Abstract
Premise
The digitization of natural history collections includes transcribing specimen label data into standardized formats. Born‐digital specimen data initially gathered in digital formats do not need to be transcribed, enabling their efficient integration into digitized collections. Modernizing field collection methods for born‐digital workflows requires the development of new tools and processes.
Methods and Results
collNotes, a mobile application, was developed for Android and iOS to supplement traditional field journals. Designed for efficiency in the field, collNotes avoids redundant data entries and does not require cellular service. collBook, a companion desktop application, refines field notes into database‐ready formats and produces specimen labels.
Conclusions
collNotes and collBook can be used in combination as a field‐to‐database solution for gathering born‐digital voucher specimen data for plants and fungi. Both programs are open source and use common file types simplifying either program's integration into existing workflows.
Keywords: biodiversity data, born digital, collection app, field work, herbarium labels, natural history collections
Biologists conducting field research, such as floristic studies, accession thousands of specimens into natural history collections. Many of these specimens’ digital records are now becoming available through online portals such as iDigBio (https://www.idigbio.org/), the Global Biodiversity Information Facility (GBIF) (Global Biodiversity Information Facility, 2018; https://www.gbif.org/), Symbiota (Gries et al., 2014; http://symbiota.org/), and regional consortia (e.g., SouthEast Regional Network of Expertise and Collections [SERNEC]; http://sernecportal.org/). One major challenge in digitizing these specimens is the accurate transcription of physical labels into digital formats. Numerous workflows have been presented to address this challenge, whereby citizen scientists, students, or professionals are tasked with transcribing these data (Hill et al., 2012; Ellwood et al., 2015; Harris and Marsico, 2017; Sweeney et al., 2018). Initially developed to digitize a backlog of historic specimens, these methods are reactive, relying on third parties (i.e., persons other than the initial collector) to transcribe the data after it has been organized onto a label. Whereas reactive digitization is necessary for historic data, new specimens are being collected every day. Among the Plantae records available on iDigBio, 77% have date of collection data and, of those, there is an average of 348,000 specimens collected per year (2006–2015). Proactively capturing data from these new specimens earlier in the process reduces the potential for transcription errors (Nelson et al., 2015) and avoids adding to the backlog of records in need of transcription. Attention to this issue has highlighted the need for born‐digital records, i.e., field data that are initially gathered in digital formats so that they are ready for upload to online data portals and label printing (Paul et al., 2015; James et al., 2018).
Here we introduce the public releases of collNotes and collBook, two open source programs that, when combined, provide a field‐to‐database solution for collections‐based research. These programs were developed with the goal of initially collecting biological specimen data in a digital format that would not contribute to the backlog of records in need of transcription. To achieve this, collNotes, a mobile application, was developed to supplement the traditional field journal. A companion desktop application, collBook, enables users to refine field notes and produce a comma‐separated values (CSV) file formatted in the Darwin Core (DwC) (Wieczorek et al., 2012) data standard. The DwC is used by many biodiversity data portals such as those built on the Symbiota framework. Adherence to this data standard makes collBook's output directly importable by any DwC‐capable portal or collection management system.
The success of migrating collections to a born‐digital workflow will depend on the adoption of these new methods by field biologists. Thus, we prioritized user experience and efficiency in the field. One time‐intensive and generally tedious step in the traditional collection process is label preparation, which requires the collector to organize and digitally transcribe field notes. To encourage adoption, a PDF file containing formatted, ready‐to‐print labels is the second output of this solution. Both collNotes and collBook are open source projects, released under the GNU General Public License v3.0. It is our hope that these programs will improve the efficiency, accuracy, and accessibility of collections‐based research.
METHODS AND RESULTS
collNotes development
A mobile application, collNotes, was developed using Microsoft's Xamarin development kit (Xamarin, San Francisco, California, USA) and is available on Android and iOS devices. collNotes was developed to supplement a traditional field journal. Although it is a mobile application, collNotes does not require cellular service to record field notes. It was designed with a minimalistic interface, prioritizing time‐saving features, especially where location data are concerned. The locality entered by the user is expected to be limited to the highest‐resolution portion of a locality string (e.g., “50 m northeast of the Illinois Monument”). There are no state, county, or municipality entry fields in collNotes. These location data are inferred later in collBook based on global positioning system (GPS) coordinates captured in the field, by collNotes. Most entry fields in collNotes are optional, and some, such as eventDate and primary collector, can be automatically populated. In the case of the entry field “reproductive condition,” where the DwC recommends controlled vocabulary, a list of terms is provided. Data from collNotes are stored in the mobile device's local storage as an SQLite database file. Exporting records produces a UTF‐8 encoded CSV file, organized (with few exceptions) under DwC terms. This resulting output may then be refined into labels and database‐ready records using collBook. Features included in collNotes are: structured data, field number generation, and coordinate capture.
Structured data
Records from a collection event often contain redundant information. To avoid repetitive manual entries, we designed a hierarchy of data categories (i.e., classes) similar to DwC classes. In collNotes, we used three classes: “trip,” “site,” and “specimen.” These classes identify which data are appropriate to duplicate across records. For example, a collection trip for a project named “Flora of Risa” might be associated with multiple sites, all of which inherit “Flora of Risa” as the project name. The classes and the data fields they are associated with are listed in Box 1. One advantage of this kind of structured data can be illustrated if numerous voucher specimens are collected from a single location. In this scenario, locality and habitat information can be entered once and propagated to pertinent records. The geographic range of a single site is left to the researcher's discretion. However, because localities and coordinates are inherited by the site class, and mobile device GPS is generally accurate to about 20 m (Tomaštík et al., 2017), a site range between 5 and 30 m is recommended.
BOX 1. The hierarchical classes used in collNotes and collBook, showing the associated Darwin Core data fields.
| Trip | Site | Specimen |
|---|---|---|
| additional | associated Taxa | catalog Number |
| Collectorsa | coordinate Uncertainty | establishment Meansc |
| event Date | In Meters | identification References |
| Label Projecta | country | identification Remarks |
| recorded By | county | individual Count |
| sampling Effort | decimal Latitude | occurrence Remarks |
| decimal Longitude | record Number | |
| habitat | reproductive Condition | |
| locality | scientific Name | |
| location Notes | scientific Name Authorship | |
| minimum Elevation In Meters | substrate | |
| municipality | ||
| pathb | ||
| state Province |
Not a Darwin Core field, used by Symbiota.
Not a Darwin Core field, used by collBook to store road or trail name.
The only possible value for “establishmentMeans” is: “cultivated” (set using the “cultivated” checkbox in either collNotes or collBook), otherwise the field is left blank.
Field number generation
When creating a new site in collNotes, a site number is automatically generated and is used by collNotes and collBook to link associated specimen records. When collections are made indiscriminately, i.e., multiple taxa from a single site are placed in the same container, the site number should be used to label the container. This allows users the option to forgo documenting specimen‐specific data in the field, and instead to generate it while refining the records in collBook. Similarly, when creating a specimen record in collNotes, a specimen number is generated that is synonymous to a traditional field number. A specimen number is formatted as two values separated by a dash, with the first value being a site number and the second being a unique value for that specimen. The starting number for specimen collection can be user defined to accommodate workers keeping lifetime numbers, although it will prepend a site number. To avoid duplicate specimen numbers, the starting site or specimen numbers can be altered in collNote's settings. For proper specimen container labeling, both site and specimen numbers are prominently displayed when generating a new record of either class (i.e., site or specimen).
Coordinate capture
GPS coordinates are the most useful data the user can capture in collNotes. Coordinates are captured in collNotes using the “SET GPS” button, which is available when creating (or editing) a site‐level record. When the user selects this feature, their phone makes a location request using the GeoLocator plugin (Montemagno, 2019). This location request includes the altitude, coordinates, and accuracy in the form of uncertainty in meters (e.g., “20,” meaning “±20 meters”). Onscreen text notifies the user of a successful location request. When uncertainty is high, successive location requests may improve it, so the font of this notification is color‐coded to reflect coordinate uncertainty: less than 20 m is green, 21–30 m is yellow, and an uncertainty over 30 m is red.
collBook development
A desktop application, collBook was written in Python 3.7 (https://www.python.org/) using the Qt5 (The Qt Company, Espoo, Finland) framework for the graphical user interface. Qt Designer 5.12 was used to design the interface layouts. Multiple custom Python modules are present in collBook's source code. A list of those modules and a brief description of their function is provided in Table 1. Available for Linux, OS X, and Windows, collBook is designed to use at the same time as specimen identification for refining field notes into database‐ready files and specimen labels. Performing data refinements in collBook, as opposed to collNotes, permits web service–dependent features without cell service dependency.
Table 1.
The Python modules written for collBook, and a brief description of their functions
| Module | Description of function |
|---|---|
| associatedtaxa.py | A dialog for selecting which associatedTaxa to include for a site. |
| collBook.py | The “Main App,” delegates commands to other modules. |
| formview.py | Manages the user entry fields in the main screen. |
| importindexdialog.py | A dialog to assist importing unrecognized data formats. |
| locality.py | Refines location related fields and calls geocoding API services. |
| pandastablemodel.py | Displays the data in a table and handles data manipulation functions. |
| pdfviewer.py | Converts PDF objects into images to display preview labels. |
| printlabels.py | Generates PDF objects for previews or output as label files. |
| progressbar.py | Status bar replacement with a progress bar and “scope of view.” |
| scinameinputdialog.py | A dialog requesting binomial names after a failed taxonomy check. |
| settingsdialog.py | A dialog for selecting and storing containing user preferences. |
| taxonomy.py | Verifies the status of binomial names and their authorities. |
Designed to be feature rich, the user interface contains four prominent panes: a label preview, form view, site navigator, and table view (Fig. 1). The label preview (Fig. 1A) presents a dynamically generated label, which is updated as edits are made. The form view is the primary method of editing or adding new records (Fig. 1B). Many of the form view's fields (e.g., date, latitude, and longitude) impose DwC‐recommended formatting. The site navigator is used to select which records are to be edited, refined, or exported (Fig. 1C). All edits made to parent classes (i.e., those of a higher class) are automatically propagated to their associated children records (i.e., lower class records). For example, in Fig. 1C, selecting “Site 1” sets the scope of records to be acted upon as all those that were collected at that site. To avoid confusion caused by changing scopes, reminder text was placed in the status bar along the bottom of the interface informing the user of the current selection type (i.e., “All records,” “Site view,” or “Specimen view”). The table view presents spreadsheet‐style access to all selected records (Fig. 1D). Contrary to the rest of the interface, the table view imposes no formatting, validation, or inheritance, providing a method to override many of the functions discussed above. Data entered using the table view may not always be visible in the form view, yet will be reflected on the label preview and in the exported data. Features included in collBook are: reverse geocoding localities, taxonomic alignments, inferred associated taxa, and creation of customizable labels that can optionally include catalog number barcodes.
Figure 1.
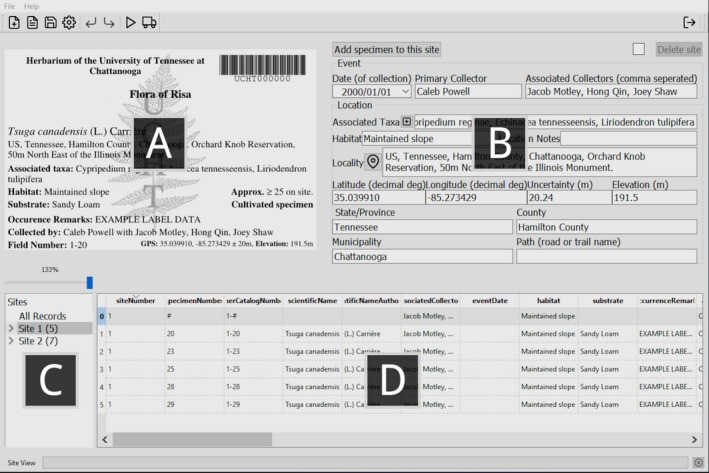
The collBook user interface, showing (A) the label preview, which presents an image of the label to be produced for a selected record; (B) the form view, where data pertinent to the selected class (e.g., “Site 1”) may be edited; (C) the site navigator, which is used to select record(s) for editing or refining; and (D) the table view, which provides an overview of the selected record(s) and allows unrestricted edits to the data.
Reverse geocoding
In collBook, location data not recorded in collNotes (i.e., “state,” “county,” “municipality”) are inferred from the GPS coordinates and prepended to the user‐entered locality string, supplementing the minimal locality data recorded in collNotes. Inference from coordinates is performed using Google's reverse geocoding web service (Google, 2019). For example, the locality string: “50 m northeast of the Illinois Monument” would become: “US, Tennessee, Hamilton County, Chattanooga, Orchard Knob Reservation, near East 4th Street, 50 m North East of the Illinois Monument.” One flaw inherent to this feature is that the user‐entered locality and the generated preamble may contain redundant terms. While testing these programs, familiarity with this feature when using collNotes was found to reduce the occurrences of such redundant terms. Nevertheless, it must remain the user's responsibility to verify labels in collBook for accuracy and redundancy.
Taxonomic alignments
When refining records, the status of the taxonomy, and the associated authority, are verified. To accomodate user preference, several sources for these alignments were included. The most recent version of the Integrated Taxonomic Information System (ITIS; https://www.itis.gov/) is bundled with the program, whereas Catalog of Life (Roskov et al., 2013) and the Taxonomic Name Resolution Service (TNRS; Boyle et al., 2013) are made available through their web services. So as not to overload web services, a one‐second delay is imposed on web service requests, making alignments through ITIS much faster. Because ITIS was packaged with collBook, it is also used to inform autofill suggestions when entering scientific names. TNRS is capable of performing partial matches, correcting minor spelling discrepancies when verifying taxonomies. In those cases, TNRS returns a score of the match's accuracy; a minimum threshold for this score can be modified at the user's preference. Alignments from these sources are applied based on user‐defined policies that delegate how recommendations should be made, and whether they should ever be automatically applied. Although not discussed in detail here, collNotes and collBook are being evaluated for groups beyond Plantae. Fungi, for example, is currently supported with a locally bundled MycoBank (Robert et al., 2013; http://www.mycobank.org/) taxonomy, as well as Catalog of Life support.
Inferred associated taxa
An additional benefit to structured data inheritance is the ability to document associations among sibling specimen records. Associated taxa information is frequently overlooked by field researchers yet may be informative for community composition, habitat, or ecosystem studies. At site level, collNotes offers an associated taxa entry field. In collBook, during record refinement, but after taxonomic alignments, the user is optionally presented with a checklist dialog box and may select some, all, or none of the taxa contemporaneously collected at the parent site. Once the taxon list is finalized, those taxa are included as comma‐separated values in the pertinent records. Associated taxa are appended after any existing user entries that may have been recorded in the field using collNotes. The determined name of each record is omitted from the inferred associated taxa for that record so that no record names itself as an associated taxon. One potential flaw in this feature occurs when a user alters a determination after the refinement steps, thereby leaving an inappropriate associated taxon in the sibling records of the altered specimen. To avoid this issue, users are encouraged to perform record refinements only after they are confident in initial determinations.
Customizable labels
Label‐containing PDF files are produced in collBook using the Python library ReportLab (https://www.reportlab.com/). There are numerous user settings for label customization, such as font type, base font size, label dimensions, and optional label elements such as: associated taxa, verified by, collection name, and collection logo. Label dimensions determine not only the resulting PDF's size but also the space available on each label. It is assumed that no label should exceed the label dimensions (i.e., there should be no multi‐page labels), and because collBook often produces information‐rich labels, space availability can become an issue. User preferences, in conjunction with dynamic placement and sizing, aid in reducing this issue. For example, the associated taxa may be omitted or restricted in item length on the label without impacting the electronic record data. By default, some label elements will share a line, but when label width is insufficient those elements may be split into separate lines. The font size of some label elements are scaled relative to the base font size. For example, GPS coordinate size is always the base font size reduced by 20%, whereas the font size used for the scientific name is usually increased somewhat. Altering the base font size therefore impacts all fonts, but does not necessarily reflect the actual font size of all elements. Another customization option we've included is the ability to load an image as a background logo, or watermark. This logo may be anchored to set locations and scaled down in either size or opacity. For best results, users should select cropped images that are larger than the target labels’ dimensions.
Catalog number barcodes
Assigning catalog numbers, usually by applying a barcode sticker, is an additional step of the digitization process. Optionally, in collBook, catalog numbers may be sequentially assigned and included on the labels as barcodes with human‐readable subtext. These barcodes are generated in the “code 39” format, using ReportLab. The catalog numbers assigned are based on a series of user inputs available in the Preferences menu. By providing a prefix (e.g., “UCHT”), a digit count (e.g., “6”), and a starting value (e.g., 12345), catalog numbers are assigned sequentially to each record (e.g., “UCHT012345,” “UCHT012346,” etc.). This feature avoids the costs of procuring and the time of applying barcode stickers, which is a significant portion of the digitization process. Nevertheless, this feature should be used with caution, as it is possible to assign non‐unique catalog numbers or to overprovision catalog numbers to specimens that are eventually not accessioned into collections. To reduce overprovisioning, catalog numbers are only assigned during the final export process; a dummy value is displayed in the preview window until those assignments are made (Fig. 1A). As these concerns do not impede the core function of the software, this feature was cautiously included. A catalog number management system that can overcome these issues remains a priority.
Interoperability with existing alternatives
We are not aware of any other solution for a complete field‐to‐database workflow, so alternatives were evaluated for collNotes and collBook independently. An alternative to collBook's label‐printing feature is to utilize Symbiota's in‐browser label‐printing option. This provides basic label formatting with some of the same features in collBook, including barcode preparation. Because those specimens have already been accessioned with catalog numbers, barcode printing is less problematic. Because collNotes and collBook maintain data in Symbiota‐friendly formats, these two platforms are not mutually exclusive. For example, a user may prepare their records with collBook, upload them to Symbiota, and use Symbiota's in‐browser label services.
There are a few alternatives to collNotes for gathering field notes directly into digital formats. Notably, the android application ColectoR (Maya‐Lastra, 2016) was developed for quick and efficient data capture in the field. ColectoR features taxonomic and location refinements and can be integrated into a Microsoft Excel template for label production. However, ColectoR does not export to a standardized format and requires mobile data service for many of its features. A clever solution for gathering digital field notes was documented in the workflow presented by Heberling and Isaac (Heberling and Isaac, 2018), where the citizen science platform iNaturalist is used to record field observations. One advantage to Heberling and Isaac's workflow is the association of iNaturalist‐hosted data with the voucher specimen's record. For example, in this process in situ photos captured using the iNaturalist mobile application are documented with the record and hosted through iNaturalist's servers. Because it is designed for citizen science observations, the iNaturalist mobile application lacks entry fields for many field notes recommended for voucher specimen labels, such as habitat or relative abundance estimates (Bridson and Forman, 1998). As noted by Heberling and Isaac, custom observation fields may be added to records made using the iNaturalist app; however, these additional data must be entered through iNaturalist's web interface, and not in the mobile application. This limitation makes either field web browser access or the eventual transcription of ancillary field notes necessary.
Preferences and priorities of individuals vary according to research needs. Therefore, while collBook has seamless integration with collNotes, we have also included functions in collBook to parse data produced by either ColectoR or iNaturalist. Additionally, a user may attempt to import into collBook any DwC‐formatted CSV file. The degree of success from such an attempt, however, will vary depending on formatting. None of these alternatives incorporate a hierarchical data structure and so lack a method to link specimens collected from the same site or during the same event. For these reasons, we chose to develop both collNotes and collBook to address the challenge of born‐digital collections‐based research.
CONCLUSIONS
Used in combination, collNotes and collBook provide a solution for field researchers and natural history collections to transition to a born‐digital process for new accessions. If adopted, we believe these applications can mitigate the continued growth of backlogged natural history data that need to be transcribed, while improving the efficiency, accuracy, and accessibility of collections‐based research. The source code for both of these works is available on GitHub (collNotes: https://github.com/j-h-m/collNotes; collBook: https://github.com/CapPow/collBook) under the GNU General Public License v3.0. The mobile application collNotes may be downloaded for free on the Google Play store and the iOS App store. The desktop program collBook is distributed for free through GitHub (https://github.com/CapPow/collBook#Installation). Feedback from early users has guided both programs through iterative improvements, prioritizing a user‐friendly experience in collNotes and a feature‐rich environment in collBook. The accessibility of these works is an invitation to the community to continue to improve, modify, or incorporate portions of them into other projects. We welcome suggestions, bug reports, and feature requests. When possible, we encourage that feedback be in the form of pull requests to our GitHub repositories.
ACKNOWLEDGMENTS
We thank Courtney Alley, Hill Craddock, Alaina Krakowiak, John Shelton, and the 2017–2019 Field Botany, Mycology, and Plant Taxonomy students at the University of Tennessee at Chattanooga for their patient testing and constructive feedback. Additionally, we would like to thank GitHub user “Zularizal” for the thoughtful logo design. This work benefited from the critical feedback of three anonymous reviewers; we appreciate their time and service. We are also thankful for internal funding from the University of Tennessee at Chattanooga, support from the department of Biology, Geology, and Environmental Science, as well as from the U.S. National Science Foundation (awards 1761839 and 1410069).
Powell, C. , Motley J., Qin H., and Shaw J.. 2019. A born‐digital field‐to‐database solution for collections‐based research using collNotes and collBook. Applications in Plant Sciences 7(8): e11284.
DATA AVAILABILITY STATEMENT
The source code for both of these works is available on GitHub (collNotes: https://github.com/j-h-m/collNotes; collBook: https://github.com/CapPow/collBook) under the GNU General Public License v3.0. The mobile application collNotes may be downloaded for free on the Google Play store and the iOS App store. The desktop program collBook is distributed for free through GitHub (https://github.com/CapPow/collBook#Installation).
LITERATURE CITED
- Boyle, B. , Hopkins N., Lu Z., Garay J. A. R., Mozzherin D., Rees T., Matasci N., et al. 2013. The taxonomic name resolution service: An online tool for automated standardization of plant names. BMC Bioinformatics 14: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridson, D. M. , and Forman L.. 1998. Herbarium handbook. Royal Botanic Gardens, Kew, Richmond, Surrey, United Kingdom. [Google Scholar]
- Ellwood, E. R. , Dunckel B. A., Flemons P., Guralnick R., Nelson G., Newman G., Newman S., et al. 2015. Accelerating the digitization of biodiversity research specimens through online public participation. BioScience 65: 383–396. [Google Scholar]
- Global Biodiversity Information Facility . 2018. What is GBIF? Website https://www.gbif.org/what-is-gbif [accessed 3 May 2019].
- Google . 2019. Google Geocoding API. Google Developers. Website https://developers.google.com/maps/documentation/geocoding [accessed 8 March 2019].
- Gries, C. , Gilbert E. E., and Franz N. M.. 2014. Symbiota – A virtual platform for creating voucher‐based biodiversity information communities. Biodiversity Data Journal 2: e1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, K. M. , and Marsico T. D.. 2017. Digitizing specimens in a small herbarium: A viable workflow for collections working with limited resources. Applications in Plant Sciences 5: 1600125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heberling, J. M. , and Isaac B. L.. 2018. iNaturalist as a tool to expand the research value of museum specimens. Applications in Plant Sciences 6: e01193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, A. , Guralnick R., Smith A., Sallans A., Gillespie R., Denslow M., Gross J., et al. 2012. The notes from nature tool for unlocking biodiversity records from museum records through citizen science. ZooKeys 209: 219–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, S. A. , Soltis P. S., Belbin L., Chapman A. D., Nelson G., Paul D. L., and Collins M.. 2018. Herbarium data: Global biodiversity and societal botanical needs for novel research. Applications in Plant Sciences 6: e1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maya‐Lastra, C. A. 2016. ColectoR, a digital field notebook for voucher specimen collection for smartphones. Applications in Plant Sciences 4: 1600035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montemagno, J. 2019. Geolocation plugin for Xamarin and Windows. Website https://github.com/jamesmontemagno/GeolocatorPlugin [accessed 15 March 2019].
- Nelson, G. , Sweeney P., Wallace L. E., Rabeler R. K., Allard D., Brown H., Carter J. R., et al. 2015. Digitization workflows for flat sheets and packets of plants, algae, and fungi. Applications in Plant Sciences 3: 1500065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul, D. , Seltmann K., Michonneau F., Masaki D., Soltis P., Ellis S., and Love K.. 2015. iDigBio Wiki: Field to Database. Website https://www.idigbio.org/wiki/index.php/Field_to_Database [accessed 5 March 2019].
- Robert, V. , Vu D., Amor A. B., van de Wiele N., Brouwer C., Jabas B., Szoke S., et al. 2013. MycoBank gearing up for new horizons. IMA Fungus 4: 371–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roskov, Y. , Kunze T., Paglinawan L., Orrell T., Nicolson D., Culham A., Bailly N., et al. 2013. Species 2000 & ITIS Catalogue of Life, 2013 Annual Checklist. Technical Report. Species 2000/ITIS, Reading, United Kingdom.
- Sweeney, P. W. , Starly B., Morris P. J., Xu Y., Jones A., Radhakrishnan S., Grassa C. J., and Davis C. C.. 2018. Large–scale digitization of herbarium specimens: Development and usage of an automated, high‐throughput conveyor system. Taxon 67: 165–178. [Google Scholar]
- Tomaštík, J. , Tomaštík J., Saloň Š., and Piroh R.. 2017. Horizontal accuracy and applicability of smartphone GNSS positioning in forests. Forestry: An International Journal of Forest Research 90: 187–198. [Google Scholar]
- Wieczorek, J. , Bloom D., Guralnick R., Blum S., Döring M., Giovanni R., Robertson T., and Vieglais D.. 2012. Darwin Core: An evolving community‐developed biodiversity data standard. PLoS ONE 7: e29715. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The source code for both of these works is available on GitHub (collNotes: https://github.com/j-h-m/collNotes; collBook: https://github.com/CapPow/collBook) under the GNU General Public License v3.0. The mobile application collNotes may be downloaded for free on the Google Play store and the iOS App store. The desktop program collBook is distributed for free through GitHub (https://github.com/CapPow/collBook#Installation).


