Abstract
The primary goals of modern genetics are to identify disease-causing mutations and to define the functions of genes in biological processes. Two complementary approaches, reverse and forward genetics, can be utilized to achieve this goal. Reverse genetics is a gene-driven approach that comprises specific gene targeting followed by phenotypic assessment. Conversely, forward genetics is a phenotype-driven approach that involves the phenotypic screening of organisms with randomly induced mutations followed by subsequent identification of the causative mutations (i.e. those responsible for phenotype). In this article, we focus on how forward genetics in mice can be utilized to explore dermatologic disease. We outline mouse mutagenesis with the chemical N-ethyl-N-nitrosourea (ENU) and the strategy utilized to instantaneously identify mutations that are causative of specific phenotypes. We also summarize the types of phenotypic screens that can be performed to explore various aspects of dermatologic disease.
Introduction
Genetic studies in mice are critical to the mechanistic understanding of human disease. In classical genetics, two common approaches are implemented to elucidate gene function: reverse and forward genetics. Reverse genetics is a gene-to-phenotype approach in which a specific gene is targeted, and the phenotypic effects of this disruption are examined. In contrast, forward genetics is a phenotype-to-gene approach that begins with a mutant phenotype of interest and proceeds to the identification of the disrupted gene. Both methods have been used to elucidate genes required for protection against dermatologic disease (DeStefanoandChristiano, 2014). Reverse genetics generally involves prior knowledge or speculation as to how a gene may function, which can limit the finding of novel and unexpected pathways leading to disease phenotype. Forward genetics makes no such assumptions. Because forward genetics does not start with any preconceived ideas as to how phenotypes arise, it can lead to the discovery of new molecules or pathways involved in a biologic process that were previously undetected by researchers. Moreover, if enough mutations are screened for a specific phenotype, the full set of genes with non-redundant function in a biological process can be defined.
ENU Mutagenesis and Breeding Strategy
The mutagen used for forward genetics in mice is N-ethyl-N-nitrosourea (ENU). While other mutation systems exist (e.g. radiation induced and transposons), ENU mutagenesis is preferred due to the high mutation rate and the ability to generate deleterious missense alleles. ENU generates the highest mutational load in the germline of any known agent, introducing approximately 3,000 mutations into each male gamete after three intraperitoneal injections of 100 mg/kg body weight, administered at weekly intervals (Arnold et al., 2012). ENU is a DNA alkylating agent that modifies nucleotide residues by the transfer of an ethyl group. ENU causes point mutations that result in A-T to T-A transversions or A-T to G-C transitions; G-C to C-G transversions are rarely observed (Arnold et al., 2012). At the protein level, 70% of ENU mutations result in nonsynonymous changes with 65% of these being missense and the remainder due to nonsense or splice mutations (Arnold et al., 2012).
Point mutations leading to missense alleles are ideal for several reasons. First, monogenic diseases, including those involving skin, are most commonly caused by coding region point mutations rather than by mutations in intronic or regulatory regions (DeStefano and Christiano, 2014, Oliver and Davies, 2012). Moreover, in addition to null alleles, ENU-induced nucleotide substitutions can generate hypomorphs, hypermorphs, and neomorphs that more closely resemble disease-causing alleles in humans (Oliver and Davies, 2012). Hypomorphic and hypermorphic correspond with mutations that result in partial loss or gain of function, respectively. Neomorphic describes a mutation that confers new function. Hypomorphic mutations can be compatible with viability even if they occur in essential genes, which comprise an estimated one-third of the genome. Indeed, viable hypomorphic mutations in essential genes are more likely to cause phenotypes as compared with hypomorphic mutations in non-essential genes (Wang et al., 2018).
ENU mutagenesis is usually carried out on a highly inbred genetic background such as C57BL/6J, in which there is homozygosity at almost all loci, and mutations induced by ENU are readily detected in the heterozygous state by high throughput DNA sequencing (e.g., with an Illumina sequencing platform). Because ENU-induced phenotypes are almost always ascribable to changes in coding sense, it is sufficient to cover coding region (whole exome sequencing) to detect the great majority of causative mutations. ENU-induced genetic variation is rather limited within a single pedigree (as compared to human genetic variation within a large population of unrelated individuals); therefore, when a phenotype is detected, it will almost always be ascribable to a single nucleotide change.
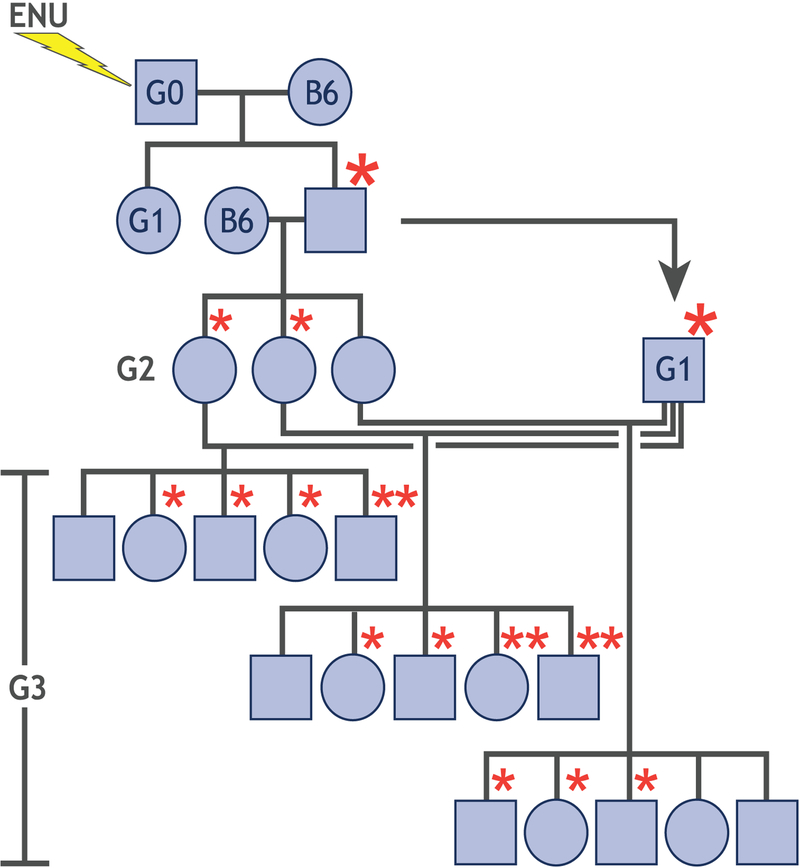
Assessment of mutations in the homozygous state is preferred over the heterozygous state because for many genes, mutations causing dominant phenotypes do not occur, or occur very rarely. Generation of mice with homozygous ENU mutations is achieved by the following breeding scheme (Figure 1). ENU-mutated male mice (G0) are bred to a wild-type female mouse, and mutations are transmitted in the heterozygous state to the G1 generation. G1 males are bred to wild-type females and mutations are again transmitted in the heterozygous state to the G2 generation. G2 daughters are backcrossed to the G1 males yielding G3 mice with mutations in the heterozygous and the homozygous states, which allows for the detection of phenotypes that are caused by both dominant and recessive models of inheritance. On average, a phenotypically neutral mutation will be transmitted to homozygosity in 12.5% of G3 mice, but if a mutation impairs survival in the homozygous state, or is linked to another mutation that does so, fewer homozygotes may be observed. This breeding scheme does not mutagenize the X chromosome, since only G1 males, bearing mutagenized Y chromosomes, are used to produce pedigrees. Alternative breeding schemes that involve breeding of G0 males with G1 females carrying germ-line mutations derived from other mutagenized males can be utilized to screen for X-linked phenotypes. Moreover, mutations on the Y chromosome rarely yield phenotypes because of the highly repetitive Y chromosome sequence.
Figure 1.
Breeding scheme for generation of G3 mice. G0 mice are bred to C57BL/6J females. G1 males are crossed to C57BL/6J females to produce G2 mice. The G3 generation results from backcrossing of G2 females to the G1 founder mouse. Asterisks represent mutations derived from the G0 male.
The size of pedigrees produced for forward genetic studies strikes a balance. On the one hand, there is a desire to detect even those mutations that do compromise survival. There is also a desire to dissociate linked mutations by meiotic recombination, and thereby resolve which mutation is causative of a particular phenotype. On the other hand, increasing the number of G3 mice analyzed increases the cost of the process per mutation screened. Typically an average number of 50–60 G3 mice are produced per pedigree, and pedigrees with fewer than 20 G3 mice are not screened.
Real-time Mapping of a Qualitative Trait
In the past, identification of the causative mutation for a phenotype involved positional cloning, a process that often required several years of breeding, outcrossing, and backcrossing to establish a critical region; then physical mapping in which the gene content of the critical region was determined; then mutation identification by Sanger sequencing of all coding regions and splice junctions. This process has been greatly accelerated, not only by the advent of massively parallel sequencing techniques, but by genotyping all G3 mice at all mutation sites, and the use of high-speed statistical computation to test the null hypothesis that each mutation has nothing to do with any phenotypic variance that might be observed in screening (Wang et al., 2015). Real-time identification of mutations is based on the premise that causation can be ascribed if all induced and/or background stock mutations are known, and if the zygosity of those mutations is known in all G3 mice (Wang et al., 2015). An average of 60 mutations in coding residues are transmitted from every ENU-mutagenized G0 progenitor to the G1 founder of each pedigree, and these mutations are identified by whole-exome sequencing. The G1 male serves as the grandsire of the pedigree. All mutations in a G1 mouse are transmitted to the G3 mice, and the G3 mice are genotyped across the mutated loci prior to phenotypic screening. As soon as phenotypic data are collected from the G3 mice, they can be combined with the genotypic information to determine the likelihood that an observed association between phenotype and genotype would occur by chance (given dominant, additive, or recessive models of inheritance).
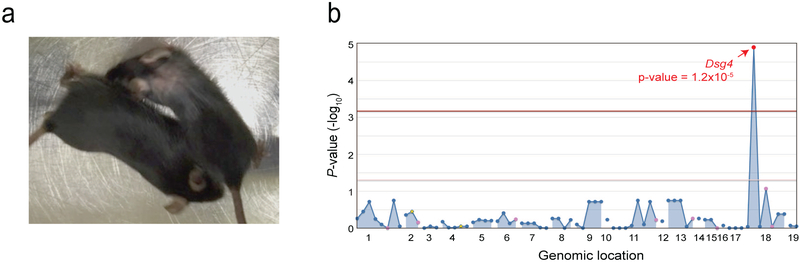
Both qualitative and quantitative phenotypes can be subjected to real-time mapping. As an example of the former, real-time mapping was used to identify a mutation in Dsg4 causative of a hair loss phenotype. Whole-exome sequencing of the pedigree revealed 72 coding region mutations. Thirty-six G3 mice were produced and assessed for a hair loss phenotype. These mice were sequenced at all 72 loci identified in the G1 founder and then assessed for various phenotypes. Upon visual inspection, four mice in this pedigree exhibited early hair loss (Figure 2A). Thirty-two appeared normal and were designated as unaffected. Automated mapping by recessive, additive, and dominant models of inheritance implicated a missense mutation in Dsg4 that results in a valine to glutamic acid change at amino acid 211 of the protein. Mapping was strongest with a recessive model of inheritance (P = 1.2 × 10−5, Figure 2B); the four affected mice were homozygous for the missense mutation in Dsg4, while the 32 mice that were unaffected were either wild-type or heterozygous at this locus. Moreover, affected mice exhibited varying zygosities at the other 71 loci that were found to be mutated with ENU, increasing the likelihood that the Dsg4 mutation is causative for the phenotype. Generally, phenotypic mappings are confirmed via generation of mice that knock-in for the ENU mutation using CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9 technology; however, in this case, mutations in Dsg4 have been reported to cause variable hair loss in lanceolate hair (lah) mouse models (Sundberg et al., 2000), making verification unnecessary. An overview of the forward genetic approach in its entirety is provided in Figure 3.
Figure 2.
Mapping of a hair loss phenotype to a Dsg4 mutation. (A) Representative image of hair loss exhibited by four mice in a pedigree. (B) Manhattan plot showing the P value of association between the phenotype and the mutations identified in the pedigree, calculated using a recessive model of inheritance. The −log10 P values were plotted versus the chromosomal positions of mutations. Horizontal red and purple lines represent thresholds of P = 0.05 with or without Bonferroni correction, respectively.
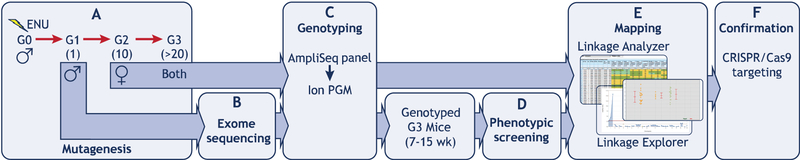
Figure 3.
Overview of the forward genetics approach conducted at the Center for Genetics of Host Defense. (A) A minimum of 20 G3 mice are produced from backcrossing of 8–10 G2 mice to their G1 father. (B) Whole exome sequencing is performed on the G1 founder. (C) G2 and G3 mice are genotyped across the mutated loci identified in the whole exome sequencing of the G1 mouse. (D) G3 mice undergo a series of phenotypic assays. (E) Genotype data and quantitative phenotype data are used for mapping by Linkage Analyzer. Calculated P values for non-linkage and scatter plots of phenotypic data for every mutant allele are displayed by Linkage Explorer. (F) Causative mutations are confirmed by observation of the mutant phenotype in mice with a second mutant allele, which may be generated by CRISPR/Cas9 targeting.
Phenotypic Screening
An incisive screen is a critical element in forward genetics. A screen should address a well-defined and realistic question. It should also be robust to the extent that few false positives are registered (i.e. minimal type 1 error), meaning that a minimum amount of resources will be dedicated to the exploration of mutations that ultimately prove to be non-causative. The less one understands about a biological phenomenon, the more one stands to gain by screening, and the less can be accomplished by other approaches. Yet it is usually a mistake to conclude at the outset that “too much is known” about a particular topic to warrant screening. Biological phenomena mediated by a small number of genes with non-redundant function will yield few causative mutations, yet these may be compellingly important. Those that depend upon large numbers of genes, perhaps operating independently in multiple cell types, will yield many causative mutations, which, however, may be more difficult to understand mechanistically. In such a situation, the mutations with greatest effect size and greatest novelty may be the ones to pursue. While some quantitative phenotypes may show a small effect size, it is important to remember that most human phenovariance results from additive or synergistic genetic differences at multiple loci. From this standpoint, ENU-induced mutations may point the way to a detailed understanding of complex phenotypes. ENU mutagenesis can and should be used to suppress disease phenotypes as well as to create them.
Mouse phenotypes can present spontaneously or only in the presence of environmental challenge. In our laboratory, G3 mice undergo a series of phenotypic assays, beginning with those that are least invasive and ending with those that are highly invasive. The pipeline provides a comprehensive assessment of visible, innate and adaptive immune, metabolic, and neurobehavioral phenotypes. The phenotypes detected during screening are catalogued and made publicly available at mutagenetix.utsouthwestern.edu. At the beginning of the phenotypic pipeline, mice are inspected for visible phenotypes, including differences in size, limb number, behavior, and skin. In total, 96,569 G3 mice that harbored over 155,957 ENU alleles have been assessed in this manner, and 32 mutations that lead to alterations in gross morphology of skin, hair, or nails have been detected. Some of these mutations were found in genes that were known to be important for cutaneous biology including Dsg4,Lyst, and Tmem79 (Barbosa et al., 1996,Perou et al., 1996, Sasaki et al., 2013,Sundberg et al., 2000), while others (e.g., Gk5, Tmprss6, Mbtps1, Dock7, Krt33a, Krt25 ((Blasius et al., 2009,Brandl et al., 2009,Crozat et al., 2010, Du et al., 2010, Du et al., 2008,Rutschmann et al., 2012, Zhang et al., 2017)) had not been previously implicated. Since all coding ENU mutations are ascertained through whole exome sequencing, the genome saturation achieved by a phenotypic assay can be determined, provided that the probability that a mutation is damaging is known. 39% of genes have been severely damaged or destroyed and assessed for visible phenotypes with at least 3 mice homozygous for each damaging mutation.
In some cases, phenotypes may only become apparent after environmental manipulation. This includes challenging mice with exogenous agents such as microbes or chemicals. Examples of dermatologic challenge assays that could be conducted include wound healing (Gradaetal., 2018) and chemically-induced psoriasis (Hawkes et al., 2018), which were recently featured in this Research Techniques Made Simple series. These two models share features with the intestinal dextran sulfate sodium (DSS) injury model, which is one of the more fruitful assays in our laboratory (Tureretal., 2017). Moreover, phenotypes observed in these assays can be scored quantitatively which offers the advantage of objectively mapping traits of even weak effect.
Almost all phenotypes detected in our pipeline are monogenic in nature, but occasionally complex phenovariance caused by mutations in the same pedigree can occur. For example, mice with white-spotted cream-colored coats were produced in the presence of a Sox10 mutation, which caused the white spots, and a Tyr mutation, which resulted in the cream color. Large numbers of G3 mice are required to detect complex traits caused by homozygosity for two unlinked recessive alleles because of the low probability (1/64) that a mouse will be homozygous at each locus. While mutations that have additive effects are rarely detected, multiple phenotypes within the same pedigree frequently occur.
Concluding Remarks
Forward genetics is an unbiased tool for elucidating the genes with non-redundant function in selected biological processes. Real time mapping technology has made it possible us to declare thousands of mutations responsible for phenotypes in a relatively short time. Moreover, the degree of saturation (percentage of genes damaged or destroyed and tested for phenotypic effect N times or more in the homozygous state) may be tracked as mutagenesis and screening progress (Wang et al., 2018). In the era of precision medicine, the ENU mutagenesis program detailed here can be envisioned for rapidly determining the significance of variants found in patients and families carrying hereditary dermatologic disease. There is a substantial opportunity to use germline mutagenesis to create or mitigate dermatologic diseases.
Summary points
Advantages:
Forward genetics is unbiased and can lead to unexpected breakthroughs.
With increased genomic damage saturation, multiple mutations effect the same pathway. The complete set of genes involved in a biologic pathway can thus be deduced by forward genetics provided that enough mice are assessed.
Hypomorphic mutations introduced by ENU can be viable, whereas early lethality may be caused by complete loss of function.
New drug targets can be ascertained from mutations that modify disease.
Limitations:
Forward genetics in mice is resource intensive and can be both expensive and laborious.
The mechanism by which mutations in some genes cause phenotype can be difficult to solve.
Questions
-
ENU mutagenesis primarily results in what type of mutations?
- Insertions
- Deletions
- Nucleotide substitutions
- Duplications
Answer: C. ENU primarily results in A→T transversions or A→G transitions.
-
Which of the following statements is correct?
- Mutations in the G2 generation can be found in the heterozygous form.
- Mutations in the G3 generation can be found in the heterozygous form.
- Mutations in the G3 generation can be found in the homozygous form.
- All of the above.
Answer: D. Mutations are found in the heterozygous form in G2 mice. Mating of G2 mice with the G1 founder can produce mutations in the heterozygous and homozygous form in the G3 generation.
-
On average, what fraction of G3 mice are homozygous for a functionally neutral mutation?
- 1/16
- 1/8
- 1/4
- 1/2
Answer: B. The probability that a G2 mouse is heterozygous for a mutation is 0.50. The probability that a heterozygous G2 mouse produces a homozygous G3 mouse is 0.25. The fraction of G3 mice homozygous for a mutation can be calculated by multiplying these values to obtain 0.125 or 1/8.
-
What is the probability that a G3 mouse is homozygous for two unlinked neutral mutations?
- 1/64
- 1/32
- 1/16
- 1/8
Answer: A. The probability can be calculated by multiplying 1/8 * 1/8 = 1/64.
-
Injection of germline mutant mice with mouse cytomegalovirus (MCMV) to determine genes required for resistance to infection is an example of a:
- Spontaneous screen
- Challenge screen
- Modifier screen
Answer: B. Challenge screens utilize an exogenous substance (e.g., chemicals or microbes) as added stressors to reveal phenotypes that may not be apparent under unperturbed conditions.
Supplementary Material
Acknowledgments
We thank Diantha Lavine for expert assistance in figure generation. This work was supported by the Immunology T32 Training Grant AI005184 at UT Southwestern Medical Center (to W.M.); NIH Grants K08 DK107886 (to E.T.), R37 GM066759 (to B.B.), R01 AI125581 (to B.B.), and U19 AI100627 (to B.B.); and by the Lyda Hill Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors report no conflicts of interest.
References
- Arnold CN, Barnes MJ, Berger M, Blasius AL, Brandl K, Croker B, et al. ENU-induced phenovariance in mice: inferences from 587 mutations. BMC research notes 2012;5:577–. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa MD, Nguyen QA, Tchernev VT, Ashley JA,Detter JC, Blaydes SM, et al. Identification of the homologous beige and Chediak-Higashi syndrome genes. Nature 1996;382(6588):262–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasius AL, Brandl K, Crozat K, Xia Y, Khovananth K, Krebs P, et al. Mice with mutations of Dock7 have generalized hypopigmentation and white-spotting but show normal neurological function. Proceedings of the National Academy of Sciences of the United States of America 2009;106(8):2706–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandl K, Rutschmann S, Li X, Du X, Xiao N, Schnabl B, et al. Enhanced sensitivity to DSS colitis caused by a hypomorphicMbtps1 mutation disrupting the ATF6-driven unfolded protein response. Proceedings of the National Academy of Sciences of the United States of America 2009;106(9):3300–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozat K, Li X-H, Smart N, Beutler B. Record for Spikey. Updated 12–23–18 2010. [Google Scholar]
- DeStefano GM, Christiano AM. The genetics of human skin disease. Cold Spring Harb Perspect Med 2014;4(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X, Li X-h, Smart NG, Beutler B. Record for Plush. 2010. [Google Scholar]
- Du X, She E, Gelbart T, Truksa J, Lee P, Xia Y, et al. The serine protease TMPRSS6 is required to sense iron deficiency. Science (New York, NY) 2008;320(5879):1088–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grada A, Mervis J, Falanga V. Research Techniques Made Simple: Animal Models of Wound Healing. The Journal of investigative dermatology 2018;138(10):2095–105.e1. [DOI] [PubMed] [Google Scholar]
- Hawkes JE, Adalsteinsson JA, Gudjonsson JE, Ward NL. Research Techniques Made Simple: Murine Models of Human Psoriasis. The Journal of investigative dermatology 2018;138(1):e1–e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver PL, Davies KE. New insights into behaviour using mouse ENU mutagenesis. Human molecular genetics 2012;21(R1):R72–R81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou CM, Moore KJ, Nagle DL, Misumi DJ, Woolf EA, McGrail SH, et al. Identification of the murine beige gene by YAC complementation and positional cloning. Nature genetics 1996;13(3):303–8. [DOI] [PubMed] [Google Scholar]
- Rutschmann S, Crozat K, Li X, Du X, Hanselman JC, Shigeoka AA, et al. Hypopigmentation and maternal-zygotic embryonic lethality caused by a hypomorphic mbtps1 mutation in mice. G3 (Bethesda, Md) 2012;2(4):499–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Shiohama A, Kubo A, Kawasaki H, Ishida-Yamamoto A, Yamada T, et al. A homozygous nonsense mutation in the gene for Tmem79, a component for the lamellar granule secretory system, produces spontaneous eczema in an experimental model of atopic dermatitis. The Journal of allergy and clinical immunology 2013;132(5):1111–20.e4. [DOI] [PubMed] [Google Scholar]
- Sundberg JP, Boggess D, Bascom C, Limberg BJ, Shultz LD, Sundberg BA, et al. Lanceolate hair-J (lahJ): a mouse model for human hair disorders. Experimental dermatology 2000;9(3):206–18. [DOI] [PubMed] [Google Scholar]
- Turer E, McAlpine W, Wang KW, Lu T, Li X, Tang M, et al. Creatine maintains intestinal homeostasis and protects against colitis. Proceedings of the National Academy of Sciences of the United States of America 2017;114(7):E1273–e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Bu CH, Hildebrand S, Jia G, Siggs OM, Lyon S, et al. Probability of phenotypically detectable protein damage by ENU-induced mutations in the Mutagenetix database. Nature communications 2018;9(1):441–. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Zhan X, Bu CH, Lyon S, Pratt D, Hildebrand S, et al. Real-time resolution of point mutations that cause phenovariance in mice. Proceedings of the National Academy of Sciences of the United States of America 2015;112(5):E440–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Tomisato W, Su L, Sun L, Choi JH, Zhang Z, et al. Skin-specific regulation of SREBP processing and lipid biosynthesis by glycerol kinase 5. Proceedings of the National Academy of Sciences of the United States of America 2017;114(26):E5197–e206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.