Abstract
Direct seeding of rice often results in poor crop establishment due to unlevelled fields, unpredicted heavy rains after sowing, and weed and pest invasion. Thus, it is important to develop varieties able to tolerate flooding during germination, also known as anaerobic germination (AG), to address these constraints. A study was conducted to identify QTLs associated with AG tolerance from an IR64/Kharsu 80A F2:3 mapping population using 190 lines phenotyped for seedling survival under the stress. Genotyping was performed using a genomewide 384-plex Indica/Indica SNP set. Four QTLs derived from Kharsu 80A providing increased tolerance to anaerobic germination were identified: three on chromosome 7 (qAG7.1, qAG7.2 and qAG7.3) and one on chromosome 3 (qAG3), with LOD values ranging from 5.7 to 7.7, and phenotypic variance explained (R2) from 8.1% to 12.6%. The QTLs identified in this study can be further investigated to better understand the genetic bases of AG tolerance in rice, and used for marker-assisted selection to develop more robust direct-seeded rice varieties.
Keywords: quantitative trait loci, anaerobic germination, direct seeding, rice, Oryza sativa
Introduction
The escalating cost of labor and increasing water crisis are the main reasons why farmers are beginning to adopt direct seeding for establishing rice in the field. Aside from these, direct-seeded rice (DSR) generally flowers earlier, leading to shorter crop duration, and matures 7–10 days earlier than when transplanted (Farooq et al. 2006a, 2006b). In spite of these advantages, DSR is constrained with lower yield because of poor crop establishment and weed infestation. Moreover, invasion of pests such as snails, birds and rats and non-germination of seeds, due to flooding caused by unlevelled fields or heavy rains, are often experienced in DSR (Pandey and Velasco 2002). One way to overcome these problems is to flood the field to a certain level right after broadcasting the seeds; however, most modern rice varieties do not have the capacity to germinate under flooded conditions. Thus, it is important to develop varieties that can withstand flooding during germination or anaerobic germination (AG).
One of the described mechanisms for AG tolerance is to escape anoxia caused by flooding through rapid shoot elongation so that seedlings can reach the water surface and thereby allowing O2 to diffuse internally to the root and endosperm (Ismail et al. 2009, Kretzschmar et al. 2015, Turner et al. 1981). Among cereals studied so far, only rice is able to extend its coleoptile and elongate its shoots rapidly under anaerobic conditions (Perata and Voesenek 2007). Additionally, Hwang et al. (1999) and Ismail et al. (2009) reported that α-amylases play a major role in starch breakdown under anaerobic conditions, thereby providing energy to the growing seedlings. The role of ethylene as a potent regulator of seedling growth under flooded conditions has also been reported. Ethylene promotes the elongation of mesocotyl and coleoptiles in rice seedlings (Ismail et al. 2009, Satler and Kende 1985). Other groups of proteins known to regulate cell wall expansion are expansins and peroxidases (Ismail et al. 2009). Expression profile analysis of anoxia-grown rice seedlings revealed that EXPA7 and EXPB12 are likely to be involved in rice coleoptile elongation (Lasanthi-Kudahettige et al. 2007). On the other hand, the activity of peroxidases is inversely related to the rate of coleoptile elongation in rice. Ismail et al. (2009) observed that seeds of tolerant rice genotypes (Khaiyan and Khao Hlan On) germinating under anoxic conditions showed substantially lower peroxidase activity than the intolerant genotypes (FR13A and IR42).
Efforts on breeding rice for tolerance to anaerobic conditions during germination and increased seed and seedling vigor have been attempted (Biswas and Yamauchi 1997, Redoňa and Mackill 1996, Yamauchi and Winn 1996). However, slow progress has been achieved over the years. This is due to the limited knowledge on the genetics of the trait, the involvement of complex mechanisms, and the lack of an effective screening method (Jiang et al. 2006, Seshu et al. 1988). However, some tolerant genotypes have been identified and used in genetic studies and breeding (Angaji et al. 2010, Septiningsih et al. 2013a, 2013b, Septiningsih and Mackill 2018, Toledo et al. 2015).
Several studies have identified quantitative trait loci (QTLs) controlling tolerance to AG. Jiang et al. (2004) found a total of five QTLs conferring AG on chromosomes 1, 2, 5 and 7 using restriction fragment length polymorphism (RFLP) markers in Kinmaze (japonica)/DV85 (indica). Furthermore, three pairs of epistatic loci were found on chromosomes 2, 3, 5 and 11. The phenotypic variance ranged from 10.5%–19.6%. In another population (USSR5/N22), Jiang et al. (2006) reported two QTLs on chromosomes 5 and 11 that explained 11–15.5% phenotypic variance. Angaji et al. (2010) identified five putative QTLs located on chromosomes 1 (qAG-1-2); 3 (qAG-3-1); 7 (qAG-7-2) and 9 (qAG-9-1) and (qAG-9-2) which explain 17.9 to 33.5% of the phenotypic variation using simple sequence repeats (SSR) in a BC2F2 population of IR64/Khao Hlan On. One of the QTLs located at the long arm of chromosome 9 (qAG-9-2) with the highest phenotypic variation (33.5%; LOD = 20.3) was fine mapped and the gene underlying the QTL was identified and characterized and used in molecular breeding (Kretzschmar et al. 2015, Toledo et al. 2015). In more recent reports, Septiningsih et al. (2013b) identified major QTLs derived from Ma-Zhan (Red), a landrace from China, while Baltazar et al. (2014) reported QTLs contributed by the aus landrace, Nanhi. The largest QTL from Ma-Zhan (Red) was located on chromosome 7, but two smaller QTLs were also detected on the same chromosome. The largest QTL derived from Nanhi was also located on chromosome 7 (Baltazar et al. 2014).
Several single nucleotide polymorphism (SNP) marker genotyping platforms are now available, with the optimal platform depending on the sample size and number of SNPs to be analyzed for each application (Saxena et al. 2012, Thomson 2014). In this study, we have taken advantage of the Illumina GoldenGate 384-plex Indica–Indica SNP set on the BeadXpress platform for mapping QTLs for AG tolerance in rice. The objective of this study was to identify and map the positions of QTLs for tolerance to AG in an F2:3 population derived from IR64/Kharsu 80A, which is an AG-tolerant aus landrace from Pakistan.
Materials and Methods
Plant materials and development of the mapping population
Kharsu 80A (IRGC 28017), an upland aus variety from Pakistan and a donor of tolerance to AG, was crossed with IR64, a non-aromatic semi-dwarf indica variety developed at the International Rice Research Institute (IRRI) which is sensitive to flooding during germination. The crosses were made in WS 2009, and F3 seeds were harvested in WS 2010. Based on our experiments, the screening of AG is affected by environmental factors, especially temperature and the amount of sunlight. However, in general, the level of AG tolerance in Kharsu 80A is comparable to or, in several field experiments, even slightly better than Khao Hlan On and Ma-Zhan (Red) (unpublished data; Angaji et al. 2010, Septiningsih et al. 2013b). The identity of F1 lines was confirmed using SSR markers, then allowed to self-pollinate to produce the F2 population for genotyping and F2:3 lines for phenotyping. The seeds used for this experiment were less than one year old, and breaking dormancy was performed at 50°C for 3 days. Prior to phenotyping, germination test was conducted at 30°C for 7 days with 30 seeds per entry, and lines having percentage germination less than 90% were discarded. A total of 190 F2:3 families were used for the QTL study; while about 50 families were discarded due to low germination.
Phenotyping AG tolerance
Phenotypic screening for AG was carried out based on the protocol of Septiningsih et al. (2013b) where 30 healthy seeds from each of the 190 lines were sown in seedling trays at about 1 cm soil depth in a greenhouse at IRRI. Each tray had 11 lines, along with Kharsu 80A and IR64 that served as the tolerant and susceptible controls, respectively. No seeds were sown in rows between lines and the outside borders. The lines were arranged according to Alpha plus randomization and the set-up was replicated twice. After sowing, the trays were submerged carefully in concrete tanks filled with 10 cm of water from the soil surface and maintained at that depth for 21 days. Seedling survival rate was assessed 21 days after sowing. This experiment was performed at IRRI greenhouse, Los Baños, Philippines, in early October 2011, where the temperature was relatively hot (average high temperature of 31°C and low temperature of 26°C) and the weather was mostly overcast (rainy season).
Genotyping using SNPs
Genomic DNA of all the F2 plants and the parents was extracted from 18 to 21-day-old seedlings using the CTAB technique of Murray and Thompson (1980) with a few modifications. The final concentration of the DNA samples was normalized to 50 ng/μl. Genotyping was performed using the Illumina BeadXpress 384-plex SNP plates with the oligo pool assay (OPA) customized for Indica–Indica (Illumina OPA ID: GS0011861-OPA; RiceOPA2.1) (Thomson et al. 2012). PCR amplification and hybridization were performed using the GoldenGate genotyping assay for VeraCode manual protocol (Illumina, San Diego, CA, USA). The VeraCode 384-plex plate was scanned using the Illumina BeadXpress Reader (Genotyping Services Lab, IRRI) and raw intensity values were exported from the GenomeStudio software V1.1.0 (Illumina). Genotype calling was performed using Alchemy software (Wright et al. 2010).
Construction of the linkage map
The rice physical map (MSU v.7) was used to order the markers for the SNP-based map. The linkage map was constructed using Map Manager QTX, vQTXb20, with Kosambi mapping function (Manly et al. 2001). To determine whether a marker followed the expected Mendelian segregation ratio (1AA:2AB:1BB). Chi square (χ2) test was performed (χ2 p < 0.05 = 6.00) using QTL Cartographer v2.5 (Wang et al. 2010). The positions of the centromeres along the chromosomes were inferred based on the rice physical map published by the International Rice Genome Sequencing Project (IRGSP) (2005).
QTL analysis
QTL analysis was performed using interval mapping (IM) and composite interval mapping (CIM) of QTL Cartographer v2.5 (Wang et al. 2010) with 1,000 permutations. The cofactors in CIM were selected automatically using forward–backward stepwise regression with F-in = 0.01 and F-out = 0.01 and window size of 10 cM (Septiningsih et al. 2012). The LOD values at p ≤ 0.05 for the permutations were used as a threshold to declare the significance of the QTLs. The data were also analyzed using Qgene 4.3.10 (Joehanes and Nelson 2008). Permutation of 10,000 iterations was used to determine the threshold of the F-value of the QTL. Two QTL mapping packages and two approaches (IM and CIM) were used to ensure that all major QTLs would be reliably detected even with minor differences in the implementation and statistical approaches by the different software packages. The relative positions of the confidence intervals and QTL peaks of the QTLs in this study were compared with those previously identified.
Results
AG tolerance in the IR64/Kharsu 80A population
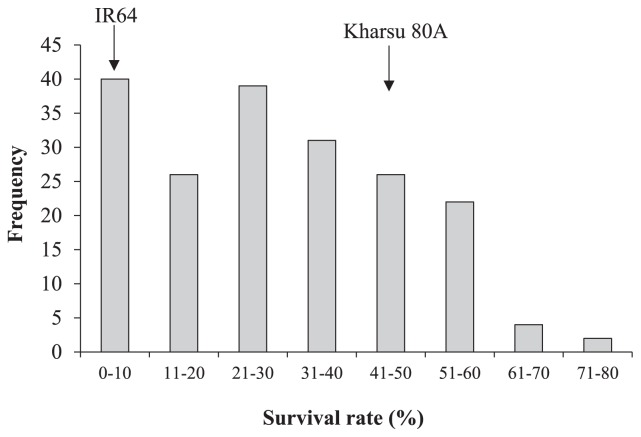
Tolerance to AG of the 190 families evaluated ranged from 0.0–73.3% with an average survival rate of 29.6% (Fig. 1). The population showed an approximately normal distribution of survival under 10 cm water after 21 days. Kurtosis and skewness were 0.27 and −0.82, respectively. The survival rates of the parents were 4.0% (IR64) and 42.8% (Kharsu 80A). There were 11 lines (5.8%) that had survival rates lower than IR64 while 48 lines (25.3%) were higher than Kharsu 80A, suggesting that transgressive segregation occurred in the population.
Fig. 1.
Phenotypic distribution of tolerance to AG in the F2:3 IR64/Kharsu 80A population as shown by survival rate. The survival rate (%) of IR64 was 4.0 ± 0.02, while that of Kharsu 80A was 42.8 ± 0.04.
Construction of the linkage map
The rice physical map (MSU v.7) was used to list the marker order, while the marker distances were calculated from the genotype data using Map-Manager QTX vQTXb20 (Manly et al. 2001). Of the 384 SNP markers used, 217 (57%) showed polymorphism and can be mapped between IR64 and Kharsu 80A. The total length of the linkage map was 1553.5 cM with an average distance of 7.6 cM between markers. Forty one percent of the polymorphic markers used showed distortion from the expected segregation distortion ratio (1AA:2AB:1BB) with χ2 (0.05) ≥ 6.00. Chromosome 10 had the highest proportion of distorted markers while the least was on chromosome 9.
QTL Analysis
Based on 1,000 permutations, the declared threshold levels for IM and CIM at p ≤ 0.05 were 3.80 and 3.97 and at p ≤ 0.01 were 4.92 and 5.05, respectively in QTL Cartographer. On the other hand, the threshold levels for IM and CIM in QGene at p ≤ 0.05 were 3.69 and 3.83 and at p ≤ 0.01 were 4.40 and 4.78, respectively. Three QTLs were detected above the QGene IM threshold level of p ≤ 0.01: qAG7.1 (LOD = 5.5), qAG7.2 (LOD = 7.7), qAG7.3 (LOD = 6.5) located on chromosome 7, with an additional QTL detected above the QGene CIM threshold at p ≤ 0.01, qAG3.1, located on chromosome 3 with a LOD of 5.9 (Table 1, Fig. 2). The Kharsu 80A allele contributed in the increase in tolerance to AG at all QTLs and was responsible for phenotypic variance of 17.5% to 23.3% (Table 1, Fig. 2). The QTL Cartographer IM results confirmed the detection of qAG7.1, qAG7.2, and qAG7.3, while QTL Cartographer CIM only detected qAG7.3 above the empirically-defined significance thresholds.
Table 1.
QTLs for tolerance to anaerobic germination detected in an F2 population derived from IR64/Kharsu 80A population
| QTL | Chr. | Flanking markers | Peak marker | QGene IM | QGene CIM | QTL Cart. IM | QTL Cart. CIM | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
||||||||||||||||
| LOD | R2 | Adda | Domb | LOD | R2 | Add | Dom | LOD | R2 | Add | Dom | LOD | R2 | Add | Dom | ||||
| qAG3 | 3 | id3002377–id3004190 | id3003215 | 5.9 | 18.4 | 8.1 | 4.9 | ||||||||||||
| qAG7.1 | 7 | id7000519–id7002260 | wd7000465 | 5.5 | 17.5 | 10.7 | −2.4 | 4.8 | 15.4 | 9.1 | −0.6 | 5.7 | 12.5 | 9.3 | 6.9 | ||||
| qAG7.2 | 7 | id7002427–id7003359 | d7003072 | 7.7 | 23.3 | 12.1 | −7.5 | 7.1 | 13.5 | 8.7 | 8.5 | ||||||||
| qAG7.3 | 7 | id7003853–id7004429 | id7004429 | 6.5 | 20.3 | 9.4 | −4.1 | 5.5 | 17.2 | 7.1 | −2.9 | 7.5 | 12.6 | 8.0 | 7.7 | 5.0 | 4.2 | 5.0 | 7.1 |
Additive effects are shown for each QTL.
Dominant effect are shown for each QTL.
Bold LOD scores were above the p = 0.01 threshold, normal font is above p = 0.05 threshold.
Fig. 2.
Linkage map of SNP markers in the IR64/Kharsu 80A F2 population showing the positions of the QTLs as black rectangles on chromosomes 3 and 7.
Discussion
Phenotyping of F2:3 progenies
The survival rates of the parents used in this population were 4% and 42.8% for IR64 and Kharsu 80A respectively. The population conforms to normal distribution, indicating that AG is a quantitative trait controlled by multiple genes. Some degree of skewness was observed. This population showed higher skewness compared with the other population (IR64/Nanhi) we investigated (Baltazar et al. 2014). For this study, the phenotypic screening of the Kharsu 80A population was performed in the greenhouse during the warm rainy season (average high of 31°C and low of 26°C) with cloudy weather compared to the Nanhi population which was screened in February when the temperature in the Philippines was cooler (average high of 29°C and low of 24°C) and with sunny weather. Cooler weather and more sunlight will enhance the growth of the germinating seeds (due to higher oxygen levels in the water and greater photosynthesis activity) increasing the ability to escape from the water which in turn will increase survival rate. Ella et al. (2010) reported that water temperature affects the survival of seeds under anaerobic conditions. Further, they found that highest seedling survival under anaerobic conditions is attained when the water temperature is 24 to 26°C, whereas temperatures above this will cause a substantial decrease in seedling survival.
The phenotypic performance of the mapping population showed several lines with more extreme phenotypes relative to the parents, indicating transgressive segregation in the population. According to Rieseberg et al. (2003), transgressive segregation is a result of recombination of two parents possessing genes with antagonistic or complementary effects. Although all the QTLs detected above the threshold were derived from the donor parent (Kharsu 80A), there potentially could be a number of smaller effect loci from the susceptible parent that fell under the significance threshold, but still contributed to transgressive segregation in the mapping population. The presence of transgressive segregants were also encountered and described both in interspecific (Septiningsih et al. 2003, Thomson et al. 2003, Xie et al. 2008) and intraspecific crosses in rice (Baltazar et al. 2014, Guo et al. 2004, Septiningsih et al. 2012, Zhou et al. 2007).
Linkage map construction
The genetic map of this population is characterized by higher marker density and had a smaller average marker distance as compared to those previously published on QTL mapping of AG (Angaji et al. 2010, Jiang et al. 2004, 2006, Septiningsih et al. 2013b). No large gaps were remaining in the map, suggesting that no QTLs were missed due to gaps in the map.
QTLs for AG
Using the software QTL Cartographer and QGene, four QTLs above the permutation threshold were identified, one positioned on chromosome 3 (qAG3.1), and the other three located on chromosome 7 (qAG7.1, qAG7.2 and qAG7.3). qAG7.2 was not detected by the CIM method, this probably is due to a close proximity with qAG7.3. Further, the AG1 (qAG-9-2) QTL or OsTPP7 gene that was previously reported on chromosome 9 (Angaji et al. 2010, Kretzschmar et al. 2015) was not detected in this population. In this case, AG1 could be missing in the genome of Kharsu 80A or this variety may have the susceptible allele of AG1. Several QTL mapping studies on AG tolerance in rice were published (Angaji et al. 2010, Baltazar et al. 2014, Jiang et al. 2004, 2006, Septiningsih et al. 2013b). Additionally, genome-wide association studies (GWAS) on AG tolerance have been reported (Hsu and Tung 2015, Zhang et al. 2017). A comparative QTL study was performed to determine if QTLs in similar locations have been previously identified from other genetic sources. One of the QTLs on chromosome 7 (qAG7.1) derived from Kharsu 80A is in a similar position with the qAG7 identified in the IR64/Nanhi population (Baltazar et al. 2014), and partially overlapped with the largest QTL (qAG7.1) derived from Ma-Zhan (Red) (Septiningsih et al. 2013b). Further, qAG7.2 is mapped in similar position as qAG7.2 from Ma-Zhan (Red); while qAG7.3 is in similar position with qAG7.3 derived from Ma-Zhan (Red) (Septiningsih et al. 2013b), qAG-7-2 from Khao Hlan On (Angaji et al. 2010), and a cluster of significant SNPs from two GWAS studies (Hsu and Tung 2015, Zhang et al. 2017). The QTL on chromosome 3, qAG3, overlaps with a significant SNP from a GWAS study by Zhang et al. (2017).
Seedling vigor plays a significant role for stable crop establishment and early vegetative growth in rice for good field emergence and crop performance and competitive ability against weeds (Foolad et al. 2007). The qAG7.2 was mapped to a similar genomic region as the largest QTL for seedling vigor, qSV-7, between markers RM214–G20–C285, identified by Zhang (2005) in a Lemont/Teqing population. Some traits associated with seedling vigor promote seedling emergence in directly broadcasted rice such as long mesocotyl and coleoptiles (Dilday et al. 1990). The QTL on chromosome 3 (qAG3) lies near the region of the QTL for coleoptile length (peak marker RZ448) identified by Cai and Morishima (2002) (peak marker CDO122) and Redoña and Mackill (1996). It is possible that these QTLs for AG tolerance also contribute to traits related to seedling vigor.
Rapid seed deterioration is a serious problem for direct seeding practice in rice (Miura et al. 2004). Ella et al. (2010) reported seedling survival decreased substantially with aging of the seeds compared with newly-harvested ones. This could be due to the increase in lipid peroxidation. Miura et al. (2004) found out that indica varieties have higher seed longevity than japonica varieties and that longevity and dormancy were found to be independent from each other. Seed dormancy is an essential trait for DSR because it entails resistance to pre-harvest sprouting (Wan et al. 2005). Interestingly, qAG7.1 and qAG7.3 are in a similar position with the largest QTLs for seed longevity or seed germination capability of seeds with different ages, qSGC7.1 and qSGC7.2, respectively, from Milyang 23/Tong 88-7 RILs and account for 11.6%–17.5% phenotypic variation explained in the study of Jiang et al. (2011). Moreover, qAG3 is located in a similar region as several seed dormancy QTLs identified in previous studies (Guo et al. 2004, Lu et al. 2011, Wan et al. 1997).
Healthy seedling growth under low temperature entails stable seedling establishment (SES). Tolerance to germination at low temperature is an important objective of temperate rice breeding programs for DSR. The QTL, qAG7.1 is in similar position with qSES7-1 (Iwata et al. 2010), while qAG7.2 resides in similar position as a low temperature vigor of germination QTL, qLVG7-2 (Han et al. 2006).
Furthermore, qAG3 is in the similar region as the qLTG3 whose peak marker was RM282 (Jiang et al. 2006). Further research needs to be done to confirm whether major QTLs derived from Kharsu 80A are the same AG tolerance QTLs as previously reported, such as by fine mapping these QTLs and identifying the underlying gene(s) controlling the QTLs. Even if the QTLs share the same locus, these studies will also provide information if the diverse alleles from different genetic donors may have varying functions. Confirmation of the gene function will also reveal whether these genes have pleiotropic effects and may govern other traits.
In summary, the major QTLs derived from Kharsu 80A present additional targets for AG tolerance improvement. It will be interesting to evaluate whether combinations of QTLs from multiple donors will provide additive effects leading to the development of high-yielding rice varieties with more robust AG tolerance under DSR system.
Acknowledgments
We thank the International Rice Research Institute for the support. The work reported here was supported in part by a grant from the Bill and Melinda Gates Foundation (BMGF) through the project “Stress-tolerant rice for Africa and South Asia (STRASA)”, by the Global Rice Science Partnership (GRiSP), and the National Institute of Food and Agriculture, U. S. Department of Agriculture, Hatch projects 1009299 and 1009300.
Literature Cited
- Angaji, S.A., Septiningsih, E.M., Mackill, D.J. and Ismail, A.M. (2010) QTLs associated with tolerance of flooding during germination in rice (Oryza sativa L.). Euphytica 172: 159–168. [Google Scholar]
- Baltazar, M.D., Ignacio, J.C.I., Thomson, M.J., Ismail, A.M., Mendioro, M.S. and Septiningsih, E.M. (2014) QTL mapping for tolerance of anaerobic germination from IR64 and the aus landrace Nanhi using SNP genotyping. Euphytica 197: 251–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas, J.K. and Yamauchi, M. (1997) Mechanism of seedling establishment of direct-seeded rice (Oryza sativa L.) under lowland conditions. Bot. Bull. Acad. Sin. 38: 29–32. [Google Scholar]
- Cai, H.W. and Morishima, H. (2002) QTL clusters reflect character associations in wild and cultivated rice. Theor. Appl. Genet. 104: 1217–1228. [DOI] [PubMed] [Google Scholar]
- Dilday, R.H., Mgonja, M.A., Amonsilpa, S.A., Collins, F.C. and Wells, B.R. (1990) Plant height vs. mesocotyl and coleoptile elongation in rice: linkage or pleiotropism? Crop Sci. 30: 815–818. [Google Scholar]
- Ella, E.S., Dionisio-Sese, M.L. and Ismail, A.M. (2010) Proper management improves seedling survival and growth during early flooding in contrasting rice genotypes. Crop Sci. 50: 1997–2008. [Google Scholar]
- Farooq, M., Basra, S.M.A. and Wahid, A. (2006a) Priming of field-sown rice seed enhances germination, seedling establishment, allometry and yield. Plant Growth Regul. 49: 285–294. [Google Scholar]
- Farooq, M., Basra, S.M.A., Tabassum, R. and Afzal, I. (2006b) Enhancing the performance of direct seeded fine rice by seed priming. Plant Prod. Sci. 9: 446–456. [Google Scholar]
- Foolad, M.R., Subbiah, P. and Zhang, L. (2007) Common QTL affect the rate of tomato seed germination under different stress and nonstress conditions. Int. J. Plant Genomics 2007: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, L., Zhu, L., Xu, Y., Zeng, D., Wu, P. and Qia, Q. (2004) QTL analysis of seed dormancy in rice (Oryza sativa L.). Euphytica 140: 155–162. [Google Scholar]
- Han, L.Z., Zhang, Y.Y., Qiao, Y.L., Cao, G.L., Zhang, S.Y., Kim, J.H. and Koh, H.J. (2006) Genetic and QTL analysis for low-temperature vigor of germination in rice. Acta Genet. Sin. 33: 998–1006. [DOI] [PubMed] [Google Scholar]
- Hsu, S.K. and Tung, C.W. (2015) Genetic mapping of anaerobic germination-associated QTLs controlling coleoptile elongation in rice. Rice 8: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, Y.S., Thomas, B.R. and Rodriguez, R.L. (1999) Differential expression of rice α-amylase genes during seedling development under anoxia. Plant Mol. Biol. 40: 911–920. [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (IRGSP) (2005) The map-based sequence of the rice genome. Nature 436: 793–800. [DOI] [PubMed] [Google Scholar]
- Ismail, A.M., Ella, E.S., Vergara, G.V. and Mackill, D.J. (2009) Mechanisms associated with tolerance to flooding during germination and early seedling growth in rice (Oryza sativa). Ann. Bot. 103: 197–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata, N., Shinada, H., Kiuchi, H., Sato, T. and Fujino, K. (2010) Mapping of QTLs controlling seedling establishment using a direct seeding method in rice. Breed. Sci. 60: 353–360. [Google Scholar]
- Jiang, L., Hou, M., Wang, C. and Wan, J. (2004) Quantitative trait loci and epistatic analysis of seed anoxia germinability in rice (Oryza sativa L.). Rice Sci. 11: 238–244. [Google Scholar]
- Jiang, L., Liu, S., Hou, M., Tang, J., Chen, L., Zhai, H. and Wan, J. (2006) Analysis of QTLs for seed low temperature germinability and anoxia germinability in rice (Oryza sativa L.). Field Crops Res. 98: 68–75. [Google Scholar]
- Jiang, W., Lee, J., Jin, Y.M., Qiao, Y., Piao, R., Jang, S.M., Woo, M.O., Kwon, S.W., Liu, X., Pan, H.Y.et al. (2011) Identification of QTLs for seed germination capability after various storage periods using two RIL populations in rice. Mol. Cells 31: 385–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joehanes, R. and Nelson, J.C. (2008) QGene 4.0, an extensible Java QTL-analysis platform. Bioinformatics 24: 2788–2789. [DOI] [PubMed] [Google Scholar]
- Kretzschmar, T., Pelayo, M.A.F., Trijatmiko, K.R., Gabunada, L.F.M., Alam, R., Jimenez, R., Mendioro, M.S., Slamet-Loedin, I.H., Sreenivasulu, N., Bailey-Serres, J.et al. (2015) A trehalose-6-phosphate phosphatase enhances anaerobic germination tolerance in rice. Nat. Plants 1: 15124. [DOI] [PubMed] [Google Scholar]
- Lasanthi-Kudahettige, R., Magneschi, L., Loreti, E., Gonzali, S., Licausi, F., Novi, G., Beretta, O., Vitulli, F., Alpi, A. and Perata, P. (2007) Transcript profiling of the anoxic rice coleoptile. Plant Physiol. 144: 218–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, B., Xie, K., Yang, C., Zhang, L., Wu, T., Liu, X., Jiang, L. and Wan, J. (2011) Genetic analysis of two weak dormancy mutants derived from strong seed dormancy wild type rice N22 (Oryza sativa). J. Integr. Plant Biol. 53: 338–346. [DOI] [PubMed] [Google Scholar]
- Manly, K.F., Cudmore, R.H. Jr and Meer, J.M. (2001) Map Manager QTX, cross-platform software for genetic mapping. Mamm. Genome 12: 930–932. [DOI] [PubMed] [Google Scholar]
- Miura, K., Lin, S.Y., Araki, H., Nagamine, T., Kuroki, M., Shimizu, H., Ando, I. and Yano, M. (2004) Genetical studies on germination of seed and seedling establishment for breeding of improved rice varieties suitable for direct seeding culture. Jpn. Agric. Res. Q. 38: 1–5. [Google Scholar]
- Murray, M.G. and Thompson, W.F. (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8: 4321–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey, S. and Velasco, L. (2002) Economics of direct seeding in Asia: patterns of adoption and research priorities. In: Pandey, S., Mortimer M., Wade L., Tuong T.P., Lopez K. and Hardy B. (eds.) Direct seeding: research issues and opportunities. Proceedings of the International Workshop on Direct Seeding in Asian Rice Systems: Strategic Research Issues and Opportunities, 25–28 Jan 2000, Bangkok, Los Baňos: International Rice Research Institute, pp. 3–14. [Google Scholar]
- Perata, P. and Voesenek, L.A. (2007) Submergence tolerance in rice requires Sub1A, an ethylene response-factor-like gene. Trends Plant Sci. 12: 43–46. [DOI] [PubMed] [Google Scholar]
- Redoňa, E.D. and Mackill, D.J. (1996) Genetic variation for seedling vigor traits in rice. Crop Sci. 36: 285–290. [DOI] [PubMed] [Google Scholar]
- Rieseberg, L.H., Widmer, A., Arntz, A.M. and Burke, J.M. (2003) The genetic architecture necessary for transgressive segregation is common in both natural and domesticated populations. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 358: 1141–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satler, S.O. and Kende, H. (1985) Ethylene and the growth of rice seedlings. Plant Physiol. 79: 194–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena, R., Elbers, C.C., Guo, Y., Peter, I., Gaunt, T.R., Mega, J.L., Lanktree, M.B., Tare, A., Castillo, B.A., Li, Y.R.et al. (2012) Large-scale gene-centric meta-analysis across 39 studies identifies type 2 diabetes loci. Am. J. Hum. Genet. 90: 410–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Septiningsih, E.M., Prasetiyono, J., Lubis, E., Tai, T.H., Tjubaryat, T., Moeljopawiro, S. and McCouch, S.R. (2003) Identification of quantitative trait loci for yield and yield components in an advanced backcross population derived from the Oryza sativa variety IR64 and the wild relative O. rufipogon. Theor. Appl. Genet. 107: 1419–1432. [DOI] [PubMed] [Google Scholar]
- Septiningsih, E.M., Sanchez, D.L., Singh, N., Sendon, P.M.D., Pamplona, A.M., Heuer, S. and Mackill, D.J. (2012) Identifying novel QTLs for submergence tolerance in rice cultivars IR72 and Madabaru. Theor. Appl. Genet. 124: 867–874. [DOI] [PubMed] [Google Scholar]
- Septiningsih, E.M., Collard, B.C.Y., Heuer, S., Bailey-Serres, J., Ismail, A.M. and Mackill, D.J. (2013a) Applying Genomics Tools for Breeding Submergence Tolerance in Rice. In: Varshney, R.K. and Tuberosa R. (eds.) Translational Genomics for Crop Breeding: Volume 2—Improvement for Abiotic Stress, Quality and Yield Improvement, Wiley-Blackwell, USA, pp. 9–30. [Google Scholar]
- Septiningsih, E.M., Ignacio, J.C.I., Sendon, P.M.D., Sanchez, D.L., Ismail, A.M. and Mackill, D.J. (2013b) QTL mapping and confirmation for tolerance of anaerobic conditions during germination derived from the rice landrace Ma-Zhan Red. Theor. Appl. Genet. 126: 1357–1366. [DOI] [PubMed] [Google Scholar]
- Septiningsih, E.M. and Mackill, D.J. (2018) Genetics and breeding of flooding tolerance in rice. In: Sasaki, T. and Ashikari M. (eds.) New Waves in Rice Genomics, Genetics, and Breeding. Springer, Singapore, pp. 275–295. [Google Scholar]
- Seshu, D.V., Krishnasamy, V. and Siddique, S.B. (1988) Seed vigor in rice. Rice seed health. International Rice Research Institute, Manila, pp. 315–329. [Google Scholar]
- Thomson, M.J., Tai, T.H., McClung, A.M., Lai, X.H., Hinga, M.E., Lobos, K.B., Xu, Y., Martinez, C.P. and McCouch, S.R. (2003) Mapping quantitative trait loci for yield, yield components and morphological traits in an advanced backcross population between Oryza rufipogon and the Oryza sativa cultivar Jefferson. Theor. Appl. Genet. 107: 479–493. [DOI] [PubMed] [Google Scholar]
- Thomson, M.J., Zhao, K., Wright, M., McNally, K.L., Rey, J., Tung, C.W., Reynolds, A., Scheffler, B., Eizenga, G., McClung, A.et al. (2012) High-throughput single nucleotide polymorphism genotyping for breeding applications in rice using the BeadXpress platform. Mol. Breed. 29: 875–886. [Google Scholar]
- Thomson, M.J. (2014) High-throughput SNP genotyping to accelerate crop improvement. Plant Breed. Biotechnol. 2: 195–212. [Google Scholar]
- Toledo, A.M.U., Ignacio, J.C.I., Casal, C. Jr, Gonzaga, Z.J., Mendioro, M.S. and Septiningsih, E.M. (2015) Development of improved Ciherang-Sub1 having tolerance to anaerobic germination conditions. Plant Breed. Biotechnol. 3: 77–87. [Google Scholar]
- Turner, F.T., Chen, C.-C. and McCauley, G.N. (1981) Morphological development of rice seedlings in water at controlled oxygen levels. Agron. J. 73: 566–570. [Google Scholar]
- Wan, J., Nakazaki, T., Kawaura, K. and Ikehashi, H. (1997) Identification of marker loci for seed dormancy in rice (Oryza sativa L.). Crop Sci. 37: 1759–1763. [Google Scholar]
- Wan, J.M., Cao, Y.J., Wang, C.M. and Ikehashi, H. (2005) Quantitative trait loci associated with seed dormancy in rice. Crop Sci. 45: 712–716. [Google Scholar]
- Wang, S., Basten, C.J. and Zeng, Z.-B. (2010) Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh: http://statgen.ncsu.edu/qtlcart/WQTLCart.htm Accessed 20 Oct 2013. [Google Scholar]
- Wright, M.H., Tung, C.W., Zhao, K., Reynolds, A., McCouch, S.R. and Bustamante, C.D. (2010) ALCHEMY: a reliable method for automated SNP genotype calling for small batch sizes and highly homozygous populations. Bioinformatics 26: 2952–2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, X.B., Jin, F.X., Song, M.H., Suh, J.P., Hwang, H.G., Kim, Y.G., McCouch, S.R. and Ahn, S.N. (2008) Fine mapping of a yield-enhancing QTL cluster associated with transgressive variation in an Oryza sativa × O. rufipogon cross. Theor. Appl. Genet. 116: 613–622. [DOI] [PubMed] [Google Scholar]
- Yamauchi, M. and Winn, T. (1996) Rice seed vigor and seedling establishment in anaerobic soil. Crop Sci. 36: 680–686. [Google Scholar]
- Zhang, M., Lu, Q., Wu, W., Niu, X., Wang, C., Feng, Y., Xu, Q., Wang, S., Yuan, X., Yu, H.et al. (2017) Association mapping reveals novel genetic loci contributing to flooding tolerance during germination in Indica rice. Front. Plant Sci. 8: 678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z.H., Qu, X.S., Wan, S., Chen, L.H. and Zhu, Y.G. (2005) Comparison of QTL controlling seedling vigour under different temperature conditions using recombinant inbred lines in rice (Oryza sativa). Ann. Bot. 95: 423–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, L., Wang, J.K., Yi, Q., Wang, Y.Z., Zhu, Y.G. and Zhang, Z.H. (2007) Quantitative trait loci for seedling vigor in rice under field conditions. Field Crops Res. 100: 294–301. [Google Scholar]