Abstract
Purpose
We report two new CAPN5 mutations associated with a phenotype of Autosomal Dominant Neovascular Inflammatory Vitreoretinopathy.
Methods
We performed next generation sequencing in two patients with ADNIV phenotype; the variants identified were explored further.
Results
Patient 1 was heterozygous for CAPN5 c.799G>A, p.(Gly267Ser). Patient 2 was heterozygous for CAPN5 c.1126G>A, p.(Gly376Ser). Both amino acids are highly conserved across species. Patient 1 had a severe phenotype and his mutation lies within the protein’s catalytic domain. Patient 2 had a mild phenotype and her mutation is the first ADNIV-causing mutation to be described in the regulatory domain of Calpain-5.
Conclusions
Our findings potentially add two new ADNIV-causing CAPN5 mutations to the three previously described. We recommend CAPN5 genetic testing in all patients with a possible ADNIV phenotype, to develop our understanding of Calpain-5; a protein which could potentially provide therapeutically accessible targets for the treatment of many leading causes of blindness.
Keywords: ADNIV, Autosomal Dominant Neovascular Inflammatory Vitreoretinopathy, CAPN5, CALPAIN-5, UVEITIS
Introduction
Inherited retinal degenerations are the leading cause of untreatable blindness in working age adults1 and the prevalence is predicted to increase with genetic drift over future generations. Gene therapy with adeno-associated viral (AAV) vectors has recently shown promise in a number of clinical trials targeting single gene replacement2–4. One of the limiting factors in selecting patients for gene therapy is establishing which mutations are pathogenic as it can be difficult to distinguish a rare polymorphism from a true mutation.
Next-generation sequencing is identifying increasing numbers of potential candidate disease alleles and analysis of these new mutations can provide useful information. For example, even identifying a new premature stop codon in a known gene could confirm which exons are assembled to generate mRNA; a number of genes, such as RPGR are known to have splice variants in the retina that are not necessarily expressed elsewhere5.
Autosomal Dominant Neovascular Inflammatory Vitreoretinopathy (ADNIV) was first described as a cause of retinal degeneration in 19906. Uniquely, it is an ocular autoimmune condition that may progress through several stages due to continued inflammation: uveitis, photoreceptor degeneration, retinal neovascularization, retinal fibrosis, retinal detachment, neovascular glaucoma, phthisis and ultimately blindness. Therefore, depending on time of presentation, it can be misdiagnosed as more common ophthalmic conditions such as autoimmune uveitis, retinitis pigmentosa (RP) and diabetic retinopathy.
The Genetic locus was mapped to chromosome 11q13 in 19927 and CAPN5 was identified as the causative gene in 20128. The encoded 73 kDa protein Calpain-5 is a calcium-dependent intracellular cysteine protease involved in signal transduction for various cellular processes, targeting multiple intracellular proteins and pathways. It comprises 640 amino acids, with 2 catalytic domains and 2 regulatory domains. CAPN5 is moderately expressed in photoreceptors9 and although it is widely expressed in other organs, no extra-ocular features are known to be associated with ADNIV.
Only three missense CAPN5 mutations have been identified to date and these mutations are in close proximity to each other in the catalytic domain encoded by Exon 68,10. Here we report 2 new cases of ADNIV caused by novel mutations in CAPN5; one of which is in the regulatory domain and has a milder phenotype.
Methods
This study was conducted in accordance with the Declaration of Helsinki (2013). Formal consent for genetic testing was obtained as per standard NHS Trust policy, peripheral blood samples were taken for next-generation DNA sequencing.
Gene Enrichment and Next Generation Sequencing
Enrichment for the CAPN5 gene was achieved as part of a customised HaloPlex enrichment system kit (Agilent technologies) designed to capture the coding exons and 10bp of the flanking introns of 111 retinal genes. HaloPlex reactions were prepared as per manufacturer’s instructions. Libraries were pooled into batches of 14 and sequenced on an Illumina MiSeq instrument (Illumina) using a MiSeq v3 kit as per manufacturer’s instructions. Reads were aligned using BWA11 and variants called using Platypus12. All variants identified by next generation sequencing were confirmed by Sanger sequencing.
Results
Patient 1 (P1)
The first patient was a 42 year-old Caucasian male with a severe phenotype and advanced disease. He had progressive visual problems from infancy with myopia, congenital nystagmus, optic nerve myelination, and progressive vitreoretinopathy with recurrent vitreous haemorrhages since 19 years old. He had multiple cryotherapy treatments, left eye ruthenium plaque brachytherapy and underwent bilateral pars plana vitrectomies. The retinal vasoproliferation continued to progress with exudation, sub-retinal fluid and fibrosis leading to bilateral tractional retinal detachments. He then developed left rubeotic glaucoma with recent insertion of a glaucoma drainage tube. His only past medical history of note was psoriasis.
At presentation to our service his visual acuity was 6/60 in the right eye and 4/60 in the left eye. On OCT imaging he had gross disturbance of normal retinal morphology due to unusual retinal nerve fibre layer (RNFL) myelination and fibrosis. Autofluorescence was decreased centrally from masking by the RNFL pathology and retinal degeneration. Wide-field laser imaging demonstrated the predominantly inferotemporal vasoproliferative abnormality. There was no immediate family history of eye problems and his mother and son were examined who both had normal ocular appearances.
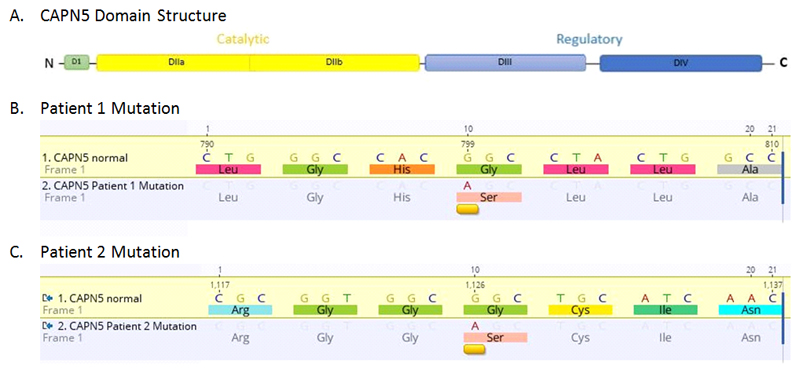
Next-generation sequencing identified that the patient was heterozygous for CAPN5 c.799G>A, p.(Gly267Ser). This variant has been identified at an allele frequency of 0.023% and 0.047% in European and Finnish populations respectively within the Exome Aggregate Consortium (ExAC) database12. p.Gly267 is highly conserved amongst vertebrates and glycine to serine is a non-polar to polar amino acid change. This mutation is in the catalytic domain of the protein encoded by exon 6, similar to previously described mutations. His mother, as the only surviving parent subsequently underwent genetic testing and was negative for the CAPN5 mutation.
Patient 2 (P2)
The second patient was a 19 year-old female of South Asian extraction with a milder phenotype and earlier disease. She was asymptomatic and an incidental finding of irregular retinal hyperpigmentation and peripheral vascular changes at a routine optometrist appointment. She was fit and well with no significant past medical history. Her visual acuity was 6/6 in the right eye and 6/5 in the left eye. Fundus fluorescein angiography (FFA) showed patchy asymmetrical pigmentary retinopathy and also a small area of periphlebitis with leakage temporally in the left eye. She was an only child with no family history of eye problems. She was initially diagnosed with retinitis pigmentosa, however her asymmetrical clinical findings, FFA and ERG were not consistent with this diagnosis. Therefore, following referral to our service, genetic testing was undertaken.
The CAPN5 variant c.1126G>A, p.(Gly376Ser) was identified in this patient. This variant is present at an allele frequency of 0.017% in the South Asian population within the Exome Aggregate Consortium (ExAC) database13. p.Gly376 is highly conserved amongst vertebrates and glycine to serine is a non-polar to polar amino acid change. Both parents were symptom free but declined undergoing ocular examination and genetic testing.
Discussion
Two new missense heterozygous mutations in CAPN5 were identified in patients with ADNIV phenotype. Both of these mutations led to glycine to serine amino acid changes at points which are highly conserved across species. The two patients have differing phenotype severity: P2 at 19 years old is asymptomatic whereas P1 at the same age already had significant visual impairment. The mutation in P1 at p.Gly267 lies within the catalytic domain similar to those previously described. The mutation in P2 at p.Gly376 however is the first to be described within a regulatory domain. ADNIV does demonstrate phenotypical heterogeneity, however it is reasonable to hypothesise that a mutation in the regulatory domain may have a less severe effect on protein function than that in the catalytic domain. The prevalence of 0.017% on the ExAc database would equate to a carrier rate of about 1 in 6,000 in the South Asian population, but it is likely that many of these patients would have subclinical disease, or other broad clinical diagnoses, such as multifocal choroiditis in which peripheral vascular lesions have been described14. Further screening of patients for variants in CAPN5, with correlation to phenotype and protein structure modelling, will provide more insight into the effect on protein function resulting in disease.
Calpain-5 targets multiple intracellular pathways15 and activates necrotic, inflammatory cell death cascades16. The pathogenic mechanism causing ADNIV is unknown as the specific protein substrates for Calpain-5 have not been identified. However, it is likely that mutations in CAPN5 could alter its activity for multiple discrete signalling cascades which could explain the array of pathological features which are unusual in occurring simultaneously in ADNIV patients. This is supported by studies which found that intraocular steroids suppress inflammatory cells and neovascularisation but not retinal fibrosis or degeneration in ADNIV patients17,18. This may indicate different pathogenic pathways for the various phenotypic characteristics of the disease.
Current opinion is that CAPN5 mutations lead to gain of function – leading to unregulated activation of potentially multiple signalling pathways and ocular inflammation – rather than haploinsufficiency8,10,19,20. This is supported by mice studies where hCAPN5-R243L-mutant mice elicit ADNIV-like disease. In a lentiviral vector transduction study hCAPN5-R243L-mutant mice demonstrated CD3+ T-cell migration into the retina, altered expression of cytokines and transcription factors involved in inflammation, reduction of ERG b-wave and photoreceptor degeneration19. A subsequent study using transgenic mutant mice additionally demonstrated upregulation of several biomarkers for uveitis including members of the Toll-like receptor pathway, chemokines and cytokines involved in innate and adaptive immune responses. Histologically, the transgenic hCAPN5-R243L mice eyes exhibited vitritis, vitreoretinal fibrosis, photoreceptor degeneration and neovascularisation20. These findings are in contrast to CAPN5 knockout mice which have no observed phenotype21.
CAPN5 mutations cause a spectrum of disease related to many significant causes of blindness; notably CAPN5 is the only identified monogenic cause for nonsyndromic uveitis. Therefore, further insight into the pathogenic mechanisms at play could improve our understanding not just of ADNIV but of autoimmune uveitis, retinitis pigmentosa, proliferative vitreoretinopathy and diabetic retinopathy.
It will be interesting to know if CAPN5 polymorphisms are risk factors for these diseases and moreover whether modifying calpain expression can alter their progression. There is already evidence that inhibition of calpain activity can rescue other types of retinal degeneration22–25. It has been shown that CAPN5 mutation expression in the eye is sufficient to cause the disease19, therefore it could be a potential candidate for gene therapy.
Our findings add to the 3 previously identified CAPN5 mutations associated with ADNIV. We would recommend CAPN5 genetic testing in all patients with potential ADNIV phenotype, particularly those with unusual inflammation or atypical retinitis pigmentosa. This may also include patients who have a phenotype of familial exudative vitreoretinopathy (FEVR) in which genetic testing for the typical FEVR genes is negative. Whilst genetic testing is becoming mainstream in RP clinics, uveitis specialists may also need to consider ADNIV in unusual cases because a clinically significant dominant history is not always evident. Identifying more ADNIV patients and their CAPN5 variants will further contribute to our understanding of Calpain-5; a protein which could potentially provide therapeutically accessible targets for the treatment of many leading causes of blindness.
Figure 1. Two Novel CAPN5 mutations identified associated with ADNIV.
A. Calpain-5 is a 640 amino acid, 73 kDa protein composed of four domains. The mutations previously described are located in catalytic domain IIb.
B. P1 mutation CAPN5 c.799G>A missense change equivalent to p.(Gly267Ser) or replacement of hydrophobic glycine for serine which is polar at the corresponding amino acid position.
C. P2 mutation CAPN5 c.1126G>A missense change equivalent to p.(Gly376Ser). This is the first ADNIV- causing mutation identified in the regulatory domain of the protein.
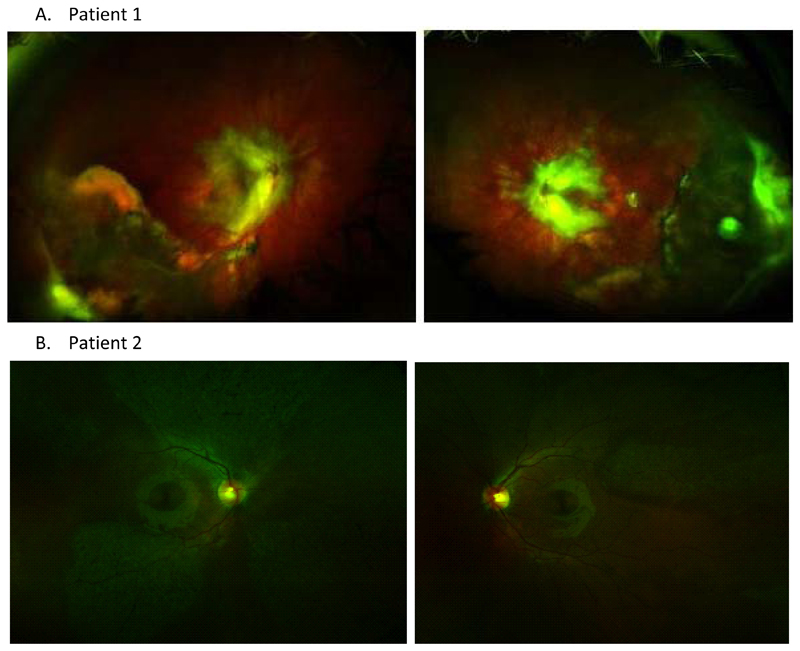
Figure 2. ADNIV Phenotype.
A. Patient 1 has a severe phenotype: optomap shows bilateral vasoproliferative vitreoretinal pathology.
B. Patient 2 has a mild phenotype: colour photographs show patchy areas of retinal pigmentary change.
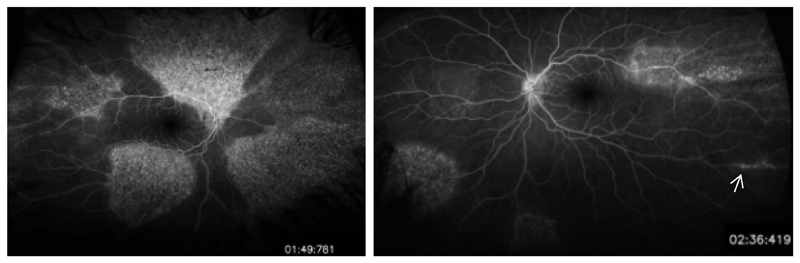
Figure 3. Patient 2 Fundus Fluorescein angiogram.
Fluorescein angiography reveals patchy asymmetrical pigmentary retinopathy and a small area of periphlebitis in the left eye (white arrow).
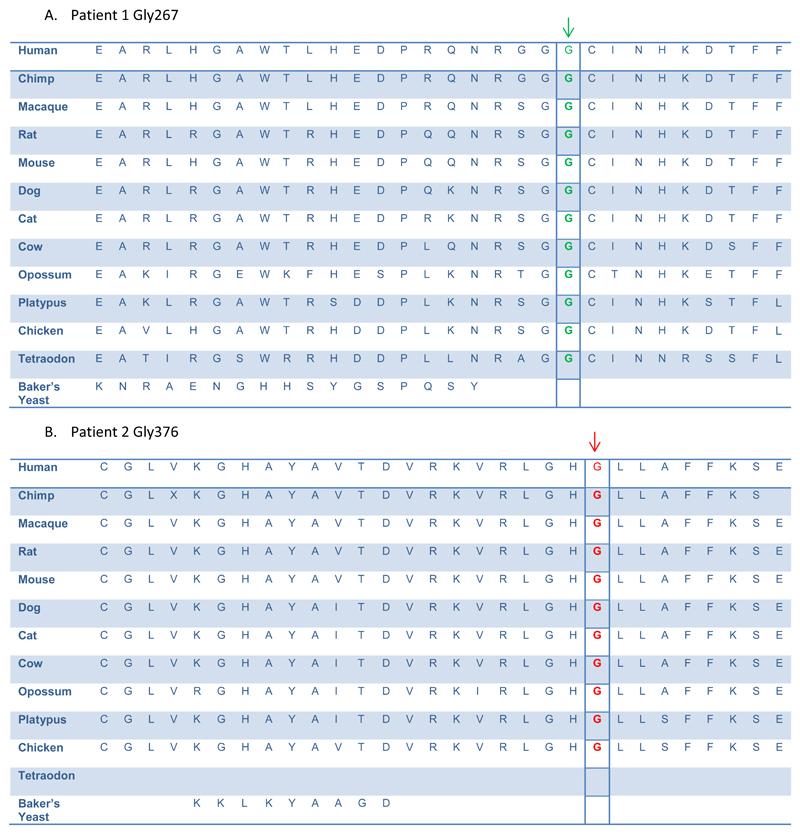
Figure 4. The novel CAPN5 mutated residues are highly conserved amongst species.
Alignment of calpain-5 orthologs show high evolutionary conservation of both patients’ CAPN5 mutations.
A. Patient 1 nucleotide change position Gly267 is indicated in green
B. Patient 2 nucleotide change position Gly 376 is indicated in red.
Funding
This work is supported by the National Institute for Health Oxford Biomedical Research Centre, Oxford University Hospitals NHS Foundation Trust and the Wellcome Trust.
Footnotes
Declaration of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- 1.Liew G, Michaelides M, Bunce C. A comparison of the causes of blindness certifications in England and Wales in working age adults (16–64 years), 1999–2000 with 2009–2010. BMJ Open. 2014;4(2):e004015. doi: 10.1136/bmjopen-2013-004015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simonelli F, Maguire AM, Testa F, et al. Gene therapy for Leber’s congenital amaurosis is safe and effective through 1.5 years after vector administration. Mol Ther. 2010;18(3):643–650. doi: 10.1038/mt.2009.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conlon TJ, Deng W-T, Erger K, et al. Preclinical Potency and Safety Studies of an AAV2-Mediated Gene Therapy Vector for the Treatment of MERTK Associated Retinitis Pigmentosa. Hum Gene Ther Clin Dev. 2013;24(1):23–28. doi: 10.1089/humc.2013.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.MacLaren RE, Groppe M, Barnard AR, et al. Retinal gene therapy in patients with choroideremia: initial findings from a phase 1/2 clinical trial. Lancet. 2014;383:1129–1137. doi: 10.1016/S0140-6736(13)62117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kirschner R, Rosenberg T, Schultz-Heienbrok R, et al. RPGR transcription studies in mouse and human tissues reveal a retina-specific isoform that is disrupted in a patient with X-linked retinitis pigmentosa. Hum Mol Genet. 1999;8(8):1571–1578. doi: 10.1093/hmg/8.8.1571. [DOI] [PubMed] [Google Scholar]
- 6.Bennett SR, Folk JC, Kimura AE, et al. Autosomal dominant neovascular inflammatory vitreoretinopathy. Ophthalmology. 1990;97:1125–1135. doi: 10.1016/s0161-6420(90)32447-8. discussion 1135-6. [DOI] [PubMed] [Google Scholar]
- 7.Stone EM, Kimura AE, Folk JC, et al. Genetic linkage of autosomal dominant neovascular inflammatory vitreoretinopathy to chromosome 11q13. Hum Mol Genet. 1992;1:685–689. doi: 10.1093/hmg/1.9.685. [DOI] [PubMed] [Google Scholar]
- 8.Mahajan VB, Skeie JM, Bassuk AG, et al. Calpain-5 Mutations Cause Autoimmune Uveitis, Retinal Neovascularization, and Photoreceptor Degeneration. PLoS Genet. 2012;8(10):e1003001. doi: 10.1371/journal.pgen.1003001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schaefer KA, Toral MA, Velez G, et al. Calpain-5 Expression in the Retina Localizes to Photoreceptor Synapses. Invest Ophthalmol Vis Sci. 2016;57(6):2509–2521. doi: 10.1167/iovs.15-18680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bassuk AG, Yeh S, Wu S, et al. Structural Modeling of a Novel CAPN5 Mutation that Causes Uveitis and Neovascular Retinal Detachment. PLoS ONE. 2015;10(4):e0122352. doi: 10.1371/journal.pone.0122352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics. 2009;25(14):1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rimmer A, Phan H, Mathieson I, et al. Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat Genet. 2014;46(8):912–8. doi: 10.1038/ng.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Exome Aggregation Consortium (ExAC) Cambridge, MA: [Accessed: 26/10/2016]. URL: http://exac.broadinstitute.org. [Google Scholar]
- 14.MacLaren RE, Lightman SL. Variable phenotypes in patients diagnosed with idiopathic multifocal chroiditis. Clin Exp Ophthalmol. 2006;34(3):233–8. doi: 10.1111/j.1442-9071.2006.01191.x. [DOI] [PubMed] [Google Scholar]
- 15.Zatz M, Starling A. Calpains and disease. N Engl J Med. 2005;352(23):2413–2423. doi: 10.1056/NEJMra043361. [DOI] [PubMed] [Google Scholar]
- 16.Syntichaki P, Xu K, Driscoll M, et al. Specific aspartyl and calpain proteases are required for neurodegeneration in C. elegans. Nature. 2002;419:939–944. doi: 10.1038/nature01108. [DOI] [PubMed] [Google Scholar]
- 17.Tlucek PS, Folk JC, Orien JA, et al. Inhibition of Neovascularization but Not Fibrosis With the Fluocinolone Acetonide Implant in Autosomal Dominant Neovascular Inflammatory Vitreoretinopathy. Arch Ophthalmol. 2012;130(11):1395–1401. doi: 10.1001/archophthalmol.2012.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tlucek PS, Folk JC, Sobol WM, et al. Surgical management of fibrotic encapsulation of the fluocinolone acetonide implant in CAPN5-associated proliferative vitreoretinopathy. Clin Ophthalmol. 2013;7:1093–1098. doi: 10.2147/OPTH.S43939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wert KJ, Skeie JM, Bassuk AG, et al. Functional validation of a human CAPN5 exome variant by lentiviral transduction into mouse retina. Hum Mol Genet. 2014;23(10):2665–77. doi: 10.1093/hmg/ddt661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wert KJ, Bassuk AG, Wu W-H, et al. CAPN5 mutation in hereditary uveitis: the R243L mutation increases calpain catalytic activity and triggers intraocular inflammation in a mouse model. Hum Mol Genet. 2015;24(16):4584–4598. doi: 10.1093/hmg/ddv189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Franz T, Winckler L, Boehm T, et al. CAPN5 is expressed in a subset of T cells and is dispensable for development. Mol Cell Biol. 2004;24:1649–1654. doi: 10.1128/MCB.24.4.1649-1654.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imai S, Shimazawa M, Nakanishi T, et al. Calpain inhibitor protects cells against light-induced retinal degeneration. J Pharmacol Exp Ther. 2010;335:645–652. doi: 10.1124/jpet.110.171298. [DOI] [PubMed] [Google Scholar]
- 23.David J, Melamud A, Kesner L, et al. A novel calpain inhibitor for treatment of transient retinal ischemia in the rat. Neuroreport. 2011;22:633–636. doi: 10.1097/WNR.0b013e32834959c5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nguyen AT, Campbell M, Kenna PF, et al. Calpain and photoreceptor apoptosis. Adv Exp Med Biol. 2012;723:547–552. doi: 10.1007/978-1-4614-0631-0_69. [DOI] [PubMed] [Google Scholar]
- 25.Ozaki T, Nakazawa M, Yamashita T, et al. Intravitreal injection or topical eye-drop application of a mu-calpain C2L domain peptide protects against photoreceptor cell death in Royal College of Surgeons’ rats, a model of retinitis pigmentosa. Biochim Biophys Acta. 2012;1822:1783–1795. doi: 10.1016/j.bbadis.2012.07.018. [DOI] [PubMed] [Google Scholar]