Abstract
To evaluate and utilize potentially valuable quantitative trait loci or genes of wild relatives in the genetic background of domesticated crop species, chromosome segment substitution lines (CSSLs) are a valuable tool. CSSLs can be constructed through the exchange of chromosome segments of AA genome species of the genus Oryza with cultivated rice, Oryza sativa L. Here we report the development of three sets of CSSLs carrying segments of AA genome species closely related to Oryza sativa—O. glaberrima (IRGC 103777 from Mali), O. rufipogon (W1962 from China), and O. nivara (IRGC 105715 from Cambodia)—in the genetic background of ssp. japonica cultivar Taichung 65 through the use of 101 to 121 simple-sequence-repeat markers in whole-genome genotyping and marker-assisted selection. The materials are available via the National Bioresource Project (Rice) Oryzabase Web page.
Keywords: experimental genetic resources, introgression lines, wild species, simple sequence repeat markers, Oryzabase
Introduction
Closed related species of domesticated crop possesses potentially valuable genetic resources that are not present in the gene pools of cultivated species. Harlan and de Wet (1971) suggested three sources of gene pools, which they called primary (via intraspecific hybridization among cultivated species: GP1), secondary (via interspecific hybridization with closely related compatible species: GP2), and tertiary (via interspecific hybridization with more distantly related species through radical artificial treatment such as embryo rescue: GP3). A breeder has made extensive efforts to exploit wild genetic resources, mainly for pest and disease resistance; more than 80% of the favorable traits conferred by gene transfer from wild species involve resistance to pests and diseases in major crops (reviewed by Hajjar and Hodgkin 2007, Prescott-Allen and Prescott-Allen 1988). Genes that improve drought and salinity tolerance, yield components, and grain and fruit quality also have been introgressed from wild germplasms (Dandan et al. 2007, Lippman et al. 2007, Nevo and Chen 2010, Zhang et al. 2014). However, unfavorable traits or genes, selected out through domestication and breeding, are frequently transferred also. Therefore, the utility of wild germplasm as a genetic resource has both benefits and drawbacks.
Cultivated rice, Oryza sativa L., a staple food in much of the world, was domesticated from the ancestral species O. rufipogon Griff. The AA genome consist of two cultivated species, O sativa L. and O. glaberrima Steud., and six wild species, O. rufipogon, O. nivara Sharma et Shastry, O. barthii A. Chev., O. longistaminata A. Chev. et Roehr., O. glumaepatula Steud., and O. meridionalis Ng (Vaughan et al. 2008). Although chromosome pairing in F1 hybrids among AA genome species is normal and gene exchange is possible in hybrid progeny, interspecific hybrids show reproductive isolation. With the progress of Next Generation Sequencing, public databases have rapidly accumulated reference sequences (Reuscher et al. 2018, Sakai et al. 2014, Schatz et al. 2014, Stein et al. 2018), haplotype maps (Alexandrov et al. 2015, Huang et al. 2012, McCouch et al. 2016, Meyer et al. 2016, Wang et al. 2014), and RNA transcription profiles (Childs et al. 2011, Sato et al. 2013, Tian et al. 2015). However, the use of wild genetic sequences in hybrid progeny is hindered by many unfavorable traits including seed dormancy, short-day requirement, lodging, seed shattering at harvest, and various maladaptive phenomena such as hybrid sterility, lethality, and breakdown. So far, genes conferring tolerance or resistance to abiotic and biotic stresses from wild species have been incorporated into cultivated species (Khush 1997, Sanchez et al. 2013). However, the vast array of allelic variations in wild germplasm has not been exploited to accelerate rice breeding and to deepen our understanding of the genetic architecture of wild species.
A chromosome segment substitution line (CSSL) is a line carrying several chromosome segments derived from a donor parent in the genetic background of a recurrent parent. A full set of CSSLs covers the whole genome. Using CSSLs, we can evaluate minor allelic differences conferred by additive quantitative trait loci (QTLs) in a uniform genetic background. Their high detection power makes it possible to manipulate a QTL as a simple Mendelian factor, subsequently allowing gene isolation by positional cloning. In addition, they offer the potential for favorable genes hidden in the genetic background of related species to be discovered in the genetic background of cultivated species (Arbelaez et al. 2015, Bessho-Uehara et al. 2017, Cheema et al. 2008, Doi et al. 1997, Furuta et al. 2014, Gutiérrez et al. 2010, He et al. 2017, Hirabayashi et al. 2010, Qiao et al. 2016, Ramos et al. 2016, Rangel et al. 2008, Shim et al. 2010, Tian et al. 2006, Yang et al. 2016). Therefore, the genetic resources of related species in GP1 or GP2 could be transferred into the genetic background of cultivated species to form a foundation for studies of genetic variation of closed related rice.
We have created chromosome segment substitution lines (CSSL) of O. glumaepatula, designated GLU-ILs, and O. meridionalis, designated MER-ILs, using the term ‘introgression lines’ (ILs) to refer to CSSLs based on intraspecific hybridization (Yoshimura et al. 2010). Here we offer new ILs of O. rufipogon, O. nivara, and O. glaberrima in the genetic background of the O. sativa ssp. japonica type cultivar Taichung 65 (T65). Applications for seed sharing are accepted through Oryzabase (https://shigen.nig.ac.jp/rice/oryzabase/).
Materials and Methods
Plant materials
The O. glaberrima, O. rufipogon, and O. nivara accessions were kindly provided by the International Rice Research Institute (IRRI), Manila, the Philippines (‘IRGC’ accessions), and the National Institute of Genetics, Mishima, Japan (‘W’ accessions). Line IRGC 103777 (O. glaberrima) originated from Mali, IRGC 105715 (O. nivara) from Cambodia, and W1962 (O. rufipogon) from China. Their derived isolates were respectively designated WK18, WK56, and WK1962. F1 hybrids carrying either T65 or O. glaberrima cytoplasm were obtained from reciprocal crosses between T65 and WK18. F1 hybrids carrying T65 cytoplasm were also obtained by pollination with either O. rufipogon or O. nivara pollen. T65 was used as the recurrent male parent to develop BC1F1, BC2F1, BC3F1, and BC4F1 plants. F1, BC1F1, and BC2F1 plants were grown in pots under short-day treatment (10 h dark, 14 h light) to promote heading and the later generation were grown in paddy field at the Harumachi farm of Kyushu University, Fukuoka, Japan.
Genotyping
Genomic DNA was extracted from freeze-dried leaves according to Dellaporta et al. (1983) with minor modifications. Simple-sequence-repeat (SSR) markers were used for genotyping of the whole genomic region (Supplemental Tables 1–3). PCR reaction mixtures (15 μL) contained 50 mM KCl, 10 mM Tris·HCl (pH 9.0), 1.5 mM MgCl2, 200 μM each dNTP, 0.2 μM each primer, 0.75 units of GoTaq polymerase (Promega), and template DNA (~5 ng) in a GeneAmp PCR system 9700 (Applied Biosystems, CA, USA). Thermal cycling for PCR started with 95°C for 5 min, followed by 35 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s. PCR products were run in 4% agarose gels (Amresco, OH, USA) in 0.5× TBE buffer to separate polymorphic DNA bands.
Results and Discussion
WK18ILs (O. glaberrima)
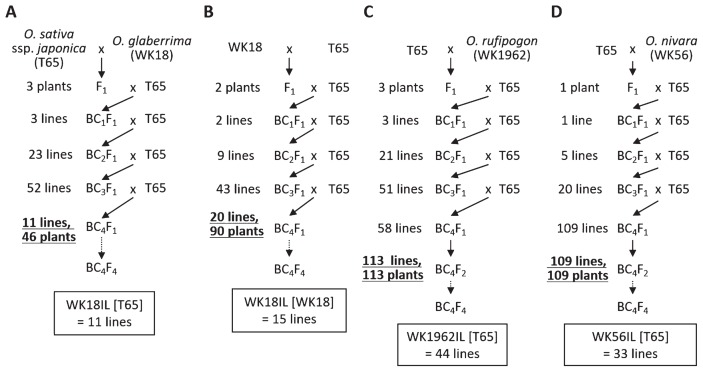
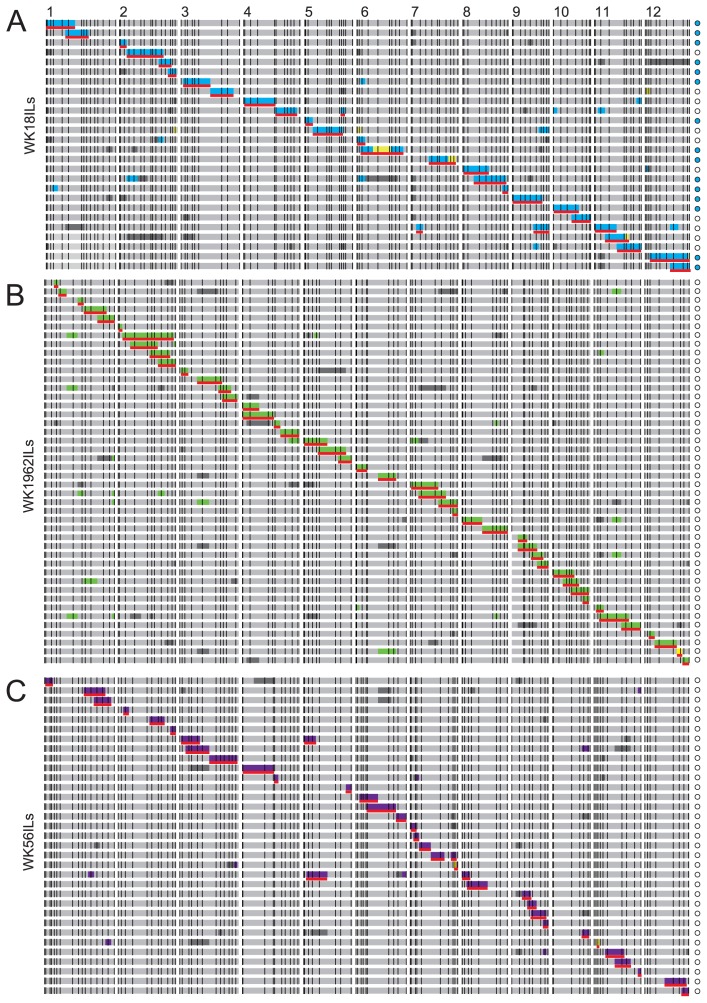
We developed 3 BC1F1, 23 BC2F1, 52 BC3F1, and 11 BC4F1 lines with T65 cytoplasm from an F1 of T65 × WK18 by recurrent backcrossing with T65 pollen (Fig. 1A). We similarly developed 2 BC1F1, 9 BC2F1, 43 BC3F1, and 20 BC4F1 lines with O. glaberrima cytoplasm from an F1 of WK18 × T65 (Fig. 1B). To develop the ILs of O. glaberrima in the T65 genetic background (WK18ILs), we conducted whole-genome genotyping using 121 SSR markers evenly distributed across the 12 chromosomes (Supplemental Table 1) with 136 plants of all 31 BC4F1 lines from both crosses. From these 31 BC4F1 lines, we selected 11 BC4F1 plants with T65 cytoplasm and 15 with O. glaberrima cytoplasm so as to select the minimum set of ILs that covers the whole genomic region (Fig. 2A, Supplemental Fig. 1). We grew BC4F2, BC4F3, and BC4F4 plants and genotyped the targeted chromosome regions so as to fix them as homozygous for O. glaberrima (red lines in Fig. 2A and Supplemental Fig. 1). Chromosome segments of O. glaberrima were not retained in the WK18 ILs on Chr. 1 (markers RM246, egt710, RM3709, RM265, RM1361, RM3362), Chr. 4 (RM567), Chr. 5 (RM3620, RM6346), Chr. 6 (RM5463), or Chr. 7 (RM3394, RM5436, RM1306). The 26 ILs cover 89.3% (108/121 markers) of the O. glaberrima genome. At the distal end of the short arm of Chr. 7 around RM1306, hybrid pollen sterility caused by S21 in heterozygous condition can reduce transmission of O. glaberrima alleles (Doi et al. 1998). This is the likely cause of non-introgression on this region in our results.
Fig. 1.
Development of introgression lines of Oryza glaberrima, O. rufipogon, and O. nivara in the genetic background of O. sativa ssp. japonica cv. Taichung 65. (A, B) Breeding of WK18ILs for O. glaberrima with (A) T65 and (B) WK18 cytoplasm. (C) Breeding of WK1962ILs for O. rufipogon. (D) Breeding of WK56ILs for O. nivara.
Fig. 2.
Graphical representation of chromosome introgression of Oryza glaberrima, O. rufipogon, and O. nivara in genetic background of O. sativa ssp. japonica cv. Taichung 65. Blue, green, and purple represent introgression of O. glaberrima, O. rufipogon, and O. nivara on homozygous condition. Missing genotypes at markers showing heterozygous genotypes at BC4F1 generations are indicated by grey. Heterozygous genotypes are indicated by yellow. The chromosome region for a minimal set of introgression for alien chromosomes indicated in red underlines were target for population maintenances. Blue circles and white circles represent the lines with WK18 and T65 cytoplams, respectively.
WK1962ILs (O. rufipogon)
We developed 3 BC1F1, 21 BC2F1, 51 BC3F1, and 58 BC4F1 lines from an F1 of T65 × WK1962 by recurrent backcrossing with T65 pollen (Fig. 1C). At the BC4F2 generation, we conducted whole-genome genotyping using 101 SSR markers (Supplemental Table 2) with 113 bulked DNA derived from BC4F1 plants (1 line per BC4F1 plants) to select a minimum set of ILs. To fix target chromosome segments and eliminate retained chromosome segments in the background of the selected lines, we performed marker-assisted selection (MAS) in the BC4F2, BC4F3, and BC4F4 generations. We selected 44 BC4F4 lines, designated ‘WK1962ILs’, with O. rufipogon segments fixed as homozygous (red lines in Fig. 2B and Supplemental Fig. 2). Chromosome segments of O. rufipogon were not retained in the WK1962 ILs on Chr. 1 (RM3148, RM5552), Chr. 6 (RM7023, RM3567, RM1031), or the short arm of Chr. 12 (RM3483). The 44 ILs cover 94.1% (95/101 markers) of the O. rufipogon genome in the T65 genetic background.
WK56ILs (O. nivara)
We developed 1 BC1F1, 5 BC2F1, 20 BC3F1, and 109 BC4F1 lines from an F1 of T65 × WK56 by recurrent backcrossing with T65 pollen. The BC4F1 plants were selfpollinated, and the bulked DNA of BC4F2 line derived from each of BC4F1 plant was genotyped using 107 SSR markers (Supplemental Table 3). In the BC4F4 generation we selected 33 lines carrying WK56 chromosome segments in the T65 genetic background, which we designated ‘WK56ILs’. Targeted chromosome segments for O. nivara introgressions were fixed in homozygous condition (red lines in Fig. 2C and Supplemental Fig. 3). The 33 ILs cover 69.2% (74/107 markers) of the O. nivara genome. Chromosome segments of O. nivara were not retained at many SSR markers on Chrs. 1 (RM272, RM3235, RM6642, RM5385, RM5638, RM3362), 2 (RM7562, RM6853, RM6611, RM5472), 4 (RM3367, RM3735, RM6089, RM3836, RM1113), 5 (RM3695, RM6841), 6 (RM7399), 7 (RM5508), 8 (RM7356, RM6976, RM3155), 10 (RM6370, WGS11, RM1375, RM4771), 11 (RM3717, RM1124, RM4504, RM4112), and 12 (RM3483, RM6296, RM7003). These omissions would have been due to the population bottleneck at BC2F1 (5 lines).
Distribution of materials via National Bioresource Project (Rice) in Japan
WK18ILs (O. glaberrima), WK1962ILs (O. rufipogon), and WK56ILs (O. nivara) are available through Oryzabase (https://shigen.nig.ac.jp/rice/oryzabase/).
Supplementary Information
Acknowledgments
This work was supported by a Grant-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT) (JP24248002 to A.Y.) and by a Grant-in-Aid from MEXT (National Bioresource Project (Rice)) and partially supported by Science and Technology Research Partnership for Sustainable Development (SATREPS).
Author Contribution Statement
YY, KTW, YM, CO, and HY conducted development of plant materials and genotyping. AY and YY design the experiment.
Literature Cited
- Alexandrov, N., Tai, S., Wang, W., Mansueto, L., Palis, K., Fuentes, R.R., Ulat, V.J., Chebotarov, D., Zhang, G., Li, Z.et al. (2015) SNP-Seek database of SNPs derived from 3000 rice genomes. Nucleic Acids Res. 43: D1023–D1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbelaez, J.D., Moreno, L.T., Singh, N., Tung, C., Maron, L.G., Ospina, Y., Martinez, C.P., Grenier, C., Lorieux, M. and McCouch, S. (2015) Development and GBS-genotyping of introgression lines (ILs) using two wild species of rice, O. meridionalis and O. rufipogon in a common recurrent parent, O. sativa cv. Curinga. Mol. Breed. 35: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessho-Uehara, K., Furuta, T., Masuda, K., Yamada, S., Angeles-Shim, R.B., Ashikari, M. and Takashi, T. (2017) Construction of rice chromosome segment substitution lines harboring Oryza barthii genome and evaluation of yield-related traits. Breed. Sci. 67: 408–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheema, K.K., Bains, N.S., Mangat, G.S., Das, A., Vikal, Y., Brar, D.S., Khush, G.S. and Singh, K. (2008) Development of high yielding IR64 × Oryza rufipogon (Griff.) introgression lines and identification of introgressed alien chromosome segments using SSR markers. Euphytica 160: 401–409. [Google Scholar]
- Childs, K.L., Davidson, R.M. and Buell, C.R. (2011) Gene coexpression network analysis as a source of functional annotation for rice genes. PLoS ONE 6: e22196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandan, L., Pfeiffer, T.W. and Cornelius, P.L. (2007) Soybean QTL for yield and yield components associated with Glycine soja alleles. Crop Sci. 48: 571–581. [Google Scholar]
- Dellaporta, S.L., Wood, J. and Hicks, J.B. (1983) A plant DNA mini-preparation: Version II. Plant Mol. Biol. Rep. 1: 19–21. [Google Scholar]
- Doi, K., Iwata, N. and Yoshimura, A. (1997) The construction of chromosome substitution lines of African rice (Oryza glaberrima Steud.) in the background of Japonica rice (O. sativa L.). Rice Genet. Newsl. 14: 39–41. [Google Scholar]
- Doi, K., Yoshimura, A. and Iwata, N. (1998) RFLP mapping and QTL analysis of heading date and pollen sterility using backcross populations between Oryza sativa L. and Oryza glaberrima Steud. Breed. Sci. 48: 395–399. [Google Scholar]
- Furuta, T., Uehara, K., Angeles-Shim, R.B., Shim, J., Ashikari, M. and Takashi, T. (2014) Development and evaluation of chromosome segment substitution lines (CSSLs) carrying chromosome segments derived from Oryza rufipogon in the genetic background of Oryza sativa L. Breed. Sci. 63: 468–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez, A.G., Carabalí, S.J., Giraldo, O.X., Martínez, C.P., Correa, F., Prado, G., Tohme, J. and Lorieux, M. (2010) Identification of a Rice stripe necrosis virus resistance locus and yield component QTLs using Oryza sativa × O. glaberrima introgression lines. BMC Plant Biol. 10: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajjar, R. and Hodgkin, T. (2007) The use of wild relatives in crop improvement: A survey of developments over the last 20 years. Euphytica 156: 1–13. [Google Scholar]
- Harlan, J.R. and de Wet, J.M.J. (1971) Toward a rational classification of cultivated plants. Taxon 20: 509–517. [Google Scholar]
- He, N., Wu, R., Pan, X., Peng, L., Sun, K., Zou, T., Zhu, H., Zeng, R., Liu, Z., Liu, G.et al. (2017) Development and trait evaluation of chromosome single-segment substitution lines of O. meridionalis in the background of O. sativa. Euphytica 213: 281. [Google Scholar]
- Hirabayashi, H., Sato, H., Nonoue, Y., Kuno-Takemoto, Y., Takeuchi, Y., Kato, H., Nemoto, H., Ogawa, T., Yano, M., Imbe, T.et al. (2010) Development of introgression lines derived from Oryza rufipogon and O. glumaepatula in the genetic background of japonica cultivated rice (O. sativa L.) and evaluation of resistance to rice blast. Breed. Sci. 60: 604–612. [Google Scholar]
- Huang, X., Kurata, N., Wei, X., Wang, Z.X., Wang, A., Zhao, Q., Zhao, Y., Liu, K., Lu, H., Li, W.et al. (2012) A map of rice genome variation reveals the origin of cultivated rice. Nature 490: 497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khush, G.S. (1997) Origin, dispersal, cultivation and variation of rice. Plant Mol. Biol. 35: 25–34. [PubMed] [Google Scholar]
- Lippman, Z.B., Semel, Y. and Zamir, D. (2007) An integrated view of quantitative trait variation using tomato interspecific introgression lines. Curr. Opin. Genet. Dev. 17: 545–552. [DOI] [PubMed] [Google Scholar]
- McCouch, S.R., Wright, M.H., Tung, C.-W., Maron, L.G., McNally, K.L., Fitzgerald, M., Singh, N., DeClerck, G., Agosto-Perez, F., Korniliev, P.et al. (2016) Open access resources for genome-wide association mapping in rice. Nat. Commun. 7: 10532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, R.S., Choi, J.Y., Sanches, M., Plessis, A., Flowers, J.M., Amas, J., Dorph, K., Barretto, A., Gross, B., Fuller, D.Q.et al. (2016) Domestication history and geographical adaptation inferred from a SNP map of African rice. Nat. Genet. 48: 1083–1088. [DOI] [PubMed] [Google Scholar]
- Nevo, E. and Chen, G. (2010) Drought and salt tolerances in wild relatives for wheat and barley improvement. Plant Cell Environ. 33: 670–685. [DOI] [PubMed] [Google Scholar]
- Prescott-Allen, R. and Prescott-Allen, C. (1988) Genes from the wild: Using wild genetic resources for food and raw materials. Earthscans Publications, London. [Google Scholar]
- Qiao, W., Qi, L., Cheng, Z., Su, L., Li, J., Sun, Y., Ren, J., Zheng, X. and Yang, Q. (2016) Development and characterization of chromosome segment substitution lines derived from Oryza rufipogon in the genetic background of O. sativa spp. indica cultivar 9311. BMC Genomics 17: 580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos, J.M., Furuta, T., Uehara, K., Chihiro, N., Angeles-Shim, R.B., Shim, J., Brar, D.S., Ashikari, M. and Jena, K.K. (2016) Development of chromosome segment substitution lines (CSSLs) of Oryza longistaminata A. Chev. & Röhr in the background of the elite japonica rice cultivar, Taichung 65 and their evaluation for yield traits. Euphytica 210: 151–163. [Google Scholar]
- Rangel, P.N., Brondani, R.P.V., Rangel, P.H.N. and Brondani, C. (2008) Agronomic and molecular characterization of introgression lines from the interspecific cross Oryza sativa (BG90-2) × Oryza glumaepatula (RS-16). Genet. Mol. Res. 7: 184–195. [DOI] [PubMed] [Google Scholar]
- Reuscher, S., Furuta, T., Bessho-Uehara, K., Cosi, M., Jena, K.K., Toyoda, A., Fujiyama, A., Kurata, N. and Ashikari, M. (2018) Assembling the genome of the African wild rice Oryza longistaminata by exploiting synteny in closely related Oryza species. Commun. Biol. 1: 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai, H., Kanamori, H., Arai-Kichise, Y., Shibata-Hatta, M., Ebana, K., Oono, Y., Kurita, K., Fujisawa, H., Katagiri, S., Mukai, Y.et al. (2014) Construction of pseudomolecule sequences of the aus rice cultivar Kasalath for comparative genomics of Asian cultivated rice. DNA Res. 21: 397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez, P.L., Wing, R.A. and Brar, D.S. (2013) The Wild Relative of Rice: Genomes and Genomics. In: Zhang, Q. and Wing R.A. (eds.) Genetics and Genomics of Rice, Springer, New York, pp. 9–25. [Google Scholar]
- Sato, Y., Takehisa, H., Kamatsuki, K., Minami, H., Namiki, N., Ikawa, H., Ohyanagi, H., Sugimoto, K., Antonio, B. and Nagamura, Y. (2013) RiceXPro Version 3.0: expanding the informatics resource for rice transcriptome. Nucleic Acids Res. 41: D1206–D1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz, M.C., Maron, L.G., Stein, J.C., Wences, A.H., Gurtowski, J., Biggers, E., Lee, H., Kramer, M., Antoniou, E., Ghiban, E.et al. (2014) Whole genome de novo assemblies of three divergent strains of rice, Oryza sativa, document novel gene space of aus and indica. Genome Biol. 15: 506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim, R.A., Angeles, E.R., Ashikari, M. and Takashi, T. (2010) Development and evaluation of Oryza glaberrima Steud. chromosome segment substitution lines (CSSLs) in the background of O. sativa L. cv. Koshihikari. Breed Sci. 60: 613–619. [Google Scholar]
- Stein, J.C., Yu, Y., Copetti, D., Zwickl, D.J., Zhang, L., Zhang, C., Chougule, K., Gao, D., Iwata, A., Goicoechea, J.L.et al. (2018) Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turnover and innovation across the genus Oryza. Nat. Genet. 50: 285–296. [DOI] [PubMed] [Google Scholar]
- Tian, F., Li, D.J., Fu, Q., Zhu, Z.F., Fu, Y.C., Wang, X.K. and Sun, C.Q. (2006) Construction of introgression lines carrying wild rice (Oryza rufipogon Griff.) segments in cultivated rice (Oryza sativa L.) background and characterization of introgressed segments associated with yield-related traits. Theor. Appl. Genet. 112: 570–580. [DOI] [PubMed] [Google Scholar]
- Tian, X., Long, Y., Wang, J., Zhang, J., Wang, Y., Li, W., Peng, Y., Yuan, Q. and Pei, X. (2015) De novo transcriptome assembly of common wild rice (Oryza rufipogon Griff.) and discovery of drought-response genes in root tissue based on transcriptomic data. PLoS ONE 10: e0131455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan, D.A., Lu, B.R. and Tomooka, N. (2008) The evolving story of rice evolution. Plant Sci. 174: 394–408. [Google Scholar]
- Wang, M., Yu, Y., Haberer, G., Marri, P.R., Fan, C., Goicoechea, J.L., Zuccolo, A., Song, X., Kudrna, D., Ammiraju, J.S.et al. (2014) The genome sequence of African rice (Oryza glaberrima) and evidence for independent domestication. Nat. Genet. 46: 982–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, D., Ye, X., Zheng, X., Cheng, C., Ye, N. and Huang, F. (2016) Development and evaluation of chromosome segment substitution lines carrying overlapping chromosome segments of the whole wild rice genome. Front Plant Sci. 7: 1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura, A., Nagayama, H., Sobrizal, Kurakazu, T., Sanchez, P.L., Doi, K., Yamagata, Y. and Yasui, H. (2010) Introgression lines of rice (Oryza sativa L.) carrying a donor genome from the wild species, O. glumaepatula Steud. and O. meridionalis Ng. Breed. Sci. 60: 597–603. [Google Scholar]
- Zhang, W., Dong, Y., Yang, L., Ma, B., Ma, R., Huang, F., Wang, C., Hu, H., Li, C., Yan, C.et al. (2014) Small brown planthopper resistance loci in wild rice (Oryza officinalis). Mol. Genet. Genomics 289: 373–382. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.