Summary
2’3’-cyclic-GMP-AMP (cGAMP) is a second messenger that activates the antiviral Stimulator of Interferon Genes (STING) pathway. We recently identified a novel role for cGAMP as a soluble, extracellular immunotransmitter that is produced and secreted by cancer cells. Secreted cGAMP is then sensed by host cells, eliciting an antitumoral immune response. Due to the antitumoral effects of cGAMP, other CDN-based STING agonists are currently under investigation in clinical trials for metastatic solid tumors. However, it is unknown how cGAMP and other CDNs cross the cell membrane to activate intracellular STING. Using a genome-wide CRISPR screen we identified SLC19A1 as the first known importer of cGAMP and other CDNs, including the investigational new drug 2’3’-bisphosphosphothioate-cyclic-di-AMP (2’3’-CDAS). These discoveries will provide insight into cGAMP’s role as an immunotransmitter and aid in the development of more targeted CDN-based cancer therapeutics.
Graphical Abstract
eTOC Blurb
2’3’-cyclic-GMP-AMP (cGAMP) is an immunotransmitter produced and secreted by cancer cells that is uptaken by host cells to elicit an antitumoral immune response. Using a CRISPR screen, Ritchie and Cordova et al. identify SLC19A1 as the first importer of cGAMP and other cyclic dinucleotides (CDNs).
Introduction
Harnessing innate immunity to treat cancer is at the cutting edge of precise and personalized cancer treatment, and there is mounting evidence that the cGAMP-STING innate immunity pathway is a potent anti-cancer target (Corrales et al., 2015; Deng et al., 2014; Wang et al., 2017). The cyclic dinucleotide (CDN) cGAMP is a second messenger that is synthesized by cyclic-GMP-AMP synthase (cGAS) after detection of double-stranded DNA (dsDNA) in the cytosol (Sun et al., 2013). cGAMP binds and activates the cytosolic domain of its ER-membrane receptor STING, which in turn activates TBK1, a kinase, and IRF3, a transcription factor, resulting in the transcription, expression, and secretion of cytokines such as interferon-beta (IFN-β). These potent antiviral and anticancer cytokines can directly neutralize threats (Apelbaum et al., 2013) and trigger downstream adaptive immunity (Iwasaki and Medzhitov, 2010). In the case of cancer clearance, IFN-β promotes cross-priming of CD8+ T cells by tumor-infiltrating antigen presenting cells (Fuertes et al., 2011). Primed CD8+ T cells can then infiltrate and kill both primary and metastatic tumors, leading to systemic tumor regression and long-term humoral memory of the tumor (Corrales et al., 2015; Woo et al., 2014).
While cytosolic dsDNA was originally discovered as a signal of viral infection (Li et al., 2013), it is now also recognized as a hallmark of cancer (Bakhoum et al., 2018; Mackenzie et al., 2017). Cancer cells often have unstable genomes that result in improper chromosome segregation during mitosis. This leads to the formation of micronuclei enclosed by leaky membranes, thereby exposing dsDNA to the cytosol and activating the cGAMP-STING pathway (Harding et al., 2017; Mackenzie et al., 2017). Instead of inactivating the pathway to escape immune detection, the vast majority of cancer cells retain the STING pathway (Bakhoum and Cantley, 2018) and exploit it to their advantage in at least two ways. First, cGAS promotes cancer progression by inhibiting DNA repair (Liu et al., 2018), thereby increasing genomic instability. Second, many cancer cells rewire the STING pathway to promote metastasis, while avoiding IFN-β production (Bakhoum and Cantley, 2018; Bakhoum et al., 2018).
While cancer cells do not typically produce type I interferons, it has been shown that cGAMP-producing cancer cells can activate the STING pathway in nearby cGAS−/− host cells (Marcus et al., 2018). Although the mechanism for this was previously unknown, we recently determined that it is due to cGAMP’s role as an immunotransmitter between cancer and host cells (Carozza et al., 2019). Cancer cells are able to efficiently export cGAMP into the extracellular space, which can then be degraded by the extracellular hydrolase ENPP1 (Li et al., 2014), or can cross the cell membrane of nearby host cells through unknown mechanisms, activating STING and the downstream anti-cancer immune response.
As cGAMP can elicit a robust antitumoral immune response, there has been great interest in developing cGAMP analogs as cancer therapeutics. In 2014, we reported stable phosphothioate analogs of cGAMP (Li et al., 2014). Subsequent studies have shown that intratumoral injections of these and other phosphothioate analogs can cure multiple tumor types in mice (Corrales et al., 2015). With these promising results, 2’3’-bisphosphothioate-cyclic-di-AMP (2’3’-CDAS) and another stable CDN analog entered phase-I clinical trials in 2017, both in combination with PD-1 checkpoint inhibitors (Trial IDs and , respectively). Despite their therapeutic potential, it is still unknown how cGAMP and other CDNs enter the cytoplasm of target cells. Because they are unable to passively diffuse across the plasma membrane due to their negative charges, they must enter cells through a facilitated mechanism. This mechanism could be either specific (e.g. through a transporter) or non-specific (e.g. pinocytosis).
Understanding how cGAMP enters cells is essential for both characterizing cGAMP as an immunotransmitter and developing CDN-based STING agonists as therapeutics. Here, we describe SLC19A1 as the first known transporter of the immunotransmitter cGAMP. In addition, we show that SLC19A1 is also an importer of several bacterial and synthetic CDNs.
Results
A Genetic Screen Identifies Putative Components of the Extracellular cGAMP-STING Pathway
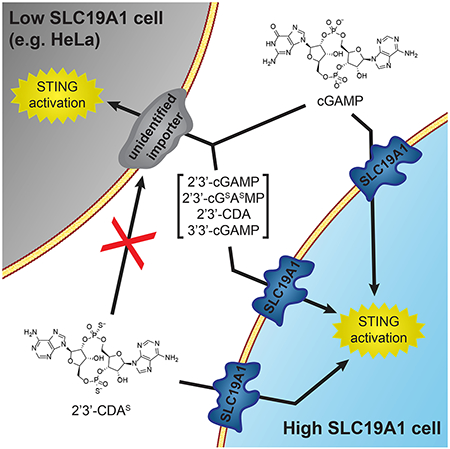
To identify important factors for cGAMP internalization, we performed a whole-genome CRISPR screen in the monocyte-derived U937 cell line. U937 cells were chosen for this screen because monocyte-lineage cells have high levels of STING and a higher response to exogenous STING agonists in vivo compared to other immune cells (Sivick et al., 2018). U937 cells express all STING pathway components (Figure S1A) and respond to extracellular cGAMP by phosphorylating the transcription factor IRF3 (Figure 1A) and producing IFN-β (Figure S1B–C). Importantly, the response is independent of the cGAMP synthase cGAS, suggesting that it is due to exogenous, extracellular cGAMP (Figure S1D). With prolonged cGAMP treatment we found that U937 cells die in a dose-dependent manner, making them well-suited for a live/dead CRISPR screen (Figure 1B).
Figure 1. A Genetic Screen Identifies Putative Components of the Extracellular cGAMP-STING Pathway.
(A) IRF3 phosphorylation in response to extracellular cGAMP. U937 cells were treated with 100 μM cGAMP for 2 h.
(B) Dose dependent cGAMP induced death in U937 cells. Cells were treated with various concentrations of cGAMP for 16 h. Cell viability was assessed using CellTiter-Glo (n = 2 biological replicates).
(C) Schematic of the CRISPR screen. A whole-genome sgRNA library was introduced into U937 cells. 250 million library cells were treated with cGAMP every 24 h for 2 weeks. The genomic DNA was sequenced and highly-enriched sgRNA sequences were identified.
(D-E) casTLE scores of individual enriched genes from (D) two replicates using LD50 doses of cGAMP and (E) two replicates using LD30 doses of cGAMP. Top hits are annotated in red.
For (B) data are shown as mean ± SD.
In previous work, we had successfully designed and introduced a custom genome-wide single-guide RNA (sgRNA) library into U937 cells to identify genes required for cell growth (Morgens et al., 2017). Additionally, we were able to synthesize hundreds of milligrams of high purity cGAMP using previously developed methods (Civril et al., 2013; Li et al., 2014). This enabled us to perform a live/dead screen by treating the library cells with a cGAMP concentration sufficient to kill 50% of cells (LD50) every 24 hours for 2 weeks. In this screen, cells containing sgRNAs that target key STING pathway members, including cGAMP importers, should be resistant to cGAMP treatment and enriched (Figure 1C). Therefore, by sequencing the sgRNAs in treated and untreated populations to calculate relative enrichment and depletion of sgRNAs, we generated a list of candidate genes involved in the extracellular cGAMP-STING pathway. We also repeated the screen with a lower cGAMP concentration (LD30) and obtained similar results. The casTLE score (a measure of statistical significance for the collective enrichment of sgRNAs targeting a particular gene (Morgens et al., 2016)) was reproducible between replicates (Figure 1D–E). The top three enriched genes in both screens were the known STING pathway members TMEM173, TBK1, and IRF3, further supporting the validity of this screening method (Figure S1E).
SLC19A1 Is Essential for Robust Extracellular cGAMP Signaling in U937 Cells
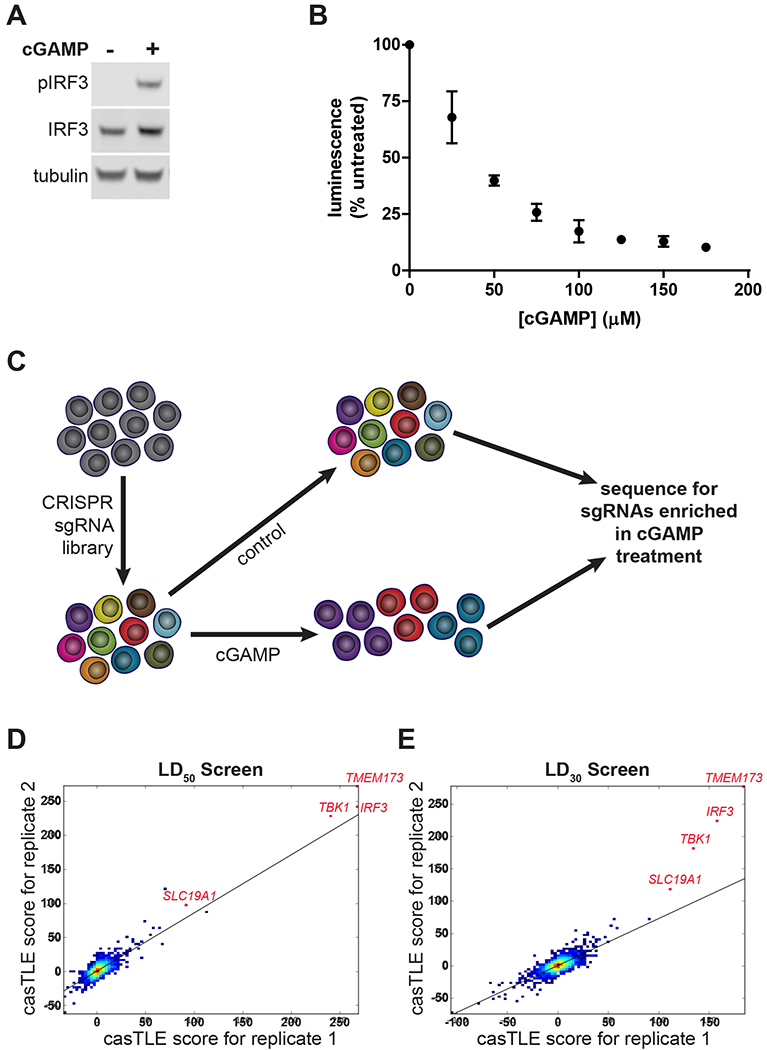
Among the top hits from our CRISPR screen, we focused on SLC19A1 as a candidate transporter of cGAMP. The SLC19A1 protein, also known as Reduced Folate Carrier 1 (RFC1), has been previously characterized as a high affinity importer of reduced folates, such as folinic acid, and antifolates, such as methotrexate (Hou and Matherly, 2014; Jansen et al., 1990). Given the structural similarity between cGAMP and these known SLC19A1 substrates (Figure 2A), we hypothesized that SLC19A1 also imports cGAMP.
Figure 2. SLC19A1 Is Essential for Robust Extracellular cGAMP Signaling in U937 Cells.
(A) Chemical structures of the known SLC19A1 substrates folinic acid and methotrexate, as compared to cGAMP.
(B) Uptake of [3H]-methotrexate in U937 SLC19A1−/− lines. U937 WT and SLC19A1−/− lines were treated with 17.5 nM [3H]-methotrexate for 5 min (n = 4 biological replicates).
(C) Effect of SLC19A1 knockout on extracellular cGAMP signaling. U937 WT and SLC19A1−/− cells were treated with 100 μM cGAMP for 2 h (n = 12 biological replicates).
(D) Effect of sulfasalazine (SSZ) on extracellular cGAMP signaling. WT and SLC19A1−/− U937 cells were pretreated with 1 mM SSZ for 20 min followed by a 100 μM cGAMP treatment for 2 h (n = 3 biological replicates).
(E) Effect of SLC19A1 overexpression on extracellular cGAMP signaling. HEK 293T cells were transfected with pcDNA3-STING-HA and either an empty pcDNA3-FLAG-HA vector or pcDNA3-FLAG-HA-SLC19A1, and then were incubated for 24 h to allow for protein production. Transfected cells were then treated with 100 μM cGAMP for 2 h (n = 6 biological replicates).
Lanes not relevant to the experiment are removed from (D) for clarity. Uncropped Western blots are available at http://dx.doi.org/10.17632/5bssvpns6h.1 For (B), (C), (D), and (E) data are shown as mean ± SD. **p < 0.01; ***p < 0.001; ****p < 0.0001
See also Figure S2
We first sought to determine if SLC19A1 is important for extracellular cGAMP signaling by using CRISPR/Cas9 to knock out SLC19A1 in U937 cells. Because there are no commercial antibodies that effectively detect SLC19A1, we sequenced the SLC19A1 locus and identified two SLC19A1−/− clones containing frameshift mutations that lead to early termination (Figure S2A–B). In contrast to the parent U937 cell line, these SLC19A1−/− clones no longer import [3H]-methotrexate, demonstrating the loss of functional SLC19A1 protein (Figure 2B). Importantly, protein levels of known essential STING pathway members were not significantly altered in these SLC19A1−/− clones (Figure S2C). When treated with extracellular cGAMP, SLC19ATa cells showed 75% less IRF3 phosphorylation than wild type cells (Figure 2C), as well as a 65% decrease in TBK1 phosphorylation (Figure S2D). This was accompanied by greatly diminished production of IFN-β mRNA (Figure S2E) and protein (Figure S2F). In addition, the noncompetitive SLC19A1 inhibitor sulfasalazine (Jansen et al., 2004) reduced phosphorylation of IRF3 in response to extracellular cGAMP by 60% in wild type U937 cells, but not in SLC19A1−/− cells (Figure 2D). Our genetic and pharmacological data together support an important role for SLC19A1 in extracellular cGAMP signaling.
To determine if SLC19A1 is sufficient to increase extracellular cGAMP signaling, HEK 293T cells (which lack endogenous cGAS and STING, and are therefore more amenable to transfection than U937 cells) were co-transfected with pcDNA3-STING-HA and either an empty pcDNA3-FLAG-HA vector or pcDNA3-FLAG-HA-SLC19A1. Overexpression of SLC19A1 increased the response to extracellular cGAMP by over 200% compared to transfection with the empty vector (Figure 2E), indicating that SLC19A1 expression is sufficient to increase extracellular cGAMP signaling.
SLC19A1 Facilitates Extracellular Uptake of cGAMP
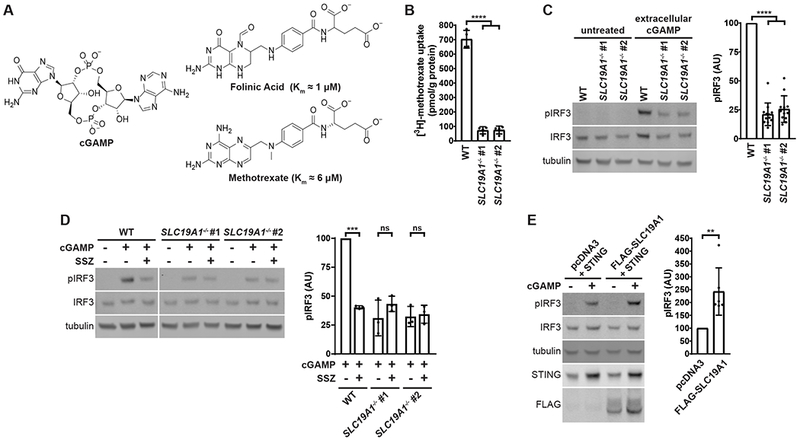
To distinguish whether SLC19A1 is required for extracellular cGAMP uptake or for downstream STING signaling, we examined its role in intracellular cGAMP signaling. While direct detection of cGAMP import would be ideal, the current technologies to measure cGAMP concentrations using 32P-radiolabeled CDNs (Li et al., 2014) or mass spectrometry (Carozza et al., 2019) are not sensitive enough to detect nanomolar concentrations of intracellular cGAMP, given the high background cGAMP in the media. However, STING binds to cGAMP with a Kd of ~5 nM (Zhang et al., 2013). Therefore, the best readout of cGAMP internalization is still STING activation (Ablasser et al., 2013; Bridgeman et al., 2015), as quantified by IRF3 phosphorylation. Intracellular cGAMP was introduced into U937 cells through cGAMP electroporation (Figure 3A). In contrast to their response to extracellular cGAMP, U937 SLC19A1−/− cells are not defective in intracellular cGAMP signaling, as evidenced by their response to electroporated cGAMP (Figure 3B). These results indicate that SLC19A1 facilitates uptake of extracellular cGAMP.
Figure 3. SLC19A1 Facilitates Extracellular Uptake of cGAMP.
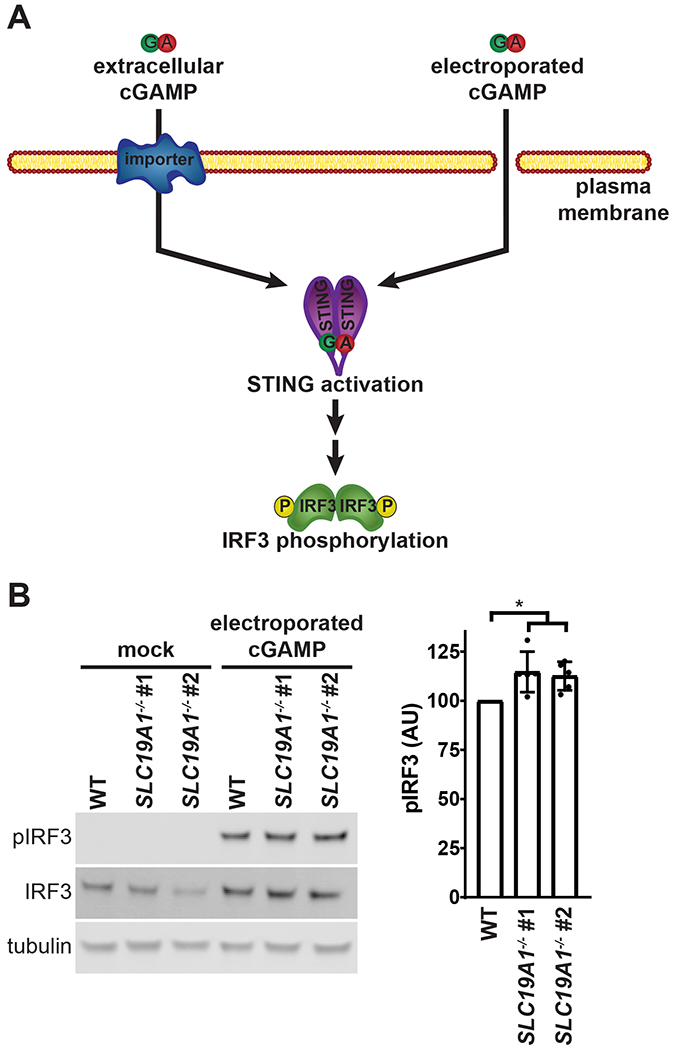
(A) Schematic of bypassing cGAMP import through cGAMP electroporation.
(B) Effect of SLC19A1 knockout on intracellular cGAMP signaling. U937 WT and SLC19A1−/− cells were electroporated with 100 nM cGAMP for 2 h (n = 5 biological replicates).
For (B) data are shown as mean ± SD. *p ≤ 0.05
SLC19A1 Is a Direct cGAMP Importer
We then tested whether SLC19A1 is a direct cGAMP importer requiring the transporter activity of the protein, rather than a receptor for endocytosis or a scaffold for the direct importer. SLC19A1 is an antiporter, and export of intracellular organic anions facilitates uptake of its substrates (Hou and Matherly, 2014). Through this mechanism, the cell-permeable compound 5-amino-4-imidazolecarboxamide riboside (AICAR) boosts import of SLC19A1 substrates. Once inside the cell, AICAR is phosphorylated to AICAR monophosphate, which serves as an organic anion for SLC19A1’s antiporter activity (Visentin et al., 2012). Treating U937 cells with AICAR increased the response to extracellular cGAMP by approximately 200% in an SLC19A1-dependent manner (Figure 4A). While AICAR is known to cause AMPK phosphorylation in some cells (Sullivan et al., 1994), which may affect STING signaling through a negative feedback loop (Konno et al., 2013), it did not have any additional effect on AMPK phosphorylation in cGAMP-treated U937 cells (Figure S3A).
Figure 4. SLC19A1 Is a Direct cGAMP Importer.
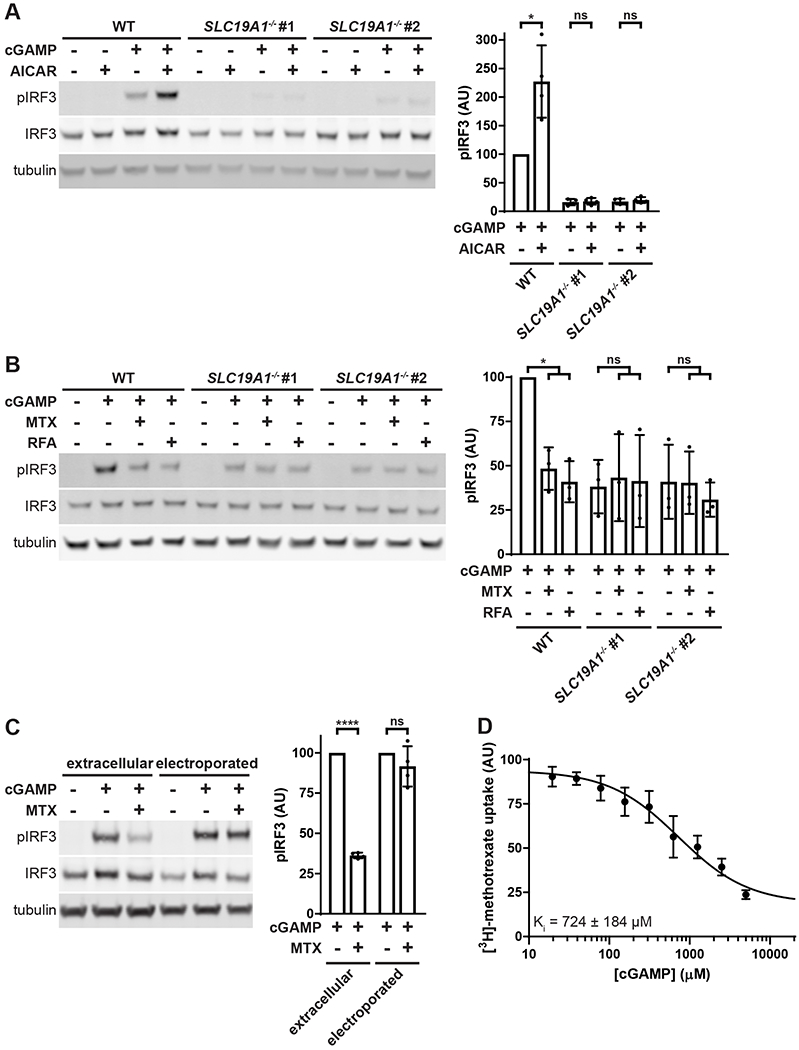
(A) Effect of AICAR on extracellular cGAMP signaling. U937 WT and SLC19A1−/− cells were pretreated with 1 mM AICAR for 20 min followed by treatment with 100 μM 2’3’-cGAMP for 2 h (n = 4 biological replicates).
(B) Effect of competitive SLC19A1 inhibitors on extracellular cGAMP signaling. U937 WT and SLC19A1−/− cells were treated with 100 μM cGAMP alone or in the presence of 500 μM methotrexate (MTX) or 500 μM folinic acid (RFA) for 2 h (n = 3 biological replicates).
(C) Bypass of MTX inhibition by electroporation. U937 WT cells were pretreated with 500 μM MTX for 5 min, and then treated with either 100 μM extracellular cGAMP or electroporated with 100 nM cGAMP for 90 min (n = 4 biological replicates).
(D) Effect of cGAMP on [3H]-methotrexate uptake. U937 cells were treated 17.5 nM [3H]-methotrexate in the presence of the indicated concentrations of cGAMP for 1 min (n = 3 biological replicates).
For (A), (B), (C), and (D) data are shown as mean ± SD. *p ≤ 0.05; ****p ≤ 0.0001
See also Figure S3
Next, we tested whether the well-characterized SLC19A1 substrates folinic acid and methotrexate could act as competitive inhibitors of cGAMP to reduce extracellular cGAMP signaling. Although folinic acid (reduced folate) and methotrexate have opposing roles in folate metabolism, they both reduced extracellular cGAMP-induced IRF3 phosphorylation by more than 50% in an SLC19A1-dependent manner (Figure 4B), with similar IC50 values for both compounds (Figure S3B). Folic acid (oxidized folate) had a much higher IC50, consistent with the previously reported lower affinity of SLC19A1 for oxidized folates (Goldman et al., 1968) (Figure S3B). In addition, methotrexate had no effect on IRF3 phosphorylation when cGAMP was electroporated into cells, indicating that methotrexate inhibits cGAMP import (Figure 4C). Furthermore, cGAMP inhibited import of [3H]-methotrexate, with a Ki of ~724 μM (Figure 4D). Together, our results demonstrate that SLC19A1 is a direct cGAMP importer and the dominant one in U937 cells. However, the presence of residual IRF3 phosphorylation in extracellular cGAMP-treated SLC19A1−/− cells suggests that there are one or more additional cGAMP import mechanisms in U937 cells.
SLC19A1 Imports Bacterial and Synthetic CDNs, Including 2’3’-CDAS
In addition to cGAMP, bacterial CDN second messengers, including 3’3’-cyclic-GMP-AMP (3’3’-cGAMP) (Zhang et al., 2013), 3’3’-cyclic-di-GMP (3’3’-CDG) (Burdette et al., 2011), and 3’3’-cyclic-di-AMP (3’3’-CDA) (Woodward et al., 2010), also bind and activate the STING pathway (Figure 5A). These CDNs are critical for the immune detection of pathogens and contribute to pathogen virulence in mice (Davies et al., 2012; Dey et al., 2015; Sauer et al., 2011; Woodward et al., 2010). It is unclear how these CDNs are exposed to STING in the cytosol since most bacterial pathogens, such as the 3’3’-cGAMP-producing Vibrio cholera, are extracellular, and many intracellular bacteria, such as the 3’3’-CDA-producing Mycobacterium tuberculosis, hide in intracellular compartments to avoid detection. Given the structural similarities shared by these bacterial CDNs and cGAMP, we hypothesized that they are also SLC19A1 substrates. As expected, extracellular 3’3’-cGAMP signaling was diminished by approximately 90% in SLC19A1−/− cells relative to wild type, indicating that 3’3’-cGAMP is an SLC19A1 substrate (Figure 5B). However, U937 cells responded poorly to extracellular 3’3’-CDA and 3’3’-CDG compared to other CDNs (Figure S4A), limiting our ability to assess their dependence on SLC19A1 for import.
Figure 5. SLC19A1 imports Bacterial and Synthetic CDNs, Including 2’3’-CDAS.

(A) Structures of bacterial (1-3), mammalian (4), and synthetic (5-7) CDNs.
(B) Importance of SLC19A1 for signaling of various extracellular CDNs. U937 WT and SLC19A1−/− cells were treated with 200 μM 3’3’-cGAMP (n = 3 biological replicates) for 3 h, or with either 100 μM cGAMP (n = 12 biological replicates), 25 μM 2’3’-cGSASMP (n = 3 biological replicates), 100 μM 2’3’-CDA (n = 2 biological replicates), or 15 μM 2’3’-CDAS (n = 3 biological replicates) for 2 h.
(C) Bypass of 2’3’-CDAS import by electroporation. U937 WT and SLC19A1−/− cells were electroporated with 100 nM 2’3’-CDAS for 2 h (n = 2 biological replicates).
(D) Effect of MTX on extracellular 2’3’-CDAS signaling. U937 cells were treated with 500 μM MTX and 15 μM 2’3’-CDAS for 90 min (n = 3 biological replicates).
(E) Comparison of extracellular and intracellular response to cGAMP and 2’3’-CDAS. U937 cells were treated with either 100 μM extracellular cGAMP or 15 μM 2’3’-CDAS, or electroporated with 100 nM cGAMP or 2’3’-CDAS for 2 h.
Vertical black bars in (B) and (E) are used to separate lanes run on different Western blots. Lanes not relevant to the experiment are removed from (E) for clarity. Uncropped Western blots are available at http://dx.doi.org/10.17632/5bssvpns6h.1
For (B), (C), and (D) data are shown as mean ± SD. ***p < 0.001
See also Figure S4
We then tested whether SLC19A1 transports synthetic cGAMP analogs, including 2’3’-bis-phosphothioate-cyclic-GMP-AMP (2’3’-cGSASMP) (Li et al., 2014), 2’3’-cyclic-di-AMP (2’3’-CDA), and the investigational new drug 2’3’-CDAS (Figure 5A). All of these 2’3’-CDNs elicited a weaker response in SLC19A1−/− cells than in wild type U937 cells, indicating that they are SLC19A1 substrates (Figure 5B). Interestingly, at the concentrations used these substrates differentially utilize SLC19A1 for import, with 2’3’-CDAS relying almost entirely on SLC19A1, followed by cGAMP, and then 2’3’-CDA and 2’3’-GSASMP. Strikingly, loss of SLC19A1 completely abolished STING signaling in response to extracellular 2’3’-CDAS but did not reduce the response to electroporated 2’3’-CDAS (Figure 5C). Consistent with this finding, extracellular 2’3’-CDAS signaling was also greatly reduced in the presence of methotrexate, demonstrating that SLC19A1 is necessary for 2’3’-CDAS import in U937 cells (Figure 5D, S4B). To determine if SLC19A1 is sufficient for 2’3’-CDAS import, we co-transfected HEK 293T cells with a STING plasmid and either FLAG-HA-SLC19A1 or an empty vector. While cells transfected with an empty vector responded poorly to extracellular 2’3’-CDAS, cells overexpressing SLC19A1 had a robust response (Figure S4C).
Interestingly, electroporation of equimolar amounts of cGAMP and 2’3’-CDAS resulted in significantly less IRF3 phosphorylation in cells electroporated with 2’3’-CDAS, as compared to cGAMP. In contrast, treatment with 15 μM extracellular 2’3’-CDAS resulted in significantly higher levels of phosphorylated IRF3 than treatment with 100 μM extracellular cGAMP (Figure 5E). This suggests that the increased response to extracellular 2’3’-CDAS is due to it being a higher-affinity substrate for SLC19A1 than cGAMP, rather than due to stronger STING activation once inside cells.
2’3’-cGAMP Import Through SLC19A1 Varies Across Cell Lines and Primary Cells
Because SLC19A1 is the dominant cGAMP importer in monocyte-derived U937 cells, we sought to characterize its role in other cell types. As U937 cells were derived from a human histiocytic lymphoma, we assayed two other human blood cancer lines, THP-1 (AML) and NU-DUL-1 (early B-cell lymphoma), for SLC19A1 dependence. As in U937 cells, SLC19A1 appears to play a dominant role in THP-1 cells, as both genetic knockout of SLC19A1 and inhibition of import with methotrexate reduced pIRF3 signal by 50% in response to extracellular cGAMP (Figure 6A). NU-DUL-1 cells also utilize SLC19A1 as a cGAMP importer, although to a lesser degree, as indicated by methotrexate inhibition (Figure 6B).
Figure 6. 2’3’-cGAMP Import Through SLC19A1 Varies Across Cell Lines and Primary Cells.
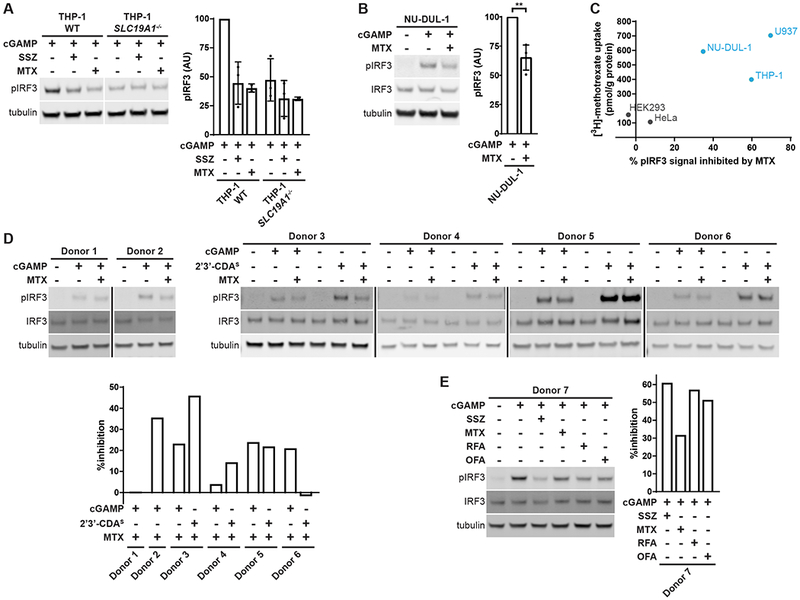
(A) Role of SLC19A1 in THP-1 cells. THP-1 WT and SLC19Ar−/− cells were pretreated with 1 mM SSZ or 500 uM MTX for 15 min, and then treated with 100 μM cGAMP for 2 h (n = 3 (SSZ) or 2 (MTX) biological replicates).
(B) Role of SLC19A1 in NU-DUL-1 cells. NU-DUL-1 cells were pretreated with 500 μM MTX for 5 min., and then treated with 100 μM cGAMP for 90 min (n = 4 biological replicates).
(C) Correlation between cellular MTX uptake and the degree to which MTX inhibits extracellular cGAMP signaling in different cell lines.
(D-E) Effect of MTX on extracellular cGAMP signaling in primary CD14+ monocytes. CD14+ monocytes were isolated from seven different healthy donors. In (D), monocytes were pretreated with 500 μM MTX for 5 min and then treated with either 50 μM cGAMP or 12.5 μM 2’3’-CDAS for 60 min. In (E), monocytes were pretreated with 1 mM sulfasalazine (SSZ), 500 μM MTX, 500 μM folinic acid (RFA), or 500 μM folic acid (OFA) for 15 min., and then treated with 50 μM cGAMP for 60 min. Response to 2’3’-CDAS was not tested in Donors 1, 2, and 7. Quantifications are shown as percent inhibition of pIRF3 signal due to MTX pretreatment relative to no pretreatment.
Vertical black bars in (D) are used to separate lanes run on different Western blots.Lanes not relevant to the experiment are removed from (A) for clarity. UncroppedWestern blots are available at http://dx.doi.org/10.17632/5bssvpns6h.1
For (A) and (B) data are shown as mean ± SD. **p ≤ 0.01
See also Figure S5
In contrast to hematologic cell lines, the epithelial cell lines HEK 293 and HeLa do not appear to use SLC19A1 as a cGAMP importer, as methotrexate inhibition and SLC19A1 loss did not affect extracellular cGAMP signaling (Figure S5A–B). Interestingly, HeLa cells did not respond to 2’3’-CDAS at concentrations that strongly activated U937 cells, suggesting that 2’3’-cGAMP and 2’3’-CDAS have different import mechanisms in these cells (Figure S5C).
Since the extent to which SLC19A1 is used as a cGAMP importer varies between cell lines, we hypothesized that this variation could be due to differential expression of SLC19A1. To this end, we measured uptake of [3H]-methotrexate in U937, THP-1, NU-DUL-1, HEK 293, and HeLa cells (Figure S5D). U937, THP-1, HEK 293, and HeLa SLC19A1−/− cells all had greatly reduced [3H]-methotrexate uptake, indicating that [3H]-methotrexate correlates with functional SLC19A1 expression (Figure S5D). As expected, higher [3H]-methotrexate uptake, and therefore higher SLC19A1 expression, correlated with a larger reliance on SLC19A1 for cGAMP import, as indicated by the degree of inhibition of extracellular cGAMP signaling in the presence of methotrexate (Figure 6C).
We next interrogated the role of SLC19A1 in primary CD14+ monocytes. As attempts to knockdown SLC19A1 using siRNA and CRISPR/Cas9 proved to be toxic to monocytes, we relied on pharmacological inhibition of SLC19A1 with methotrexate. CD14+ monocytes were isolated from the whole blood of six individual donors. The inhibitory effect of methotrexate on the response to extracellular cGAMP and 2’3’-CDAS varied between donors, ranging from 0% inhibition to 35% (for cGAMP) and 45% (for 2’3’-CDAS) inhibition (Figure 6D). Monocytes were isolated from one additional healthy donor, and was then treated with sulfasalazine, methotrexate, folinic acid, or folic acid. All treatments resulted in a reduction in pIRF3 ranging from 30-60% in response to extracellular cGAMP (Figure 6E). These data suggest that SLC19A1 may also play a role in CDN import in primary monocytes, and that its role varies among donors.
Discussion
In this study we performed a whole-genome CRISPR screen in the monocyte-derived U937 cell line to identify key regulators of the response to extracellular cGAMP. From the screen we identified SLC19A1 as a cGAMP importer, providing the first evidence of a specific import mechanism for cGAMP. In addition to importing cGAMP, we showed that SLC19A1 also imports other CDNs, such as the investigational new drug 2’3’-CDAS We found that the selectivity of CDNs for SLC19A1 is dependent on the identity of nucleotide bases and linkages. For example, whereas 3’3’-cGAMP is selective for SLC19A1, the other 3’3’-linked CDNs, 3’3’-CDA and 3’3’-CDG, do not appear to be imported efficiently by SLC19A1. Similarly, while the phosphodiester-linked 2’3’-CDA is only moderately selective for SLC19A1 in U937 cells, the phosphothioate linked 2’3’-CDAS is highly selective for SLC19A1 in this cell line.
Interestingly, different cell types use different sets of CDN transport mechanisms. While U937 and THP-1 cells largely depend on SLC19A1 to import cGAMP, CD14+ primary monocytes also use SLC19A1 independent, unidentified import mechanisms. Furthermore, HEK 293 and HeLa cells do not use SLC19A1 to import cGAMP and 2’3’-CDAS at the concentrations tested. Although the identity of these additional importer(s) remains unknown, it is unlikely that import is due to a nonspecific mechanism, such as pinocytosis, as this would not explain the observation that HeLa cells respond efficiently to extracellular cGAMP but not 2’3’-CDAS. It warrants further investigation to determine the extent to which SLC19A1 contributes to cGAMP uptake in primary monocytes and other cell types across the human population. Regardless, the fact that humans have more than one importer for cGAMP is another supporting evidence that extracellular cGAMP signaling plays important roles and the system cannot afford to fail.
It has been shown that both endogenous and exogenous extracellular cGAMP promotes immune cell recruitment and tumor shrinkage in a STING-dependent manner (Corrales et al. 2015; Woo et al. 2014; Carozza et al., 2019). However, it is currently unknown what cell type(s) is directly responding to extracellular cGAMP to promote an anti-tumoral immune response. Further studies are necessary to identify these responder cell types and determine to what extent SLC19A1 and other cGAMP importers are necessary for the anti-tumor effect of extracellular cGAMP.
Due to the strong anti-tumoral effects of non-hydrolysable CDN analogs in mice (Corrales et al., 2015), two CDN-based STING agonists have entered and finished phase 1b clinical trials. Unfortunately, these CDNs showed less efficacy in humans as both monotherapies and in combination with anti-PD1 antibodies (Harrington et al., 2018; Meric-Bernstam et al., 2018). Given that our data indicate that CDNs are differentially imported by different cell types, it is possible that the CDNs used in these trials were not efficiently imported by cell types that would promote an anti-tumoral response. Identification of which cell types in humans are important to target for STING mediated anti-tumor immunity and their import mechanisms will aid in the design of more effective CDN-based therapeutics.
STAR Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Lingyin Li (lingyinl@stanford.edu).
Experimental Model and Subject Details
Mammalian cell lines
HEK-Blue IFN-α/β Cells, HEK 293, HEK 293T, and HeLa cells were maintained in DMEM (Cellgro) supplemented with 10% FBS (Atlanta Biologicals) and 1% penicillin-streptomycin (Gibco). U937 and NU-DUL-1 cells were maintained in RPMI (Cellgro) supplemented with 10% heat-inactivated FBS (Atlanta Biologicals) and 1% penicillin-streptomycin (Gibco). THP-1 cells were maintained in RPMI (Cellgro) supplemented with 10% heat-inactivated FBS (Atlanta Biologicals), 1% penicillin-streptomycin (Gibco), and 50 μM 2-mercaptoethanol (Sigma). All cell lines were maintained in a 5% CO2 incubator at 37 °C.
Primary human cells
CD14+ primary cells were maintained in RPMI (Cellgro) supplemented with 10% heat-inactivated FBS (Atlanta Biologicals), 1 mM sodium pyruvate (Gibco), 50 μM 2-mercaptoethanol (Sigma), and 10 ng/mL M-CSF (PeproTech). Cells were maintained in a 5% CO2 incubator at 37 °C.
Method Details
Reagents and antibodies
2’3’-cyclic-GMP-AMP (cGAMP) was synthesized in house as described below. 3’3’-cyclic-GMP-AMP (3’3’-cGAMP), 3’3’-cyclic-di-AMP (3’3’-CDA), 3’3’-cyclic-di-GMP (3’3’-CDG), 2’3’-cyclic-di-AMP (2’3’-CDA), 2’3’-bisphosphothioate-cyclic-di-AMP (2’3’-CDAS), and 2’3’-bisphosphothioate-cyclic-GMP-AMP (2’3’-cGSASMP) were purchased from Invivogen. 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR), sulfasalazine, folinic acid, and methotrexate were purchased from Sigma Aldrich. Sulfasalazine was dissolved in 50 mM NaHCO3 and methotrexate was dissolved in 100 mM NaHCO3. CellTiter-Glo luminescent cell viability assay was purchased from Promega. Rabbit polyclonal antibodies against TBK1 (1:1000), IRF3 (1:1000), pTBK1 (S172, 1:1000), pIRF3 (S396, 1:1000), AMPKa (1:1000), pAMPKa (THr172, 1:1000), STING (1:1000), and cGAS (1:1000) were purchased from Cell Signaling Technology. Mouse monoclonal anti-a-tubulin (1:1000) was purchased from Cell Signaling Technology.
Recombinant DNA
For creation of pcDNA3-FLAG-HA-SLC19A1, the DNA sequence encoding human SLC19A1 was amplified from a U937 cell cDNA library using SLC19A1 FWD and SLC19A1 REV primers (Supplemental Table 1) and insert into the XbaI-BamHI sites of pcDNA3-FLAG-HA (Addgene). For creation of pDB-His-MBP-sscGAS, the DNA sequence encoding porcine cGAS (residues 135-497) was amplified from a porcine cDNA library using the primer pair sscGAS FWD and sscGAS REV (Supplemental Table 1) and inserted into pDB-His-MBP (a generous gift from Qian Yin, Florida State University) via Gibson assembly.
Expression and purification of recombinant cGAS
Rosetta cells expressing pDB-His-MBP-sscGAS were grown in 2xYT medium with 100 μg/mL kanamycin and induced with 0.5 mM IPTG when OD600 reached 1, and then were grown overnight at 16°C. All subsequent proced ures using proteins and cell lysates were performed at 4°3. Cells were pelleted and lysed in 20 mM HEPES pH 7.5, 400 mM NaCl, 10% glycerol, 10 mM imidazole, 1 mM DTT, and protease inhibitors (cOmplete, EDTA-free protease inhibitor cocktail, Roche). Cell lysate was then cleared by ultracentrifugation at 50,000 × g for 1 h. The cleared supernatant was incubated with HisPur cobalt resin (ThermoFisher Scientific; 1 mL resin per 1 L bacterial culture) for 30 min. Cobalt resin was then washed with 20 mM HEPES pH 7.5, 1 M NaCI, 10% glycerol, 10 mM imidazole, and 1 mM DTT. Protein was eluted from resin with 300 mM imidazole in 20 mM HEPES pH 7.5, 400 mM NaCl, and 1 mM DTT. Fractions containing His-MBP-cGAS were pooled, concentrated, and dialyzed against 20 mM HEPES pH 7.5, 400 mM NaCl, 1 mM DTT, and then snap-frozen in aliquots for future use.
Synthesis and purification of cGAMP
To enzymatically synthesize cGAMP, 1 μM purified cGAS was incubated with 50 mM Tris-HCl pH 7.4, 2 mM ATP, 2 mM GTP, 20 mM MgCl2, and 100 μg/mL herring testis DNA (Sigma) for 24 h. The reaction was then heated at 95 °C for 3 min and filtered through a 3-kDa filter. cGAMP was purified from the reaction mixture using a PLRP-S polymeric reversed phase preparatory column (100 A, 8 μm, 300 × 25 mm; Agilent Technologies) on a preparatory HPLC (1260 Infinity LC system; Agilent Technologies) connected to UV-vis detector (ProStar; Agilent Technologies) and fraction collector (440-LC; Agilent Technologies). The flow rate was set to 25 mL/min. The mobile phase consisted of 10 mM triethylammonium acetate in water and acetonitrile. The mobile phase started as 2% acetonitrile for first 5 min. Acetonitrile was then ramped up to 30% from 5-20 min, then to 90% from 20-22 min, maintained at 90% from 22-25 min, and then ramped down to 2% from 25-28 min. Fractions containing cGAMP were lyophilized and resuspended in water. The concentration was determined by measuring absorbance at 280 nm.
U937 CRISPR knockout library generation
The methods used to create the U937 CRISPR knockout library line were previously described by Morgens et al. (2017). Briefly, a whole-genome library of exon-targeting sgRNAs were designed, with the goal of minimizing off-target effects and maximizing gene disruption. The top 10 sgRNA sequences for each gene were included in the library, along with thousands of safe-targeting and non-targeting negative controls. The library was cloned into a lentiviral vector, pMCB320, which also expresses mCherry and a puromycin resistance cassette. The cells were infected with the lentiviral library, and then selected with puromycin.
CRISPR screen
The U937 CRISPR knockout library line was grown in 4 spinner flasks (1 L), with 2 flasks serving as untreated controls and 2 flasks receiving cGAMP treatment. Throughout the screen all of the samples were split daily to keep the cell density at 250 million cells per 500 mL, which corresponded to 1,000 cells per guide in the untreated samples. The experimental samples were treated daily with enough cGAMP to reduce cell viability by 50% as compared to the control samples. The initial treatment was 20 μM and was increased steadily over the 2-week screen, ultimately reaching 30 μM on the final treatment day. At the end of 2 weeks, the genomic DNA was extracted using a Qiagen Blood Maxi Kit. The library was sequenced using a NextSeq 500/550 Mid Output v2 kit (Illumina). The experimental and control samples were compared using casTLE (Morgens et al., 2016), available at https://bitbucket.org/dmorgens/castle. The algorithm determines the likely effect size for each gene, as well as the statistical significance of this effect.
PBMC isolation
Buffy coat (Stanford Blood Center) was diluted 1:3 with PBS supplemented with 2 mM EDTA. Diluted buffy coat was layered on top of 50% Percoll (GE Healthcare) containing 140 mM NaCI and centrifuged at 600 × g for 30 min. The separated PBMC layer was collected and washed once with PBS and once with RPMI before MACS CD14+ isolation.
CD14+ primary cell MACS Isolation
CD14+ cells in bulk PBMCs were labeled using CD14 MicroBeads (Miltenyi Biotec) and purified using a MACS LS Column on a MidiMACS Separator (Miltenyi Biotec) following the manufacturer’s instructions.
IFNβ qPCR
For detection of !ΡΝβ transcript, U937 cells (5 × 105 cells in 1 mL) were first treated with 100 μM cGAMP for 6 h. Cells were then pelleted and lysed with 1 mL TRIzol (Invitrogen). Total RNA from cells was extracted following manufacturer’s instructions. This RNA was reverse transcribed in 20 μL reactions containing 500 ng total RNA, 100 pmol Random Hexamer Primers (Thermo Scientific), 0.5 mM dNTPs (NEB), 20 U RNaseOUT (Invitrogen), 1× Maxima RT Buffer (Thermo Scientific), and 200 U Maxima Reverse Transcriptase (Thermo Scientific). Reverse transcription reactions were incubated first for 10 min at 37 Ό, then for 30 mi n at 50 C Reactions were then terminated by incubating for 5 min at 85 C To qua ntify transcript levels, 10 μL reactions were set up containing 1× GreenStar Master Mix (Bioneer), 10× ROX dye (Bioneer), 100 nM forward and reverse primers, and 0.7 μL of reverse transcription reactions. To determine Ct values, reactions were run on a ViiA 7 Real-Time PCR System (Applied Biosystems) using the following program: ramp up to 50 “C (1.6 O/s) and incubate for 2 min, ramp up to 95 “C (1.6 “C/s) and incubate for 10 min; then 40 cycles of ramp up to 95 °C (1.6 °C/s) and incubate for 15 sec, ramp down to 60 (1.6 °C/s) and incubate for 1 min. Primers used for qPCR can be found in Table S1.
IFN-β Protein Detection
U937 cells (5 × 105 cells in 1 mL) were treated with 100 μM cGAMP for 6 h. Then, cells were pelleted and supernatant containing secreted IFN-β was collected. 20 μL of supernatant were added to 180 μL HEK-Blue IFN-α/β cells (InvivoGen) (2.8 × 105 cells/mL) and incubated for 20 h in a 5% CO2 incubator at 37 °C. Supernatant from HEK-Blue IFN-α/β cells was then assayed for alkaline phosphatase activity using QUANTI-Blue (InvivoGen), following manufacturer’s instructions.
Cell Viability Measurements
U937 cells were treated with the indicated concentrations of cGAMP for 16 hours. 100 μL of CellTiter-Glo (Promega) were added directly to 100 μL of cells. The cells were shaken gently for 2 min, and then allowed to rest at room temperature for 8 min. The luminescence was measured on a Tecan Spark with a 500 ms integration time.
SLC19A1−/− and cGAS’−/− cell line generation
LentiCRISPR v2 (Addgene) was used as the 3rd-generation lentiviral backbone for all knockout lines. Guide sequences targeting SLC19A1 and cGAS (Table S1) were cloned into the lentiviral backbone using the Lentiviral CRISPR Toolbox protocol from the Zhang Lab at MIT(Sanjana et al., 2014; Shalem et al., 2014). Lentiviral packaging plasmids (pHDM-G, pHDM-Hgmp2, pHDM-tat1b, and RC/CMV-rev1b) were purchased from Harvard Medical School. 500 ng of the lentiviral backbone plasmid containing the guide sequence and 500 ng of each of the packaging plasmids were transfected into HEK 293T cells using FuGENE 6 transfection reagent (Promega). The viral media was exchanged after 24 hours and was harvested after 48 hours. The virus-containing media was passed through a 0.45 μη filter and added to U937 cells in media containing 8 μg/mL polybrene (Sigma Aldrich). The cells were spun at 1000 × g for 1 hour, and then the cells were resuspended in fresh virus-free media. The cells were put under selection with 1 μg/mL puromycin (Sigma Aldrich) for 1 week.
Electroporation of STING agonists
For some experiments, cells were pretreated with 500 μM methotrexate for 5 min. 1 × 106 cells were pelleted and resuspended in 100 μΐ electroporation solution (90 mM Na2HPO4, 90 mM NaH2PO4, 5 mM KCl, 10 mM MgCl2, 10 mM sodium succinate) with the appropriate concentration of STING agonist. Cells were then transferred to a cuvette with a 0.2 cm electrode gap (Bio-Rad) and electroporated using program U-013 on a Nucleofector II device (Lonza). Following electroporation, cells were transferred to the appropriate culture media and cultured for 90 min - 2 h (depending on the experiment) before processing for Western blot analysis.
SLC19A1 Overexpression
To prepare for transfection, 7 × 106 HEK 293T cells were split into a 12-well plate the day before transfection. These cells were then transfected with 50 ng of pcDNA3-STING-HA and 500 ng of either pcDNA3-FLAG-HA or pcDNA3-FLAG-HA-SLC19A1 using FuGENE 6 transfection reagent (Promega). After 24 h, transfected cells were treated with 100 μM cGAMP for 2 h. Following this, cells were directly lysed in Laemmli Sample Buffer and run on an SDS-PAGE gel for Western blot analysis.
[3H]-Methotrexate uptake assay
2-4 × 106 cells were treated with 17.5 nM [3H]-methotrexate in MHS buffer (20 mM HEPES, 225 mM sucrose, pH to 7.4 with MgO) for 5 min. Cold PBS was then added to cells to halt import. Following two more washes in cold PBS, cells were lysed in 500 μL 200 mM NaOH and heated at 65 °C for 45 min to compl etely dissolve the lysate. Then, 400 μL of cell lysate was used to count 3H signal on a scintillation counter and 25 μL were used in a bicinchoninic acid assay (Thermo Scientific) to quantify protein levels for normalization.
Quantification and Statistical Analysis
All statistical analyses were performed using GraphPad Prism 7.4. For [3H]-methotrexate import in U937 cell lines, p values were calculated using an unpaired t-test assuming a Gaussian distribution. For all experiments involving Western blots, densitometric measurements of protein bands were made using ImageJ 1.51g. Measurements of total IRF3 or tubulin bands were used to normalize samples that were run on the same gel, and p values were calculated using a ratio paired t-test assuming a Gaussian distribution.
Data and Software Availability
Sequencing data are available in Table S2. Complete, uncropped versions of all Western blots appearing in this paper are accessible from Mendeley Data at http://dx.doi.org/10.17632/5bssvpns6h.1.
Supplementary Material
Table S2. Guide Counts from CRISPR Screen, Related to Figure 1
Highlights.
A CRISPR screen identifies the Reduced Folate Carrier SLC19A1 as a cGAMP importer Other cyclic dinucleotides (CDNs) also utilizes SLC19A1 with varying selectivity Other CDN import mechanisms exist and vary by cell type
Acknowledgements
We thank J. Carozza, K. Shaw, and S. L. Ergun for synthesizing cGAMP. We also thank all Li Lab members for their insightful comments and discussion throughout the course of this study. C.R. was supported by NIH 5T32GM007276. A.F.C. was supported by NIH 2T32GM007365. This work is supported by the National Institute of Health (5R00CA19089 and DP2CA228044 to L.L., and DP2HD084069 to M.C.B.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References
- Ablasser A, Schmid-Burgk JL, Hemmerling I, Horvath GL, Schmidt T, Latz E, and Hornung V (2013). Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature 503, 530–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apelbaum A, Yarden G, Warszawski S, Harari D, and Schreiber G (2013). Type I interferons induce apoptosis by balancing cFLIP and caspase-8 independent of death ligands. Mol. Cell. Biol. 33, 800–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhoum SF, and Cantley LC (2018). The Multifaceted Role of Chromosomal Instability in Cancer and Its Microenvironment. Cell 174, 1347–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhoum SF, Ngo B, Laughney AM, Cavallo J-A, Murphy CJ, Ly P, Shah P, Sriram RK, Watkins TBK, Taunk NK, et al. (2018). Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 553, 467–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridgeman A, Maelfait J, Davenne T, Partridge T, Peng Y, Mayer A, Dong T, Kaever V, Borrow P, and Rehwinkel J (2015). Viruses transfer the antiviral second messenger cGAMP between cells. Science 349, 1228–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, and Vance RE (2011). STING is a direct innate immune sensor of cyclic di-GMP. Nature 478, 515–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carozza JA, Bohnert V, Shaw KE, Nguyen KC, Skariah G, Brown JA, Rafat M, von Eyben R, Graves EE, Glenn JS, et al. (2019). 2’3’-cGAMP is an immunotransmitter produced by cancer cells and regulated by ENPP1. bioRxiv https://www.biorxiv.org/content/10.1101/539312v1. [DOI] [PMC free article] [PubMed]
- Civril F, Deimling T, de Oliveira Mann CC, Ablasser A, Moldt M, Witte G, Hornung V, and Hopfner K-P (2013). Structural mechanism of cytosolic DNA sensing by cGAS. Nature 498, 332–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrales L, Glickman LH, McWhirter SM, Kanne DB, Sivick KE, Katibah GE, Woo S-RR, Lemmens E, Banda T, Leong JJ, et al. (2015). Direct Activation of STING in the Tumor Microenvironment Leads to Potent and Systemic Tumor Regression and Immunity. Cell Rep. 11, 1018–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies BW, Bogard RW, Young TS, and Mekalanos JJ (2012). Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell 149, 358–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Liang H, Xu M, Yang X, Burnette B, Arina A, Li X-DD, Mauceri H, Beckett M, Darga T, et al. (2014). STING-dependent cytosolic DNA sensing promotes radiation-induced type I interferon-dependent antitumor immunity in immunogenic tumors. Immunity 41, 543–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey B, Dey RJ, Cheung LS, Pokkali S, Guo H, Lee J-H, and Bishai WR (2015). A bacterial cyclic dinucleotide activates the cytosolic surveillance pathway and mediates innate resistance to tuberculosis. Nat. Med. 21, 401–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuertes MB, Kacha AK, Kline J, Woo S-R, Kranz DM, Murphy KM, and Gajewski TF (2011). Host type I IFN signals are required for antitumor CD8+ T cell responses through CD8{alpha}+ dendritic cells. J. Exp. Med. 208, 2005–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman ID, Lichtenstein NS, and Oliverio VT (1968). Carrier-mediated transport of the folic acid analogue, methotrexate, in the L1210 leukemia cell. J. Biol. Chem. 243, 5007–5017. [PubMed] [Google Scholar]
- Han K, Jeng EE, Hess GT, Morgens DW, Li A, and Bassik MC (2017). Synergistic drug combinations for cancer identified in a CRISPR screen for pairwise genetic interactions. Nat. Biotechnol. 35, 463–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding SM, Benci JL, Irianto J, Discher DE, Minn AJ, and Greenberg RA (2017). Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature 548, 466–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington KJ, Brody J, Ingham M, Strauss J, Cemerski S, Wang M, Tse A, Khilnani A, Marabelle A, and Golan T (2018). Preliminary results of the first-inhuman (FIH) study of MK-1454, an agonist of stimulator of interferon genes (STING), as monotherapy or in combination with pembrolizumab (pembro) in patients with advanced solid tumors or lymphomas. In ESMO 2018 Congress, p. [Google Scholar]
- Hou Z, and Matherly LH (2014). Biology of the major facilitative folate transporters SLC19A1 and SLC46A1. Curr. Top. Membr. 73, 175–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A, and Medzhitov R (2010). Regulation of adaptive immunity by the innate immune system. Science 327, 291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen G, Westerhof GR, Jarmuszewski MJ, Kathmann I, Rijksen G, and Schornagel JH (1990). Methotrexate transport in variant human CCRF-CEM leukemia cells with elevated levels of the reduced folate carrier. Selective effect on carrier-mediated transport of physiological concentrations of reduced folates. J. Biol. Chem. 265, 18272–18277. [PubMed] [Google Scholar]
- Jansen G, van der Heijden J, Oerlemans R, Lems WF, Ifergan I, Scheper RJ, Assaraf YG, and Dijkmans BAC (2004). Sulfasalazine is a potent inhibitor of the reduced folate carrier: implications for combination therapies with methotrexate in rheumatoid arthritis. Arthritis Rheum. 50, 2130–2139. [DOI] [PubMed] [Google Scholar]
- Konno H, Konno K, and Barber GN (2013). Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell 155, 688–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Yin Q, Kuss P, Maliga Z, Millan JL, Wu H, and Mitchison TJ (2014). Hydrolysis of 2’3’-cGAMP by ENPP1 and design of nonhydrolyzable analogs. Nat. Chem. Biol. 10, 1043–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X-D, Wu J, Gao D, Wang H, Sun L, and Chen ZJ (2013). Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science 341, 1390–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Zhang H, Wu X, Ma D, Wu J, Wang L, Jiang Y, Fei Y, Zhu C, Tan R, et al. (2018). Nuclear cGAS suppresses DNA repair and promotes tumorigenesis. Nature 563, 131–136. [DOI] [PubMed] [Google Scholar]
- Mackenzie KJ, Carroll P, Martin C-A, Murina O, Fluteau A, Simpson DJ, Olova N, Sutcliffe H, Rainger JK, Leitch A, et al. (2017). cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 548, 461–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus A, Mao AJ, Lensink-Vasan M, Wang L, Vance RE, and Raulet DH (2018). Tumor-Derived cGAMP Triggers a STING-Mediated Interferon Response in Non-tumor Cells to Activate the NK Cell Response. Immunity 49, 754–763. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meric-Bernstam F, Werner TL, Hodi FS, Messersmith W, Lewis N, Talluto C, Dostalek M, Tao A, McWhirter SM, Trujillo D, et al. (2018). Phase I dose-finding study of MIW815 (ADU-S100), an intratumoral STING agonist, in patients with advanced solid tumors or lymphomas. In 33rd Annual SITC Meeting, p. [Google Scholar]
- Morgens DW, Deans RM, Li A, and Bassik MC (2016). Systematic comparison of CRISPR/Cas9 and RNAi screens for essential genes. Nat. Biotechnol. 34, 634–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgens DW, Wainberg M, Boyle EA, Ursu O, Araya CL, Tsui CK, Haney MS, Hess GT, Han K, Jeng EE, et al. (2017). Genome-scale measurement of off-target activity using Cas9 toxicity in high-throughput screens. Nat. Commun. 8, 15178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, and Zhang F (2014). Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer JD, Sotelo-Troha K, Von Moltke J, Monroe KM, Rae CS, Brubaker SW, Hyodo M, Hayakawa Y, Woodward JJ, Portnoy DA, et al. (2011). The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect. Immun. 79, 688–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, Heckl D, Ebert BL, Root DE, Doench JG, et al. (2014). Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivick KE, Desbien AL, Glickman LH, Reiner GL, Corrales L, Surh NH, Hudson TE, Vu UT, Francica BJ, Banda T, et al. (2018). Magnitude of Therapeutic STING Activation Determines CD8+ T Cell-Mediated Anti-tumor Immunity. Cell Rep. 25, 3074–3085.e5. [DOI] [PubMed] [Google Scholar]
- Sullivan JE, Brocklehurst KJ, Marley AE, Carey F, Carling D, and Beri RK (1994). Inhibition of lipolysis and lipogenesis in isolated rat adipocytes with AICAR, a cell-permeable activator of AMP-activated protein kinase. FEBS Lett. 353, 33–36. [DOI] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, and Chen ZJ (2013). Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visentin M, Zhao R, and Goldman ID (2012). Augmentation of Reduced Folate Carrier-Mediated Folate/Antifolate Transport through an Antiport Mechanism with 5-Aminoimidazole-4-Carboxamide Riboside Monophosphate. Mol. Pharmacol. 82, 209–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Hu S, Chen X, Shi H, Chen C, Sun L, and Chen ZJ (2017). cGAS is essential for the antitumor effect of immune checkpoint blockade. Proc. Natl. Acad. Sci. U. S. A. 114, 1637–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo S-R, Fuertes MB, Corrales L, Spranger S, Furdyna MJ, Leung MYK, Duggan R, Wang Y, Barber GN, Fitzgerald KA, et al. (2014). STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity 41, 830–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward JJ, lavarone AT, and Portnoy DA (2010). c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science 328, 1703–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Shi H, Wu J, Zhang X, Sun L, Chen C, and Chen ZJ (2013). Cyclic GMP-AMP containing mixed Phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol. Cell 51, 226–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S2. Guide Counts from CRISPR Screen, Related to Figure 1
Data Availability Statement
Sequencing data are available in Table S2. Complete, uncropped versions of all Western blots appearing in this paper are accessible from Mendeley Data at http://dx.doi.org/10.17632/5bssvpns6h.1.


