to non-progressors, the latter had higher levels of Streptococcus thermophilus and Lactococcus lactis, whereas the progressors had increased levels of Bifidobacterium pseudocatenulatum, Roseburia hominis, and Alistipes shahii species. The intestinal microbiomes of the control children had also more genes related to fermentation and to biosynthesis of short-chain fatty acids [31]. Other recent analyses on the host-microbiome interactions have also revealed that factors promoting the host's mucosal barrier, adhesion of the microvilli, and exocrine function of the pancreas are depleted in T1D patients [21,25]. Some seemingly contradictory findings observed in man (Table 2), may be at least partly explained by non-optimal selection of controls, or by the differences in sample collection and handling as well as in sequencing methods.
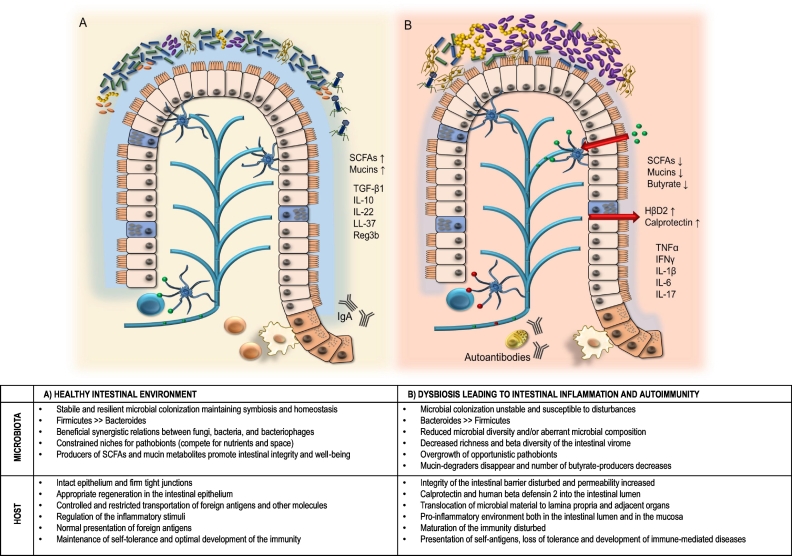
It appears that in man, a prolonged or repetitive deviation from the optimal microbial homeostasis (dysbiosis) may lead to loss of self-tolerance and systemic spreading of the proinflammatory signals and effector cells. Thus far it has been suggested that a health-promoting intestinal microbiota (i) has a certain level of microbial abundance, the optimal quantity of which depends on the functional net effects of the co-occurring microbial networks, (ii) has a composition that is resilient to transient perturbations, such as antibiotic treatment or gastrointestinal infections, (iii) provides the host with protection against pathogen invasions, and (iv) provides the host and the co-occurring commensal microbes with nutritional metabolites they are otherwise unable to obtain, e.g. certain vitamins, degraded complex polysaccharides, butyrate, and mucin metabolites (Fig. 1) [13,48].
Fig. 1.
Healthy intestinal environment (A) compared to dysbiosis (B) that leads to intestinal inflammation and emerging of autoimmunity.
Abbreviations: SCFA, short-chain fatty acids; TGF-β, transforming growth factor beta; IL, interleukin; LL-37,cathelicidin-related antimicrobial peptide 37; Ref3b, regenerating islet-derived protein 3-beta; HβD2, human beta defensin-2 (HβD2); TNFα , tumor necrosis factor alfa; IFNγ, interferon gamma.
Symbols:  dendritic cell;
dendritic cell;  T cell;
T cell;  B cell;
B cell;  plasma cell;
plasma cell;  Firmicutes;
Firmicutes;  Bacteroides;
Bacteroides;  opportunistic pathobionts;
opportunistic pathobionts;  mucin metabolisers like Akkermansia muciniphila;
mucin metabolisers like Akkermansia muciniphila;  fungi like Candida albicans;
fungi like Candida albicans;  Bacteriophage;
Bacteriophage;  foreign antigens;
foreign antigens;  self antigens.
self antigens.
3. Intestinal virome
3.1. Virus and T1D: From infections to virome
Viral infections have been associated with diabetes since 1864, when mumps was for the first time implicated as a cause of diabetes [49]. In the 1920's, the observed seasonal variation in the acute diabetes onset created the hypothesis that diabetes might have an infectious origin [50,51]. The technical developments in the next three decades rendered further findings possible: (i) viral infections can induce diabetes in mice [[52], [53], [54]], (ii) some patients with newly-diagnosed insulin-dependent diabetes have neutralizing Coxsackievirus B4 antibodies [55], and (iii) Coxsackievirus B4 isolated from a patients who died at the diagnosis of diabetes caused pancreatic islet inflammation and beta cell death when inoculated into mice [56].
Thereafter, the pathomechanisms behind the virus-induced insulitis and beta-cell autoimmunity leading to T1D have been intensively studied and discussed [e.g. [57], [58]]. In addition to Coxsackie B4, other enteroviruses and related viruses have been shown to induce diabetes in animals. In humans, enterovirus PCR-positivity has been correlated with subsequent development of T1D-associated autioantibodies in at-risk individuals, and post-mortem examinations have shown that unlike controls, patients with newly diagnosed T1D have beta-cell tropic destructive Coxsackie viruses in their pancreas [[59], [60], [61], [62]]. The latest advancement in the field is the endeavour to develop a vaccine for prevention of T1D by inducing Coxsackievirus B immunity [63].
Studies on associations between viral infections and T1D have underlined the highly variable nature of the viruses, e.g. in some mice models inoculation with assumedly diabetogenic variants of encephalomyocarditis virus or Coxsackie B caused paradoxical protection against insulitis and diabetes [64,65]. Subsequently, it has been suggested that certain latent infections might bring immune benefits for the host and thus represent a part of the host's beneficial virome [66,67].
3.2. Early life dynamics of the intestinal virome in man
Lim et al. have described the dynamics of the early human intestinal virome in eight twins followed from birth until the age of two years [68]. For all virus groups, interpersonal variations were significantly lower within the twin pairs than between different pairs of twins. The main findings were that in man, populations of eukaryotic viruses have their nadir at the birth of the child, after which the abundance and the diversity of both RNA and DNA viruses increase by age. Entero-, parecho-, tombamo- and sapoviruses were the most common eukaryotic RNA viruses, while anelloviruses were the most common eukaryotic DNA viruses. Dynamics of the anelloviruses seemed to reflect the host's immunocompetence: they were rarely seen in breastfed infants before the age of 3 months but peaked at the age of 6–12 months, when the immune protection provided by the mother faded, and the infant's own immunity gradually prevailed.
Bacteriophages, viruses that infect bacteria, were observed in all samples from these twins and comprised the dominant viral component of the normal intestinal virome. Contrary to eukaryotic RNA and DNA viruses, richness and diversity of the bacteriophages peaked at birth and decreased therafter. Among the bacteriophages, Caudovirales (Siphoviridae, Inoviridae, Myoviridae and Podoviridae families) and Microviridae families were the most abundant ones. The abundance of the two groups of bacteriophages showed an inverse correlation, and the balance of the bacteriophage populations shifted towards Microviridae dominance by the age of 2 years. The otherwise commonly observed human intestinal bacteriophage, CrAssphage, appeared only in one sample taken at the age of 2 years, indicating that these bacteriophages peak at a later age. Predator-prey relationships between bacteriophages and their targets showed clear age-dependent trajectories: after high abundance of bacteriophages and low number of intestinal bacteria at birth, a decrease in Caudovirales and a shift towards Microviridae dominance allowed a physiological increase in the abundance and diversity of the intestinal bacterial colonies by the age of 2 years [68].
3.3. Human intestinal virome and islet autoimmunity
Thus far, there are few clinical studies exploring the role of the intestinal virome in the development of T1D in man [[69], [70], [71]]. In the first report, stool samples from 19 case children with early advanced islet autoimmunity and subsequent progression to clinical T1D, and their tightly matched controls, were analysed with next-generation sequencing. The detected human viruses were verified by using real-time PCR [69]. Samples in focus were gathered 3, 6, and 9 months before the first signs of islet autoimmunity. No dramatic changes in the intestinal virome preceded the appearance of the T1D-associated autoantibodies and no associations between islet autoimmunity and any of the detected human viruses were observed. In the second report from the same DIPP Study cohort, associations between islet autoimmunity and the intestinal bacteriome (16S rDNA profiles) and virome (both DNA and RNA viruses) were assessed [70]. Compared to controls, four bacterial operational taxonomic units were less abundant in children who progressed to islet autoimmunity. The concurrent quantitative relation between bacteriophage CrAssphage and prevalent species of the Bacteroides genus suggested that CrAssphage might possibly modify the intestinal bacteriome towards dysbiosis characteristic of islet autoimmunity [70].
In another small longitudinal study on young Finns and Estonians, comprising 11 children with T1D-associated autoantibodies and 22 controls, intestinal viromes of the controls had higher diversity and higher frequency of Circoviridae-related sequences, when compared to cases [71]. Human enterovirus, kobuvirus, parechovirus, parvovirus, and rotavirus sequences were frequently detected in the samples, but were not associated with islet autoimmunity. After adjusting for age, specific T1D-associated viral bacteriophage contigs could be identified. T1D-associated contigs were statistically linked to specific components of the bacterial microbiome, thus indicating an interplay between the two kingdoms. Results from this study confirmed that certain viral signatures, remarkably consistent across individuals, reflect normal intestinal development, and that the observed age-discriminative contigs are predictive of the developmental changes occurring in the gut microbiome [71]. The partly contradictory findings in these three early studies may, again, be explained by differences in sampling and handling methods, sequencing techniques, or in the identification of the viral sequences. Future larger studies on the role of the intestinal virome in the development of islet autoimmunity will show whether T1D-associated alterations in the intestinal bacterial microbiome are independent phenomena, or a result from earlier changes in the intestinal virome. As bacteriophages can modulate bacterial abundance and colonization, changes in their abundance and diversity might well be able to disturb the intestinal homeostasis and lead to bacterial dysbiosis. On the other hand, there are already indications that certain eukaryotic viruses modify mucosal immunity similarly to commensal bacteria, thus supporting the intestinal homeostasis [72].
4. Intestinal fungal community (mycobiota)
4.1. Mycobiota is an elementary part of the human microbiota
Recent studies of the intestinal microbiome in relation to T1D have largely focused on the role of alterations in the bacterial community. The human intestine, however, harbours a diverse range of bacteria, fungi, viruses, and protista [73,74]. Mycobiota is an important part of the human microbiota, but its role in various diseases is scarcely known. As all human barrier surfaces, including mucosa, are colonized with fungi, alterations in fungal communities may have profound effects on human health. The intestinal mycobiome may modulate the composition and functions of the microbiota either directly by interacting with microbial organisms or indirectly by modulating the immune system of the host [75]. The development of culture-independent methods for the identification of the fungi has increased the knowledge of fungal colonization in humans and revealed associations between organ-specific diseases and the mycobiome. The modern methods for identifying fungal communities is based on sequencing the 18 s and 28 s sub-units of ribosomal DNA or so called internal transcribed spacer regions (ITS) [76].
4.2. Intestinal mycobiota in healthy humans
Several fungal species have been identified in the gastrointestinal tract of healthy humans, representing approximately 0.1% of the intestinal microbiota [77,78]. Stability of the intestinal mycobiota appears limited, as few fungal species have constantly been observed across different studies. Within the 36 studies analysed for a recent review, only 15 of the identified 267 fungal species were detected in more than five studies [77]. Fungal species most commonly detected belonged to the following genera: Candida, Saccharomyces, Penicillium, Aspergillus, Cryptococcus, Malassezia, Cladosporium, Galactomyces, Debaryomyces, and Trichosporon. The majority (~75%) of the fungal species were detected only in one study [77].
4.3. Disease associations of intestinal fungi
Even though it seems plausible that mycobiota plays an active role in the maintenance of the intestinal integrity, data on associations between mycobiota and human diseases is scarce. The main findings thus far relate to Saccharomyces cerevisiae and Candida species. Overgrowth of Candida spp. has been observed in humans with T1D or inflammatory bowel diseases. Poor glycaemic control may promote fungal overgrowth, and patients with established diabetes are more susceptible to have frequent and prolonged fungal infections [79]. The same holds true for immune compromised individuals [80,81]. In Crohn's disease, increased colonization with Candida species, elevated levels of antifungal-antibodies against Saccharomyces cerevisiae (ASCA Ig), and decreased abundance of Saccharomyces cerevisiae in the intestinal mycobiome have been observed [82]. Increased Candida colonization in patients with inflammatory bowel disease (IBD) may arise from alterations in the patients' immune system and from the frequent use of anti-inflammatory medications and antibiotics [83]. In addition to patients with IBD or autism spectrum disorders, overgrowth of Candida spp., especially that of C. albicans, has been observed in children with clinical T1D [11,[83], [84], [85]]. The idea that intestinal mycobiota has a modifying effect on extra-intestinal immunity has been supported by findings from a mouse model of allergy in which antibiotic-induced intestinal overgrowth of Candida promoted airway inflammation [86,87]. Mycobiota may also play its own important role in promoting the well-being of the mucosal barrier, especially when the bacterial microbiome is disturbed [88,89].
4.4. Mycobiome-bacteriome interactions
Inter-kingdom crosstalk between bacteria and fungi has been observed in patients with Crohn's disease, among whom fungal genera correlated with several bacterial taxa [90,91]. The potency of the mycobiota to regulate the bacterial microbiota is emphasised by findings showing that after antibiotic treatment, the restoration of the bacterial compartment is considerably influenced by colonization with C. albicans [91]. Animals treated with cefoperazone and subsequently fed with C. albicans had increased level of yeasts accompanied with other alterations in their intestinal bacterial microbiota [92]. In a mouse model of liver injury, administration of Saccharomyces cerevisiae var. boulardii changed the composition of the intestinal bacterial compartment by increasing the relative abundance of Bacteroidetes and decreasing the Firmicutes [93]. The observation that fungal and bacterial communities interact with each other has been established, but the detailed crosstalk networks and their relevance regarding the health of the human host are yet to be more thoroughly characterized.
5. Microbiota in other sites
In addition to the gut, microbes colonize a series of other mucosal and epithelial barriers, such as the oral cavity, airways, skin, and genitalia. Data on the immunomodulatory role of the microbiota residing in other compartments than the intestine is limited. Hu et al. reported that in the NOD mice, specific islet autoimmunity related changes in the intestinal microbiota were predictive for future diabetes, and that changes in the microbiota were more obvious in the intestine than other sites [27]. A Dutch study compared the oral and the intestinal microbiota between adult patients with established T1D and healthy controls [94]. Marked differences were observed in the oral microbiota between patients and control subjects. In the oral microbiota, the abundance of the genera Streptococcus, Rothia, and Actinomyces were higher in cases, among whom there was also a decreased abundance of butyrate producing intestinal bacteria.
Microbial communities residing in other niches than the intestine may have a significant role in other immune-mediated diseases, such as anelloviruses in the development of asthma [95] or skin microbiota in the management of atopic dermatitis [96], but regarding the development of islet autoimmunity, gut with its microbiota seems to be the major early modifier of the human immune system.
6. Crosstalk between microbiota and the host
Data on microbiota and its modulatory effects on the host has expanded rapidly during the last five years and will be discussed elsewhere. However, to summarize the big picture, current discussions circle around three main categories: the known known, the known unknown, and the unknown unknown. The first category comprise data on the crosstalk between microbiota and various organs and tissues, for example brain [97], adipose tissue and gut [98], muscles [99], and liver [100]. Until future studies provide further knowledge, the unknown can only be deduced by exclusion. For example, we are still lacking data on the importance of the unidentified reads commonly detected in the microbiota studies; the interactions between all molecules in a particular cell (interactomes) and in wider systems of interacting molecules and cells (meta-interactomes); the crosstalk between various kingdoms of the microbiota, and details on communication networks between various organs and their microbiota.
7. Dysbiosis and diabetes –contemporary or causal phenomena?
Even though reports on the T1D-related changes in the gut microbiota are accumulating, proving the causality has remained challenging. Causality requires that when all possible confounders have been controlled for, ceteris paribus (other things being equal), change in one variable should lead to change in another variable in a repeatable and generalizable way [101]. As long as no randomized, controlled, placebo-controlled intervention studies showing long-term changes in the intestinal microbial colonization and in the immunological maturation that lead repeatedly to comparable changes in the T1D-associated outcomes have been performed in man, causality between dysbiosis and T1D remains open.
8. Hypothesis and future directions
Based on current knowledge, we hypothesize that intestinal microbiota may contribute to the development of T1D via a two-phased process. The first phase of the process starts at birth and ends with the appearance of the first T1D-associated autoantibodies. During that phase, a successful training of the developing immune system is required in order to establish self-tolerance and to control inflammatory responses. If during that phase the gut microbiome tilts towards disproportion between the abundance of Bacteroides, Bifidobacteria, and Eschericia coli, the maturation of the immune system becomes distorted and susceptibility to immune-mediated diseases increases. The second phase from seroconversion to overt T1D seems to be characterized by a reduced microbial diversity and a proinflammatory intestinal dysbiosis. The mechanisms behind the spreading of the relatively local intestinal inflammation towards extra-intestinal autoimmunity remains to be explored in the future.
The challenge for the near future is to identify the causal relationships between intestinal microbiota and T1D in man. This will require extensive longitudinal studies with optimal methodology for collecting, processing and storage of the stool samples. In addition, standardized and reproducible methods for DNA and RNA extraction and sequencing, for stool transcriptomics, metabolomics and metaproteomics, and for bioinformatics are needed. We are only at the beginning of learning about the role of the intestinal microbiome in the development of T1D and other immune-mediated diseases. Further efforts should be directed to studies on the role of the intestinal mycobiota and virome, their interplay, and their interaction with the host and other microbes.
9. Search strategy and selection criteria
Data for this review were identified through PubMed and references from relevant articles using the search terms “microbiome”, “microbiota”, “mycobiota”, “virome”,” type 1 diabetes”, and” dysbiosis”. Articles between 2014 and 2018 were included in the initial analysis, with the exceptions of mycobiota and other topics for which recent data were scarcely available.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Acknowledgements
The authors' research that is relevant for this review is and has been supported by the following grants: Academy of Finland (Centre of Excellence in Molecular Systems Immunology and Physiology Research 2012-2017, Decision No. 525 250114), Novo Nordisk Foundation, Sigrid Juselius Foundation, Finland, Finska Läkaresällskapet, and Medicinska understödsföreningen Liv och Hälsa.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.06.031.
Appendix A. Supplementary data
Supplementary material
References
- 1.Atkinson M.A., Eisenbarth G.S., Michels A.W. Type 1 diabetes. Lancet. 2014;383(9911):69–82. doi: 10.1016/S0140-6736(13)60591-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Knip M., Luopajärvi K., Härkönen T. Early life origin of type 1 diabetes. Semin Immunopathol. 2017;39(6):653–667. doi: 10.1007/s00281-017-0665-6. [DOI] [PubMed] [Google Scholar]
- 3.Harjutsalo V., Sund R., Knip M., Groop P.H. Incidence of type 1 diabetes in Finland. JAMA. 2013;310(4):427–428. doi: 10.1001/jama.2013.8399. [DOI] [PubMed] [Google Scholar]
- 4.Yatsunenko T., Rey F.E., Manary M.J., Trehan I., Dominguez-Bello M.G., Contreras M. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yassour M., Vatanen T., Siljander H., Hämäläinen A.-M., Härkönen T., Ryhänen S.J. Natural history of the infant gut microbiome and impact of antibiotic treatments on bacterial strain diversity and stability. Sci Transl Med. 2016;8(343):343ra81. doi: 10.1126/scitranslmed.aad0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belkaid Y., Hand T.W. Role of the microbiota in immunity and inflammation. Cell. 2014;157(1):121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knip M., Siljander H. The role of the intestinal microbiota in type 1 diabetes mellitus. Nat Rev Endocrinol. 2016;12(3):154–167. doi: 10.1038/nrendo.2015.218. [DOI] [PubMed] [Google Scholar]
- 8.Suzuki T., Yamada T., Takao T., Fujimura T., Kawamura E., Shimizu Z.M. Diabetogenic effects of lymphocyte transfusion on the NOD or NOD nude mouse. In: Rygaard J., Sprang-Thomsen M., editors. Immune-deficient Animals in Biomedical Research. Karger; Basel: 1987. [Google Scholar]
- 9.Mejía-León M.E., Petrosino J.F., Ajami N.J., Domínguez-Bello M.G., de la Barca A.M. Fecal microbiota imbalance in Mexican children with type 1 diabetes. Sci Rep. 2014;4:3814. doi: 10.1038/srep03814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kemppainen K.M., Ardissone A.N., Davis-Richardson A.G., Fagen J.R., Gano K.A. Early childhood gut microbiomes show strong geographic differences among subjects at high risk for type 1 diabetes. Diabetes Care. 2015;38(2):329–332. doi: 10.2337/dc14-0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soyucen E., Gulcan A., Aktuglu-Zeybek A.C., Onal H., Kiykim E., Aydin A. Differences in the gut microbiota of healthy children and those with type 1 diabetes. Pediatr Int. 2014;56(3):336–343. doi: 10.1111/ped.12243. [DOI] [PubMed] [Google Scholar]
- 12.Kostic A.D., Gevers D., Siljander H., Vatanen T., Hyötyläinen T., Hämäläinen A.M. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe. 2015;17(2):260–273. doi: 10.1016/j.chom.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Endesfelder D., Engel M., Davis-Richardson A.G., Ardissone A.N., Achenbach P., Hummel S. Towards a functional hypothesis relating anti-islet cell autoimmunity to the dietary impact on microbial communities and butyrate production. Microbiome. 2016;4:17. doi: 10.1186/s40168-016-0163-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heintz-Buschart A., May P., Laczny C.C., Lebrun L.A., Bellora C., Krishna A. Integrated multi-omics of the human gut microbiome in a case study of familial type 1 diabetes. Nat Microbiol. 2016;2:16180. doi: 10.1038/nmicrobiol.2016.180. [DOI] [PubMed] [Google Scholar]
- 15.Maffeis C., Martina A., Corradi M., Quarella S., Nori N., Torriani S. Association between intestinal permeability and faecal microbiota composition in Italian children with beta cell autoimmunity at risk for type 1 diabetes. Diabetes Metab Res Rev. 2016;32(7):700–709. doi: 10.1002/dmrr.2790. [DOI] [PubMed] [Google Scholar]
- 16.Qi C.J., Zhang Q., Yu M., Xu J.P., Zheng J., Wang T. Imbalance of fecal microbiota at newly diagnosed type 1 diabetes in Chinese children. Chin Med J (Engl) 2016;129(11):1298–1304. doi: 10.4103/0366-6999.182841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vatanen T., Kostic A.D., d'Hennezel E., Siljander H., Franzosa E.A., Yassour M. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell. 2016;165(5):842–853. doi: 10.1016/j.cell.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cinek O., Kramna L., Lin J., Oikarinen S., Kolarova K., Ilonen J. Imbalance of bacteriome profiles within the Finnish diabetes prediction and prevention study: parallel use of 16S profiling and virome sequencing in stool samples from children with islet autoimmunity and matched controls. Pediatr Diabetes. 2017;18(7):588–598. doi: 10.1111/pedi.12468. [DOI] [PubMed] [Google Scholar]
- 19.de Groot P.F., Belzer C., Aydin Ö., Levin E., Levels J.H., Aalvink S. Distinct fecal and oral microbiota composition in human type 1 diabetes, an observational study. PLoS One. 2017;12(12) doi: 10.1371/journal.pone.0188475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pellegrini S., Sordi V., Bolla A.M., Saita D., Ferrarese R., Canducci F. Duodenal mucosa of patients with type 1 diabetes shows distinctive inflammatory profile and microbiota. J Clin Endocrinol Metab. 2017;102(5):1468–1477. doi: 10.1210/jc.2016-3222. [DOI] [PubMed] [Google Scholar]
- 21.Pinto E., Anselmo M., Calha M., Bottrill A., Duarte I., Andrew P.W. The intestinal proteome of diabetic and control children is enriched with different microbial and host proteins. Microbiology. 2017;163(2):161–174. doi: 10.1099/mic.0.000412. [DOI] [PubMed] [Google Scholar]
- 22.Stewart C.J., Nelson A., Campbell M.D., Walker M., Stevenson E.J., Shaw J.A. Gut microbiota of type 1 diabetes patients with good glycaemic control and high physical fitness is similar to people without diabetes: an observational study. Diabet Med. 2017;34(1):127–134. doi: 10.1111/dme.13140. [DOI] [PubMed] [Google Scholar]
- 23.Cinek O., Kramna L., Mazankova K., Odeh R., Alassaf A., Ibekwe M.U. The bacteriome at the onset of type 1 diabetes: a study from four geographically distant African and Asian countries. Diabetes Res Clin Pract. 2018;144:51–62. doi: 10.1016/j.diabres.2018.08.010. [DOI] [PubMed] [Google Scholar]
- 24.Gao X., Huynh B.T., Guillemot D., Glaser P., Opatowski L. Inference of significant microbial interactions from longitudinal metagenomics data. Front Microbiol. 2018;9:2319. doi: 10.3389/fmicb.2018.02319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gavin P.G., Mullaney J.A., Loo D., Cao K.L., Gottlieb P.A., Hill M.M. Intestinal metaproteomics reveals host-microbiota interactions in subjects at risk for type 1 diabetes. Diabetes Care. 2018;41(10):2178–2186. doi: 10.2337/dc18-0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Higuchi B.S., Rodrigues N., Gonzaga M.I., Paiolo J.C.C., Stefanutto N., Omori W.P. Intestinal dysbiosis in autoimmune diabetes is correlated with poor glycemic control and increased interleukin-6: a pilot study. Front Immunol. 2018;9:1689. doi: 10.3389/fimmu.2018.01689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang Y., Li S.C., Hu J., Ruan H.B., Guo H.M., Zhang H.H. Gut microbiota profiling in Han Chinese with type 1 diabetes. Diabetes Res Clin Pract. 2018;141:256–263. doi: 10.1016/j.diabres.2018.04.032. [DOI] [PubMed] [Google Scholar]
- 28.Leiva-Gea I., Sánchez-Alcoholado L., Martín-Tejedor B., Castellano-Castillo D., Moreno-Indias I., Urda-Cardona A. Gut microbiota differs in composition and functionality between children with type 1 diabetes and MODY2 and healthy control subjects: a case-control study. Diabetes Care. 2018;41(11):2385–2395. doi: 10.2337/dc18-0253. [DOI] [PubMed] [Google Scholar]
- 29.Mejía-León M.E., López-Domínguez L., Aguayo-Patrón S.V., Caire-Juvera G. Calderón de la Barca AM. Dietary changes and gut dysbiosis in children with type 1 diabetes. J Am Coll Nutr. 2018;37(6):501–507. doi: 10.1080/07315724.2018.1444519. [DOI] [PubMed] [Google Scholar]
- 30.Stewart C.J., Ajami N.J., O'Brien J.L., Hutchinson D.S., Smith D.P., Wong M.C. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature. 2018;562(7728):583–588. doi: 10.1038/s41586-018-0617-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vatanen T., Franzosa E.A., Schwager R., Tripathi S., Arthur T.D., Vehik K. The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature. 2018;562(7728):589–594. doi: 10.1038/s41586-018-0620-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mullen Y. Development of the nonobese diabetic mouse and contribution of animal models for understanding type 1 diabetes. Pancreas. 2017;46(4):455–466. doi: 10.1097/MPA.0000000000000828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.International Diabetes Federation . 8th ed. 2017. IDF Diabetes Atlas.https://diabetesatlas.org/ Available from: [Google Scholar]
- 34.Mordes J.P., Bortell R., Blankenhorn E.P., Rossini A.A., Greiner D.L. Rat models of type 1 diabetes: genetics, environment, and autoimmunity. ILAR J. 2004;45(3):278–291. doi: 10.1093/ilar.45.3.278. [DOI] [PubMed] [Google Scholar]
- 35.Bonder M.J., Kurilshikov A., Tigchelaar E.F., Mujagic Z., Imhann F., Vila A.V. The effect of host genetics on the gut microbiome. Nat Genet. 2016;48(11):1407–1412. doi: 10.1038/ng.3663. [DOI] [PubMed] [Google Scholar]
- 36.Alkanani A.K., Hara N., Gottlieb P.A., Ir D., Robertson C.E. Alterations in intestinal microbiota correlate with susceptibility to type 1 diabetes. Diabetes. 2015;64(10):3510–3520. doi: 10.2337/db14-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knip M., Åkerblom H.K., Al Taji E., Becker D., Bruining J., Castano L. Effect of hydrolyzed infant formula vs conventional formula on risk of type 1 diabetes: the TRIGR randomized clinical trial. JAMA. 2018;319(1):38–48. doi: 10.1001/jama.2017.19826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beyerlein A., Chmiel R., Hummel S., Winkler C., Bonifacio E., Ziegler A.G. Timing of gluten introduction and islet autoimmunity in young children: updated results from the BABYDIET study. Diabetes Care. 2014;37(9):e194–e195. doi: 10.2337/dc14-1208. [DOI] [PubMed] [Google Scholar]
- 39.Michels A.W., Gottlieb P.A. Learning from past failures of oral insulin trials. Diabetes. 2018;67(7):1211–1215. doi: 10.2337/dbi17-0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hedman M., Ludvigsson J., Faresjö M.K. Nicotinamide reduces high secretion of IFN-gamma in high-risk relatives even though it does not prevent type 1 diabetes. J Interferon Cytokine Res. 2006;26(4):207–213. doi: 10.1089/jir.2006.26.207. [DOI] [PubMed] [Google Scholar]
- 41.Uusitalo U., Liu X., Yang J., Aronsson C.A., Hummel S., Butterworth M. Association of early exposure of probiotics and islet autoimmunity in the TEDDY study. JAMA Pediatr. 2016;170(1):20–28. doi: 10.1001/jamapediatrics.2015.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Groele L., Szajewska H., Szypowska A. Effects of lactobacillus rhamnosus GG and Bifidobacterium lactis Bb12 on beta-cell function in children with newly diagnosed type 1 diabetes: protocol of a randomised controlled trial. BMJ Open. 2017;7(10) doi: 10.1136/bmjopen-2017-017178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ho J., Reimer R.A., Doulla M., Huang C. Effect of prebiotic intake on gut microbiota, intestinal permeability and glycemic control in children with type 1 diabetes: study protocol for a randomized controlled trial. Trials. 2016;17(1):347. doi: 10.1186/s13063-016-1486-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Milani C., Duranti S., Bottacini F., Casey E., Turroni F., Mahony J. The first microbial colonizers of the human gut: Composition, activities, and health implications of the infant gut microbiota. Microbiol Mol Biol Rev. 2017;81(4) doi: 10.1128/MMBR.00036-17. (pii:e00036–17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ding T., Schloss P.D. Dynamics and associations of microbial community types across the human body. Nature. 2014;509(7500):357–360. doi: 10.1038/nature13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Opazo M.C., Ortega-Rocha E.M., Coronado-Arrázola I., Bonifaz L.C., Boudin H., Neunlist M. Intestinal microbiota influences non-intestinal related autoimmune diseases. Front Microbiol. 2018;9:432. doi: 10.3389/fmicb.2018.00432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kosiewicz M.M., Zirnheld A.L., Alard P. Gut microbiota, immunity, and disease: a complex relationship. Front Microbiol. 2011;2:180. doi: 10.3389/fmicb.2011.00180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomas S., Izard J., Walsh E., Batich K., Chongsathidkiet P., Clarke G. The host microbiome regulates and maintains human health: a primer and perspective for non-microbiologists. Cancer Res. 2017;77(8):1783–1812. doi: 10.1158/0008-5472.CAN-16-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gale E.A. The discovery of type 1 diabetes. Diabetes. 2001;50(2):217–226. doi: 10.2337/diabetes.50.2.217. [DOI] [PubMed] [Google Scholar]
- 50.Adams S.F. The seasonal variation in the onset of acute diabetes: the age and sex factors in 1000 patients. Arch Intern Med. 1926;37:861–864. [Google Scholar]
- 51.Gundersen E. Is diabetes of infectious origin? J Infect Dis. 1927;41:197–202. [Google Scholar]
- 52.Pappenheimer A.M., Kunz L.J., Richardson S. Passage of Coxsackie virus (Connecticut-5 strain) in adult mice with production of pancreatic disease. J Exp Med. 1951;94:45–64. doi: 10.1084/jem.94.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Craighead J., McLane M.F. Diabetes mellitus: induction in mice by encephalomyelitis virus. Science. 1968;162:913–914. doi: 10.1126/science.162.3856.913. [DOI] [PubMed] [Google Scholar]
- 54.Coleman T.J., Gamble D.R., Taylor K.W. Diabetes in mice after Coxsackie B4 virus infection. Br Med J. 1973;3:25–27. doi: 10.1136/bmj.3.5870.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gamble D.R., Kinsley M.L., FitzGerald M.G., Bolton R., Taylor K.W. Viral antibodies in diabetes mellitus. Br Med J. 1969;3:627–630. doi: 10.1136/bmj.3.5671.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yoon J.W., Austin M., Onodera T., Notkins A.L. Isolation of a virus from the pancreas of a child with diabetic ketoacidosis. N Engl J Med. 1979;300:1173–1179. doi: 10.1056/NEJM197905243002102. [DOI] [PubMed] [Google Scholar]
- 57.Horwitz M.S., Bradley L.M., Harbertson J., Krahl T., Lee J., Sarvetnick N. Diabetes induced by Coxsackie virus: initiation by bystander damage and not molecular mimicry. Nat Med. 1998;4(7):781–785. doi: 10.1038/nm0798-781. [DOI] [PubMed] [Google Scholar]
- 58.Filippi C.M., von Herrath M.G. Viral trigger for type 1 diabetes: pros and cons. Diabetes. 2008;57:2863–2871. doi: 10.2337/db07-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stene L.C., Oikarinen S., Hyöty H., Barriga K.J., Norris J.M., Klingensmith G. Enterovirus infection and progression from islet autoimmunity to type 1 diabetes: the diabetes and autoimmunity study in the young (DAISY) Diabetes. 2010;59:3174–3180. doi: 10.2337/db10-0866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sioofy-Khojine A.B., Lehtonen J., Nurminen N., Laitinen O.H., Oikarinen S., Huhtala H. Coxsackievirus B1 infections are associated with the initiation of insulin-driven autoimmunity that progresses to type 1 diabetes. Diabetologia. 2018;61(5):1193–1202. doi: 10.1007/s00125-018-4561-y. [DOI] [PubMed] [Google Scholar]
- 61.Sioofy-Khojine A.B., Oikarinen S., Honkanen H., Huhtala H., Lehtonen J.P., Briese T. TEDDY Study Group. Molecular epidemiology of enteroviruses in young children at increased risk of type 1 diabetes. PLoS One. 2018;13(9):e0201959. doi: 10.1371/journal.pone.0201959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Richardson S.J., Willcox A., Bone A.J., Foulis A.K., Morgan N.G. The prevalence of enteroviral capsid protein vp1 immunostaining in pancreatic islets in human type 1 diabetes. Diabetologia. 2009;52(6):1143–1151. doi: 10.1007/s00125-009-1276-0. [DOI] [PubMed] [Google Scholar]
- 63.Hyöty H., Leon F., Knip M. Developing a vaccine for type 1 diabetes by targeting coxsackievirus B. Expert Rev Vaccines. 2018;17(12):1071–1083. doi: 10.1080/14760584.2018.1548281. [DOI] [PubMed] [Google Scholar]
- 64.Hermitte L., Vialettes B., Naquet P., Atlan C., Payan M.J., Vague P. Paradoxical lessening of autoimmune processes in non-obese diabetic mice after infection with the diabetogenic variant of encephalomyocarditis virus. Eur J Immunol. 1990;20:1297–1303. doi: 10.1002/eji.1830200615. [DOI] [PubMed] [Google Scholar]
- 65.Tracy S., Drescher K.M., Chapman N.M., Kim K.S., Carson S.D., Pirruccello S. Toward testing the hypothesis that group B coxsackieviruses (CVB) trigger insulin-dependent diabetes: inoculating nonobese diabetic mice with CVB markedly lowers diabetes incidence. J Virol. 2002;76:12097–12111. doi: 10.1128/JVI.76.23.12097-12111.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Barton E.S., White D.W., Cathelyn J.S., Brett-McClellan K.A., Engle M., Diamond M.S. Herpesvirus latency confers symbiotic protection from bacterial infection. Nature. 2007;447(7142):326–329. doi: 10.1038/nature05762. [DOI] [PubMed] [Google Scholar]
- 67.Virgin H.W. The virome in mammalian physiology and disease. Cell. 2014;157:142–150. doi: 10.1016/j.cell.2014.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lim E., Zhou Y., Zhao G., Bauer I.K., Droit L., Ndao I.M. Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat Med. 2015;21(10):1228–1234. doi: 10.1038/nm.3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kramná L., Kolářová K., Oikarinen S., Pursiheimo J.-P., Ilonen J., Simell O. Gut virome sequencing in children with early islet autoimmunity. Diabetes Care. 2015;38(5):930–933. doi: 10.2337/dc14-2490. [DOI] [PubMed] [Google Scholar]
- 70.Cinek O., Kramná L., Lin J., Oikarinen S., Kolarova K., Ilonen J. Imbalance of bacteriome profiles within the Finnish diabetes prediction and prevention study: parallel use of 16S profiling and virome sequencing in stool samples from children with islet autoimmunity and matched controls. Pediatr Diabetes. 2017;18(7):588–598. doi: 10.1111/pedi.12468. [DOI] [PubMed] [Google Scholar]
- 71.Zhao G., Vatanen T., Droit L., Park A., Kostic A.D., Poon T.W. Intestinal virome changes precede autoimmunity in type 1 diabetes-susceptible children. Proc Natl Acad Sci U S A. 2017;114(7):E6166–E6175. doi: 10.1073/pnas.1706359114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kernbauer E., Ding Y., Cadwell K. An enteric virus can replace the beneficial function of commensal bacteria. Nature. 2014;516:94–98. doi: 10.1038/nature13960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rajilić-Stojanović M., Smidt H., de Vos W.M. Diversity of the human gastrointestinal tract microbiota revisited. Environ Microbiol. 2007;9(9):2125–2136. doi: 10.1111/j.1462-2920.2007.01369.x. [DOI] [PubMed] [Google Scholar]
- 74.Scanlan P.D., Marchesi J.R. Micro-eukaryotic diversity of the human distal gut microbiota: qualitative assessment using culture-dependent and -independent analysis of faeces. ISME J. 2008;2(12):1183–1193. doi: 10.1038/ismej.2008.76. [DOI] [PubMed] [Google Scholar]
- 75.Oever J.T., Netea M.G. The bacteriome-mycobiome interaction and antifungal host defense. Eur J Immunol. 2014;44(11):3182–3191. doi: 10.1002/eji.201344405. [DOI] [PubMed] [Google Scholar]
- 76.Underhill D.M., Iliev I.D. The mycobiota: interactions between commensal fungi and the host immune system. Nat Rev Immunol. 2014;14(6):405–416. doi: 10.1038/nri3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hallen-Adams H.E., Suhr M.J. Fungi in the healthy human gastrointestinal tract. Virulence. 2017;8(3):352–358. doi: 10.1080/21505594.2016.1247140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Qin J., Li R., Raes J., Arumugam M., Burgdorf K.S., Manichanh C. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nowakowska D., Kurnatowska A., Stray-Pedersen B., Wilczyński J. Species distribution and influence of glycemic control on fungal infections in pregnant women with diabetes. J Infect. 2004;48(4):339–346. doi: 10.1016/j.jinf.2004.01.017. [DOI] [PubMed] [Google Scholar]
- 80.Li Q., Wang C., Tang C., He Q., Li N., Li J. Dysbiosis of gut fungal microbiota is associated with mucosal inflammation in Crohn's disease. J Clin Gastroenterol. 2014;48(6):513–523. doi: 10.1097/MCG.0000000000000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chehoud C., Albenberg L.G., Judge C., Hoffmann C., Grunberg S., Bittinger K. Fungal signature in the gut microbiota of pediatric patients with inflammatory bowel disease. Inflamm Bowel Dis. 2015;21(8):1948–1956. doi: 10.1097/MIB.0000000000000454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gosiewski T., Salamon D., Szopa M., Sroka A., Malecki M.T., Bulanda M. Quantitative evaluation of fungi of the genus Candida in the feces of adult patients with type 1 and 2 diabetes - a pilot study. Gut Pathog. 2014;6(1):43. doi: 10.1186/s13099-014-0043-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Plantinga T.S., van der Velden W.J., Ferwerda B., van Spriel A.B., Adema G., Feuth T. Early stop polymorphism in human DECTIN-1 is associated with increased candida colonization in hematopoietic stem cell transplant recipients. Clin Infect Dis. 2009;49(5):724–732. doi: 10.1086/604714. [DOI] [PubMed] [Google Scholar]
- 84.Thanyasrisung P., Kesakomol P., Pipattanagovit P., Youngnak-Piboonratanakit P., Pitiphat W., Matangkasombut O. Oral Candida carriage and immune status in Thai human immunodeficiency virus-infected individuals. J Med Microbiol. 2014;63(Pt 5):753–759. doi: 10.1099/jmm.0.069773-0. [DOI] [PubMed] [Google Scholar]
- 85.Strati F., Cavalieri D., Albanese D., De Felice C., Donati C., Hayek J. New evidences on the altered gut microbiota in autism spectrum disorders. Microbiome. 2017;5(1):24. doi: 10.1186/s40168-017-0242-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Noverr M.C., Falkowski N.R., McDonald R.A., McKenzie A.N., Huffnagle G.B. Development of allergic airway disease in mice following antibiotic therapy and fungal microbiota increase: role of host genetics, antigen, and interleukin-13. Infect Immun. 2005;73(1):30–38. doi: 10.1128/IAI.73.1.30-38.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Noverr M.C., Noggle R.M., Toews G.B., Huffnagle G.B. Role of antibiotics and fungal microbiota in driving pulmonary allergic responses. Infect Immun. 2004;72(9):4996–5003. doi: 10.1128/IAI.72.9.4996-5003.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hoarau G., Mukherjee P.K., Gower-Rousseau C., Hager C., Chandra J., Retuerto M.A. Bacteriome and mycobiome interactions underscore microbial dysbiosis in familial Crohn's disease. MBio. 2016;7(5) doi: 10.1128/mBio.01250-16. (pii: e01250–16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sokol H., Leducq V., Aschard H., Pham H.P., Jegou S., Landman C. Fungal microbiota dysbiosis in IBD. Gut. 2017;66(6):1039–1048. doi: 10.1136/gutjnl-2015-310746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Huang Y.J., Erb-Downward J.R., Dickson R.P., Curtis J.L., Huffnagle G.B., Han M.K. Understanding the role of the microbiome in chronic obstructive pulmonary disease: principles, challenges, and future directions. Transl Res. 2017;179(1):71–83. doi: 10.1016/j.trsl.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Huffnagle G.B., Noverr M.C. The emerging world of the fungal microbiome. Trends Microbiol. 2013;21(7):334–341. doi: 10.1016/j.tim.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Erb Downward J.R., Falkowski N.R., Mason K.L., Muraglia R., Huffnagle G.B. Modulation of post-antibiotic bacterial community reassembly and host response by Candida albicans. Sci Rep. 2013;3:2191. doi: 10.1038/srep02191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yu L., Zhao X.K., Cheng M.L., Yang G.Z., Wang B., Liu H.J. Saccharomyces boulardii administration changes gut microbiota and attenuates D-Galactosamine-induced liver injury. Sci Rep. 2017;7(1):1359. doi: 10.1038/s41598-017-01271-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.de Groot P.F., Belzer C., Aydin Ö., Levin E., Levels J.H., Aalvink S. Distinct fecal and oral microbiota composition in human type 1 diabetes, an observational study. PLoS One. 2017;12(12) doi: 10.1371/journal.pone.0188475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Freer G., Maggi F., Pifferi M., Di Cicco M.E., Peroni D.G., Pistello M. The Virome and its major component, Anellovirus, a convoluted system molding human immune defenses and possibly affecting the development of asthma and respiratory diseases in childhood. Front Microbiol. 2018;9:686. doi: 10.3389/fmicb.2018.00686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kim J.E., Kim H.S. Microbiome of the skin and gut in atopic dermatitis (AD): understanding the pathophysiology and finding novel management strategies. J Clin Med. 2019;8(4) doi: 10.3390/jcm8040444. (pii: E444) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bessac A., Cani P.D., Meunier E., Dietrich G., Knauf C. Inflammation and gut-brain Axis during type 2 diabetes: focus on the crosstalk between intestinal immune cells and enteric nervous system. Front Neurosci. 2018;12:725. doi: 10.3389/fnins.2018.00725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Geurts L., Neyrinck A.M., Delzenne N.M., Knauf C., Cani P.D. Gut microbiota controls adipose tissue expansion, gut barrier and glucose metabolism: novel insights into molecular targets and interventions using prebiotics. Benef Microbes. 2014;5(1):3–17. doi: 10.3920/BM2012.0065. [DOI] [PubMed] [Google Scholar]
- 99.Grosicki G.J., Fielding R.A., Lustgarten M.S. Gut microbiota contribute to age-related changes in skeletal muscle size, composition, and function: biological basis for a gut-muscle Axis. Calcif Tissue Int. 2018;102(4):433–442. doi: 10.1007/s00223-017-0345-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Delzenne N.M., Knudsen C., Beaumont M., Rodriguez J., Neyrinck A.M., Bindels L.B. Contribution of the gut microbiota to the regulation of host metabolism and energy balance: a focus on the gut-liver axis. Proc Nutr Soc. 2019:1–10. doi: 10.1017/S0029665118002756. [DOI] [PubMed] [Google Scholar]
- 101.Chambliss D.F., Schutt R.K. 5th ed. Vol. 6. Sage Publications; 2016. Making Sense of the Social World; pp. 111–138. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material