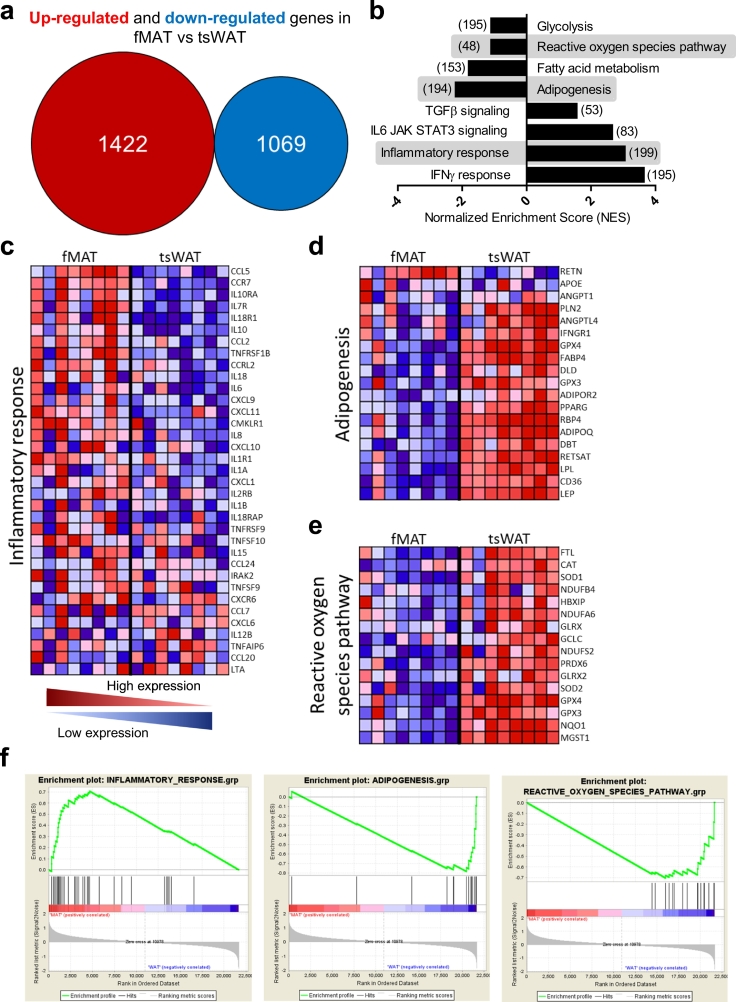
Fig. 2.
Identification of genes and pathways differentially expressed in fMAT versus tsWAT adipocytes. (a) Overall expression of up-regulated (red) and down-regulated (blue) genes in fMAT compared to tsWAT measured by GeneChip® HuGene 2.1 microarray. (b) Inversely up- and down-regulated pathways in fMAT vs tsWAT analysed with Gen Set Enrichment analysis. The number of genes of each gene set is shown within brackets. Highlighted enrichment gene sets show an FDR of <25%, p < .0001. (c - e) Heatmap of genes for selected pathways in fMAT and tsWAT. Heatmap colours indicate red, high; pink, moderate; light blue, low; and dark blue, lowest expression. Samples of eight donors pooled from at least three independent experiments. (c) Inflammatory response genes are shown. (d) Adipogenesis and fatty acid metabolism genes are shown. (e) Genes involved in the reactive oxygen species pathway are shown. (f) Corresponding enrichment plots of the pathways inflammatory response (left), adipogenesis (middle), and reactive oxygen species (right). The enrichment score (ES) shows the degree to which a gene set is overrepresented at the top (positive correlated) or at the bottom (negative correlated) of the ranked list of genes. The ranked list metric (SignalNoise) shows positive values that correlate with the first phenotype (fMAT) and negative values that correlate with the second phenotype (tsWAT) at least 2-fold up- and down-regulated. n = eight female individuals. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)