Abstract
Four major clades of Candida auris have been described, and all infections have clustered in these 4 clades. We identified an isolate representative of a potential fifth clade, separated from the other clades by >200,000 single-nucleotide polymorphisms, in a patient in Iran who had never traveled outside the country.
Keywords: Candida auris, whole-genome sequencing, typing, clades, clonal, genotyping, fungi, fungal infections, multidrug-resistant infections, antimicrobial resistance
In the past decade, Candida auris has emerged in healthcare facilities as a multidrug-resistant pathogen that can cause outbreaks of invasive infections (1). C. auris has now been identified in >35 countries, many of which have documented healthcare-associated person-to-person spread (2). Transmission of this yeast is facilitated by its ability to colonize skin and other body sites, as well as its ability to persist for weeks on surfaces and equipment (3).
Whole-genome sequencing of C. auris has identified 4 major populations in which isolates cluster by geography (4). These populations are commonly referred to as the South Asian (I), East Asian (II), African (III), and South American (IV) clades. Worldwide, C. auris isolates continue to cluster in 1 of the 4 clades (Figure; 5–7). We report an isolate representative of a fifth clade in Iran from a patient who never traveled outside that country. The patient was a 14-year-old girl in whom C. auris otomycosis had been diagnosed; her case was the first known C. auris case in Iran (8).
Figure.
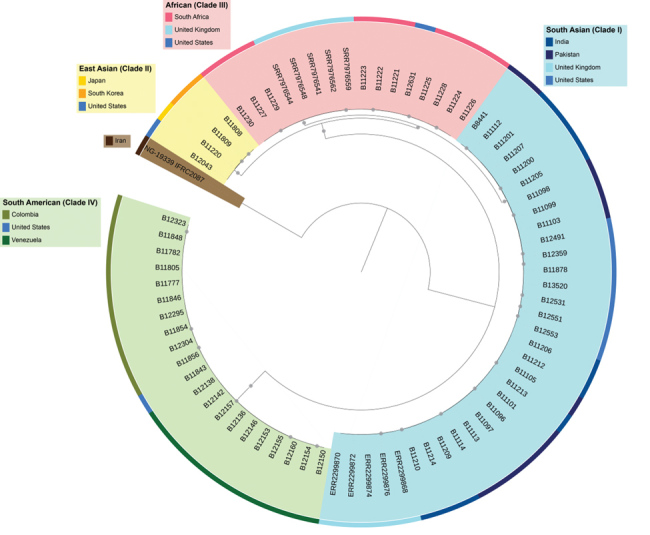
Major clades of Candida auris. Maximum-likelihood phylogenetic tree shows isolates from C. auris cases from 10 countries. Circles at nodes indicate separations with a bootstrap value >99%.
We conducted whole-genome sequencing of the isolate from Iran and 74 isolates from other countries (Appendix) and confirmed that the isolate from Iran was genetically distinct from the 4 existing clades, having a difference of >200,000 single-nucleotide polymorphisms compared with the other 4 clades. Isolates from the East Asian clade were its closest neighbors. Within the South Asian clade, isolates from C. auris cases in India, Pakistan, the United Kingdom, and the United States clustered together; within the East Asian clade, isolates from cases in Japan, South Korea, and the United States clustered together; within the African clade, isolates from cases in South Africa, the United Kingdom, and the United States clustered together; and within the South American clade, isolates from cases in Colombia, the United States, and Venezuela clustered together (Figure).
The C. auris isolate from Iran appears to represent a fifth major clade. Although this case was reported in 2018, additional cases of C. auris infections and colonization are thought to exist in Iran, given that challenges in diagnostic capacity in the country have probably limited the identification of more C. auris cases. The patient in this case was reported to have never traveled outside Iran (8), suggesting that this population structure might not be a result of a recent C. auris introduction into the country and that it might have emerged in Iran some time ago. Determining whether additional C. auris cases exist in Iran and whether such strains are related will help shed light on how C. auris emerged in Iran.
The isolate from Iran was susceptible to the 3 major classes of antifungal drugs and was cultured from ear swab specimens from the patient (8). C. auris of the East Asian clade is thought to have a propensity for the ear that is uncharacteristic of the other major clades (9). A recent study showed that, of 61 C. auris isolates obtained from 13 hospitals across South Korea during a 20-year period, 57 (93%) came from ear cultures (10). Although a systematic analysis has not been conducted, there are limited reports of ear infections or colonization caused by C. auris of the South Asian, African, or South American clades, so it is of interest that the isolate from Iran was most closely related to isolates of the East Asian clade, albeit with a difference of hundreds of thousands of single nucleotide polymorphisms. Ultimately, our discovery is a reminder that much about C. auris remains to be learned and underscores the need for vigilance in areas where C. auris has not yet emerged.
Additional information regarding a potential fifth clade of Candida auris, Iran, 2018.
Acknowledgments
We thank the Canisius-Wilhelmina Hospital for providing the research funds for the molecular analysis in this study.
Authors’ contributions: N.A.C. and J.F.M. designed the study; T.dG., H.B., and A.M. were involved in laboratory investigations; N.A.C. and T.M.C. performed the bioinformatics; N.A.C. drafted the manuscript; and all authors read, revised, and approved the final manuscript.
Biography
Dr. Chow is a molecular epidemiologist in the Mycotic Diseases Branch of the Division of Foodborne, Waterborne, and Environmental Diseases, National Center for Emerging and Zoonotic Infectious Diseases, Centers for Disease Control and Prevention. Her primary research interests include application of whole-genome sequencing and metagenomics for outbreak investigations as well as integrating and visualizing epidemiologic and laboratory data sources.
Footnotes
Suggested citation for this article: Chow NA, de Groot T, Badali H, Abastabar M, Chiller TM, Meis JF. Potential fifth clade of Candida auris, Iran, 2018. Emerg Infect Dis. 2019 Sep [date cited]. https://doi.org/10.3201/eid2509.190686
References
- 1.Chowdhary A, Sharma C, Meis JF. Candida auris: A rapidly emerging cause of hospital-acquired multidrug-resistant fungal infections globally. PLoS Pathog. 2017;13:e1006290. 10.1371/journal.ppat.1006290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saris K, Meis JF, Voss A. Candida auris. Curr Opin Infect Dis. 2018;31:334–40. [DOI] [PubMed] [Google Scholar]
- 3.Welsh RM, Bentz ML, Shams A, Houston H, Lyons A, Rose LJ, et al. Survival, persistence, and isolation of the emerging multidrug-resistant pathogenic yeast Candida auris on a plastic healthcare surface. J Clin Microbiol. 2017;55:2996–3005. 10.1128/JCM.00921-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lockhart SR, Etienne KA, Vallabhaneni S, Farooqi J, Chowdhary A, Govender NP, et al. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis. 2017;64:134–40. 10.1093/cid/ciw691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chow NA, Gade L, Tsay SV, Forsberg K, Greenko JA, Southwick KL, et al. ; US Candida auris Investigation Team. Multiple introductions and subsequent transmission of multidrug-resistant Candida auris in the USA: a molecular epidemiological survey. Lancet Infect Dis. 2018;18:1377–84. 10.1016/S1473-3099(18)30597-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Escandón P, Chow NA, Caceres DH, Gade L, Berkow EL, Armstrong P, et al. Molecular epidemiology of Candida auris in Colombia reveals a highly related, countrywide colonization with regional patterns in amphotericin B resistance. Clin Infect Dis. 2019;68:15–21. [DOI] [PubMed] [Google Scholar]
- 7.Rhodes J, Abdolrasouli A, Farrer RA, Cuomo CA, Aanensen DM, Armstrong-James D, et al. Genomic epidemiology of the UK outbreak of the emerging human fungal pathogen Candida auris. Emerg Microbes Infect. 2018;7:43. 10.1038/s41426-018-0045-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abastabar M, Haghani I, Ahangarkani F, Rezai MS, Taghizadeh Armaki M, Roodgari S, et al. Candida auris otomycosis in Iran and review of recent literature. Mycoses. 2019;62:101–5. 10.1111/myc.12886 [DOI] [PubMed] [Google Scholar]
- 9.Welsh RM, Sexton DJ, Forsberg K, Vallabhaneni S, Litvintseva A. Insights into the unique nature of the East Asian clade of the emerging pathogenic yeast Candida auris. J Clin Microbiol. 2019;57:e00007–00019. 10.1128/JCM.00007-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwon YJ, Shin JH, Byun SA, Choi MJ, Won EJ, Lee D, et al. Candida auris clinical isolates from South Korea: Identification, antifungal susceptibility, and genotyping. J Clin Microbiol. 2019;57:e01624–18. 10.1128/JCM.01624-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional information regarding a potential fifth clade of Candida auris, Iran, 2018.


