Abstract
The calcium-activated chloride channel TMEM16A is intimately linked to cancers. Over decades, TMEM16A over-expression and contribution to prognosis have been widely studied for multiple cancers strengthening the idea that TMEM16A could be a valuable biomarker and a promising therapeutic target. Surprisingly, from the survey of the literature, it appears that TMEM16A has been involved in multiple cancer-related functions and a large number of molecular targets of TMEM16A have been proposed. Thus, TMEM16A appears to be an ion channel with a multifaceted role in cancers.
In this review, we summarize the latest development regarding TMEM16A contribution to cancers. We will survey TMEM16A contribution in cancer prognosis, the origins of its over-expression in cancer cells, the multiple biological functions and molecular pathways regulated by TMEM16A. Then, we will consider the question regarding the molecular mechanism of TMEM16A in cancers and the possible basis for the multifaceted role of TMEM16A in cancers.
Graphical Abstract.
1. Introduction
Ion channels are critical for regulating ion homeostasis in any cells and thus are involved in a wide variety of physiological processes. In recent decades, a growing literature describes a contribution of ion channels in all aspects or hallmarks of cancer [1,2]. Thus, ion channels could represent an unprecedented reservoir of unused therapeutic targets. Investigating their molecular mechanisms in cancer cells is crucial to both improve our understanding of the contribution of ion channels in their cancer-related mechanisms and to better define drugs and conditions for using ion channels as therapeutic targets in cancers.
TMEM16A, also known as ANO1, DOG1, ORAOV2, or TAOS2, is a calcium-activated chloride channel (CaCC) [3–5] with physiological functions in epithelial tissues, exocrine glands, dorsal root ganglion neurons and smooth muscle [6–8]. Before its molecular identification as CaCC in 2008, TMEM16A/DOG1 was described as a biomarker for gastro-intestinal squamous cancer (GIST) [9]. Since then, a number of investigations has commented its over-expression in various cancers including Head and Neck Squamous Cancer Cells (HNSCC), Breast, Prostate, Pancreatic, Gastric, Parotid and Colorectal (CRC) cancers.
However, despite the consensus regarding its over-expression in cancers, a surprising range of the biological and molecular functions associated to TMEM16A emerged from the literature suggesting that TMEM16A has a multifaceted role in cancers. In this review, we will summarize evidence regarding TMEM16A expression in cancers and its associated biological and molecular functions. We will then review our understanding of the molecular mechanism of TMEM16A in cancers and propose hypothesis that could explain the multifaceted role observed for TMEM16A in cancers.
2. TMEM16A expression in cancer
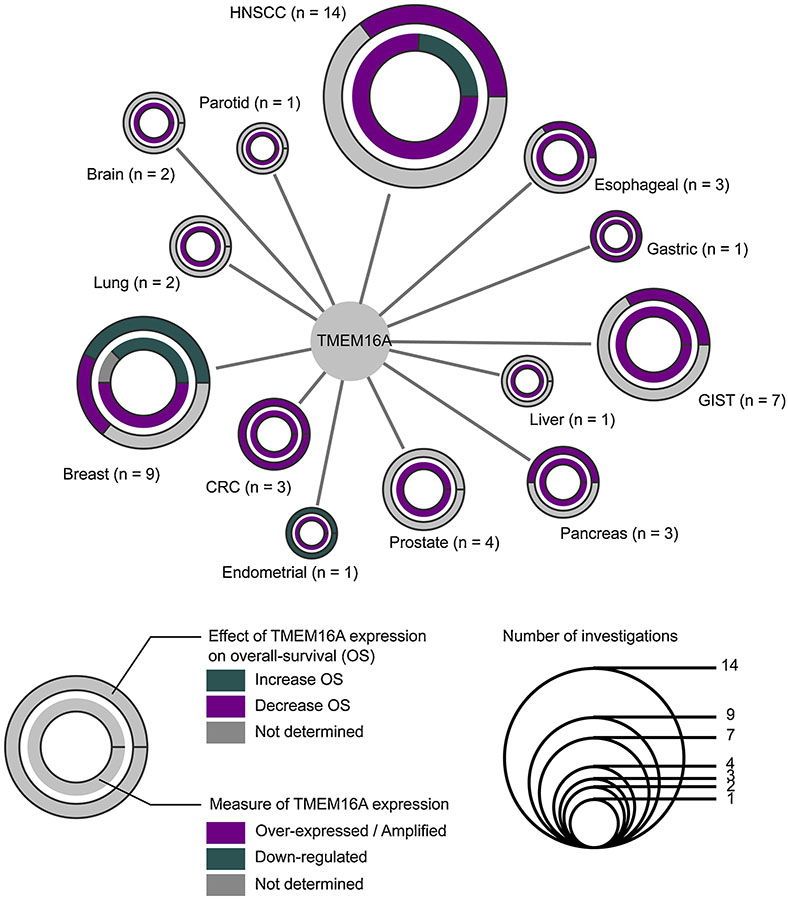
TMEM16A is intimately associated to cancer. Historically known as DOG1 – a biomarker of Gastro Intestinal Squamous Carcinoma (GIST) [9], TMEM16A has now been investigated in multiple different cancers including HNSCC [10–23], Breast [14,24–32], Brain [33,34], CRC [35–37], Esophageal [38–40], Endometrial [41], Gastric [42], GIST [10,43–47], Liver [48], Lung [49,50], Pancreatic [51–54], Parotid [55] and Prostate cancers [56–58] (Figure 1). In the wide majority of these investigations,TMEM16A is over-expressed or amplified compared to its expression in healthy tissues (Figure 1) suggesting that TMEM16A over-expression could represent a conserved mechanism in oncogenesis.
Figure 1. Expression and contribution to the overall survival of TMEM16A in cancers.
Node size is defined by the number of different articles investigating TMEM16A expression or amplification and/or TMEM16A contribution to the overall survival. The number of different articles per cancer is noted in parenthesis. Inner circle represents the distribution of results obtained from the literature per cancer types regarding TMEM16A expression. TMEM16A is either over-expressed or amplified (magenta), down-regulated (green) or not affected (grey). Outer circle represents the distribution of results obtained from the literature per cancer types regarding the contribution of a high TMEM16A expression to the overall survival (OS). A high TMEM16A expression could either reduce the OS (magenta), increase the OS (green) or have no effect or have not been determined (grey).
Interestingly, TMEM16A over-expression is not homogeneous and detailed investigations have found that TMEM16A is not over-expressed in some subtypes of various cancers such as Human Papilloma Positive (HPV)-positive HNSCC, Estrogen Receptor (ER)-positive and Human Epidermal Growth Factor Receptor 2 (HER2)-positive breast cancers and Pancreatic Neuro-Endocrine tumors [13,18,30,31,52]. Additionally, Shiwarski and colleagues have demonstrated that TMEM16A expression can be modulated depending on the progression of the tumor. They show that primary tumors exhibit a high level of TMEM16A whereas metastasis from lymph nodes have a low expression of TMEM16A [22]. This interesting observation has to be counterbalanced by other studies demonstrating high expression of TMEM16A in liver metastasis of CRC [35] or an amplification of the TMEM16A gene in metastasis from HNSCC [23]. It is also notable that TMEM16A over-expression in HNSCC could be predictive of the presence of distant metastasis [11].
At the clinical level, TMEM16A over-expression has been associated to multiple clinical parameters. The most recurrent one is the overall survival. In most investigations, TMEM16A over-expression is correlated with a poor prognosis [13,14,16,19–21,25,27,32,35,39,42,44,45] with the exception of endometrial cancer, HER2-positive and ER-positive breast cancers [30,31,41] (Figure 1). In addition, TMEM16A over-expression has also been correlated to the tumor size [10,45], the presence of distant metastasis [11,23], the recurrence rate [45,49], the improvement of the clinical outcomes by the chemotherapy [32] or the clinical stage of cancer [34,41,44,56]. These findings suggest that TMEM16A represents a promising biomarker for various cancers. TMEM16A could also represent a valuable therapeutic target as its pharmacological inhibition of TMEM16A reduces tumor growth in vivo [59] and promotes the sensitivity of other chemotherapy [60,61] and its expression could be correlated to the improvement of clinical outcomes by chemotherapy [32].
Thus, over-expression of TMEM16A is a frequent feature observed in multiple cancers suggestive of a conserved mechanism for the promotion of carcinogenesis. However, further investigations will be required to better characterize TMEM16A expression and its contribution to clinical prognosis in specific cancer subtypes in order to improve future therapeutic strategies targeting TMEM16A.
3. A multifaceted biological role of TMEM16A in cancer
In addition to its over-expression in multiple cancers and its contribution to several clinical features, TMEM16A has been associated to multiple biological functions in cancer cells. Hanahan and colleagues have proposed several features or essential functions that could define a cancer cell [62]. While these functions could be seen as reducing the complexity of cancer cells, we found it interesting to overlap some of these hallmarks with different cancer functions attributed to TMEM16A in the literature.
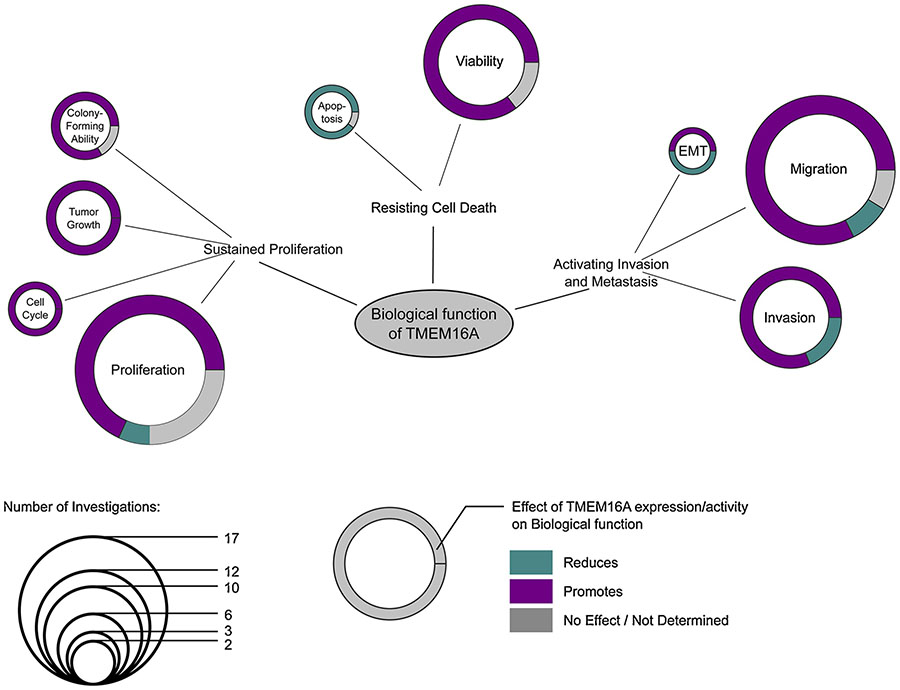
The contribution of TMEM16A to the hallmark “Sustained Proliferation” is the most represented in the literature. Thus, molecular or pharmacological silencing of TMEM16A in cancer cells could impair proliferation [30,32,34,36,40,48,50,54,58,63–68], cell cycle [37,48,63], tumor growth in vivo [14,25,32,48,50,56,58], or the ability to form colony [25,50,56,63] (Figure 2).
Figure 2. Association of TMEM16A expression/activity with hallmarks of cancer.
For each hallmark of cancer as defined by Hanahan et al. [62], we associated biological functions reported in the literature to be modulated by TMEM16A expression/activity. Node size of each biological function is relative to the number of different cancer cell lines for which the contribution of TMEM16A expression/activity has been reported. The outer colored circle represents the distribution of conclusions obtained for each biological functions. TMEM16A expression/activity could either promotes (magenta), reduces (green) or have no effect (grey) on the biological function.
The contribution of TMEM16A has also been investigated with respect to the resistance to cell death of cancer cells [69]. Most of investigations across multiple cancer types have demonstrated that TMEM16A expression sustains cancer cell viability [13,22,25,30,32,37,37,56,57,61,63,64,66,70] and impair apoptosis [58,63] (Figure 2).
The contribution of TMEM16A to the hallmark “Invasion and Metastasis” has also been extensively investigated. Most investigations conclude that TMEM16A expression promotes cancer cell migration and invasion [11,17,21,33,34,36,37,42,48,50,52,54,56,63,64,66,68,70–72] (Figure 2). However, one report suggests a different role of TMEM16A on this hallmark [22]. In this investigation, Shiwarski and colleagues show that in HNSCC cancer cells, TMEM16A expression inhibits cancer cell migration and invasion while also preventing the epithelial-to-mesenchymal transition (EMT), a critical process for cancer cells to develop metastasis and secondary tumors [22]. A recent study, however found that TMEM16A expression is required for EMT, migration and invasion of gastric cancer cells [42]. Several other investigations have concluded that TMEM16A expression promotes HNSCC cancer cell migration or invasion [11,21].
Thus, it seems reasonable to conclude that TMEM16A is involved in cancer cell proliferation, apoptosis resistance, migration and invasion. However, there is heterogeneity for TMEM16A over-expression in various cancer subtypes as well as heterogeneity could be observed for the role of TMEM16A in cancer cells.
4. Multiple origins of TMEM16A over-expression in cancer
The multifaceted role of TMEM16A in cancers may reflect the origins of TMEM16A over-expression in cancer cells. Several mechanisms have been suggested to explain the over-expression of TMEM16A.
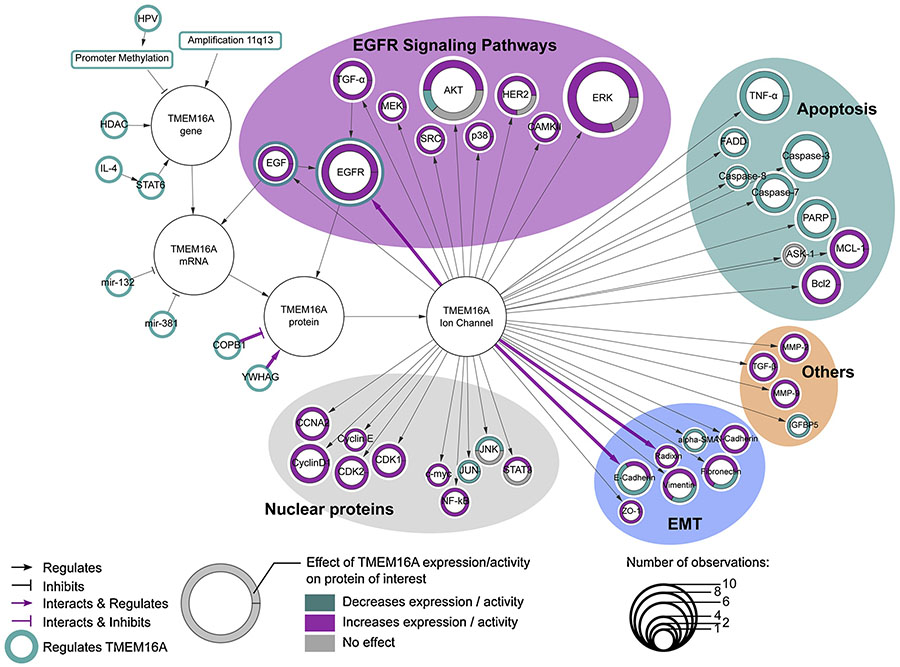
At the genomic level, amplification of the locus 11q13 is the most frequently mechanism associated to TMEM16A over-expression in cancers (Figure 3). This amplicon has been observed in various cancers such as GIST, HNSCC, breast cancers, esophageal carcinoma and lung cancers and is correlated to an increase of the number of copies of TMEM16A gene [11,13,16,18,20,23,25,38,40,49]. Hypermethylation of the promoter region of the TMEM16A gene has been observed in HPV-positive HNSCC and in distant metastasis of HNSCC and it correlates with a low expression of TMEM16A [13,22] suggesting that hypermethylation of TMEM16A promoter could repress TMEM16A transcription. Thus, even if direct evidence of the hypomethylation of this particular genomic region in cancer has not been reported, it is conceivable that hypomethylation of TMEM16A promoter could be a putative mechanism contributing to its over-expression in cancer. In prostate cancer cells, Histone Deacetylase 3 (HDAC3) promotes the expression of TMEM16A [57] (Figure 3). In non-cancerous cells, TMEM16A expression is induced by extracellular ligands such as Epidermal Growth Factor (EGF) or interleukin-4 (IL-4) [73,74]. Analysis of TMEM16A promoter revealed that Signal Transducer and Activator of Transcription 6 (STAT6) mediates the IL-4 induced TMEM16A expression (Figure 3) [75]. Recently, the same group has demonstrated that TMEM16A gene transcription is regulated by Glioma-associated-oncogenes (Gli) proteins [76]. In a recent report, EGF promotes TMEM16A expression in breast cancer cells through the Epidermal Growth Factor Receptor (EGFR)-STAT3 pathway [32]. Thus, TMEM16A expression could also be modulated by soluble factors in the tumor micro-environment.
Figure 3. Regulation of TMEM16A and its molecular targets in cancers.
Each node correspond to a protein described as either a regulator or a downstream target of TMEM16A expression/activity in cancers. Proteins represented with a thin green bordered node have been described as regulators of TMEM16A expression or activity. Proteins represented as a node with a thick colored border have been described as molecular targets of TMEM16A expression/activity. The node size of target proteins is relative to the number of different cell lines in which the contribution of TMEM16A on the expression/activity of the protein of interest has been investigated. The thick outer colored circle represents the distribution of conclusions made for each targets of TMEM16A. TMEM16A expression/activity could either promotes (magenta), inhibits (green) or has no effect (grey) on the protein expression/activity.
At the mRNA levels, miRNA-132 and 381 repress TMEM16A expression (Figure 3). These miRNA species have been found to be down-regulated in gastric and colorectal cancers respectively [35,42] and their down-regulation correlates with a higher expression of TMEM16A expression.
TMEM16A expression is also be regulated at the post-translational level. In glioblastoma cells, TMEM16A has been found to interact with the Coatomer protein complex subunit beta 1 (COPB1) and tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein gamma (14-3-3γ) (Figure 3). Such interaction could inhibit or promote the trafficking of TMEM16A to the plasma membrane [33,77]. Additionally, TMEM16A interaction with EGFR has been found to improve the stability of TMEM16A protein [61].
Thus, it appears that TMEM16A over-expression in cancer has multiple origins supporting the idea that TMEM16A over-expression is an important feature for cancer development that could be achieved through multiple ways.
5. Molecular targets of TMEM16A in cancers
Over the years, the modulation of TMEM16A expression or activity in cancer cells has been associated with the up- or down-regulation of different proteins, making it possible to create a network of protein targets of TMEM16A in cancer cells (Figure 3). While most of these targets are probably indirectly modulated by TMEM16A, it is interesting to observe that they fall into several groups based on the biological functions they are associated with.
Alteration of TMEM16A expression affects proteins dedicated to transcription or cell division (Figure 3). For example, cell cycle proteins such as Cyclin A2 (CCNA2), Cyclin D1, Cyclin E, Cyclin-dependent Kinases 1 and 2 (CDK1 and CDK2) are up-regulated in cancer cells with increased TMEM16A expression [34,63]. In addition, the activation of some transcription factors such as NF-κB, c-myc and STAT3 are positively correlated with TMEM16A expression whereas the activity of transcription factors JUN and JNK are negatively correlated with TMEM16A expression [32,34,58,78]. Thus, these group of targets modulated in cancer cells with increased expression of TMEM16A could contribute to the biological role of TMEM16A in cancer cell proliferation.
An other group of targets includes proteins related to apoptosis (Figure 3). Indeed, the activity of proapoptotic proteins such as caspase-3, 7, 8, Fas-associated protein with death domain (FADD) and Tumor Necrosis Factor-α (TNF-α) are down-regulated while those of anti-apoptotic proteins B Cell Lymphoma 2 (Bcl-2) and Myeloid Cell Leukemia-1 (MCL-1) are up-regulated in cancer cells with increased expression of TMEM16A [25,58], indicating that TMEM16A promotes cancer cell viability and inhibits apoptosis.
At the molecular level, variety of TMEM16A functions have been implicated (Figure 3). For example, TMEM16A could either promote or repress E-Cadherin, vimentin or fibronectin [22,42]. In addition, while in HNSCC, TMEM16A expression promotes the expression of the pro-epithelial marker Zonula Occludens-1 (ZO-1) and represses the mesenchymal marker α-Smooth Muscle Actin (α-SMA) [22], in gastric cancer cells, TMEM16A promotes the expression of the mesenchymal marker N-Cadherin [42]. It is clear that TMEM16A has an impact on several of proteins involved in EMT [22,42]. Several proteins such as radixin, Tumor Growth Factor-β (TGF-β), matrix metallopeptidases 2 and 9 (MMP2 and MMP9) associated with cancer cell migration or invasion are affected in cancer cells with increased TMEM16A expression (Figure 3) [22,34,42].
Epidermal growth factor receptor (EGFR) is a tyrosine kinase that has been extensively investigated in various cancers [79–83]. In cancer cells, most recurrent molecular targets associated with TMEM16A are also associated with EGFR signaling pathways (Figure 3). Initially, Britschgi and colleagues report that TMEM16A expression is able to regulate EGFR constitutive phosphorylation and associated signaling pathways such as SRC, Akt, ERK and CamKII in HNSCC and breast cancer cells [25]. They also demonstrated that TMEM16A promotes autocrine secretion of the EGFR ligands EGF and TGF-α [25]. Then, while elucidating the interactome of TMEM16A in HNSCC cancer cells, Bill and colleagues found interaction of TMEM16A and EGFR independent of either EGFR or TMEM16A activities [61]. They also reported that EGFR and TMEM16A show mutual regulation of their expression levels promoting their stability [61]. Interestingly, the pharmacological inhibition of TMEM16A enhanced the sensitivity to EGFR-based therapeutic strategies in HNSCC cancer cells strengthening the relationship between EGFR and TMEM16A [60,61]. In breast cancer, EGF-induced EGFR-STAT3 signaling pathway promotes TMEM16A expression [32]. Recently, we found that TMEM16A is required for promoting EGF-induced EGFR signaling in a pancreatic cancer cell line [52]. Interestingly, while confirming the close relationship between TMEM16A and EGFR, we also found that Akt and ERK signaling are not affected by TMEM16A expression suggesting the involvement of a different EGFR-related signaling pathway [52]. Moreover, TMEM16A expression has a profound effect on the phosphoproteome of pancreatic cancer cells and impairs the EGFR-related signaling pathways [52]. TMEM16A can also modulate the expression of HER2, an other member of human epidermal growth factor receptor (HER), thereby enlarging the role of TMEM16A in signaling involving tyrosine kinases receptors [78,84]. Thus, at the molecular level, TMEM16A also has multifaceted roles on a variety of molecular targets that converge to the multiple oncogenic function regulated by TMEM16A.
6. Understanding the mechanism of TMEM16A in cancers
The multifaceted molecular and cellular roles of TMEM16A in cancer cells raise the question regarding the molecular mechanism for TMEM16A.
As TMEM16A is an ion channel, it is tempting to speculate that TMEM16A exerts its pro-oncogenic function through chloride transport. However, TMEM16A-related chloride conduction may not be mandatory for its pro-oncogenic effects [13,54,63], as indicated by the difference observed between the use pharmacological blockers of TMEM16A and the molecular silencing of TMEM16A protein expression. Thus, TMEM16A blockers T16Ainh-A01 or CaCCinh-A01 does not reproduce the effect observed using siRNA on the migration of the pancreatic cancer cell line BxPC-3 [54]. Similarly, the use of T16Ainh-A01 does not reproduce the effect observed with the silencing of TMEM16A expression on the viability of the colorectal cancer cell line HCT-116 [63]. Conversely, the reduction of HNSCC cell viability by the use of CaCCinh-A01 is not observed when silencing TMEM16A expression [13]. However, other investigations observed that TMEM16A blockers inhibit cancer cell proliferation and/or migration to the same extent as the molecular silencing of TMEM16A [11,25,33,37,58–60,64]. Moreover, over-expression of non-conductive TMEM16A mutants (R621E, K668E and K610A) failed to reproduce the increase of proliferation observed with wild-type TMEM16A [14,25,32].
Taken together, these studies suggest that both the protein expression and the ion conduction of TMEM16A may contribute to its pro-oncogenic functions, consistent with the idea of a very multifaceted role of TMEM16A in cancers.
6.1. TMEM16A-interacting partners
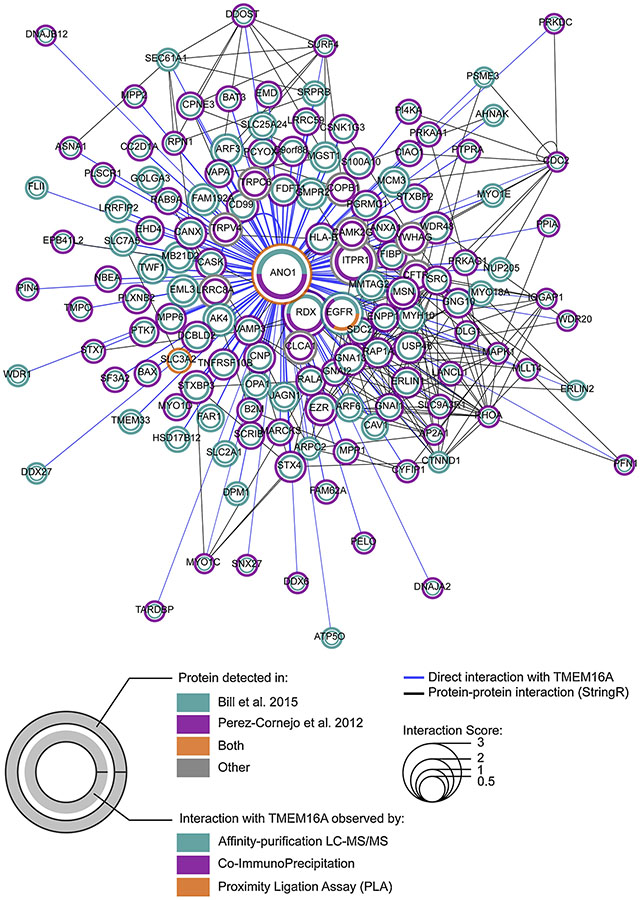
Independent or synergistic to its ion channel function, TMEM16A protein may modulate cancer cell function by interacting with other proteins as described for other ion channels [85–91]. In an attempt to identify TMEM16A-interacting partners, two independent investigations have been conducted to determine the TMEM16A interactome [61,92].
TMEM16A has been found to interact with the Ezrin-Radixin-Moesin (ERM) network in HEK cells over-expressing TMEM16A (Figure 4) [92]. The ERM network is an intermediate in the connection between the actin cytoskeleton and the plasma membrane. This network may regulate cancer cell migration, invasion or adhesion, raising the possibility that the TMEM16A interaction with components of ERM network could modulate cancer cell migration [93,94]. TMEM16A interaction with radixin was observed by co-immunoprecipitation in HNSCC cell lines and has been found to be mediated by the phosphorylation of S970 in the TMEM16A C-terminal domain [22]. Interestingly, while the over-expression of TMEM16A inhibits EMT and promotes proliferation in T24 cell line, over-expressing TMEM16A S970A (a mutant not interacting with radixin) fails to inhibit EMT but still promotes T24 proliferation [22]. This suggest that TMEM16A interaction with the ERM network could contribute to one aspect of its biological functions in cancer cells. The relationship between TMEM16A and other components of the ERM network (ezrin and moesin) has not been elucidated yet in the context of cancer.
Figure 4. TMEM16A-interacting partners.
Each TMEM16A interacting partner found in the literature has been represented as a single node connected to TMEM16A by a blue edge. The node size and edge width are relative to the interaction score calculated as the relative ascending rank order obtained in each of interactome published [61,92] + 1 for each data demonstrating a direct interaction between TMEM16A and the partner of interest by either co-immunoprecipitation (co-IP) or proximity-ligation-assay (PLA). Network obtained has been enriched for high confident protein-protein interaction (PPI) (confidency = 0.8, additional interactors = 0) using StringApp (version 1.4.2) integrated in Cytoscape 3.7.0. Then, the network layout have been obtained by using the integrated prefuse-direct force layout algorithm. The outer circle of each node represents the detection of TMEM16A-interacting protein in the interactome published by Bill et al. (green), the one published by Perez et al. (magenta), both (orange) or neither of them (grey). The inner circle represent the method used to demonstrate the interaction between TMEM16A and the protein of interest (proteomic approaches = green, co-IP = magenta or PLA =orange).
In HNSCC cancer cells, characterization of the TMEM16A interactome has revealed that TMEM16A interacts with EGFR (Figure 4) [61], supporting the notion that TMEM16A is strongly associated to EGFR signaling pathways in cancer cells [25,52,61]. The TMEM16A/EGFR interaction was also observed by proximity-ligation assay (PLA) in pancreatic cancer cells (Figure 4) [52]. Mutagenesis experiments revealed that TMEM16A/EGFR interaction is independent of TMEM16A ion channel or EGFR kinase activities and is mediated by the juxta-membrane domain of EGFR. While the direct binding site of EGFR on TMEM16A has not been identified, the C-terminal domain of TMEM16A has been found to be non-essential suggesting a different binding site than that for radixin [61]. EGFR interaction with TMEM16A is associated to proliferation of HNSCC and pancreatic cancer cell migration [52,61]. Compiling studies of TMEM16A interaction with EGFR and radixin, we can thus associate TMEM16A protein with two different partners with distinct and unique binding sites for the modulation of at least two different cancer functions, indication that the multifaceted role of TMEM16A in cancer may be due to multiple interactions on multiple locations on the TMEM16A protein.
In addition of the ERM network, Perez and colleagues found that TMEM16A interacts with the SNARE protein complex (including Syntaxin-4, Syntaxin-7, VAMP3 and STXBP3) (Figure 4) [92]. While these proteins might be associated with TMEM16A biogenesis and trafficking, the interaction of TMEM16A with this particular protein complex could hypothetically be associated with the TMEM16A-dependent EGF and TGFα secretion observed in breast and HNSCC cancer cells [25]. So far, no experiments have been performed to elucidate the relationship between the vesicle trafficking network and TMEM16A.
Survey of the literature reveals that additional proteins interact with TMEM16A. In cancer cells, COPB1 and 14-3-3γ have been found to interact with TMEM16A (Figure 4) and to inhibit or promote TMEM16A trafficking at the plasma membrane respectively [33,77]. 14-3-3γ interacts with the residue T9 of TMEM16A [33], thus providing additional evidence of multiple protein binding sites on TMEM16A.
In non-cancer cells, TRPC6, TRPV4, CFTR, LRRC8A, CLCA1, CAMK2G and inositol triphosphate receptor 1 (IP3R1) interact with TMEM16A and regulate its activity (Figure 4) [8,95–102].
It is interesting to note that from TMEM16A interactomes defined by a proteomic approach, only TMEM16A itself and SLC3A2 have been found in both studies [61,92]. This absence of overlap between both proteomics approach could be attributed to a difference in the techniques, the methodology, or the biological model. Moreover, from the list of TMEM16A-interacting partners obtained by the proteomics approach, only radixin and EGFR have also been observed as TMEM16A-interacting partners in other investigations conducted in cancer cells [22,52], suggesting that great effort will be required to validate all of these putative partners of TMEM16A and to characterize their interaction and function in cancer cells.
6.2. TMEM16A ion conduction
As discussed above, the pharmacological inhibition of TMEM16A using various molecules inhibits several cancer cell functions such as proliferation, migration or apoptosis resistance [13,25,33,37,52,54,58–60,63–66,68,70,72,103]. This suggests that TMEM16A channel activity contributes to the multifaceted role of TMEM16A in cancer. How might TMEM16A-mediated chloride current mediate pro-oncogenic functions?
Ion fluxes are essential to maintain osmolarity and intracellular water content and ion composition in every single cell. In the context of cancer, ion fluxes and associated changes in the intracellular water content regulate cell volume and thus are critical for morphological changes occurring during cell division, migration, invasion, EMT or for preventing cell death [104,105]. Thus, osmoregulation represents a mechanism that associates ion fluxes and different cancer cell functions and could, at least in part, explain the multifaceted role of TMEM16A. In agreement with a contribution of TMEM16A to cancer cell osmoregulation, several reports have observed that both molecular silencing and TMEM16A inhibition are able to affect cancer cell volume or morphology [11,22,71].
Lysine deficient kinases (WNK) are considered to be intracellular chloride sensors [106,107] and regulate cell volume, proliferation and EMT [108,109]. In cancer cells, WNK kinases regulate PI3KAkt, TGF-β and NF-kB signaling and autophagy [110,111]. In pancreatic cancer cells, we have found that both TMEM16A expression and EGFR activation modulate the phosphorylation of WNK1 kinases (Crottes et al. Unpublished data). This suggest that TMEM16A-related chloride current may mediate a part of its pro-oncogenic effects through WNK kinases signaling. Further experiments are required to validate this molecular mechanism.
Recently, our group observed that TMEM16A-related chloride currents are critical for the distribution and clustering of Phosphatidylinositol 4,5-bisphosphate (PIP2) at the plasma membrane (PM) [112,113]. In cancer cells, lipid distribution in microdomains at the PM is critical for cancer cell proliferation and migration [80]. It is also a critical determinant for the interaction of ion channels with various receptors such as EGFR at the surface of cancer cells [80]. PIP2 is the precursor of inositol triphosphate (IP3) obtained by the cleavage of PIP2 by phospholipase C (PLC) (which can be activated by EGFR). IP3 is a well characterized second messenger that activates IP3R which initiates intracellular calcium (Ca2+) release known to be a regulator of TMEM16A activation (discussed below) [98].
Our recent study linked TMEM16A activity, EGFR and IP3-dependent intracellular Ca2+ release to pancreatic cancer cell migration [52]. Combined with the known interaction of TMEM16A with IP3R [97,98], these results allowed us to propose the following mechanism in which TMEM16A-related chloride currents promote PIP2 clustering at the PM in close vicinity to the EGFR/TMEM16A/IP3R macro-complex. This restricted spatial localization of all three molecules will promote the release of IP3 from the PM and subsequently intracellular Ca2+ release in response to EGFR activation. The spatial localization and the dynamics of these interactions need to be further investigated, to test this hypothesis as the basis for unifying the ion conduction and physical interaction of TMEM16A into one molecular mechanism to define the multifaceted role of TMEM16A in cancer cells.
Similar to the multiple roles observed for TMEM16A protein interactions in cancer cells, TMEM16A-mediated ion conduction may have multiple roles in cancer cells by modulating osmoregulation, WNK kinases activities or lipids distribution. Thus, both the TMEM16A physical interaction and its biophysical properties contribute to the multifaceted role observed for TMEM16A in cancer.
6.3. Regulation of TMEM16A chloride transport
As we discussed above, TMEM16A protein could be modulated at the transcriptional, translational or post-transcriptional levels in cancer cells. TMEM16A ion conduction could also be modulated by multiples mechanisms.
Since its identification as a CaCC [4,5,73], several regulatory mechanisms of TMEM16A chloride channel have been identified [114]. TMEM16A ion conduction can be affected by direct binding of calcium [115–118], membrane potential [115,119,120], lipid environment [112,113,121,122], phosphorylation of serine residues, [99,100,122], the presence of protons or temperature alteration [123,124]. As discussed previously, its interaction with other ion channels and kinases also modulates its ion conduction [8,95–98,101,102].
Taken individually, each of these regulatory mechanisms of TMEM16A is relevant in cancer. For instance, membrane potential changes in tumors cells has been observed since the late 1950’s [125]. Lipid environment is essential for the proper function of ion channels in cancer cells [80]. As in non-cancerous tissues, finely tuned regulation of Ca2+ homeostasis is critical for cancer cells [126]. Local extracellular acidification is regulated by ion channels to promote extracellular matrix digestion and cancer cell invasion [127,128].
However, although pharmacological approaches have emphasized the importance of TMEM16A-dependent chloride transport for cancer-related functions (as discussed above), the investigation of the molecular mechanisms regulating TMEM16A chloride currents in cancer cells remains largely unknown.
Importantly, one cannot entirely rely on extrapolation of regulatory mechanisms of TMEM16A observed in non-cancerous cells to the context of cancer. For example, while the activation of TMEM16A-mediated chloride current occurs downstream of either IP3R-dependent intracellular Ca2+ release or extracellular Ca2+ fluxes mediated by TRPC6, TRPV4 or ORAI1 in non-cancerous cells [8,98,129–131], TMEM16A is upstream of the calcium signaling and controls both intracellular Ca2+ release and store-operated Ca2+ entry induced by EGFR activation in pancreatic cancer cells [52].
Investigating the different known activators of TMEM16A in the context of cancer will provide valuable inputs to improve our understanding of the function and regulation of TMEM16A in cancer cells.
7. Elucidating the multifaceted functions of TMEM16A in cancers.
We have discussed above how the multifaceted role of TMEM16A in cancers could be explained by either protein-protein interaction, ion conduction or both. However, it is interesting to speculate whether, in a single cancer cell, TMEM16A could contribute to all of these functions simultaneously or asynchronously. The resolution of this question has not yet been achieved as several reports indicate that TMEM16A was unable to concomitantly regulate both proliferation and migration in the same cancer cells [11,21,22,42,54] while other reports demonstrate that TMEM16A could regulate both proliferation and migration in the same cancer cells [34,48,50,56,63,68].
7.1. A cell-specific mechanism
In their recent review, Wang and colleagues proposed that multifaceted functions of TMEM16A could be explained by a cell-specific role of TMEM16A that depends on the cellular environment [132]. Thus, the TMEM16A oncogenic function is dependent on the cancer cell type under investigation and its protein expression profile. While it is an interesting hypothesis that could explain why TMEM16A could promotes proliferation, migration or cell death depending on the cancer cell types, this hypothesis may not account for the multiple function that TMEM16A could exert in a single cancer cell.
7.2. TMEM16A and the plasticity of cancer cells
Composition of cellular membranes is highly heterogeneous. In any cellular membrane, micro-domains such as lipid rafts may have different lipid and protein compositions. These micro-domains are involved in intracellular signaling and inter-organelle communications.
In cancer cells, ion channels may exert intriguing functions on lipid rafts, thus promoting the formation and the stability of macro-complexes via their interaction with various receptors, other ion channels and signaling molecules [80,86,91]. By doing so, ion channels contribute to sustained downstream signaling pathways associated to these receptors or proteins. Disturbing ion channel function or expression or lipid composition will abrogate the formation of such macro-complexes and inhibit the associated downstream signaling pathways. These ion channels-dependent complexes could form in response to the sensing of extracellular cues such as extracellular matrix or growth factors. Thus, ion channels could promote cancer cell adaptive response by the formation of such macro-complexes [86,90,133].
As discussed above, TMEM16A promotes PIP2 clustering at the plasma membrane [112] and interacts with various receptors (EGFR), signaling molecules (the ERM network) or other ion channels expressed in different sub-cellular compartments (IP3R) [22,52,61,92,98]. Thus, we hypothesize that TMEM16A may contribute to the formation of such macro-complexes associated to lipid rafts (enriched in PIP2) to promote the adaptive response of cancer cells to various stimuli.
Thus, either by being involved in signaling confined to multiple lipid rafts at different sub-cellular locations or by contributing to adaptive response to extra- or intra-cellular cues, TMEM16A may regulate various signaling pathways and biological functions in a single cancer cell thus leading to the multifaceted role of TMEM16A in cancer cells.
Supporting this hypothesis, our recent investigation in pancreatic cancer cells observed that TMEM16A interacts with EGFR and sustains ligand-dependent EGFR activation and signaling pathways by initiating intracellular Ca2+ release and store-operated calcium entry (SOCE) [52]. This study provides an example of a TMEM16A-related macro-complexes (TMEM16A/EGFR) involved in the sensing of extracellular growth factor (EGF) and involving the intracellular inter-organelle communication (Intracellular Ca2+ release + SOCE). Further experiments will be required to define each aspect of this particular molecular mechanism of TMEM16A and determine if by this mechanism, TMEM16A can indeed regulate multiple oncogenic functions in the same cancer cell.
8. Perspectives and Future directions
In this review, we summarize the multifaceted role of TMEM16A in cancer. TMEM16A over-expression is a common feature of multiple cancers suggesting that TMEM16A is a promising biomarker.
However, a survey of the literature suggests that the role of TMEM16A could not be attributed to a single function or molecular mechanism. Whether by the origins of TMEM16A over-expression, signaling pathways, proteins or cancer cell functions regulated by TMEM16A, all of these converge toward a complex and multifaceted role of TMEM16A. Aggregating evidence from the literature suggests that both TMEM16A protein and TMEM16A-related ion conduction contribute to its cancer-related functions. TMEM16A could thus be engaged in a large variety of different signaling pathways by either its ability to interact with multiple proteins or its ion conduction that regulates ion homeostasis in cancer cells thereby contributing to critical functions in cancer cells such as osmoregulation, lipid distribution or calcium homeostasis.
To explain the multifaceted role observed for TMEM16A in cancers, Wang and colleagues recently proposed that TMEM16A could be associated to a cell-type specific mechanism [132]. However, this hypothesis suggests that TMEM16A modulates a single function in a unique cancer cell line.
In this review, we propose a complementary hypothesis that may explain the multifaceted role of TMEM16A in cancer. Our hypothesis proposes that TMEM16A contributes to the formation of macro-complexes of proteins in specialized micro-domains such as lipid rafts. Such specialized macro- complexes will promote the adaptive response to various stimuli thus associating TMEM16A to the regulation of multiple functions in cancer cells.
This hypothesis is partially supported by current evidence, it will require further investigation to characterize each aspect of the molecular mechanism of TMEM16A to elucidate its multifaceted role in cancer.
Highlights:
TMEM16A is a prognosis biomarker of several cancers
TMEM16A expression and function is regulated by multiples factors
TMEM16A contributes to multiple biological function of cancer cells
TMEM16A forms macro-complexes that defined its multifaceted role in cancer cells
Integration of lipid, ion and protein signaling by TMEM16A may explain its multifaceted role
Acknowledgment:
The authors thank Drs. Chin Fen Teo and Tina Han for helpful discussions and for editing the article.
Funding Sources:
D.C. is supported by Philippe Foundation. L.Y.J is supported by R01CA185039–04. L.Y.J. is Howard Hughes Medical Institute Investigator.
Abbreviation tables:
- 14-3-3γ
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein gamma (YWHAG)
- α-SMA
α-Smooth Muscle Actin
- ANO1
Anoctamin-1
- Bcl-2
B Cell Lymphoma 2
- CaCC
Calcium Activated Chloride Channel
- CDK
Cyclin-dependent Kinase
- COPB1
Coatomer protein complex subunit beta 1
- CRC
Colorectal Cancer
- DOG1
Discovered on gastrointestinal stromal tumors protein 1
- EGF
Epidermal Growth Factor
- EGFR
Epidermal Growth Factor Receptor
- EMT
Epithelial-to-Mesenchymal Transition
- ER
Estrogen Receptors
- ERM
Ezrin-Radixin-Moesin
- FADD
Fas-associated protein with death domain
- GIST
Gastro Intestinal Squamous Tumors
- Gli
Glioma-associated oncogenes
- HDAC3
Histone Deacetylase 3
- HER2
Human Epidermal Growth Factor Receptor 2
- HNSCC
Head and Neck Squamous Carcinoma Cells
- HPV
Human Papilloma Virus
- IL-4
Interleukin-4
- IP3
Inositol triphosphate
- IP3R
inositol triphosphate receptor
- MCL-1
Myeloid Cell Leukemia 1
- MMP
matrix metallopeptidase
- PIP2
Phosphatidylinositol 4,5-bisphosphate
- PLA
Proximity-Ligation Assay
- PLC
Phospholipase C
- SOCE
Store-Operated Calcium Entry
- STAT3
Signal Transducer and Activator of Transcription 3 TGF-β: Tumor Growth Factor-β
- TNF-α
Tumor Necrosis Factor-α
- WNK
(With-No-Lysine) WNK lysine deficient protein kinase
- ZO-1
Zonula Occludens-1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest:
No conflict of interest to declare
References:
- [1].Prevarskaya N, Skryma R, Shuba Y, Ion Channels in Cancer: Are Cancer Hallmarks Oncochannelopathies?, Physiol. Rev 98 (2018) 559–621. doi: 10.1152/physrev.00044.2016. [DOI] [PubMed] [Google Scholar]
- [2].Prevarskaya N, Skryma R, Shuba Y, Ion channels and the hallmarks of cancer, Trends Mol Med. 16 (2010) 107–121. doi: 10.1016/j.molmed.2010.01.005. [DOI] [PubMed] [Google Scholar]
- [3].Caputo A, Caci E, Ferrera L, Pedemonte N, Barsanti C, Sondo E, Pfeffer U, Ravazzolo R, Zegarra-Moran O, Galietta LJV, TMEM16A, a membrane protein associated with calcium-dependent chloride channel activity, Science. 322 (2008) 590–594. doi: 10.1126/science.1163518. [DOI] [PubMed] [Google Scholar]
- [4].Schroeder BC, Cheng T, Jan YN, Jan LY, Expression Cloning of TMEM16A as a Calcium-Activated Chloride Channel Subunit, Cell. 134 (2008) 1019–1029. doi: 10.1016/j.cell.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Yang YD, Cho H, Koo JY, Tak MH, Cho Y, Shim W-S, Park SP, Lee J, Lee B, Kim B-M, Raouf R, Shin YK, Oh U, TMEM16A confers receptor-activated calcium-dependent chloride conductance, Nature. 455 (2008) 1210–1215. doi: 10.1038/nature07313. [DOI] [PubMed] [Google Scholar]
- [6].Huang F, Wong X, Jan LY, International Union of Basic and Clinical Pharmacology. LXXXV: Calcium-Activated Chloride Channels, Pharmacological Reviews. 64 (2012) 1–15. doi: 10.1124/pr.111.005009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Pedemonte N, Galietta LJV, Structure and Function of TMEM16 Proteins (Anoctamins), Physiological Reviews. 94 (2014) 419–459. doi: 10.1152/physrev.00039.2011. [DOI] [PubMed] [Google Scholar]
- [8].Wang Q, Leo MD, Narayanan D, Kuruvilla KP, Jaggar JH, Local coupling of TRPC6 to ANO1/TMEM16A channels in smooth muscle cells amplifies vasoconstriction in cerebral arteries, American Journal of Physiology-Cell Physiology. 310 (2016) C1001–C1009. doi: 10.1152/ajpcell.00092.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].West RB, Corless CL, Chen X, Rubin BP, Subramanian S, Montgomery K, Zhu S, Ball CA, Nielsen TO, Patel R, Goldblum JR, Brown PO, Heinrich MC, van de Rijn M, The Novel Marker, DOG1, Is Expressed Ubiquitously in Gastrointestinal Stromal Tumors Irrespective of KIT or PDGFRA Mutation Status, The American Journal of Pathology. 165 (2004) 107–113. doi: 10.1016/S0002-9440(10)63279-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Abd El-Rehim DM, Gayyed MF, Does Immunohistochemistry for Discovered on GIST1 and Minichromosome Maintenance Protein7 Provide Additional Clinicopathological Value in Gastrointestinal Stromal Tumors?, World Journal of Oncology. 6 (2015) 355–363. doi: 10.14740/wjon918w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ayoub C, Wasylyk C, Li Y, Thomas E, Marisa L, Robé A, Roux M, Abecassis J, de Reyniès A, Wasylyk B, ANO1 amplification and expression in HNSCC with a high propensity for future distant metastasis and its functions in HNSCC cell lines, British Journal of Cancer. 103 (2010) 715–726. doi: 10.1038/sj.bjc.6605823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Carles A, Millon R, Cromer A, Ganguli G, Lemaire F, Young J, Wasylyk C, Muller D, Schultz I, Rabouel Y, Dembélé D, Zhao C, Marchal P, Ducray C, Bracco L, Abecassis J, Poch O, Wasylyk B, Head and neck squamous cell carcinoma transcriptome analysis by comprehensive validated differential display, Oncogene. 25 (2006) 1821–1831. doi: 10.1038/sj.onc.1209203. [DOI] [PubMed] [Google Scholar]
- [13].Dixit R, Kemp C, Kulich S, Seethala R, Chiosea S, Ling S, Ha PK, Duvvuri U, TMEM16A/ANO1 is differentially expressed in HPV-negative versus HPV-positive head and neck squamous cell carcinoma through promoter methylation, Scientific Reports. 5 (2015). doi: 10.1038/srep16657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Duvvuri U, Shiwarski DJ, Xiao D, Bertrand C, Huang X, Edinger RS, Rock JR, Harfe BD, Henson BJ, Kunzelmann K, Schreiber R, Seethala RS, Egloff AM, Chen X, Lui VW, Grandis JR, Gollin SM, TMEM16A Induces MAPK and Contributes Directly to Tumorigenesis and Cancer Progression, Cancer Research. 72 (2012) 3270–3281. doi: 10.1158/0008-5472.CAN-12-0475-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Friedrich RE, Wunder T, Schumacher U, Bartel-Friedrich S, Zustin J, Expression of DOG1 (Using SP31) in Poorly Differentiated Carcinoma of the Head and Neck, Anticancer Res. 36 (2016) 3117–3122. [PubMed] [Google Scholar]
- [16].Hermida-Prado F, Menéndez S, Albornoz-Afanasiev P, Granda-Diaz R, Álvarez-Teijeiro S, Villaronga M, Allonca E, Alonso-Durán L, León X, Alemany L, Mena M, del-Rio-Ibisate N, Astudillo A, Rodríguez R, Rodrigo J, García-Pedrero J, Distinctive Expression and Amplification of Genes at 11q13 in Relation to HPV Status with Impact on Survival in Head and Neck Cancer Patients, Journal of Clinical Medicine. 7 (2018) 501. doi: 10.3390/jcm7120501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Li Y, Zhang J, Hong S, ANO1 as a marker of oral squamous cell carcinoma and silencing ANO1 suppresses migration of human scc-25 cells, Medicina Oral Patología Oral y Cirugia Bucal. (2014) e313–e319. doi: 10.4317/medoral.19076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Pattle SB, Utjesanovic N, Togo A, Wells L, Conn B, Monaghan H, Junor E, Johannessen I, Cuschieri K, Talbot S, Copy number gain of 11q13.3 genes associates with pathological stage in hypopharyngeal squamous cell carcinoma: ANO1, MOLECULAR MARKER OF METASTATIC DISEASE, Genes, Chromosomes and Cancer. 56 (2017) 185–198. doi: 10.1002/gcc.22425. [DOI] [PubMed] [Google Scholar]
- [19].Reddy RB, Bhat AR, James BL, Govindan SV, Mathew R, Ravindra DR, Hedne N, Illiayaraja J, Kekatpure V, Khora SS, Hicks W, Tata P, Kuriakose MA, Suresh A, Meta-Analyses of Microarray Datasets Identifies ANO1 and FADD as Prognostic Markers of Head and Neck Cancer, PLoS ONE. 11 (2016) e0147409. doi: 10.1371/journal.pone.0147409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Rodrigo JP, Menéndez ST, Hermida-Prado F, Álvarez-Teijeiro S, Villaronga MÁ, Alonso-Durán L, Vallina A, Martínez-Camblor P, Astudillo A, Suárez C, María García-Pedrero J, Clinical significance of Anoctamin-1 gene at 11q13 in the development and progression of head and neck squamous cell carcinomas, Scientific Reports. 5 (2015). doi: 10.1038/srep15698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ruiz C, Martins JR, Rudin F, Schneider S, Dietsche T, Fischer CA, Tornillo L, Terracciano LM, Schreiber R, Bubendorf L, Kunzelmann K, Enhanced Expression of ANO1 in Head and Neck Squamous Cell Carcinoma Causes Cell Migration and Correlates with Poor Prognosis, PLoS ONE. 7 (2012) e43265. doi: 10.1371/journal.pone.0043265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Shiwarski DJ, Shao C, Bill A, Kim J, Xiao D, Bertrand CA, Seethala RS, Sano D, Myers JN, Ha P, Grandis J, Gaither LA, Puthenveedu MA, Duvvuri U, To “Grow” or “Go”: TMEM16A Expression as a Switch between Tumor Growth and Metastasis in SCCHN, Clinical Cancer Research. 20 (2014) 4673–4688. doi: 10.1158/1078-0432.CCR-14-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Sugahara K, Michikawa Y, Ishikawa K, Shoji Y, Iwakawa M, Shibahara T, Imai T, Combination effects of distinct cores in 11q13 amplification region on cervical lymph node metastasis of oral squamous cell carcinoma, Int. J. Oncol 39 (2011) 761–769. doi: 10.3892/ijo.2011.1094. [DOI] [PubMed] [Google Scholar]
- [24].Baghai F, Yazdani F, Etebarian A, Garajei A, Skalova A, Clinicopathologic and molecular characterization of mammary analogue secretory carcinoma of salivary gland origin, Pathol. Res. Pract 213 (2017) 1112–1118. doi: 10.1016/j.prp.2017.07.017. [DOI] [PubMed] [Google Scholar]
- [25].Britschgi A, Bill A, Brinkhaus H, Rothwell C, Clay I, Duss S, Rebhan M, Raman P, Guy CT, Wetzel K, George E, Popa MO, Lilley S, Choudhury H, Gosling M, Wang L, Fitzgerald S, Borawski J, Baffoe J, Labow M, Gaither LA, Bentires-Alj M, Calcium-activated chloride channel ANO1 promotes breast cancer progression by activating EGFR and CAMK signaling, Proceedings of the National Academy of Sciences. 110 (2013) E1026–E1034. doi: 10.1073/pnas.1217072110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Cheng H, Yang S, Qu Z, Zhou S, Ruan Q, Novel Use for DOG1 in Discriminating Breast Invasive Carcinoma from Noninvasive Breast Lesions, Disease Markers. 2016 (2016) 1–8. doi: 10.1155/2016/5628176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Choi E, Yun JA, Jabeen S, Jeon E, Won H, Ko Y, Kim S, Prognostic significance of TMEM16A, PPFIA1, and FADD expression in invasive ductal carcinoma of the breast, World Journal of Surgical Oncology. 12 (2014) 137. doi: 10.1186/1477-7819-12-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kuwabara H, Yamamoto K, Terada T, Kawata R, Nagao T, Hirose Y, Hemorrhage of MRI and Immunohistochemical Panels Distinguish Secretory Carcinoma From Acinic Cell Carcinoma: Hemorrhage of MRI, Laryngoscope Investigative Otolaryngology. 3 (2018) 268–274. doi: 10.1002/lio2.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ubby I, Bussani E, Colonna A, Stacul G, Locatelli M, Scudieri P, Galietta L, Pagani F, TMEM16A alternative splicing coordination in breast cancer, Molecular Cancer. 12 (2013) 75. doi: 10.1186/1476-4598-12-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wu H, Wang H, Guan S, Zhang J, Chen Q, Wang X, Ma K, Zhao P, Zhao H, Yao W, Jin F, Xiao Q, Wei M, Cell-specific regulation of proliferation by Ano1/TMEM16A in breast cancer with different ER, PR, and HER2 status, Oncotarget. 8 (2017). doi: 10.18632/oncotarget.18662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wu H, Guan S, Sun M, Yu Z, Zhao L, He M, Zhao H, Yao W, Wang E, Jin F, Xiao Q, Wei M, Ano1/TMEM16A Overexpression Is Associated with Good Prognosis in PR-Positive or HER2-Negative Breast Cancer Patients following Tamoxifen Treatment, PLOS ONE. 10 (2015) e0126128. doi: 10.1371/journal.pone.0126128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wang H, Yao F, Luo S, Ma K, Liu M, Bai L, Chen S, Song C, Wang T, Du Q, Wu H, Wei M, Fang Y, Xiao Q, A mutual activation loop between the Ca2+-activated chloride channel TMEM16A and EGFR/STAT3 signaling promotes breast cancer tumorigenesis, Cancer Lett. (2019). doi: 10.1016/j.canlet.2019.04.027. [DOI] [PubMed] [Google Scholar]
- [33].Lee Y-S, Lee JK, Bae Y, Lee B-S, Kim E, Cho C-H, Ryoo K, Yoo J, Kim C-H, Yi G-S, Lee S-G, Lee CJ, Kang SS, Hwang EM, Park J-Y, Suppression of 14-3-3γ-mediated surface expression of ANO1 inhibits cancer progression of glioblastoma cells, Sci Rep. 6 (2016) 26413. doi: 10.1038/srep26413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Liu J, Liu Y, Ren Y, Kang L, Zhang L, Transmembrane protein with unknown function 16A overexpression promotes glioma formation through the nuclear factor-κB signaling pathway, Mol Med Rep. 9 (2014) 1068–1074. doi: 10.3892/mmr.2014.1888. [DOI] [PubMed] [Google Scholar]
- [35].Mokutani Y, Uemura M, Munakata K, Okuzaki D, Haraguchi N, Takahashi H, Nishimura J, Hata T, Murata K, Takemasa I, Mizushima T, Doki Y, Mori M, Yamamoto H, Down-Regulation of microRNA-132 is Associated with Poor Prognosis of Colorectal Cancer, Ann. Surg. Oncol 23 (2016) 599–608. doi: 10.1245/s10434-016-5133-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Park YR, Lee ST, Kim SL, Zhu SM, Lee MR, Kim SH, Kim IH, Lee SO, Seo SY, Kim SW, Down-regulation of miR-9 promotes epithelial mesenchymal transition via regulating anoctamin-1 (ANO1) in CRC cells, Cancer Genetics. 231–232 (2019) 22–31. doi: 10.1016/j.cancergen.2018.12.004. [DOI] [PubMed] [Google Scholar]
- [37].Sui Y, Sun M, Wu F, Yang L, Di W, Zhang G, Zhong L, Ma Z, Zheng J, Fang X, Ma T, Inhibition of TMEM16A expression suppresses growth and invasion in human colorectal cancer cells, PLoS ONE. 9 (2014) e115443. doi: 10.1371/journal.pone.0115443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Carneiro A, Isinger A, Karlsson A, Johansson J, Jönsson G, Bendahl P-O, Falkenback D, Halvarsson B, Nilbert M, Prognostic impact of array-based genomic profiles in esophageal squamous cell cancer, BMC Cancer. 8 (2008) 98. doi: 10.1186/1471-2407-8-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Shang L, Hao J-J, Zhao X-K, He J-Z, Shi Z-Z, Liu H-J, Wu L-F, Jiang Y-Y, Shi F, Yang H, Zhang Y, Liu Y-Z, Zhang T-T, Xu X, Cai Y, Jia X-M, Li M, Zhan Q-M, Li E-M, Wang L-D, Wei W-Q, Wang M-R, ANO1 protein as a potential biomarker for esophageal cancer prognosis and precancerous lesion development prediction, Oncotarget. 7 (2016) 24374–24382. doi: 10.18632/oncotarget.8223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Shi Z-Z, Shang L, Jiang Y-Y, Hao J-J, Zhang Y, Zhang T-T, Lin D-C, Liu S-G, Wang B-S, Gong T, Zhan Q-M, Wang M-R, Consistent and Differential Genetic Aberrations between Esophageal Dysplasia and Squamous Cell Carcinoma Detected By Array Comparative Genomic Hybridization, Clinical Cancer Research. 19 (2013) 5867–5878. doi: 10.1158/1078-0432.CCR-12-3753. [DOI] [PubMed] [Google Scholar]
- [41].Wang F, Wang B, Long J, Wang F, Wu P, Identification of candidate target genes for endometrial cancer, such as ANO1, using weighted gene co-expression network analysis, Exp Ther Med. 17 (2019) 298–306. doi: 10.3892/etm.2018.6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Cao Q, Liu F, Ji K, Liu N, He Y, Zhang W, Wang L, MicroRNA-381 inhibits the metastasis of gastric cancer by targeting TMEM16A expression, J. Exp. Clin. Cancer Res 36 (2017) 29. doi: 10.1186/s13046-017-0499-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Kosemehmetoglu K, Kaygusuz G, Fritchie K, Aydin O, Yapicier O, Coskun O, Karatayli E, Boyacigil S, Guler G, Dervisoglu S, Kuzu I, Clinical and pathological characteristics of gastrointestinal stromal tumor (GIST) metastatic to bone, Virchows Archiv. 471 (2017) 77–90. doi: 10.1007/s00428-017-2138-7. [DOI] [PubMed] [Google Scholar]
- [44].Li Q, Zhi X, Zhou J, Tao R, Zhang J, Chen P, Røe OD, Sun L, Ma L, Circulating tumor cells as a prognostic and predictive marker in gastrointestinal stromal tumors: a prospective study, Oncotarget. 7 (2016) 36645–36654. doi: 10.18632/oncotarget.9128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Rizzo FM, Palmirotta R, Marzullo A, Resta N, Cives M, Tucci M, Silvestris F, Parallelism of DOG1 expression with recurrence risk in gastrointestinal stromal tumors bearing KIT or PDGFRA mutations, BMC Cancer. 16 (2016) 87. doi: 10.1186/s12885-016-2111-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Simon S, Grabellus F, Ferrera L, Galietta L, Schwindenhammer B, Mühlenberg T, Taeger G, Eilers G, Treckmann J, Breitenbuecher F, Schuler M, Taguchi T, Fletcher JA, Bauer S, DOG1 regulates growth and IGFBP5 in gastrointestinal stromal tumors, Cancer Res. 73 (2013) 3661–3670. doi: 10.1158/0008-5472.CAN-12-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Xu C, Han H, Wang J, Zhang B, Shao Y, Zhang L, Wang H, Wang H, Wu Y, Li X, Li R, Tian Y, Diagnosis value of CD117 and PDGFRA, alone or in combination DOG1, as biomarkers for gastrointestinal stromal tumors, Ann Transl Med. 3 (2015) 308. doi: 10.3978/j.issn.2305-5839.2015.10.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Deng L, Yang J, Chen H, Ma B, Pan K, Su C, Xu F, Zhang J, Knockdown of TMEM16A suppressed MAPK and inhibited cell proliferation and migration in hepatocellular carcinoma, Onco Targets Ther. 9 (2016) 325–333. doi: 10.2147/OTT.S95985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].He Y, Li H, Chen Y, Li P, Gao L, Zheng Y, Sun Y, Chen J, Qian X, Expression of anoctamin 1 is associated with advanced tumor stage in patients with non-small cell lung cancer and predicts recurrence after surgery, Clin Transl Oncol. 19 (2017) 1091–1098. doi: 10.1007/s12094-017-1643-0. [DOI] [PubMed] [Google Scholar]
- [50].Jia L, Liu W, Guan L, Lu M, Wang K, Inhibition of Calcium-Activated Chloride Channel ANO1/TMEM16A Suppresses Tumor Growth and Invasion in Human Lung Cancer, PLoS ONE. 10 (2015) e0136584. doi: 10.1371/journal.pone.0136584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Ardeleanu C, Arsene D, Hinescu M, Andrei F, Gutu D, Luca L, Popescu LM, Pancreatic expression of DOG1: a novel gastrointestinal stromal tumor (GIST) biomarker, Appl. Immunohistochem. Mol. Morphol 17 (2009) 413–418. doi: 10.1097/PAI.0b013e31819e4dc5. [DOI] [PubMed] [Google Scholar]
- [52].Crottès D, Lin, Yu-Hsiu T, Peters CJ, Gilchrist, John M, Wiita, Arun P, Jan YN, Jan LY, TMEM16A controls EGF-induced calcium signaling implicated in pancreatic cancer prognosis, Proceedings of the National Academy of Sciences. (2019). doi: 10.1073/pnas.1900703116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Nielsen MFB, Mortensen MB, Detlefsen S, Typing of pancreatic cancer-associated fibroblasts identifies different subpopulations, World J. Gastroenterol 24 (2018) 4663–4678. doi: 10.3748/wjg.v24.i41.4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Sauter DRP, Novak I, Pedersen SF, Larsen EH, Hoffmann EK, ANO1 (TMEM16A) in pancreatic ductal adenocarcinoma (PDAC), Pflugers Arch. 467 (2015) 1495–1508. doi: 10.1007/s00424-014-1598-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Chênevert J, Duvvuri U, Chiosea S, Dacic S, Cieply K, Kim J, Shiwarski D, Seethala RR, DOG1: a novel marker of salivary acinar and intercalated duct differentiation, Mod. Pathol 25 (2012) 919–929. doi: 10.1038/modpathol.2012.57. [DOI] [PubMed] [Google Scholar]
- [56].Liu W, Lu M, Liu B, Huang Y, Wang K, Inhibition of Ca(2+)-activated Cl(−) channel ANO1/TMEM16A expression suppresses tumor growth and invasiveness in human prostate carcinoma, Cancer Lett. 326 (2012) 41–51. doi: 10.1016/j.canlet.2012.07.015. [DOI] [PubMed] [Google Scholar]
- [57].Matsuba S, Niwa S, Muraki K, Kanatsuka S, Nakazono Y, Hatano N, Fujii M, Zhan P, Suzuki T, Ohya S, Downregulation of Ca2+-activated Cl- channel TMEM16A by the inhibition of histone deacetylase in TMEM16A-expressing cancer cells, J. Pharmacol. Exp. Ther 351 (2014) 510–518. doi: 10.1124/jpet.114.217315. [DOI] [PubMed] [Google Scholar]
- [58].Song Y, Gao J, Guan L, Chen X, Gao J, Wang K, Inhibition of ANO1/TMEM16A induces apoptosis in human prostate carcinoma cells by activating TNF-α signaling, Cell Death Dis. 9 (2018) 703. doi: 10.1038/s41419-018-0735-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Guo S, Chen Y, Pang C, Wang X, Shi S, Zhang H, An H, Zhan Y, Matrine is a novel inhibitor of the TMEM16A chloride channel with antilung adenocarcinoma effects, J. Cell. Physiol 234 (2019) 8698–8708. doi: 10.1002/jcp.27529. [DOI] [PubMed] [Google Scholar]
- [60].Kulkarni S, Bill A, Godse NR, Khan NI, Kass JI, Steehler K, Kemp C, Davis K, Bertrand CA, Vyas AR, Holt DE, Grandis JR, Gaither LA, Duvvuri U, TMEM16A/ANO1 suppression improves response to antibody-mediated targeted therapy of EGFR and HER2/ERBB2, Genes Chromosomes Cancer. 56 (2017) 460–471. doi: 10.1002/gcc.22450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Bill A, Gutierrez A, Kulkarni S, Kemp C, Bonenfant D, Voshol H, Duvvuri U, Gaither LA, ANO1/TMEM16A interacts with EGFR and correlates with sensitivity to EGFR-targeting therapy in head and neck cancer, Oncotarget. 6 (2015) 9173–9188. doi: 10.18632/oncotarget.3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Hanahan D, Weinberg RA, Hallmarks of cancer: the next generation, Cell. 144 (2011) 646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- [63].Guan L, Song Y, Gao J, Gao J, Wang K, Inhibition of calcium-activated chloride channel ANO1 suppresses proliferation and induces apoptosis of epithelium originated cancer cells, Oncotarget. 7 (2016) 78619–78630. doi: 10.18632/oncotarget.12524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Hu C, Zhang R, Jiang D, TMEM16A as a Potential Biomarker in the Diagnosis and Prognosis of Lung Cancer, Arch Iran Med. 22 (2019) 32–38. [PubMed] [Google Scholar]
- [65].Mazzone A, Eisenman ST, Strege PR, Yao Z, Ordog T, Gibbons SJ, Farrugia G, Inhibition of cell proliferation by a selective inhibitor of the Ca(2+)-activated Cl(−) channel, Ano1, Biochem. Biophys. Res. Commun 427 (2012) 248–253. doi: 10.1016/j.bbrc.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Seo Y, Park J, Kim M, Lee HK, Kim J-H, Jeong J-H, Namkung W, Inhibition of ANO1/TMEM16A Chloride Channel by Idebenone and Its Cytotoxicity to Cancer Cell Lines, PLoS ONE. 10 (2015) e0133656. doi: 10.1371/journal.pone.0133656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Wanitchakool P, Wolf L, Koehl GE, Sirianant L, Schreiber R, Kulkarni S, Duvvuri U, Kunzelmann K, Role of anoctamins in cancer and apoptosis, Philos. Trans. R. Soc. Lond., B, Biol. Sci 369 (2014) 20130096. doi: 10.1098/rstb.2013.0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Zhang X, Li H, Zhang H, Liu Y, Huo L, Jia Z, Xue Y, Sun X, Zhang W, Inhibition of transmembrane member 16A calcium-activated chloride channels by natural flavonoids contributes to flavonoid anticancer effects, Br. J. Pharmacol 174 (2017) 2334–2345. doi: 10.1111/bph.13841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Kunzelmann K, Ousingsawat J, Benedetto R, Cabrita I, Schreiber R, Contribution of Anoctamins to Cell Survival and Cell Death, Cancers (Basel). 11 (2019). doi: 10.3390/cancers11030382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Seo Y, Ryu K, Park J, Jeon D-K, Jo S, Lee HK, Namkung W, Inhibition of ANO1 by luteolin and its cytotoxicity in human prostate cancer PC-3 cells, PLoS ONE. 12 (2017) e0174935. doi: 10.1371/journal.pone.0174935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Jacobsen KS, Zeeberg K, Sauter DRP, Poulsen KA, Hoffmann EK, Schwab A, The role of TMEM16A (ANO1) and TMEM16F (ANO6) in cell migration, Pflugers Arch. 465 (2013) 1753–1762. doi: 10.1007/s00424-013-1315-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Sui Y, Wu F, Lv J, Li H, Li X, Du Z, Sun M, Zheng Y, Yang L, Zhong L, Zhang X, Zhang G, Identification of the Novel TMEM16A Inhibitor Dehydroandrographolide and Its Anticancer Activity on SW620 Cells, PLoS ONE. 10 (2015) e0144715. doi: 10.1371/journal.pone.0144715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Caputo A, Caci E, Ferrera L, Pedemonte N, Barsanti C, Sondo E, Pfeffer U, Ravazzolo R, Zegarra-Moran O, Galietta LJV, TMEM16A, a membrane protein associated with calcium-dependent chloride channel activity, Science. 322 (2008) 590–594. doi: 10.1126/science.1163518. [DOI] [PubMed] [Google Scholar]
- [74].Mroz MS, Keely SJ, Epidermal growth factor chronically upregulates Ca(2+)-dependent Cl(−) conductance and TMEM16A expression in intestinal epithelial cells, J. Physiol. (Lond.) 590 (2012) 1907–1920. doi: 10.1113/jphysiol.2011.226126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Mazzone A, Gibbons SJ, Bernard CE, Nowsheen S, Middha S, Almada LL, Ordog T, Kendrick ML, Reid Lombardo KM, Shen KR, Galietta LJV, Fernandez-Zapico ME, Farrugia G, Identification and characterization of a novel promoter for the human ANO1 gene regulated by the transcription factor signal transducer and activator of transcription 6 (STAT6), FASEB J. 29 (2015) 152–163. doi: 10.1096/fj.14-258541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Mazzone A, Gibbons SJ, Eisenman ST, Strege PR, Zheng T, D’Amato M, Ordog T, Fernandez-Zapico ME, Farrugia G, Direct repression of anoctamin 1 (ANO1) gene transcription by Gli proteins, The FASEB Journal. (2019) fj.201802373R. doi: 10.1096/fj.201802373R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Lee Y-S, Bae Y, Park N, Yoo JC, Cho C-H, Ryoo K, Hwang EM, Park J-Y, Surface expression of the Anoctamin-1 (ANO1) channel is suppressed by protein-protein interactions with β-COP, Biochem. Biophys. Res. Commun 475 (2016) 216–222. doi: 10.1016/j.bbrc.2016.05.077. [DOI] [PubMed] [Google Scholar]
- [78].Fujimoto M, Kito H, Kajikuri J, Ohya S, Transcriptional repression of human epidermal growth factor receptor 2 by ClC-3 Cl-/H+ transporter inhibition in human breast cancer cells, Cancer Sci. 109 (2018) 2781–2791. doi: 10.1111/cas.13715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Caldieri G, Malabarba MG, Di Fiore PP, Sigismund S, EGFR Trafficking in Physiology and Cancer, Prog. Mol. Subcell. Biol 57 (2018) 235–272. doi: 10.1007/978-3-319-96704-2_9. [DOI] [PubMed] [Google Scholar]
- [80].Guéguinou M, Gambade A, Félix R, Chantôme A, Fourbon Y, Bougnoux P, Weber G, Potier-Cartereau M, Vandier C, Lipid rafts, KCa/ClCa/Ca2+ channel complexes and EGFR signaling: Novel targets to reduce tumor development by lipids?, Biochim. Biophys. Acta 1848 (2015) 2603–2620. doi: 10.1016/j.bbamem.2014.10.036. [DOI] [PubMed] [Google Scholar]
- [81].Hsu JL, Hung M-C, The role of HER2, EGFR, and other receptor tyrosine kinases in breast cancer, Cancer Metastasis Rev. 35 (2016) 575–588. doi: 10.1007/s10555-016-9649-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Sigismund S, Avanzato D, Lanzetti L, Emerging functions of the EGFR in cancer, Mol Oncol. 12 (2018) 3–20. doi: 10.1002/1878-0261.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Wang Z, ErbB Receptors and Cancer, Methods Mol. Biol 1652 (2017) 3–35. doi: 10.1007/978-1-4939-7219-7_1. [DOI] [PubMed] [Google Scholar]
- [84].Fujimoto M, Inoue T, Kito H, Niwa S, Suzuki T, Muraki K, Ohya S, Transcriptional repression of HER2 by ANO1 Cl- channel inhibition in human breast cancer cells with resistance to trastuzumab, Biochem. Biophys. Res. Commun 482 (2017) 188–194. doi: 10.1016/j.bbrc.2016.11.033. [DOI] [PubMed] [Google Scholar]
- [85].Becchetti A, Crescioli S, Zanieri F, Petroni G, Mercatelli R, Coppola S, Gasparoli L, D’Amico M, Pillozzi S, Crociani O, Stefanini M, Fiore A, Carraresi L, Morello V, Manoli S, Brizzi MF, Ricci D, Rinaldi M, Masi A, Schmidt T, Quercioli F, Defilippi P, Arcangeli A, The conformational state of hERG1 channels determines integrin association, downstream signaling, and cancer progression, Sci Signal. 10 (2017). doi: 10.1126/scisignal.aaf3236. [DOI] [PubMed] [Google Scholar]
- [86].Crottès D, Rapetti-Mauss R, Alcaraz-Perez F, Tichet M, Gariano G, Martial S, Guizouarn H, Pellissier B, Loubat A, Popa A, Paquet A, Presta M, Tartare-Deckert S, Cayuela ML, Martin P, Borgese F, Soriani O, SIGMAR1 Regulates Membrane Electrical Activity in Response to Extracellular Matrix Stimulation to Drive Cancer Cell Invasiveness, Cancer Res. 76 (2016) 607–618. doi: 10.1158/0008-5472.CAN-15-1465. [DOI] [PubMed] [Google Scholar]
- [87].Gueguinou M, Crottès D, Chantôme A, Rapetti-Mauss R, Potier-Cartereau M, Clarysse L, Girault A, Fourbon Y, Jézéquel P, Guérin-Charbonnel C, Fromont G, Martin P, Pellissier B, Schiappa R, Chamorey E, Mignen O, Uguen A, Borgese F, Vandier C, Soriani O, The SigmaR1 chaperone drives breast and colorectal cancer cell migration by tuning SK3-dependent Ca2+ homeostasis, Oncogene. 36 (2017) 3640–3647. doi: 10.1038/onc.2016.501. [DOI] [PubMed] [Google Scholar]
- [88].Guéguinou M, Harnois T, Crottes D, Uguen A, Deliot N, Gambade A, Chantôme A, Haelters JP, Jaffrès PA, Jourdan ML, Weber G, Soriani O, Bougnoux P, Mignen O, Bourmeyster N, Constantin B, Lecomte T, Vandier C, Potier-Cartereau M, SK3/TRPC1/Orai1 complex regulates SOCE-dependent colon cancer cell migration: a novel opportunity to modulate anti-EGFR mAb action by the alkyl-lipid Ohmline, Oncotarget. 7 (2016) 36168–36184. doi: 10.18632/oncotarget.8786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Pillozzi S, Masselli M, De Lorenzo E, Accordi B, Cilia E, Crociani O, Amedei A, Veltroni M, D’Amico M, Basso G, Becchetti A, Campana D, Arcangeli A, Chemotherapy resistance in acute lymphoblastic leukemia requires hERG1 channels and is overcome by hERG1 blockers, Blood. 117 (2011) 902–914. doi: 10.1182/blood-2010-01-262691. [DOI] [PubMed] [Google Scholar]
- [90].Pillozzi S, Brizzi MF, Bernabei PA, Bartolozzi B, Caporale R, Basile V, Boddi V, Pegoraro L, Becchetti A, Arcangeli A, VEGFR-1 (FLT-1), beta1 integrin, and hERG K+ channel for a macromolecular signaling complex in acute myeloid leukemia: role in cell migration and clinical outcome, Blood. 110 (2007) 1238–1250. doi: 10.1182/blood-2006-02-003772. [DOI] [PubMed] [Google Scholar]
- [91].Pillozzi S, Arcangeli A, Physical and functional interaction between integrins and hERG1 channels in cancer cells, Adv. Exp. Med. Biol 674 (2010) 55–67. [DOI] [PubMed] [Google Scholar]
- [92].Perez-Cornejo P, Gokhale A, Duran C, Cui Y, Xiao Q, Hartzell HC, Faundez V, Anoctamin 1 (Tmem16A) Ca2+-activated chloride channel stoichiometrically interacts with an ezrinradixin-moesin network, Proceedings of the National Academy of Sciences. 109 (2012) 10376–10381. doi: 10.1073/pnas.1200174109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Clucas J, Valderrama F, ERM proteins in cancer progression, J. Cell. Sci 127 (2014) 267–275. doi: 10.1242/jcs.133108. [DOI] [PubMed] [Google Scholar]
- [94].He J, Ma G, Qian J, Zhu Y, Liang M, Yao N, Ding Q, Chen L, Liu X, Xia T, Wang S, Interaction Between Ezrin and Cortactin in Promoting Epithelial to Mesenchymal Transition in Breast Cancer Cells, Medical Science Monitor. 23 (2017) 1583–1596. doi: 10.12659/MSM.904124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Benedetto R, Ousingsawat J, Wanitchakool P, Zhang Y, Holtzman MJ, Amaral M, Rock JR, Schreiber R, Kunzelmann K, Epithelial Chloride Transport by CFTR Requires TMEM16A, Scientific Reports. 7 (2017). doi: 10.1038/s41598-017-10910-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Benedetto R, Sirianant L, Pankonien I, Wanitchakool P, Ousingsawat J, Cabrita I, Schreiber R, Amaral M, Kunzelmann K, Relationship between TMEM16A/anoctamin 1 and LRRC8A, Pflügers Archiv - European Journal of Physiology. 468 (2016) 1751–1763. doi: 10.1007/s00424-016-1862-1. [DOI] [PubMed] [Google Scholar]
- [97].Cabrita I, Benedetto R, Fonseca A, Wanitchakool P, Sirianant L, Skryabin BV, Schenk LK, Pavenstädt H, Schreiber R, Kunzelmann K, Differential effects of anoctamins on intracellular calcium signals, The FASEB Journal. 31 (2017) 2123–2134. doi: 10.1096/fj.201600797RR. [DOI] [PubMed] [Google Scholar]
- [98].Jin X, Shah S, Liu Y, Zhang H, Lees M, Fu Z, Lippiat JD, Beech DJ, Sivaprasadarao A, Baldwin SA, Zhang H, Gamper N, Activation of the Cl- channel ANO1 by localized calcium signals in nociceptive sensory neurons requires coupling with the IP3 receptor, Sci Signal. 6 (2013) ra73. doi: 10.1126/scisignal.2004184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Jung J, Nam JH, Park HW, Oh U, Yoon J-H, Lee MG, Dynamic modulation of ANO1/TMEM16A HCO3− permeability by Ca2+/calmodulin, Proceedings of the National Academy of Sciences. 110 (2013) 360–365. doi: 10.1073/pnas.1211594110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Lin C-X, Lv X-F, Yuan F, Li X-Y, Ma M-M, Liu C-Z, Zhou J-G, Wang G-L, Guan Y-Y, Ca2+/Calmodulin-Dependent Protein Kinase II γ-Dependent Serine727 Phosphorylation Is Required for TMEM16A Ca2+-Activated Cl- Channel Regulation in Cerebrovascular Cells, Circ. J 82 (2018) 903–913. doi: 10.1253/circj.CJ-17-0585. [DOI] [PubMed] [Google Scholar]
- [101].Sala-Rabanal M, Yurtsever Z, Nichols CG, Brett TJ, Secreted CLCA1 modulates TMEM16A to activate Ca(2+)-dependent chloride currents in human cells, Elife. 4 (2015). doi: 10.7554/eLife.05875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Takayama Y, Tominaga M, Involvement of TRPV1-ANO1 Interactions in Pain-Enhancing Mechanisms, Adv. Exp. Med. Biol 1099 (2018) 29–36. doi: 10.1007/978-981-13-1756-9_3. [DOI] [PubMed] [Google Scholar]
- [103].Berglund E, Akcakaya P, Berglund D, Karlsson F, Vukojević V, Lee L, Bogdanović D, Lui W-O, Larsson C, Zedenius J, Fröbom R, Bränström R, Functional role of the Ca2+-activated Cl− channel DOG1/TMEM16A in gastrointestinal stromal tumor cells, Exp. Cell Res 326 (2014) 315–325. doi: 10.1016/j.yexcr.2014.05.003. [DOI] [PubMed] [Google Scholar]
- [104].Cuddapah VA, Sontheimer H, Ion channels and transporters [corrected] in cancer. 2. Ion channels and the control of cancer cell migration, Am. J. Physiol., Cell Physiol 301 (2011) C541–549. doi: 10.1152/ajpcell.00102.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Hoffmann EK, Sørensen BH, Sauter DPR, Lambert IH, Role of volume-regulated and calcium-activated anion channels in cell volume homeostasis, cancer and drug resistance, Channels (Austin). 9 (2015) 380–396. doi: 10.1080/19336950.2015.1089007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Chen J-C, Lo Y-F, Lin Y-W, Lin S-H, Huang C-L, Cheng C-J, WNK4 kinase is a physiological intracellular chloride sensor, Proc. Natl. Acad. Sci. U.S.A (2019). doi: 10.1073/pnas.1817220116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Shekarabi M, Zhang J, Khanna AR, Ellison DH, Delpire E, Kahle KT, WNK Kinase Signaling in Ion Homeostasis and Human Disease, Cell Metab. 25 (2017) 285–299. doi: 10.1016/j.cmet.2017.01.007. [DOI] [PubMed] [Google Scholar]
- [108].Dbouk HA, Weil LM, Perera GKS, Dellinger MT, Pearson G, Brekken RA, Cobb MH, Actions of the protein kinase WNK1 on endothelial cells are differentially mediated by its substrate kinases OSR1 and SPAK, Proc. Natl. Acad. Sci. U.S.A 111 (2014) 15999–16004. doi: 10.1073/pnas.1419057111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Rodan AR, Jenny A, WNK Kinases in Development and Disease, Curr. Top. Dev. Biol 123 (2017) 1–47. doi: 10.1016/bs.ctdb.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Gallolu Kankanamalage S, Karra AS, Cobb MH, WNK pathways in cancer signaling networks, Cell Commun. Signal 16 (2018) 72. doi: 10.1186/s12964-018-0287-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Gallolu Kankanamalage S, Lee A-Y, Wichaidit C, Lorente-Rodriguez A, Shah AM, Stippec S, Whitehurst AW, Cobb MH, WNK1 is an unexpected autophagy inhibitor, Autophagy. 13 (2017) 969–970. doi: 10.1080/15548627.2017.1286431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].He M, Ye W, Wang W-J, Sison ES, Jan YN, Jan LY, Cytoplasmic Cl- couples membrane remodeling to epithelial morphogenesis, Proc. Natl. Acad. Sci. U.S.A 114 (2017) E11161–E11169. doi: 10.1073/pnas.1714448115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Ye W, Han TW, Nassar LM, Zubia M, Jan YN, Jan LY, Phosphatidylinositol-(4, 5)-bisphosphate regulates calcium gating of small-conductance cation channel TMEM16F, Proc. Natl. Acad. Sci. U.S.A 115 (2018) E1667–E1674. doi: 10.1073/pnas.1718728115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Ji Q, Guo S, Wang X, Pang C, Zhan Y, Chen Y, An H, Recent advances in TMEM16A: Structure, function, and disease, J. Cell. Physiol (2018). doi: 10.1002/jcp.27865. [DOI] [PubMed] [Google Scholar]
- [115].Contreras-Vite JA, Cruz-Rangel S, De Jesús-Pérez JJ, Figueroa IAA, Rodríguez-Menchaca AA, Pérez-Cornejo P, Hartzell HC, Arreola J, Revealing the activation pathway for TMEM16A chloride channels from macroscopic currents and kinetic models, Pflugers Arch. 468 (2016) 1241–1257. doi: 10.1007/s00424-016-1830-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Dang S, Feng S, Tien J, Peters CJ, Bulkley D, Lolicato M, Zhao J, Zuberbühler K, Ye W, Qi L, Chen T, Craik CS, Jan YN, Minor DL, Cheng Y, Jan LY, Cryo-EM structures of the TMEM16A calcium-activated chloride channel, Nature. 552 (2017) 426–429. doi: 10.1038/nature25024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Paulino C, Kalienkova V, Lam AKM, Neldner Y, Dutzler R, Activation mechanism of the calcium-activated chloride channel TMEM16A revealed by cryo-EM, Nature. 552 (2017) 421–425. doi: 10.1038/nature24652. [DOI] [PubMed] [Google Scholar]
- [118].Tien J, Peters CJ, Wong XM, Cheng T, Jan YN, Jan LY, Yang H, A comprehensive search for calcium binding sites critical for TMEM16A calcium-activated chloride channel activity, Elife. 3 (2014). doi: 10.7554/eLife.02772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Peters CJ, Gilchrist JM, Tien J, Bethel NP, Qi L, Chen T, Wang L, Jan YN, Grabe M, Jan LY, The Sixth Transmembrane Segment Is a Major Gating Component of the TMEM16A Calcium-Activated Chloride Channel, Neuron. 97 (2018) 1063–1077.e4. doi: 10.1016/j.neuron.2018.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Peters CJ, Yu H, Tien J, Jan YN, Li M, Jan LY, Four basic residues critical for the ion selectivity and pore blocker sensitivity of TMEM16A calcium-activated chloride channels, Proc. Natl. Acad. Sci. U.S.A 112 (2015) 3547–3552. doi: 10.1073/pnas.1502291112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Schreiber R, Ousingsawat J, Wanitchakool P, Sirianant L, Benedetto R, Reiss K, Kunzelmann K, Regulation of TMEM16A/ANO1 and TMEM16F/ANO6 ion currents and phospholipid scrambling by Ca2+ and plasma membrane lipid, J. Physiol. (Lond.) 596 (2018) 217–229. doi: 10.1113/JP275175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Tian Y, Schreiber R, Wanitchakool P, Kongsuphol P, Sousa M, Uliyakina I, Palma M, Faria D, Traynor-Kaplan AE, Fragata JI, Amaral MD, Kunzelmann K, Control of TMEM16A by INO-4995 and other inositolphosphates, Br. J. Pharmacol 168 (2013) 253–265. doi: 10.1111/j.1476-5381.2012.02193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Cho H, Yang YD, Lee J, Lee B, Kim T, Jang Y, Back SK, Na HS, Harfe BD, Wang F, Raouf R, Wood JN, Oh U, The calcium-activated chloride channel anoctamin 1 acts as a heat sensor in nociceptive neurons, Nat. Neurosci 15 (2012) 1015–1021. doi: 10.1038/nn.3111. [DOI] [PubMed] [Google Scholar]
- [124].Cruz-Rangel S, De Jesús-Pérez JJ, Aréchiga-Figueroa IA, Rodríguez-Menchaca AA, Pérez-Cornejo P, Hartzell HC, Arreola J, Extracellular protons enable activation of the calcium-dependent chloride channel TMEM16A, J. Physiol. (Lond.) 595 (2017) 1515–1531. doi: 10.1113/JP273111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Tokuoka S, Morioka H, The membrane potential of the human cancer and related cells. I, Gan. 48 (1957) 353–354. [PubMed] [Google Scholar]
- [126].Prevarskaya N, Skryma R, Shuba Y, Calcium in tumour metastasis: new roles for known actors, Nat. Rev. Cancer 11 (2011) 609–618. doi: 10.1038/nrc3105. [DOI] [PubMed] [Google Scholar]
- [127].Brisson L, Driffort V, Benoist L, Poet M, Counillon L, Antelmi E, Rubino R, Besson P, Labbal F, Chevalier S, Reshkin SJ, Gore J, Roger S, NaV1.5 Na+ channels allosterically regulate the NHE-1 exchanger and promote the activity of breast cancer cell invadopodia, J. Cell. Sci 126 (2013) 4835–4842. doi: 10.1242/jcs.123901. [DOI] [PubMed] [Google Scholar]
- [128].Brisson L, Gillet L, Calaghan S, Besson P, Le Guennec J-Y, Roger S, Gore J, Na(V)1.5 enhances breast cancer cell invasiveness by increasing NHE1-dependent H(+) efflux in caveolae, Oncogene. 30 (2011) 2070–2076. doi: 10.1038/onc.2010.574. [DOI] [PubMed] [Google Scholar]
- [129].Concepcion AR, Vaeth M, Wagner LE, Eckstein M, Hecht L, Yang J, Crottes D, Seidl M, Shin HP, Weidinger C, Cameron S, Turvey SE, Issekutz T, Meyts I, Lacruz RS, Cuk M, Yule DI, Feske S, Store-operated Ca2+ entry regulates Ca2+-activated chloride channels and eccrine sweat gland function, J. Clin. Invest 126 (2016) 4303–4318. doi: 10.1172/JCI89056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Sharma A, Ramena G, Yin Y, Premkumar L, Elble RC, CLCA2 is a positive regulator of store-operated calcium entry and TMEM16A, PLoS ONE. 13 (2018) e0196512. doi: 10.1371/journal.pone.0196512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Sun Y, Birnbaumer L, Singh BB, TRPC1 regulates calcium-activated chloride channels in salivary gland cells, J. Cell. Physiol 230 (2015) 2848–2856. doi: 10.1002/jcp.25017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Wang H, Zou L, Ma K, Yu J, Wu H, Wei M, Xiao Q, Cell-specific mechanisms of TMEM16A Ca2+-activated chloride channel in cancer, Mol. Cancer 16 (2017) 152. doi: 10.1186/s12943-017-0720-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Crottès D, Guizouarn H, Martin P, Borgese F, Soriani O, The sigma-1 receptor: a regulator of cancer cell electrical plasticity?, Front Physiol. 4 (2013) 175. doi: 10.3389/fphys.2013.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]