Abstract
Studies on glucocorticoid receptor (GR) action typically assess gene responses by long-term stimulation with synthetic hormones. As corticosteroids are released from adrenal glands in a circadian and high-frequency (ultradian) mode, such treatments may not provide an accurate assessment of physiological hormone action. Here we demonstrate that ultradian hormone stimulation induces cyclic GR-mediated transcriptional regulation, or gene pulsing, both in cultured cells and in animal models. Equilibrium receptor-occupancy of regulatory elements precisely tracks the ligand pulses. Nascent RNA transcripts from GR-regulated genes are released in distinct quanta, demonstrating a profound difference between the transcriptional programs induced by ultradian and constant stimulation. Gene pulsing is driven by rapid GR exchange with response elements and by GR recycling through the chaperone machinery, which promotes GR activation and reactivation in response to the ultradian hormone release, thus coupling promoter activity to the naturally occurring fluctuations in hormone levels. The GR signalling pathway has been optimized for a prompt and timely response to fluctuations in hormone levels, indicating that biologically accurate regulation of gene targets by GR requires an ultradian mode of hormone stimulation.
Corticosteroids are important signalling molecules in a large variety of human physiological responses. Their action is mediated by two members of the nuclear receptor family, the glucocorticoid and the mineralocorticoid receptors. These hormones (cortisol in humans and corticosterone in rodents) are released from the adrenal glands in a daily, circadian cycle. Although not widely recognized, the actual pattern of hormone secretion is highly pulsatile (ultradian), with a periodicity of approximately 1 h; this pattern is also maintained in extracellular fluid1–5. Potential effects of the ultradian mode of hormone secretion on the GR-and MR-mediated transcriptional programs are unknown. Studies on gene regulation by GR typically assess gene responses using long-term stimulation with synthetic hormones. Considering the pulsatile mode of glucocorticoid secretion, such treatments may not provide an accurate assessment of physiological hormone action.
The GR has been shown to exchange rapidly with response elements in living cells6. The residence time for GR on regulatory elements is measured in seconds. It is important to evaluate the implications of these complex aspects of steroid action (rapid fluctuation of serum hormone levels; fast dynamics of GR interactions with chromatin) for the transcriptional programs that are induced by these ligands.
To address these questions, we developed protocols to examine the potential effects of ultradian hormone release on gene responses, both in cultured cell systems and in live animal models. We used live-cell imaging, photobleaching techniques and biochemical analysis of gene function to characterize the real-time action of corticosteroids during treatment with ligand pulses. We show that ultradian hormone stimulation induces cyclic GR-mediated transcriptional regulation, or gene pulsing. We also demonstrate the importance of naturally occurring hormones in the functioning of this cycle, and provide a mechanistic model that integrates three key aspects of dynamic GR action: (1) rapid GR exchange with response elements, (2) reassociation of the receptor with its ligand through the chaperone cycle and (3) extracellular fluctuation in hormone concentration.
These findings suggest potentially important consequences of ultradian secretion. The transcriptional program induced by hormone pulses is significantly different from that produced by constant hormone treatment. Thus, the physiological response to GR activation is probably significantly modulated by the naturally occurring ultradian secretion of glucocorticoids.
RESULTS
Real-time interaction of GR with gene targets
The potential effects of ultradian hormone release (Fig. 1a) on GR interactions with gene targets are unknown. Using the 3617 mouse cell line6–8, which contains an amplified array of GR responsive promoter structures (the MMTV array), GFP (green fluorescent protein)–GR interactions with GR response elements (GREs) can be directly visualized in living cells6,9,10. When corticosterone (a naturally occurring hormone in rodents) is applied to cells in a periodic manner (consecutive additions and withdrawals), the equilibrium concentration of GFP–GR associated with GREs in the array responds rapidly to the serum ligand concentration (Fig. 1b, c; Supplementary Information, Fig. S1 and Movie S1).
Figure 1.
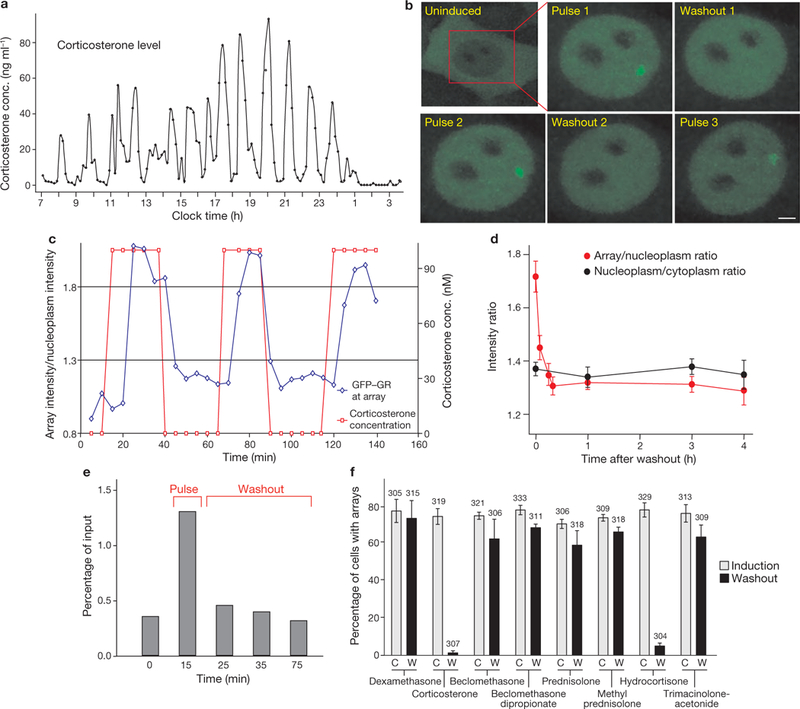
GR-MMTV promoter interactions in response to ultradian corticosterone fluctuations. (a) Corticosterone plasma levels in a freely moving rat show ultradian (pulsatile) fluctuations. (b) Pulsatile corticosterone treatment induces cyclic GR loading at the MMTV promoter array (green) in the 3617 cell line (single cell analysis). Scale bar, 2 μm. (c) GFP-GR intensity fluctuations at the array in response to the changing hormone concentration (in the cell presented in b). (d) GFP-GR dissociation from the array in response to hormone withdrawal (measured by the GFP-GR fluorescent intensity at the ChRFP-NF1-marked locus) is completed in < 10 min, whereas GR remains nuclear for > 3 h. Error bars represent the mean ± s.e.m., n = 25 cells. (e) Representative ChIP experiment demonstrates a similar rate of dissociation of endogenous GR from the MMTV promoter array on hormone withdrawal. (f) GFP-GR dissociation from the MMTV promoter is completed in <15 min after hormone withdrawal when the activating hormone is a naturally occurring glucocorticoid (corticosterone or hydrocortisone), but is incomplete for all of the synthetic glucocorticoids tested, indicating stronger and longer-lived interactions between the synthetic hormones and the GR. Error bars represent the mean ± s.e.m., n ≥ 300 cells.
To facilitate array localization, we generated a new cell line, 3617-ChRFP-NF1 (a derivative of the 3617 cell line), which in addition to GFP-GR expresses cherry red fluorescent protein-tagged nuclear factor 1 (ChRFP-NF1) under tetracycline regulation (Tet-off; Fig. 3a, e). NF1 associates constitutively with several sites in the array locus (Supplementary Information, Fig. S2), and serves as a convenient marker for array identification in hormone-free cells. Using this approach, we characterized the rate of GFP-GR dissociation from the array in response to hormone withdrawal (washout), and found that the GR-array binding and exchange equilibrium shifted away from the GREs less than 10 min after the washout (Fig. 1d; Supplementary Information, Movie S1). Consistent with previous studies11,12, the total nuclear GFP-tagged or endogenous GR levels remained relatively unchanged (Supplementary Information, Fig. S3). Using chromatin immunoprecipitation (ChIP), we observed a similar shift in equilibrium association of endogenous GR with the MMTV array in the parental 3134 cell line (Fig. 1e). Thus, rapid changes in GR affinity for its regulatory elements in response to hormone fluctuations imply that GR interactions with natural ligands are also highly dynamic. In contrast, when cells are treated with the synthetic hormone dexamethasone in the pulsatile ultradian mode, the cyclic pattern of GR-array interaction was attenuated (Supplementary Information, Figs S4a, b, S6 and Movie S2). We hypothesized that slow disassociation of the high affinity dexamethasone ligand from the receptor10,13 prevents the receptor from responding adequately to the extracellular hormone level. Interestingly, of the eight separate GR agonists examined, only the naturally occurring ligands, hydrocortisone and corticosterone, could result in rapid (less than 15 min) redistribution of the receptor-response element equilibrium after hormone withdrawal (Fig. 1f). This previously unappreciated property of ligand-dependent receptor dynamics probably contributes to the biological selectivity of the ligands.
Figure 3.
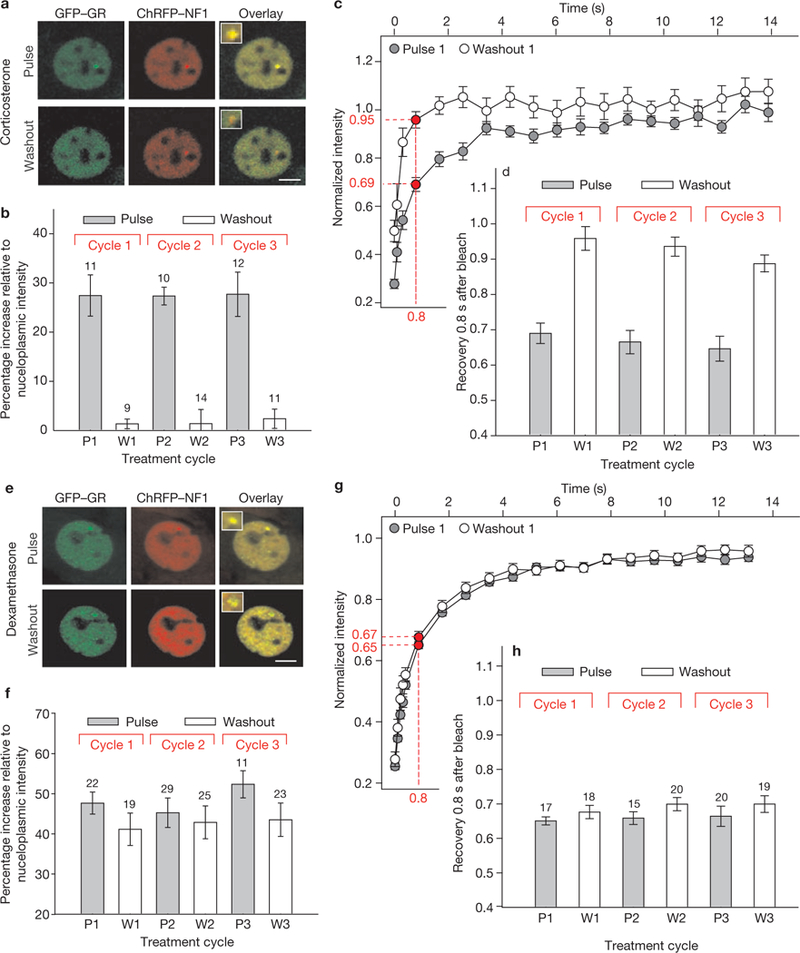
GFP-GR loading and dynamics at the MMTV promoter array during ultradian stimulation with natural and synthetic glucocorticoids. (a) A 3617 ChRFP-NF1 cell line was used to determine the GFP-GR loading at, and dissociation from, the MMTV array (marked by ChRFP-tagged NF1) during corticosterone induction (Pulse) and withdrawal (Washout). Scale bar, 5 μm. (b) 3617-ChRFP-NF1 cells were subjected to one, two or three cycles of 100 nM corticosterone induction (20 min) and withdrawal (40 min) and fixed either after the induction (P1, 2 and 3) or on hormone withdrawal (W1, 2 and 3). GR loading at the array (measured as GFP-GR intensity at the ChRFP-NF1-marked locus relative to the nucleoplasmic GFP-GR intensity) precisely followed the hormone addition and withdrawal cycles. Error bars represent the mean ± s.e.m., n = 9–14 cells. (c)GFP-GR dynamics at the array was measured by fluorescent recovery after photobleaching (FRAP) on corticosterone addition and withdrawal. Recovery during hormone stimulation, reflective of productive GR-MMTV promoter interactions, was slower than recovery on hormone withdrawal. Error bars represent the mean ± s.e.m., n ≥ 15 cells. (d)Similar fluctuations in GFP-GR dynamics were detected during subsequent cycles 2 and 3. Error bars represent the mean ± s.e.m., n ≥ 15 cells. (e) GFP-GR associates with the ChRFP-NFl-marked MMTV array after dexamethasone treatment, but remains associated with this locus even after dexamethasone removal from the growth media, indicating stable GR-dexamethasone interactions. Scale bar, 5 μm. (f) Only small fluctuations of the GFP-GR intensity at the array site are observed over three consecutive cycles of dexamethasone addition and withdrawals. Error bars represent the mean ± s.e.m., n = 17–29 cells. (g) A minor change in GFP-GR dynamics at the array is observed after dexamethasone washout. Fluorescent recovery 0.8 s after the bleach period is 65% ± 0.01 and 67% ± 0.02 for the pulsed and washout sample, respectively. Error bars represent the mean ± s.e.m., n ≥ 15 cells. (h) Comparable changes in the GFP-GR dynamics are observed during the second and third subsequent cycles. Error bars represent the mean ± s.e.m., n ≥ 15 cells.
GR-chaperone cycle
From the results of our single cell (Fig. 1b, c) and washout experiments (Fig. 1d–f), we infer that corticosterone dissociates from the entire nuclear GR pool within 10 min of hormone withdrawal. Reassociation of the hormone with unliganded steroid receptors requires the formation of a large, multicomponent chaperone protein complex11,14 (Fig. 2a). Thus, ligand-free receptors must be ‘re-chaperoned’ in the nucleus before binding free hormone and initiating new chromatin interactions15,16. If this chaperone cycle is blocked by inhibition of Hsp90 (heat shock protein 90) with geldanamycin, the equilibrium of GR association is shifted away from the array within 5–10 min after drug treatment, even in the continued presence of corticosterone10 (Fig. 2b, c). Failure to ‘re-ligand’ the receptor prevents the receptor from re-entering the rapid exchange cycle on chromatin. Hsp90 inhibition also accelerates proteasome-mediated degradation of GR (Fig. 2). Thus, nuclear GR recycling is critical for GR stability, GR activation and reactivation, and subsequent gene regulation in response to fluctuating hormone levels.
Figure 2.
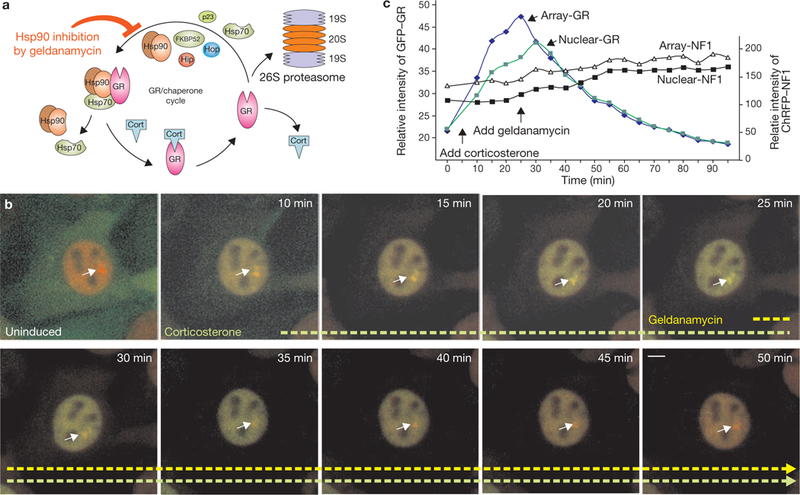
GR-chaperone cycle. (a) GR is involved in frequent hormone disassociation-reassociation cycles. After hormone release, GR must reassociate with the chaperone machinery to regain its hormone-binding affinity in the nucleus. Hsp90 inhibition (with the selective Hsp90 inhibitor geldanamycin) in the constant presence of hormone (Cort), induces rapid GR disassociation from the array (within 5–10 min) before accelerated proteasome-mediated degradation. (b) Images of a single cell expressing GFP-GR and ChRFP-NF1. The cell was induced with corticosterone (100 nM), before being challenged with 2.5 μg ml−1 geldanamycin in the continuing presence of hormone. The equilibrium association of GFP-GR with the array decreases rapidly during inhibition, and total nuclear concentration of GR decreases afterwards due to accelerated degradation. The effect of geldanamycin treatment is confined to GR, as the intensity of the ChRFP-tagged NF1 at the array (and its concentration in the nucleus) is unaffected by it. Images were recorded at 5 min intervals. Geldanamycin was added 25 min after the start of the experiment, 20 min after corticosterone stimulation. Scale bar, 5 μm. (c) The average intensities (in relative units) of GFP-GR and ChRFP-NF1 associated with the array, or in the nucleus, are plotted as a function of time.
GR mobility and equilibrium occupancy at promoter elements
The characterization of GR response-element dynamics using photobleaching techniques in living cells led to the discovery that GR exchanges rapidly with its regulatory sites6, and that the GR exchange rate at the array is ligand- and ATP-dependent10. We also observed10,17,18 slower receptor exchange rates at response elements, compared with the general nucleoplasmic space, reflecting specific and productive interactions at regulatory sites. Thus, GR mobility at the array during a corticosterone pulse should be slower than GR mobility after hormone withdrawal. We therefore characterized exchange rates for GR at the array during the course of three pulsatile corticosterone cycles. As expected, GFP-GR mobility at the array locus varied considerably during the ultradian hormone cycle. Lower mobility (69 ± 0.02% fluorescent recovery at 0.8 s after photobleaching) was observed during the peak of GR localization to the array, reflecting specific GFP-GR interactions with the promoter, whereas an increased exchange rate (95 ± 0.03% fluorescent recovery at 0.8 s after photobleaching) was found on hormone withdrawal, indicative of nonspecific interactions (Fig. 3c, d; Supplementary Information, Fig. S5). Thus, productive receptor action at the GREs is limited to the peak of an ultradian pulse. Dexamethasone failed to induce significant ultradian cycling of receptor-GRE interactions (Fig. 3e, f) or fluctuations in receptor-chromatin dynamics (Fig. 3g, h).
We also examined GR loading at multiple GR response elements in the genome using ChIP analysis (Fig. 4). Ultradian hormone treatment induced cyclic GR loading at GR-binding elements associated with the Glul locus, the Mt1 locus and the MMTV promoter19. Thus, as observed at the single cell level by GFP-GR fluorescence (Fig. 3b), pulsatile hormone stimulation induced cyclic GR loading at endogenous GR response elements in cell populations. These findings demonstrate that the phenomenon of ultradian cycling is a general property of GR response elements throughout the genome.
Figure 4.
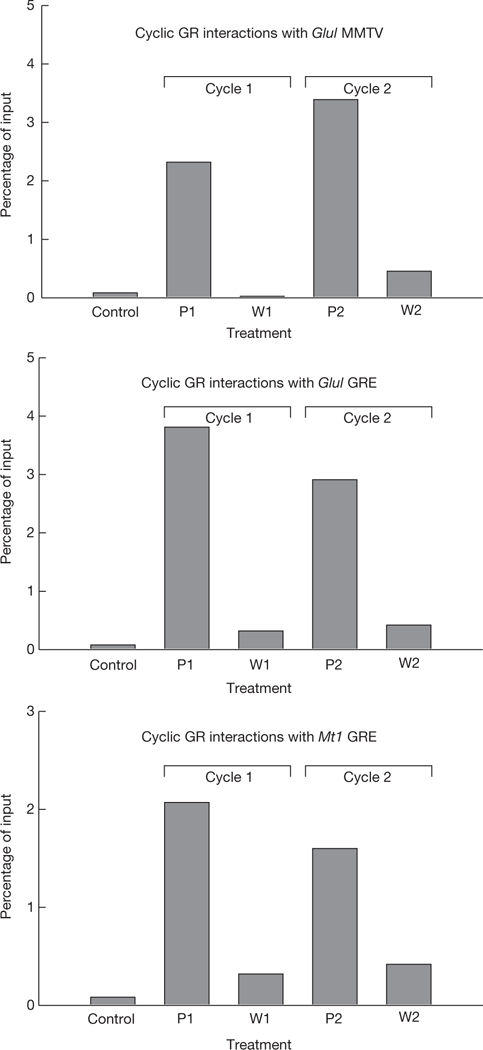
Equilibrium association of GR with endogenous GR response elements (GREs). GR associated with GREs was determined by chromatin immunoprecipitation (ChIP) during ultradian stimulation. Cyclic loading of GR is observed for the MMTV nucleosome B GR response element, the Glul response element, and the Mt1 response element. The data are from three independent experiments. P, induction; W, withdrawal.
RNA polymerase II residency at the MMTV array
We examined the recruitment and dynamics of polymerase II (Pol II) at the MMTV array in response to ultradian hormone stimulation in live cells expressing endogenous GR. Using GFP fused to the largest Pol II subunit, hRBP1 (ref. 20), we characterized GFP-Pol II exchange rates by FRAP (fluorescence recovery after photobleaching) analysis. As seen in Fig. 5a–d and Supplementary Information, Fig. S7, the rate of Pol II exchange was significantly reduced during the periods of maximal GR binding, indicating productive engagement of the Pol II9. In contrast, Pol II mobility during withdrawal periods was substantially higher. Thus, Pol II promoter residency is also affected by the ultradian mode of hormone stimulation.
Figure 5.
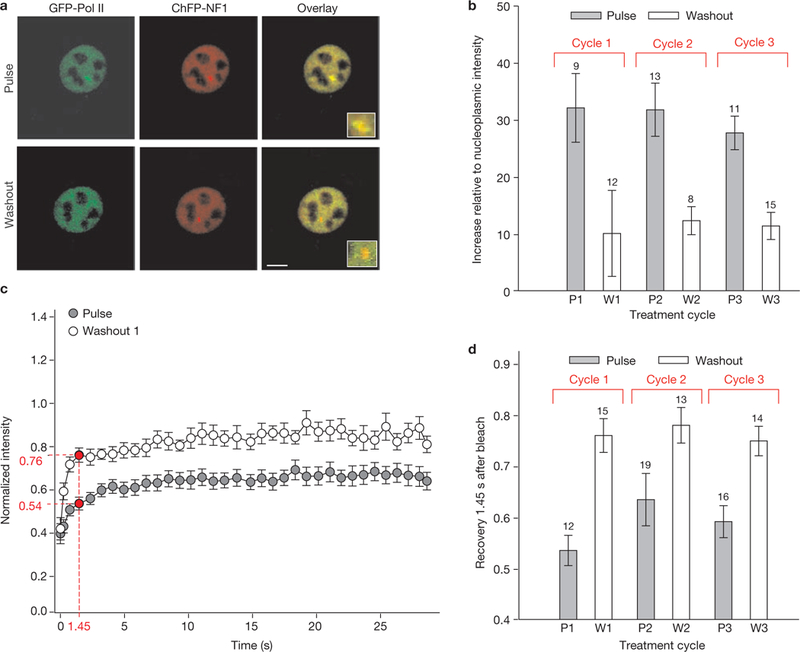
Cyclic RNA Polymerase II loading and dynamics in response to ultradian corticosterone treatment. (a) GFP-Pol II associates with the ChRFP-NF1-marked MMTV array locus in response to corticosterone induction (Pulse) and dissociates from the array on corticosterone withdrawal (Washout). Scale bar, 5 μm. (b) Cyclic Pol II loading at the MMTV promoter array, measured by GFP-Pol II intensity relative to nucleoplasmic intensity, during three consecutive cycles of hormone addition and withdrawal. Error bars represent the mean ± s.e.m., n = 8–15 cells. (c) GFP-Pol II dynamics at the array as measured by FRAP is slower during corticosterone induction, compared with withdrawal (54% and 76% recovery at 1.45 s after the bleach, respectively). Error bars represent the mean ± s.e.m., n ≥ 12 cells. Reduced Pol II mobility on hormone stimulation indicates that more Pol II molecules are productively engaged in transcription and elongation. (d) Comparable fluctuations in the GFP-Pol II dynamics are observed over several cycles of ultradian corticosterone treatment. P, induction; W, withdrawal. Error bars represent mean ± s.e.m., n = 12–19 cells.
Effects of ultradian hormone stimulation on GR-mediated transcription
A critical question is whether ultradian dynamics modulates the transcriptional program by GR. To test the effect of pulsatile stimulation on gene expression, we measured transcription levels from a number of GR-upregulated genes in the 3134 cell line by real time quantitative PCR (rtqPCR), using primers with recognition specificities limited to nascent mRNA transcripts (intron-exon junction). For each gene examined (Figs 6 and 7a), the level of nascent transcript increased rapidly during the hormone pulse, then decayed to very low levels during the washout period. When the cells were treated with dexamethasone, its impaired dissociation from GR prevented efficient transcription downregulation on dexamethasone withdrawal (Supplementary Information, Fig. S8), uncoupling hormone-level fluctuations from the transcription response. In contrast, the lower affinity of corticosterone for GR promoted its fast dissociation and rapid transcriptional downregulation on hormone withdrawal (Fig. 6; Supplementary Information, Fig. S8). Varying corticosterone concentrations influenced the level, but not the pulsatile nature, of nascent RNA release (Supplementary Information, Fig. S9). These data demonstrate that ultradian hormone stimulation induces coordinated cyclic downstream events leading to the pulsatile release of nascent RNA from GR-regulated genes. We designate this phenomenon as a form of gene pulsing.
Figure 6.
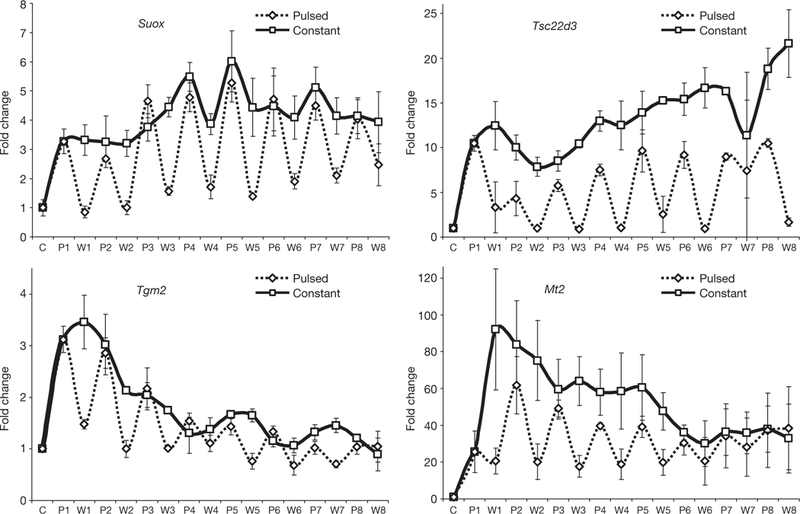
Effects of ultradian corticosterone treatment on transcription of GR-regulated genes in 3134 cells. Nascent transcripts from several GR-upregulated genes in the 3134 cell line are synthesized in distinct quanta during the ultradian hormone induction (eight consecutive pulses), whereas constant corticosterone treatment induces continuous RNA release. Error bars represent mean ± s.e.m. from three independent experiments. C, control; P, induction; W, withdrawal.
Figure 7.
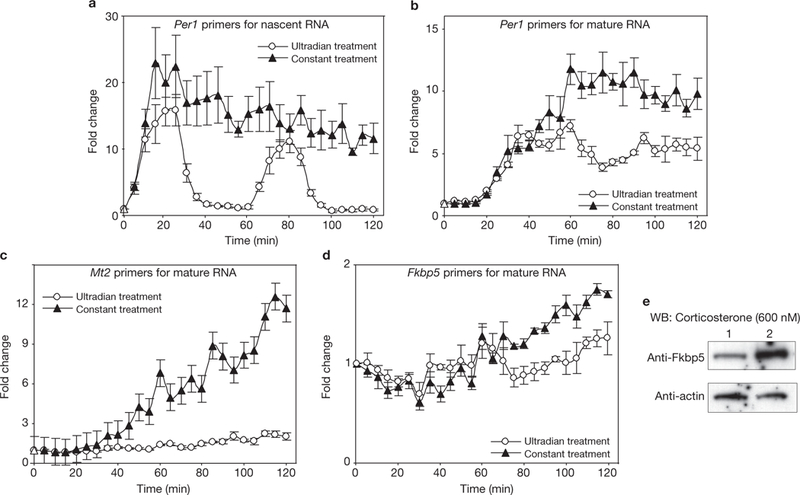
Differential transcription outputs of ultradian and constant hormone treatments. (a) Two pulses of corticosterone induce characteristic pulsatile release of nascent RNA, whereas constant treatment induces continuous release of Per1 transcript. (b–d) Constant treatment leads to accumulation of higher levels of mature RNA from GR-regulated genes, Per1 (b), Mt2 (c) and Fkbp5 (d), than pulsatile treatment. Error bars represent the mean ± s.e.m. n = 4. (e) Western blot (WB) experiments demonstrate that the Fkbp5 protein level after constant treatment is higher than the protein level after pulsatile treatment (600 nM corticosterone for 8 h). Lane 1, pulsatile treatment; Lane 2, constant treatment.
We hypothesize that gene pulsing is required for the biologically accurate regulation of gene targets. Mature RNA levels from samples treated constantly with corticosterone are significantly higher than those from samples treated in an ultradian manner (Fig. 7b–d). This difference is apparent as early as 1–2 h after stimulation starts and is preserved at the protein level (Fig. 7e). These findings suggest that constant hormone stimulation may lead to inappropriate protein expression and altered body physiology.
Gene pulsing in live animals
To ascertain whether the striking functional response to pulsatile hormone action occurs at the whole animal level, we examined the effects of ultradian stimulation on live rats. Sprague-Dawley rats were first adrenalectomized to remove all circulating corticosterone. Freely behaving animals were then treated with pulses of corticosterone through an indwelling intravenous cannula to establish a pattern of circulating natural ligand that mimicked the natural ultradian profile, as described previously21 (Fig. 8a; Supplementary Information, Fig. S10). To test whether these brief pulses of circulating corticosterone in vivo could elicit a functional response, we focused on an important metabolic glucocorticoid target organ, the liver. Hepatic GR activation was measured by binding to synthetic GRE-containing oligonucleotides in the TransAM assay (see Methods). As observed in the cultured cell models, each pulse of corticosterone induced a ‘wave’ of GR activation which decreased to the baseline level within 60 min of each pulse. GR association with regulatory regions in the target clock gene Period1 (Per1) was then assessed by a ChIP assay over the duration of the first pulse (Supplementary Information, Fig. S10c). GR association with chromatin was rapid and transient, with significant dissociation from the chromatin template occurring within 30 min of the corticosterone pulse. Functional output was assessed using quantitative (qPCR) to measure nascent RNA transcripts from the Per1 gene. Strikingly, the kinetics of Per1 transcription were highly transitory, correlating well with the duration of GR-DNA association. These results indicate that the in vivo system for glucocorticoid signalling is highly dynamic, and intrinsically dependent on the secretion pattern of corticosteroid released from the adrenal gland.
Figure 8.
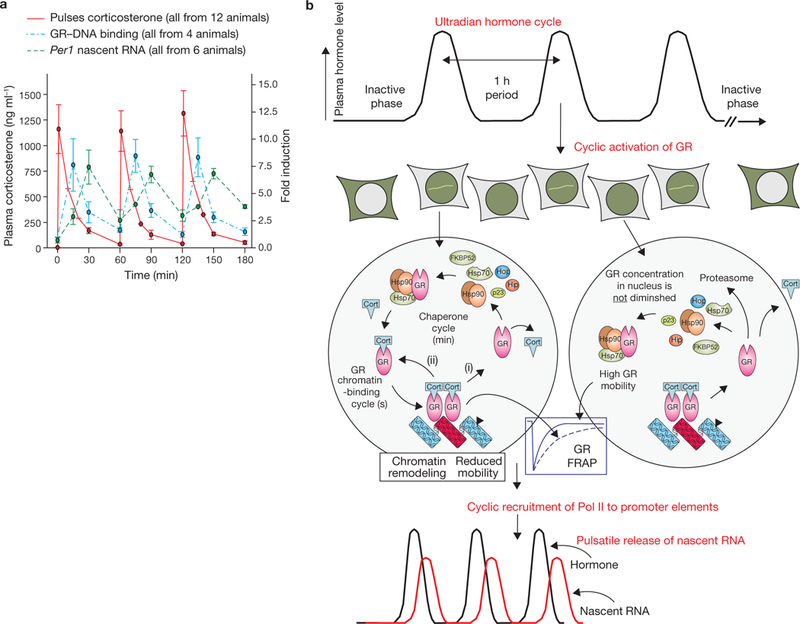
Pulsatile corticosterone treatment of adrenalectomized (ADX) rats and a model for gene pulsing. (a) Pulsatile corticosterone replacement in freely ambulating ADX rats was achieved with hourly bolus injections through an indwelling venous cannula. Plasma corticosterone levels, measured by an enzyme immunoassay (EIA), were undetectable in the ADX rats before corticosterone injection (time 0 min), and rose to a maximal value 1 min after injection before clearing according to the approximate 10 min half-life of corticosterone in blood (red trace). Subsequent injections at 60 min and 120 min resulted in similar profiles. GR activation and DNA association-dissociation kinetics reflected the pulsatile pattern of circulating corticosterone. GR activation and binding to GREs containing synthetic oligonucleotides were determined in a quantitative ELISA-based assay (TransAM) using nuclear extracts prepared from rat liver at the indicated times (blue trace). Chromatin immunoprecipitation (ChIP) assays (shown in Supplementary Information, Fig. S10) revealed GR association-dissociation with regulatory regions of the Period 1 gene promoter shows a similar profile to the results obtained in the TransAM assay. The functional output of this dynamic activation profile is the pulsatile release of nascent Per 1 RNA, measured by qPCR analysis of heterogeneous nuclear RNA (hnRNA) during the time course (green trace). Error bars represent mean ± s.e.m., n = 4–12 animals. (b) Gene pulsing: a schematic representation of cyclic GR interactions with response elements leading to pulsatile release of nascent RNA. Ultradian hormone fluctuations (periodicity ~1h) induce GR translocation to the nucleus, before transient interactions (timescale of seconds) with GREs (chromatin-binding cycle). In addition, GR is involved in the nuclear chaperone cycle (timescale of min), through which unliganded GR regains its hormone-binding affinity and re-enters the chromatin-binding cycle. Receptors recently disengaged from chromatin may lose ligand (i), or retain ligand (ii) and rebind chromatin. During hormone withdrawal periods, the unliganded GR fraction increases rapidly (timescale of min), leading to loss of GR from the array, faster GR mobility and transcription downregulation. Cort, ligand.
DISCUSSION
We explored the effects of ultradian hormone secretion on the GR-mediated transcriptional program and discovered that, in contrast to constant treatment, the ultradian mode of hormone secretion induces cyclic GR-mediated transcriptional regulation, or gene pulsing. We also showed that the dynamics of GR-template interaction, as well as RNA Pol II loading and exchange, fluctuate together with the changes in extracellular hormone concentration. As a result, nascent RNA transcripts from a number of GR regulated genes are released in distinct quanta during ultradian treatment, demonstrating profound differences in the transcriptional program induced by pulsatile ligand-stimulation from that induced by constant stimulation. We conclude that dynamic GR-promoter interactions in live cells6, together with the chaperone cycle, are critical for rapid ‘sensing’ of the ligand environment by GR. Our findings suggest that dynamic receptor exchange is crucial to elicit biologically appropriate actions of the receptor.
We propose that gene pulsing in the GR system is necessary for correct transcriptional programming. As modelled in Fig. 8b, ligand-bound receptor complexes exchange rapidly and continuously with response elements in chromatin (in the time scale of seconds). During each exchange event, a receptor will either lose its ligand (Fig. 8b), requiring entry into the chaperone cycle to re-acquire a ligand, or retain its ligand and return to the template (Fig. 8b). During periods between ultradian pulses, intracellular ligand levels fall below the KD required to maintain GR occupancy, and the equilibrium for GRE occupancy shifts largely to nucleoplasmic receptors.
The chromatin and chaperone cycles are much faster than the ultradian circulating hormone cycle. The dynamic action of these processes enables accurate signal transduction, leading to time-efficient ‘translation’ of information regarding hormone bioavailability into gene activity. Synthetic glucocorticoids, having much higher receptor affinity, fail to disengage from nuclear receptors with sufficient speed to support the ultradian cycles, thus uncoupling extracellular hormone fluctuations from appropriate receptor function at response elements. Therefore, even low doses of synthetic glucocorticoids (such as dexamethasone) will significantly alter the transcription program set by ultradian hormone release, resulting in dramatically altered RNA accumulation profiles (Supplementary Information, Fig. S9). This effect can be quite dramatic, as seen for the Mt2 locus (Fig. 7c). For this locus, very little mature transcript is accumulated during pulsed hormone treatment, whereas significant levels continue to increase during constant treatment. Moreover, a longer dexamethasone half-life in the plasma (3–4 h) will further exacerbate those effects in vivo.
Cycling of other nuclear receptors, including the oestrogen (ER) and androgen (AR) receptors, on specific binding sites have been described previously22,23. However, these cycling events are intrinsic properties of ER and AR signalling, and depend on proteasome function. Most importantly, the oscillations observed for these receptors occur in the constant presence of hormone. In contrast, the GR cycles described here reflect pulsed release of glucocorticoids from the adrenal glands and thus are externally driven.
It is likely that the pulsatile release of glucocorticoids has significant biological consequences, as it has been conserved during evolution. Furthermore, transient ligand release has been described for several other mammalian hormones24–28, and in some cases is known to be absolutely required for correct physiological action24. Considering previous observations of oscillatory action in other systems (p53, refs 29, 30; NfκB31; Ace1, ref. 32; calcium signalling33,34 and eukaryotic developmental genes35) as well as data regarding the pulsatile release of other hormones24–26–28, it is likely that pulsatility represents a common mechanism for target gene regulation.
Except for prohormones, all hormones that have been appropriately investigated seem to be released in pulses. The relevance of this pulsatility has been particularly well described for the G protein coupled receptor GnRH28, for which pulsatile administration is used clinically for the treatment of infertility. Constant receptor activation (using GnRH superagonists) is used to induce chemical castration for the treatment of conditions such as prostatic carcinoma36. The lack of effective ultradian rhythmicity in corticosteroid replacement regiments for patients with hypocortisolaemia may well contribute to the lack of energy often described by patients undergoing treatment for Addison’s disease. Furthermore, attention to the kinetics of therapy in patients with glucocorticoid-responsive diseases such as rheumatoid arthritis could provide a new approach to decreasing glucocorticoid side effects37, while maximizing the therapeutic response.
Our data provide the basis for a more complex view of gene regulation by GR. Glucocorticoids are released as a result of activity of the highly dynamic hypothalamic-pituitary-adrenal (HPA) axis. Temporal HPA axis activity drives a pattern of GR action that leads to gene stimulation that is reflective of the HPA axis profile. Physical and psychologically stressful events induce a rise in glucocorticoid plasma levels, super-imposed on the ultradian and circadian rhythm. Due to its plasticity, the HPA axis integrates many internal and external stimuli, which in turn have a direct impact on GR-regulated transcriptional programs. Rapid exchange of GR with chromatin, a phenomenon described for several transcription factors6,38,39 (reviewed in ref. 40), is a key element in the rapid response of this critical endocrine system. These insights open new approaches for the development of synthetic glucocorticoids. Considering the frequency with which these drugs are administered for multiple indications, further studies are clearly needed to understand the potential of ultradian therapy in GR therapeutic regiments. □
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/naturecellbiology/.
Methods summary.
The 3134 and 3617 cell lines were described previously8,41. The 3617-ChRFP-NF1 cell line is a derivative of 3617, and expresses both GFP-GR and ChRFP-NF1 under tetracycline regulation. FRAP experiments were carried out on a Zeiss 510 confocal microscope with 100× 1.3-N.A. oil immersion objective and the cells were kept at 37 °C using an air stream stage incubator (Nevtek). Initial images were acquired in red fluorescence to independently locate the MMTV arrays marked by ChRFP-NF1. Bleaching was then performed at these sites with the 488- and 514-nm lines from a 45-mW argon laser operating at 90% laser power. A single iteration was used for the bleach, and fluorescent recovery was monitored at low laser intensity (0.2% for a 45-mW laser). All time-lapse imaging experiments were carried out on a Zeiss-Live Duoscan microscope equipped with a chamber allowing media exchange over the course of the experiments, and with automated temperature, humidity and CO2 control. Gene-expression studies were performed in a specially adapted incubator, allowing media replacement under conditions of stable CO2 and temperature levels throughout the duration of an experiment. The 3134 cells were treated with pulses (20 min each) of 100 nM corticosterone, interspaced with 40 min periods of incubation with hormone-free media for 8 h consecutively. Total RNA was extracted, reverse transcribed (iScript cDNA) and used in a qPCR reaction applying SyBr green and Bio-Rad IQ system (Biorad). Primer sequences designed to amplify nascent RNA (amplicons that cross an exon-intron boundary) were used. Chromatin immunoprecipitation (ChIP) was performed according to the standard protocol (Upstate Biotechnology). All animal procedures were carried out in accordance with the UK Home Office animal welfare regulations.
Plasmids and cell lines.
GFP-Pol II (hRPB1) was described previously20. The cherryRFP-NF1A1.1 (ChRFP-NF1) construct was generated by ligating NF1A1.1 cDNA (provided by R. M. Gronostajski, The State University of NY, USA) to a ChRFP-C1 vector as an EcoRI-SalI fragment. The ChRFP-C1 vector was generated by replacing GFP in the pEGFP-C1(Clontech) vector with Cherry red fluorescent protein (ChRFP) (a gift from R. Tsien, University of California, San Diego, USA). The 3134 cell line is a mouse mammary adenocarcinoma cell line. It contains a large tandem array (~200 copies) of a mouse mammary tumour virus, Harvey viral ras (MMTV-v-Ha-ras), reporter. The 3617 cell line is a derivative of 3134 cell line expressing a green fluorescent protein (GFP)-tagged version of GR (GFP-GR) from a chromosomal locus under control of the tetracycline-repressible promoter. Both cell lines were described previously8,41. The derivative cell line, 3617-ChRFP-NF1, expresses both GFP-GR and ChRFP-NF1 under tetracycline regulation. Commercially available pRev-TRE vector (Clontech) was opened using BamHI and SalI restriction enzymes, and a synthetic linker oligonucleotide containing the BamHI, AgeI, NotI, MfeI and SalI restriction sites was inserted to produce the pRev-TRE-Link Age-Mfe vector. The Clontech pEGFP-C1 and the related ChRFP-C1 vector contain unique AgeI-MfeI sites, allowing transfer of the ChRFP-NF1 coding sequence into the pRev-TRE-Link vector as an Age-Mfe fragment. The resulting pRev-TRE-Link Age-Mfe ChRFP-NF1 vector was introduced into Phoenix retroviral packaging cells (Orbigen, Inc.) to produce replication deficient virions. Medium containing virions from the Phoenix cells was then collected and used to infect 3617 cells, which express the Tet-regulator protein. Infected 3617 cells were then treated with hygromycin for 10 days to select cells with stable integration of the pRev-TRE-Link ChRFP-NF1 DNA. All cell lines were grown in Dulbeccos modified Eagle’s medium (DMEM, Gibco BRL) supplemented with 10% fetal bovine serum (FBS) (Hyclone). Tetracycline (5mg ml−1) was added to the medium to suppress GFP-GR and CherryRFP-NF1expression in 3617 and 3617-ChRFP-NF1 cells.
FRAP and live-cell imaging.
Before live-cell imaging and FRAP, 3617 cells, 3617-ChRFP-NF1 cells, or 3617 cells transiently expressing GFP-Pol II and ChRFP-NF1 were transferred to 35-mm glass bottom dishes (MatTek Corporation) at a density of 2 × 105 in Phenol Red-free DMEM medium containing 10% charcoal-stripped fetal bovine serum (Hyclone) for at least 18 h. Tetracycline was omitted from the media of the 3617 and 3617-ChRFP-NF1 cells to allow the expression of GFP-GR and ChRFP-NF1. The 3617 cells transiently expressing GFP-PolII and ChRFP-NF1 were kept with 5 mg ml−1 tetracycline to suppress GFP-GR expression. Ultradian experiments were conducted with 20 min treatment with 100 nM corticosterone or dexamethasone, followed by washout and incubation in hormone-free medium for 40 min (unless stated otherwise). All cells were induced simultaneously and at each time point one of the dishes was used for the FRAP experiments and discarded afterwards. To allow time for the collection of the 3–5 FRAP curves per dish, data collection began 10 min after the hormone induction for the pulse samples and was terminated 10–15 min later. Collection of FRAP data for the washout samples began 20 min after hormone withdrawal and was terminated 10–15 min later. Data from at least three independent experiments were collected and used to generate corresponding average FRAP curves (± s.e.m.). Fluorescent recovery at a single time point (0.8 sec and 1.45 sec after the bleach for GFP-GR and GFP-Pol II, respectively) was selected to represent the changes in fluorescent recovery during the three consecutive induction and hormone withdrawals. FRAP and time-lapse imaging experiments were carried as described in the Methods Summary.
Fixed cell experiments.
To detect the effect of hormone withdrawal on GFP-GR loading at the array, 3617 cells were grown overnight on 22-mm2 coverslips in DMEM medium containing 10% charcoal-stripped serum (Hyclone) without tetracycline (to allow the expression of the GFP-GR), and induced for 15 min with 100 nM hormone (corticosterone, dexamethasone, prednisolone, hydrocortisone, methylprednisolone, beclomethasone dipropionate, betamethasone or trimacinolone acetonite; all from Sigma). After hormone withdrawal, cells were kept in hormone-free medium for an additional 15 min before fixation with 3.5% paraformaldehyde in phosphate-buffered saline (PBS) and mounted in PBS. In parallel, 3617 cells were treated with the selected glucocorticoids for 30 min before fixation (with 3.5% paraformaldehyde in PBS) and mounted in PBS before examination with a Leica 100× 1.3-N.A. oil immersion objective. The percentage of cells with visible arrays under both conditions was determined. The difference between the two numbers indicates the efficiency of the hormone washout.
To determine the equilibrium association of GFP-GR with the array, as well as GFP-GR localization on corticosterone withdrawal, 3617-ChRFP-NF1 cells were grown overnight on 22-mm2 coverslips in DMEM medium containing 10% charcoal-stripped serum (Hyclone) without tetracycline (to induce the expression of the GFP-GR and ChRFP-NF1) and treated with 100 nM hormone for 20 min. Two of the coverslips were fixed immediately (with 3.5% paraformaldehyde in PBS), while the remainder were fixed at different times after the hormone withdrawal and mounted in PBS. Over 100 cells from each slide were examined with the Zeiss 510 confocal microscope, using a 100× 1.3-N.A. oil immersion objective. Images were acquired in green (GFP-GR) and red (ChRFP-NF1) fluorescence and the ratio of the GFP-GR intensity of the array (determined by the ChRFP-NF1 staining) to the nucleoplasm intensity was determined using the Zeiss 510 software. The same method was also used to determine the GFP-GR array-nucleoplasm intensity ratio during subsequent hormone treatments and withdrawals. The experiment was repeated three times.
Ultradian gene expression.
For gene expression studies, 3134 cells were plated in charcoal-stripped fetal bovine serum (Hyclone) in 6-well dishes 24 h before the experiment. Cells were treated with eight pulses (20 min each) of 100 nM corticosterone, interspaced with 40 min periods of incubation with hormone-free media. To prevent cell stress, these experiments were performed in a specially adapted incubator, allowing media replacement under conditions of stable CO2 and temperature levels throughout the duration of an experiment. To ensure complete hormone withdrawal during the wash period, cells were washed once with an excess of hormone-free media and replaced with fresh media. Cells were collected before hormone treatment (NH) and after each hormone pulse (P1, P2 and P3), as well as at the end of each wash period (W1, W2 and W3). Control cells were treated constantly with 100 nM corticosterone and collected at the matching times.
Cells were lysed in 600 μl of RLT buffer (with β-mercaptoethanol added) followed by syringe-needle shearing. Total RNA was extracted using the RNeasy Mini Kit (Qiagen), including a DNaseI digestion step (RNase free DNase Set, Qiagen). RNA (1 μg) was reverse transcribed (iScript cDNA Synthesis Kit, BioRad) in 20 μl reaction volume, and 0.5 μl was used per qPCR reaction using SyBr green and a Bio-Rad IQ system (Biorad). Primer sequences were designed to amplify nascent RNA (amplicons that cross an exon-intron boundary). The primer sequences are shown in Supplementary Information, Table S1. PCR was performed as recommended by the manufacturer. Standard curves were created by 10-fold serial dilutions of the template. The expression data from three independent experiments were normalized to the expression of a control gene Mcm3, the mean values and s.e.m. were calculated and displayed as a fold change in relation to the untreated (NH) sample.
Western blot analyses.
Cells were treated constantly (600 nM corticosterone for 8 h) or with eight 20 min-pulses of 600 nM corticosterone followed by 40 min hormone withdrawal periods. After treatment, cells were washed in phosphate-buffered saline, lysed in whole-cell lysis buffer (20 mM Hepes at pH 7.6, 20% Glycerol, 1.5 mM MgCl2, 0.2 mM EDTA, 0.1% Triton-X100, 1 mM DTT and 1× complete protease inhibitors cocktails, Roche) containing 500 mM NaCl for 30 min, and pelleted by microcentrifugation at top speed. The supernatants were diluted to 140 mM NaCl with lysis buffer without salt and proteins and were then separated by SDS-PAGE and transferred to PVDF membranes. Blots were probed with primary anti-Fkbp5 (sc-11518; 1:1000) or anti-actin (SC01615; 1:800) antibodies (Santa Cruz Biotechnology) in Tris-buffered saline (TBS) containing 5% nonfat dry milk, followed by incubation with horseradish peroxidase (HRP)-conjugated anti-goat antibody (Santa Cruz Biotechnology). All blots were visualized with the ECL kit (Supersignal).
Chromatin immunoprecipitation (ChIP).
3134 cells were seeded in 150mm tissue culture plates and collected the next day for ChIP experiments after the indicated times of 100 nM corticosterone treatment or washout. Media was changed under the conditions, as described above for the RNA expression experiments. ChIP was performed according to the standard protocol (Upstate Biotechnology) with a crosslinking step (1% formaldehyde at 37 °C), followed by a quenching step with 150 mM glycine. Chromatin was sonicated by using the Bioruptor sonicator (Diagenode) with 45 sec ‘on’ and 15 sec ‘off’ for 30 min. Sonication efficiency was monitored by 1% agarose gel electrophoresis. Sonicated chromatin (500 μg) was immunoprecipitated with antibodies against GR (Affinity BioReagents; PA1–510 and PA1–511). DNA isolated from immunoprecipitates, as well as input DNA, was used as a template for real-time PCR (rtPCR). Primers for the promoter region of MMTV (Nuc B), were used for amplification: sense 5′-TTAAGTAAGTTTTTGGTTACAAACT-3′ and antisense 5′-TCAGAGCTCAGATCAGAACCTTTGATACC-3′. Other primers used for amplification for the GR binding sites in Glul and Mt1 promoter region were: sense 5′-CACTTGGGCAAACATGGACGGT-3′ and antisense 5′-CACAAGAGGAAATGCCCCCCT-3′ for Glul, and sense 5′-TAGGGACATG ATGTTCCACACGTC-3′and 5′-TTTTCGGGCGGAGTGCAGAG-3′ for Mt1. Standard curves were created by 10-fold serial dilution of an input template. The data presented are representative of three independent experiments.
Live animal experiments.
Adult Sprague-Dawley rats (250–300g; Harlan) were maintained under standard housing conditions with a 14:10 light-dark cycle (lights on 05:15, lights off 19:15). Food and water (or saline when specified) were available ad libitum. All animal procedures were carried out in accordance with the UK Home Office animal welfare regulations.
Rats (n = 6/timepoint/group) were anaesthetised with IM injection of Hypnorm (fentanyl citrate 0.252 mg per kg and fluanisone 8 mg per kg) after IP injection of Diazepam (4 mg per kg) then subjected to bilateral adrenalectomy and jugular cannulation. After surgery, the rats were recovered for 5 days, with ADX rats receiving corticosterone replacement (15μg ml−1) in 0.1% ethanol saline for drinking.
Corticosterone was withdrawn 24 h before the experiment. On the day of the experiment, the rats received up to three bolus IV injections of corticosterone-HBC, in the form of a preformed water-soluble complex of corticosterone and 2-hydroxypropyl-β-cyclodextrin (corticosterone-HBC; C-174; Sigma-Aldrich) administered at times 0, 60 and 120 min. Each IV injection contained the equivalent of 100 μg of corticosterone. At defined times, the rats were killed by decapitation after an overdose of pentabarbital. A section of liver was rapidly removed, sliced into 3-mm sections, rapidly frozen in liquid nitrogen and stored at −80 °C until nuclear extracts were prepared or RNA extracted. Blood was collected from the trunk into heparinised tubes and the plasma obtained by centrifugation before being stored at −20 °C until measurement of corticosterone by enzyme immunoassasays (EIA - Vinci Biochem).
Nuclear extract preparation.
Nuclear extracts were prepared according to the method used by Vallone et al.42 as previously described for rat tissue21. All procedures were performed at 4 °C and on ice. Each 50 mg piece of liver was homogenized in 1 ml S1 buffer (10 mM hepes at pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.1 mM EDTA at pH 8, supplemented with 0.5 mM DTT, 0.2 mM Na orthovanadate, 2 mM NaF and Complete protease inhibitor, Roche) using a Dounce homogenizer. Nuclear proteins were extracted in 1.2-pellet volumes of S2 buffer (10 mM hepes at pH 7.9, 400 mM NaCl, 1.5 mM MgCl2, 0.1 mM EDTA at pH 8 and 5% glycerol, supplemented with 0.5 mM DTT, 0.2 mM sodium orthovanadate, 2 mM NaF and Complete protease inhibitor, Roche) then stored at −80 °C. The integrity of nuclear fractions was verified by western blotting with antibodies specific to either the nuclear (anti-Histone H1 sc-8030, Santa Cruz Biotechnology) or cytoplasmic fraction (anti-RAS 05–516, Upstate Biotechnology; results not shown).
Corticosterone enzyme immunoassay (EIA).
Total corticosterone concentration in plasma was quantified by EIA using the Correlate-EIA kit (cat# 900–097; Assay Designs, Inc.).
TransAM GR:DNA-binding ELISA.
A commercially available ELISA-based transcription factor binding assay kit (TransAM GR; cat #45496; Active Motif) was used to measure GRE-binding activity for each nuclear sample. The protein concentration of each sample was determined with a bicinchoninic acid (BCA) assay (cat #23225; Thermo Scientific Rockford), and as an additional normalization control to more accurately measure the integrity of each sample, an aliquot of nuclear extract from each sample was also processed for NF-YA DNA binding activity (cat #40396; Active Motif). NF-YA is a ubiquitous transcription factor that showed no significant difference between the treatment groups. DNA-binding assay kits were used in accordance with the manufacturer’s instructions. Briefly, nuclear extracts (20 μg for GR, 5 μg for NF-Y) were incubated 96-well plates coated with GR or NF-Y binding consensus oligonucleotide sequence for 1 h, before being incubated with the supplied primary anti-GR or NF-Y antibody (1:1000) for 1 h, then with a peroxidase-conjugated secondary antibody (1:1000) for 1 h. After the substrate was added, colour development was read at 450 nm, and the optical density (OD) of both GR and NF-Y were recorded. The OD results obtained from the GR assay were first normalized to the results obtained from the NF-Y assay. Data were then expressed as fold induction relative to time zero.
Rat liver ChIP.
Buffers from the EZ ChIP kit (Upstate Biotechnology) were used in ChIP experiments, although the protocol was modified for use of animal tissue. Fresh liver samples of approximately 400 mg were cut into 3–4-mm3 cubes on ice. Cubes were fixed in 1% (v/v) formaldehyde in PBS (10 min, room temperature) and cross-linking was quenched by addition of glycine to 125 mM for 5 min. Fixing solution was removed fully by aspiration, followed by three washes in ice cold PBS supplemented with 2 mM NaF, 0.2 mM sodium orthovanadate and Complete protease inhibitor (Roche). Samples were blotted dry and homogenized in S1 buffer (described above) to release the nuclear fraction. Nuclei were recovered by centrifugation and lysed in SDS lysis buffer supplemented with Complete protease inhibitor, NaF and sodium orthovandate. Chromatin was sonicated with a Branson Sonifier 450, to an average fragment size of 1.5 kb, applying 10 s pulses at 10% output. Sheared chromatin (50 μg) was immunoprecipitated at 4 °C over-night with 2 μg M-20 anti-GR antibody (Santa Cruz Inc.). Antibody-GR-DNA complexes were collected on 60 μl protein A agarose beads (2 h at 4 °C), then washed once with low salt buffer, twice with high salt buffer, once with LiCl buffer and twice with TE buffer (pH 8.0) to remove non-specific binding. Following elution of antibody-GR-DNA complexes, RNase (Roche) and proteinase K (Sigma) digestion removed specific contaminants before samples were extracted once in 25:24:1 phenol-chloroform-isoamyl alcohol and once in 24:1 chloroform-isoamyl alcohol (Sigma). DNA was ethanol precipitated overnight at −20 °C, washed in 70% ethanol and finally dissolved in nuclease-free water (Ambion), ready for PCR amplification. Unlabelled PCR primers (FOR sequence 5′-CCAAGGCTGAGTGCATGTC-3′; REV sequence 5′-GCGGCCAGCGCACTA-3′) and a FAM-labelled probe (sequence 5′-CAAGAGAACACGATGTTCC-3′) were designed to amplify across the single GRE in the rat Per1 gene promoter (Custom Taqman Gene Expression Assay, Applied Biosystems; plate ID 619710). Absolute quantification was achieved by comparing samples with a standard curve generated from known quantities of sonicated rat genomic DNA. Samples and standards were amplified on an ABI PRISM 7500 Detection System (Applied Biosystems), according to the manufacturer’s instructions, with Universal Taqman PCR master mix (Applied Biosystems, cat # 4304437).
Quantitative PCR.
Total RNA was extracted from approximately 50 mg of liver with the phenol-guanidine isothiocyanate-chloroform extraction method using Trizol reagent (cat #15596026; Invitrogen Hopkinton) then further purified through RNeasy columns (cat #74104; Qiagen). Genomic DNA contamination was removed from the samples with DNase digestion (cat #79254; Qiagen). Complementary DNA was reverse transcribed from 1 μg of total RNA using the cloned AMV first strand synthesis kit (cat #12328–040; Invitrogen). Rat Period 1 (NM_001034125.1 GI:86477154) nascent transcript was quantified with a Taqman custom designed expression assay, with unlabelled primers and a FAM-dye-labelled probe designed within intron 21; primer sequences 5′-ATGTGTGCCTGGTGTCTGT-3′, 5′-ATGGTGGCTCAACTCCTTAACTC-3′; FAM probe 5′-CCTTCTTC-AGATTTCC-3′ (ABI plate ID 656011; Applied Biosystems). Briefly, PCR primer-probe mix was added to 50 ng cDNA and Taqman PCR master mix (cat #4304437; Applied Biosystems), and a comparative CT method was used to detect relative expression curves on an ABI PRISM 7500 detection system (Applied Biosystems). Data were normalized to the expression of the endogenous control, rat p-Actin (ABI assay #4352931E; Applied Biosystems).
Supplementary Material
ACKNOWLEDGEMENTS
We thank T. Karpova and J. McNally for assistance, C. Zeiss Inc. for the opportunity to carry out live cell experiments on a Zeiss Duoscan microscope, D. Shatti for technical assistance with nuclear extract preparation and Y. Kershaw for assistance with the in vivo experiments. This research was supported (in part) by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. The animal work was supported by a BBSRC grant (BB/C51297X/1) and a Wellcome Trust Programme Grant (074112/Z/04/Z) to S.L. The qPCR was performed on an Applied Biosystems 7500 System funded by a Wellcome Trust Equipment Grant (075548/Z/04/Z).
Footnotes
Note: Supplementary Information is available on the Nature Cell Biology website.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Lightman SL et al. Hypothalamic-pituitary-adrenal function. Arch. Physiol Biochem. 110, 90–93 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Lightman SL Patterns of exposure to glucocorticoid receptor ligand. Biochem. Soc. Trans. 34, 1117–1118 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Droste SK et al. Corticosterone levels in the brain show a distinct ultradian rhythm but a delayed response to forced swim stress. Endocrinology 149, 3244–3253 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Young EA, Abelson J & Lightman SL Cortisol pulsatility and its role in stress regulation and health. Front Neuroendocrinol. 25, 69–76 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Lewis JG, Bagley CJ, Elder PA, Bachmann AW & Torpy DJ Plasma free cortisol fraction reflects levels of functioning corticosteroid-binding globulin. Clin. Chim. Acta 359, 189–194 (2005). [DOI] [PubMed] [Google Scholar]
- 6.McNally JG, Mueller WG, Walker D, Wolford RG & Hager GL The glucocorticoid receptor: Rapid exchange with regulatory sites in living cells. Science 287, 1262–1265 (2000). [DOI] [PubMed] [Google Scholar]
- 7.Kramer P et al. Transcriptional state of the mouse mammary tumor virus promoter can effect topological domain size in vivo. J. Biol. Chem. 274, 28590–28597 (1999). [DOI] [PubMed] [Google Scholar]
- 8.Walker D, Htun H & Hager GL Using inducible vectors to study intracellular trafficking of GFP-tagged steroid/nuclear receptors in living cells. Methods 19, 386–393 (1999). [DOI] [PubMed] [Google Scholar]
- 9.Becker M et al. Dynamic behavior of transcription factors on a natural promoter in living cells. EMBO Rep. 3, 1188–1194 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stavreva DA, Muller WG, Hager GL, Smith CL & McNally JG Rapid glucocorticoid receptor exchange at a promoter is coupled to transcription and regulated by chaperones and proteasomes. Mol. Cell. Biol. 24, 2682–2697 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu J & DeFranco DB Chromatin recycling of glucocorticoid receptors: implications for multiple roles of heat shock protein 90. Mol. Endocrinol. 13, 355–365 (1999). [DOI] [PubMed] [Google Scholar]
- 12.Qi M, Stasenko LJ & DeFranco DB Recycling and desensitization of glucocorticoid receptors in v-mos transformed cells depend on the ability of nuclear receptors to modulate gene expression. Mol. Endocrinol. 4, 455–464 (1990). [DOI] [PubMed] [Google Scholar]
- 13.Munck A & Foley R Kinetics of glucocorticoid-receptor complexes in rat thymus cells. J. Steroid Biochem. 7, 1117–1122 (1976). [DOI] [PubMed] [Google Scholar]
- 14.Pratt WB, Galigniana MD, Harrell JM & DeFranco DB Role of hsp90 and the hsp90-binding immunophilins in signalling protein movement. Cell Signal. 16, 857–872 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Freeman BC & Yamamoto KR Disassembly of transcriptional regulatory complexes by molecular chaperones. Science 296, 2232–2235 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Freeman BC & Yamamoto KR Continuous recycling: a mechanism for modulatory signal transduction. Trends Biochem. Sci. 26, 285–290 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Rayasam GV et al. Ligand specific dynamics of the progesterone receptor in living cells and during chromatin remodeling in vitro. Mol. Cell. Biol. 25, 2406–2418 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klokk TI et al. Ligand-specific dynamics of the androgen receptor at its response element in living cells. Mol. Cell Biol. 27, 1823–1843 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.John S et al. Interaction of the glucocorticoid receptor with the global chromatin landscape. Mol. Cell 29, 611–624 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Sugaya K, Vigneron M & Cook PR Mammalian cell lines expressing functional RNA polymerase II tagged with the green fluorescent protein. J. Cell Sci. 113 ( Pt 15), 2679–2683 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Conway-Campbell BL et al. Proteasome-dependent down-regulation of activated nuclear hippocampal glucocorticoid receptors determines dynamic responses to corticosterone. Endocrinology 148, 5470–5477 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Metivier R et al. Estrogen receptor-α directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115, 751–763 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Kang Z, Pirskanen A, Janne OA & Palvimo JJ Involvement of proteasome in the dynamic assembly of the androgen receptor transcription complex. J. Biol. Chem. 277, 48366–48371 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Rothman MS & Wierman ME The role of gonadotropin releasing hormone in normal and pathologic endocrine processes. Curr. Opin. Endocrinol. Diabetes Obes. 14, 306–310 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Thompson NM, Davies JS, Mode A, Houston PA & Wells T Pattern-dependent suppression of growth hormone (GH) pulsatility by ghrelin and GH-releasing peptide-6 in moderately GH-deficient rats. Endocrinology 144, 4859–4867 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Chadwick D & Goode JA Mechanisms and Biological Significance of Pulsatile Hormone Secretion (John Wiley & Sons Ltd, 2000). [Google Scholar]
- 27.Belchetz PE, Plant TM, Nakai Y, Keogh EJ & Knobil E Hypophysial responses to continuous and intermittent delivery of hypopthalamic gonadotropin-releasing hormone. Science 202, 631–633 (1978). [DOI] [PubMed] [Google Scholar]
- 28.Gore AC GnRH: The master molecule of reproduction (Kluwer Academic Publishers, 2002). [Google Scholar]
- 29.Geva-Zatorsky N et al. Oscillations and variability in the p53 system. Mol. Syst. Biol. 2, 2006 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Batchelor E, Mock CS, Bhan I, Loewer A & Lahav G Recurrent initiation: a mechanism for triggering p53 pulses in response to DNA damage. Mol. Cell 30, 277–289 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nelson DE et al. Oscillations in NF-kappaB signaling control the dynamics of gene expression. Science 306, 704–708 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Karpova TS et al. Concurrent fast and slow cycling of a transcriptional activator at an endogenous promoter. Science 319, 466–469 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Dolmetsch RE, Xu K & Lewis RS Calcium oscillations increase the efficiency and specificity of gene expression. Nature 392, 933–936 (1998). [DOI] [PubMed] [Google Scholar]
- 34.Cai L, Dalal CK & Elowitz MB Frequency-modulated nuclear localization bursts coordinate gene regulation. Nature 455, 485–490 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chubb JR, Trcek T, Shenoy SM & Singer RH Transcriptional pulsing of a developmental gene. Curr. Biol. 16, 1018–1025 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mezo G, Manea M, Szabi I, Vincze B & Kovacs M New derivatives of GnRH as potential anticancer therapeutic agents. Curr. Med. Chem. 15, 2366–2379 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Schacke H, Docke WD & Asadullah K Mechanisms involved in the side effects of glucocorticoids. Pharmacol. Ther. 96, 23–43 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Sharp ZD et al. Estrogen-receptor-alpha exchange and chromatin dynamics are ligand-and domain-dependent. J Cell Sci. 119, 4101–4116 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Bosisio D et al. A hyper-dynamic equilibrium between promoter-bound and nucleoplasmic dimers controls NF-kB-dependent gene activity. EMBO J. 25, 798–810 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hager GL et al. Chromatin dynamics and the evolution of alternate promoter states. Chromosome. Res. 14, 107–116 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Muller WG, Walker D, Hager GL & McNally JG Large-scale chromatin decondensation and recondensation regulated by transcription from a natural promoter. J. Cell Biol. 154, 33–48 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vallone D et al. Neoplastic transformation of rat thyroid cells requires the junB and fra-1 gene induction which is dependent on the HMGI-C gene product. EMBO J. 16, 5310–5321 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


