Abstract
Background
Esophageal squamous cell carcinoma (ESCC) is one of the most lethal malignancies with poor prognosis. Cancer-testis genes (CTGs) have been vigorously pursued as targets for cancer immunotherapy, but the expressive patterns and functional roles of CTGs remain unclear in ESCC.
Methods
A systematic screening strategy was adopted to screen CTGs in ESCC by integrating multiple public databases and RNA expression microarray data from 119 ESCC subjects. For the newly identified ESCC prognosis-associated CTGs, an independent cohort of 118 patients with ESCC was recruited to validate the relationship via immunohistochemistry. Furthermore, functional assays were performed to determine the underlying mechanisms.
Findings
21 genes were recognized as CTGs, in particular, CDCA5 was aberrantly upregulated in ESCC tissues and significantly associated with poor prognosis (HR = 1.85, 95%CI: 1.14–3.01, P = .013). Immunohistochemical staining confirmed that positive CDCA5 expression was associated with advanced TNM staging and a shorter overall survival rate (45.59% vs 28.00% for CDCA5−/+ subjects, P = 1.86 × 10−3). H3K27 acetylation in CDCA5 promoter might lead to the activation of CDCA5 during ESCC tumorigenesis. Functionally, in vitro assay of gain- and loss-of-function of CDCA5 suggested that CDCA5 could promote ESCC cells proliferation, invasion, migration, apoptosis resistance and reduce chemosensitivity to cisplatin. Moreover, in vivo assay showed that silenced CDCA5 could inhibit tumor growth. Mechanistically, CDCA5 knockdown led to an arrest in G2/M phase and changes in the expression of factors that played fundamental roles in the cell cycle pathway.
Interpretation
CDCA5 contributed to ESCC progression and might serve as an attractive target for ESCC immunotherapy.
Fund
This work was supported by the Natural Science Foundation of Jiangsu Province (No. BK20181083 and BK20181496), Jiangsu Top Expert Program in Six Professions (No. WSW-003 and WSW-007), Major Program of Science and Technology Foundation of Jiangsu Province (No. BE2016790 and BE2018746), Jiangsu Medical Young Talent Project (No. QNRC2016566), the Program of Jiangsu Medical Innovation Team (No. CXTDA2017006), Postgraduate Research & Practice Innovation Program of Jiangsu Province (KYCX18_1487) and Jiangsu Province 333 Talents Project (No. BRA2017545).
Keywords: Esophageal squamous cell cancer, Cancer-testis gene, CDCA5, Immunotherapy, Biomarker
Research in context.
Evidence before this study
Cancer-testis antigens (CTAs) are promising immunotherapeutic targets of malignancies given their resilient immunogenicity and tumor-restricted expression pattern. However, systematic analysis of the specific CTAs involvement in ESCC has not been conducted.
Added value of this study
In this study, we performed a systematic screening for cancer-testis genes (CTGs) in ESCC by integrating multiple public databases with our own data. As a result, 21 CTGs were identified, 13 of which were novel. CDCA5 was aberrantly expressed in ESCC tumor tissues and showed significant association with poor ESCC prognosis. Mechanistically, we found that CDCA5 might be activated by the gain of H3K27ac. Furthermore, knockdown of CDCA5 inhibited tumor growth both in vitro and in vivo through the cell cycle pathway.
Implications of all the available evidence
These findings expanded our understanding of the systematic expression of CTGs in ESCC and how CTGs drove ESCC progression. Moreover, this study proposed novel CTGs as potential targets for ESCC immunotherapy for use in the clinics.
Alt-text: Unlabelled Box
1. Introduction
Esophageal cancer is the sixth leading cause of cancer-related death and the ninth most frequently diagnosed cancer worldwide [1]. Esophageal squamous cell carcinoma (ESCC) is the main histology subtype and accounts for >95% of all esophageal cancer cases in China [2]. Although the prognosis of ESCC has profited from the development of diagnostic techniques and therapeutic modalities over the past decades, it remains poor with a 5-year overall survival (OS) rate ranging from 10% to 30% [3]. Therefore, it is extremely important to identify effective novel therapeutic strategies to improve the survival rate of patients with ESCC, particularly when current therapies are exhausted.
In recent years, novel therapies for the treatment of malignant tumors have been proposed and developed due to an improved understanding of the fundamental mechanisms underlying tumor genomics and biology [4,5]. Immunotherapy is a novel treatment strategy that has emerged as an effective and promising option for various types of cancers [6]. The targeting of immune checkpoints and agonists of T-cell activation in melanoma and lung cancer have made their way into clinical practice; however, data regarding ESCC remain immature, and immunotherapy should be used within the framework of the clinical trial [7]. Nevertheless, ESCC might be excellent candidate disease for immunotherapy, in light of the abundant somatic mutations found in tumors, which might make the cancer cells more susceptible to recognition by the immune system due to neoepitope presentation on their surfaces that enhances tumor immunogenicity [7,8].
Cancer testis antigens (CTAs) are a large family of tumor-associated and immunogenic antigens that are highly expressed in cancer cells but limited in normal cells, except for cells in reproductive tissues, such as testis, ovary, and placenta [9,10]. The specific expression patterns and immunogenicity of CTAs make them perfect molecular target candidates for cancer immunotherapy [[11], [12], [13]]. Over the past decades, clinical trials using CTA-targeted therapeutic vaccines (such as MAGE-A and NY-ESO-1 antigens) have shown positive clinical efficacy, well-established safety and tolerability in various cancers [[13], [14], [15]]. However, the immunogenicity of different CTAs and their distribution in heterogeneous tumors vary significantly [13,16]. Previous studies have identified several CTAs that not only participate in the development of ESCC but also exhibit potential as therapeutic targets for ESCC based on a candidate gene strategy [[17], [18], [19], [20], [21]]. For example, the induction of NY-ESO-1 immunity and preferable outcomes were observed in a clinical trial of patients with ESCC vaccinated with NY-ESO-1 [22]. Similarly, a cancer vaccine therapy using three HLA-A24-restricted epitope peptides derived from three CTAs (TTK, LY6K and IMP-3) demonstrated satisfactory safety, strong immunogenicity and a high rate of disease control for patients with advanced ESCC [23]. These promising findings motivated us to explore the specific CTAs in ESCC and to provide effective immunotherapies for ESCC.
In our previous study, we established a maneuver to systematically explore the molecular landscape of cancer testis genes (CTGs) and successfully identified 876 novel CTGs across 19 cancer types using transcriptomics data from multiple independent public-available databases; unfortunately, the ESCC was not included [24]. In the present study, we performed a systematic analysis based on public-available databases and our published RNA microarray data from 119 paired ESCC samples to identify specific CTAs in ESCC. Associations between our identified CTGs and ESCC prognosis were evaluated, followed by validation in an independent ESCC cohort with 118 specimens. Furthermore, functional assays were conducted to evaluate the biological functions and potential therapeutic value of novel CTGs in ESCC. Overall, this study would provide us a deeper insight into the roles of CTGs in ESCC and prospective immunotherapeutic targets for ESCC treatment.
2. Materials and methods
2.1. Databases used in this study
Several public-available databases were used in this study, including GTEx, HBM and HPM. Transcriptomics data from GTEx, HBM and our NJMU-seq were analyzed to screen the testis-specific genes (TSGs). We used the proteomic data from the HPM database to define the testis-specific proteins (TSPs). RNA expression microarray data from 119 paired ESCC samples were further adopted to determinate candidate CTGs in ESCC. Detailed information about these databases and samples have been described in our previous papers [24,25].
2.2. Study subjects
A total of 118 ESCC patients were recruited from the First Affiliated Hospital of Nanjing Medical University. All patients received esophagectomy between January 2002 and December 2003. Patients were diagnosed as having primary ESCC by a pathological examination and received no chemo-or radio-therapy prior to surgery. This study was approved by the medical ethics committees of the First Affiliated Hospital of Nanjing Medical University.
2.3. Immunohistochemistry
Tissue microarrays (TMAs) of the 118 subjects were constructed from paraffin-embedded tissue blocks. For each tumor, a representative area was carefully selected from a hematoxylin and eosin-stained section. For each case, normal tissue and cancer tests were repeated twice. We used the avidin-biotin complex method to perform the immunohistochemistry (IHC) analysis. In brief, xylene was used for deparaffinization followed by rehydrating (ethanol) and rinsing (0.1 M PBS, pH 7.4). After incubating at 95 °C for 20 min in a citrate buffer (10 mM, pH 6.0), the sections were cooled to 30 °C and rinsed using 0.1 M PBS. Then, 3% hydrogen peroxide was used to inactivate the endogenous peroxidase and binding sites were blocked by incubating in 10% normal animal serum for 30 min. Sections were then incubated at 4 °C for 24 h with a primary antibody for CDCA5 (Abcam, ab210610; 1/200) and followed by incubation with the 2-step Polymer Detection System (Polink-2 Plus, GBI, USA) at room temperature. Dako Envision system was used for the detection with diaminobenzidine (DAB) as the chromogen. Finally, the specimens were lightly counterstained using Mayer's hematoxylin and then dehydrated and mounted. We established the negative controls by replacing the specific primary antibody with animal serum.
Two experienced pathologists who were blind to these subjects were responsible for the final IHC scores, which were calculated by combining both staining intensity and extent. Staining intensity was divided into four grades: 0 (no), 1 (weak), 2 (moderate), and 3 (strong), and staining extent was graded as 0 (≤10%), 1 (11–25%), 2 (26–50%), 3 (51–75%), and 4 (>75%). Samples with an IHC score ≥ 3 were considered as positive; otherwise, they were designated as negative.
2.4. Cell culture and transfection
The human normal esophageal epithelial cell line HET-1a and the ESCC cell lines KYSE30, ECA109, KYSE70, KYSE150, KYSE180, KYSE410, KYSE450, KYSE510 and TE-1 were purchased from Genechem Co. Ltd. (Shanghai, China). HET-1a cells were cultured in DMEM (Gibco, USA), and other cells were cultured in RPMI-1640 medium (Gibco, USA); 10% fetal bovine serum (FBS, Gibco, USA) and 1% penicillin G sodium/streptomycin sulphate were also used with a humidified atmosphere consisting of 95% air and 5% CO2 at 37 °C.
Short hairpin (shRNA) RNAs for human CDCA5 were cloned into a phU6-MCS-Ubiquitin-EGFP-IRES-puromycin plasmid (GV280, Genechem, China). To knock down endogenous CDCA5, we used two lentivirus vector sequences 5′-GCAGTTTGATCTCCTGGTT-3′ (named KD1), and 5′-AGAAACAGAAACGTAAGAA-3′ (named KD2) and one scrambled control sequence 5′-TTCTCCGAACGTGTCACGT-3′ (named NC). The transfection of the plasmids was performed using Lipofectamine 3000 reagent (Invitrogen, USA). Lentiviral infection was used to obtain stable cell lines with a low expression of sh-CDCA5 in KYSE30 and Eca109 cells. For the chemosensitivity array to cisplatin and mouse xenograft assay, only ESCC cells of KD2 group were used because of its optimum knockdown efficiency.
KYSE150 cells, which have a low CDCA5 expression, were transfected with pcDNA-CDCA5 (seq: 5′-GAGGATCCCCGGGTACCGGTCGCCACCATGTCTGGGAGGCGAACGCG-3′, named OE) or a negative control (named NC) sequence as before. The qualified ones were selected with 0.5 mg/l puromycin for 10 days.
2.5. RNA extraction, reverse transcription, and real-time PCR
The Total RNA Isolation Reagent - SuperfecTRI (Pufei Biotech, China) was used to extract the total RNA. The concentration and quality of RNA were evaluated using Nano Drop 2000C Spectrophotometer (Thermo, USA) and agarose gel electrophoresis. Complementary DNA (cDNA) was generated by using the M-MLV Reverse Transcriptase Kit (Promega, China) according to the manufacturer's protocol. The primers (RiboBio, Guangzhou, China) for human CDCA5 were as follows: F: 5′-AGAAAGTCAGGCGTTCCTACAG-3′ and R: 5′-GGGAGATTCCAGGGAGAGTCAT-3′. Real-time PCR was performed on the LightCycler480 System (Roche, USA) using SYBR Prime Script RT-PCR Kits (Takara, Japan). The expression of CDCA5 was calculated using the 2-ΔΔCt method with GAPDH as the reference gene.
2.6. Cell function tests
Cell viability was assessed via 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT) assay (Genview, USA) and colony formation assay. Flow cytometry analysis (Guava easyCyte HT, Millipore, USA) was used to detect the cell cycle distribution. Cell apoptosis was assessed by an Annexin V-FITC early apoptosis detection kit (eBioscience, USA) with Annexin V and PI double staining according to the manufacturer's instructions. Tumor cell invasion and migration activities were performed using Transwell plates (8 μm; Corning, USA). Exactly 40 μL ECM gel (Sigma, USA) was added to each Transwell for cell invasion assay. The effect of CDCA5 on the chemosensitivity of the ESCC cells to cisplatin (DDP) was assessed by MTT assay. Cells were treated with 10 μM DDP for 48 h. All assays were performed in triplicate.
2.7. Chromatin immunoprecipitation
Formaldehyde treatment crosslinks proteins to DNA to ensure co-precipitation. Cells were lysed and sonication was performed to shear the chromatin to manageable size. The average fragment size of DNA was confirmed by gel electrophoresis to ensure fragment sizes of 200–1000 bp. We used One-Day Chromatin Immunoprecipitation Kits (Cat#17-408, EZ-Magna ChIP, Millipore) for the Chromatin immunoprecipitation (ChIP) assay according to the manufacturer's protocol. Anti-Histone H3 (acetyl K27) antibody-ChIP Grade was obtained from Abcam (ab177178). Protein-DNA crosslinks were reversed and DNA was purified to remove the chromatin proteins and prepare the DNA for real-time PCR analysis. We designed 2 pairs of primers and the sequences were: Primer 1: F: 5′-AGGAAGCCAATACCGCCTTG −3′ and R: 5′-ACTGCCTGGTAGCCAATCAC-3′; Primer 2: F: 5′-TATCACCCCAAGGTCCGACT-3′ and R: 5′-ACTGCCTGGTAGCCAATCAC-3′.
2.8. Mouse xenograft assay
A total of 4 × 106 KYSE30 or Eca109 cells (in which CDCA5 was stably expressed or knocked down, respectively) in 0.1 ml phosphate-buffered saline were subcutaneously injected into the right or left oxter of female BALB/c nude mice at 4 weeks of age (SLAC Laboratory Animal Co., Ltd., Shanghai, China). Tumor size was measured with calipers every three days. Mice were euthanized by anesthesia 21 days after injection. The tumors were removed from the mice and then measured. All animal studies were approved by the Institutional Animal Care and Use Committee of the First Affiliated Hospital of Nanjing Medical University.
2.9. Western blot
Proteins from the KYSE30 and Eca109 cells were extracted using a lysis buffer (P0028, Beyotime) after being lysed in a RIPA lysis buffer (P0013, Beyotime). The cell lysates were boiled in 5× SDS-PAGE loading buffer for 10 min and then resolved by 8% SDS-PAGE. These lysates were transferred to a nitrocellulose membrane. Antibodies for CCNA2, CCNB1, CDC25A, PCNA and CDCA5 were purchased from Abcam. Tubulin (Maixin, China) was used as the internal reference and bound antibodies were visualized using an ECL kit (P0018, Beyotime).
2.10. Statistics
χ2 test was used to compare the difference distributions of categorical variables between subgroups. Associations between clinicopathological factors and the prognosis of ESCC patients were evaluated based on Cox proportional hazards regression analysis. The OS or disease-free survival was defined as from the time of operation to the date of death or disease relapse. Age, sex, TNM staging, tumor differentiation grade and tumor location were adjusted when appropriate. Differential gene expression analysis was performed using paired Student's t-test. Pearson correlation was adopted to conduct the gene co-expression analysis using the RNA expression microarray data of 119 ESCC samples. A correlation coefficient > 0.5 and a P value < .05 were defined as significantly correlated. Gene Ontology (GO) analysis was performed based on the DAVID Bioinformatics Resources 6.8 database (https://david.ncifcrf.gov/home.jsp). All analyses were performed using R 3.5.1 software.
3. Results
3.1. Characteristics of study subjects
In this study, multiple public databases were combined with the RNA expression microarray data from 119 ESCC samples to screen for CTGs in ESCC, followed by IHC validation in 118 recruited ESCC cases. The characteristics of study subjects are shown in Table 1. The OS rate of the patients in the screening dataset was 38.66% (46/119), which was similar to that in the IHC validation dataset (38.14%, 45/118). Age, sex, tumor differentiation grade and tumor location showed no significant association with the prognosis of ESCC patients, whereas lymph node stage was significantly associated with ESCC prognosis. Samples with positive lymph node metastasis had lower OS rate (55.56% vs 24.62% for N−/N+ subjects, respectively, Cox proportional hazards regression analysis, P = .002) in the screening dataset. A consistent result was also observed in the validation dataset (52.17% vs 18.37% for N−/N+ patients, respectively, Cox proportional hazards regression analysis, P = 1.85 × 10−5).
Table 1.
Characteristics of subjects in screening and validation stages.
| Characteristics | RNA expression array samples: 119 |
IHC Validation: 118 |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Dead |
Alive |
OS (%) | HR (95%CI)a | Pa | Dead |
Alive |
OS (%) | HR (95%CI)a | Pa | |
| (N = 73) | (N = 46) | (N = 73) | (N = 45) | |||||||
| Gender | 0.505 | 0.652 | ||||||||
| Male | 58 (79.45%) | 40 (86.96%) | 40.82% | 1.00 | 46 (63.01%) | 30 (66.67%) | 39.47% | 1.00 | ||
| Female | 15 (20.55%) | 6 (13.04%) | 28.57% | 1.22 (0.69–2.15) | 27 (36.99%) | 15 (33.33%) | 35.71% | 1.12 (0.69–1.80) | ||
| Age | 0.059 | 0.886 | ||||||||
| ≤60 years | 38 (52.05%) | 31 (67.39%) | 44.93% | 1.00 | 34 (46.58%) | 22 (48.89%) | 39.29% | 1.00 | ||
| >60 years | 35 (47.95%) | 15 (32.61%) | 30.00% | 1.56 (0.99–2.48) | 39 (53.42%) | 23 (51.11%) | 37.10% | 1.03 (0.65–1.64) | ||
| T stage | 0.898 | 0.035 | ||||||||
| T1-2 | 18 (24.66%) | 10 (21.74%) | 35.71% | 1.00 | 10 (13.70%) | 13 (28.89%) | 56.52% | 1.00 | ||
| T3-4 | 55 (75.34%) | 36 (78.26%) | 39.56% | 0.97 (0.57–1.64) | 63 (86.30%) | 32 (71.11%) | 33.68% | 1.95 (1.00–3.79) | ||
| N stage | 0.002 | 1.85 × 10−5 | ||||||||
| N0 | 24 (32.88%) | 30 (65.22%) | 55.56% | 1.00 | 33 (45.21%) | 36 (80.00%) | 52.17% | 1.00 | ||
| N+ | 49 (67.12%) | 16 (34.78%) | 24.62% | 2.16 (1.32–3.53) | 40 (54.79%) | 9 (20.00%) | 18.37% | 2.76 (1.74–4.40) | ||
| Differentiation | ||||||||||
| G1 | 14 (19.18%) | 9 (19.57%) | 39.13% | 1.00 | 27 (36.99%) | 19 (42.22%) | 41.30% | 1.00 | ||
| G2 | 36 (49.32%) | 28 (60.86%) | 43.75% | 0.87 (0.47–1.61) | 0.655 | 37 (50.68%) | 23 (51.11%) | 38.33% | 1.05 (0.64–1.72) | 0.849 |
| G3 | 23 (31.50%) | 9 (19.57%) | 28.13% | 1.36 (0.70–2.66) | 0.360 | 9 (12.33%) | 3 (6.67%) | 25.00% | 1.98 (0.93–4.21) | 0.078 |
| Location | ||||||||||
| Upper | 11 (15.07%) | 3 (6.52%) | 21.43% | 1.00 | 4 (5.48%) | 1 (2.22%) | 20.00% | 1.00 | ||
| Middle | 40 (54.79%) | 29 (63.04%) | 42.03% | 0.64 (0.33–1.25) | 0.193 | 18 (24.66%) | 12 (26.67%) | 40.00% | 0.47 (0.16–1.41) | 0.179 |
| Lower | 22 (30.14%) | 14 (30.43%) | 38.89% | 0.71 (0.35–1.48) | 0.365 | 51 (69.86%) | 32 (71.11%) | 38.50% | 0.48 (0.17–1.34) | 0.161 |
| CDCA5 | 0.013 | 1.86 × 10−3 | ||||||||
| Negative (Low expression) | 30 (41.10%) | 30 (65.22%) | 50.00% | 1.00 | 37 (50.68%) | 31 (68.89%) | 45.59% | 1.00 | ||
| Positive (High expression) | 43 (58.90%) | 16 (34.78%) | 27.12% | 1.85 (1.14–3.01) | 36 (49.32%) | 14 (31.11%) | 28.00% | 2.27 (1.36–3.82) | ||
Based on cox proportional hazards regression analysis, age, gender, TNM stage, tumor differentiation grades and tumor location were adjusted when appropriate.
3.2. Systematic screening for CTGs in ESCC
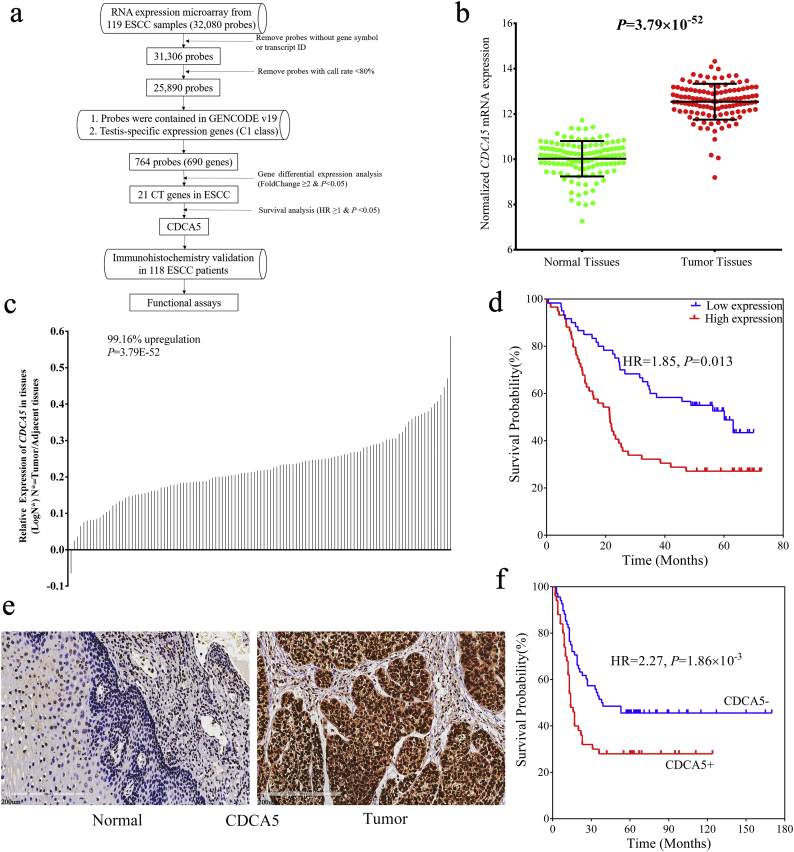
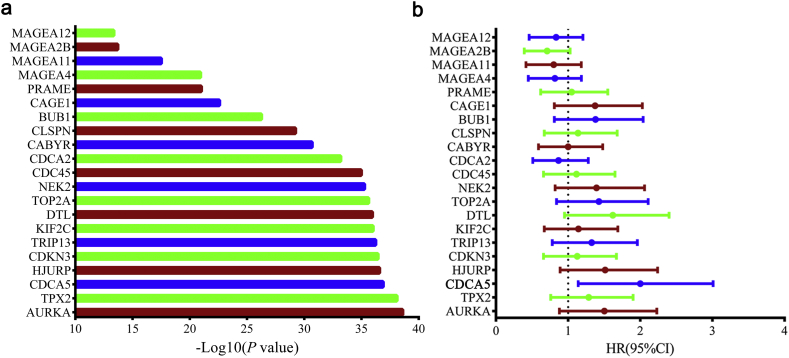
A flow chart of this study is shown in Fig. 1a. Briefly, 32,080 probes were included in the RNA expression microarray. Probes without gene symbols or transcript IDs, or probes with a detection rate ≤ 80% in all samples were excluded. As a result, 25,890 probes were initially retained. Of these probes, genes that were not contained in the Gencode v19 database were further removed. Then, the genes that belonged to the 1336 testis-specific genes (C1 class) identified by our previous study were selected [24]. Consequently, 764 probes (corresponding to 690 TSGs) were conserved as candidate CTGs in ESCC (Table S1). Of these genes, 21 genes showed aberrant expression (Paired Student's t-test, fold change ≥ 2 & P < .05) in ESCC tumor tissues and were therefore considered as CTGs in ESCC (Table S2 and Fig. S1a). Among them, 8 genes (MAGEA4, KIF2C, CABYR, CAGE1, MAGEA2B, MAGEA11, PRAME and MAGEA12) were recognized as CTGs in previous studies [11,[26], [27], [28], [29]], however, 13 genes were newly identified as ESCC specific CTGs in this study.
Fig. 1.
CDCA5 was aberrantly upregulated in ESCC tumor tissues and significantly associated with poor ESCC prognosis. (a) Flowchart of this study; (b–c) CDCA5 was significantly elevated in ESCC tumor tissues based on the RNA expression microarray data from 119 paired ESCC samples (Mean ± SD, Paired Student's t-test, P < .001); (d) ESCC patients with higher CDCA5 mRNA expression showed poorer prognosis (Log-rank test, P = .013); (e) Expression of CDCA5 protein was increased in ESCC tumor tissues compared with adjacent normal tissues (Bar represents 200 μm); (f) Positive CDCA5 expression was significantly associated with poor prognosis of ESCC in the IHC validation dataset (Log-rank test, P = 1.86 × 10−3).
3.3. Higher CDCA5 mRNA expression was associated with poorer ESCC prognosis
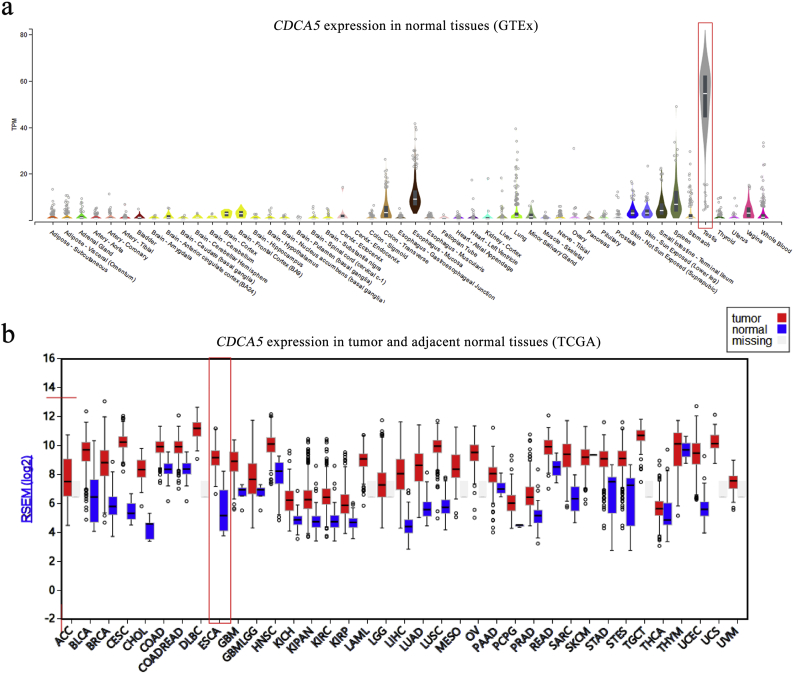
To gain insight into the roles of the identified 21 CTGs in the prognosis of ESCC, survival analysis was performed. Notably, CDCA5 was the unique one that was overexpressed in ESCC tumor tissues and associated with the poor ESCC prognosis (Fig. S1b). As shown in Fig. 1b & c, CDCA5 was upregulated in 99.16% (118/119) of all ESCC tumor tissues compared with adjacent normal tissues (Paired Student's t-test, P = 3.79 × 10−52). Furthermore, we identified the restricted pattern of CDCA5 over-expression in testis and tumor tissues by analyzing public-available databases, and the expression of CDCA5 in various normal tissues and tumor tissues was shown in Fig. S2. Patients with higher CDCA5 expression (the median expression value was set as the cut-off) showed a significantly increased mortality risk (Cox proportional hazards regression analysis, HR = 1.85, 95%CI: 1.14–3.01, P = .013) and reduced OS rate (27.12% vs 50.00%, Table 1, Fig. 1d) compared with those with lower CDCA5 expression.
3.4. CDCA5 expression was validated in an independent cohort of ESCC patients
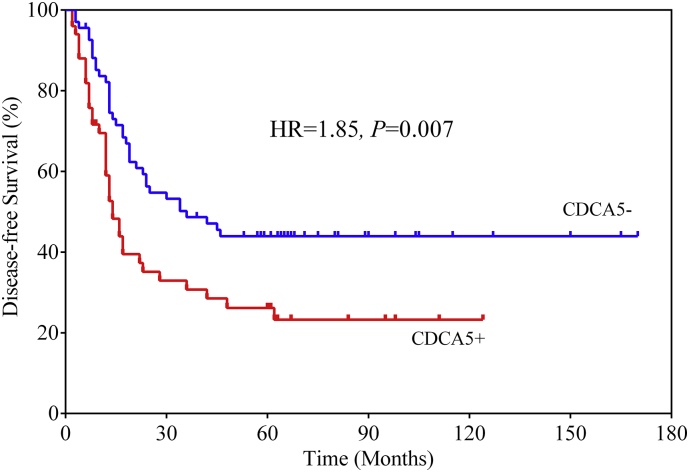
For the validation of CDCA5 at the protein level, 118 ESCC patients were recruited from our hospital. As shown in Table 1, the medium survival time was 23 months with 73 deaths in the validation dataset. IHC detection showed that CDCA5 was aberrantly elevated in ESCC tumor tissues (Fig. 1e). As a result, 50 of the 118 ESCC samples showed positive expression of CDCA5 and were correlated with poorer prognosis than the negative cases (Cox proportional hazards regression analysis, HR = 2.27, 95%CI: 1.36–3.82, P = 1.86 × 10−3, Table 1 and Fig. 1f). The OS rate was 45.59% for CDCA5- subjects, while 28.00% for the CDCA5+ patients. In addition, CDCA5 expression was significantly associated with the disease-free survival and the cumulative relapse rate of ESCC (Cox proportional hazards regression analysis, HR = 1.85, 95%CI: 1.17–2.94, P = .007, Fig. S3). The disease-free survival rate was 44% for the CDCA5- subjects, while 23.2% for the CDCA5+ patients.
3.5. Positive expression of CDCA5 was associated with advanced TNM stages
To uncover the possible mechanism by which CDCA5 modulated ESCC prognosis, we analyzed the associations between CDCA5 protein expression and clinicopathological characteristics. Positive expression of CDCA5 was significantly associated with T stage (χ2 test, P = .007), N stage (χ2 test, P = .018) and TNM stage (χ2 test, P = .008, Table 2). Patients with positive CDCA5 expression had a higher percentage of advanced T stage (92.0% vs 72.1%), N stage (54.0% vs 32.4%) and TNM stage (52.0% vs 27.9%). These findings suggested that CDCA5 might modulate the prognosis of ESCC patients by promoting tumor cells proliferation and migration.
Table 2.
Associations between CDCA5 protein expression and clinicopathological factors.
| Factors | N = 118 | Negative (n = 68) | Positive (n = 50) | Pa |
|---|---|---|---|---|
| Gender | 0.937 | |||
| Male | 76 | 44 (64.7%) | 32 (64.0%) | |
| Female | 42 | 24 (35.3%) | 18 (36.0%) | |
| Age | 0.397 | |||
| ≤60 years | 56 | 30 (44.1%) | 26 (52.0%) | |
| >60 years | 62 | 38 (55.9%) | 24 (48.0%) | |
| T stage | 0.007 | |||
| T1-2 | 23 | 19 (27.9%) | 4 (8.0%) | |
| T3-4 | 95 | 49 (72.1%) | 46 (92.0%) | |
| N stage | 0.018 | |||
| N0 | 69 | 46 (67.6%) | 23 (46.0%) | |
| N+ | 49 | 22 (32.4%) | 27 (54.0%) | |
| Differentiation | 0.604 | |||
| G1 | 46 | 29 (42.6%) | 17 (34.0%) | |
| G2 | 60 | 33 (48.5%) | 27 (54.0%) | |
| G3 | 12 | 6 (8.9%) | 6 (12.0%) | |
| Location | 0.397 | |||
| Upper | 5 | 2 (2.9%) | 3 (6.0%) | |
| Middle | 30 | 15 (22.1%) | 15 (30.0%) | |
| Lower | 83 | 51 (75.0%) | 32 (64.0%) | |
| TNM stage | 0.008 | |||
| I–II | 73 | 49 (72.1%) | 24 (48.0%) | |
| III–IV | 45 | 19 (27.9%) | 26 (52.0%) |
Based on χ2 test.
3.6. CDCA5 was epigenetically activated in ESCC cells
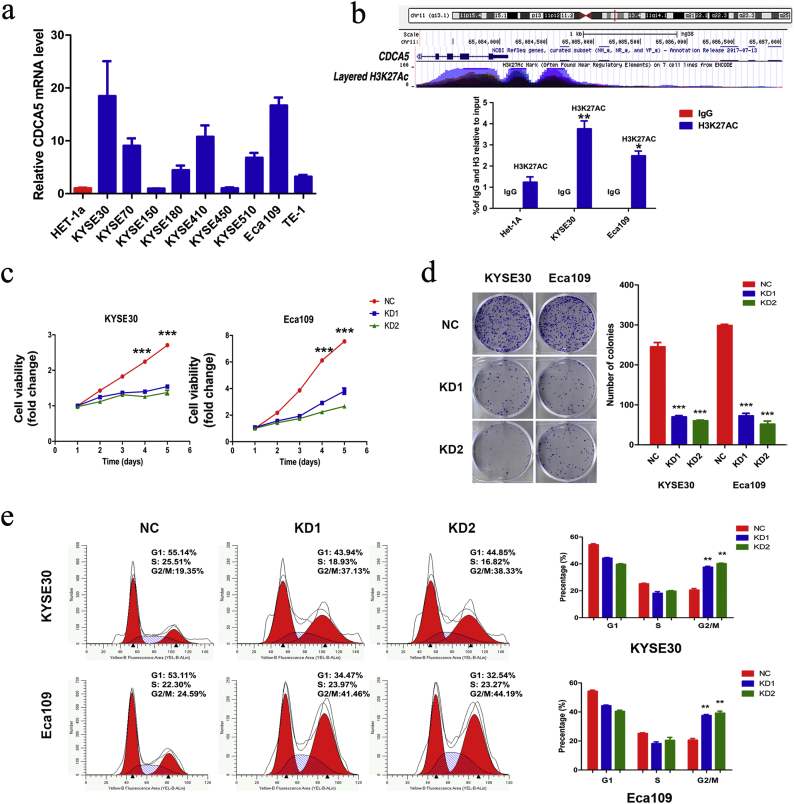
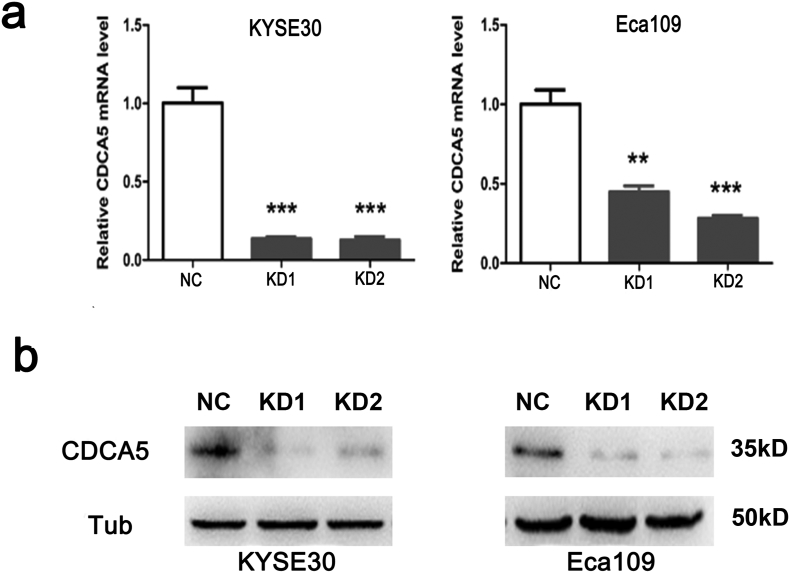
Previous studies have revealed the roles of CDCA5 in other cancer types [[30], [31], [32]]. However, the role of CDCA5 in ESCC is not well understood. CDCA5 expression was aberrantly elevated in ESCC cells (Fig. 2a). Of these cells, KYSE30 and ECA109 cells showed the most abundant expression of CDCA5 and were therefore selected as the model cells for the following functional assays. Epigenetic modifications such as methylation, acetylation and phosphorylation were one of the most common mechanisms of regulation of the CTGs expression [13,33,34]. To suggest the possible activation mechanism of CDCA5 in ESCC, we performed functional annotations based on the ENCODE database. As shown in Fig. 2b, the high enrichment of H3K27Ac (acetylation of histone H3 at lysine 27), which represents an active enhancer marker, was observed at the promoter of CDCA5, which suggested that the acetylation of the CDCA5 promoter might enhance the promoter activity of CDCA5. As we expected, the anti-H3K27Ac levels in both KYSE30 (Student's t-test, P < .01) and Eca109 (Student's t-test, P < .05) cells were significantly higher than that in Het-1A cells. These findings suggested that CDCA5 promoter acetylation might at least partially account for the activation and upregulation of CDCA5 during ESCC tumorigenesis.
Fig. 2.
CDCA5 promotes tumor proliferation, cell cycle arrest and its potential regulation. (a) Expression of CDCA5 mRNA in HET-1a and ESCC cell lines; (b) H3K27Ac (acetylation of histone H3 at lysine 27) was enriched at the promoter of CDCA5 gene (Mean ± SD, Student's t-test, *P < .05, **P < .01); (c) CDCA5 knockdown inhibited proliferation in both KYSE30 and Eca109 cells (Mean ± SD, Student's t-test, ***P < .001); (d) The amount of colony formation of ESCC cells was decreased in the CDCA5 knockdown group compared with the control (Mean ± SD, Student's t-test, ***P < .001); (e) KYSE30 and Eca109 cells transfected with shRNAs-CDCA5 exhibited a G2/M phase arrest (Mean ± SD, Student's t-test, **P < .01).
3.7. Knockdown of CDCA5 inhibits ESCC cells proliferation, migration and invasion, cell cycle arrest and promotes cell apoptosis
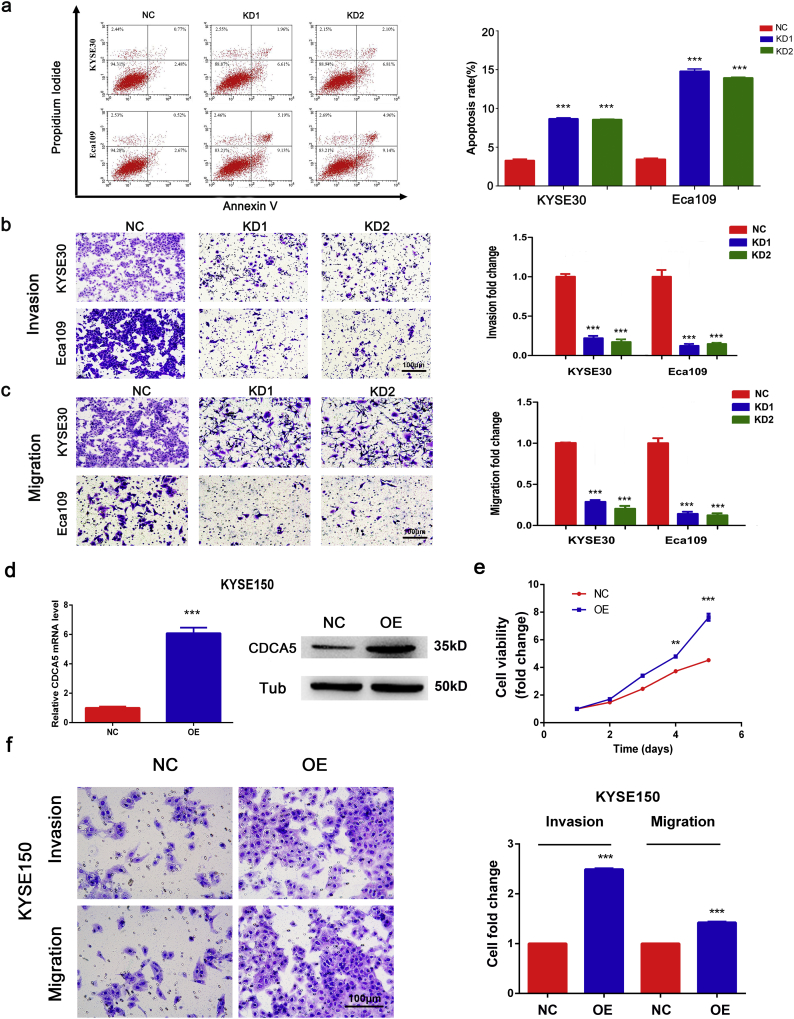
ESCC cells with suppressed CDCA5 expression were obtained by using two lentivirus vectors (Fig. S4). As shown in Fig. 2c–d, the knockdown of CDCA5 significantly reduced cell viability and proliferation in both KYSE30 and ECA109 cells (Student's t-test, P < .001). In addition, results from the flow-cytometry analysis indicated that cells (both KYSE30 and ECA109) with CDCA5 knockdown had higher percentage in G2/M phase (Student's t-test, P < .01, Fig. 2e), suggesting that the knockdown of CDCA5 could induce cell cycle arrest in ESCC. Furthermore, inhibition of CDCA5 in KYSE30 and ECA109 cells significantly promoted the cell apoptosis (Student's t-test, P < .001, Fig. 3a). These findings provided biological evidence that CDCA5 was associated with tumor proliferation. Moreover, consistent with results shown in Table 2, knockdown of CDCA5 significantly repressed the invasion (Student's t-test, P < .001, Fig. 3b) and migration (Student's t-test, P < .001, Fig. 3c) of ESCC cells.
Fig. 3.
CDCA5 promotes ESCC cells proliferation, migration and invasion. (a) CDCA5 knockdown induced apoptosis in both KYSE30 and Eca109 cells according to the flow cytometric analysis (Mean ± SD, Student's t-test, ***P < .001); (b) CDCA5 knockdown inhibited ESCC cells invasion in KYSE30 and Eca109 cells, (Mean ± SD, Student's t-test, ***P < .001); (c) Knockdown of CDCA5 inhibited the migration of tumor cells in KYSE30 and Eca109 cells, (Mean ± SD, Student's t-test, ***P < .001); (d) The efficiency of CDCA5 overexpression in KYSE150 (Mean ± SD, Student's t-test, ***P < .001); (e) Overexpression of CDCA5 promoted the proliferation of KYSE150 cells (Mean ± SD, Student's t-test, ***P < .001); (f) CDCA5 overexpression promoted KYSE150 cells invasion and migration (Mean ± SD, Student's t-test, ***P < .001).
3.8. Overexpression of CDCA5 promotes ESCC cells proliferation, invasion and migration
CDCA5 overexpression assays were performed to further confirm the effect of CDCA5 on the proliferation, invasion and migration of ESCC cells. As shown in Fig. 3d, CDCA5 was successfully overexpressed in KYSE150 cells (Student's t-test, P < .001). The MTT assay showed that the overexpression of CDCA5 promoted ESCC cells proliferation (Fig. 3e, Student's t-test, P < .001). In addition, the invasion and migration of ESCC cells with CDCA5 overexpression were significantly enhanced compared with the NC group (Fig. 3f, Student's t-test, P < .001). Both the knockdown and overexpression of CDCA5 assays proved that CDCA5 functioned as an oncogene and could promote the progression of ESCC.
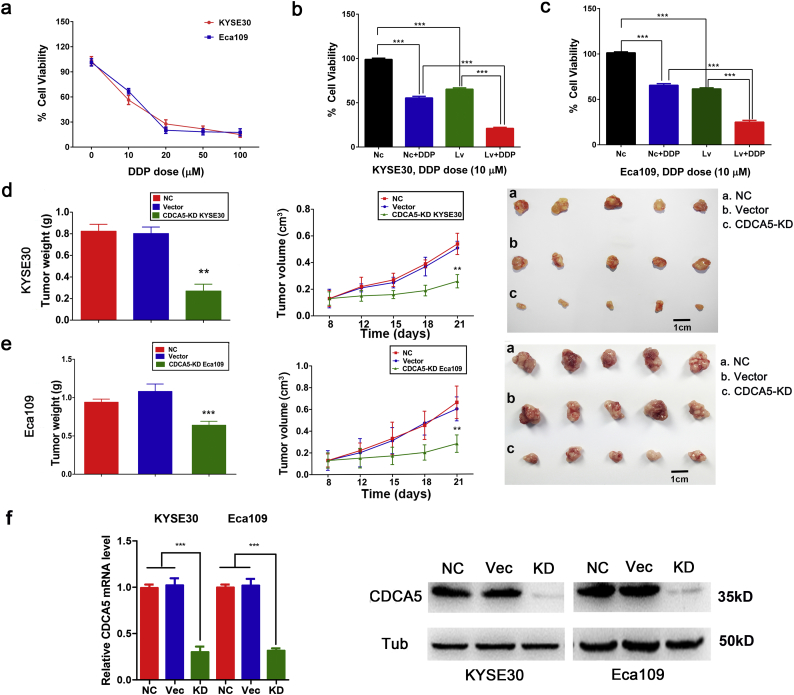
3.9. CDCA5 knockdown enhances the chemosensitivity of ESCC cells to cisplatin
Furthermore, we explored the effect of CDCA5 on the sensitivity of ESCC cells to cisplatin. As shown in Fig. 4a, KYSE30 and Eca109 cells were cultured in 10 μM, 20 μM, 50 μM and 100 μM DDP for 48 h. The concentration of 10 μM DDP was chosen to perform the following assays. The cell viability of ESCC cells with CDCA5 knockdown by using lentivirus was significantly decreased compared with the viability of the NC cells (Fig. 4b–c, Student's t-test, P < .001). Furthermore, CDCA5 knockdown cells treated with DDP showed a significantly reduced cell viability than CDCA5 knockdown cells or DDP culture cells alone (both KYSE30 and Eca109 cells, Student's t-test, P < .001). This finding suggested that CDCA5 could induce cisplatin resistance and that the inhibition of CDCA5 could enhance the sensitivity of ESCC cells to cisplatin.
Fig. 4.
Knockdown of CDCA5 enhances the chemosensitivity of ESCC cells to cisplatin and inhibits the tumor growth in vivo. (a) Dose-response curves showing the effect of DDP at different concentrations (0, 10 μM, 20 μM, 50 μM and 100 μM) on the cells viability of KYSE30 and Eca109; (b–c) Inhibition of CDCA5 enhanced the chemosensitivity of KYSE30 (b) and Eca109 (c) cells to cisplatin (Mean ± SD, Student's t-test, ***P < .001); Lv: lentivirus transfection; (d–e) CDCA5 knockdown significantly inhibited tumor growth in vivo (both Eca109 and KYSE30 cells, (Mean ± SD, Student's t-test, **P < .01, ***P < .001)); (f) The CDCA5 knockdown efficiency in established xenograft tumors.
3.10. Inhibition of CDCA5 suppresses tumor growth in vivo
As described above, CDCA5 has been proven to promote the progression of ESCC in vitro. Furthermore, we evaluated the effects of CDCA5 on tumor in vivo by a mouse xenograft assay. Three models were constructed: (1) NC: without transfection; (2) Vector: transfected with an empty vector; (3) CDCA5-KD: transfected with cells that CDCA5 was stable knockdown (KYSE30 or ECA109). As shown in Fig. 4d–e, tumor size was measured every three days and the CDCA5-KD group (both KYSE30 and ECA109 cells) had smaller tumor size than the NC or Vector group (Student's t-test, P < .01). Similarly, tumor weight in CDCA5-KD group was significantly lower compared with that in NC or Vector group (Student's t-test, P < .01). The knockdown efficiency of CDCA5 in established xenograft tumors is shown in Fig. 4f. All these findings suggested that knockdown of CDCA5 could inhibit tumor growth in vivo and CDCA5 might be a potential therapeutic target for the treatment of ESCC.
3.11. CDCA5 exerted its activity via influencing the cell cycle pathway
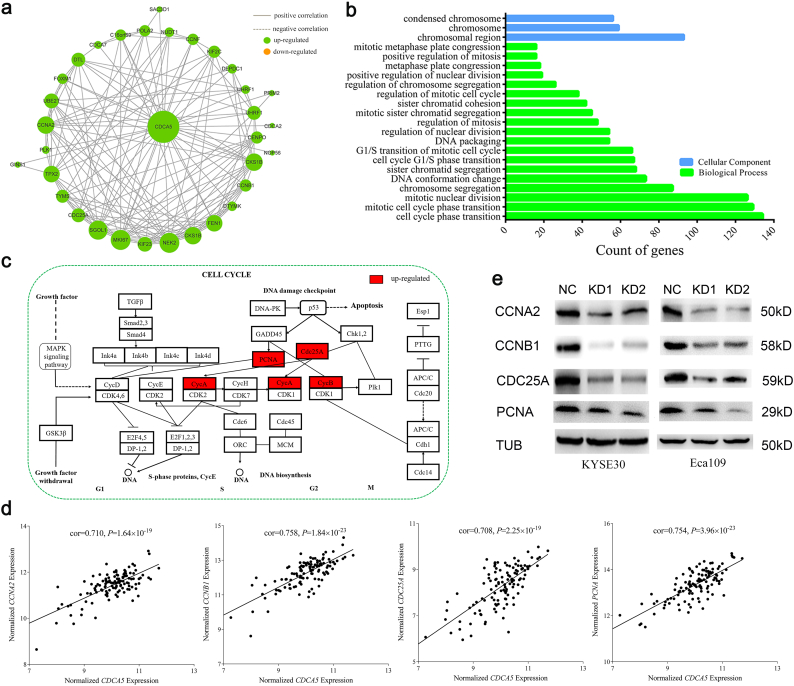
To explore the underlying mechanisms and the potential pathway through which CDCA5 promoted ESCC progression. As shown in Fig. 5a, 30 genes were aberrantly expressed (Fold change ≥ 2 & P < .05) and showed significant correlation with the expression of CDCA5 in ESCC tumor tissues. GO analysis suggested that CDCA5 might mainly participate in cell cycle related biological processes according to the KEGG database (Fig. 5b). Specifically, CCNA2, CCNB1, CDC25A and PCNA were considered as the key members in the cell cycle pathway and showed closely co-expression with CDCA5 (Fig. 5c and e). Western blot assay confirmed that the expression of CCNA2, CCNB1, CDC25A and PCNA was greatly decreased in CDCA5-knockdown cells (both KYSE30 and ECA109, Fig. 5e). These findings suggested that CDCA5 could promote the progression of ESCC by activating the cell cycle pathway.
Fig. 5.
CDCA5 promotes ESCC progression through the cell cycle pathway. (a) Genes co-expressed with CDCA5 in the RNA microarray data; (b) Gene Ontology analysis (GO) of differentially expressed genes in the RNA microarray based on DAVID Bioinformatics Resources 6.8; (c) CycA (CCNA2), CycB (CCNB1), CDC25A and PCNA participates in the cell cycle pathway according to the KEGG database; (d) Expression of CCNA2, CCNB1, CDC25A and PCNA was positively correlated with CDCA5 mRNA expression in the RNA microarray data (Pearson correlation analysis, P < .001); (e) Western blot assay showed that knockdown of CDCA5 inhibited the expression of CCNA2, CCNB1, CDC25A and PCNA in both KYSE30 and Eca109 cells.
4. Discussion
Recent prominent advances in the identification and characterization of novel specific molecular targeting has enhanced the development of innovative cancer treatment strategies, including new types of therapeutic agents or antibodies and cancer vaccines [35,36]. Molecular targeted drugs are expected to be highly specific to malignant cells, with minimal adverse effects due to their well-defined mechanisms. One attractive strategy to achieve this goal is to combine the power of a comprehensive analysis to effectively screen for genes that are overexpressed in cancer cells but minimally expressed in normal organs. CTAs are multifunctional protein group with specific expression patterns in various types of cancer cells; they are considered as unique and promising cancer biomarkers and targets for cancer therapy [13]. Using our systematic approach, 21 CTGs were identified. Of these CTGs, 8 have been recognized, while the other 13 CTGs were considered as novel in ESCC.
Of the 8 reported CTGs, MAGEA4, MAGEA2B, MAGEA11 and MAGEA12 belong to the MAGEA family, which is a well-known highly conserved family of CTGs located on the human X-chromosome q28 [13]. MAGEA proteins are processed by the intracellular proteasome and their peptides are presented by the MHC class I molecules on the surface of cancer cells thereby making them ideal cancer specific antigens. Growing evidence supports MAGEA protein involvement in the regulation of the processes that underlie cancer cell survival, tumor formation, and metastasis [37,38]. For instance, MAGEA4 inhibits the apoptosis of cancer cells by suppressing endogenousp53, or by enhancing malignant progression via p53-independent pathways [39]. Simultaneously, the overexpression of TWIST1, a bHLH transcription factor, may transcriptionally up-regulate the expression of MAGEA4 to activate cancer cell migration-invasion program [40]. In addition, MAGEA11 can activate androgen receptor signaling by forming a molecular bridge between transcriptionally active androgen receptor dimers to contribute to cancer cell growth [41]. Most recently, MAGEA11 was found to promote oncogenesis through interactions with retinoblastoma-related protein p107 and the E2F1 transcription factor, which is important for cell cycle progression and apoptosis [42]. Regarding the other 4 reported CTGs (KIF2C, CABYR, CAGE1 and PRAME), although previous studies have discovered that they may play a critical role in tumorigenesis [[43], [44], [45], [46]], the potential molecular mechanisms, especially those in ESCC, are not yet well understood. More studies are warranted to further explore their functions in ESCC.
Of the thirteen identified novel CTGs, several CTGs have been reported as having essential roles in cancers. For example, AURKA encodes a cell cycle-regulated kinase and participates in the regulation of the cell cycles in multiple cancers, including esophageal cancer [[47], [48], [49]]. Similarly, TPX2 encodes microtubule-associated protein and takes part in the normal assembly of the microtubules involved in the tumorigenesis of colorectal cancer, bladder cancer and hepatocellular carcinoma during apoptosis [[50], [51], [52]]. Importantly, CDCA5 expression was significantly increased in ESCC tumor tissues and was correlated with poor clinical outcomes. The CDCA5 gene, located on chromosome11q, is a critical regulator of sister-chromatid cohesion and separation during cell division [53,54]. The aberrant upregulation of CDCA5 has been observed in human cancers during tumor progression, and indicated poor prognosis such as lung cancer, hepatocellular carcinoma, and breast cancer [31,55,56]. For the first time, we found that the expression of CDCA5 was aberrantly elevated in ESCC tumor tissues and higher CDCA5 expression was significantly associated with the poorer prognosis of ESCC patients.
Regarding the underlying mechanisms, CDCA5 was identified as an evolutionarily conserved small molecular protein that contributes to maintenance of sister chromatid cohesion from the S phase to the onset of anaphase [57]. It has been reported that high levels of CDCA5 expression promoted lung cancer cells proliferation interaction with ERK kinase through the activation of the MAPK pathway [31]. In another study, the upregulation of CDCA5 was confirmed to increase cell viability and proliferation in gastric cancer malignant progression via influencing cyclin E1 [58]. However, the tumorigenic functions of CDCA5 in ESCC have remained largely unclear thus far. The present study found that the knockdown of CDCA5 strongly inhibited the proliferation, migration and invasion of ESCC cells. Consistently, the overexpression of CDCA5 promoted ESCC cells proliferation, invasion and migration. What's more, CDCA5 knockdown enhanced the chemosensitivity of ESCC cells to cisplatin and led to a significant reduction in the tumor volumes and weights of mice tumors. All of these findings suggested that CDCA5 might serve as a potential immunotherapeutic target for ESCC intervention. Recent evidence indicates that the regulation of the cell cycle is extremely important to the cell because a dysregulated cell cycle leads the cell to grow autonomously, which is thought to be a fundamental hallmark of cancer, especially the checkpoint pathways [30,59]. In this study, we present the first evidence that CDCA5 knockdown induced G2/M phases arrest and apoptosis in ESCC. The results support the putative role of CDCA5 in chromatid separation during the G2/M transition [60]. Notably, the GO enrichment analysis demonstrated that the term “cell cycle” ranks first among CDCA5-related potential pathways. Moreover, the key members of cell cycle pathway (CCNA2, CCNB1, CDC25A and PCNA) showed significant co-expression with CDCA5. Consistent with this finding, in vitro experiments revealed that inhibition of CDCA5 significantly suppressed the expression of CCNA2, CCNB1, CDC25A and PCNA, and these genes appear to function with cell cycle pathway to govern ESCC development. Regarding the regulation of CDCA5, we found that the CDCA5 promoter was highly acetylated in ESCC cells. Highlighting the finding, we speculated that the acetylation modification of the CDCA5 promoter might be the potential activation mechanism of CDCA5 in ESCC. Further functional assays are warranted to support our speculation.
In conclusion, we outline a schematic of CTGs in ESCC by integrating public databases and our data. A total of 13 novel CTGs were successfully identified in ESCC. Moreover, CDCA5 was characterized as a novel tumor-promoting gene and was significantly associated with unfavorable ESCC prognosis, suggesting that it might be a favorite prognostic biomarker in the clinic and a prospective immunotherapeutic target for ESCC vaccines.
The following are the supplementary data related to this article.
Fig. S1.
Differential expression analysis and prognosis analysis of the 21 identified CT genes in ESCC. (a) A total of 21 genes were significantly elevated in ESCC tumor tissues (Paired Student's t-test, Fold change ≥ 2.00, P < .05); (b) Associations between 21 CT genes expression and the prognosis of ESCC patients. CDCA5 was the only gene that showed significant association with ESCC prognosis (Cox proportional hazards regression analysis, HR > 1.00, P < .05).
Fig. S2.
The expression of CDCA5 in various normal and tumor tissues.
(a) CDCA5 expression in various normal tissues based on the GTEx v7 database; CDCA5 showed the highest expression in normal testis tissues (marked in red box) but limited expression in other normal tissues; (b) The expression of CDCA5 in a variety of tumor tissues and adjacent normal tissues based on the TCGA database; CDCA5 showed aberrantly upregulated expression in most of tumor tissues, including esophageal cancer (marked in red box).
Fig. S3.
CDCA5 was significantly associated with the disease-free survival of ESCC patients. ESCC patients with positive CDCA5 expression had a higher risk of relapse and a poorer prognosis (Cox proportional hazards regression analysis, HR = 1.85, P = .007).
Fig. S4.
CDCA5 knockdown efficiency of shRNAs in KYSE30 and Eca109 cells. (a) The knockdown efficiency of KD1 and KD2 in KYSE30 and Eca109 cells, Mean ± SD, Student's t-test, **P < .01, ***P < .001; (b) The western blot of knockdown efficiency of KD1 and KD2 in KYSE30 and Eca109 cells.
Table S1. List of 764 testis-specific expression probes (690 genes, C1 class). Table S2. 21 CT genes in ESCC identified in this study.
Author contributions
J. Xu, C. Zhu, Y. Yu and W. Wu participated in data analysis and drafting of the manuscript; J. Cao, Z. Li, J. Dai and C. Wang contributed to the analysis and the making of tables and figures; Y. Tang, Q. Zhu, J. Wang, W. Wen and L. Xue performed the functional assays; F. Zhen, J. Liu, C. Huang, F. Zhao, Y. Zhou and Z. He were involved in material and technical support; X. Pan, H. Wei, Y. Zhu, Y. He and J. Que took part in the collection of samples; J. Luo, L. Chen and W. Wang designed this study and revised the manuscript. All authors approved the final version of the manuscript.
Declaration of Competing Interest
None.
Acknowledgments
Acknowledgments
We thank the study participants and research staff for their contributions and commitment to this study.
Funding
This work was supported by the Natural Science Foundation of Jiangsu Province (No. BK20181083 and BK20181496), Jiangsu Top Expert Program in Six Professions (No. WSW-003 and WSW-007), Major Program of Science and Technology Foundation of Jiangsu Province (No. BE2016790 and BE2018746), Jiangsu Medical Young Talent Project (No. QNRC2016566), the Program of Jiangsu Medical Innovation Team (No. CXTDA2017006), Postgraduate Research & Practice Innovation Program of Jiangsu Province (KYCX18_1487) and Jiangsu Province 333 Talents Project (No. BRA2017545).
Contributor Information
Jinghua Luo, Email: ljh1966@126.com.
Liang Chen, Email: clbright0909@njmu.edu.cn.
Wei Wang, Email: wangwei15261883958@163.com.
References
- 1.Lagergren J., Smyth E., Cunningham D., Lagergren P. Oesophageal cancer. Lancet. 2017;390(10110):2383–2396. doi: 10.1016/S0140-6736(17)31462-9. [DOI] [PubMed] [Google Scholar]
- 2.Arnold M., Soerjomataram I., Ferlay J., Forman D. Global incidence of oesophageal cancer by histological subtype in 2012. Gut. 2015;64(3):381–387. doi: 10.1136/gutjnl-2014-308124. [DOI] [PubMed] [Google Scholar]
- 3.Allemani C., Matsuda T., Di Carlo V., Harewood R., Matz M., Niksic M. Global surveillance of trends in cancer survival 2000-14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet. 2018;391(10125):1023–1075. doi: 10.1016/S0140-6736(17)33326-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kirkwood J.M., Butterfield L.H., Tarhini A.A., Zarour H., Kalinski P., Ferrone S. Immunotherapy of cancer in 2012. CA Cancer J Clin. 2012;62(5):309–335. doi: 10.3322/caac.20132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mun E.J., Babiker H.M., Weinberg U., Kirson E.D., Von Hoff D.D. Tumor-treating fields: a fourth modality in cancer treatment. Clin Cancer Res. 2018;24(2):266–275. doi: 10.1158/1078-0432.CCR-17-1117. [DOI] [PubMed] [Google Scholar]
- 6.Myint Z.W., Goel G. Role of modern immunotherapy in gastrointestinal malignancies: a review of current clinical progress. J Hematol Oncol. 2017;10(1):86. doi: 10.1186/s13045-017-0454-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lawrence M.S., Stojanov P., Polak P., Kryukov G.V., Cibulskis K., Sivachenko A. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499(7457):214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lesokhin A.M., Callahan M.K., Postow M.A., Wolchok J.D. On being less tolerant: enhanced cancer immunosurveillance enabled by targeting checkpoints and agonists of T cell activation. Sci Transl Med. 2015;7(280):280sr1. doi: 10.1126/scitranslmed.3010274. [DOI] [PubMed] [Google Scholar]
- 9.Scanlan M.J., Gure A.O., Jungbluth A.A., Old L.J., Chen Y.T. Cancer/testis antigens: an expanding family of targets for cancer immunotherapy. Immunol Rev. 2002;188:22–32. doi: 10.1034/j.1600-065x.2002.18803.x. [DOI] [PubMed] [Google Scholar]
- 10.Simpson A.J., Caballero O.L., Jungbluth A., Chen Y.T., Old L.J. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer. 2005;5(8):615–625. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- 11.Yao J., Caballero O.L., Yung W.K., Weinstein J.N., Riggins G.J., Strausberg R.L. Tumor subtype-specific cancer-testis antigens as potential biomarkers and immunotherapeutic targets for cancers. Cancer Immunol Res. 2014;2(4):371–379. doi: 10.1158/2326-6066.CIR-13-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li B., Severson E., Pignon J.C., Zhao H., Li T., Novak J. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17(1):174. doi: 10.1186/s13059-016-1028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gordeeva O. Cancer-testis antigens: unique cancer stem cell biomarkers and targets for cancer therapy. Semin Cancer Biol. 2018;53:75–89. doi: 10.1016/j.semcancer.2018.08.006. [DOI] [PubMed] [Google Scholar]
- 14.Dreno B., Thompson J.F., Smithers B.M., Santinami M., Jouary T., Gutzmer R. MAGE-A3 immunotherapeutic as adjuvant therapy for patients with resected, MAGE-A3-positive, stage III melanoma (DERMA): a double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol. 2018;19(7):916–929. doi: 10.1016/S1470-2045(18)30254-7. [DOI] [PubMed] [Google Scholar]
- 15.Yoshitake Y., Fukuma D., Yuno A., Hirayama M., Nakayama H., Tanaka T. Phase II clinical trial of multiple peptide vaccination for advanced head and neck cancer patients revealed induction of immune responses and improved OS. Clin Cancer Res. 2015;21(2):312–321. doi: 10.1158/1078-0432.CCR-14-0202. [DOI] [PubMed] [Google Scholar]
- 16.Aubry F., Satie A.P., Rioux-Leclercq N., Rajpert-De Meyts E., Spagnoli G.C., Chomez P. MAGE-A4, a germ cell specific marker, is expressed differentially in testicular tumors. Cancer. 2001;92(11):2778–2785. doi: 10.1002/1097-0142(20011201)92:11<2778::aid-cncr10125>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 17.Ishikawa N., Takano A., Yasui W., Inai K., Nishimura H., Ito H. Cancer-testis antigen lymphocyte antigen 6 complex locus K is a serologic biomarker and a therapeutic target for lung and esophageal carcinomas. Cancer Res. 2007;67(24):11601–11611. doi: 10.1158/0008-5472.CAN-07-3243. [DOI] [PubMed] [Google Scholar]
- 18.Chen Y.T., Panarelli N.C., Piotti K.C., Yantiss R.K. Cancer-testis antigen expression in digestive tract carcinomas: frequent expression in esophageal squamous cell carcinoma and its precursor lesions. Cancer Immunol Res. 2014;2(5):480–486. doi: 10.1158/2326-6066.CIR-13-0124. [DOI] [PubMed] [Google Scholar]
- 19.Forghanifard M.M., Gholamin M., Farshchian M., Moaven O., Memar B., Forghani M.N. Cancer-testis gene expression profiling in esophageal squamous cell carcinoma: identification of specific tumor marker and potential targets for immunotherapy. Cancer Biol Ther. 2011;12(3):191–197. doi: 10.4161/cbt.12.3.15949. [DOI] [PubMed] [Google Scholar]
- 20.Okabayashi K., Fujita T., Miyazaki J., Okada T., Iwata T., Hirao N. Cancer-testis antigen BORIS is a novel prognostic marker for patients with esophageal cancer. Cancer Sci. 2012;103(9):1617–1624. doi: 10.1111/j.1349-7006.2012.02355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.da Silva V.L., Fonseca A.F., Fonseca M., da Silva T.E., Coelho A.C., Kroll J.E. Genome-wide identification of cancer/testis genes and their association with prognosis in a pan-cancer analysis. Oncotarget. 2017;8(54):92966–92977. doi: 10.18632/oncotarget.21715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wada H., Sato E., Uenaka A., Isobe M., Kawabata R., Nakamura Y. Analysis of peripheral and local anti-tumor immune response in esophageal cancer patients after NY-ESO-1 protein vaccination. Int J Cancer. 2008;123(10):2362–2369. doi: 10.1002/ijc.23810. [DOI] [PubMed] [Google Scholar]
- 23.Kono K., Mizukami Y., Daigo Y., Takano A., Masuda K., Yoshida K. Vaccination with multiple peptides derived from novel cancer-testis antigens can induce specific T-cell responses and clinical responses in advanced esophageal cancer. Cancer Sci. 2009;100(8):1502–1509. doi: 10.1111/j.1349-7006.2009.01200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang C., Gu Y., Zhang K., Xie K., Zhu M., Dai N. Systematic identification of genes with a cancer-testis expression pattern in 19 cancer types. Nat Commun. 2016;7 doi: 10.1038/ncomms10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J., Chen Z., Tian L., Zhou C., He M.Y., Gao Y. LncRNA profile study reveals a three-lncRNA signature associated with the survival of patients with oesophageal squamous cell carcinoma. Gut. 2014;63(11):1700–1710. doi: 10.1136/gutjnl-2013-305806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Specht E., Kaemmerer D., Sanger J., Wirtz R.M., Schulz S., Lupp A. Comparison of immunoreactive score, HER2/neu score and H score for the immunohistochemical evaluation of somatostatin receptors in bronchopulmonary neuroendocrine neoplasms. Histopathology. 2015;67(3):368–377. doi: 10.1111/his.12662. [DOI] [PubMed] [Google Scholar]
- 27.Nishikawa H., Maeda Y., Ishida T., Gnjatic S., Sato E., Mori F. Cancer/testis antigens are novel targets of immunotherapy for adult T-cell leukemia/lymphoma. Blood. 2012;119(13):3097–3104. doi: 10.1182/blood-2011-09-379982. [DOI] [PubMed] [Google Scholar]
- 28.Nettersheim D., Arndt I., Sharma R., Riesenberg S., Jostes S., Schneider S. The cancer/testis-antigen PRAME supports the pluripotency network and represses somatic and germ cell differentiation programs in seminomas. Br J Cancer. 2016;115(4):454–464. doi: 10.1038/bjc.2016.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H., Fang L., Xiao X., Shen L. The expression and effects the CABYR-c transcript of CABYR gene in hepatocellular carcinoma. Bull Cancer. 2012;99(3):E26–E33. doi: 10.1684/bdc.2011.1538. [DOI] [PubMed] [Google Scholar]
- 30.Chang I.W., Lin V.C., He H.L., Hsu C.T., Li C.C., Wu W.J. CDCA5 overexpression is an indicator of poor prognosis in patients with urothelial carcinomas of the upper urinary tract and urinary bladder. Am J Transl Res. 2015;7(4):710–722. [PMC free article] [PubMed] [Google Scholar]
- 31.Nguyen M.H., Koinuma J., Ueda K., Ito T., Tsuchiya E., Nakamura Y. Phosphorylation and activation of cell division cycle associated 5 by mitogen-activated protein kinase play a crucial role in human lung carcinogenesis. Cancer Res. 2010;70(13):5337–5347. doi: 10.1158/0008-5472.CAN-09-4372. [DOI] [PubMed] [Google Scholar]
- 32.Xu T., Ma M., Dai J., Yu S., Wu X., Tang H. Gene expression screening identifies CDCA5 as a potential therapeutic target in acral melanoma. Hum Pathol. 2018;75:137–145. doi: 10.1016/j.humpath.2018.02.009. [DOI] [PubMed] [Google Scholar]
- 33.Yawata T., Nakai E., Park K.C., Chihara T., Kumazawa A., Toyonaga S. Enhanced expression of cancer testis antigen genes in glioma stem cells. Mol Carcinog. 2010;49(6):532–544. doi: 10.1002/mc.20614. [DOI] [PubMed] [Google Scholar]
- 34.Wischnewski F., Pantel K., Schwarzenbach H. Promoter demethylation and histone acetylation mediate gene expression of MAGE-A1, -A2, -A3, and -A12 in human cancer cells. Mol Cancer Res. 2006;4(5):339–349. doi: 10.1158/1541-7786.MCR-05-0229. [DOI] [PubMed] [Google Scholar]
- 35.Kawaguchi H., Ohno S., Miyazaki M., Hashimoto K., Egashira A., Saeki H. CYFRA 21-1 determination in patients with esophageal squamous cell carcinoma: clinical utility for detection of recurrences. Cancer. 2000;89(7):1413–1417. doi: 10.1002/1097-0142(20001001)89:7<1413::aid-cncr1>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 36.Hirata D., Yamabuki T., Miki D., Ito T., Tsuchiya E., Fujita M. Involvement of epithelial cell transforming sequence-2 oncoantigen in lung and esophageal cancer progression. Clin Cancer Res. 2009;15(1):256–266. doi: 10.1158/1078-0432.CCR-08-1672. [DOI] [PubMed] [Google Scholar]
- 37.Lian Y., Meng L., Ding P., Sang M. Epigenetic regulation of MAGE family in human cancer progression-DNA methylation, histone modification, and non-coding RNAs. Clin Epigenetics. 2018;10(1):115. doi: 10.1186/s13148-018-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schooten E., Di Maggio A., van Bergen En Henegouwen P.M.P., Kijanka M.M. MAGE-A antigens as targets for cancer immunotherapy. Cancer Treat Rev. 2018;67:54–62. doi: 10.1016/j.ctrv.2018.04.009. [DOI] [PubMed] [Google Scholar]
- 39.Fujiwara-Kuroda A., Kato T., Abiko T., Tsuchikawa T., Kyogoku N., Ichinokawa M. Prognostic value of MAGEA4 in primary lung cancer depends on subcellular localization and p53 status. Int J Oncol. 2018;53(2):713–724. doi: 10.3892/ijo.2018.4425. [DOI] [PubMed] [Google Scholar]
- 40.Forghanifard M.M., Rad A., Farshchian M., Khaleghizadeh M., Gholamin M., Moghbeli M. TWIST1 upregulates the MAGEA4 oncogene. Mol Carcinog. 2017;56(3):877–885. doi: 10.1002/mc.22541. [DOI] [PubMed] [Google Scholar]
- 41.Meek D.W., Marcar L. MAGE-A antigens as targets in tumour therapy. Cancer Lett. 2012;324(2):126–132. doi: 10.1016/j.canlet.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 42.Su S., Minges J.T., Grossman G., Blackwelder A.J., Mohler J.L., Wilson E.M. Proto-oncogene activity of melanoma antigen-A11 (MAGE-A11) regulates retinoblastoma-related p107 and E2F1 proteins. J Biol Chem. 2013;288(34):24809–24824. doi: 10.1074/jbc.M113.468579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gnjatic S., Cao Y., Reichelt U., Yekebas E.F., Nolker C., Marx A.H. NY-CO-58/KIF2C is overexpressed in a variety of solid tumors and induces frequent T cell responses in patients with colorectal cancer. Int J Cancer. 2010;127(2):381–393. doi: 10.1002/ijc.25058. [DOI] [PubMed] [Google Scholar]
- 44.Luo C., Xiao X., Liu D., Chen S., Li M., Xu A. CABYR is a novel cancer-testis antigen in lung cancer. Clin Cancer Res. 2007;13(4):1288–1297. doi: 10.1158/1078-0432.CCR-06-1742. [DOI] [PubMed] [Google Scholar]
- 45.Qian Z., Li M., Wang R., Xiao Q., Wang J., Li M. Knockdown of CABYR-a/b increases chemosensitivity of human non-small cell lung cancer cells through inactivation of Akt. Mol Cancer Res. 2014;12(3):335–347. doi: 10.1158/1541-7786.MCR-13-0391. [DOI] [PubMed] [Google Scholar]
- 46.Oehler V.G., Guthrie K.A., Cummings C.L., Sabo K., Wood B.L., Gooley T. The preferentially expressed antigen in melanoma (PRAME) inhibits myeloid differentiation in normal hematopoietic and leukemic progenitor cells. Blood. 2009;114(15):3299–3308. doi: 10.1182/blood-2008-07-170282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prigent C., Giet R. Aurora A and mitotic commitment. Cell. 2003;114(5):531–532. doi: 10.1016/s0092-8674(03)00685-8. [DOI] [PubMed] [Google Scholar]
- 48.Shah K.N., Bhatt R., Rotow J., Rohrberg J., Olivas V., Wang V.E. Aurora kinase A drives the evolution of resistance to third-generation EGFR inhibitors in lung cancer. Nat Med. 2019;25(1):111–118. doi: 10.1038/s41591-018-0264-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tanaka E., Hashimoto Y., Ito T., Okumura T., Kan T., Watanabe G. The clinical significance of Aurora-A/STK15/BTAK expression in human esophageal squamous cell carcinoma. Clin Cancer Res. 2005;11(5):1827–1834. doi: 10.1158/1078-0432.CCR-04-1627. [DOI] [PubMed] [Google Scholar]
- 50.Sillars-Hardebol A.H., Carvalho B., Tijssen M., Belien J.A., de Wit M., Delis-van Diemen P.M. TPX2 and AURKA promote 20q amplicon-driven colorectal adenoma to carcinoma progression. Gut. 2012;61(11):1568–1575. doi: 10.1136/gutjnl-2011-301153. [DOI] [PubMed] [Google Scholar]
- 51.Yan L., Li Q., Yang J., Qiao B. TPX2-p53-GLIPR1 regulatory circuitry in cell proliferation, invasion, and tumor growth of bladder cancer. J Cell Biochem. 2018;119(2):1791–1803. doi: 10.1002/jcb.26340. [DOI] [PubMed] [Google Scholar]
- 52.Huang Y., Guo W., Kan H. TPX2 is a prognostic marker and contributes to growth and metastasis of human hepatocellular carcinoma. Int J Mol Sci. 2014;15(10):18148–18161. doi: 10.3390/ijms151018148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nishiyama T., Ladurner R., Schmitz J., Kreidl E., Schleiffer A., Bhaskara V. Sororin mediates sister chromatid cohesion by antagonizing Wapl. Cell. 2010;143(5):737–749. doi: 10.1016/j.cell.2010.10.031. [DOI] [PubMed] [Google Scholar]
- 54.Nishiyama T., Sykora M.M., Huis in 't Veld P.J., Mechtler K., Peters J.M. Aurora B and Cdk1 mediate Wapl activation and release of acetylated cohesin from chromosomes by phosphorylating Sororin. Proc Natl Acad Sci U S A. 2013;110(33):13404–13409. doi: 10.1073/pnas.1305020110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tian Y., Wu J., Chagas C., Du Y., Lyu H., He Y. CDCA5 overexpression is an Indicator of poor prognosis in patients with hepatocellular carcinoma (HCC) BMC Cancer. 2018;18(1):1187. doi: 10.1186/s12885-018-5072-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Phan N.N., Wang C.Y., Li K.L., Chen C.F., Chiao C.C., Yu H.G. Distinct expression of CDCA3, CDCA5, and CDCA8 leads to shorter relapse free survival in breast cancer patient. Oncotarget. 2018;9(6):6977–6992. doi: 10.18632/oncotarget.24059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang N., Pati D. Sororin is a master regulator of sister chromatid cohesion and separation. Cell Cycle. 2012;11(11):2073–2083. doi: 10.4161/cc.20241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Z., Shen M., Zhou G. Upregulation of CDCA5 promotes gastric cancer malignant progression via influencing cyclin E1. Biochem Biophys Res Commun. 2018;496(2):482–489. [Google Scholar]
- 59.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 60.Borton M.T., Rashid M.S., Dreier M.R., Taylor W.R. Multiple levels of regulation of sororin by Cdk1 and aurora B. J Cell Biochem. 2016;117(2):351–360. doi: 10.1002/jcb.25277. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. List of 764 testis-specific expression probes (690 genes, C1 class). Table S2. 21 CT genes in ESCC identified in this study.