Abstract
Background
Most studies on regenerative medicine focus on cell-based therapies and transplantations. Small-molecule therapeutics, though proved effective in different medical conditions, have not been extensively investigated in regenerative research. It is known that healing potential decreases with development and developmental changes are driven by epigenetic mechanisms, which suggests epigenetic repression of regenerative capacity.
Methods
We applied zebularine, a nucleoside inhibitor of DNA methyltransferases, to stimulate the regenerative response in a model of ear pinna injury in mice.
Findings
We observed the regeneration of complex tissue that was manifested as improved ear hole repair in mice that received intraperitoneal injections of zebularine. Six weeks after injury, the mean hole area decreased by 83.2 ± 9.4% in zebularine-treated and by 43.6 ± 15.4% in control mice (p < 10−30). Combined delivery of zebularine and retinoic acid potentiated and accelerated this effect, resulting in complete ear hole closure within three weeks after injury. We found a decrease in DNA methylation and transcriptional activation of neurodevelopmental and pluripotency genes in the regenerating tissues.
Interpretation
This study is the first to demonstrate an effective induction of complex tissue regeneration in adult mammals using zebularine. We showed that the synergistic action of an epigenetic drug (zebularine) and a transcriptional activator (retinoic acid) could be effectively utilized to induce the regenerative response, thus delineating a novel pharmacological strategy for regeneration. The strategy was effective in the model of ear pinna regeneration in mice, but zebularine acts on different cell types, therefore, a similar approach can be tested in other tissues and organs.
Keywords: Regeneration; Epigenetics; Ear pinna; Mouse; Zebularine; Wound healing, regenerative medicine
Research in context.
Evidence before this study
Most research efforts on regenerative medicine are focused on transplantation and cell based therapies. Pharmacological activation of endogenous regenerative potential has not been extensively studied. Epigenetic basis of regeneration are poorly recognized but these are the epigenetic mechanisms that determine cell differentiation and dedifferentiation. High regenerative capacity characteristic of foetal and neonatal periods declines with development, which is connected with epigenetic repression of multiple genes. Therefore, epigenetic inhibitors appear as attractive tools that can be used to release the suppressed regenerative potential.
In order to investigate as to whether epigenetic derepression can activate regenerative responses, we decided to apply a simple but efficient model of ear pinna injury. In normal laboratory strains of mouse, 2- mm holes made in the ear pinna, remain for life, but pharmacological stimulation can induce ear pinna hole closure. The model of ear punch wound in mice has been used in several studies to test pro-regenerative activities of small molecules. As the epigenetic inhibitor, we selected zebularine, a DNA methyltransferase inhibitor, known for its low toxicity. To potentiate the effect of transcriptional activation, we chose retinoic acid, a regulatory molecule, reported for its role in wound healing and regeneration responses in different tissues.
Added value of this study
This study is a successful attempt to induce regeneration of complex tissue in the model of ear pinna injury in mice using an epigenetic inhibitor, zebularine. Further, we report a novel pharmacological regenerative strategy based on epigenetic derepression using zebularine combined with transcriptional activation induced with retinoic acid. The result indicates the importance of epigenetic aspects of regeneration.
Implications of all the available evidence
Zebularine treatment was proved effective to activate regeneration in the ear pinna, but it is important to translate the epigenetic therapy to other models of clinical significance. Although murine models are not perfect, they are useful tools of preclinical studies owing to the resemblance of mice and humans on genetic, biochemical and anatomical levels. Of note, animal models of injury allow the examination of not only tissue responses in the site of injury but also the reactions of immune, nervous and vascular systems. In addition, the molecular targets of zebularine and retinoic acid are ubiquitous in different species, and therefore, they are likely to act in a similar manner in mice and humans. The proposed regenerative strategy can be applied to different tissues and organs using other combinations of epigenetic inhibitors and transcriptional activators than zebularine and retinoic acid. Further research is necessary to investigate the mechanisms of pro-regenerative processes activated by zebularine.
Alt-text: Unlabelled Box
1. Introduction
At present, the main direction of regenerative medicine is towards ex vivo strategies based on stem cells and tissue engineering. Another possibility, the in vivo pharmacological stimulation of endogenous regeneration potential has not attracted much attention to date.
Regeneration potential is known to decrease with organism development. The spectacular regenerative abilities in the embryonic and neonatal stages, such as scarless skin wound healing in mammalian foetuses [1], cardiac repair in neonatal mice [2] and spinal cord regeneration following complete transection in opossum pups [3], are lost in adulthood. Development is driven by epigenetic reprogramming, while epigenetic reprogramming is critical for cells to acquire pluripotency [4]. Several observations in animal models indicate the significance of the epigenetic status for regeneration capability [1,[5], [6], [7]].
The transgenic delivery of transcription factors have been successfully applied to induce cell pluripotency, and thus activate massive epigenetic reprogramming [8]. However, small molecule epigenetic inhibitors, as e.g. those of DNA methyltransferases, are more convenient tools to modify the epigenome.
Zebularine is a cytidine analogue and DNA methyltransferase inhibitor. Similar to 5-azacitidine, zebularine inhibits DNA methyltransferases after it is incorporated into DNA during replication. Metabolic activation of zebularine consists of several steps: phosphorylation to zebularine monophosphate by uridine cytidine kinase, phosphorylation to zebularine diphosphate by nucleoside-phosphate kinase, reduction to deoxyzebularine diphosphate by ribonucleotide reductase, and finally phosphorylation to zebularine triphosphate by nucleoside-diphosphate kinase [9]. Deoxyzebularine triphosphate is a substrate in DNA synthesis. DNA methyltransferase forms a stable covalent adduct with zebularine integrated into DNA, which leads to passive demethylation during DNA replication [10]. Zebularine but not 5-azacitidine shows minimal toxicity in cell culture [9] and animal models [11,12]. No toxic effects were observed in mice treated with high doses (400 mg/kg) of zebularine for 78 consecutive days [12].
Intrinsic regeneration ability has been investigated in different animal models [3] [2]. However, testing whether the regenerative response is induced by pharmacological stimulation in such organs as the heart, spinal cord or limb would require a sophisticated experimental setup. Ear punch wound is a simple model of mammalian tissue regeneration. It is worth noting that the research on ear pinna regeneration dates back to as early as the 1950's. As mentioned by Williams-Boyce and Daniel [13], Markelova was the first to demonstrate the phenomenon in the rabbit. Varied outcomes of ear pinna injuries were observed in different mammalian species [13]. Natural inborn ability of perfect ear pinna regeneration was well characterized in the MRL/MpJ mouse (1998) [14] and in the African spiny mouse (2012) [15]. In the MRL/MpJ, an inbred laboratory strain, the regenerative phenotype was found to be a multigenic trait [16], but enhanced tissue repair can be a result of a monogenic mutation as it is in the case of the Foxn1 gene. The nude mutation in Foxn1 is known to induce a hairless phenotype as well as improved ear hole closure and skin wound healing in mice of different genetic backgrounds (2004) [17]. While in most laboratory strains, 2-mm-diameter through-and-through ear holes in ear pinnae remain for life, they close completely within 30 days in the MRL/MpJ [14]. Not only skin but also muscles, blood vessels, cartilage [14], and peripheral nerves [18] are restored; thus, the phenomenon could be regarded as an example of complex tissue and epimorphic regeneration. Further, enhanced regenerative abilities are not limited to ear pinnae but seem to extend to the whole body, as they were reported in tendons [19], joints [20], cornea [21], retina [22], digit tips [23], spinal cord [24] and heart [25] of the MRL/MpJ mouse. Due to its experimental simplicity and convenient quantitation, ear pinna injury appears to be a compelling model to test the pro-regenerative activity of chemical compounds [26,27].
As the decline in regenerative capacity with development is likely to be linked to epigenetic repression [1,[5], [6], [7],28,29], the question arises as to whether epigenetic inhibitors can restore this lost ability. Epigenetic derepression alone may be insufficient to activate silenced genes without positive induction. Therefore, we decided to examine the combined action of zebularine, an epigenetic inhibitor, and retinoic acid, a transcriptional activator, to stimulate the regenerative response. We chose zebularine for its low toxicity, retinoic acid for its established role in regeneration processes [[30], [31], [32], [33], [34]], and the ear punch wound model for its efficiency.
2. Materials and methods
2.1. Animals
The experiments on mice of the BALB/c strain were conducted at the Tri-City Academic Laboratory Animal Centre where the animals were bred and maintained. The experiments on C57BL/6 J mice were performed at the animal facility of the Nencki Institute in Warsaw. The C57BL/6 J mice were purchased from the Center for Experimental Medicine of the Medical University of Białystok, Poland. The animal study protocols were approved by the Local Ethics Committee for Animal Experimentation in Bydgoszcz (permit No. 5/2015) and the First Local Ethics Committee in Warsaw (permit No. 491/2013) for the experiments in the BALB/c and the C57BL/6 J mouse, respectively. Animal experimentation was carried out in accordance with EU directive 2010/63/EU. The experiments were performed using 2-month-old animals. Unlike female mice, male mice are more prone to aggressive behaviour, which may complicate wound healing. Therefore, most research was conducted on females but experiments on males were also carried out in order to test the effects of zebularine treatment on both sexes. Prior to tissue collection, the mice were euthanized in a CO2 chamber.
2.2. Ear pinna injury
Through-and-through holes of 2 mm in diameter were made in the mouse ear pinna using a scissor-style ear punch. The mice were randomly divided into two groups of six animals each. The treatment group received seven intraperitoneal injections of zebularine (TCI, Cat. No. Z0022) dissolved in saline (50 mg/mL, 1000 mg/kg body weight on the day of injury directly after punching and at 1, 2, 3, 4, 7, and 10 days after injury), while saline was administered to the control mice. All-trans-retinoic acid for injection (TCI, Cat. No. R0064) was dissolved in rape seed oil and administered intraperitoneally (16 mg/kg) directly after punching and at 2, 4, 7, 9, and 11 days after injury, while pure rape seed oil and saline were administered to the control mice. Lower zebularine doses and the doses of the other tested nucleosides are indicated in the legend of Fig. 2. To track healing progress, ears were photographed, and ear hole areas were measured using computer-assisted image analysis (ImageJ [35]). In the replicate experiment in C57BL/6 J mice, a biopsy punch of 2 mm in diameter was used to produce ear holes.
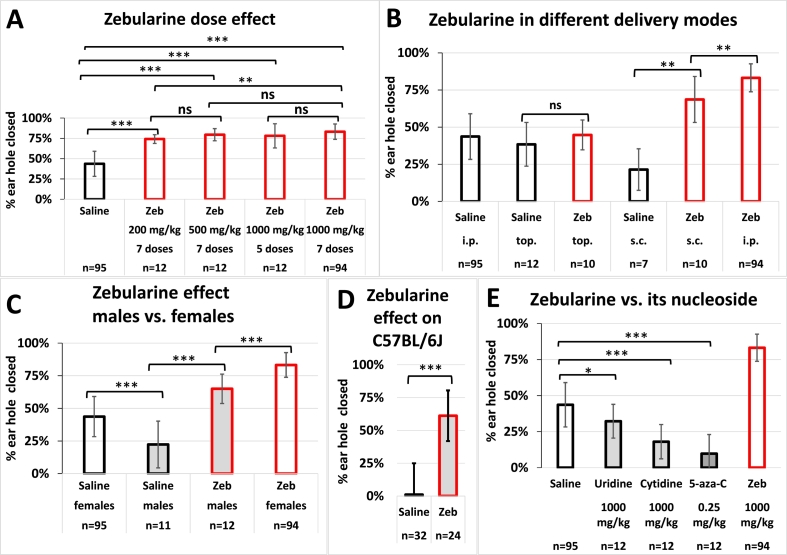
Fig. 2.
Zebularine, unlike its nucleotide analogues, promotes ear hole closure in mice at different doses and delivery modes and of different sexes and strains.(A) Similar effects of lower zebularine doses (200 and 500 vs. 1000 mg/kg) and shortened treatment periods (5 vs. 7 doses). (B) Results of topical, subcutaneous and intraperitoneal zebularine delivery (C) Zebularine effects in male and female mice. (D) Effective zebularine treatment in C57BL/6 J mice. (E) Effects of zebularine nucleoside analogues. Histograms show the mean ear hole area on day 42 post injury. Error bars represent the SD. If not indicated otherwise, the experiments were performed in BALB/c female mice that received 7 intraperitoneal injections of zebularine according to the time schedule presented in Fig. 1a. “n” refers to the number of ears from n/2 animals.
2.3. Histology
After collection, the ears were fixed on a cork with metal pins and preserved for a week in 4% formalin buffered with PBS. The tissues were then embedded in paraffin, sectioned to 5-μm thickness, and stained with Masson's trichrome or haematoxylin and Alcian blue. Image acquisition was performed with a Leica DM IL microscope at 100× magnification.
2.4. Quantification of collagen density and blood vessels
The assessment of collagen density within wound area was performed from the images of histological tissue sections stained with Masson's trichrome using colour deconvolution method for the ImageJ software [36]. Blood vessels in newly formed tissue were quantified as average number of occurrences per sq. mm. The results were presented as mean values for 7–9 photographs representing 2–3 mice.
2.5. Transcript levels
Tissue samples were preserved in RNAlater (Qiagen) and stored at −80 °C. The rings surrounding ear pinna holes were excised with a 3-mm biopsy punch. RNA was extracted using an RNeasy Kit (Qiagen). cDNA synthesis was performed in a reaction mix containing 200 ng of RNA, 100 pmoles of oligo dT20, 4 μL of 5× reaction buffer (250 mM Tris-HCl, 375 mM KCl, 15 mM MgCl2, 50 mM DTT), and 200 units of Maxima Reverse Transcriptase (ThermoScientific Bio, Cat. No. EP0742) in a final volume of 20 μL. Real-time PCR was carried out in a final volume of 10 μL containing 5 μL of FastStart Essential DNA Green Master (Roche, Cat. No. 06402712001), 1 μL of cDNA, and 0.25 μL each of forward and reverse primers (10 μM) on a LightCycler LC96 (Roche). The transcript levels were calculated using the 2-∆∆Ct method relative to Actb and Tbp (mouse templates) and ACTB, GAPDH and TBP (human templates). The primer sequences are listed in Supplemental Information, Table S1. PCR was performed in triplicate.
2.6. Global CpG methylation levels
Tissue samples were preserved in RNAlater (Qiagen) and stored at −80 °C. The rings surrounding ear pinna holes were excised with a 3-mm biopsy punch. Genomic DNA was extracted using a DNeasy Blood and Tissues Kit (Qiagen) with RNase A treatment.
CpG methylation levels were estimated using an ELISA-based assay (ZYMO, Cat. No. D5325) in 100-ng aliquots of genomic DNA according to the manufacturer's instructions.
A mass spectrometry approach (LC/MS) was applied to verify the ELISA results. Aliquots of 100 ng of genomic DNA were digested to nucleosides in a 12.5 μL total volume containing 2.5 units of DNA Degradase Plus (ZYMO, E2020) and 1.25 μL of reaction buffer at 37 °C for 2 h, followed by heat-inactivation at 70 °C for 20 min. 5-Methyl-2′-deoxycytidine (5-mC) (TCI, Cat. No. D3610) and 2′-deoxyguanosine (dG) (Sigma, Cat. No. D7145) diluted in water (1 nM) were used as references. The samples were diluted to 30 μL with water, loaded onto an Eksigent HALO C18 column (100 × 0.5 mm, 2.7 μm, 90 Å), and developed using an Eksigent MicroLC 200 Plus (Milford, MA) LC system at 40 °C. The mobile phases consisted of water (A) and acetonitrile (B), both with the addition of 0.1% formic acid. The mobile phase was delivered using isocratic elution with 0% B for 2 min, followed by gradient elution of 0–20% B for 3 min. Detection was carried out using a triple quadrupole time-of-flight mass spectrometer (ABSciex TripleTOF 5600+) operating in positive electrospray ionization (ESI) mode. The analyses were conducted by monitoring the [M + H]+ parent ion to product ion transitions (generated by loss of the deoxyribose moiety): 5-mC m/z 242.13 → 126.05 and dG m/z 268.14 → 152.05. The 5-mC content was calculated by dividing the 5-mC peak area by the dG peak area. All analyses were performed in triplicate.
2.7. Gene-specific DNA methylation
Methylation levels of selected DNA loci were estimated using a CpG methylation-dependent restriction enzyme. A 100-ng aliquot of DNA extracted as described above was treated with McrBC (New England Biolabs, Cat. No. M0272S) in a final volume of 10 μL. Two microliters of 10-fold diluted DNA was used as a template for PCR quantitation as described above in the Transcript levels section. The value r = 1-(McrBC digested DNA/input DNA) represented the DNA methylation level. The primer sequences are listed in Supplemental Information, Table S1. PCR was performed in triplicate.
2.8. Cell culture conditions
Immortalized human keratinocytes (HaCaT, (DKFZ Heidelberg, Germany) [37] and human dermal fibroblasts (46BR.1 N, Sigma-Aldrich) were grown in DMEM-HG (Sigma–Aldrich, Cat. No. D6429) supplemented with 10% foetal bovine serum (FBS Sigma-Aldrich, Cat. No. F9665), 100 units/mL penicillin, and 100 μg/mL streptomycin. The cells were routinely cultured in a humidified atmosphere with 5% CO2 at 37 °C.
2.9. Zebularine stimulation of cell cultures for RNA-seq
For zebularine stimulation, the medium was exchanged for DMEM supplemented with 2% FBS, 16 μM thymidine, and zebularine (1 μg/mL). After 48 h, medium was exchanged and cells were stimulated with zebularine for a second time. After another 48 h of incubation, cells were detached, counted, washed in PBS, centrifuged and stored as pellets at −80 °C. Three independent experiments were performed for each cell line.
2.10. XTT cell proliferation and LDH cell cytotoxicity assay
Briefly, HaCaT and 46BR.1 N cells were incubated with zebularine at the concentrations indicted in Fig. 6 for 48 h or 72 h. The XTT assay was carried out according to the manufacturer's instructions (Roche, Cat. No. 11465015001). Cell proliferation was normalized with respect to the negative control (100%). In the LDH assay, cell death was quantified by measurement of lactate dehydrogenase activity in cell supernatants according to the manufacturer's protocol (Takara, Cat. No. MK401). Cell death was normalized with respect to the negative control (0%) and 1% (v/v) Triton X-100 for the maximum LDH release indicating the maximum cytotoxicity (100%). Three independent cell culture experiments were conducted for each assay and cell line. The LDH and XTT assays were performed in triplicates.
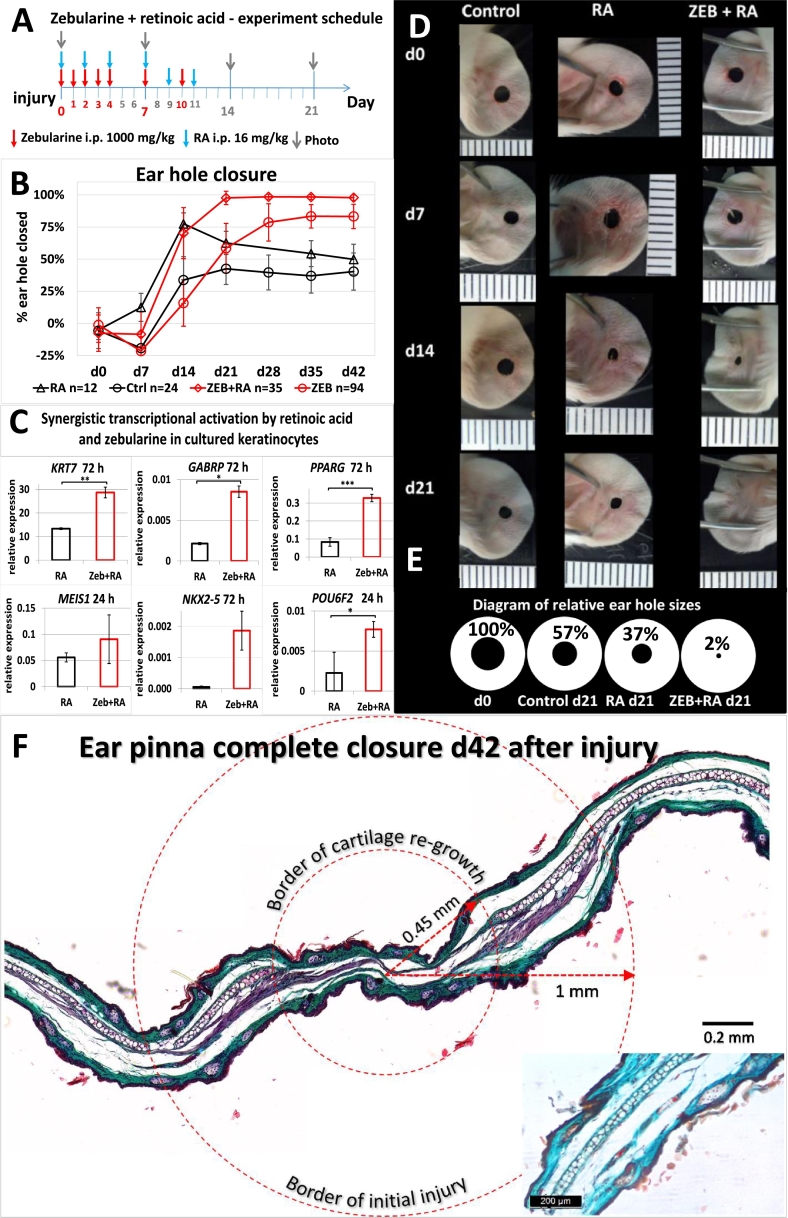
Fig. 6.
Combined intraperitoneal delivery of zebularine and retinoic acid results in accelerated and complete ear pinna hole closure in mice. (A) Experiment time schedule. (B) Ear wound closure demonstrated as the mean hole area for n ears from n/2 mice receiving zebularine and retinoic acid (ZEB + RA), retinoic acid alone (RA), zebularine alone (ZEB), and vehiculum only: the injection of saline and the injection of pure rape seed oil (Ctrl); error bars represent the SD. (C) Zebularine-mediated augmentation of retinoic acid target genes expression in cultured human keratinocytes; the results are presented as the fold change of gene expression in the keratinocytes treated with zebularine and retinoic acid (ZEB + RA) or retinoic acid alone (RA) as compared to the untreated control cells; 24 and 72 h indicate the time of incubation with RA. (D) Representative photographs showing ear holes in mice that received both zebularine and retinoic acid (ZEB + RA), retinoic acid alone (RA) or vehiculum only (Control); the scale is calibrated in mm. (E) Diagram presenting mean ear hole areas for zebularine and retinoic acid-treated mice and controls. (F) Histological analysis of ear pinna after complete hole closure in response to combined treatment with zebularine and retinoic acid. Masson's trichrome staining shows cartilage re-growth within a 1-mm radius of the initial wound and restored tissue architecture and on the day 42 after injury; the inset presents non-injured ear pinna for comparison.
2.11. Stimulation of keratinocytes with retinoic acid and zebularine
For zebularine and all-trans-Retinoic acid (RA) stimulation, the HaCaT cells were seeded in EpiLife medium (Gibco, Cat. No. MEPI500CA) medium supplemented with EDGS (EpiLife™ Defined Growth Supplement, Life Technologies, Gibco, Cat. No. S0125) and 10% FBS. After 24 h, the medium was exchanged for serum free EpiLife with EDGS and the cells were stimulated with zebularine (1 μg/mL). After another 48 h of incubation, RA (TCI, Cat. No. R0064, 1 μg/mL) was added either together with zebularine or 48 h after zebularine was applied. After another 24–h of incubation, the cells were counted, washed in PBS, centrifuged and stored as pellets at −80 °C.
2.12. Transcriptome profiling (RNA-seq)
Total RNA was isolated from cultured keratinocytes (HaCaT) and fibroblasts (46BR.1 N) using the miRNeasy Mini Kit (Qiagen) according to the manufacturer's instructions, except 1-bromo-3-chloropropane was used for extraction. RNA extracted from cell culture replicates was pooled in equal concentrations. The RIN (RNA integrity number) was determined using Eukaryote Total RNA Nano assay for Agilent 2100 Bioanalyzer. The measured RIN values were > 8.0 in the scale from 1 (most degraded profile) to 10 (intact RNA). A Ribosome Reduction Kit (Arraystar Inc., Rockville, MD, USA) and a SureSelect Strand-Specific mRNA Library Kit (Agilent, Santa Clara, CA, USA) were used according to standard procedures. The cDNA libraries were quantitated using qPCR on a Roche LightCycler LC480 with the Kapa Library Quantification Kit for Illumina (Kapa Biosystems, Woburn, MA, USA). Onboard cluster generation and 50-nt paired-end RNA sequencing were performed on the Illumina HiSeq2500 using Rapid Run v2 sequencing chemistry and flow cells as recommended by the manufacturer (Illumina Inc., San Diego, CA, USA). as a service provided by the Genomics Core of Heflin Center for Genomic Science (University of Alabama at Birmingham, U.S.A.). Paired end 50 bp sequencing runs were completed and the data was converted to the FASTQ Sanger format Illumina's with the bcl2fastq converter. Trim Galore was used to remove residual adapter sequences from the reads (https://www.bioinformatics.babraham.ac.uk). Sequencing reads were aligned to the human reference genome assembly (hg19) using TopHat [38]. Transcript assembly and estimation of the relative abundances were carried out with Cufflinks [39] with the following settings: quartile normalization method for the libraries, blind dispersion method for replicates, and biased correction using the canonical hg19 as reference. Cuffmerge and Cuffquant were then used followed by a final comparison analysis in Cuffdiff according to the workflow for Cufflinks version 2.2.0 and higher (http://cole-trapnell-lab.github.io/cufflinks/manual). The RNA-seq data has been deposited in ArrayExpress database under accession number E-MTAB-6832.
2.13. Gene ontology analyses
First, the transcripts showing no or very low expression levels determined in RNA-seq analyses (FPKM values 〈100) for both zebularine-treated and control cells were filtered out. Next, the genes displaying a minimum twofold change in expression were selected for subsequent analyses. The Gene Ontology analysis was carried out with ClueGO (version 2.3.2) using the Biological Process and Molecular Function tools [40]. The statistical significance was calculated by a two-sided hypergeometric test with the Benjamini-Hochberg correction.
2.14. Statistical analyses
Unless otherwise indicated, statistical significance was determined using the Mann-Whitney U test. Two-tailed statistics and a significance level of 5% were applied. P-values were computed using the asymptotic method with continuity correction. The statistical power values were calculated for animal experiments and listed in Supplemental Information Table S2. The analyses were carried out in XLSTAT software (Addinsoft). Statistical significance is indicated by one, two, or three asterisks indicating p < .05, p < .01, or p < .001, respectively.
3. Results
3.1. Zebularine-stimulated regenerative response in murine ear pinnae
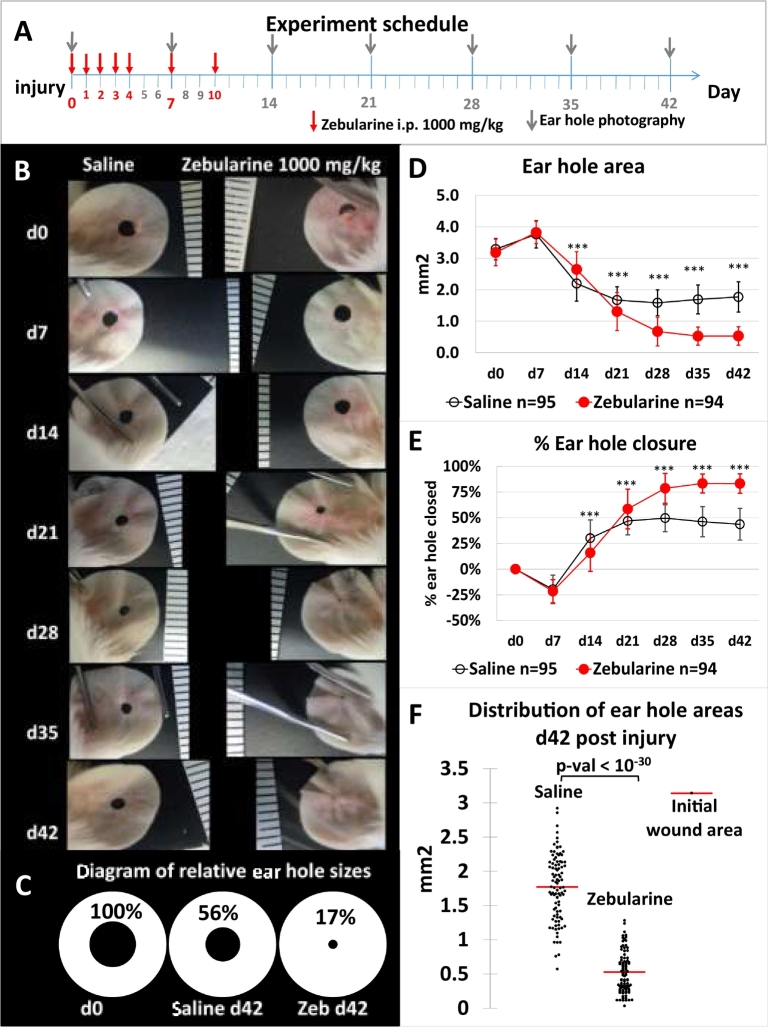
To investigate whether the pharmacological induction of epigenetic reprogramming stimulates regeneration in adult mammals, we examined the effect of zebularine, a nucleoside DNA methyltransferase inhibitor, on ear pinna wound healing in 8-week-old mice. Punches of 2 mm in diameter were made in the centres of ear auricles following intraperitoneal zebularine injections (1000 mg/kg) (Fig. 1a). Healing progress was evaluated weekly for six weeks in experimental groups, each consisting of six zebularine-treated mice and six saline-treated mice; collectively, the study included 47 treated and 48 control mice. Extensive tissue restoration occurred in the zebularine-treated but not in the control mice (Fig. 1b, c). Initially, as measured on the day 14 after injury, zebularine treatment slightly impeded ear hole closure, whereas the mean ear hole area was significantly smaller in zebularine-treated animals at later time points (Fig. 1d). Ear hole closure in mice receiving zebularine plateaued on the day 35 after injury and significantly exceeded that in the controls (Fig. 1e). On the day 42 after injury, the mean hole area had decreased to 0.53 ± 0.3 sq. mm in zebularine-treated and 1.77 ± 0.48 sq. mm in control mice (p < 10−30), corresponding to 83.2 ± 9.4% and 43.6 ± 15.4% closure, respectively (Fig. 1f). Not only better closure but also less variation was observed in the treated than in the control group (Fig. 1f).
Fig. 1.
Zebularine stimulates ear pinna hole closure in the mouse. (A) Experimental time schedule. (B) Representative photographs showing ear hole closure for the same zebularine-treated and control female mice; the scale is calibrated in mm. (C) Diagram showing the mean ear hole area at day 0 (d0) and day 42 (d42) for zebularine-treated and control female mice. (D) Ear wound closure demonstrated as the mean hole area for 47 zebularine-treated and 48 control female mice; “n” indicates the number of ears, and the error bars represent the SD. (E) Mean percentage of closed area throughout wound healing; “n” indicates the number of ears, and the error bars represent the SD. (F) Distribution of ear hole areas at d42 post injury; each dot represents a single ear. The calculated p-values were as follows d14–2.42E-07, d21–3.01E-06, d28–0.00E+00, d35–0.00E+00, d42.
The applied dose of pharmacological agent of 1000 mg/kg is unusually high, but much lower zebularine doses of 500 and 200 mg/kg proved effective, as did the administration of five instead of seven injections (Fig. 2a). While trials of topical zebularine delivery through rubbing 10-μL droplets of zebularine solution (50 mg/mL) with gloved fingers were unsuccessful, subcutaneous injections resulted in significant ear hole closure but were less effective than intraperitoneal administration (Fig. 2b). The above described experiments were done on females. To determine whether the zebularine effect is restricted to females, we carried out an analogous experiment in males. We observed less effective closure in males than in females, but treated males had significantly better closure than control males (Fig. 2c). In addition, we confirmed that the ear hole closure in response to zebularine treatment was not limited to the BALB/c strain but was also significantly stimulated in C57BL/6 J mice (Fig. 2d). More detailed ear hole closure data demonstrated as dot plots representing ear hole areas are included in Supplemental Information Fig. S1.
3.2. Zebularine vs. its nucleoside analogues
5-Azacitidine, a close structural analogue of zebularine, is a more potent but more toxic DNA methyltransferase inhibitor [41]. Intraperitoneal injections of 5-azacitidine (0.25 mg/kg), on the same schedule as that followed for zebularine (Fig. 1a) did not induce regeneration and, moreover, significantly inhibited the closure process (Fig. 2e). Due to 5-azacitidine toxicity, a dose 4000-times lower than that of zebularine was applied.
Zebularine is rapidly converted to uridine in the cytosol [42], which prompts the question of whether enhanced levels of this metabolite could be responsible for the observed regenerative response. Neither high doses (1000 mg/kg) of uridine, nor cytidine, another zebularine analogue, promoted the regenerative response, and on the contrary, they even markedly suppressed ear hole closure (Fig. 2e).
3.3. Histological examination of restored tissues
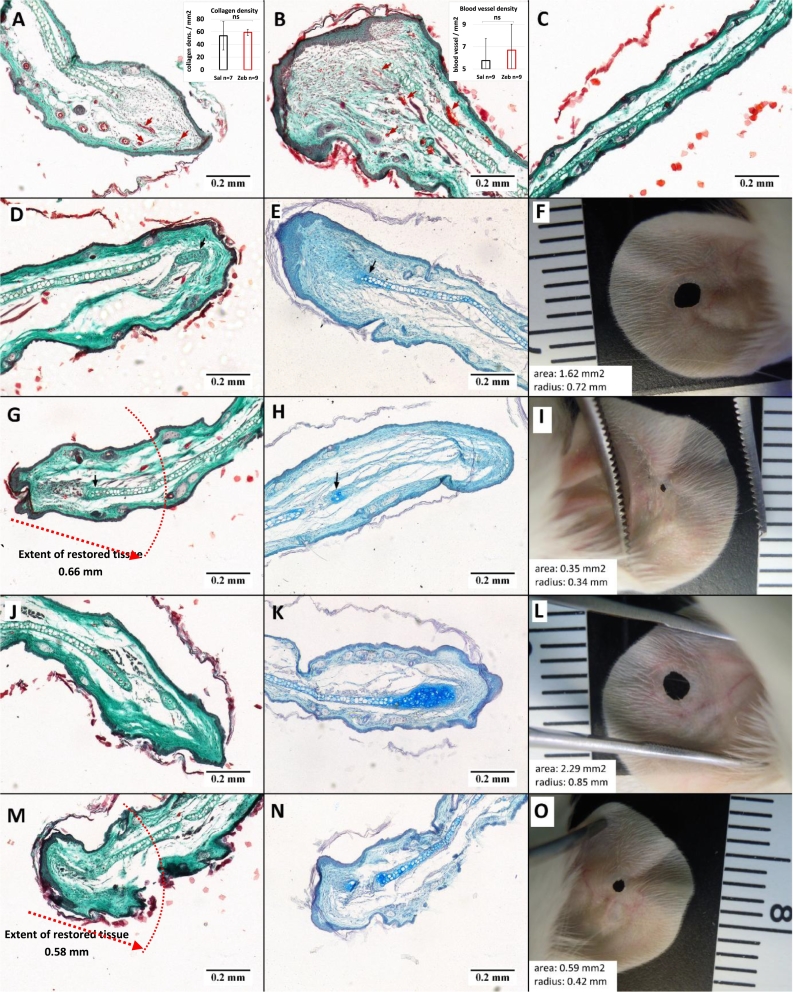
To describe tissue architecture, collagen deposition, and cartilage formation, we stained ear pinna tissue sections collected from the wound area using Masson's trichrome, and Alcian blue, respectively (Fig. 3). In both control (Fig. 3a, d, e, f, j, k, l) and zebularine-treated tissues (Fig. 3b, g, h, i, m, n, o), we observed cartilage formation, fibroblasts, and detected no collagen deposition within the restoration zone. Furthermore, we detected blood vessels within the regenerating area but we found no significant differences in blood vessel and collagen densities between the control and zebularine-treated ear pinnae (Fig. 3a, b, insets). We were unable to identify any distinctive traits of the tissue architecture in mice that received zebularine. Rather, it was the extent of restoration within ear pinnae that distinguished the zebularine-treated animals from the controls (Fig. 3g, m). The comparison of regenerated tissues from experimental mice with those from naïve controls (Fig. 3c) showed no remarkable structural differences.
Fig. 3.
Histological examination reveals similar tissue architecture in the restored areas of ear-pinnae in zebularine treated and control mice. (A) control and (B) zebularine treated ears at d21; insets: blood vessel and collagen densities in zebularine-treated and control sections at day 21; (C) normal, uninjured ear; (D, E, F) control ears at d42; (G, H, I) zebularine treated ears at d42; (J, K, L) control ears at d70; (M, N, O) zebularine treated ears at d70; (A, B, C, D, G, J, M) were stained with Masson's trichrome and (E, H, K, N) were stained with haematoxylin and Alcian Blue. (F, I, L, O) ears from which the sections were taken for histological staining. Blood vessels and cartilage are indicated with short red and black arrows, respectively. The red dotted lines indicate the extension of the restored tissue where newly formed cartilage is found The zone of restoration (0.66 mm) was calculated by deducing the radius of ear hole at day 42 (0.34 mm) (I) from the radius of initial ear hole (1 mm) (G). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.4. Decreased DNA methylation in the wound area
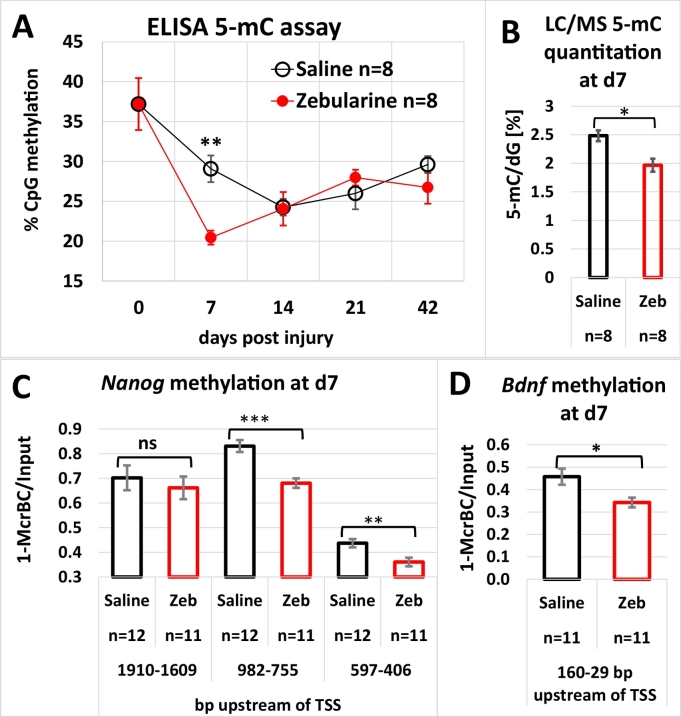
Zebularine inhibits DNA methyltransferases after incorporation into DNA, and the resultant demethylation occurs primarily after replication. Extensive DNA replication is more likely to occur in proliferating cells in the wound area than in non-injured tissue. Therefore, we determined the levels of CpG methylation in the tissues surrounding ear pinna holes of zebularine-treated and control animals using an antibody specific for 5-methylcytosine. The results showed that wounding itself resulted in decreased DNA methylation. The change was significantly greater in zebularine treated than in control mice on the day 7 after injury, which was the second day after 5 consecutive daily injections (Fig. 4a). Enhanced DNA demethylation in response to zebularine in tissues surrounding ear pinna wounds was confirmed using mass spectrometry (Fig. 4b). As determined using digestion with a CpG methylation-dependent restriction enzyme, decreased DNA methylation following zebularine treatment was also found in the promoter regions of the Nanog gene, a key pluripotency marker, and the Bdnf gene, encoding a canonical neurotrophic factor (Fig. 4c, d). The demethylation effects were accompanied by transcriptional activation (Fig. 5).
Fig. 4.
Global and gene-specific demethylation in regenerating ear pinnae follows intraperitoneal zebularine delivery. (A) ELISA analysis of 5-methylcytosine (5-mC) content in the wound area and uninjured tissue (day 0, d0). (B) LC-MS quantitation of 5-mC at d7 after injury. (C, D) Promoter demethylation of the Nanog and Bdnf genes in the wound area after zebularine treatment determined using a CpG methylation-sensitive restriction enzyme. Plots show the mean value, and error bars represent the SEM; “n” indicates the number of ears.
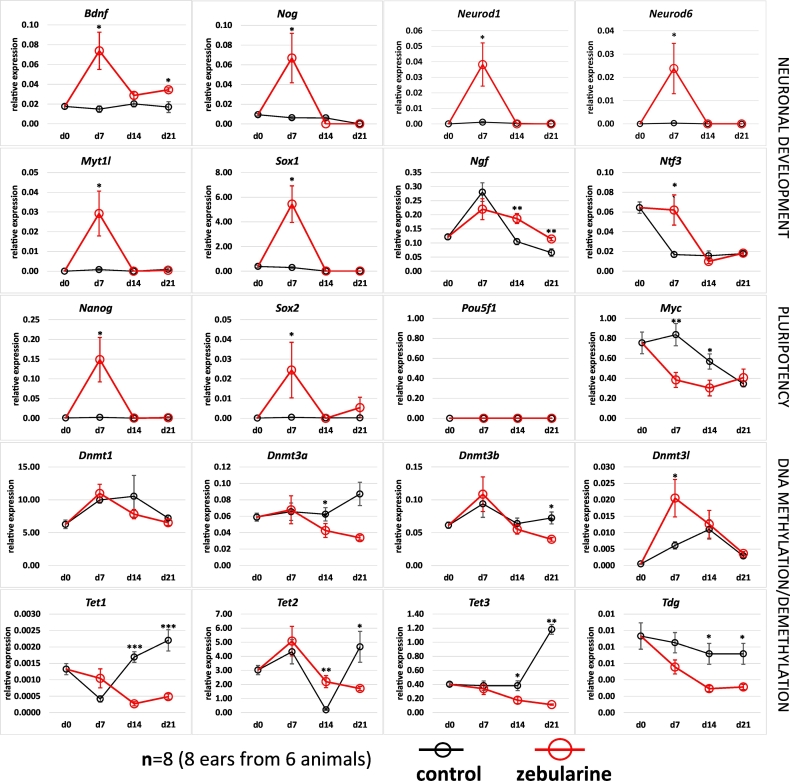
Fig. 5.
Genes involved in neuronal development, pluripotency and DNA methylation show transcriptional changes in the wound area after intraperitoneal zebularine injections. D0 indicates uninjured tissue. The transcript levels were determined using qPCR. Plots show the mean value, error bars represent the SEM.
3.5. Gene expression changes under zebularine treatment
To investigate the transcriptional consequences of zebularine delivery, we quantitated the transcript levels of a panel of genes related to pluripotency, neuronal development, and DNA methylation in the ear pinna tissues collected from the injury area (Fig. 5). Several neurodevelopmental genes, including Bdnf, Nog, Neurod1, Neurod6, Myt1l, and Sox1, displayed significantly higher expression levels at day 7 after injury in zebularine-treated than in control mice. Remarkably, the pluripotency markers, Nanog and Sox2 were also induced. Genes involved in de novo (Dnmt3a, Dnmt3b) and maintenance DNA methylation (Dnmt1) and in demethylation processes (Tet1, Tet2, Tet3, Tdg) showed decreases in expression at some time points after zebularine treatment, with the exception of Dnmt3l, which is thought to stimulate de novo DNA methylation.
In addition, we performed RNA-seq transcriptome profiling of cultured human keratinocytes (HaCaT) and fibroblasts (46BR.1 N) to observe the extensive alterations following zebularine stimulation. A substantial proportion of genes (over 10%) displayed over a two-fold change in expression after zebularine treatment (Supplemental Information, Fig. S2). In the context of transcriptional changes observed in ear pinnae after zebularine treatment, neuronal development genes constituted one of the top functional categories enriched among the transcripts upregulated in cultured keratinocytes and fibroblasts in response to zebularine stimulation (Supplemental Information, Fig. S3).
3.6. Potentiation of the regenerative effect of zebularine by retinoic acid
Considering that DNA methyltransferase inhibition leads to the demethylation of gene promoters, zebularine action could enable the activation of silenced genes. However, promoter demethylation is insufficient to induce gene expression without an additional activator. To potentially stimulate gene expression towards regenerative response, we decided to try retinoic acid. Retinoic acid has an established role as a signalling molecule in development [34], and regeneration in amphibians but also in mammals [[30], [31], [32]]. Retinoic acid is a transcriptional regulator which targets a number of genes including the homeobox genes [43] which are known for their activity during embryonic development and regeneration. Combined delivery (Fig. 6a) of retinoic acid (16 mg/kg) and zebularine (1000 mg/kg) resulted in accelerated and complete ear hole closure within three weeks (Fig. 6b, d, e). The experiment was repeated three times, including a total of 35 treated animals. Six weeks post injury, histological examination showed the restoration of ear tissue architecture and cartilage re-growth into the regenerated area (Fig. 6f).
3.7. Zebularine-mediated augmentation of retinoic acid target genes expression
In order to investigate whether zebularine supports the action of retinoic acid as the transcriptional activator we chose a cell culture model in order to avoid potential complications expected in heterogeneous tissue samples collected from the wound area. We performed gene expression profiling of several retinoic acid target genes in human keratinocytes cells (HaCaT) cultured with zebularine and stimulated with retinoic acid. We determined enhanced transcript levels of three homeobox genes (MEIS1, NKX2–5, POU6F2) and three other known retinoic acid target genes (KRT7, GABRP, PPARG) in the cells treated with both zebularine and retinoic acid as compared to those exposed to retinoic acid alone (Fig. 6c).
3.8. Zebularine toxicity and effect on cell proliferation
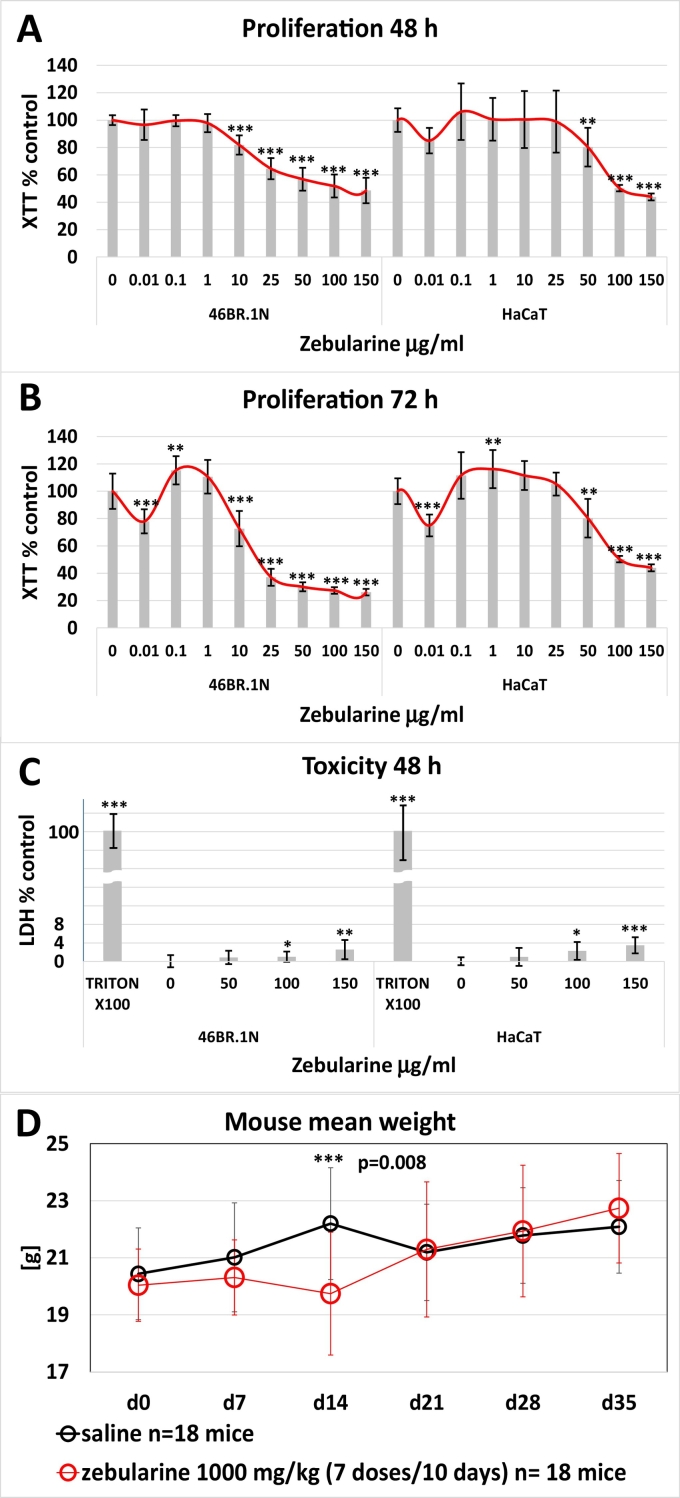
The effect of zebularine on proliferation was examined in cell line models of cultured human keratinocytes (HaCaT) and fibroblasts (46BR.1 N) (Fig. 7a, b). Zebularine slightly stimulated proliferation after 72 h at low concentrations (0.1 and 1.0 μg/mL) but inhibited keratinocyte and fibroblast proliferation at the lowest concentration used (0.01 μg/mL). Stronger inhibition was observed at concentrations within the range of 10–150 μg/mL and 50–150 μg/mL for fibroblasts and keratinocytes, respectively (Fig. 7b). Weaker inhibitory effects were observed after 48 h of incubation (Fig. 7a). Zebularine displayed low cytotoxicity, with the greatest toxicity at 100 and 150 μg/mL (Fig. 7c). The low toxicity in cell culture models corresponded with our observations that mice receiving high doses of zebularine showed only a small and transient decrease in body weight (Fig. 7d).
Fig. 7.
Experiments on cell lines and animals indicate limited toxicity of zebularine. (A, B) Effect of increasing zebularine concentrations on the proliferation of cultured fibroblasts (46BR.1 N) and keratinocytes (HaCaT) assessed using the XTT assay after 48 and 72 h of incubation and normalized to the negative control (100%). (C) Toxicity of increasing zebularine concentrations estimated using the LDH assay after a 48-h incubation and normalized to the negative control (0%) and 1% Triton X-100 (100%). (D) Effect of zebularine injections on mouse weight. Error bars represent the SD.
4. Discussion
In the present study, we demonstrated that zebularine, a nucleoside inhibitor of DNA methyltransferases, effectively promoted the regenerative response in mouse ear pinnae. While limited ear hole closure occurred in vehicle-treated mice (43.6%), zebularine-treated animals showed extensive restoration of lost tissue (83.2%) within six weeks after injury. As revealed by histological examination, the restoration was not limited to the formation of connective tissue but it involved the growth of cartilage and fibroblasts. Combined application of zebularine and retinoic acid displayed a synergistic effect, resulting in accelerated and complete ear hole closure within three weeks of injury.
4.1. Hypothesized mechanisms
The remarkable regenerative effect of zebularine in animals we demonstrated raises the question about the mechanism of this activity. The inhibitory action of zebularine on DNA methyltransferases, suggests the potential role of DNA demethylation in the activation of regenerative response. Zebularine inhibits DNA methyltransferases after it is incorporated into DNA leading to DNA demethylation. Zebularine causes the depletion of both the maintenance and de novo DNA methyltransferases [44]. Active cell proliferation in the wound area could promote both zebularine integration into DNA and passive DNA demethylation. DNA demethylation is likely to re-activate silenced developmental genes such as those of pluripotency factors that may be critical for regeneration. Indeed, DNA demethylating agents induce pluripotency in cell lines [45], and passive demethylation has been reported to preferentially activate pluripotency genes [46]. The decreased DNA methylation and increased transcriptional activation we found in the wound area of zebularine-treated mice support the above line of reasoning.
In addition, different cell types may show varied responses to zebularine stimulation. Various cell subsets within the wound and penetrating the injury area could reveal diverse sensitivities to zebularine, as cultured fibroblasts and keratinocytes did (Fig. 7a, b). Notably, zebularine displayed an inhibitory effect on proliferation (Fig. 7a, b) that did not lead to cell damage (Fig. 7c). Accordingly, we cannot exclude the possibility that zebularine suppresses certain cell lines involved in the normal response to wounding, thus promoting regenerative repair. This concept could be supported by the observation that zebularine initially (at two weeks after injury) slowed down the rate of wound closure (Fig. 1d, e).
4.2. Minimal zebularine toxicity and its pro-regenerative activity
Low toxicity of zebularine in cell culture [9] and mice (no toxicity symptoms at 400 mg/kg i.p. for 78 consecutive days [12]) is an important feature that appears to have been decisive in the successful use of the epigenetic inhibitor to promote regeneration. This is particularly interesting in the context of high toxicity of 5-azacitidine, another nucleoside inhibitor of DNA methylotransferases with LD50 in mice of 2.48 mg/kg i.p. for five days [47]. Zebularine and 5-azacitidine undergo similar metabolic activation and have a similar mechanism of action, but zebularine displays much better water solubility [11]. In contrast to 5-aza-2′-deoxycytidine, 2′-deoxyzebularine is unlikely to become methylated while exhibiting inhibitory activity [48]. Zebularine has several other features that may explain its low toxicity. First, all zebularine metabolites are endogenous substances [49]. Second, a substantial fraction of zebularine is oxidized to uridine by liver aldehyde oxidase, thus preventing DNA incorporation [9]. DNA incorporation is also challenged by that of RNA, which is estimated as to be 7-fold higher [9], and by competitive inhibition of zebularine phosphorylation by cytidine [9]. Zebularine functions as a cytidine deaminase and thymidylate synthase inhibitor, which may temper its inhibitory action on DNA methyltransferase. While cytidine deaminase has been conjectured to bind a significant amount of zebularine [50], it is possible that the inhibition of thymidylate synthase [51] reduces DNA synthesis, thus preventing passive DNA demethylation. Cellular metabolism could explain not only the requirement of high effective doses in animals (500–1000 mg/kg) [11] but also the low toxicity of zebularine. In addition to metabolism, the biological activity of nucleoside inhibitors relies on active transport that is counter-acted by efflux through protein transporters [52]. Consequently, sensitivity to zebularine may be determined by the cell type and, in particular, by the expressed transporter proteins.
4.3. Synergistic effect of zebularine and retinoic acid
The central role of retinoic acid signalling in development and organogenesis has been explored in different animal models [34]. Retinoids regulate amphibian limb regeneration [53] and stimulate repair in diverse tissues in mammals [30] such as the lungs [31], axons [32] and the skin [54]. Interestingly, the adult MRL/MpJ mouse, known for its regenerative potential, shows a distinctive expression profile of retinol metabolism genes [55]. As a transcriptional activator, retinoic acid targets a number of genes [34]. These clues led us to the concept of using this molecule to activate the genes demethylated by zebularine. Based on this idea, we predict that zebularine could synergize with other transcriptional activators to induce selected pathways. The augmentation of retinoic acid target genes activation by zebularine we found in cultured keratinocytes supports this prediction (Fig. 6c).
4.4. Critical remarks
Although regenerative results are promising, there are several concerns regarding the use of zebularine, including the very high doses and the mutagenicity risk. These complications could be avoided if zebularine was replaced by another DNA methyltransferase inhibitor, preferably a non-nucleoside compound, that is more resistant to metabolic degradation. Nevertheless, the use of zebularine can be regarded as a promising solution. In most trials, we administered seven injections of a very high dose of 1000 mg/kg body weight. However, we determined that five doses of zebularine were almost equally effective as seven doses and that a fivefold lower dose produced a significant regenerative response (Fig. 2a).
Zebularine mutagenicity has not been extensively investigated, except for tests using bacterial cells [56,57]. Zebularine is mainly incorporated instead of cytidine and mispairs with adenine [57]. However, its analogues, 5-azacytidine and 5-aza-2′-deoxycytidine are approved as anticancer drugs, despite the potential risk of secondary neoplasms. Considering that regenerative therapies, unlike anticancer treatment, may involve short-period administration and local delivery, the use of zebularine in regenerative medicine does not seem unthinkable.
General doubts regarding epigenetic therapies could be raised. Epigenetic alterations harbour the risk of promoting cancer [58], and epigenetic reprogramming is difficult to control. Although these concerns appear substantiated, a number of epigenetic drugs are in clinical studies [59], suggesting that their potential outweighs the related risks.
4.5. Final conclusions
We demonstrated that epigenetic-based pharmacological strategy using zebularine, a DNA methyltransferase inhibitor, was effective to promote regeneration in a mouse ear punch model. We observed significant DNA demethylation and the transcriptional activation of several pluripotency and neurodevelopmental genes in the tissues collected from the regenerating area. Noteworthy, a stimulating example of a therapy based on epigenetic derepression of neuronal genes has been recorded. The activation of neuronal genes in sensory cells of ear induced by treatment with a histone deacetylase inhibitor SAHA (suberanilohydroxamic acid) has been reported to rescue hearing in deaf mice [60].
The key question is whether zebularine will be active as a regenerative therapy in models more complex than ear pinnae. Solving this problem will require further research.
Importantly, the main achievement of the present study is not the selection of drug candidates but the delineation of a novel successful strategy for regenerative medicine. Zebularine could be replaced by a compound with better pharmacodynamic properties, and different transcriptional regulators may be used to activate desired gene sets. The principle of the proposed strategy involves the combination of epigenetic derepression and transcriptional activation in order to stimulate endogenous regenerative potential.
Funding sources
This study was supported by the grant of the National Centre for Research and Development of Poland No. STRATEGMED1/235077/9/NCBR/2014. The funding source had no role in the writing of the manuscript or in the decision to submit it for publication. Nor did they have any role in the study design, experiments, data collection, analysis, interpretation of findings, or in any other aspect of the study. As the corresponding author Dr. Paweł Sachadyn had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Author contributions
PiS designed and performed animal experiments and collected tissue samples, designed and performed transcriptional analysis for pluripotency and cell cycle genes, analysed histological results; PSo designed and performed animal experiments and collected tissue samples, designed and performed transcriptional analysis for neurodevelopmental genes; JPP significantly contributed to the study concept and the design of preliminary experiments; BG determined global DNA methylation levels with ELISA and processed DNA for LC/MS 5-mC quantitation, performed transcriptional analysis for genes responsible for DNA methylation, and carried out gene ontology analyses; JK determined gene specific DNA methylation levels and performed transcriptional profiling in human keratinocytes; MD conducted cell culture experiments and analysed results; EN - performed histological staining; AW conducted cell culture experiments and analysed results; PM participated in the study concept design and data interpretation; carried out LC/MS 5-mC quantitation; JR carried out LC/MS 5-mC quantitation; PRe carried out LC/MS 5-mC quantitation; PR performed experiments on C57BL/6 J mice; AC participated in the study concept design and data interpretation, supervised experiments on C57BL/6 J mice; GPS contributed significantly to the design of animal experiments; NF conducted RNA-seq transcriptome profiling; AM conducted RNA-seq transcriptome profiling; AP participated in the study concept design and data interpretation, designed RNA-seq analysis; LJ participated in the study concept design; PSk participated in the study concept design; SRM participated in the study concept design, and provided financial support; MP participated in the study concept design; conceived, designed and supervised cell culture experiments and histological examinations, analysed and interpreted the results, revised the manuscript, and provided financial support; PSa created the study concept, participated in the design of experiments, supervised animal and molecular experiments, analysed and interpreted the results, performed statistical analyses; drafted and revised the manuscript, and provided financial support.
Declaration of Competing Interest
Patent applications (PCT/PL2018/000027, EP18000264, P-423672) to protect novel use of zebularine in regenerative therapies have been filed (PiS, PSo, JPP, BG, ŁJ, PSk, AC, PM, AP, SRM, MP, PSa,). The authors declare no other competing interests.
Acknowledgements
This study was conducted within the “REGENNOVA” project on novel technologies for pharmacological stimulation of regeneration under the “STRATEGMED” program of the National Centre for Research and Development of Poland (grant no. STRATEGMED1/235077/9/NCBR/2014). We thank all researchers participating in the “REGENNOVA” consortium.
We thank Prof. Mirosława Cichorek (Dept. of Embryology, Medical University of Gdańsk), Prof. Marek Grzybiak and Prof. Adam Kosiński (Dept. of Clinical Anatomy, Medical University of Gdańsk) for providing access to laboratory equipment. We thank Monika Dmochowska, Agnieszka Jakubiak, Anna Żyłko, and Beata Muszyńska, DVM, from the Tri-City Academic Laboratory Animal Centre, Medical University of Gdańsk for their technical assistance in animal experiments and Dr. Rafał Płatek from the Nencki Institute for introducing us to image analyses tools.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.07.010.
Contributor Information
Michał Pikuła, Email: pikula@gumed.edu.pl.
Paweł Sachadyn, Email: psach@pg.edu.pl.
Appendix A. Supplementary data
Supplemental information
References
- 1.Podolak-Popinigis J., Ronowicz A., Dmochowska M., Jakubiak A., Sachadyn P. The methylome and transcriptome of fetal skin: implications for scarless healing. Epigenomics. 2016;8(10):1331–1345. doi: 10.2217/epi-2016-0068. [DOI] [PubMed] [Google Scholar]
- 2.Porrello E.R., Mahmoud A.I., Simpson E. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331(6020):1078–1080. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fry E.J., Stolp H.B., Lane M.A., Dziegielewska K.M., Saunders N.R. Regeneration of supraspinal axons after complete transection of the thoracic spinal cord in neonatal opossums (Monodelphis domestica) J Comp Neurol. 2003;466(3):422–444. doi: 10.1002/cne.10904. [DOI] [PubMed] [Google Scholar]
- 4.Hochedlinger K., Jaenisch R. Induced pluripotency and epigenetic reprogramming. Cold Spring Harb Perspect Biol. 2015;7(12) doi: 10.1101/cshperspect.a019448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Powell C., Grant A.R., Cornblath E., Goldman D. Analysis of DNA methylation reveals a partial reprogramming of the Muller glia genome during retina regeneration. Proc Natl Acad Sci U S A. 2013;110(49):19814–19819. doi: 10.1073/pnas.1312009110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sim C.B., Ziemann M., Kaspi A. Dynamic changes in the cardiac methylome during postnatal development. FASEB J. 2015;29(4):1329–1343. doi: 10.1096/fj.14-264093. [DOI] [PubMed] [Google Scholar]
- 7.Gornikiewicz B., Ronowicz A., Krzeminski M., Sachadyn P. Changes in gene methylation patterns in neonatal murine hearts: implications for the regenerative potential. BMC Genomics. 2016;17:231. doi: 10.1186/s12864-016-2545-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 9.Ben-Kasus T., Ben-Zvi Z., Marquez V.E., Kelley J.A., Agbaria R. Metabolic activation of zebularine, a novel DNA methylation inhibitor, in human bladder carcinoma cells. Biochem Pharmacol. 2005;70(1):121–133. doi: 10.1016/j.bcp.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 10.Cheng J.C., Yoo C.B., Weisenberger D.J. Preferential response of cancer cells to zebularine. Cancer Cell. 2004;6(2):151–158. doi: 10.1016/j.ccr.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 11.Cheng J.C., Matsen C.B., Gonzales F.A. Inhibition of DNA methylation and reactivation of silenced genes by zebularine. J Natl Cancer Inst. 2003;95(5):399–409. doi: 10.1093/jnci/95.5.399. [DOI] [PubMed] [Google Scholar]
- 12.Herranz M., Martín-Caballero J., Fraga M.F. The novel DNA methylation inhibitor zebularine is effective against the development of murine T-cell lymphoma. Blood. 2006;107(3):1174–1177. doi: 10.1182/blood-2005-05-2033. [DOI] [PubMed] [Google Scholar]
- 13.Williams-Boyce P.K., Daniel J.C., Jr. Comparison of ear tissue regeneration in mammals. J Anat. 1986;149:55. [PMC free article] [PubMed] [Google Scholar]
- 14.Clark L.D., Clark R.K., Heber-Katz E. A new murine model for mammalian wound repair and regeneration. Clin Immunol Immunopathol. 1998;88(1):35–45. doi: 10.1006/clin.1998.4519. [DOI] [PubMed] [Google Scholar]
- 15.Seifert A.W., Kiama S.G., Seifert M.G., Goheen J.R., Palmer T.M., Maden M. Skin shedding and tissue regeneration in African spiny mice (Acomys) Nature. 2012;489(7417):561. doi: 10.1038/nature11499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blankenhorn E.P., Troutman S., Clark L.D., Zhang X.M., Chen P., Heber-Katz E. Sexually dimorphic genes regulate healing and regeneration in MRL mice. Mamm Genome. 2003;14(4):250–260. doi: 10.1007/s00335-002-2222-3. [DOI] [PubMed] [Google Scholar]
- 17.Gawronska-Kozak B. Regeneration in the ears of immunodeficient mice: identification and lineage analysis of mesenchymal stem cells. Tissue Eng. 2004;10(7–8):1251–1265. doi: 10.1089/ten.2004.10.1251. [DOI] [PubMed] [Google Scholar]
- 18.Buckley G., Metcalfe A.D., Ferguson M.W. Peripheral nerve regeneration in the MRL/MpJ ear wound model. J Anat. 2011;218(2):163–172. doi: 10.1111/j.1469-7580.2010.01313.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lalley A.L., Dyment N.A., Kazemi N. Improved biomechanical and biological outcomes in the MRL/MpJ murine strain following a full-length patellar tendon injury. J Orthop Res. 2015;33(11):1693–1703. doi: 10.1002/jor.22928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fitzgerald J., Rich C., Burkhardt D., Allen J., Herzka A.S., Little C.B. Evidence for articular cartilage regeneration in MRL/MpJ mice. Osteoarthr Cartil. 2008;16(11):1319–1326. doi: 10.1016/j.joca.2008.03.014. [DOI] [PubMed] [Google Scholar]
- 21.Ueno M., Lyons B.L., Burzenski L.M. Accelerated wound healing of alkali-burned corneas in MRL mice is associated with a reduced inflammatory signature. Invest Ophthalmol Vis Sci. 2005;46(11):4097–4106. doi: 10.1167/iovs.05-0548. [DOI] [PubMed] [Google Scholar]
- 22.Xia H., Krebs M.P., Kaushal S., Scott E.W. Enhanced retinal pigment epithelium regeneration after injury in MRL/MpJ mice. Exp Eye Res. 2011;93(6):862–872. doi: 10.1016/j.exer.2011.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chadwick R.B., Bu L., Yu H. Digit tip regrowth and differential gene expression in MRL/Mpj, DBA/2, and C57BL/6 mice. Wound Repair Regen. 2007;15(2):275–284. doi: 10.1111/j.1524-475X.2007.00216.x. [DOI] [PubMed] [Google Scholar]
- 24.Thuret S., Thallmair M., Horky L.L., Gage F.H. Enhanced functional recovery in MRL/MpJ mice after spinal cord dorsal hemisection. PLoS One. 2012;7(2) doi: 10.1371/journal.pone.0030904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leferovich J.M., Bedelbaeva K., Samulewicz S. Heart regeneration in adult MRL mice. Proc Natl Acad Sci U S A. 2001;98(17):9830–9835. doi: 10.1073/pnas.181329398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bastakoty D., Saraswati S., Cates J., Lee E., Nanney L.B., Young P.P. Inhibition of Wnt/β-catenin pathway promotes regenerative repair of cutaneous and cartilage injury. FASEB J. 2015;29(12):4881–4892. doi: 10.1096/fj.15-275941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y., Strehin I., Bedelbaeva K. Drug-induced regeneration in adult mice. Sci Transl Med. 2015;7(290):290ra92. doi: 10.1126/scitranslmed.3010228. [-ra92] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gornikiewicz B., Ronowicz A., Podolak J., Madanecki P., Stanislawska-Sachadyn A., Sachadyn P. Epigenetic basis of regeneration: analysis of genomic DNA methylation profiles in the MRL/MpJ mouse. DNA Res. 2013;20(6):605–621. doi: 10.1093/dnares/dst034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yakushiji N., Suzuki M., Satoh A. Correlation between Shh expression and DNA methylation status of the limb-specific Shh enhancer region during limb regeneration in amphibians. Dev Biol. 2007;312(1):171–182. doi: 10.1016/j.ydbio.2007.09.022. [DOI] [PubMed] [Google Scholar]
- 30.Maden M., Hind M. Retinoic acid, a regeneration-inducing molecule. Dev Dyn. 2003;226(2):237–244. doi: 10.1002/dvdy.10222. [DOI] [PubMed] [Google Scholar]
- 31.Hind M., Maden M. Retinoic acid induces alveolar regeneration in the adult mouse lung. Eur Respir J. 2004;23(1):20–27. doi: 10.1183/09031936.03.00119103. [DOI] [PubMed] [Google Scholar]
- 32.Puttagunta R., Di Giovanni S. Retinoic acid signaling in axonal regeneration. Front Mol Neurosci. 2012;4:59. doi: 10.3389/fnmol.2011.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gudas L.J. Emerging roles for retinoids in regeneration and differentiation in normal and disease states. Biochim Biophys Acta (BBA)-Mol Cell Biol Lipids. 2012;1821(1):213–221. doi: 10.1016/j.bbalip.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cunningham T.J., Duester G. Mechanisms of retinoic acid signalling and its roles in organ and limb development. Nat Rev Mol Cell Biol. 2015;16(2):110–123. doi: 10.1038/nrm3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ruifrok A.C., Johnston D.A. Quantification of histochemical staining by color deconvolution. Anal Quant Cytol Histol. 2001;23(4):291–299. [PubMed] [Google Scholar]
- 37.Boukamp P., Popp S., Altmeyer S. Sustained nontumorigenic phenotype correlates with a largely stable chromosome content during long-term culture of the human keratinocyte line HaCaT. Genes Chromosomes Cancer. 1997;19(4):201–214. doi: 10.1002/(sici)1098-2264(199708)19:4<201::aid-gcc1>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 38.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trapnell C., Williams B.A., Pertea G. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bindea G., Mlecnik B., Hackl H. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25(8):1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Billam M., Sobolewski M.D., Davidson N.E. Effects of a novel DNA methyltransferase inhibitor zebularine on human breast cancer cells. Breast Cancer Res Treat. 2010;120(3):581–592. doi: 10.1007/s10549-009-0420-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klecker R.W., Cysyk R.L., Collins J.M. Zebularine metabolism by aldehyde oxidase in hepatic cytosol from humans, monkeys, dogs, rats, and mice: influence of sex and inhibitors. Bioorg Med Chem. 2006;14(1):62–66. doi: 10.1016/j.bmc.2005.07.053. [DOI] [PubMed] [Google Scholar]
- 43.Marshall H., Morrison A., Studer M., Pöpperl H., Krumlauf R. Retinoids and Hox genes. FASEB J. 1996;10(9):969–978. [PubMed] [Google Scholar]
- 44.Marquez V.E., Barchi J.J., Kelley J.A. Zebularine: a unique molecule for an epigenetically based strategy in cancer chemotherapy. The magic of its chemistry and biology. Nucleosides Nucleotides Nucleic Acids. 2005;24(5–7):305–318. doi: 10.1081/ncn-200059765. [DOI] [PubMed] [Google Scholar]
- 45.De Carvalho D.D., You J.S., Jones P.A. DNA methylation and cellular reprogramming. Trends Cell Biol. 2010;20(10):609–617. doi: 10.1016/j.tcb.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.He S., Sun H., Lin L. Passive DNA demethylation preferentially up-regulates pluripotency-related genes and facilitates the generation of induced pluripotent stem cells. J Biol Chem. 2017;292(45):18542–18555. doi: 10.1074/jbc.M117.810457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.European Medicines Agency . 2008. European public assessment report Vidaza. EMEA/593162/2008. [Google Scholar]
- 48.Zhou L., Cheng X., Connolly B.A., Dickman M.J., Hurd P.J., Hornby D.P. Zebularine: a novel DNA methylation inhibitor that forms a covalent complex with DNA methyltransferases. J Mol Biol. 2002;321(4):591–599. doi: 10.1016/S0022-2836(02)00676-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beumer J.H., Joseph E., Egorin M.J. A mass balance and disposition study of the DNA methyltransferase inhibitor zebularine (NSC 309132) and three of its metabolites in mice. Clin Cancer Res. 2006;12(19):5826–5833. doi: 10.1158/1078-0432.CCR-06-1234. [DOI] [PubMed] [Google Scholar]
- 50.Yoo C.B., Cheng J.C., Jones P.A. Zebularine: a new drug for epigenetic therapy. Biochem Soc Trans. 2004;32:910–912. doi: 10.1042/BST0320910. Pt 6. [DOI] [PubMed] [Google Scholar]
- 51.Yoo C.B., Valente R., Congiatu C. Activation of p16 gene silenced by DNA methylation in cancer cells by phosphoramidate derivatives of 2′-deoxyzebularine. J Med Chem. 2008;51(23):7593–7601. doi: 10.1021/jm8005965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arimany-Nardi C., Errasti-Murugarren E., Minuesa G. Nucleoside transporters and human organic cation transporter 1 determine the cellular handling of DNA-methyltransferase inhibitors. Br J Pharmacol. 2014;171(16):3868–3880. doi: 10.1111/bph.12748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stocum D.L. Mechanisms of urodele limb regeneration. Regeneration. 2017;4(4):159–200. doi: 10.1002/reg2.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Riahi R.R., Bush A.E., Cohen P.R. Topical retinoids: therapeutic mechanisms in the treatment of photodamaged skin. Am J Clin Dermatol. 2016;17(3):265–276. doi: 10.1007/s40257-016-0185-5. [DOI] [PubMed] [Google Scholar]
- 55.Podolak-Popinigis J., Gornikiewicz B., Ronowicz A., Sachadyn P. Transcriptome profiling reveals distinctive traits of retinol metabolism and neonatal parallels in the MRL/MpJ mouse. BMC Genomics. 2015;16:926. doi: 10.1186/s12864-015-2075-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ang J., Song L.Y., D'Souza S. Mutagen synergy: hypermutability generated by specific pairs of base analogs. J Bacteriol. 2016;198(20):2776–2783. doi: 10.1128/JB.00391-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee G., Wolff E., Miller J.H. Mutagenicity of the cytidine analog zebularine in Escherichia coli. DNA Repair. 2004;3(2):155–161. doi: 10.1016/j.dnarep.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 58.Priestley C.C., Anderton M., Doherty A.T. Epigenetics–relevance to drug safety science. Toxicol Res. 2012;1(1):23–31. [Google Scholar]
- 59.Esteller M. Epigenetic drugs: more than meets the eye. Epigenetics. 2017;12(5):307. doi: 10.1080/15592294.2017.1322881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nakano Y., Kelly M.C., Rehman A.U. Defects in the alternative splicing-dependent regulation of REST cause deafness. Cell. 2018;174(3):536–548. doi: 10.1016/j.cell.2018.06.004. [e21] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental information