Abstract
Collaboration can be challenging; nevertheless, the emerging successes of large, multi-partner, multi-national cooperatives and research networks in the biomedical sector have sustained the appetite of academics and industry partners for developing and fostering new research consortia. This model has percolated down to national funding agencies across the globe, leading to funding for projects that aim to realise the true potential of genomic medicine in the 21st century and to reap the rewards of ‘big data’. In this Perspectives article, the experiences of the RA-MAP consortium, a group of more than 140 individuals affiliated with 21 academic and industry organizations that are focused on making genomic medicine in rheumatoid arthritis a reality are described. The challenges of multi-partner collaboration in the UK are highlighted and wide-ranging solutions are offered that might benefit large research consortia around the world.
Over the past few years, the relative failure by scientists to reap the benefits of the genomics revolution, along with the pressing challenges and perceived opportunities that accompany the analysis of ‘big data’, have led to a concerted drive towards the development of cooperative academia–industry initiatives across a range of diseases1,2. This move towards consortia acknowledges the need to advance health care initiatives in a systematic way and places emphasis on the collective harnessing of knowledge, resources and expertise in ways that are both complementary and mutually beneficial to all parties3–6. Central to these initiatives has been the creation of nonexclusive consortia in pre-competitive areas of research (research aimed at the generation of new knowledge) that capitalize on expertise from multiple sources and reward all partners for their contributions7,8. In this Perspectives article, we describe the experience of setting up the RA-MAP consortium, a multi-partner academia–industry partnership, and highlight some of the challenges we faced and solutions we adopted to successfully direct a collaborative consortium focused on rheumatoid arthritis (RA).
Stratified medicine
Stratified medicine has been defined in a wide variety of ways9: the Association of the British Pharmaceutical Industry (ABPI) defines it as “the ability to classify individuals into subpopulations that differ in their susceptibility to a particular disease or their response to a particular treatment”10. The term has also been used interchangeably with precision, personalised or P4 medicine9,11. In line with these definitions, and in an effort to realise the full potential of stratified medicine12, funding bodies have sought to support research that provides new insights into disease mechanisms, enabling the tailoring of existing treatments to individuals and paving the way for the development of new treatments, diagnostic methods and care pathways13,14.
Arguably, physicians have been practising precision medicine for centuries, individualizing therapy on the basis of personalized clinical assessment in combination with rudimentary investigations such as haematological and biochemical profiles, as well as radiographic imaging and histopathological investigations. Contemporary concepts of tailoring therapy to specific patient subgroups have been driven by a growing appreciation of pathway biology, in which common clinical syndromes are underpinned by aberrations in specific molecular and cellular processes, and the development of sophisticated laboratory tools to define these distinct pathways15,16. Sequencing and annotation of the human genome, coupled with advances in next generation sequencing technology, have been at the forefront of stratified medicine, enabling researchers to uncover molecular associations with specific disease phenotypes17,18, drug responses and drug toxicities19, as well as to define novel pathogenic molecular pathways that underpin disease risk20. Genomic fingerprinting, along with transcriptomics, epigenomics, proteomics and metabolomics, are just a few of the ‘omics’ technologies that enable a truly systematic and unbiased approach to understanding the molecular basis of disease. The omics revolution is generating data on an unprecedented scale21, leading to the need for major advances in informatics, data integration, data science and methods for analysing big data, a set of disciplines that are often captured under the umbrella term of ‘systems biology and bioinformatics’22. The overriding goal of stratified medicine is early, precise diagnosis of disease and early therapeutic intervention, applying ‘the five rights’ of medication use (a concept adapted from standards for safe medication practices): the right patient, the right drug, the right time, the right dose and the right route of administration23. A future goal of stratified medicine would be to use these data to define the preclinical disease state with a view to personalized preventive medicine. Such big data approaches are underpinned by the belief that the classical clinical phenotype of a disease such as RA is actually composed of a variety of distinct molecular endotypes24, each one predicated on inherited, environmental and stochastic differences between patients.
Nowhere has stratified medicine had a greater effect to date than in cancer; genotyping patients for BRCA mutations25, screening patients for gene translocations26,27 and analysis of expression of ERBB2 combined with in situ tissue typing in patients with breast cancer28,29, for example, have transformed therapy through a deeper understanding of oncogenesis at the molecular level. This deeper knowledge of oncogenesis has led to cancer prevention and to the rational design of small molecule tyrosine kinase inhibitors and monoclonal antibodies, with proof-of-concept being established during clinical trials30,31. The stratification of patients according to their immune phenotype is also progressing rapidly in the field of checkpoint inhibitor therapy32–34. On the basis of these advances, there has been considerable interest in the past few years in applying these principles to other diseases that might benefit from a similar experimental approach. An academia–industry collaboration designed along the lines of the contemporary concepts outlined above would provide a strong platform from which to deliver such an ambitious programme of work.
MRC–ABPI-funded programmes
In 2008, the UK Medical Research Council (MRC) published a strategic review of human immunology, which provided a roadmap for building capacity, for the creation of an interdisciplinary environment and for an increase in connectivity between institutions and sectors35. In 2009, in response to the last of these points, the MRC Human Immunology and Inflammation Initiative identified obstacles to closer academia–industry interaction, solutions to which included improved networking, improved access to human tissue samples and improved support for clinical researchers. Two disease-focused workshops, covering RA and chronic obstructive pulmonary disease, were held in 2010 to begin to address these important issues. The rationale for selecting RA as a model disease for this approach was driven by a combination of UK expertise in the field and specific unmet clinical needs and knowledge gaps for the disease. These unmet needs included robust strategies for the stratification of patients and suitable biomarkers to inform such stratification, technology to predict responses to specific therapies and molecular and cellular signatures to identify a state of true biological remission. At these workshops, the discussions focused on approaches to stratified medicine and placed particular emphasis on prioritising research into disease pathways and on how an ambitious and incisive programme of research might best be delivered. Key requirements for establishing a successful consortium were highlighted during these discussions and are summarised in BOX 1. In 2011, the MRC–ABPI Inflammation and Immunity Initiative was formally launched in an attempt to address some of the specific unmet needs of patients with RA.
Box 1. Establishing a successful stratified medicine consortium.
Several key elements are required when setting up an academia–industry partnership.
A consensus on the importance of identifying common disease pathways.
Engaged industrial partners with emerging drug pipelines.
Existing efficacious therapies that might be suitable for repurposing.
An urgent need for disease phenotyping and biomarker-based patient stratification.
The need for a better understanding of the relationship between clinical and pathological phenotypes.
The availability of emerging technologies to redefine disease subtypes at a molecular and cellular level.
Regional or national colocalisation of partners.
A rich patient bioresource.
Access to clinical research infrastructures, for example the National Health Service and National Institute for Health Research in the UK.
Enthusiastic support from patient groups.
The immunological concept
After considering the requirements listed in BOX 1, the RA-focused working group concluded that the missing element was a full understanding of the immune dysregulation that underpins RA. If the immunology of the disease could be better characterized, it followed that biomarkers could then be developed to stratify patients with the disease and to inform therapy choices. Theoretically, these cellular and molecular tools could be integrated into an immunological toolkit that would consist of a combination of clinical and laboratory parameters measured in patients with early RA that could be used to predict clinical responses to DMARDs, to monitor biological responses to therapy and to define a true state of biological remission. This proposal was predicated on the following principles: the healthy immune system is associated with an immunological fingerprint that can be defined by serum, cellular and/or molecular signatures in peripheral blood; RA is associated with detectable perturbations of the immune system at very early stages of disease36 that can be used to distinguish subsets of patients; restoration of immune health in patients with RA might be inducible by therapies that target these perturbations; and clinical remission is associated with a biological state that might have similarities to a healthy immune system. It was thought that, if successful, such an approach could have an immediate effect on our understanding of a broad range of immune-mediated inflammatory diseases.
The RA-MAP Consortium
In 2012, following a successful funding application focused on the principles described above, the Rheumatoid Arthritis MRC–ABPI (RA-MAP) Consortium was conceived. The consortium has since expanded to include 11 industry partners and 10 UK academic partners who share a deep-rooted enthusiasm for translational science in the field of immunology and inflammation in the pre-competitive space (Supplementary information S1 (figure)). Membership of the consortium reflects contributions and commitments by various partners to genomic medicine, genetics and immunology and inflammation biology; expertise in immune phenotyping, metabolomics and proteomics; clinical expertise in assembling and curating patient cohorts and deep clinical phenotyping; and centres of excellence in experimental medicine with a focus on early inflammatory arthritis. Unusually, the consortium was established with a close relationship between the funding body and the researchers, which created a new paradigm for collaborative working.
The RA-MAP Consortium has similarities to other research networks that focus on research into rheumatic diseases (TABLE 1), including the Accelerating Medicines Partnership (AMP) RA and systemic lupus erythematosus network, a partnership that was launched in 2014. This US network seeks to define new therapies and diagnostic technologies for rheumatic autoimmune diseases by utilizing a systems-level understanding of transcriptomic signatures derived from synovial, kidney and skin tissues. Along similar lines, the European Union (EU)-funded PRECISESADS consortium focuses on redefining autoimmune diseases at a molecular level (TABLE 1). In operational terms, EU consortia have benefited considerably from the experiences of previous academia–industry partnerships, such as AutoCure, MASTERSWITCH and Be the Cure (BTCure) (TABLE 1). The longevity of these programmes has served to fuel the productivity of research and to facilitate collaborations between public sector and private sector organizations. Since its inception, the MRC Stratified Medicine strategic initiative has also supported several other consortia that focus on immune-mediated inflammatory diseases (TABLE 1).
Table 1. Academia–industry consortia in immune-mediated inflammatory diseases.
| Consortium | Contributors | Website |
|---|---|---|
| International consortia | ||
| AMP RA and SLE network |
|
https://amp-ralupus.stanford.edu/ |
| PRECISESADS consortium |
|
http://www.precisesads.eu/ |
| AutoCure |
|
http://www.crb.uu.se/research/projects/autocure/ |
| MASTERSWITCH |
|
http://cordis.europa.eu/result/rcn/147588_en.html |
| Be the Cure |
|
http://btcure.eu |
| Rheuma Tolerance for Cure |
|
http://cordis.europa.eu/project/rcn/211964_en.html |
| MRC Stratified Medicine consortia | ||
| MATURA |
|
http://www.matura.whri.qmul.ac.uk |
| PSORT |
|
http://www.psort.org.uk |
| MASTERPLANS |
|
http://www.lupusmasterplans.org/home.html |
AMP, Accelerating Medicines Partnership; ARUK, Arthritis Research UK; EFPIA, European Federation of Pharmaceutical Industries and Associations; FDA, US Food and Drug Administration; NIH, National Institutes of Health; NHS, National Health Service; RA, rheumatoid arthritis; SLE, systemic lupus erythematosus; SME, small or medium sized enterprise.
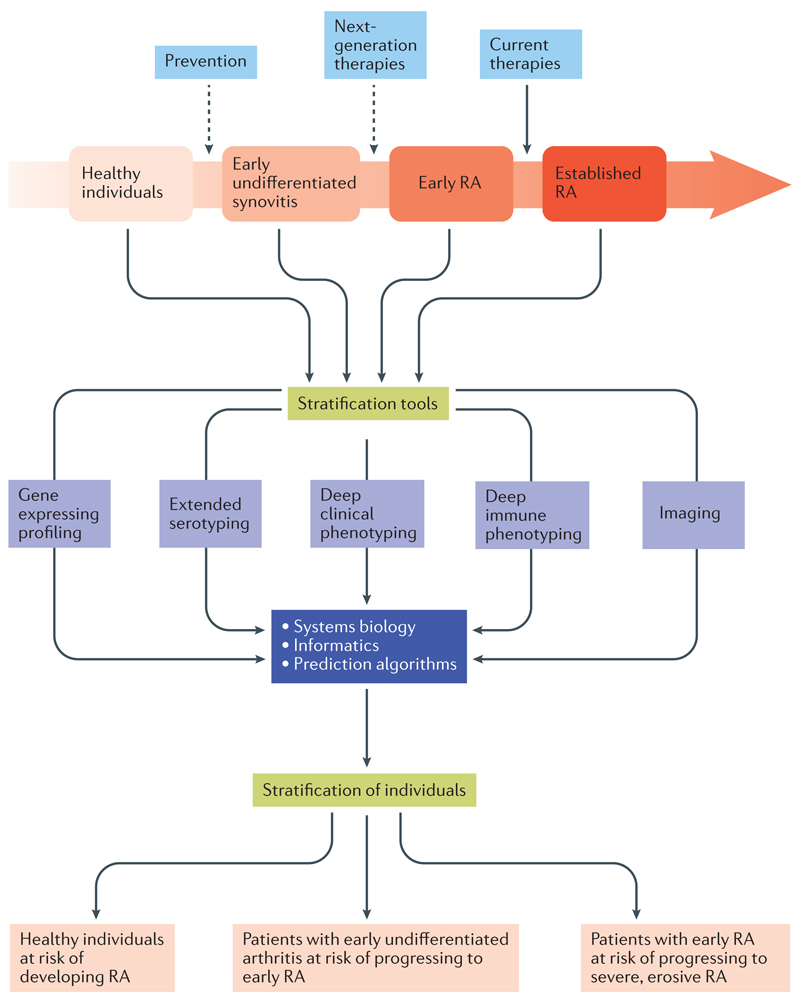
A key challenge for the RA-MAP Consortium was to harness the synergistic skill sets of pharmaceutical companies, biotechnology companies and academic partners to develop a programme of activities that would address each specific scientific goal. To do so required the establishment of a sizeable new inception cohort of treatment-naive patients with RA who had a relatively short duration of symptoms and were willing to provide biological samples. This cohort of patients was called Towards a Cure for Early RA (TACERA), and the samples from these patients provided the substrate for cutting-edge analytical techniques. The next step was to apply innovative systems approaches to analyse and assemble the data from multiple omics platforms into predictive algorithms, with the ultimate aim being the development of a set of informative assays that would provide a toolkit to facilitate patient stratification in a clinical setting (FIG. 1). A cohort of healthy individuals who were followed longitudinally following vaccination with a neoantigen was enrolled to provide a suitable control population with which to compare the signatures of immune dysregulation identified in patients with RA.
Figure 1. Stratification of patients with rheumatoid arthritis.
Stratification of patients with rheumatoid arthritis (RA) can occur at several points during the natural history of the disease. Stratification describes a process of characterising subgroups of patients according to distinct clinical, cellular and molecular features (or endotypes) using any combination of parameters. Multiple platforms can be adopted to stratify patients throughout the disease course, including serotyping, clinical and immunological phenotyping, genotyping and imaging.
Although each industry partner had their own strategic reasons for joining the consortium, the overriding motivation of these companies to partner with academia was the shared recognition that this study would generate data in a real-world population of patients with RA that could improve our understanding of the subsets of disease and associated immunological phenotypes that characterize the early phase of RA. Working collaboratively with companies and various academic centres was thought to increase the chances of producing clinically relevant knowledge about opportunities for intervention and indicators of response in these patients. To achieve these goals, the RA-MAP Consortium divided its tasks into various research work packages (see Supplementary information S2 (figure)).
For the remainder of this Perspectives article we aim to describe some of the operational and scientific challenges that are faced by large research consortia and to highlight solutions that can be adopted to overcome such challenges with reference to specific examples from our experience with the RA–MAP Consortium.
Challenges and solutions
Some of the key challenges that are faced by academia–industry consortia are summarised in BOX 2. Further insights and suggested solutions derived from the experience of the RA-MAP Consortium are described in detail below.
Box 2. Challenges faced by research consortia and possible solutions.
Agreement as to the terms of reference and ground rules for consortium operations
Generate a contract or consortium agreement with input from the contract and legal teams of all partners from the outset.
Data ownership
In any pre-competitive project, data can be shared and intellectual property arrangements can be addressed directly in the consortium agreement.
Industry contributions
Contributions from industry partners should be agreed from the start of the project. Examples of contributions should be provided that cover the areas of specific interest or expertise of each partner.
Project management
Management structures are essential and part of ‘normal business’ for industry partners. Capitalize on private sector expertise to establish lean, functional committees with clear terms of reference. Invest in a project manager, ideally with both academic and industry experience.
Managing staff turnover
Anticipate and redistribute resources to support the training of incoming technical and research staff; close liaison with industry partners to identify new colleagues with relevant skills and experience is essential.
Building a strong collaborative ethos
Identify areas of expertise and establish working groups made up of individuals from across all sectors who share common goals and who will commit to regular teleconference meetings.
Recruiting, site approval and set-up
Engage contract research organizations to support activities such as coordinating the acquisition of documentation for timely site-specific regulatory approval.
Quality control
Quality control applies as much to study protocols and standard operating procedures as it does to sample acquisition, processing and storage and to data analysis; procurement should be robust and outward-looking if the necessary expertise does not exist within the consortium.
Data analysis
Invest in state-of-the-art data warehouse capabilities and facilitate access by all parties. Define research priorities and construct a mutually agreed data analysis plan. Frequent opportunities for all partners to discuss results are essential to maintain momentum.
Publication
Agree to a publication policy and plan that provides shared authorship, where appropriate, and recognizes the contributions of the extended network of investigators.
Scientific review of milestones
Project reviews should be agreed with the funding organization, as appropriate, but should be regular, robust and led by an independent expert advisory committee and chair.
The contract
A major challenge for any consortium is one of scale. In any group of academic and industry partners who each have distinct agendas, experiences and governance structures, individual partners will have different expectations. This discrepancy requires sympathetic management so that the ambitions of all parties can be met. Agreement of the scientific goals of the consortium provides a common purpose, for which each partner can identify their potential contributions and resource provision. Tangible benefits for industry partners are central to success and to the sustainable engagement of such partners; each company will value research ‘currency’ in a different way, but good examples might include access to deeply phenotyped cohorts of patients, access to downstream data and sharing of samples among partners. Interactions between and operations involving multiple institutions require a set of clear ground rules that go beyond a ‘terms of reference’ template. One possible solution is the consortium agreement, which provides an operating framework that emphasizes the obligations and responsibilities of leadership and membership and contains guidelines about the transfer and use of materials, liabilities and indemnity of each party, details of project management and data management practices including data protection and, importantly, publicity, publication and intellectual property rights. In essence, the agreement needs to be simple, pragmatic and a point of reference for the lifetime of the consortium and beyond.
Who owns the data?
Reaching agreement over data protection and ownership can be a major challenge for research consortia because priorities and expectations can vary between the private and public sectors, notwithstanding the nuances that research in the pre-competitive space can offer. Nonetheless, this is an area in which the experience of industry can add value to a consortium, by helping to define relevant background to the project, supporting registration and protection of intellectual property rights arising from the data, filing and prosecuting patent applications or assisting in actions relating to infringement of intellectual property rights. In return, academic partners might agree to grant worldwide non-exclusive licenses to any industry partner to use the results of experiments and intellectual property for commercial purposes, taking into account the relative contribution made to the consortium by that industry partner. Members of the RA-MAP Consortium learned that much time can be saved, and barriers promptly overcome, by facilitating frequent, robustly managed communication between the intellectual property and technology transfer offices of each partner from the very outset.
How can industry partners contribute?
Resource frameworks differ greatly depending on the scale and context of the research programme and the funding agency involved. For example, industry partners might be required to pledge specific levels of support, such as in-kind contributions, contributions of skilled personnel, funding for specific research projects or provision of access to technology platforms. Such has been the approach of the EU Framework 7 and Horizon 2020 programmes with respect to matched contributions from European Federation of Pharmaceutical Industries and Associations (EFPIA) partners37. Commitment to provide matched-funding from the outset has obvious advantages but, although these ground rules might not apply to all consortia, there are other imaginative ways that industry partners can support the research agenda. The RA-MAP Consortium has benefited greatly from the patient-level data, advice on the setup of and study operations for the TACERA study, omics platforms, advice on the management of informatics and bioinformatics and statistical analysis that were provided by industry partners.
Consortium operations
Concepts of project management differ widely across sectors, yet robust management can determine the success or failure of a project. So, what are the options? Experience suggests that oversight of multi-partner projects can be greatly facilitated by a small executive Consortium Management Board that is co-chaired by industry and academia principal investigators. This board might take responsibility for coordinating activities and for reporting progress to the funder. A larger Project Steering Group, comprising representatives of all consortium partners, can operate as the decision-making body, using a legally binding consortium agreement as its terms of reference. Investment in full-time project managers with experience in both academia and industry can reap dividends. As the ‘operators of operations’, project managers are essential for organizing meetings and maintaining a sharp focus on project timelines, deliverables and milestones, as well as for the robust management of high-risk work packages, and are increasingly appreciated as vital assets in the academic setting. Infusing a project with a momentum that will last for its lifetime can be critical to success — an exemplar operating structure is illustrated in Supplementary information S3 (figure).
Coordinating biological sampling at multiple sites
Traditionally, the acquisition of an extended portfolio of samples, including intensive sampling over short periods of time, has been the remit of small, single-centre experimental medicine studies. Accredited centres specializing in phase I clinical trials and contract research organizations have streamlined this process over several decades, facilitated by the proximity of patients to the lab, short times from venesection to processing of samples and tried and tested standard operating procedures (SOPs) for processing, storing and analysing fresh samples. Large, multi-centre studies present a challenge in this regard, necessitating sizeable efforts to harmonize the acquisition, processing and storage of prospectively acquired biological samples, and compromises in terms of sample range and assay complexity. Sampling is often limited in such multi-centre studies to the monitoring of drug safety using local accredited clinical laboratories.
To address the challenge of collecting samples from multiple sites, the RA-MAP Consortium established a hub-and-spoke network of seven academic laboratory hubs across England and Scotland that serve 28 patient-recruiting centres. This approach enabled the transportation of study samples from any patient-recruiting site to a lab within 4 hours of venesection. The requirements for sample transport, and for subsequent processing and storage, were clearly documented in study SOPs and protocols, with each step of the sample transport process carefully logged by study staff. Specifically designed sample tracking and logging software was placed in each of the hub laboratories along with the necessary hardware, including barcode scanners. SOPs for complex sample processing were developed by the relevant partners, scrutinized by industry partners, and refined before participant recruitment. This approach enabled high quality, barcoded aliquots of serum, peripheral blood cells, whole-blood RNA, RNA from lymphocyte and monocyte subsets purified in each laboratory, genomic DNA and urine to be processed and stored (Supplementary information S4 (figure)).
Combined input from academic and industry partners can ensure that sampling protocols are optimized to support immuno-phenotyping, as well as metabolomic, proteomic and transcriptomic analyses. In addition, sample procurement of this magnitude requires sample storage that facilitates long-term access to samples by the wider research community. Well-funded national repositories are ideally suited to provide this platform; in the UK, the UK Biobank provides such a resource.
Quality control
By centralizing sample analysis, single-centre studies can ensure the consistency and quality of sample processing and analysis of fresh material. However, when a broad portfolio of analytical platforms, analysis and expertise is required, there are several pragmatic approaches that can be adopted. Analysing all samples at a single sitting has obvious advantages, especially for transcriptomics, proteomics and metabolomics; when performing such assays at scale (for example, RNA extraction and microarray analysis), outsourcing can prove to be both cost effective and scientifically justifiable. A particular challenge for multi-centre studies is flow cytometric analysis, because cell staining protocols vary widely and hardware and machine settings can dramatically alter immune phenotypes, not to mention the varying expression profiles generated by different antibodies and fluorophores. To address this challenge, aliquots of cryopreserved peripheral blood cells can be distributed to designated laboratories that have expertise in the deep phenotyping of a single leukocyte subset. Flow cytometer configurations can be harmonized and batches of fluorescence-conjugated monoclonal antibodies can be purchased in bulk and distributed to each centre to minimize experimental variability across sites and between assays. In cases when samples are evaluated by flow cytometry at multiple time-points, additional measures can be adopted to minimize batch effects (for example, by applying corrections using standard tools such as COMBAT38).
Curating the data
Data are one of the defining metrics for determining the success of a consortium. Study participant data is often derived from multiple sources, especially when combining clinical, laboratory, imaging and omics datasets. As an example, the RA-MAP Consortium oversaw the recruitment of an inception cohort of patients with RA (participants in the TACERA study), who were followed from first presentation for up to 18 months, accumulating >1,280 baseline and follow-up visits from 275 study participants. The scale of the programme and the breadth and depth of data acquired necessitated investment in data cleaning, curating and storage, in accordance with data protection guidelines and sharing and communication policies, which needed to comply with requirements for patient confidentiality on the one hand while facilitating data analysis on the other. For the TACERA study, data were securely transferred and pseudo-anonymised using the OpenPseudonymiser package before undergoing a curation process, which included data integrity checks and semantic normalization. The curated and reformatted data were uploaded to TranSMART, a data warehouse that enables data access, visualisation, exploration and download to all members of the consortium (Supplementary information S5 (figure)). The local platform of TranSMART belonging to the RA-MAP Consortium has provided service to 82 users from multiple organizations and stores 37GB of data on the MRC eMedLab cloud computing facility, offering high performance computing capacity, a solution for long-term data sustainability and an appropriate environment for future meta-analyses by the rheumatology and immune-mediated inflammatory disease research communities.
Analysis of multi-omic data
When dealing with large volumes of data, challenges arise beyond storage. The RA-MAP Consortium’s portfolio of studies generated approximately 40 million analysis-ready data points from approximately 1 billion raw data points derived from more than 5,721 patient samples. The results of each omics platform investigation were stored in the TranSMART data warehouse, which provided an integrated view of omics platforms and linked clinical phenotypes, alongside a highly curated selection of pre-existing public data. TranSMART was chosen as it provided the RA-MAP Consortium and their partners with a unified, secure and, critically, sustainable research environment that offered on-board analytical capacity (including additional plugins such as SmartR39), data export and an R application programming interface, which enabled the use of a broad range of systems biology and machine learning methods for biomarker discovery.
Encouraging a sense of ownership of the data among all members of a consortium and overseeing the analysis by multiple parties require robust management. Agreement between partners and a clear alignment of goals between clinicians and the analytical teams, which might comprise biostatisticians, bioinformaticians and systems biologists from multiple partners, are essential for sustaining research momentum, maximizing output and for maintaining focus on pre-defined clinical questions. The RA-MAP Consortium found the adoption of a series of ‘lab meeting’-style teleconferences to be particularly productive. During these meetings, bioinformaticians could discuss the analysis of data on individual platforms and systems biologists could direct overall data integration while at the same time retaining a sharp focus on immunologically relevant research questions.
Publication policy
Communicating the outcome of large-scale consortia-driven projects is extremely important. The research community is familiar with manuscripts that are co-authored by large numbers of investigators; however, authorship requires further consideration when multiple parties have contributed equally. Discussions with publishers indicate that assigning authorship collectively to a consortium is generally acceptable; however, for operational and pragmatic reasons, either one or a few lead investigators can be designated as named and/or corresponding authors. To appropriately acknowledge the contributions of the consortium members in general, and the work of specific investigators in particular (such as graduate students, postdoctoral researchers, statisticians and bioinformaticians), separate documents listing specific contributions can be submitted to the relevant journal as supplementary information in accordance with journal policy. In addition, this approach provides a process whereby credentials for a larger number of academic investigators can be evaluated as part of the UK government’s Research Excellence Framework, a process whereby higher education institutions are allocated resources on the basis of research excellence. It is prudent for publication policies that address issues of authorship and author contributions to be defined from the outset of collaborative projects and included in the consortium agreement.
Meeting the milestones
Strategies for monitoring progress and outputs from large collaborative groups can vary from a remote approach (for example, annual written reports), which is typical of large EU consortia, to a more intense and actively managed relationship between funder and researcher. The latter option is the chosen method for the stratified medicine consortia funded by the MRC (TABLE 1), who opted for a formal and engaging face-to-face method of review. Members of the Consortium Management Board were requested to attend face-to-face reviews of milestones and deliverables by an independent panel of experts convened by the MRC on a 6-monthly basis. Progress was robustly and critically reviewed and additional targets established or revised when required and, on occasion, suggestions for additional analyses were given. Although challenging and highly supportive, this review process was uncompromising in its expectations of milestone delivery. During each review session, the panel of experts sought to challenge the science and experimental approach of the consortium, seeking solutions at every opportunity and strategies to mitigate risk. The funding body also gained from these review sessions through a deeper understanding of the steps required to develop operational and functional research consortia.
Future directions
Using the TACERA early RA cohort, the RA-MAP Consortium set out to stratify patients with RA on the basis of clinical findings (mapping patients to distinct trajectories), whole-blood transcriptomic profiles (uncovering major disease endotypes) and clusters of serum analytes that might guide treatment choices at the time of disease onset. At the time of writing, data from the TACERA study that fulfil these aims have been submitted for publication. In the near future, the RA-MAP Consortium aims to focus on integration of these stratification tools into clinical practice. The multi-omics approach of the RA-MAP Consortium strongly indicates that disease stratification might be multi-dimensional and require stratification of patients by use of an immunological toolkit, depending on the specific clinical question being asked. Once validated, the priority will be to apply the discovered stratification algorithms in a clinical trial setting.
Conclusions
The RA-MAP Consortium, comprising more than 140 investigators, has embarked on a stimulating journey, negotiating its way through difficulties at various points along the way. The successful operation of a large consortium of academic and industry investigators relies on several key factors: the development of a functional multi-partner research infrastructure; a strong pre-competitive collaborative ethos; an uncompromising emphasis on the generation of high-quality data; the nurturing of relationships for a productive research community; the sharing of insights about understanding the disease and its treatment; and the sharing of outputs through delivery of a publication plan that targets high-impact journals. Under the existing framework of project approvals, the RA-MAP Consortium will offer the wider research community access to data and samples as soon as our own investigations have been completed. We anticipate that access to samples might be granted as early as February 2018, and to data the following year. This process will be actively managed by a dedicated Data and Sample Access Committee in a transparent manner, facilitated by a structured application form.
Supplementary Material
Acknowledgements
The programme of research described in this Perspectives article was funded by the Medical Research Council (MRC), UK. The RA-MAP Consortium would particularly like to thank members of the MRC Immunity and Inflammation Stratified Medicine Steering Group and officers of the MRC, who have supported the work of the RA-MAP Consortium with unbridled enthusiasm.
Footnotes
Author contributions
A.P.C., M.R.B. and J.D.I. wrote the manuscript. A.P.C., S.B., F.B.-C. and A.W.P. researched data for the article. A.P.C., M.R.B., A.B., M.B., S.B., F.B.-C., B.A.F., C.S.G., P.E., M.E.R., N.G., R.H., S.H., M.F.M., I.B.M., S.R., A.W.P., F.P., D.P., R.R., A.R., M.A.S., D.S., B.T. and J.D.I. provided substantial contributions to discussions of content. A.P.C., M.R.B., A.B., S.B., F.B.-C., C.C., B.A.F., C.S.G., M.E.R., N.G., R.H., S.H., S.K., M.L., C.L., M.F.M., I.B.M., C.M.M., G.P., S.R., A.W.P., F.P., D.P., A.R., P.S.-K., M.A.S., D.S., P.C.T., B.T., W.T., D.V. and J.D.I. reviewed and/or edited the manuscript before submission.
Competing interests statement
A.P.C. declares that he has acted as a consultant for or received honoraria from BMS, Eisai, GSK, Janssen and Roche. For a full list of competing interests for all co-authors, see Supplementary information S6 (table).
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Further Information
EU Framework 7: https://ec.europa.eu/research/fp7/index_en.cfm
Horizon 2020: https://ec.europa.eu/programmes/horizon2020/
MRC eMedLab: www.emedlab.ac.uk
OpenPseudonymiser: https://www.openpseudonymiser.org/
Research Excellence Framework: http://www.ref.ac.uk/
TranSMART: http://transmartfoundation.org/overview-of-platform/
UK Biobank: http://www.ukbiobank.ac.uk/resources/
Contributor Information
Andrew P. Cope, Centre for Inflammation Biology and Cancer Immunology, School of Immunology and Microbial Sciences, Faculty of Life Sciences and Medicine, King’s College London, Great Maze Pond, London, SE1 1UL, UK
Michael R. Barnes, Queen Mary University of London, UK
Sarah Brockbank, Newcastle University, UK.
Francisco Bonachela-Capdevila, Janssen.
Claudio Carini, Pfizer.
Benjamin A. Fisher, University of Birmingham, UK
Carl S. Goodyear, University of Glasgow, UK
Paul Emery, University of Leeds, UK.
Michael R. Ehrenstein, University College London, UK
Neil Gozzard, UCB Pharma.
Ray Harris, Elsai.
Sally Hollis, AstraZeneca.
Catharina Lindholm, AstraZeneca.
Michael F. McDermott, University of Leeds, UK
Iain B. McInnes, University of Glasgow, UK
Christopher M. Mela, Roche
Gerry Parker, UCB Pharma.
Simon Read, Grunenthal.
Ayako Wakatsuki Pedersen, Newcastle University, UK.
Frederique Ponchel, University of Leeds, UK.
Duncan Porter, University of Glasgow, UK.
Ravi Rao, GlaxoSmithKline.
Anthony Rowe, Janssen.
Peter Schulz-Knappe, Protagen AG.
Matthew A. Sleeman, MedImmune
Deborah Symmons, University of Manchester, UK.
Peter C. Taylor, University of Oxford, UK
Brian Tom, University of Cambridge, UK.
Wayne Tsuji, Amgen.
Denny Verbeeck, Janssen.
John D. Isaacs, Newcastle University, UK
References
- 1.Jones A, Clifford L. Drug discovery alliances. Nat Rev Drug Discov. 2005;4:807–808. doi: 10.1038/nrd1856. [DOI] [PubMed] [Google Scholar]
- 2.Barnes MR, et al. Lowering industry firewalls: pre-competitive informatics initiatives in drug discovery. Nat Rev Drug Discov. 2009;8:701–708. doi: 10.1038/nrd2944. [DOI] [PubMed] [Google Scholar]
- 3.Yildirim O, Gottwald M, Schüler P, Michel MC. Opportunities and challenges for drug development: public-private partnerships, adaptive designs and big data. Front Pharmacol. 2016;7:461. doi: 10.3389/fphar.2016.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmeck B, Bertrams W, Lai X, Vera J. Systems medicine for lung diseases: phenotypes and precision medicine in cancer, infection, and allergy. Methods Mol Biol. 2016;1386:119–133. doi: 10.1007/978-1-4939-3283-2_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stingone JA, et al. Big and disparate data: considerations for pediatric consortia. Curr Opin Pediatr. 2017;29:231–239. doi: 10.1097/MOP.0000000000000467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morrison M. “A good collaboration is based on unique contributions from each side”: assessing the dynamics of collaboration in stem cell science. Life Sci Soc Policy. 2017;13:7. doi: 10.1186/s40504-017-0053-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilan KH. Opening up the ivory tower. Cell. 2007;129:847–850. doi: 10.1016/j.cell.2007.05.032. [DOI] [PubMed] [Google Scholar]
- 8.Melese T, Lin SM, Chang JL, Cohen NH. Open innovation networks between academia and industry: an imperative for breakthrough therapies. Nat Med. 2009;15:502–507. doi: 10.1038/nm0509-502. [DOI] [PubMed] [Google Scholar]
- 9.Schleidgen S, Klingler C, Bertram T, Rogowski WH, Marckmann G. What is personalized medicine: sharpening a vague term based on a systematic literature review. BMC Med Ethics. 2013;14:55. doi: 10.1186/1472-6939-14-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Association of the British Pharmaceutical Industry. Stratified medicine in the NHS: an assessment of the current landscape and implementation challenges for non-cancer applications. ABPI; 2014. http://www.abpi.org.uk/our-work/library/medical-disease/Documents/stratified_med_nhs.pdf. [Google Scholar]
- 11.Association of the British Pharmaceutical Industry. The stratification of disease for personalised medicines. Research driven recommendations to strengthen a unified UK strategy through a stakeholder alliance. ABPI; 2014. http://www.abpi.org.uk/our-work/library/medical-disease/Documents/strat_med.pdf. [Google Scholar]
- 12.Academy of Medical Sciences. Stratified, personalised or P4 medicine: a new direction for placing the patient at the centre of healthcare and health education. Academy of Medical Sciences; 2015. https://acmedsci.ac.uk/download?f=file&i=32644. [Google Scholar]
- 13.UK Trade and Investment. Unlock your global business potential UK stratified medicine. GOV.UK; 2013. https://www.gov.uk/government/uploads/system/uploads/attachment_data/file/301775/UK_Stratified_Medicine.pdf. [Google Scholar]
- 14.Medical Research Council. The Medical Research Council stratified medicine research initiatives. MRC; 2017. https://www.mrc.ac.uk/research/initiatives/stratified-medicine/ [Google Scholar]
- 15.Peltonen L, McKusick VA. Genomics and medicine. Dissecting human disease in the postgenomic era. Science. 2001;291:1224–1229. doi: 10.1126/science.291.5507.1224. [DOI] [PubMed] [Google Scholar]
- 16.Loscalzo J, Kohane I, Barabasi AL. Human disease classification in the postgenomic era: a complex systems approach to human pathobiology. Mol Syst Biol. 2007;3:124. doi: 10.1038/msb4100163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Botstein D, Risch N. Discovering genotypes underlying human phenotypes: past successes for Mendelian disease, future approaches for complex disease. Nat Genet. 2003;33:228–237. doi: 10.1038/ng1090. [DOI] [PubMed] [Google Scholar]
- 18.Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- 19.Giacomini KM, et al. Genome-wide association studies of drug response and toxicity: an opportunity for genome medicine. Nat Rev Drug Discov. 2017;16:70. doi: 10.1038/nrd.2016.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chatterjee N, Shi J, García-Closas M. Developing and evaluating polygenic risk prediction models for stratified disease prevention. Nat Rev Genet. 2016;17:392–406. doi: 10.1038/nrg.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berger B, Peng J, Singh M. Computational solutions for omics data. Nat Rev Genet. 2013;14:333–346. doi: 10.1038/nrg3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hood L, Balling R, Auffray C. Revolutionizing medicine in the 21st century through systems approaches. Biotechnol J. 2012;7:992–1001. doi: 10.1002/biot.201100306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Institute for Safe Medication Practices. The five rights: a destination without a map. Institute for Safe Medication Practices; 2007. http://www.ismp.org/newsletters/acutecare/articles/20070125.asp. [Google Scholar]
- 24.McInnes IB, Buckley CD, Isaacs JD. Cytokines in rheumatoid arthritis — shaping the immunological landscape. Nat Rev Rheumatol. 2016;12:63–68. doi: 10.1038/nrrheum.2015.171. [DOI] [PubMed] [Google Scholar]
- 25.Baretta Z, Mocellin S, Goldin E, Olopade OI, Huo D. Effect of BRCA germline mutations on breast cancer prognosis: a systematic review and meta-analysis. Medicine (Baltimore) 2016;95:e4975. doi: 10.1097/MD.0000000000004975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bunting SF, Nussenzweig A. End-joining, translocations and cancer. Nat Rev Cancer. 2013;13:443–454. doi: 10.1038/nrc3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lieber MR. Mechanisms of human lymphoid chromosomal translocations. Nat Rev Cancer. 2016;16:387–398. doi: 10.1038/nrc.2016.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Slamon DJ, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244:707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 29.Pauletti G, Godolphin W, Press MF, Slamon DJ. Detection and quantitation of HER-2/neu gene amplification in human breast cancer archival material using fluorescence in situ hybridization. Oncogene. 1996;13:63–72. [PubMed] [Google Scholar]
- 30.Maemondo M, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 31.Slamon DJ, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 32.Khagi Y, Kurzrock R, Patel SP. Next generation predictive biomarkers for immune checkpoint inhibition. Cancer Metastasis Rev. 2017;36:179–190. doi: 10.1007/s10555-016-9652-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siniard RC, Harada S. Immunogenomics: using genomics to personalize cancer immunotherapy. Virchows Arch. 2017;471:209–219. doi: 10.1007/s00428-017-2140-0. [DOI] [PubMed] [Google Scholar]
- 34.Ramamurthy C, Godwin JL, Borghaei H. Immune checkpoint inhibitor therapy: what line of therapy and how to choose? Curr Treat Options Oncol. 2017;18:33. doi: 10.1007/s11864-017-0476-y. [DOI] [PubMed] [Google Scholar]
- 35.Medical Research Council. MRC Strategic Review of Human Immunology. MRC; 2007. https://www.mrc.ac.uk/publications/browse/strategic-review-of-human-immunology/ [Google Scholar]
- 36.Gerlag D, et al. EULAR recommendations for terminology and research in individuals at risk of rheumatoid arthritis: report from the Study Group for Risk Factors for Rheumatoid Arthritis. Ann Rheum Dis. 2012;71:638–641. doi: 10.1136/annrheumdis-2011-200990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Innovative Medicines Initiative. The IMI funding model. IMI; 2017. http://www.imi.europa.eu/about-imi/imi-funding-model [Google Scholar]
- 38.Johnson WE, Rabinovic A, Li C. Adjusting batch effects in microarray expression data using Empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 39.Herzinger S, et al. SmartR: an open-source platform for interactive visual analytics for translational research data. Bioinformatics. 2017;33:2229–2231. doi: 10.1093/bioinformatics/btx137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.