Abstract
DNA methylation is an epigenetic modification that can affect gene expression and transposable element (TE) activities. Because cytosine DNA methylation patterns are inherited through both mitotic and meiotic cell divisions, differences in these patterns can contribute to phenotypic variability. Advances in high-throughput sequencing technologies have enabled the generation of abundant DNA sequence data. Integrated analyses of genome-wide gene expression patterns and DNA methylation patterns have revealed the underlying mechanisms and functions of DNA methylation. Moreover, associations between DNA methylation and agronomic traits have also been uncovered. The resulting information may be useful for future applications of natural epigenomic variation, for crop breeding. Additionally, artificial epigenome editing may be an attractive new plant breeding technique for generating novel varieties with improved agronomic traits.
Keywords: DNA methylation, epigenome, transposable element, small RNA
Introduction
Cytosine DNA methylation is a chemical modification of the fifth position of the cytosine base. In plants, DNA methylation occurs in three distinct sequence contexts, symmetrical CG and CHG as well as asymmetrical CHH, where H is either C, A, or T (Law and Jacobsen 2010). These DNA methylation patterns are stably inherited through cell division. Changes in DNA methylation can occur spontaneously and may be induced by genetic factors and environmental stimuli. Additionally, stress conditions can alter DNA methylation patterns (Dowen et al. 2012, Hossain et al. 2017, Secco et al. 2015, Wibowo et al. 2016). There are two types of DNA methylation patterns in plants, namely the CG-only gene body methylation (gbM), which is DNA methylation within transcriptional regions, and non-CG as well as CG transposable element (TE)-like methylation (teM). gbM is associated with mild constitutive expression levels, whereas teM is associated with the repression of TE activities (TE silencing) and gene expression (gene silencing) (Coleman-Derr and Zilberman 2012, Tran et al. 2005, Zhang et al. 2006, Zilberman et al. 2007). Transposable element activities increase genomic diversity, which is applicable for breeding as well as functional genomics investigations. However, TE silencing is important for stable crop cultivation and production because excessive TE activities can cause variable phenotypes, including deleterious ones. Moreover, changes in gene expression can lead to visible phenotypic alterations. Therefore, DNA methylation must be appropriately regulated for effective crop breeding.
High-throughput DNA sequencing has enabled transcriptome and epigenome profiling at single-base resolution, as well as genome re-sequencing (Lister et al. 2008). Integrating these omics-based data has resulted in the accumulation of information regarding the biological roles of epigenomes. Importantly, there are considerable intra- and inter-species variabilities in DNA methylation patterns (Kawakatsu et al. 2016a, Niederhuth et al. 2016). The associated data have not been restricted to model plant species with compact genomes, but have been extended to agronomically important crops with large and complex genomes (Daccord et al. 2017, Regulski et al. 2013, Schmitz et al. 2013a, Turco et al. 2017, Zhong et al. 2013). Natural genomic variation, such as single nucleotide variants and structural variations, has been exploited for plant breeding (Morrell et al. 2011). Recent studies suggest that it may also be possible to exploit natural epigenomic variation as a new tool for breeding.
In this review, we describe the DNA methylation machinery, diversity, and dynamics in the model plant Arabidopsis (Arabidopsis thaliana) as well as the agronomic traits associated with DNA methylation. Heterosis is one of the best-known agronomic traits associated with DNA methylation. Please refer to Fujimoto et al. (2018) in the same review series for details regarding heterosis and related epigenetics, including the transgenerational inheritance of DNA methylation or the epigenetics of recombinant inbred lines (epiRILs) derived from hybrids between DNA methylation deficient mutants and wild type.
DNA methylation machinery
CG methylation is maintained by MET1 and VIM1 (Kankel et al. 2003, Woo et al. 2007). VIM1 recognizes hemimethylated DNA and recruits MET1 to replication foci. The recruited MET1 catalyzes DNA methylation on newly synthesized hemimethylated DNA strands. Thus, CG methylation is maintained in a semi-conservative manner during DNA replication. CHG methylation is catalyzed by CMT3, which binds to methylated histone 3 lysine 9 (H3K9) (Bartee et al. 2001, Lindroth et al. 2001). Additionally, CHG and CHH methylation within heavily heterochromatic regions is regulated by CMT2, which binds to dimethylated H3K9 (H3K9me2) (Stroud et al. 2014, Zemach et al. 2013).
DNA methylation within heterochromatic regions also depends on the chromatin remodeling factor DDM1, which removes histone H1 linker proteins from densely packed chromatin to enable MET1, CMT3, and CMT2 to methylate the DNA in heterochromatic regions (Zemach et al. 2013). Furthermore, RNA-directed DNA methylation (RdDM) mediates all types of cytosine methylation within short TEs in euchromatin and along the edges of long TEs in heterochromatin (Zemach et al. 2013). In canonical RdDM, two plant-specific RNA polymerases Pol IV and Pol V, which are the result of Pol II duplications, play critical roles in small interfering RNA (siRNA) biogenesis and de novo methylation during RdDM, respectively (Matzke and Mosher 2014). Pol IV is recruited to target regions through a direct association with the SHH1, which recognizes H3K9me2, and CLSY proteins (Law et al. 2013, Zhou et al. 2018).
Pol V is recruited to target regions through an indirect association with the inactive histone methyltransferases SUVH2 and SUVH9, which recognize methylated DNA (Johnson et al. 2014). DDR (DRD1-DMS3-RDM1) complex mediates the association between Pol V and SUVH2/9 (Matzke and Mosher 2014). Pol IV synthesizes short RNAs [approximately 30–40 nucleotides (nt)] that are converted to double-stranded RNA (dsRNA) by RDR2 (Blevins et al. 2015, Zhai et al. 2015). The dsRNAs are diced into 24-nt siRNAs by DCL3 (Xie et al. 2004). AGO4 binds to these siRNAs, and the resulting AGO4-siRNA complex is guided to Pol V target loci, with Pol V transcripts as scaffolds (Gao et al. 2010, Havecker et al. 2010). DRM2 is recruited to target regions through an indirect association with AGO4 and catalyzes methylation reactions in all contexts (Gao et al. 2010).
There are several non-canonical RdDM pathways (Cuerda-Gil and Slotkin 2016). Once RdDM is initiated by non-canonical RdDM pathways, it is then established by canonical RdDM pathways (McCue et al. 2015, Stroud et al. 2014). Additionally, the histone methyltransferases KYP/SUVH4, SUVH5, and SUVH6 recognize methylated CHG and CHH via the SRA domain and catalyze the dimethylation of H3K9 (Du et al. 2014, Ebbs et al. 2005, Ebbs and Bender 2006, Rajakumara et al. 2011). Hence, non-CG DNA methylation, histone modification, and nucleosome positioning form self-reinforcing loops.
DNA demethylation machinery
DNA demethylation is initiated by a bi-functional DNA glycosylase that exhibits both DNA glycosylase activity and apurinic/apyrimidinic (AP) lyase activity, through a base excision repair mechanism (Zhang and Zhu 2012). Specifically, DNA glycosylase excises methylated cytosines, while AP lyase nicks the AP site. Additionally, DNA phosphatase ZDP, AP endonuclease APE1L, DNA polymerases, and the DNA ligase AtLIG1 cooperatively fill the single nucleotide gap with unmethylated cytosine (Li et al. 2015, Martínez-Macías et al. 2012). Arabidopsis has four DNA glycosylases: DME, ROS1/DML1, DML2, and DML3. The DME gene is predominantly expressed in the central cell of the female gametophyte before fertilization, where DME induces global hypomethylation, leading to maternal allele-specific demethylation and expression of imprinting genes as well as some transposons (Choi et al. 2002, Hsieh et al. 2009). These imprinting genes include FWA, FIS2 and MEA (Kinoshita et al. 1999, 2004, Luo et al. 2000, Vielle-Calzada et al. 1999). FWA encodes a homeodomain-containing transcription factor that controls flowering. FIS2 and MEA encode components of the PRC2 that catalyzes the repressive H3K27me3 modification. Because PRC2 is required for endosperm cellularization, DME-dependent demethylation in the central cell is indispensable (Köhler et al. 2003). DME is also expressed in the vegetative cell of the male gametophyte, and is required for demethylation of imprinting genes and transposons (Ibarra et al. 2012). Moreover, ROS1, DML2, and DML3 are expressed in vegetative tissues, and are required for the demethylation of thousands of discrete loci, including TEs within the promoters of stress-responsive genes (Calarco et al. 2012, Le et al. 2014, Tang et al. 2016). The overlap between RdDM target regions and ROS1 target regions reveals the antagonism between active DNA methylation and demethylation. Interestingly, a TE located in the ROS1 promoter region is a target of RdDM and ROS1. DNA methylation in this TE promotes the expression of ROS1. Therefore, the balance between DNA methylation and demethylation in the TE may be critical for fine-tuning the genome-wide methylation level (Lei et al. 2015, Williams et al. 2015).
Finally, because nascent DNA being synthesized during DNA replication is not methylated, cell division itself can induce “passive” demethylation by diluting DNA methylation in the absence of maintenance DNA methylation or de novo DNA methylation. Nucleoside analogs of cytidine, such as 5-azacitidine and zebularine, can be incorporated into DNA and substituted for cytosine. The 5-azacitidine- or zebularine-substituted DNA inhibits DNA methyltransferase activity, leading to genomic DNA demethylation.
Lessons from the reference plant Arabidopsis
Cell type-specific DNA methylation in Arabidopsis
Mature pollen grains are the final form of male sexual lineage cells. Meiosis produces haploid microspores from diploid meiocytes. Two rounds of mitosis result in the production of mature pollen grains comprising two sperm cells and a vegetative cell. Sperm cells initiate a simultaneous “double fertilization” process: one fusing with haploid egg cell and the other with the diploid central cell. The vegetative cell which supports growth of the pollen tube does not transmit its genomic information to the next generation. Active RdDM induces locus-specific hypermethylation in the sperm cell genomes, but not in the vegetative cell genome. The male sexual lineage-specific methylation within intron 9 of MPS1/PRD2, which is crucial for meiosis, is required for the proper splicing of this intron (Walker et al. 2018). In RdDM-deficient drm1 drm2 meiocytes, approximately 30% of MPS1/PRD2 mRNAs retain intron 9, which introduces a premature stop codon, suggesting that proper MPS1 expression (and meiosis) is regulated by RdDM. Indeed, the meiocytes of drm1 drm2 and rdr2 mutants do not undergo normal meiosis, forming triad, tetrad, and pentad microspores.
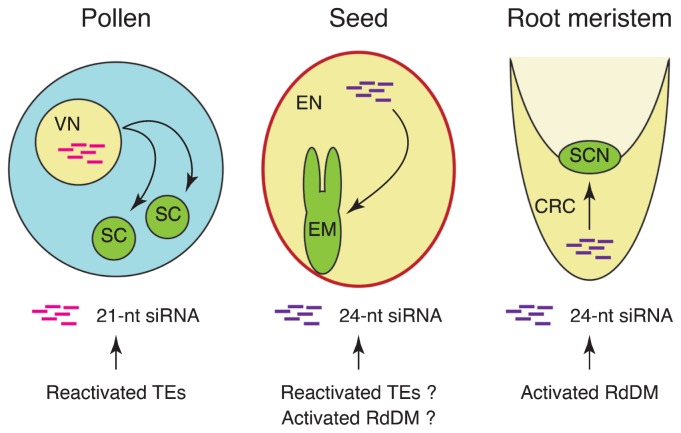
Transposable elements are typically silenced by DNA methylation; however, they are explicitly reactivated in the vegetative cell nucleus (Slotkin et al. 2009). The chromatin in the vegetative nucleus is decondensed, whereas the sperm nuclei are compact (Slotkin et al. 2009). The DNA glycosylase genes DME, ROS1, DML2, and DML3 are expressed in the vegetative nucleus, but not in the sperm (Schoft et al. 2011). These DNA glycosylases mediate the demethylation of small AT-rich euchromatic TEs in the vegetative nucleus, which reactivates these TEs (Calarco et al. 2012, Ibarra et al. 2012). In contrast, in sperm, these TEs undergo DME-dependent hypermethylation suggesting there is a link between demethylation and de-condensation in the vegetative nucleus and hypermethylation along with compact chromatin in the sperm (Calarco et al. 2012, Ibarra et al. 2012). The transcripts of reactivated TEs are degraded in the RNAi pathway, like trans-acting siRNA-generating TAS transcripts (Creasey et al. 2014). The vegetative nucleus-specific expression of a truncated GFP gene fused to miRNA173 or an endogenous 21-nt transposon siRNA target site produces a 21-nt siRNA that can target GFP, leading to non-cell autonomous silencing of the sperm cell-specific expression of GFP (Grant-Downton et al. 2013, Martínez et al. 2016, Slotkin et al. 2009). These results suggest that the 21-nt siRNAs produced from reactivated TEs in the vegetative nucleus are transported to the sperm (Fig. 1). Such non-canonical RdDM 21-nt siRNAs may reinforce TE silencing in the sperm germline. It is noteworthy that several protein-coding genes silenced by DNA methylation are also reactivated specifically in the vegetative nucleus, and are important for pollen tube growth and development (Schmitz et al. 2013b).
Fig. 1.
Epigenetic silencing of transposable elements (TEs) reinforced by siRNAs produced in companion cells. In developing pollen grains, TEs are reactivated via demethylation in the vegetative nucleus. The TE transcripts are converted to dsRNAs, then degraded into 21-nt siRNAs. These 21-nt siRNAs are transported to sperm cells and reinforce the DNA methylation within TEs. In developing seeds, 24-nt siRNAs are produced by activated RdDM or from reactivated TEs in the endosperm. These 24-nt siRNAs are transported to the embryo and reinforce TE silencing in the embryo. In the root meristem, 24-nt siRNAs are over-produced in the columella cells by activated RdDM. These 24-nt siRNAs may be transported to the stem cell niche, where they reinforce TE silencing. VN: vegetative nucleus, SC: sperm cell, EN: endosperm, EM: embryo, CRC: columella root cap, SCN: stem cell niche.
During double fertilization, the sperm cells fertilize the egg cell and the central cell to produce the embryo and the endosperm. The endosperm genome is globally hypomethylated in all contexts, relative to the embryo genome (Gehring et al. 2009, Hsieh et al. 2009). The hypomethylation of the endosperm genome occurs only in maternal chromosomes. Additionally, the hypomethylation in the endosperm depends on DME activity, similar to that in pollen grains (Hsieh et al. 2009, Ibarra et al. 2012). The DME gene is expressed in the central cell before fertilization, and the demethylation is initiated in the central cells (Park et al. 2016). Maternal-specific demethylation contributes to the maternal-specific expression of imprinted genes. Hypomethylation in the endosperm has also been observed in rice and maize (Wang et al. 2015, Zemach et al. 2010). In rice, the both CG and CHG hypomethylation in the endosperm are associated with the endosperm-specific expression of some seed storage protein genes and starch synthase genes. As described above, DME targets AT-rich euchromatic TEs. Hypomethylated TEs in the endosperm are hypermethylated in the embryo, suggesting the non-cell autonomous regulation of TE silencing in the embryo is due to siRNAs produced in the endosperm (Ibarra et al. 2012). The expression of an endosperm-specific artificial microRNA targeting GFP results in silencing of embryo-specific expression of GFP, suggests transfer of siRNAs from the endosperm to the embryo. Some TEs are weakly reactivated in the developing seed suggesting that, like in pollen grains, epigenetically activated TE transcripts are the source of 21-nt siRNAs (Hsieh et al. 2009, Slotkin et al. 2009). However, 24-nt Pol IV-dependent siRNAs, rather than 21-nt siRNAs, are explicitly produced from the maternal chromosomes in the endosperm suggesting that, like in pollen grains, endosperm-derived 24-nt siRNAs are transported to the embryo and reinforce TE silencing (Lu et al. 2012, Mosher et al. 2009). A link between the weak reactivation of TEs and the increased production of 24-nt siRNAs in the endosperm has not been established.
Plants have stem cell niches in the shoot apical meristem (SAM) and root apical meristem (RAM). These stem cells are responsible for shoot and root architecture patterning and are affected by TE activities. Hypermethylation in the SAM and the RAM due to the increased abundance of RdDM factors and DDM1 likely reinforces the TE silencing in the meristems (Baubec et al. 2014). To elucidate the dynamics underlying the TE silencing mediated by DNA methylation in the RAM, DNA methylation patterns for specific cell types within the RAM needs to be clarified. Manually collecting specific cell types from somatic tissues is not feasible. Thus, dissociating single cells from somatic tissues by enzymatically digesting the cell wall, and a subsequent fluorescence-activated cell sorting analysis enables researchers to distinguish cells producing fluorescent proteins (e.g., GFP) from the various cells of somatic tissues. Among the six major cell types of the RAM (epidermis, cortex, endodermis, stele, whole columella root cap, and lower columella), the whole columella root cap is globally hypermethylated in all contexts, but especially in the CHH context (Kawakatsu et al. 2016b). Global CHH hypermethylation has also been observed in the lower columella, indicating that CHH hypermethylation is a signature of columella cells.
Among all of the Arabidopsis cell types that have been analyzed, CHH hypermethylation is greatest in columella cells. Columella CHH hypermethylation occurs primarily in the pericentromeric regions, in which TEs are abundant, but also occurs in chromosomal arms. Local DNA methylation changes were identified as CG-only differentially methylated regions (CG-DMRs), non-CG only DMRs (CH-DMRs), and CG and non-CG DMRs (C-DMRs). The CH-DMRs are the major DMRs among root cell types and more than 70% of the CH-DMRs overlap with all classes of TEs. The DNA methylation patterns in CG- and C-DMRs and the transcriptional profiles are more similar between cell types originating from the same initial cells than between cell types derived from different initial cells. However, DNA methylation patterns in CH-DMRs are more dependent on the physical position in the RAM, suggesting that positional information or cell-to-cell communication also influence the regulation of DNA methylation patterns. The CHH hypermethylation in the columella is accompanied by an over-accumulation of 24-nt siRNAs, likely due to the upregulated expression of siRNA biogenesis machinery genes. The DNA methylation within TE bodies is primarily dependent on either RdDM or CMT2, and, in leaves, 24-nt siRNAs do not accumulate within CMT2-dependent TE bodies. In the columella, RdDM-dependent TEs as well as CMT2-dependent TEs exhibit CHH hypermethylation with an over-accumulation of 24-nt siRNAs, indicating that an enhanced RdDM is responsible for a genome-wide CHH hypermethylation in the columella. The downregulated expression of heterochromatin-related component genes may suggest that heterochromatin is loosened in the columella. Decondensed heterochromatin in the columella may be responsible for the enhanced production of 24-nt siRNAs within heterochromatin, where CMT2, but not RdDM, is responsible for DNA methylation. The biological importance of enhanced RdDM in the columella is unclear because these cells are sloughed into the soil soon after differentiating from initial cells but likely does not involve extensive TE silencing. Columella cells are adjacent to the stem cell niches in the RAM, which are presumably vulnerable to TE activities. One attractive hypothesis is that excessive amounts of 24-nt siRNAs produced in the columella are transported into the stem cell niches to reinforce TE silencing, analogous to the cell non-autonomous TE silencing in the reproductive cells (Fig. 1).
Developmentally regulated DNA methylation
During embryogenesis, plants form the basis of their architecture with two apical meristems and a few leaves. Meanwhile, the embryo and/or endosperm store energy and amino acid reserves for germination. After maturing, dry seeds can remain dormant for an extended period until conditions are favorable for germination. In developing seeds, CHH methylation of TEs increases, but not CG or CHG methylations (Bouyer et al. 2017, Kawakatsu et al. 2017, Lin et al. 2017, Narsai et al. 2017).
Additionally, CHH hypermethylation decreases in the dry seeds of the drm1 drm2 cmt3 triple mutant, and is absent in the dry seeds of the drm1 drm2 cmt2 cmt3 quadruple mutant, suggesting that both RdDM and CMT2 are responsible for the CHH methylation occurring in developing seeds. In contrast, the CHH methylation of TEs drastically decreases during germination. The global demethylation resets the hypermethylation in dry seeds. A lack of DNA demethylases does not affect the global demethylation during germination. Therefore, the global demethylation during germination likely occurs passively, in which methylation is diluted because of repeated cell divisions. Intriguingly, both RdDM components and CMT2 are produced, and 24-nt siRNA levels are relatively unchanged during germination. This suggests that unknown factor(s) inhibit de novo re-methylation or that cells are dividing so quickly that de novo re-methylation cannot compensate for the passive demethylation.
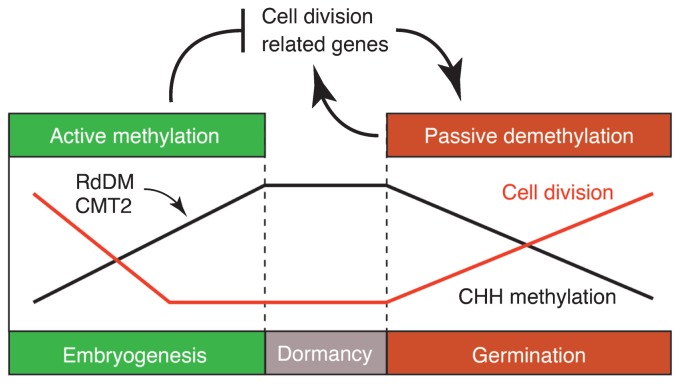
Many of the genes exhibiting upregulated expression upon germination are associated with cell division and cell wall organization. These genes tend to have nearby DMRs that are methylated during seed development and demethylated during germination. This raises the possibility of the epigenetic regulation of germination and the existence of a positive feedback loop between passive demethylation and induction of cell division-related genes (Fig. 2; Kawakatsu et al. 2017). These dynamic changes to CHH methylation have also been observed in rice and soybean, implying that the epigenomic reconfiguration during seed development and germination is widely conserved in the plant kingdom. The drm1 drm2 cmt2 cmt3 quadruple mutant exhibits normal seed development, with minor transcriptome changes, although TEs are reactivated, suggesting that in Arabidopsis, CHH hypermethylation during seed development is a failsafe mechanism for TE silencing (Lin et al. 2017). A maternally transmitted defect in RdDM increases the seed abortion rate and severely decreases seed size in Brassica rapa, which produces seeds that are much larger than those of the related Arabidopsis (Grover et al. 2018). Closer examination of an Arabidopsis mutant with defective RdDM, also revealed a decrease in the weight of seeds, although the extent was much smaller than that observed for B. rapa seeds. The diversity in the embryo and endosperm sizes in aborted seeds may reflect an asynchronous seed abortion in B. rapa RdDM mutants, suggesting that a sudden increase in TE activities due to TE reactivation terminates normal seed development at random times.
Fig. 2.
Hypothesized epigenetic regulation of seed germination. During seed development, global CHH methylation levels (black line) increase, whereas the cell division rate (red line) decreases toward maturation. Under conditions that are favorable for germination, cell division is induced, leading to passive CHH demethylation. Cell division-related genes whose expression levels are upregulated in response to germination are often located near regions affected by CHH methylation reconfigurations. According to this model, reconfiguration of CHH methylation may help to regulate the seed germination process.
Population-wide DNA methylation diversity
In addition to genetic variations, natural epigenetic variation might also be shape phenotypic diversity and adaptation. Analyses of DNA methylomes from more than 1000 Arabidopsis accessions revealed extensive epigenomic variation with 38% of the reference genome differentially methylated among these accessions (Dubin et al. 2015, Kawakatsu et al. 2016a, Schmitz et al. 2013b). While CG-DMRs mainly overlap with protein-coding genes related to housekeeping processes, the CH-DMRs overlap with TEs and intergenic regions, and the C-DMRs overlap with TEs and/or protein-coding genes whose expression levels vary across tissues or environments.
Earlier investigations involving the reference Arabidopsis accession Col-0 indicated that gbM may be associated with the exclusion of the histone variant H2A.z from gene bodies, leading to constitutive gene expression (Tran et al. 2005, Zhang et al. 2006, Zilberman et al. 2007). Across accessions, genes that undergo gbM tend to be more highly expressed than unmethylated genes or genes that undergo teM. However, gbM variation is significantly larger than transcriptome variation among accessions. Additionally, global gene expression levels in accessions that nearly lack gbM are similar to those of accessions with higher gbM levels, which is consistent with the observation that the loss of gbM in the met1 mutant does not affect gbM gene expression patterns (Bewick et al. 2016). Moreover, gbM is conserved between orthologous genes, but two Brassicaceae species completely lack gbM, presumably because of the absence of CMT3 (Bewick et al. 2016). Similar global expression patterns have been observed for orthologs in Arabidopsis and these two Brassicaceae species. Therefore, gbM is associated with mild constitutive gene expression, but there is no clear evidence of its impact on transcriptomes, at least under normal growth conditions.
Arguably the most striking finding from the 1000 epigenomes population study is that one-quarter of all protein-coding genes (7,524 genes) are poly-epiallelic (PE) genes, which undergo gbM in some accessions and teM in other accessions (Kawakatsu et al. 2016a). The ratio of teM epialleles of PE genes is much lower than that of gbM epialleles, and only one accession has teM epialleles in approximately 30% of the PE genes, suggesting that many teM epialleles of PE genes are newly formed in gbM genes. There are several possible explanations for the emergence of poly-epialleles. First, RdDM may have spread from nearby newly inserted TEs. Second, siRNAs produced from the newly formed inverted repeats at unlinked loci (e.g., PAI loci) may reinforce DNA methylation (Luff et al. 1999). Third, aberrant mRNA from gbM genes may be subjected to non-canonical RdDM (Cuerda-Gil and Slotkin 2016). Last, purely spontaneous reversions may occur, as gbM may have evolved from teM (Bewick et al. 2016). The PE genes are enriched with genes involved in signaling and metabolic pathways, with an emphasis on phosphorylation-related and immune response-related genes. Among the analyzed PE genes that have gbM and teM epialleles in at least five accessions, 10% of the genes exhibit an association between DNA methylation and gene expression, with the expression levels of teM epialleles significantly lower than those of gbM epialleles under normal growth conditions. Thus, the epigenetic regulation of PE genes may provide a mechanism for increasing phenotypic diversity and plant adaptation.
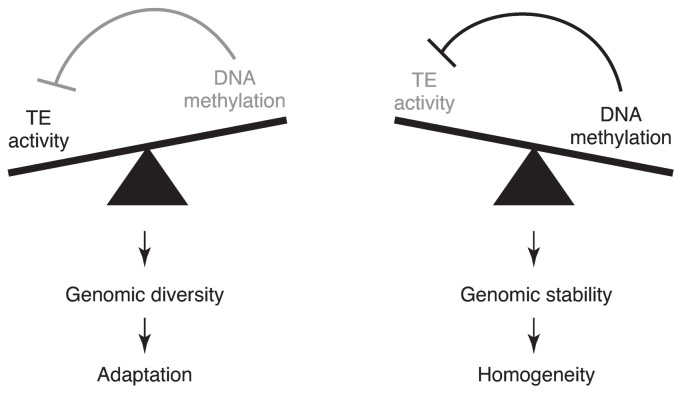
The correlation between genome-wide methylation and the place of origin suggests there is a genetic basis for methylation variation (Kawakatsu et al. 2016a). A genome-wide association study revealed the associations between RNA silencing or DNA methyltransferase activities and genome-wide methylation levels. The methylation levels within RdDM-targeted TEs are associated with SNPs linked with AGO1, NRPD1B, and AGO9, whereas those of CMT2-targeted TEs are associated with SNPs linked with CMT2 and AGO9. Natural variation in AGO9 expression patterns may help to regulate TE methylation (Rodriguez-Leal et al. 2015). Additionally, gbM levels are reasonably associated with SNPs linked with MET1. Relatively high DNA methylation levels within TEs likely repress TE expression, resulting in increased genomic integrity and homogeneity within the population. However, relatively low DNA methylation levels may allow an increase in TE expression and transposition. When TEs are expressed, mobilized and inserted into genes, the affected gene may be knocked out, or the expression of nearby genes may be positively or negatively altered. In some cases, TE insertions may cause nearby genes to become responsive to stress (Naito et al. 2009). Therefore, decreasing DNA methylation levels potentially leads to genomic diversity and possibly enable adaptations to environmental changes. Natural variation in identified genes may provide a balance between adaptation and population homogeneity (Fig. 3).
Fig. 3.
The balance between global DNA methylation levels and transposable element (TE) activities potentially influences population diversity. Natural variation in several components involved in the DNA methylation pathway are associated with global DNA methylation levels. Lower DNA methylation levels are conducive for TE activation, whereas higher DNA methylation levels can silence TEs. Because TE activities are associated with genomic integrity, such natural variation may function as biological rheostat.
DNA methylation and adaptation
Both abiotic and biotic stress can change DNA methylation patterns globally or locally. However, transgenerational inheritance of stress-induced epigenetic variation is controversial, because there have been few comprehensive analyses. Phosphate starvation increases DNA methylation around highly induced genes, especially in the adjacent TEs in rice (Secco et al. 2015). Interestingly, this process unlikely depends on RdDM, because DCL3 is dispensable for TE hypermethylation. These DNA methylation changes occur after transcriptional changes, possibly reflecting the failsafe system to inactivate TEs in the vicinity of accessible chromatin. However, the hypermethylation is recovered in the following generations.
Repeated hyper-osmotic stress for over 5 generations confers tolerance against osmotic stress in Arabidopsis (Wibowo et al. 2016). This stress memory is transmitted to the next generation, suggesting epigenetic regulation. Indeed, repetitive hyper-osmotic stress induces both hypermethylation and hypomethylation within TEs and 2-kb upstream regions of protein coding genes in non-CG contexts, although hypermethylation is dominant. One stress-induced hyper-DMR locates upstream of MYB20, that is involved in abscisic acid signaling and stress tolerance (Wibowo et al. 2016). Progenies with the stress memory show the decreased expression of MYB20 under salt stress condition, whereas the down regulation of MYB20 is not observed in progenies without stress memory. On the other hand, one stress-induced hypo-DMR locates downstream of CNI1 (Wibowo et al. 2016). Salt stress induces the expression of lncRNA including antisense CNI1, which downregulates the expression of CNI1 under hyperosmotic stress. DNA methylation at the CNI1-downstream hypo-DMR represses the expression of the lncRNA under normal condition, but hypermethylation allows its expression under salt stress. Reciprocal crossing revealed the stress memory is transmitted through the female germline (Wibowo et al. 2016). This biased sexual transmission is likely caused by DME-dependent resetting of stress-induced DNA methylation changes in the male germline, because repeatedly osmotic stressed dme is more tolerant than stressed wild type. Stress-induced DNA methylation changes are inherited to the next generation, however, they are not further inherited to their offspring without stress. Another study also identified repetitive hyper-osmotic stress for over 10 generations induced DNA methylation changes, especially in CG context (Jiang et al. 2014). The rate of accumulated epimutation is significantly higher in progenies of repeatedly salt stress-treated plants than progenies of control plants. Among CG-DMRs accumulated during 10 generations with stress and one generation without stress, over 75% of CG-DMRs are inherited to the next generation, suggesting that some stress-induced DMRs can be inherited to subsequent multi-generational progenies even without stress.
Growing seeds in discreate patches and collecting only dispersed seeds creates the artificial dynamic landscape, therefore simulates selection (Fakheran et al. 2010). Starting with 19 RILs between Cvi and Ler, selected populations after five rounds of selection experiments showed later flowering and increased number of branches and siliques, accompanied by significantly reduced genetic variations, in which only 2 genotypes dominated the all populations (Schmid et al. 2018). Epigenetic variation, that is, single DNA methylation polymorphisms, among these populations were also reduced, compared to their ancestors, suggesting that epigenetic variation is also subjected to selection. Although there is no global correlation between differentially methylated cytosines accumulated during selection and gene expression, the expression of a lncRNA At2g06002 was negatively associated with DNA methylation (Schmid et al. 2018). Selected populations tended to lose DNA methylation within At2g06002 and showed the higher expression of At2g06002. Association of lower DNA methylation, higher gene expression and delayed flowering time was also observed among natural accessions. At2g06002 locates upstream of FIP1, and the expression levels of At2g06002 and FIP1 were correlated. Since FIP1 interacts with FRIGIDA involved in flowering time regulation, this association may suggest a possible link between selection-associated hypomethylation of At2g06002 and FIP1-FRIGIDA-mediated delayed flowering, thus can contribute to rapid adaptation (Schmid et al. 2018).
DNA methylation-associated gene activation
As described above, DNA methylation is associated with gene silencing. However, in some cases, DNA methylation can also be associated with gene activation (Harris et al. 2018). A forward genetic screen identified SUVH1 as an anti-silencing factor (Li et al. 2016). SUVH1 is required for both transgene and endogenous genes with methylated promoters. SUVH1 and its close homolog SUVH3 were also identified as methylated DNA binding proteins (Harris et al. 2018). SUVH1 and SUVH3 colocalize with RdDM target regions. SUVH1 and SUVH3 form a protein complex with chaperone proteins DNAJ1 and DNAJ2. DNAJ1 and DNAJ2 are essential for SUVH1/SUVH3 anti-silencing activities. Additionally, recruiting DNAJ1 to promoters enhanced reporter gene expression. Finally, constitutive expression of DNAJ1 upregulated the expression of genes proximal to DNAJ1 binding sites. Interestingly, FWA, which is stably silenced in the wild type plants, could not be reactivated by DNAJ1, suggesting that DNAJ1 activity is effective only on expressed genes. The underlying mechanisms of DNAJ1’s preference for expressed genes are unknown, but are keys to enhanced expression of proximal genes, whereas TE expression remains silent.
Agronomic traits associated with DNA methylation
Previous studies have characterized the association between DNA methylation and observable phenotypes that are potentially important for crop yield or quality. Some of these phenotypes were initially described several decades ago. The peloric toadflax (Linaria vulgaris), which is a naturally occurring epi-mutant that was initially described in 1744, has radially symmetrical flowers, whereas the wild-type plant produces bilaterally symmetrical flowers (Gustafsson 1979). Mutations in the homeobox gene CYC lead to a similar phenotype in snapdragon (Antirrhinum majus), suggesting that a mutation in Lcyc, which is a toadflax homolog of CYC, is responsible for the peloric phenotype (Cubas et al. 1999, Luo et al. 1999). Indeed, Lcyc is transcriptionally silenced in the peloric mutant; however, no mutation was observed in the coding region or in the approximately 1-kb upstream region. Interestingly, the link between the peloric phenotype and Lcyc was initially identified by a restriction fragment length polymorphism, likely not by a genetic variant but because of the use of a DNA methylation-sensitive enzyme Sau3AI. Thus, a heavily methylated Lcyc is likely the basis for the peloric phenotype. Indeed, the occasional reversion of the peloric phenotype appears to be correlated with the demethylation of Lcyc.
Paramutation
Paramutation, which is also a classical epigenetic phenomenon (Chandler 2007), refers to an interaction between a paramutagenic allele and a paramutable allele. Both alleles can have identical DNA sequences. The paramutagenic allele induces a heritable change in the paramutable allele, which becomes paramutagenic. The paramutagenic state is heritable even after the original paramutagenic allele is lost during segregation. The paramutagenic R-stippled (R-st) allele at the r1 locus confers the spotted pigmentation of pericarps, whereas the paramutable R-r allele is responsible for full pigmentation (Kermicle et al. 1995). Although F1 pericarps with R-r/R-st are fully pigmented, the pericarps of their progenies with the R-r allele exhibit decreased pigmentation. Loss of pigmentation is associated with the downregulated expression of the r gene and increased DNA methylation at the r locus (Walker 1998). A silenced R-r allele (R-r’) is inherited by the progenies, which revert to R-r phenotypes in a few generations (Brown and Brink 1960). The paramutagenic B’ allele at the b1 gene, involved in anthocyanin biosynthesis, confers the light pigmentation of the whole plant body, whereas the paramutable Booster-Intense (B-I) allele confers the dark pigmentation (Stam et al. 2002b). Seven tandem repeats of approximately 850 bp spanning about 6 kb located 100 kb upstream of b1 are required for the paramutation of b1 (Stam et al. 2002b). A transcriptionally active B-I allele is associated with increased chromatin accessibility and DNA methylation, compared to those in silenced B’ allele (Stam et al. 2002a). In contrast to the frequent reversion from R-st to R-r, a newly established B’ from B-I is stable, with no reports describing the reversion from B’ to B-I. Several other paramutations have been reported in maize and in other plant species (Das and Messing 1994, Hollick et al. 1995, Pilu et al. 2009). A forward genetic screen identified genes required for paramutations in maize, including MOP genes and RMR genes (Chandler 2007). These genes are required for siRNA production, and contribute to RdDM, except for RMR2, implying that RdDM regulates paramutation. However, the transcriptional gene silencing (TGS) induced by RdDM in transgenic plants is less stable than that induced by paramutation when the trigger T-DNA is lost during segregation. Additionally, the alleles silenced by RdDM are not paramutagenic. Therefore, DNA methylation cannot solely explain paramutation.
Incompatibility
Sterility and inviability may result from crossing of distinct accessions (hybrid incompatibility) or the same accessions (self-incompatibility). Hybrid incompatibility produces reproductive barriers, whereas self-incompatibility leads to outcrossing. In Arabidopsis, the hybrid incompatibility between Col-0 and Shandara (Sha) is caused by the combined effects of the duplicated genes FOLT1 and FOLT2 (Durand et al. 2012). Both Col-0 and Sha possess FOLT1, whereas only Sha carries FOLT2 along with two truncated FOLT2 copies. The complete FOLT2 sequence and the rearranged truncated FOLT2 copies produce siRNAs that induce RdDM at the FOLT1 locus that silences FOLT1 in Sha. In contrast, the Col-0 FOLT1 allele is actively expressed. Additionally, FOLT2 is actively expressed in Sha. Recombinant inbred lines with insufficient FOLT transcripts (i.e. silenced FOLT1 and lack of FOLT2) are not viable.
Histidine biosynthesis is essential for viability. Arabidopsis possesses two histidinol-phosphate aminotransferase genes (HISN6A and HISN6B). The Col-0 HISN6A allele is actively expressed, whereas the Cvi HISN6A allele is mutated and non-functional. The Col-0 HISN6B allele is silenced by CG and CHG methylation, whereas the Cvi HISN6B allele is actively expressed (Blevins et al. 2017). Recombinant inbred lines with the non-functional Cvi HISN6A allele and the silenced Col-0 HISN6B allele are not viable because of inhibited histidine biosynthesis.
In Brassica species, the recognition of self or non-self is controlled by haplotypes of the S locus encoding SP11/SCR and SRK (Kitashiba and Nasrallah 2014). The encoded SP11/SCR and SRK proteins function as the pollen-derived ligand and the stigmatic receptor, respectively, and the identical haplotype at the S locus causes self-incompatibility (Fujii et al. 2016). Additionally, SP11/SCR is expressed in the tapetum, and the dominance relationships between SP11/SCR determine the self-incompatibility phenotype in pollen grains (Kusaba et al. 2002, Shiba et al. 2002). In heterozygotes with both dominant and recessive SP11/SCR, the recessive SP11/SCR is silenced by the methylation within the promoter region, leading to the monoallelic expression of the dominant SP11/SCR allele (Shiba et al. 2006). The dominant S haplotype includes the inverted repeat(s) similar to those in the promoter region of the recessive SP11/SCR allele in the vicinity of the dominant SP11/SCR allele (Tarutani et al. 2010). The inverted repeat encoded by the dominant S haplotype is also expressed in the tapetum and produces 24-nt siRNAs targeting the promoter region of the recessive SP11/SCR allele in trans.
Sex determination
Most flowering plants, including crops, produce bisexual flowers with pistils and stamens. In addition to self-incompatibility, unisexual flowers that have either pistils or stamens enhance outcrossing. Consequently, sex determination is related to the expansion of genetic diversity. In the female flowers of melon (Cucumis melo), ethylene produced in the carpel primordia by the 1-aminocyclopropane-1-carboxylate synthase CmACS-7 represses stamen development (Boualem et al. 2008). In male flowers, the C2H2 zinc-finger transcription factor CmWIP1 arrests carpel development and indirectly represses CmACS-7 expression. Moreover, the insertion of a hAT DNA transposon into the CmWIP1 promoter converts a male flower to a female flower because of the dispersion of DNA methylation due to the hAT transposon and the subsequent silencing of CmWIP1 (Martin et al. 2009).
Diploid persimmon (Diospyros lotus) is a dioecious species, in which an individual plant has either male or female flowers, whereas hexaploid persimmon (Diospyros kaki) is a monoecious species, in which an individual plant has both male and female flowers. Homeodomain transcription factor genes MeGI and OGI help mediate the sex determination in persimmon (Akagi et al. 2014). The encoded MeGI protein represses anther development in female flowers. The Y-chromosome-encoded pseudogene OGI includes inverted repeats and produces 21-nt siRNAs targeting MeGI. In D. lotus, these 21-nt siRNAs post-transcriptionally silence MeGI, resulting in male flowers with fertile stamens. In D. kaki, OGI expression is suppressed by the insertion of a Kali-type SINE retrotransposon in the promoter region. Sex determination in D. kaki depends on the expression of MeGI (Akagi et al. 2016). Specifically, MeGI is silenced in male flowers because of DNA methylation at the MeGI locus, and the spontaneous conversion to female flowers is associated with demethylation at this locus. Additionally, zebularine treatment inhibits anther development in D. kaki male flower buds, likely because of the associated re-activation of MeGI expression.
Fruit ripening
Many fruits are edible and are important components of human/animal diets. Ripening alters fruit texture, flavor, taste, color, and nutrition. In addition to the plant hormone ethylene, the SQUAMOSA promoter-binding protein-like transcription factor CNR and the MADS-box transcription factor RIN are essential for fruit ripening in tomato (Solanum lycopersicum) (Eriksson et al. 2004, Thompson et al. 1999, Vrebalov et al. 2002). The dominant mutant Cnr and the semi-dominant mutant rin exhibit pleiotropic phenotypes, including the production of colorless fruits and delayed softening. In mature Cnr fruits, the CNR promoter is methylated in CG and CHG contexts, which silences CNR (Manning et al. 2006). In contrast, the same region is demethylated during ripening in wild-type fruits. Rare, but occasional revertant sectors in Cnr fruits are consistent with the epigenetic regulation of CNR (Zhong et al. 2013). Artificial global demethylation induced by a 5-azacitidine treatment causes premature ripening. During fruit ripening, the promoters of various ripening-related genes that are directly targeted by RIN are frequently demethylated by the DNA demethylase SlDML2 (Lang et al. 2017, Liu et al. 2015). In the fruits of SlDML2 knock-down/knock-out transgenic plants, the demethylation of ripening-related genes is inhibited, which results in downregulated expression.
Vitamin E (VTE) is a valuable nutrient for humans. The VTE content in ripe fruits is higher for the wild tomato species Solanum pennellii than for the cultivated tomato S. lycopersicum. A 2-methyl-6-phytylquinol methyltransferase, VTE3(1), catalyzes the final steps of tocopherol biosynthesis, and VTE3(1) expression is correlated with VTE content (Almeida et al. 2011). In S. lycopersicum, down-regulated VTE3(1) expression has been associated with the insertion of a SINE retrotransposon in the promoter and hypermethylation of the inserted SINE (Quadrana et al. 2014). The spontaneous demethylation of the VTE3(1) promoter leads to the upregulated expression of VTE3(1) and an increase in fruit VTE content.
Harvested tomato fruits are stored under cool conditions to extend their shelf-life, leading to a loss of flavor due to altered volatile synthesis. Cold storage downregulates the expression of RIN and some of its targets involved in fruit maturation and volatile synthesis. The expression of these genes resumes when plants are exposed to normal temperatures. The transient repression of these genes induced by a cold treatment is accompanied by increased DNA methylation within their promoters (Zhang et al. 2016). The expression of SlDML2 is also transiently downregulated during cold storage but is immediately upregulated at normal temperatures, suggesting that SlDML2 contributes to changes to chilling-responsive DNA methylation and gene expression.
Somaclonal variation
Tissue cultures are widely used for clonal propagations and the generation of transgenic crops. However, abnormal phenotypes often arise after tissue culture processes related to dedifferentiation and regeneration. These phenomena are called somaclonal variations and have been applied for mutagenesis studies aimed at improving agronomic traits. Single nucleotide variants and small insertions/deletions are sources of somaclonal variations. In rice, reactivation of the Copia-type LTR retrotransposon Tos17 located on chromosome 7 reportedly results in somaclonal variations (Hirochika et al. 1996). Additionally, Tos17 is methylated throughout the life cycle but is demethylated by the DNA demethylase DNG701 in calli (La et al. 2011). The knockout of DNG701 decreases Tos17 activity, while the overexpression of DNG701 has the opposite effect, indicating that DNA methylation has important effects on Tos17 silencing.
Aberrant DNA methylation reprogramming is also a source of somaclonal variation. In rice calli grown under tissue culture conditions, loss of DNA methylation may occur stochastically. The loss of DNA methylation phenotype is randomly inherited by regenerated plants and can affect the expression of nearby genes. Notably, there are rice genome regions particularly susceptible to the loss of DNA methylation. In the dominant df mutant, hypomethylation in the promoter region of the rice homolog of FIE1, which encodes an Esc-like component of PRC2, induces the ectopic expression of FIE1 (Zhang et al. 2012). Indeed, the constitutive expression of FIE1 results in the same phenotype as that of the df mutant. Another representative example of the link between aberrant DNA methylations and somaclonal variations is the mantled phenotype of African oil palm (Ong-Abdullah et al. 2015), which is important for the production of edible oils and biofuels. Clonal propagation is widely used to improve yields. However, tissue culture techniques often induce the hypomethylation of a LINE retrotransposon in the intron of the homolog of the B-class MADS box gene DEFICIENS, resulting in alternative splicing and premature termination of expression. These changes result in the conversion of stamens and staminodes to pseudocarpels, leading to the production of parthenocarpic flowers and decreased oil yields.
Conclusions and perspectives
Plants potentially employ epigenome regulatory processes as a survival strategy, including for the maintenance of genome stability in germline cells and adaptation during cell differentiation and under long-term or transient stress conditions. Studies mainly involving Arabidopsis have revealed the mechanisms underlying DNA methylation regulation and dynamics. However, DNA methylation patterns and the set of DNA methylation associated genes are different among plant species, suggesting the importance of methylome analysis in individual crops (Bewick et al. 2016, Li et al. 2014, Niederhuth et al. 2016, Stroud et al. 2013). Advances in high-throughput sequencing techniques have enabled the identification of agronomic traits controlled by epigenetic regulation. Applying methods for creating even greater epigenomic variation may help breeders develop crops with new properties such as better qualities or those better enable to adapt to global environmental changes. Both targeted and global methods for epigenome editing represent an attractive new approach for plant breeding. Targeted de novo DNA methylation and gene silencing can be induced by expressing siRNAs. In addition, tethering SUVH2 to target gene promoters by an engineered zinc-finger induces DNA methylation and results in gene silencing (Johnson et al. 2014). Conversely, recruiting human TET1 by an artificial zinc-finger cause DNA demethylation of target genes and their reactivation (Gallego-Bartolomé et al. 2018). Other genome editing tools, such as transcription activator-like effector and dead Cas9 (dCas9) that loses nuclease activity, might also be applied to targeted epigenome editing in crops (Luo et al. 2018). Although DNA methylation deficit mutants are viable in Arabidopsis, they are often lethal in crops (Hu et al. 2014, Li et al. 2014, Moritoh et al. 2012, Yamauchi et al. 2014). Constitutive expression of TET1 randomly induces DNA demethylation so that resulting individual transgenic plants have distinct methylomes, leading to phenotypic variation (Ji et al. 2018). Since induced DNA demethylation is relatively mild, it is feasible to apply TET1-mediated epimutagenesis to crops. One anticipated problem of epigenome-edited crops involves the reversion of methylation status. Further characterizing the mechanisms underlying DNA methylation and demethylation may help researchers overcome the problems associated with epigenome-edited crops to improve sustainable agricultural practices.
Acknowledgments
This work was supported by JSPS KAKENHI grants (17H05851, 17H03753 and 19H04873) (to T.K.). J.R.E. is an investigator at the Howard Hughes Medical Institute.
Abbreviations
- ACS
1-Aminocyclopropane-1-carboxylate synthase
- AGO4/6
ARGONAUTE 4/6
- AtLIG1
ARABIDOPSIS DNA LIGASE 1
- b1
booster 1
- CLSY1–4
CLASSY 1–4
- CMT2/3
CHROMOMETHYRASE 2/3
- CNI1
CARBON/NITROGEN INSENSITIVE 1
- CNR
COLORLESS NON-RIPENING
- CYC
CYCLOIDEA
- DCL3
DICER-LIKE 3
- DDM1
DECREASED IN DNA METHYLATION 1
- df
DWARF AND FLOWER ABERRANT
- DME
DEMETER
- DML1–3
DEMETER LIKE 1–3
- DMR
Differentially methylated region
- DMS3
DEFECTIVE IN MERISTEM SILENCING 3
- DNAJ
DnaJ-domain containing chaperone protein
- DRD1
DEFECTIVE IN RNA-DIRECTED DNA METHYLATION 1
- DRM2
DOMAINS OF REARRANGED METHYLTRANSFERASE 2
- dsRNA
Double-stranded RNA
- FIE1
FERTILIZATION-INDEPENDENT ENDOSPERM1
- FIS2
FERTILIZATION-INDEPENDENT SEED 2
- FIP1
FRIGIDA INTERACTING PROTEIN1
- FOLT1
FOLATE TRANSPORTER1
- FWA
FLOWERING WAGENINGEN
- gbM
Gene body methylation
- GFP
Green fluorescent protein
- H3K4/9/27
Histone H3 lysine 4/9/27
- KYP
KRYPTONITE
- lncRNA
Long non-coding RNA
- MEA
MEDEA
- MeGI
MALE GROWTH INHIBITOR
- MET1
DNA METHYLTRANSFERASE 1
- MOP
MEDIATOR OF PARAMUTATION
- MPS1/PRD2
MULTIPOLAR SPINDLE 1/PUTATIVE RECOMBINATION INITIATION DEFECTS 2
- MYB20
MYB DOMAIN PROTEIN 20
- OGI
OPPRESSOR OF MEGI
- PAI
Phosphoribosylanthranilate isomerase
- PE gene
Poly-epiallelic gene
- PRC2
Polycomb repressive complex 2
- r1
red color 1
- RdDM
RNA-directed DNA methylation
- RDM1
RNA-DIRECTED METHYLATION 1
- RDR2
RNA-DEPENDENT RNA POLYMERASE 2
- RIL
Recombinant inbred line
- RIN
RIPENING INHIBITOR
- RMR
REQUIRED TO MAINTAIN REPRESSION
- ROS1
REPRESSOR OF SILENCING 1
- SHH1
SAWADEE HOMEODOMAIN HOMOLOGUE 1
- siRNA
Small interfering RNA
- SNP
Single nucleotide polymorphism
- SP11/SCR
S-locus protein 11/S-locus cysteine-rich
- SRK
S-receptor kinase
- SUVH2/4/5/6/9
SU(VAR) HOMOLOGUE 2/4/5/6/9
- TE
Transposable element
- teM
TE-like methylation
- TET1
Ten-eleven translocation methylcytosine dioxygenase 1
- VIM1
VARIATION IN DNA METHYLATION 1
- VTE
Vitamin E
Literature Cited
- Akagi, T., Henry, I.M., Tao, R. and Comai, L. (2014) Plant genetics. A Y-chromosome-encoded small RNA acts as a sex determinant in persimmons. Science 346: 646–650. [DOI] [PubMed] [Google Scholar]
- Akagi, T., Henry, I.M., Kawai, T., Comai, L. and Tao, R. (2016) Epigenetic regulation of the sex determination gene MeGI in polyploid persimmon. Plant Cell 28: 2905–2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida, J., Quadrana, L., Asís, R., Setta, N., de Godoy, F., Bermúdez, L., Otaiza, S.N., Corrêa da Silva, J.V., Fernie, A.R., Carrari, F.et al. (2011) Genetic dissection of vitamin E biosynthesis in tomato. J. Exp. Bot. 62: 3781–3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartee, L., Malagnac, F. and Bender, J. (2001) Arabidopsis cmt3 chromomethylase mutations block non-CG methylation and silencing of an endogenous gene. Genes Dev. 15: 1753–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baubec, T., Finke, A., Mittelsten Scheid, O. and Pecinka, A. (2014) Meristem-specific expression of epigenetic regulators safeguards transposon silencing in Arabidopsis. EMBO Rep. 15: 446–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewick, A.J., Ji, L., Niederhuth, C.E., Willing, E.M., Hofmeister, B.T., Shi, X., Wang, L., Lu, Z., Rohr, N.A., Hartwig, B.et al. (2016) On the origin and evolutionary consequences of gene body DNA methylation. Proc. Natl. Acad. Sci. USA 113: 9111–9116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blevins, T., Podicheti, R., Mishra, V., Marasco, M., Wang, J., Rusch, D., Tang, H. and Pikaard, C.S. (2015) Identification of Pol IV and RDR2-dependent precursors of 24 nt siRNAs guiding de novo DNA methylation in Arabidopsis. Elife 4: e09591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blevins, T., Wang, J., Pflieger, D., Pontvianne, F. and Pikaard, C.S. (2017) Hybrid incompatibility caused by an epiallele. Proc. Natl. Acad. Sci. USA 114: 3702–3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boualem, A., Fergany, M., Fernandez, R., Troadec, C., Martin, A., Morin, H., Sari, M.A., Collin, F., Flowers, J.M., Pitrat, M.et al. (2008) A conserved mutation in an ethylene biosynthesis enzyme leads to andromonoecy in melons. Science 321: 836–838. [DOI] [PubMed] [Google Scholar]
- Bouyer, D., Kramdi, A., Kassam, M., Heese, M., Schnittger, A., Roudier, F. and Colot, V. (2017) DNA methylation dynamics during early plant life. Genome Biol. 18: 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, D. and Brink, R. (1960) Paramutagenic action of paramutant Rr and Rg alleles in maize. Genetics 45: 1313–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarco, J.P., Borges, F., Donoghue, M.T., Van Ex, F., Jullien, P.E., Lopes, T., Gardner, R., Berger, F., Feijo, J.A., Becker, J.D.et al. (2012) Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell 151: 194–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler, V.L. (2007) Paramutation: from maize to mice. Cell 128: 641–645. [DOI] [PubMed] [Google Scholar]
- Choi, Y., Gehring, M., Johnson, L., Hannon, M., Harada, J.J., Goldberg, R.B., Jacobsen, S.E. and Fischer, R.L. (2002) DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in arabidopsis. Cell 110: 33–42. [DOI] [PubMed] [Google Scholar]
- Coleman-Derr, D. and Zilberman, D. (2012) Deposition of histone variant H2A.Z within gene bodies regulates responsive genes. PLoS Genet. 8: e1002988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creasey, K.M., Zhai, J., Borges, F., Van Ex, F., Regulski, M., Meyers, B.C. and Martienssen, R.A. (2014) miRNAs trigger widespread epigenetically activated siRNAs from transposons in Arabidopsis. Nature 508: 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubas, P., Vincent, C. and Coen, E. (1999) An epigenetic mutation responsible for natural variation in floral symmetry. Nature 401: 157–161. [DOI] [PubMed] [Google Scholar]
- Cuerda-Gil, D. and Slotkin, R.K. (2016) Non-canonical RNA-directed DNA methylation. Nat. Plants 2: 16163. [DOI] [PubMed] [Google Scholar]
- Daccord, N., Celton, J.M., Linsmith, G., Becker, C., Choisne, N., Schijlen, E., van de Geest, H., Bianco, L., Micheletti, D., Velasco, R.et al. (2017) High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat. Genet. 49: 1099–1106. [DOI] [PubMed] [Google Scholar]
- Das, O.P. and Messing, J. (1994) Variegated phenotype and developmental methylation changes of a maize allele originating from epimutation. Genetics 136: 1121–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowen, R.H., Pelizzola, M., Schmitz, R.J., Lister, R., Dowen, J.M., Nery, J.R., Dixon, J.E. and Ecker, J.R. (2012) Widespread dynamic DNA methylation in response to biotic stress. Proc. Natl. Acad. Sci. USA 109: E2183–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, J., Johnson, L.M., Groth, M., Feng, S., Hale, C.J., Li, S., Vashisht, A.A., Gallego-Bartolome, J., Wohlschlegel, J.A., Patel, D.J.et al. (2014) Mechanism of DNA methylation-directed histone methylation by KRYPTONITE. Mol. Cell 55: 495–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubin, M.J., Zhang, P., Meng, D., Remigereau, M.S., Osborne, E.J., Paolo Casale, F., Drewe, P., Kahles, A., Jean, G., Vilhjalmsson, B.et al. (2015) DNA methylation in Arabidopsis has a genetic basis and shows evidence of local adaptation. Elife 4: e05255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand, S., Bouché, N., Perez Strand, E., Loudet, O. and Camilleri, C. (2012) Rapid establishment of genetic incompatibility through natural epigenetic variation. Curr. Biol. 22: 326–331. [DOI] [PubMed] [Google Scholar]
- Ebbs, M.L., Bartee, L. and Bender, J. (2005) H3 lysine 9 methylation is maintained on a transcribed inverted repeat by combined action of SUVH6 and SUVH4 methyltransferases. Mol. Cell Biol. 25: 10507–10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbs, M.L. and Bender, J. (2006) Locus-specific control of DNA methylation by the Arabidopsis SUVH5 histone methyltransferase. Plant Cell 18: 1166–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson, E.M., Bovy, A., Manning, K., Harrison, L., Andrews, J., De Silva, J., Tucker, G.A. and Seymour, G.B. (2004) Effect of the Colorless non-ripening mutation on cell wall biochemistry and gene expression during tomato fruit development and ripening. Plant Physiol. 136: 4184–4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakheran, S., Paul-Victor, C., Heichinger, C., Schmid, B., Grossniklaus, U. and Turnbull, L.A. (2010) Adaptation and extinction in experimentally fragmented landscapes. Proc. Natl. Acad. Sci. USA 107: 19120–19125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii, S., Kubo, K. and Takayama, S. (2016) Non-self- and self-recognition models in plant self-incompatibility. Nat. Plants 2: 16130. [DOI] [PubMed] [Google Scholar]
- Fujimoto, R., Uezono, K., Ishikura, S., Osabe, K., Peacock, W.J. and Dennis, E.S. (2018) Recent research on the mechanism of heterosis is important for crop and vegetable breeding systems. Breed. Sci. 68: 145–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Bartolomé, J., Gardiner, J., Liu, W., Papikian, A., Ghoshal, B., Kuo, H.Y., Zhao, J.M., Segal, D.J. and Jacobsen, S.E. (2018) Targeted DNA demethylation of the Arabidopsis genome using the human TET1 catalytic domain. Proc. Natl. Acad. Sci. USA 115: E2125–E2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Z., Liu, H., Daxinger, L., Pontes, O., He, X., Qian, W., Lin, H., Xie, M., Lorkovic, Z., Zhang, S.et al. (2010) An RNA polymerase II- and AGO4-associated protein acts in RNA-directed DNA methylation. Nature 465: 106–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring, M., Bubb, K.L. and Henikoff, S. (2009) Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science 324: 1447–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant-Downton, R., Kourmpetli, S., Hafidh, S., Khatab, H., Le Trionnaire, G., Dickinson, H. and Twell, D. (2013) Artificial microRNAs reveal cell-specific differences in small RNA activity in pollen. Curr. Biol. 23: R599–601. [DOI] [PubMed] [Google Scholar]
- Grover, J.W., Kendall, T., Baten, A., Burgess, D., Freeling, M., King, G.J. and Mosher, R.A. (2018) Maternal components of RNA-directed DNA methylation are required for seed development in Brassica rapa. Plant J. 94: 575–582. [DOI] [PubMed] [Google Scholar]
- Gustafsson, A. (1979) Linnaeus’ Peloria: The history of a monster. Theor. Appl. Genet. 54: 241–248. [DOI] [PubMed] [Google Scholar]
- Harris, C.J., Scheibe, M., Wongpalee, S.P., Liu, W., Cornett, E.M., Vaughan, R.M., Li, X., Chen, W., Xue, Y., Zhong, Z.et al. (2018) A DNA methylation reader complex that enhances gene transcription. Science 362: 1182–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havecker, E.R., Wallbridge, L.M., Hardcastle, T.J., Bush, M.S., Kelly, K.A., Dunn, R.M., Schwach, F., Doonan, J.H. and Baulcombe, D.C. (2010) The Arabidopsis RNA-directed DNA methylation argonautes functionally diverge based on their expression and interaction with target loci. Plant Cell 22: 321–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirochika, H., Sugimoto, K., Otsuki, Y., Tsugawa, H. and Kanda, M. (1996) Retrotransposons of rice involved in mutations induced by tissue culture. Proc. Natl. Acad. Sci. USA 93: 7783–7788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollick, J.B., Patterson, G.I., Coe, E.H., Cone, K.C. and Chandler, V.L. (1995) Allelic interactions heritably alter the activity of a metastable maize pl allele. Genetics 141: 709–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain, M.S., Kawakatsu, T., Kim, K.D., Zhang, N., Nguyen, C.T., Khan, S.M., Batek, J.M., Joshi, T., Schmutz, J., Grimwood, J.et al. (2017) Divergent cytosine DNA methylation patterns in single-cell, soybean root hairs. New Phytol. 214: 808–819. [DOI] [PubMed] [Google Scholar]
- Hsieh, T.F., Ibarra, C.A., Silva, P., Zemach, A., Eshed-Williams, L., Fischer, R.L. and Zilberman, D. (2009) Genome-wide demethylation of Arabidopsis endosperm. Science 324: 1451–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, L., Li, N., Xu, C., Zhong, S., Lin, X., Yang, J., Zhou, T., Yuliang, A., Wu, Y., Chen, Y.R.et al. (2014) Mutation of a major CG methylase in rice causes genome-wide hypomethylation, dysregulated genome expression, and seedling lethality. Proc. Natl. Acad. Sci. USA 111: 10642–10647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra, C.A., Feng, X., Schoft, V.K., Hsieh, T.F., Uzawa, R., Rodrigues, J.A., Zemach, A., Chumak, N., Machlicova, A., Nishimura, T.et al. (2012) Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science 337: 1360–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, L., Jordan, W.T., Shi, X., Hu, L., He, C. and Schmitz, R.J. (2018) TET-mediated epimutagenesis of the Arabidopsis thaliana methylome. Nat. Commun. 9: 895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, C., Mithani, A., Belfield, E.J., Mott, R., Hurst, L.D. and Harberd, N.P. (2014) Environmentally responsive genome-wide accumulation of de novo Arabidopsis thaliana mutations and epimutations. Genome Res. 24: 1821–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, L.M., Du, J., Hale, C.J., Bischof, S., Feng, S., Chodavarapu, R.K., Zhong, X., Marson, G., Pellegrini, M., Segal, D.J.et al. (2014) SRA- and SET-domain-containing proteins link RNA polymerase V occupancy to DNA methylation. Nature 507: 124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankel, M.W., Ramsey, D.E., Stokes, T.L., Flowers, S.K., Haag, J.R., Jeddeloh, J.A., Riddle, N.C., Verbsky, M.L. and Richards, E.J. (2003) Arabidopsis MET1 cytosine methyltransferase mutants. Genetics 163: 1109–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakatsu, T., Huang, S.S., Jupe, F., Sasaki, E., Schmitz, R.J., Urich, M.A., Castanon, R., Nery, J.R., Barragan, C., He, Y.et al. (2016a) Epigenomic diversity in a global collection of Arabidopsis thaliana accessions. Cell 166: 492–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakatsu, T., Stuart, T., Valdes, M., Breakfield, N., Schmitz, R.J., Nery, J.R., Urich, M.A., Han, X., Lister, R., Benfey, P.N.et al. (2016b) Unique cell-type-specific patterns of DNA methylation in the root meristem. Nat. Plants 2: 16058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakatsu, T., Nery, J.R., Castanon, R. and Ecker, J.R. (2017) Dynamic DNA methylation reconfiguration during seed development and germination. Genome Biol. 18: 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermicle, J.L., Eggleston, W.B. and Alleman, M. (1995) Organization of paramutagenicity in R-stippled maize. Genetics 141: 361–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita, T., Yadegari, R., Harada, J.J., Goldberg, R.B. and Fischer, R.L. (1999) Imprinting of the MEDEA polycomb gene in the Arabidopsis endosperm. Plant Cell 11: 1945–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita, T., Miura, A., Choi, Y., Kinoshita, Y., Cao, X., Jacobsen, S.E., Fischer, R.L. and Kakutani, T. (2004) One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science 303: 521–523. [DOI] [PubMed] [Google Scholar]
- Kitashiba, H. and Nasrallah, J.B. (2014) Self-incompatibility in Brassicaceae crops: lessons for interspecific incompatibility. Breed. Sci. 64: 23–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler, C., Hennig, L., Spillane, C., Pien, S., Gruissem, W. and Grossniklaus, U. (2003) The Polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1. Genes Dev. 17: 1540–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusaba, M., Tung, C.W., Nasrallah, M.E. and Nasrallah, J.B. (2002) Monoallelic expression and dominance interactions in anthers of self-incompatible Arabidopsis lyrata. Plant Physiol. 128: 17–20. [PMC free article] [PubMed] [Google Scholar]
- La, H., Ding, B., Mishra, G.P., Zhou, B., Yang, H., del Bellizzi, M. R., Chen, S., Meyers, B.C., Peng, Z., Zhu, J.K.et al. (2011) A 5-methylcytosine DNA glycosylase/lyase demethylates the retrotransposon Tos17 and promotes its transposition in rice. Proc. Natl. Acad. Sci. USA 108: 15498–15503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang, Z., Wang, Y., Tang, K., Tang, D., Datsenka, T., Cheng, J., Zhang, Y., Handa, A.K. and Zhu, J.K. (2017) Critical roles of DNA demethylation in the activation of ripening-induced genes and inhibition of ripening-repressed genes in tomato fruit. Proc. Natl. Acad. Sci. USA 114: E4511–E4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law, J.A. and Jacobsen, S.E. (2010) Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11: 204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law, J.A., Du, J., Hale, C.J., Feng, S., Krajewski, K., Palanca, A.M., Strahl, B.D., Patel, D.J. and Jacobsen, S.E. (2013) Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1. Nature 498: 385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le, T.N., Schumann, U., Smith, N.A., Tiwari, S., Au, P.C., Zhu, Q.H., Taylor, J.M., Kazan, K., Llewellyn, D.J., Zhang, R.et al. (2014) DNA demethylases target promoter transposable elements to positively regulate stress responsive genes in Arabidopsis. Genome Biol. 15: 458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei, M., Zhang, H., Julian, R., Tang, K., Xie, S. and Zhu, J.K. (2015) Regulatory link between DNA methylation and active demethylation in Arabidopsis. Proc. Natl. Acad. Sci. USA 112: 3553–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q., Eichten, S.R., Hermanson, P.J., Zaunbrecher, V.M., Song, J., Wendt, J., Rosenbaum, H., Madzima, T.F., Sloan, A.E., Huang, J.et al. (2014) Genetic perturbation of the maize methylome. Plant Cell 26: 4602–4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S., Liu, L., Li, S., Gao, L., Zhao, Y., Kim, Y.J. and Chen, X. (2016) SUVH1, a Su(var)3–9 family member, promotes the expression of genes targeted by DNA methylation. Nucleic Acids Res. 44: 608–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., Córdoba-Cañero, D., Qian, W., Zhu, X., Tang, K., Zhang, H., Ariza, R.R., Roldán-Arjona, T. and Zhu, J.K. (2015) An AP endonuclease functions in active DNA demethylation and gene imprinting in Arabidopsis. PLoS Genet. 11: e1004905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, J.Y., Le, B.H., Chen, M., Henry, K.F., Hur, J., Hsieh, T.F., Chen, P.Y., Pelletier, J.M., Pellegrini, M., Fischer, R.L.et al. (2017) Similarity between soybean and Arabidopsis seed methylomes and loss of non-CG methylation does not affect seed development. Proc. Natl. Acad. Sci. USA 114: E9730–E9739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroth, A.M., Cao, X., Jackson, J.P., Zilberman, D., McCallum, C.M., Henikoff, S. and Jacobsen, S.E. (2001) Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 292: 2077–2080. [DOI] [PubMed] [Google Scholar]
- Lister, R., O’Malley, R.C., Tonti-Filippini, J., Gregory, B.D., Berry, C.C., Millar, A.H. and Ecker, J.R. (2008) Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133: 523–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, R., How-Kit, A., Stammitti, L., Teyssier, E., Rolin, D., Mortain-Bertrand, A., Halle, S., Liu, M., Kong, J., Wu, C.et al. (2015) A DEMETER-like DNA demethylase governs tomato fruit ripening. Proc. Natl. Acad. Sci. USA 112: 10804–10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, J., Zhang, C., Baulcombe, D.C. and Chen, Z.J. (2012) Maternal siRNAs as regulators of parental genome imbalance and gene expression in endosperm of Arabidopsis seeds. Proc. Natl. Acad. Sci. USA 109: 5529–5534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luff, B., Pawlowski, L. and Bender, J. (1999) An inverted repeat triggers cytosine methylation of identical sequences in Arabidopsis. Mol. Cell 3: 505–511. [DOI] [PubMed] [Google Scholar]
- Luo, C., Hajkova, P. and Ecker, J.R. (2018) Dynamic DNA methylation: In the right place at the right time. Science 361: 1336–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, D., Carpenter, R., Copsey, L., Vincent, C., Clark, J. and Coen, E. (1999) Control of organ asymmetry in flowers of Antirrhinum. Cell 99: 367–376. [DOI] [PubMed] [Google Scholar]
- Luo, M., Bilodeau, P., Dennis, E.S., Peacock, W.J. and Chaudhury, A. (2000) Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc. Natl. Acad. Sci. USA 97: 10637–10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning, K., Tör, M., Poole, M., Hong, Y., Thompson, A.J., King, G.J., Giovannoni, J.J. and Seymour, G.B. (2006) A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat. Genet. 38: 948–952. [DOI] [PubMed] [Google Scholar]
- Martin, A., Troadec, C., Boualem, A., Rajab, M., Fernandez, R., Morin, H., Pitrat, M., Dogimont, C. and Bendahmane, A. (2009) A transposon-induced epigenetic change leads to sex determination in melon. Nature 461: 1135–1138. [DOI] [PubMed] [Google Scholar]
- Martínez, G., Panda, K., Köhler, C. and Slotkin, R.K. (2016) Silencing in sperm cells is directed by RNA movement from the surrounding nurse cell. Nat. Plants 2: 16030. [DOI] [PubMed] [Google Scholar]
- Martínez-Macías, M.I., Qian, W., Miki, D., Pontes, O., Liu, Y., Tang, K., Liu, R., Morales-Ruiz, T., Ariza, R.R., Roldán-Arjona, T.et al. (2012) A DNA 3′ phosphatase functions in active DNA demethylation in Arabidopsis. Mol. Cell 45: 357–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke, M.A. and Mosher, R.A. (2014) RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat. Rev. Genet. 15: 394–408. [DOI] [PubMed] [Google Scholar]
- McCue, A.D., Panda, K., Nuthikattu, S., Choudury, S.G., Thomas, E.N. and Slotkin, R.K. (2015) ARGONAUTE 6 bridges transposable element mRNA-derived siRNAs to the establishment of DNA methylation. EMBO J. 34: 20–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritoh, S., Eun, C.H., Ono, A., Asao, H., Okano, Y., Yamaguchi, K., Shimatani, Z., Koizumi, A. and Terada, R. (2012) Targeted disruption of an orthologue of DOMAINS REARRANGED METHYLASE 2, OsDRM2, impairs the growth of rice plants by abnormal DNA methylation. Plant J. 71: 85–98. [DOI] [PubMed] [Google Scholar]
- Mosher, R.A., Melnyk, C.W., Kelly, K.A., Dunn, R.M., Studholme, D.J. and Baulcombe, D.C. (2009) Uniparental expression of PolIV-dependent siRNAs in developing endosperm of Arabidopsis. Nature 460: 283–286. [DOI] [PubMed] [Google Scholar]
- Naito, K., Zhang, F., Tsukiyama, T., Saito, H., Hancock, C.N., Richardson, A.O., Okumoto, Y., Tanisaka, T. and Wessler, S.R. (2009) Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature 461: 1130–1134. [DOI] [PubMed] [Google Scholar]
- Narsai, R., Gouil, Q., Secco, D., Srivastava, A., Karpievitch, Y.V., Liew, L.C., Lister, R., Lewsey, M.G. and Whelan, J. (2017) Extensive transcriptomic and epigenomic remodelling occurs during Arabidopsis thaliana germination. Genome Biol. 18: 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederhuth, C.E., Bewick, A.J., Ji, L., Alabady, M.S., Kim, K.D., Li, Q., Rohr, N.A., Rambani, A., Burke, J.M., Udall, J.A.et al. (2016) Widespread natural variation of DNA methylation within angiosperms. Genome Biol. 17: 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong-Abdullah, M., Ordway, J.M., Jiang, N., Ooi, S.E., Kok, S.Y., Sarpan, N., Azimi, N., Hashim, A.T., Ishak, Z., Rosli, S.K.et al. (2015) Loss of Karma transposon methylation underlies the mantled somaclonal variant of oil palm. Nature 525: 533–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, K., Kim, M.Y., Vickers, M., Park, J.S., Hyun, Y., Okamoto, T., Zilberman, D., Fischer, R.L., Feng, X., Choi, Y.et al. (2016) DNA demethylation is initiated in the central cells of Arabidopsis and rice. Proc. Natl. Acad. Sci. USA 113: 15138–15143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilu, R., Panzeri, D., Cassani, E., Cerino Badone, F., Landoni, M. and Nielsen, E. (2009) A paramutation phenomenon is involved in the genetics of maize low phytic acid1-241 (lpa1-241) trait. Heredity (Edinb) 102: 236–245. [DOI] [PubMed] [Google Scholar]
- Quadrana, L., Almeida, J., Asís, R., Duffy, T., Dominguez, P.G., Bermúdez, L., Conti, G., Corrêa da Silva, J.V., Peralta, I.E., Colot, V.et al. (2014) Natural occurring epialleles determine vitamin E accumulation in tomato fruits. Nat. Commun. 5: 3027. [DOI] [PubMed] [Google Scholar]
- Rajakumara, E., Law, J.A., Simanshu, D.K., Voigt, P., Johnson, L.M., Reinberg, D., Patel, D.J. and Jacobsen, S.E. (2011) A dual flip-out mechanism for 5mC recognition by the Arabidopsis SUVH5 SRA domain and its impact on DNA methylation and H3K9 dimethylation in vivo. Genes Dev. 25: 137–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regulski, M., Lu, Z., Kendall, J., Donoghue, M.T., Reinders, J., Llaca, V., Deschamps, S., Smith, A., Levy, D., McCombie, W.R.et al. (2013) The maize methylome influences mRNA splice sites and reveals widespread paramutation-like switches guided by small RNA. Genome Res. 23: 1651–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Leal, D., Leon-Martinez, G., Abad-Vivero, U. and Vielle-Calzada, J.P. (2015) Natural variation in epigenetic pathways affects the specification of female gamete precursors in Arabidopsis. Plant Cell 27: 1034–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, M.W., Heichinger, C., Coman Schmid, D., Guthörl, D., Gagliardini, V., Bruggmann, R., Aluri, S., Aquino, C., Schmid, B., Turnbull, L.A.et al. (2018) Contribution of epigenetic variation to adaptation in Arabidopsis. Nat. Commun. 9: 4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz, R.J., He, Y., Valdes-Lopez, O., Khan, S.M., Joshi, T., Urich, M.A., Nery, J.R., Diers, B., Xu, D., Stacey, G.et al. (2013a) Epigenome-wide inheritance of cytosine methylation variants in a recombinant inbred population. Genome Res. 23: 1663–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz, R.J., Schultz, M.D., Urich, M.A., Nery, J.R., Pelizzola, M., Libiger, O., Alix, A., McCosh, R.B., Chen, H., Schork, N.J.et al. (2013b) Patterns of population epigenomic diversity. Nature 495: 193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoft, V.K., Chumak, N., Choi, Y., Hannon, M., Garcia-Aguilar, M., Machlicova, A., Slusarz, L., Mosiolek, M., Park, J.S., Park, G.T.et al. (2011) Function of the DEMETER DNA glycosylase in the Arabidopsis thaliana male gametophyte. Proc. Natl. Acad. Sci. USA 108: 8042–8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secco, D., Wang, C., Shou, H., Schultz, M.D., Chiarenza, S., Nussaume, L., Ecker, J.R., Whelan, J. and Lister, R. (2015) Stress induced gene expression drives transient DNA methylation changes at adjacent repetitive elements. Elife 4: e09343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba, H., Iwano, M., Entani, T., Ishimoto, K., Shimosato, H., Che, F.S., Satta, Y., Ito, A., Takada, Y., Watanabe, M.et al. (2002) The dominance of alleles controlling self-incompatibility in Brassica pollen is regulated at the RNA level. Plant Cell 14: 491–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba, H., Kakizaki, T., Iwano, M., Tarutani, Y., Watanabe, M., Isogai, A. and Takayama, S. (2006) Dominance relationships between selfincompatibility alleles controlled by DNA methylation. Nat. Genet. 38: 297–299. [DOI] [PubMed] [Google Scholar]
- Slotkin, R.K., Vaughn, M., Borges, F., Tanurdzic, M., Becker, J.D., Feijo, J.A. and Martienssen, R.A. (2009) Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136: 461–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam, M., Belele, C., Dorweiler, J.E. and Chandler, V.L. (2002a) Differential chromatin structure within a tandem array 100 kb upstream of the maize b1 locus is associated with paramutation. Genes Dev. 16: 1906–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam, M., Belele, C., Ramakrishna, W., Dorweiler, J.E., Bennetzen, J.L. and Chandler, V.L. (2002b) The regulatory regions required for B′ paramutation and expression are located far upstream of the maize b1 transcribed sequences. Genetics 162: 917–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud, H., Greenberg, M.V., Feng, S., Bernatavichute, Y.V. and Jacobsen, S.E. (2013) Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 152: 352–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud, H., Do, T., Du, J., Zhong, X., Feng, S., Johnson, L., Patel, D.J. and Jacobsen, S.E. (2014) Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat. Struct. Mol. Biol. 21: 64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, K., Lang, Z., Zhang, H. and Zhu, J.K. (2016) The DNA demethylase ROS1 targets genomic regions with distinct chromatin modifications. Nat. Plants 2: 16169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarutani, Y., Shiba, H., Iwano, M., Kakizaki, T., Suzuki, G., Watanabe, M., Isogai, A. and Takayama, S. (2010) Trans-acting small RNA determines dominance relationships in Brassica self-incompatibility. Nature 466: 983–986. [DOI] [PubMed] [Google Scholar]
- Thompson, A.J., Tor, M., Barry, C.S., Vrebalov, J., Orfila, C., Jarvis, M.C., Giovannoni, J.J., Grierson, D. and Seymour, G.B. (1999) Molecular and genetic characterization of a novel pleiotropic tomato-ripening mutant. Plant Physiol. 120: 383–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, R.K., Henikoff, J.G., Zilberman, D., Ditt, R.F., Jacobsen, S.E. and Henikoff, S. (2005) DNA methylation profiling identifies CG methylation clusters in Arabidopsis genes. Curr. Biol. 15: 154–159. [DOI] [PubMed] [Google Scholar]
- Turco, G.M., Kajala, K., Kunde-Ramamoorthy, G., Ngan, C.Y., Olson, A., Deshphande, S., Tolkunov, D., Waring, B., Stelpflug, S., Klein, P.et al. (2017) DNA methylation and gene expression regulation associated with vascularization in Sorghum bicolor. New Phytol. 214: 1213–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vielle-Calzada, J.P., Thomas, J., Spillane, C., Coluccio, A., Hoeppner, M.A. and Grossniklaus, U. (1999) Maintenance of genomic imprinting at the Arabidopsis medea locus requires zygotic DDM1 activity. Genes Dev. 13: 2971–2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrebalov, J., Ruezinsky, D., Padmanabhan, V., White, R., Medrano, D., Drake, R., Schuch, W. and Giovannoni, J. (2002) A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296: 343–346. [DOI] [PubMed] [Google Scholar]
- Walker, E.L. (1998) Paramutation of the r1 locus of maize is associated with increased cytosine methylation. Genetics 148: 1973–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, J., Gao, H., Zhang, J., Aldridge, B., Vickers, M., Higgins, J.D. and Feng, X. (2018) Sexual-lineage-specific DNA methylation regulates meiosis in Arabidopsis. Nat. Genet. 50: 130–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P., Xia, H., Zhang, Y., Zhao, S., Zhao, C., Hou, L., Li, C., Li, A., Ma, C. and Wang, X. (2015) Genome-wide high-resolution mapping of DNA methylation identifies epigenetic variation across embryo and endosperm in Maize (Zea may). BMC Genomics 16: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibowo, A., Becker, C., Marconi, G., Durr, J., Price, J., Hagmann, J., Papareddy, R., Putra, H., Kageyama, J., Becker, J.et al. (2016) Hyperosmotic stress memory in Arabidopsis is mediated by distinct epigenetically labile sites in the genome and is restricted in the male germline by DNA glycosylase activity. Elife 5: e13546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, B.P., Pignatta, D., Henikoff, S. and Gehring, M. (2015) Methylation-sensitive expression of a DNA demethylase gene serves as an epigenetic rheostat. PLoS Genet. 11: e1005142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo, H.R., Pontes, O., Pikaard, C.S. and Richards, E.J. (2007) VIM1, a methylcytosine-binding protein required for centromeric heterochromatinization. Genes Dev. 21: 267–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Z., Johansen, L.K., Gustafson, A.M., Kasschau, K.D., Lellis, A.D., Zilberman, D., Jacobsen, S.E. and Carrington, J.C. (2004) Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2: E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi, T., Johzuka-Hisatomi, Y., Terada, R., Nakamura, I. and Iida, S. (2014) The MET1b gene encoding a maintenance DNA methyltransferase is indispensable for normal development in rice. Plant Mol. Biol. 85: 219–232. [DOI] [PubMed] [Google Scholar]
- Zemach, A., Kim, M.Y., Silva, P., Rodrigues, J.A., Dotson, B., Brooks, M.D. and Zilberman, D. (2010) Local DNA hypomethylation activates genes in rice endosperm. Proc. Natl. Acad. Sci. USA 107: 18729–18734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach, A., Kim, M.Y., Hsieh, P.H., Coleman-Derr, D., Eshed-Williams, L., Thao, K., Harmer, S.L. and Zilberman, D. (2013) The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 153: 193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai, J., Bischof, S., Wang, H., Feng, S., Lee, T.F., Teng, C., Chen, X., Park, S.Y., Liu, L., Gallego-Bartolome, J.et al. (2015) A one precursor one siRNA model for Pol IV-dependent siRNA biogenesis. Cell 163: 445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, B., Tieman, D.M., Jiao, C., Xu, Y., Chen, K., Fei, Z., Giovannoni, J.J. and Klee, H.J. (2016) Chilling-induced tomato flavor loss is associated with altered volatile synthesis and transient changes in DNA methylation. Proc. Natl. Acad. Sci. USA 113: 12580–12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. and Zhu, J.K. (2012) Active DNA demethylation in plants and animasl. Cold Spring Harb. Symp. Quant. Biol. 77: 161–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Cheng, Z., Qin, R., Qiu, Y., Wang, J.L., Cui, X., Gu, L., Zhang, X., Guo, X., Wang, D.et al. (2012) Identification and characterization of an epi-allele of FIE1 reveals a regulatory linkage between two epigenetic marks in rice. Plant Cell 24: 4407–4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., Yazaki, J., Sundaresan, A., Cokus, S., Chan, S.W., Chen, H., Henderson, I.R., Shinn, P., Pellegrini, M., Jacobsen, S.E.et al. (2006) Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126: 1189–1201. [DOI] [PubMed] [Google Scholar]
- Zhong, S., Fei, Z., Chen, Y.R., Zheng, Y., Huang, M., Vrebalov, J., McQuinn, R., Gapper, N., Liu, B., Xiang, J.et al. (2013) Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat. Biotechnol. 31: 154–159. [DOI] [PubMed] [Google Scholar]
- Zhou, M., Palanca, A.M.S. and Law, J.A. (2018) Locus-specific control of the de novo DNA methylation pathway in Arabidopsis by the CLASSY family. Nat. Genet. 50: 865–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman, D., Gehring, M., Tran, R.K., Ballinger, T. and Henikoff, S. (2007) Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 39: 61–69. [DOI] [PubMed] [Google Scholar]