Abstract
During metamorphosis, holometabolous insects completely replace the larval gut and must control the microbiota to avoid septicaemia. Rapid induction of bactericidal activity in the insect gut at the onset of pupation has been described in numerous orders of the Holometabola and is best-studied in the Lepidoptera where it is under control of the 20-hydroxyecdysone (20E) moulting pathway. Here, using RNAseq, we compare the expression of immune effector genes in the gut during metamorphosis in a holometabolous (Galleria mellonella) and a hemimetabolous insect (Gryllus bimaculatus). We find that in G. mellonella, the expression of numerous immune effectors and the transcription factor GmEts are upregulated, with peak expression of three antimicrobial peptides (AMPs) and a lysozyme coinciding with delamination of the larval gut. By contrast, no such upregulation was detectable in the hemimetabolous Gr. bimaculatus. These findings support the idea that the upregulation of immune effectors at the onset of complete metamorphosis is an adaptive response, which controls the microbiota during gut replacement.
This article is part of the theme issue ‘The evolution of complete metamorphosis’.
Keywords: gut metamorphosis, antimicrobial peptides, lysozyme, Gryllus bimaculatus, Galleria mellonella, RNAseq
1. Introduction
Complete metamorphosis is the defining trait of holometabolous insects and entails a radical reorganization of the anatomy during a pupal stage [1]. The adaptive value of such a drastic event is to decouple the larval and adult stages [2], which separates growth and differentiation [3,4]. However, the addition of the sessile pupal stage, which seems to have evolved only once [5], also comes with costs, such as increased predation rates [6], parasitoid attacks [7] and infections [8]. Another challenge that needed to be resolved during the evolution of complete metamorphosis is the control of the microbiota during gut metamorphosis, which entails the replacement of the entire larval gut epithelium. The insect host must control the gut microbiota to avoid septicaemia, but complete eradication of the gut microbiota would risk the loss of potential mutualists [9]. Early work in Diptera used experimental infections of the house fly Musca domestica to demonstrate the persistence of bacteria during metamorphosis [10]. Such persistence was subsequently described in Coleoptera [11], Diptera [12], Lepidoptera [13] and Hymenoptera [14]. In the lepidopteran Manduca sexta, Russell & Dunn [15] showed that lysozyme and antimicrobial peptides (AMPs), best-studied as immune effectors produced in the fat body in response to microbial infections, are secreted by the replacement adult gut epithelium into the gut lumen en masse in a manner analogous to that of vertebrate Paneth cells. Importantly, the synthesis of immune effectors within the replacement gut epithelium begins before delamination of the larval epithelium and hence before the replacement epithelium becomes directly exposed to the microbiota [15]. Thus, it was proposed that the release of immune effectors upon delamination of the larval epithelium constitutes a prophylactic response under endocrine control that functions to protect the host from infection in the absence of the peritrophic matrix [16]. Recent work has described the same phenomenon in the silkworm Bombyx mori and the tobacco cutworm Spodoptera litura, where lysozyme and the lepidopteran AMP lebocin are induced in the pupal gut [17,18] in addition to their roles in other tissues as canonical immune defences [19]. In B. mori, this dual use has been achieved by co-option of lysozyme and lebocin expression by the 20-hydroxyecdysone (20E) moulting pathway [17]. In the case of lebocin, this permits Toll-mediated induction in the fat body in response to infection [19] in addition to 20E-mediated induction via the Broad complex and the transcription factor BmEts in the gut during metamorphosis [17]. Taken together, these results support the notion that the induction of immune effectors in the gut prior to metamorphosis is under endocrine control and independent of microbial recognition [17]. The study of immune gene induction across the entirety of the larval–pupal moult is hampered by the difficulties inherent in the specification of the stages of gut metamorphosis. Delamination of the larval gut epithelium occurs in the prepupa and precedes ecdysis of the external cuticle. Thus, it is not possible to directly observe the ultrastructural changes in the prepupal gut in a non-destructive manner. In Galleria mellonella, the stage of gut replacement can be precisely specified by observing the migration of pigments in the stemmata (larval eyes) [20], which allows sampling of guts at each stage of the larval–pupal moult. We have previously used this classification system to enable the quantification and manipulation of lysozyme expression in the gut during the larval–pupal moult, demonstrating that changes in the composition of the gut microbiota during metamorphosis is under partial host immunological control [9].
Metamorphosis in hemimetabolous insects contrasts with holometaboly, in that it entails much less physiological and morphological reorganization. While development is under the control of the same major pathways [21], little is known about the regulation of immune genes during moults and especially during the nymphal–adult moult. Indirect evidence of differences in the regulation of immunity in the gut is provided by studies that quantify the changes in the gut microbiota during development. Sudukaran et al. [22] reported a constant increase in microbial density and diversity during the development of the hemimetabolous fire bug. By contrast, complete metamorphosis drives strong reductions in density, diversity and changes in composition [9,13,23], with some bacterial species being driven to extinction [9,23].
Here we compare, using RNAseq, the temporal dynamics of immune effector gene expression in the midgut throughout the final larval moult in the holometabolous insect G. mellonella with the hemimetabolous insect Gryllus bimaculatus, taking these as examples of the different developmental modes [24]. We hypothesize that the induction of AMP and lysozyme expression observed in holometabolous insects should coincide with delamination of the larval gut epithelium. By contrast, given that the gut does not undergo extensive remodelling or replacement during hemimetabolous metamorphosis, we expect no immune effector induction in the absence of infection.
2. Material and methods
(a). Gryllus bimaculatus rearing and sampling
A laboratory culture of Gr. bimaculatus (two-spotted cricket) was established with insects purchased from a commercial supplier (Der Terraristikladen, Düsseldorf, Germany) and reared at the Freie Universität Berlin. They were held in plastic containers with a 16 L : 8 D cycle at 27 ± 1°C. Crickets were fed a diet of insect pellets and were supplied with water ad libitum via a cotton plugged tube. Under these conditions, a complete generation cycle included eight moults over a period of approximately 62 days, with the final larval instar lasting for 9 days [25,26]. Crickets were reared individually from the seventh instar and checked daily to determine their development into the final instar. Stage I samples were collected 7 days after crickets moulted into the final nymphal instar and individuals stopped feeding. Individuals of stage II were sampled 24 h later and crickets representing stage III were collected after an additional 12 h. Adults were sampled within 1 h after the imaginal moult. RNA was isolated from dissected midguts as described below and used to create 3 independent replicate pools per stage, each representing 8 individual insects, resulting in 12 sample pools.
(b). Galleria mellonella rearing and sampling
Final-instar larvae were purchased from a commercial supplier (Livefoods Direct, Sheffield, UK) and used to establish a laboratory colony. Larvae were reared in the dark at 30°C on a grain-honey diet as described previously [9].
The staging system of Kühn & Piepho [27], as described by Uwo et al. [20], was used to specify the stages of midgut metamorphosis in larvae and pupae (see Uwo et al. [20] and Ellis et al. [28] for illustrations):
-
1.
Stage I, a wandering larva that has ceased feeding and started spinning. Pigments remain entirely within the stemmata.
-
2.
Stage II, a spinning larva. Pigments have partially migrated from the stemmata.
-
3.
Stage III, a spinning larva. Pigments have left the stemmata completely but are still in contact with the cuticle. The larval gut epithelium has delaminated and floats freely in the lumen.
-
4.
Stage IV, a mature spinning larva. Pigments have merged and sunk beneath the cuticle but are still visible. This stage is very brief and could not be adequately sampled.
-
5.
Stage V, a prepupa that has ceased spinning. Stemmatal pigments are no longer visible. The midgut is laterally flattened and the detached larval gut has formed the yellow body that undergoes apoptosis.
-
6.
Stage VI, a new pupa. The cuticle has not sclerotized and is completely white.
-
7.
Stage VII, a sclerotized pupa. Approximately 24 h after the larval–pupal moult.
The migration of stemmatal pigments was monitored under a stereo microscope. For each of six stages (I–III and V–VII), RNA was isolated (as described below) from dissected midguts and combined in 6 (stages II and VI), 7 (stage III) or 8 (stages I, V and VII) independent pools, each representing 10 individuals.
(c). RNA isolation and library preparation
Midguts were homogenized in Trizol (Sigma, Taufkirchen, Germany) with two sterile 5 mm steel beads (Qiagen, Hilden, Germany) using a TissueLyzer (Qiagen) at 30 Hz for 2 × 3 min. Total RNA was recovered according to the manufacturer's instructions using chloroform extraction and isopropanol precipitation and re-dissolved in RNA Storage Solution (Ambion). Samples were incubated with 2 units of TurboDNase (Ambion) for 30 min at 37°C before a phenol/chloroform extraction was performed in order to purify the RNA. Equal quantities of total RNA were used to create independent replicate pools for each stage. RNA pools were quantified using a Qubit 2.0 fluorometer (Life Technologies, Carlsbad, California) and an RNA 6000 Nano chip on a BioAnalyzer 2100 (Aligent, California). Gr. bimaculatus libraries were prepared using the NEXTflex™ Rapid Directional mRNA-Seq Kit (Bioo Scientific, Austin, Texas) and sequenced on an Illumina NextSeq550 platform (Illumina, San Diego, CA, USA) at the Berlin Center for Genomics in Biodiversity Research (BeGenDiv) for 150 cycles to yield 15–41 million read pairs per library (mean 28 million). G. mellonella libraries were prepared using an illumina TruSeq mRNA kit and sequenced for 200 cycles on a HiSeq 2000 at the Earlham Institute (Norwich, UK) to yield 23–45 million read pairs per library (mean 32 million). Sequencing data are available under BioProject accessions PRJNA268796 and PRJNA547710.
(d). De novo assembly and annotation
Assemblies for both species were produced using Trinity v. 2.8.4 [29], incorporating quality and adapter filtering via Trimmomatic [30] and subsequent in silico normalization. Assemblies were annotated with the Trinotate annotation pipeline [31].
(e). Inference of orthologous gene groups
OrthoFinder v. 2.2.7 [32] was used to infer orthology between predicted peptide sequences from Gr. bimaculatus, G. mellonella and the proteomes of other insect species with sequenced genomes. These included Acyrthosiphon pisum, Apis mellifera, Drosophila melanogaster, Tribolium castaneum, Zootermopsis nevadensis, as well as all Lepidopteran genomes from lepbase release 4 [33].
(f). Immune effector gene identification
Immune effectors of Gr. bimaculatus and G. mellonella were identified from orthologue groups containing annotated immune genes from previously published insect genome projects. Additionally, blast and HMM homology searches were performed using previously described insect immune effector proteins as queries against each de novo assembly.
(g). Differential gene expression
For both species, transcript abundances were quantified by pseudo-aligning RNAseq reads to de novo assemblies using Salmon v. 0.1.2.0 [34]. tximport [35] was used in conjunction with DESeq2 [36] to model gene-level estimated counts while correcting for changes in transcript usage across samples. Specifically, to identify differential expression as a function of developmental stage, likelihood-ratio tests were performed between full and intercept-only negative binomial GLMs. Differential expression was considered to be significant when fold changes were greater than 2 for pairwise Wald contrasts of developmental stages, with a false discovery rate (FDR)-corrected p-value of less than 0.05. The mean of the normalized counts for each gene was used as the informative covariate for independent hypothesis weighting [37] in order to optimize the power of multiple testing.
3. Results
(a). Galleria mellonella
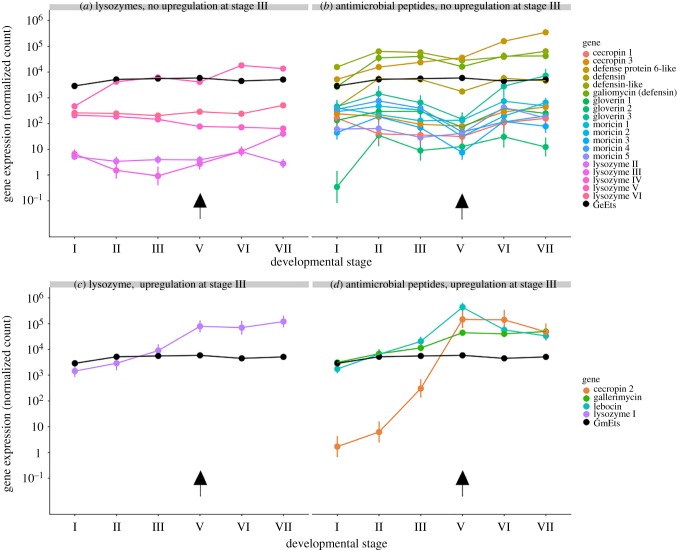
A total of 6 lysozymes and 23 AMPs were identified in the G. mellonella de novo assembly, the majority of which have been previously described ([38,39] figure 1a). These included three gloverins, five moricins, three cecropins, three defensins and one antifungal peptide (gallerimycin). A previously described lebocin-like proline-rich protein [39,40] was predicted to belong to an orthologue group together with all annotated lebocins from the lepbase v4 and is here referred to as lebocin. Lysozyme I and lebocin were predicted to be orthologues of B. mori lysozyme and lebocin, respectively, which were previously shown to be under control of 20E via the Broad complex [41] and the BmEts transcription factor [17]. The expression of all immune effectors varied significantly as a function of developmental stage; however, only four (lysozyme I, cecropin 2, lebocin and gallerimycin) were identified as being induced at the onset of the larval–pupal moult in pairwise Wald contrasts of stage III versus stage V (figure 1a). A putative orthologue of the BmEts transcription factor (here GmEts) was identified as part of a single-copy Ets orthologue group. GmEts showed significant induction between stage I and stage II in a pairwise Wald contrast (Wald statistic = 214.7199, adjusted p-value < 0.001), figure 1).
Figure 1.
Immune effector gene expression in the midgut during the larval–pupal moult of Galleria mellonella. Roman numerals correspond to precise developmental stages (§2). Plotted values represent the coefficients and 95% confidence intervals from negative binomial generalized linear models. (a,b) Lysozyme and antimicrobial peptide genes with no upregulation between stages III and V. (c,d) Lysozyme and antimicrobial peptide genes with significant upregulation between stages III and V. Upregulation was considered to be significant when fold changes were greater than 2 in a pairwise Wald contrast at an FDR-corrected p-value of less than 0.05. Arrows denote the onset of the larval–pupal moult.
(b). Gryllus bimaculatus
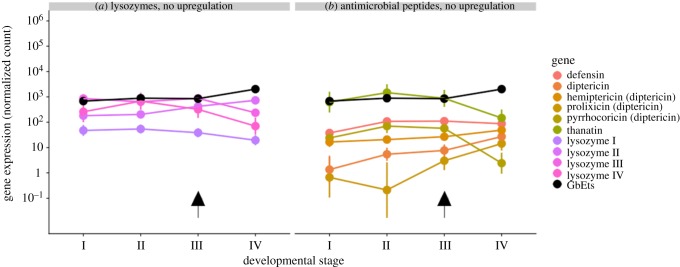
Orthologues of 10 immune effector genes were identified, comprising 4 inducible lysozymes and 6 AMPs. We identified peptides with similarities to the classical AMPs defensin and diptericin, as well as to 3 other diptericin-like peptides: pyrrhocoricin [42], prolixicin [43] and hemiptericin [44]. Thanatin was first isolated from the hemipteran insect Podisus maculiventris and does not show similarities to the conventional families of insect AMPs but exhibits sequence similarities with the brevinine family of AMPs from frog skin [45]. It shows activity against Gram-positive, Gram-negative bacteria as well as against fungi [45]. There was no significant upregulation of any of the annotated immune effector genes during the nymphal–adult moult (figure 2). A putative orthologue of the transcription factor BmEts [17] (here GbETS) also showed no significant change in expression (figure 2).
Figure 2.
(a,b) Immune effector gene expression in the midgut during the nymphal–adult moult of Gryllus bimaculatus. Plotted values represent the coefficients and 95% confidence intervals from negative binomial generalized linear models. Arrows denote the onset of the nymph–adult moult.
4. Discussion
Taking G. mellonella and Gr. bimaculatus as representatives of holo- and hemimetabolous insects, we found stark differences in immune effector gene expression between both species during metamorphosis consistent with the hypothesis that only complete metamorphosis elicits a prophylactic gut immune response [16]. Clearly, further comparisons of representative species from multiple orders from holo- and hemimetabolous groups are needed to confirm whether this finding reflects a general pattern. As previously observed in other lepidoptera, G. mellonella shows strong upregulation of a subset of immune effector genes at the onset of the larval–pupal moult, whereas in Gr. bimaculatus, no significant induction was detected. Our precise specification of the stages of gut metamorphosis in G. mellonella prepupae [20] shows that peak immune gene expression coincides with delamination of the larval gut epithelium. Work in Bombyx has demonstrated that transcription factor BmEts mediated by 20E can induce the expression of immune effectors independent of microbe-associated molecular patterns (MAMPs) [17]. BmEts' upregulation occurs in the prepupa approximately 12 h prior to immune effector induction [14]. A similar pattern is evident in our own data, with significant induction of GmEts occurring at stage II, the spinning larva, prior to both delamination of the larval gut epithelium and significant induction of immune effector gene expression (figure 1b,c). MAMP-independent 20E-mediated upregulation of immune effectors has not been reported in hemimetabolous insects and we detected no such upregulation during gut metamorphosis in Gr. bimaculatus. This is likely explained by the less drastic changes to gut anatomy of hemimetabolous insects during metamorphosis, which does not entail gut replacement [46] and therefore does not necessitate prophylaxis. However, experimental support for a prophylactic effect in holometabolous insects is lacking, and our own work manipulating lysozyme expression during gut metamorphosis via RNAi knockdown did not detect any difference in pupal mortality [9]. We also cannot exclude the possibility that in either species there may be differentially expressed immune genes which were not successfully annotated or that are expressed in other parts of the gut (e.g. the hindgut, which may be an important site of bacterial colonization) and are therefore unintentionally excluded from our analysis. The present data show that in G. mellonella, the cocktail of induced immune effectors is more complex than previously described, with three AMPs (lebocin, cecropin 2 and gallerimycin) showing induction, in addition to lysozyme I. Future work could use RNAi knockdown to test whether functional redundancy in the effector cocktail has hindered the detection of a protective effect. This is likely given that synergistic interactions between insect AMPs are common [47,48] and are known to occur between G. mellonella immune effectors, including lysozyme I [49]. Alternatively, immune induction may serve to control the proliferation of the microbiota rather than to specifically protect against opportunistic infection. We observed that immune effector upregulation persists into the pupal stage (figure 1, stages VI and VII), during which time the degenerating larval gut epithelium forms the ‘yellow body’ within the gut lumen and undergoes apoptosis and necrosis to release breakdown products that are recycled by the replacement gut [20,50]. This raises the possibility that immune induction may function to suppress bacterial growth that might otherwise disrupt the complex trophic relationship between the autolytic larval gut and the replacement adult gut [51].
A third explanation for the observed immune induction is that it may function to drive changes in microbial community composition, for example, in order to facilitate ontogenetic shifts in habitat and/or diet. This has recently been suggested as one possible explanation for changes in microbial community composition during amphibian gut metamorphosis [52], where development is coupled with a shift from aquatic detritivore to a terrestrial insectivore.
5. Conclusion
The replacement of the larval gut during metamorphosis in holometabolous insects is associated with the induction of an immune response in the gut. Future work should investigate the adaptive value of this response and whether the upregulation of gut immunity functions either as a simple form of prophylaxis with collateral effects on the composition of the gut microbiota, or to suppress the growth of microbes that could otherwise compete with the host for nutrients released by the autolytic larval gut, or to actively regulate shifts in the composition of the microbiota, for example, to facilitate ontogenetic diet shifts.
Data accessibility
Sequencing data are available under BioProject accessions PRJNA268796 and PRJNA547710.
Authors' contributions
P.R.J., V.P. and J.R. conceived and designed the experiments, analysed the data and wrote the paper. P.R.J. and V.P. performed the experiments.
Competing interests
We declare we have no competing interests.
Funding
This research was supported by European Research Council (http://erc.europa.eu/) grant no. 260986 to J.R., by the BBSRC Genome Analysis Centre Capacity & Capability Challenge programme to J.R. and P.R.J. (http://www.bbsrc.ac.uk/) and by Deutsche Forschungsgemeinschaft (DFG) project 389139730 to J.R. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hall MJR, Martín-Vega D. 2019. Visualization of insect metamorphosis. Phil. Trans. R. Soc. B 374, 20190071 ( 10.1098/rstb.2019.0071) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moran NA. 1994. Adaptation and constraint in the complex life cycles of animals. Annu. Rev. Ecol. Syst. 25, 573–600. ( 10.1146/annurev.es.25.110194.003041) [DOI] [Google Scholar]
- 3.Arendt JD. 1997. Adaptive intrinsic growth rates: an integration across taxa. Q. Rev. Biol. 72, 149–177. ( 10.1086/419764) [DOI] [Google Scholar]
- 4.Berlese A. 1913. Intorno alle metamorfosi degli insetti. Redia 9, 9. [Google Scholar]
- 5.Misof B, et al. 2014. Phylogenomics resolves the timing and pattern of insect evolution. Science 346, 763–767. ( 10.1126/science.1257570) [DOI] [PubMed] [Google Scholar]
- 6.Lindstedt C, Murphy L, Mappes J. 2019. Antipredator strategies of pupae: how to avoid predation in an immobile life stage? Phil. Trans. R. Soc. B 374, 20190069 ( 10.1098/rstb.2019.0069) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Godfray HCJ. 1994. Parasitoids: behavioral and evolutionary ecology. Princeton, NJ: Princeton University Press. [Google Scholar]
- 8.Critchlow JT, Norris A, Tate AT. 2019. The legacy of larval infection on immunological dynamics over metamorphosis. Phil. Trans. R. Soc. B 374, 20190066 ( 10.1098/rstb.2019.0066) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnston PR, Rolff J. 2015. Host and symbiont jointly control gut microbiota during complete metamorphosis. PLoS Pathog. 11, e1005246 ( 10.1371/journal.ppat.1005246) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bacot AW. 1911. The persistence of Bacillus pyocyaneus in pupae and imagines of Musca domestica raised from larvae experimentally infected with the bacillus. Parasitology 4, 68–74. ( 10.1017/S0031182000002468) [DOI] [Google Scholar]
- 11.Delalibera I, Vasanthakumar A, Klepzig KD, Raffa KF. 2007. Composition of the bacterial community in the gut of the pine engraver, Ips pini (Say)(Coleoptera) colonizing red pine. Symbiosis 43, 97–104. [Google Scholar]
- 12.Wong CNA, Ng P, Douglas AE. 2011. Low-diversity bacterial community in the gut of the fruitfly Drosophila melanogaster: bacterial community in Drosophila melanogaster. Environ. Microbiol. 13, 1889–1900. ( 10.1111/j.1462-2920.2011.02511.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammer TJ, McMillan WO, Fierer N. 2014. Metamorphosis of a butterfly-associated bacterial community. PLoS ONE 9, e86995 ( 10.1371/journal.pone.0086995) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brucker RM, Bordenstein SR. 2012. The roles of host evolutionary relationships (genus: Nasonia) and development in structuring microbial communities. Evolution 66, 349–362. ( 10.1111/j.1558-5646.2011.01454.x) [DOI] [PubMed] [Google Scholar]
- 15.Russell VW, Dunn PE. 1991. Lysozyme in the midgut of Manduca sexta during metamorphosis. Arch. Insect. Biochem. Physiol. 17, 67–80. ( 10.1002/arch.940170202) [DOI] [PubMed] [Google Scholar]
- 16.Russell V, Dunn PE. 1996. Antibacterial proteins in the midgut of Manduca sexta during metamorphosis. J. Insect. Physiol. 42, 65–71. ( 10.1016/0022-1910(95)00083-6) [DOI] [PubMed] [Google Scholar]
- 17.Mai T, Chen S, Lin X, Zhang X, Zou X, Feng Q, Zheng S. 2017. 20-hydroxyecdysone positively regulates the transcription of the antimicrobial peptide, lebocin, via BmEts and BmBR-C Z4 in the midgut of Bombyx mori during metamorphosis. Dev. Comp. Immunol. 74, 10–18. ( 10.1016/j.dci.2017.04.002) [DOI] [PubMed] [Google Scholar]
- 18.Xu Q, et al. 2012. Transcriptional profiling of midgut immunity response and degeneration in the wandering silkworm, Bombyx mori. PLoS ONE 7, e43769 ( 10.1371/journal.pone.0043769) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanaka H, Yamamoto M, Moriyama Y, Yamao M, Furukawa S, Sagisaka A, Nakazawa H, Mori H, Yamakawa M. 2005. A novel Rel protein and shortened isoform that differentially regulate antibacterial peptide genes in the silkworm Bombyx mori. Biochim. Biophys. Acta BBA Gene Struct. Exp. 1730, 10–21. ( 10.1016/j.bbaexp.2005.05.007) [DOI] [PubMed] [Google Scholar]
- 20.Uwo M, Ui-Tei K, Park P, Takeda M. 2002. Replacement of midgut epithelium in the greater wax moth, Galleria mellonela, during larval–pupal moult. Cell Tissue Res. 308, 319–331. ( 10.1007/s00441-002-0515-1) [DOI] [PubMed] [Google Scholar]
- 21.Belles X. 2019. The innovation of the final moult and the origin of insect metamorphosis. Phil. Trans. R. Soc. B 374, 20180415 ( 10.1098/rstb.2018.0415) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sudakaran S, Salem H, Kost C, Kaltenpoth M. 2012. Geographical and ecological stability of the symbiotic mid-gut microbiota in European firebugs, Pyrrhocoris apterus (Hemiptera, Pyrrhocoridae). Mol. Ecol. 21, 6134–6151. ( 10.1111/mec.12027) [DOI] [PubMed] [Google Scholar]
- 23.Greenberg B. 1969. Salmonella suppression by known populations of bacteria in flies. J. Bacteriol. 99, 629–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berenbaum MR, Isman MB. 1989. Herbivory in holometabolous and hemimetabolous insects: contrasts between Orthoptera and Lepidoptera. Experientia 45, 229–236. ( 10.1007/BF01951808) [DOI] [Google Scholar]
- 25.Gerstenlauer B, Hoffmann KH. 1995. Ecdysteroid release and ecdysteroid titer during larval–adult development of the Mediterranean field cricket, Gryllus bimaculatus (Ensifera: Gryllidae). Eur. J. Entomol. 92, 81–92. ( 10.1016/s0022-1910(98)00057-2) [DOI] [Google Scholar]
- 26.Sturm R. 2008. Morphology and ultrastructure of the female accessory sex glands in various crickets (Orthoptera, Saltatoria, Gryllidae). Dtsch Entomol. Z. 49, 185–195. ( 10.1002/mmnd.20020490203) [DOI] [Google Scholar]
- 27.Kühn A, Piepho H. 1936. Über hormonale Wirkungen bei der Verpuppung der Schmetterlinge, von Alfred Kühn und Hans Piepho. Nachrichten Von Ges Wiss Zu Gött. 2, 141–154. [Google Scholar]
- 28.Ellis JD, Graham JR, Mortensen A. 2013. Standard methods for wax moth research. J. Apic. Res. 52, 1–17. ( 10.3896/IBRA.1.52.1.10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haas BJ, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512. ( 10.1038/nprot.2013.084) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. ( 10.1093/bioinformatics/btu170) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. ( 10.1038/nbt.1883) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emms DM, Kelly S. 2015. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16, 157 ( 10.1186/s13059-015-0721-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Challis RJ, Kumar S, Dasmahapatra KKK, Jiggins CD, Blaxter M. 2016. Lepbase: the Lepidopteran genome database. bioRxiv [cited 2019 Apr 2]; see http://biorxiv.org/lookup/doi/10.1101/056994.
- 34.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. 2017. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419. ( 10.1038/nmeth.4197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soneson C, Love MI, Robinson MD. 2015. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res. 4, 1521 ( 10.12688/f1000research.7563.1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 ( 10.1186/s13059-014-0550-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ignatiadis N, Klaus B, Zaugg JB, Huber W. 2016. Data-driven hypothesis weighting increases detection power in genome-scale multiple testing. Nat. Methods 13, 577–580. ( 10.1038/nmeth.3885) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vogel H, Altincicek B, Glöckner G, Vilcinskas A. 2011. A comprehensive transcriptome and immune-gene repertoire of the lepidopteran model host Galleria mellonella. BMC Genomics 12, 308 ( 10.1186/1471-2164-12-308) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brown SE, Howard A, Kasprzak AB, Gordon KH, East PD. 2009. A peptidomics study reveals the impressive antimicrobial peptide arsenal of the wax moth Galleria mellonella. Insect. Biochem. Mol. Biol. 39, 792–800. ( 10.1016/j.ibmb.2009.09.004) [DOI] [PubMed] [Google Scholar]
- 40.Cytryńska M, Mak P, Zdybicka-Barabas A, Suder P, Jakubowicz T. 2007. Purification and characterization of eight peptides from Galleria mellonella immune hemolymph. Peptides 28, 533–546. ( 10.1016/j.peptides.2006.11.010) [DOI] [PubMed] [Google Scholar]
- 41.Ma H, Abbas MN, Zhang K, Hu X, Xu M, Liang H, Kausar S, Yang L, Cui H. 2019. 20-Hydroxyecdysone regulates the transcription of the lysozyme via broad-complex Z2 gene in silkworm, Bombyx mori. Dev. Comp. Immunol. 94, 66–72. ( 10.1016/j.dci.2019.01.014) [DOI] [PubMed] [Google Scholar]
- 42.Kragol G, et al. 2002. Identification of crucial residues for the antibacterial activity of the proline-rich peptide, pyrrhocoricin: multifunctional antimicrobial peptides. Eur. J. Biochem. 269, 4226–4237. ( 10.1046/j.1432-1033.2002.03119.x) [DOI] [PubMed] [Google Scholar]
- 43.Ursic-Bedoya R, Buchhop J, Joy JB, Durvasula R, Lowenberger C. 2011. Prolixicin: a novel antimicrobial peptide isolated from Rhodnius prolixus with differential activity against bacteria and Trypanosoma cruzi. Insect. Mol. Biol. 20, 775–786. ( 10.1111/j.1365-2583.2011.01107.x) [DOI] [PubMed] [Google Scholar]
- 44.Cociancich S, Dupont A, Hegy G, Lanot R, Holder F, Hetru C, Hoffmann JA, Bulet P. 1994. Novel inducible antibacterial peptides from a hemipteran insect, the sap-sucking bug Pyrrhocoris apterus. Biochem. J. 300, 567–575. ( 10.1042/bj3000567) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fehlbaum P, Bulet P, Chernysh S, Briand JP, Roussel JP, Letellier L, Hetru C, Hoffmann JA. 1996. Structure-activity analysis of thanatin, a 21-residue inducible insect defense peptide with sequence homology to frog skin antimicrobial peptides. Proc. Natl Acad. Sci. USA 93, 1221–1225. ( 10.1073/pnas.93.3.1221) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teixeira AdD, Fialho MdC, Zanuncio JC, Ramalho FdS, Serrão JE. 2013. Degeneration and cell regeneration in the midgut of Podisus nigrispinus (Heteroptera: Pentatomidae) during post-embryonic development. Arthropod. Struct. Dev. 42, 237–246. ( 10.1016/j.asd.2013.02.004) [DOI] [PubMed] [Google Scholar]
- 47.Zanchi C, Johnston PR, Rolff J. 2017. Evolution of defence cocktails: antimicrobial peptide combinations reduce mortality and persistent infection. Mol. Ecol. 26, 5334–5343. ( 10.1111/mec.14267) [DOI] [PubMed] [Google Scholar]
- 48.Yu G, Baeder DY, Regoes RR, Rolff J. 2016. Combination effects of antimicrobial peptides. Antimicrob. Agents Chemother. 60, 1717–1724. ( 10.1128/AAC.02434-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zdybicka-Barabas A, Mak P, Klys A, Skrzypiec K, Mendyk E, Fiołka MJ, Cytryńska M. 2012. Synergistic action of Galleria mellonella anionic peptide 2 and lysozyme against Gram-negative bacteria. Biochim. Biophys. Acta BBA Biomembr. 1818, 2623–2635. ( 10.1016/j.bbamem.2012.06.008) [DOI] [PubMed] [Google Scholar]
- 50.Tettamanti G, Casartelli M. 2019. Cell death during complete metamorphosis. Phil. Trans. R. Soc. B 374, 20190065 ( 10.1098/rstb.2019.0065) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Franzetti E, et al. 2015. The midgut of the silkmoth Bombyx mori is able to recycle molecules derived from degeneration of the larval midgut epithelium. Cell Tissue Res. 361, 509–528. ( 10.1007/s00441-014-2081-8) [DOI] [PubMed] [Google Scholar]
- 52.Warne RW, Kirschman L, Zeglin L. 2017. Manipulation of gut microbiota reveals shifting community structure shaped by host developmental windows in amphibian larvae. Integr. Comp. Biol. 57, 786–794. ( 10.1093/icb/icx100) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sequencing data are available under BioProject accessions PRJNA268796 and PRJNA547710.