Summary
The most widespread form of malaria is caused by Plasmodium vivax. To replicate, this parasite must invade immature red blood cells, through a process which requires interaction of the Plasmodium vivax Duffy binding protein, PvDBP with its human receptor, the Duffy antigen receptor for chemokines, DARC. Naturally acquired antibodies that inhibit this interaction associate with clinical immunity, suggesting PvDBP as a leading candidate for inclusion in a vaccine to prevent malaria due to Plasmodium vivax. Here, we isolated a panel of monoclonal antibodies from human volunteers immunised in a clinical vaccine trial of PvDBP. We screened their ability to prevent PvDBP from binding to DARC, and their capacity to block red blood cell invasion by a transgenic Plasmodium knowlesi parasite genetically modified to express PvDBP and to prevent reticulocyte invasion by multiple clinical isolates of Plasmodium vivax. This identified a broadly neutralising human monoclonal antibody which inhibited invasion of all tested strains of Plasmodium vivax. Finally, we determined the structure of a complex of this antibody bound to PvDBP, indicating the molecular basis for inhibition. These findings will guide future vaccine design strategies and open up possibilities for testing the prophylactic use of such an antibody.
Introduction
In large parts of the world, Plasmodium vivax is the dominant species causing human malaria 1,2. An effective blood-stage vaccine would reduce morbidity, lower blood-stage asexual and sexual parasite densities and aid progress towards elimination. The symptoms of malaria occur as parasites invade, replicate within and burst out of red blood cells (RBC) of infected individuals. Plasmodium vivax parasites invade immature RBC, selecting reticulocytes expressing transferrin receptor CD71 3,4. Plasmodium vivax Duffy-binding protein, PvDBP, interacts with the Duffy antigen receptor for chemokines, DARC/Fy, on the surface of reticulocytes, and is essential for invasion by most strains 5,6. Indeed, genetic knockout of the orthologous DBPα gene from the closely-related simian malaria Plasmodium knowlesi also prevents invasion of Duffy-positive erythrocytes in vitro 7 and antibodies that target PkDBP can block invasion of erythrocytes by Plasmodium knowlesi 8. Moreover, a human polymorphism which results in Duffy-negative erythrocytes 9 is widespread across Africa and is associated with protection from Plasmodium vivax infection 10, leading to low levels of this form of malaria across much of the continent 11. PvDBP is therefore the most promising candidate for inclusion in a vaccine to prevent Plasmodium vivax 12.
PvDBP has been divided into six distinct regions, with the DARC binding site mapped to a ~350 amino acid residue Duffy-binding-like (DBL) domain known as Region II, PvDBPII6. This domain is split into three regions, known as subdomains 1-3 5. PvDBPII binds to the sixty-residue extracellular N-terminal ectodomain of DARC, in an interaction which requires DARC to be post-translationally modified by sulphation of two tyrosine residues, Tyr30 and Tyr41 13,14. Nevertheless, the interaction between PvDBPII and DARC is only partially understood. Structural studies of PvDBPII show it to form dimers in both solution and in a crystal 13. In addition, structures of PvDBPII crystallised in the presence of the ectodomain of DARC, reveal an ordered 11-residue helical peptide (DARC19-30), which lies close to the PvDBPII dimer interface in subdomain 2 15. However, the DARC protein used in these studies was not tyrosine-sulphated and sulphated Tyr41, which cannot be seen in this study, was previously shown to be essential for high affinity PvDBPII binding 14. Indeed, mapping residues identified by mutagenesis onto the structure of PvDBPII reveals an additional patch on subdomain 2 proposed to contribute to DARC binding 16,17 and it is likely that the DARC binding site spans much of the surface of subdomain 2, including both this patch and the region that interacts with the helical DARC19-30 peptide.
Development of PvDBPII-based vaccine candidates has progressed through pre-clinical studies to two recent Phase I human clinical trials18,19. Immunisation of mice, rabbits and non-human primates using PvDBPII-based vaccines induces inhibitory antibodies that block binding of recombinant PvDBPII to DARC 20,21. Challenges in sustaining Plasmodium vivax in long-term culture 22 have prevented these antibodies from being robustly tested in standardised functional growth inhibition assays, of the type traditionally performed for vaccines targeting Plasmodium falciparum 12. Nevertheless, in humans, high-titres of naturally-acquired antibodies that target PvDBPII and prevent DARC binding in vitro are associated with reduced risk of Plasmodium vivax infection 23, lower parasite densities following invasion and decreased risk of clinical malaria 24,25. Encouragingly, in a recent Phase Ia clinical trial, immunisation of human volunteers using recombinant viral vectors expressing PvDBPII induced strain-transcending antibodies which prevented recombinant PvDBPII from binding to DARC 19, confirming that such antibodies could be raised by human vaccination.
Several studies have investigated the molecular basis for antibody-mediated inhibition of DARC binding by PvDBPII 13,15,26,27. Screening a linear peptide array with non-inhibitory and inhibitory human serum identified peptides which recognise antibodies found specifically in inhibitory serum 27. These peptides are located within subdomain 2, in regions involved in PvDBPII dimerisation and DARC19-30 peptide binding 15, suggesting that antibodies which directly block dimerisation and DARC binding are desirable. In addition, a study of monoclonal antibodies (mAbs) derived from PvDBPII-immunised mice used structural studies and mass-spectrometry based mapping approaches to identify three epitopes for antibodies shown to block DARC binding in vitro 26. Surprisingly, these epitopes are located on subdomain 3 of PvDBPII, distant from the DARC binding site.
To understand the epitopes for human inhibitory antibodies, and to guide future structure-guided immunogen development, we have now cloned a panel of antibodies from PvDBPII immunised human volunteers. We determined their ability to inhibit binding of recombinant PvDBPII to DARC in vitro, as well as their capacity to neutralise parasite invasion in two distinct parasite-based functional assays. This provides important insight into these different in vitro readouts of antibody function and led to the structural characterisation of an epitope for an antibody that shows broadly neutralising activity against parasite invasion.
Results
Cloning of a panel of vaccine-induced human monoclonal antibodies that bind to PvDBPII
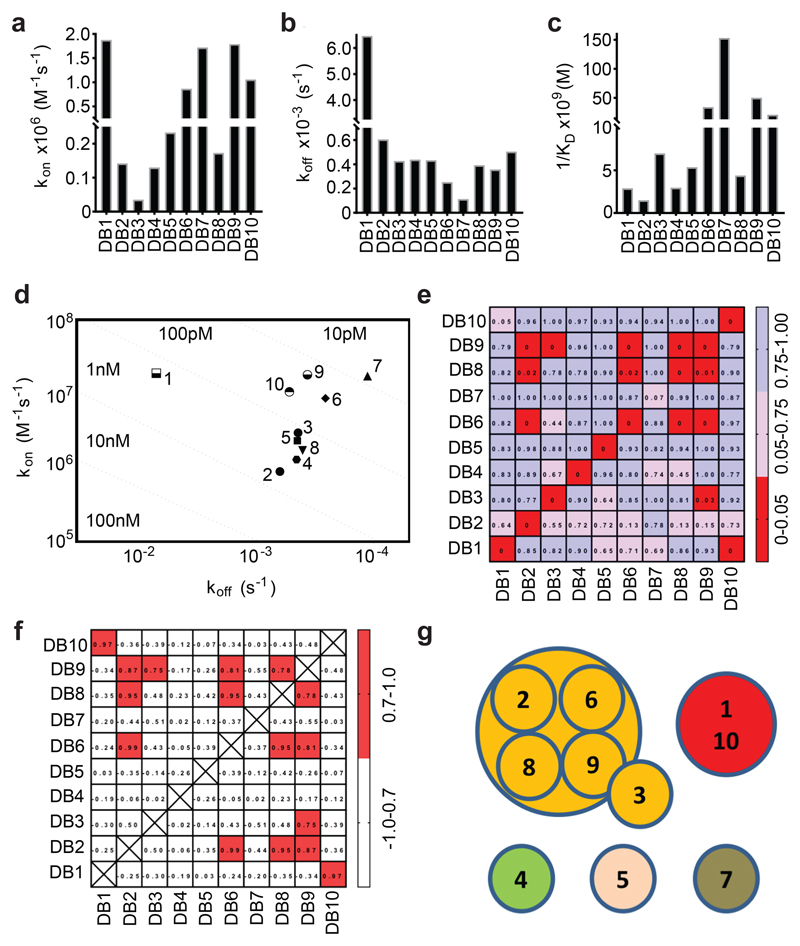
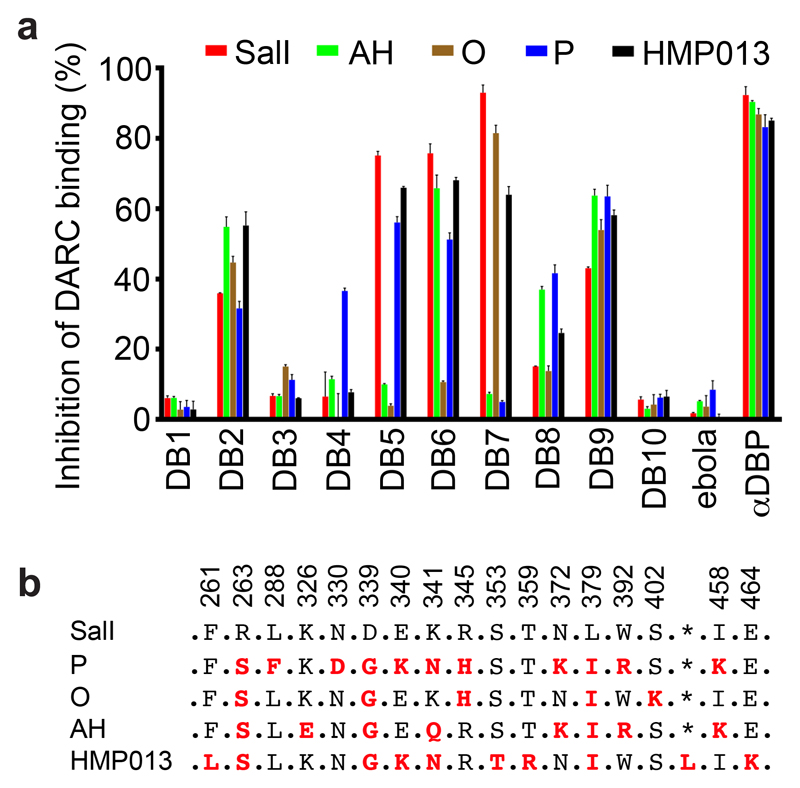
A panel of anti-PvDBPII mAbs was isolated from antibody-secreting cells of immunised volunteers enrolled in a human Phase Ia clinical trial of a PvDBPII-based vaccine delivered using recombinant chimpanzee adenovirus and poxvirus viral vectors 19. RT-PCR and PCR were used to isolate variable region (VR)-coding genes and these were cloned into a human IgG1 scaffold. Cognate heavy-chain and light-chain plasmids were co-expressed in HEK293 cells and vaccine antigen-specificity confirmed by ELISA reactivity of culture supernatants to recombinant PvDBPII. Ten genetically distinct mAbs were isolated (Supplementary Table 1), which recognised parasite-derived PvDBP from culture supernatant (Supplementary Figure 1). These were aligned with the most similar germline VR genes in IgBLAST 28, showing them to contain little non-germline sequence. Their monovalent binding affinities for PvDBPII, as determined by surface plasmon resonance (SPR) (Figure 1A-D and Supplementary Table 2), reveal low nanomolar to high picomolar affinities. Assessment of their binding to biotinylated PvDBPII by Biolayer Interferometry (BLI) (Figure 1E,F) defined two distinct epitope bins containing mAbs which bind overlapping epitopes, and three mAbs with distinct non-overlapping epitopes (Figure 1G). An ELISA-based binding inhibition assay 29 was used to assess their inhibition of binding of five sequence-divergent PvDBPII proteins 19 to DARC (Figure 2, Supplementary Figure 2). Most mAbs showed differential binding inhibition, with only two (DB2 and DB9) inhibiting the DARC binding of all five PvDBPII variant alleles (Figure 2).
Figure 1. Binding kinetics and epitope bins for the human anti-PvDBPII mAb panel.
Kinetic rate constants of binding for ten human mAbs (DB1-DB10) to PvDBPII as determined by SPR. (A) Association rate constant (kon); (B) Dissociation rate constant (koff); (C) Association constant (1/KD); and (D) Iso-affinity plot of kon against koff. (E) A “relative binding” matrix showing the fraction of a mAb bound to PvDBPII in the presence of a second bound mAb. Assays were conducted in both orientations. Boxes are colour-coded with ≥ 0.75 in blue, 0.75 > X > 0.05 in pink and ≤ 0.05 in red. Negative values were normalised to 0 and values >1 were normalised to 1. (F) A “binding profile correlation” matrix showing the Pearson product-moment correlation values of each mAb pair. The correlation threshold was set at 0.7; values equal to or above this are coloured in red as the threshold chosen to represent competition. (G) Epitope bins determined from E and F.
Figure 2. Inhibition of the binding of recombinant PvDBPII to DARC ectodomain.
(A) Assessment of the % binding of five naturally occurring variants of PvDBPII to the DARC ectodomain in vitro in the presence of 100 μg/mL concentration of each mAb (DB1-DB10). Individual titration curves are shown in Supplementary Figure 2. “αDBP” is polyclonal human anti-PvDBPII serum at 1:5 dilution while ‘ebola’ is an anti-Ebolavirus recombinant human IgG1 mAb included as a negative control. Data points represent the mean of three technical replicates, while the error bars represent the standard deviation. (B) Sequence polymorphisms of PvDBPII variants used in the assay. Numbering is according to the SalI reference sequence. Amino acid polymorphisms are indicated for the PvDBPII variants (P, O, AH and HMP013). Amino acids that are the same as the SalI reference sequence are black, those divergent from SalI are red and * indicates the absence of a leucine insertion between V429 and P430 in HMP013.
Assessing the neutralisation of parasite invasion by the antibody panel
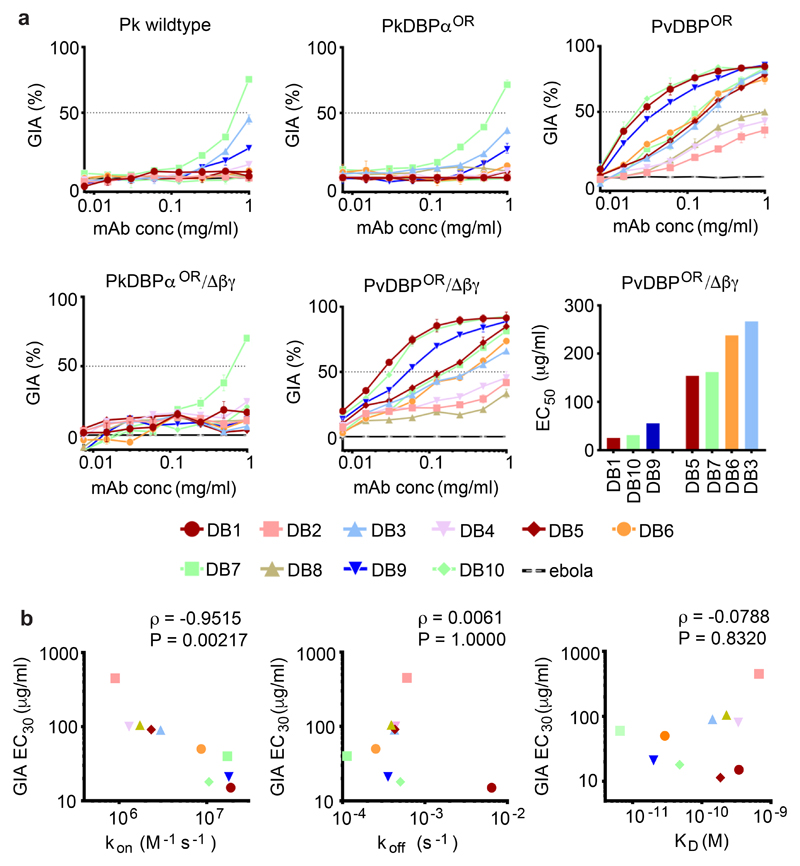
To characterise the ability of the human mAbs to block merozoite entry into RBC, assays of growth inhibition activity (GIA) were performed. A Plasmodium knowlesi line was used that has been adapted to long-term in vitro culture in human RBC 30 and in which the native PkDBPα gene had been replaced by the PvDBP gene 31. Plasmodium knowlesi is dependent on DARC for RBC invasion, but unlike Plasmodium vivax, has three DBP genes; PkDBPα, PkDBPβ and PkDBPγ 32. Both PkDBPβ and PkDBPγ are thought to be required for invasion of rhesus RBC whereas PkDBPα is essential for invasion of human RBC 7,33,34. The ten mAbs were assayed for growth inhibition against five Plasmodium knowlesi lines. First, the original human-RBC adapted PkA1-H.1 strain 30. Second, two transgenic PkA1-H.1 lines, PvDBPOR in which the PkDBPα gene had been replaced with the PvDBP gene (this transgenic insert was the full-length PvDBP Salvador I (SalI) strain vaccine-homologous sequence), and PvDBPOR/Δβγ, in which the same gene swap had taken place and additionally the native PkDBPβ and PkDBPγ genes had been deleted. PvDBPOR/Δβγ was considered the best model for Plasmodium vivax as the only DBP gene present is that for PvDBP. Finally, were two transgenic control lines, PkDBPαOR and PkDBPαOR/Δβγ in which native PkDBP genes had been deleted and replaced with re-codonised versions of themselves.
One anti-PvDBPII mAbs, DB7, at high concentration, inhibited growth of the three Plasmodium knowlesi control lines (Figure 3A), suggesting an epitope cross-reactive with PkDBPα. In contrast, when assayed against the transgenic PvDBP-expressing Plasmodium knowlesi lines, three mAbs showed high levels of inhibition of parasite growth, with four showing intermediate levels and the remaining three showing modest activity. A control human mAb against Ebolavirus showed no detectable GIA (Figure 3A). The three most neutralising anti-PvDBPII mAbs (DB1, DB9 and DB10) had EC50 values comparable to mouse-derived mAbs which potently prevent erythrocyte invasion by Plasmodium falciparum 35,36. A strong correlation (P = 0.002, ρ=-0.951) was observed between the association-rate (kon) and GIA (Figure 3B), suggesting that the opportunity for PvDBP blockade in the context of merozoite invasion is time-limited 36,37. In contrast, no correlation was observed between GIA and either dissociation rate or affinity (Figure 3B).
Figure 3. Growth inhibition of transgenic Plasmodium knowlesi (Pk) lines expressing PvDBP.
(A) Assays of growth Inhibitory activity (GIA) by the ten anti-PvDBPII human mAbs against five different Pk lines: ‘Wild type’ Pk (A1-H.1); PkDBPαOR; PkDBPαOR/Δβγ; PvDBPOR and PvDBPOR/Δβγ. Inhibition was tested in a two-fold dilution series starting at 1 mg/mL. Data points represent the mean of three technical replicates, while the error bars represent the standard deviation. The EC50 values (interpolated by non-linear regression) are shown in ascending rank order for the seven mAbs which reached >50 % GIA against PvDBPOR/Δβγ at the maximum concentration (1 mg/mL). (B) kon (on-rate), koff (off-rate) and KD (dissociation constant), plotted against GIA EC30 for PvDBPOR/Δβγ. EC30 values were used to include weaker-neutralising mAbs and were interpolated from non-linear regression curves. Kinetic data are as Figure 1 and Supplementary Table 2. Spearman’s rank correlation coefficient (ρ) and P value are shown.
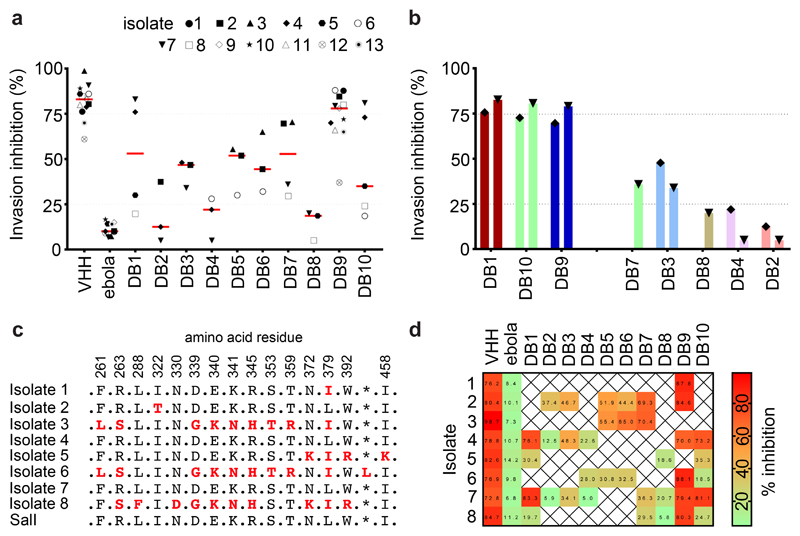
To assess the degree of strain transcendence of mAb inhibition across naturally occurring isolates, we next tested the mAbs directly for their capacity to prevent reticulocyte invasion by Plasmodium vivax parasites derived from thirteen clinical isolates originating from Thai patients, using ex-vivo short-term culture invasion inhibition assays (Figure 4A,B). Limited availability of these clinical samples prevented us from assessing the inhibitory potential of every antibody against every isolate. We were able to sequence the PvDBPII gene region from eight of these isolates, revealing multiple polymorphisms (Figure 4C). Some isolates had PvDBPII sequences identical to the SalI reference strain used in the vaccine (#4 and #7) while others were polymorphic, with as many as ten amino acid substitutions in region II of the DBP gene (#6 and #8).
Figure 4. Inhibition of invasion of reticulocytes by Thai Plasmodium vivax clinical isolates.
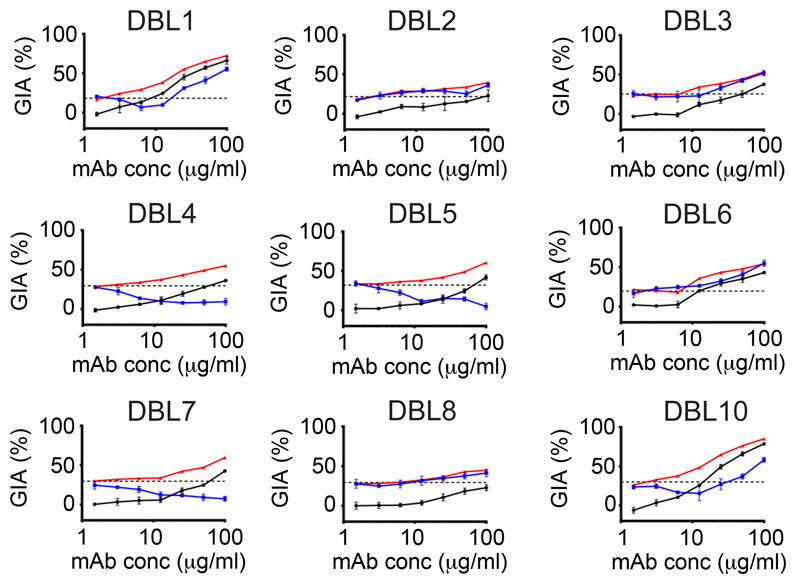
(A) Plasmodium vivax ex-vivo invasion assays were performed with thirteen separate isolates of infected blood from local patients. Each data point represents the % inhibition of each antibody against one of the thirteen isolates. All antibodies were tested at a final concentration of 1 mg/mL, except the positive control anti-DARC VHH (VHH) which was assayed at 25 μg/mL. The red bars represent the median % inhibition for each antibody. A recombinant human IgG1 anti-Ebolavirus mAb (ebola) was used as a negative control at 1mg/mL. (B) Percentage invasion inhibition by mAbs tested against the two Thai Plasmodium vivax isolates which share the PvDBPII gene sequence of SalI; isolate #4 (left hand bar of each pair) and isolate #7 (right hand bar of each pair). This demonstrates the degree of correlation with the hierarchy of mAb inhibition in the transgenic Plasmodium knowlesi GIA assays (Figure 3A). (C) Amino acid polymorphisms found within the PvDBPII gene segment of the eight Thai Pv isolates for which we obtained sequence information. The vaccine homologous SalI reference sequence is shown in the bottom row. Amino acids that are the same as the reference sequence black, divergent residues are red and * represents absence of a leucine insertion between V429 and P430 in the SalI reference sequence. (D) Summary matrix showing the percentage inhibition of invasion by each mAb for each of the sequenced strains. The positive control was the camelid anti-DARC nanobody (VHH) at 25 μg/mL and the negative control was a recombinant human IgG1 anti-Ebolavirus mAb (ebola) at a concentration of 1 mg/mL.
These assays revealed marked strain-dependent differences in the potency of the anti-PvDBPII mAbs. For example, DB1 and DB10, the two most inhibitory mAbs in the transgenic Plasmodium knowlesi GIA assay (containing the SalI PvDBP transgene), inhibited invasion of Thai isolates with matching SalI PvDBPII sequences (isolates #4 and #7) but not those with heterologous PvDBPII sequences (isolates #5 and #8) (Figure 4B,D). Notably, the hierarchy of mAb potency for the two Plasmodium vivax parasite isolates which possessed the homologous SalI PvDBPII sequence mirrored that observed in the transgenic Plasmodium knowlesi model (Figure 4B), suggesting that this model is highly predictive of Plasmodium vivax neutralisation. DB3, 5, 6 and 7 showed intermediate median levels of inhibition (~40-60 %), whilst DB2, 4 and 8 showed low median levels (~10-20 %). Only one of the ten mAbs (DB9) potently inhibited invasion (~65-90 %) of 10/11 isolates. The inhibition of invasion by isolate 12 was lower but a lack of sequence information, due to the challenges of working with small quantities of samples from field isolates, makes it challenging to speculate on the reasons for this. DB9 also showed potent growth inhibition in the transgenic Plasmodium knowlesi assays of GIA (Figure 3A) and inhibited binding of all five variant alleles of PvDBPII to DARC (Figure 2), highlighting it as an antibody with broadly neutralising activity against a wide range of Plasmodium vivax variants.
Antagonism of DB9-mediated inhibitory activity
We next sought to assess whether the efficacy of DB9 would be enhanced or diminished by the other nine mAbs. A BLI binding-competition assay (Figure 1E-G), showed that half of the mAbs, including DB9, compete with each other for binding sites on recombinant PvDBPII. To test for evidence of synergy, additivity or antagonism 38, we ran GIA assays using the PvDBPOR line of transgenic Plasmodium knowlesi in which DB9 was present at 25 μg/mL, together with a dilution series of a second mAb (Figure 5). While no synergy was detected, there was antagonism between DB9 and five other mAbs, with addition of a second mAb leading to growth inhibition lower than that predicted from adding the inhibition due to the individual mAbs at equivalent concentrations. Surprisingly, these were the five mAbs (DB1, DB4, DB5, DB7 and DB10) which did not compete with DB9 for binding to PvDBPII in the BLI competition assay (Figure 1E-G). The other four mAbs (DB2, DB3, DB6 and DB8) with epitopes overlapping that of DB9, showed an additive growth inhibitory effect with DB9 in this assay. Therefore, antibodies that bind in and around the DB9 epitope function in an independent and additive manner, whilst antibodies that bind to distinct epitope regions elsewhere on the PvDBPII molecule are mutually antagonistic. Given DB9 was able to show strain-transcending high-level neutralising activity, this raised the imperative to understand the epitope of DB9 and thus enable future design of immunogens which specifically induce DB9-like antibodies.
Figure 5. Assessment of synergy, additivity and antagonism by anti-PvDBPII human mAb combinations.
Assays of growth inhibitory activity (GIA) were performed to assess the inhibitory activity of DB9 in combination with the other mAbs against the PvDBPOR transgenic Plasmodium knowlesi line. DB9 was held at a fixed concentration of 25 μg/mL, while the other mAbs are in a two-fold dilution series starting at 100 μg/mL. The black line shows the % inhibition of each mAb in the absence of DB9. The red line shows the predicted additive inhibition (‘Bliss additivity’ as calculated in the equation in Methods) of the indicated mAb plus DB9 at 25 μg/mL. The blue line shows the actual observed % inhibition of the two combined mAbs. The dotted black line gives the % inhibition of DB9 alone at 25 μg/mL. Data points represent the mean of three technical replicates, while the error bars represent the standard deviation.
The molecular basis for the action of DB9
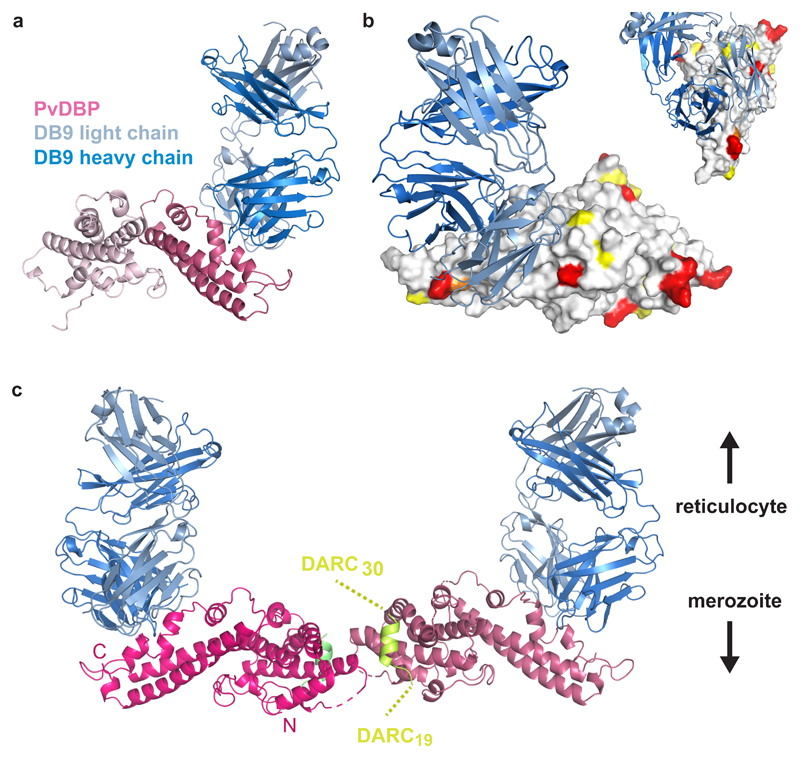
To obtain structural insight, we crystallised a complex of PvDBPII bound to the Fab fragment of DB9. Crystals diffracted to 3.0 Å resolution, allowing structure determination by molecular replacement (Figure 6 and Supplementary Table 3). Comparison of the structure of PvDBPII in complex with DB9 with that of un-liganded PvDBPII (PDB: 4NUV) revealed no change in PvDBPII conformation (with an RMSD of 0.523Å).
Figure 6. The structural basis for inhibition of PvDBPII by antibody DB9.
(A) The structure of PvDBPII (pink) bound to the Fab fragment of DB9 (blue). PvDBPII is shown in two shades of pink, with subdomains 1 and 2 in light pink and subdomain 3 in dark pink. DB9 is in two shades with the light chain in light blue and the heavy chain in dark blue. (B) PvDBPII is shown in surface representation in grey, with residues known to be polymorphic highlighted according to their sequence entropy (yellow = 0.15-0.3, orange = 0.3-0.45 and red > 0.45). DB9 is in blue, and binds to a conserved region of PvDBPII. (C) The structure of PvDBPII:DB9 complex superimposed on the structure of the PvDBPII dimer structure bound to a peptide from the DARC ectodomain, indicating that DB9 may prevent the binding of PvDBP to DARC in the context of the reticulocyte membrane.
The epitope of DB9 is contained within subdomain 3 of PvDBPII (Figure 6A). This region consists of two long α-helices (H1 and H2), which form a platform on which three small helices (H3-H5) and their intervening loops are arranged (Supplementary Figure 3). All three CDR loops of the heavy chain and CDR1 and CDR2 of the light chain directly contact PvDBPII (Supplementary Figure 3 and Supplementary Table 4). The epitope includes a single residue from H1 (K412) of PvDBPII and residues from each of helices H3, H4 and H5, and the intervening loops. Contacts are predominantly mediated by hydrogen bonds between DB9 and hydrophilic residues from PvDBPII.
We next compared the position of the epitope with locations of known polymorphic residues on PvDBPII, as alignment of globally disparate sequences of PvDBPII revealed 120 polymorphic sites with nucleotide diversity varying between 0.006 and 0.0109 39. We retrieved 383 amino acid sequences of PvDBPII and calculated their sequence entropies. This revealed that the surface of PvDBPII contacted by DB9 is one of the most conserved regions of the domain, with low sequence variation explaining its broadly reactive nature (Figure 6B and Supplementary Figure 3).
In the membrane context, PvDBPII is thought to form a dimer which interacts with the extracellular domains of DARC. A crystal structure is available for a dimer of PvDBPII bound to a small helix, consisting of residues 19-30 of the extracellular ectodomain of DARC 15. We next superimposed our PvDBPII:DB9 structure onto this structure, suggesting a model for how DB9 might inhibit invasion by Plasmodium vivax (Figure 6C). While DB9 binds at a distance from the known region of the DARC binding site, it protrudes from subdomain 3 in the same direction as the C-terminus of the DARC19-30 peptide. This end of the DARC peptide will face the transmembrane region of DARC, located in the reticulocyte membrane. Although the arrangement of DARC31-60 is currently unknown, this model suggests that DB9 can prevent the PvDBPII dimer from approaching the reticulocyte membrane in an orientation which is compatible with DARC binding, thereby preventing initiation of the invasion process.
With the epitope for DB9 contained solely within subdomain 3, we next assessed binding of the other mAbs bind to this region. Subdomain 3 was produced by refolding material expressed in E. coli. An ELISA-based binding assay showed that 6 of the 10 mAbs bound subdomain 3 (Supplementary Figure 4). With the exception of DB4, these were the mAbs that competed for binding on PvDBPII (Figure 1E-G). Indeed, with the exception of DB4, the mAbs that bind to subdomain 3 are not those that antagonise the effect of DB9. Therefore, despite the fact that antibodies that target subdomain 3 differ in their inhibitory potential, these data identify subdomain 3 of PvDBPII as a promising potential target for broadly inhibitory antibodies, and identify the epitope of one such antibody.
Discussion
The Duffy-binding protein is the most promising candidate for inclusion in a vaccine to prevent infection by Plasmodium vivax due to its essential interaction with human DARC during reticulocyte invasion. Antibodies which bind to PvDBP are found in individuals from malaria endemic regions, and their presence associates with protection from clinical vivax malaria 23–25. However, no human monoclonal antibody has been reported which targets PvDBP and displays broadly inhibitory potential. We have therefore vaccinated human volunteers with PvDBPII to induce a humoral response and isolated a monoclonal antibody which can inhibit all tested isolates of Plasmodium vivax from invading blood cells. This opens the possibility for testing the efficacy of this monoclonal as a prophylactic to prevent vivax malaria, while knowledge of its epitope will guide future vaccine development.
The mAbs generated here have been analysed through a series of complementary approaches and the differing outcomes of these assays illustrate the need for caution in attributing broadly inhibitory potential (Supplementary Table 5). The most accessible assay involves analysis of purified protein binding in an ELISA-based format, with antibodies assessed for prevention of recombinant PvDBPII from binding to N-terminal DARC ectodomain, with the assumption that direct prevention of DARC binding is the goal for a protective antibody. This assay allows assessment of a range of PvDBPII variants to analyse breadth of inhibitory potential and identified two antibodies that broadly inhibited DARC binding (DB9 and DB2), and three with vaccine-homologous (SalI sequence) binding-inhibition activity (DB5, DB6 and DB7).
With the blood-stage of Plasmodium vivax currently impossible to sustain in long-term in vitro culture conditions, we also used a culture-compatible transgenic parasite-based assay involving the closely related simian malaria, Plasmodium knowlesi. Here Duffy-binding protein, PkDBPα was replaced by PvDBP, generating parasite lines in which human RBC invasion inhibition can be studied in a laboratory setting using long-term culture. This showed a clear hierarchy of mAb potency against transgenic parasites in vitro, whether or not the PkDBPβ and PkDBPγ genes were removed. Most informatively, the outcomes of this study only partially overlapped with those of the protein-based study, with some antibodies (i.e. DB1 and DB10) among those with the greatest potency to block parasite invasion but proving ineffective at blocking the PvDBPII-DARC interaction in a protein-protein assay. The failure of the binding-inhibition ELISA, which only uses fragments of DARC and PvDBP, to predict parasite neutralisation in both subsequent functional assays, calls into question its utility as a method of screening antibodies in this context, suggesting that future studies should instead use parasite-based growth inhibition studies.
Finally, we used the most authentic in vitro assay available, by assessing the invasion of reticulocytes by ex-vivo Plasmodium vivax taken directly from patient isolates. When studying parasite isolates which expressed the SalI PvDBPII variant incorporated into the transgenic Plasmodium knowlesi strain, these studies give a very similar outcome to the transgenic Plasmodium knowlesi assays (Figures 3A and 4B). However, when studying isolates with heterologous PvDBPII sequences, the outcomes were different. Here, just one antibody, DB9, inhibited reticulocyte invasion by all tested strains. Comparison of data from these three assays suggests that a protein-protein analysis will miss valuable inhibitory antibodies and highlights the applicability of a transgenic Plasmodium knowlesi-based assay as an accessible in vitro proxy for the ex-vivo analysis of Plasmodium vivax. In the future, panels of transgenic parasites expressing different PvDBPII variants will allow assessment of broadly-inhibitory potential of therapeutic interventions.
Various properties have been suggested as desirable for inhibitory antibodies that target PvDBP, including capacity to directly block DARC binding or to inhibit dimerisation 6,13. In contrast, DB9 binds distantly from either the DARC-binding or dimerisation interfaces, on a non-polymorphic site on the distal surface of subdomain 3. Remarkably, it also prevents recombinant PvDBPII from binding to DARC immobilised onto an ELISA plate. A composite model, in which DB9 is docked onto that of a dimer of PvDBPII bound to the 19-30 peptide from DARC shows that DB9 emerges from the same side of PvDBPII as the C-terminus of the DARC peptide. DB9 may therefore sterically prevent dimeric PvDBPII from approaching a DARC coated surface and thereby prevent receptor engagement and invasion. A previous study identified antibodies from immunised mice which also bind to subdomain 3 26. The inhibitory potential of these antibodies was tested using the protein-based binding assay, which we found to be only partially indicative of efficacy in parasite-based neutralisation assays. Nevertheless, one of these antibodies, 2C6, has an epitope close to that of DB9 (Supplementary Figure 5) and could be operating through the same mechanism.
These findings have significant consequences for vaccine development, identifying subdomain 3 of PvDBPII as an important site of broadly-inhibitory epitopes for human antibodies. Not all antibodies that bind to subdomain 3 are inhibitory. However, our discovery of antibodies which bind to PvDBPII and antagonise the function of DB9 also raises the imperative to avoid inducing these deleterious antibodies. While further studies are needed to understand the molecular basis for this antagonism, perhaps involving analysis of their effect on the interaction between full length PvDBP and DARC in the membrane context, it is encouraging to note that 80% of the antibodies that antagonise the function of DB9 do not recognise subdomain 3. This suggests future vaccination strategies in which the surface of subdomain 3 containing the DB9 epitope is specifically presented to the immune system in the form of a vaccine immunogen to produce a strong and protective immune response as part of the malaria vaccines of the future.
Methods
Generation of monoclonal antibodies
Plasmablast isolation and sorting
Volunteers from a Phase Ia clinical trial were bled seven days after the second immunisation using MVA (modified vaccinia virus Ankara) encoding PvDBPII 19. Blood was collected from volunteers in heparinised tubes and centrifuged in Leucosep tubes (Greiner Bio-One) to separate the peripheral blood mononuclear cells (PBMC). The PBMC were enriched for B cells using a human pan-B cell enrichment kit (Easysep) and re-suspended in DMEM before staining with a CD19+, CD10–, CD21–, CD27+, CD20–, CD38+, IgG+ fluorophore-conjugated antibody panel. Plasmablasts were single-cell sorted using a MoFlo cell sorter (DakoCytomation) into 96-well PCR plates containing 10 μL 10 mM Tris HCl buffer containing 40 U/mL of RNase inhibitor (Promega). The study received ethical approval from the Oxfordshire Research Ethics Committee A in the UK (REC reference 13/SC/0001) and was registered with ClinicalTrials.gov NCT01816113. The volunteers signed consent forms and consent was verified before each vaccination.
Antibody variable gene amplification
In wells of a 96-well plate, each containing a single antibody-secreting cell (ASC), a two-step RT-PCR was carried out with a first reverse transcription (RT) step using a Sensiscript RT kit (Qiagen) and degenerate primers 1-17 (modified from 40, see Supplementary Table 6). Next, a PCR was performed on 1 μL of the RT reaction product using the same set of primers used before (1-17) which cover the diversity of all Vγ, Vκ and Vλ sequences using Phusion HF master mix (New England Biolabs). Following this, a nested PCR was performed with primers 18-51, also using Phusion HF master mix, (Supplementary Table 6) on 1 μL of the previous product diluted 1:100 to amplify inserts which contain plasmid-homologous extensions designed for circular polymerase extension cloning 41.
Cloning
The AbVec-hIgG1/AbVec-hIgKappa/AbVec-hIgLambda expression plasmids were a kind gift from Patrick C. Wilson (University of Chicago) 42. These plasmids were 5’ digested using BshTI and at the 3’ using SalI (AbVec-hIgG1), XhoI (AbVec-hIgLambda) and Pfl23II (AbVec-hIgKappa) to yield linear products. CPEC assembly was performed by mixing 100 ng of a 1:1 molar ratio of insert:plasmid in 20 μL containing 1x Phusion HF polymerase master mix and assembled using an 8-cycle CPEC protocol (8 cycles: 98 °C 10 s, slow ramp anneal from 70 °C to 55 °C at 0.1 °C/s, 72 °C 35 s). Full nicked plasmids were then transformed into Zymo 5α Mix & go competent E. coli (Zymo Research) according to manufacturer’s instructions, streaked on LB agar petri dishes containing 100 μg/mL carbenicillin and grown at 37°C overnight in a static incubator. Colonies were screened by PCR for correctly sized inserts.
Screening
Exponential growth-phase adherent HEK293 cells were re-suspended in DMEM (Sigma-Aldrich) supplemented with 2 mM L-glutamine, 100 U/mL penicillin, 1 mM sodium pyruvate, 0.1 mg/mL streptomycin and 10 % ultra-low IgG foetal bovine serum (FBS) (Thermo Fischer Scientific) and seeded at 4 x 104 cells/well in 100 μL 24 h prior to transfection in Costar 96-well cell culture plates (Corning). On the day of transfection, for each well, 50 μL of 60 μg/mL linear 25 kDa PEI (Alfa Aesar) was mixed with 200 ng of cognate heavy- and light-chain coding plasmid in a volume of 50 μL and shaken at 20°C for 30 min. The DNA-PEI complexes were added to the HEK293 cells. The following day, an additional 50 μL of supplemented DMEM (as described above) was added to each well. Supernatants were screened for PvDBPII (produced in S2 cells as described below) binding by indirect ELISA. Over 100 mAb supernatants were screened in this assay and of these, ten bound PvDBPII.
Recombinant protein constructs, expression and purification
The production of the recombinant PvDBPII used in the IgG screening assay described above, the DARC-binding inhibition assays (Figure 2) and the SPR assays (Figure 1A-D), from Drosophila S2 cells has been previously described 19. In brief; the sequence used was identical to the vaccine sequence; D194–T521 of PvDBPII (SalI), with T257A, S353A and T422A substitutions to remove sites of possible N-linked glycosylation, followed by a shortened Pk/V5 epitope tag (IPNPLLGLD) and a C-tag (EPEA) 43 for detection and purification, respectively. Purification was performed on an AKTA Pure 25 system (GE Healthcare, UK), consisting of an affinity step with CaptureSelect™ C-tag column (Thermo Fisher Scientific, UK) and a polishing size exclusion chromatography (SEC) using Superdex 200 16/60 PG (GE Healthcare, UK) in 20 mM Tris-HCl, 150 mM NaCl, pH 7.4 (TBS). Purified protein was quantified by Nanodrop (Thermo Fisher Scientific, UK) and stored at -80°C until further use. A gene encoding the PvDBPII HMP013 allele 19 (aa 194-521) was codon optimised for human expression and synthesised (Genewiz, USA). This sequence, with a 5’ KpnI site and 3’ XbaI site, was cloned into a mammalian expression plasmid in frame with an N-terminal mouse IgK light chain leader sequence and a C-terminal C-tag prior to the stop codon. Suspension EXPI293F cells (Thermo Fisher Scientific, UK) were transiently transfected and culture supernatants were harvested after 4 days. Purification was performed on an AKTA Pure 25 system (GE Healthcare, UK), consisting of an affinity step with CaptureSelect™ C-tag column (Thermo Fisher Scientific, UK) and a polishing SEC step using Superdex 200 Increase 10/300 GL (GE Healthcare, UK) in 20 mM Tris-HCl, 150 mM NaCl, 0.1% Tween 20, pH 7.4 (TBS). Purified protein was quantified by Nanodrop (Thermo Fisher Scientific, UK) and stored at -80°C until further use.
The PvAH, PvO and PvP alleles of PvDBPII 44 used in the DARC binding-inhibition assays (Figure 2) were a kind gift from Chetan Chitnis (Institute Pasteur, France).
Monobiotinylated PvDBPII supernatant was produced for use in the BLI experiments (Figures 1E-G) by transient transfection of suspension EXPI293F cells, using a plasmid encoding PvDBPII (SalI) with C-terminal rat CD4 domains 3 and 4 followed by a biotin acceptor peptide obtained from Addgene (plasmid #68529) and courtesy of Dr Julian Rayner (Wellcome Trust Sanger Institute, Hinxton, UK) 45. This PvDBPII plasmid was co-transfected with another plasmid encoding E. coli biotin ligase (BirA). Supernatant was harvested after 4 days, clarified, dialysed against PBS using snakeskin and concentrated ~10-fold using spin columns.
The production of the PvDBPII and PvDBPII subdomain 3 proteins used in the crystal complex (Figure 6) and the subdomain 3 binding ELISA (Supplementary Figure 4) is described in ‘Structural Methods’ below.
The production of recombinant N-terminal DARC, used in the DARC-binding inhibition assays, has been previously described 19. In brief; a gene encoding the first 60 amino acids of DARC Fyb allele (GenBank Accession #ABA10433.1) followed by a thrombin cleavage site (LVPRGS) and an AviTag (GLNDIFEAQKIEWHE) was codon-optimised for E. coli expression and synthesised (GeneArt, Life Technologies). Cysteines 4, 51 and 54 of DARC were mutated to alanine. This sequence with 5’ KpnI site and 3’ BamHI site was cloned into a mammalian expression plasmid in-frame with an N-terminal human tissue plasminogen activator (tPA) leader sequence 20 and a C-terminal hexa-histidine (His6) tag prior to the stop codon. Suspension HEK293E cells grown in EXPI293 expression medium (Thermo Fisher Scientific, UK) were transiently transfected with plasmid and allowed to grow for three days before the supernatant was harvested, purified using a HisTrap Excel column (GE Healthcare, UK) and buffer exchanged into PBS. Purified protein was quantified by Nanodrop (Thermo Fisher Scientific, UK) and stored at -80°C until further use.
Recombinant monoclonal antibodies were transiently expressed in HEK293F cells using the Expi293™ Expression System (Thermo Fischer Scientific) according to the manufacturer’s recommendations. Cognate heavy and light chain-coding plasmids were co-transfected at a 1:1 ratio. Supernatants were harvested by centrifuging the culture at 2500 xg for 15 min and filtering the supernatant with a 0.22 μm vacuum filter. All mAbs were purified using a 5 mL Protein G HP column (GE Healthcare) on an ÄKTA start FPLC system or an ÄKTA Pure FPLC system (both GE Healthcare). Equilibration and wash steps were performed with Dulbecco’s PBS and mAbs were eluted in 0.1 M glycine pH 2.7. The eluates were pH equilibrated to 7.4 using 1.0 M Tris HCl pH 9.0 and immediately buffer-exchanged into Dulbecco’s PBS and concentrated using an Amicon® ultra centrifugal concentrator (Millipore) with a molecular weight cut-off of 30 kDa.
Bio-Layer Interferometry (BLI)
BLI was carried out on an OctetRED384 (Pall FortéBio) using streptavidin-coated biosensors (Pall FortéBio) to immobilise PvDBPII enzymatically monobiotinylated on a C-terminal AviTag™. Assays were carried out in 96-well format in black plates (Greiner). For epitope binning studies (Figure 1E-G), a six-step sequential assay was performed: Baseline (PBS, 30 s); Protein immobilisation (neat supernatant, 120 s); Wash (PBS, 60 s); first mAb (mAb1) binding (300 nM mAb1, 120 s); Wash (PBS, 60 s); second mAb (mAb2) binding (150 nM mAb2, 120 s). “Relative binding” in Figure 1E shows the ratio (SignalmAb2 with mAb1 bound)/(SignalmAb2 with no mAb1) where “SignalmAb2” was normalised for the amount of PvDBPII bound to the biosensor, such that “SignalmAb2” = the raw signal in “mAb2 binding” divided by the raw signal in the “Protein immobilisation” phase. The resulting “binding profile” for any given mAb corresponds to the column of “relative binding values” under that mAb in the “relative binding” table. To establish the epitope bins, binding profiles between each mAb pair were correlated using a Pearson product-moment correlation coefficient, the values of which are shown in the “binding profile correlation” matrix in Figure 1F. mAb pairs whose binding profile correlation was > 0.7 were grouped into the same epitope bin (Figure 1G).
Measurement of binding by ELISA
Qualitative mAb binding ELISAs such as those used in Supplementary Figure 4 were carried out by coating PvDBPII or PvDBPII subdomain 3, produced as described in ‘Structural Methods’, on Maxisorp flat-bottom 96-well ELISA plates (Nunc) at 2 μg/mL in 50 μL at 4 °C overnight. Plates were then washed twice with PBS and 0.05% Tween 20 (PBS/T) and blocked with 200 μL of Blocker™ Casein (Thermo Fischer Scientific) for 1 h. Next, wells were incubated with 10 μg/mL of mAb for approximately 45 min at 20 °C then washed 4 times with PBS/T before the addition of 50 μL of 1:1000 dilution of goat anti-human gamma-chain alkaline phosphatase-conjugated secondary antibody (Sigma-Aldrich) for 45 min at 20 °C. Wells were then washed 6 times with PBS/T and developed with 100 μL of p-nitrophenyl phosphate substrate at 1 mg/mL (Sigma-Aldrich) and optical density read at 405 nm (OD405) using a Model 550 Microplate Reader (Bio-Rad, UK).
Affinity determination by SPR
Data were collected on a Biacore X100 (GE Healthcare). Experiments were performed at 25 °C in Dulbecco’s PBS + 0.005 % Polysorbate-20 (GE Healthcare). In Figure 1 and Supplementary Table 2 a sensor chip protein A (GE Healthcare) was used to capture 50-100 RU of purified mAb diluted in SPR running buffer at a flow rate of 5 μL/min on flow cell 2. Next, an appropriate range (typically 20 nM-0.625 nM) of six 2-fold dilutions, with one replicate, of PvDBPII (expressed in S2 cells as described above) was injected for 90 s at 60 μL/min and dissociation was measured for 1600 s (7200 s when necessary). Specific binding of the PvDBPII protein to mAb was obtained by reference-subtracting the response of a blank surface from that of the mAb-coated surface. The sensor surface was regenerated with a 60 s pulse of 10 mM glycine-HCl pH 1.5 (GE Healthcare). Sensorgrams were fitted to a global Langmuir 1:1 interaction model, allowing determination of the kinetic association and dissociation rate constants using Biacore X100 evaluation software.
PvDBPII-DARC binding inhibition assays
This assay methodology has been previously reported 19,29. In brief; recombinant N-terminal DARC protein was coated onto Nunc-Immuno Maxisorp plates at 1 μg/mL. mAb samples were diluted down in a 2-fold series (starting at 100 μg/mL) and pre-incubated with PvDBPII protein for 30 min at room temperature (RT). The pre-incubated PvDBPII protein plus mAb mixture was then added to the DARC-coated plates (in duplicate wells). The plates were incubated for 1 h at 37 °C, washed and then incubated with anti-PvDBPII polyclonal rabbit serum 20. After a further wash step, the plates were incubated with a 1:1000 dilution of anti-rabbit IgG alkaline phosphatase (Sigma Aldrich, UK) and then developed with 1 mg/mL p-nitrophenyl phosphate in diethanolamine buffer (Pierce, UK). OD405 was read using a Model 550 Microplate Reader (Bio-Rad, UK) when control wells containing PvDBPII protein and buffer only reached a value of 1.0. In one column of each 96 well plate, wells contained only mAb (100 μg/ml) and buffer (no PvDBPII protein) and these ‘background’ OD405 values were subtracted from all test values. To calculate % binding-inhibition for each mAb sample the formula below was used:
Five allelic variants of recombinant PvDBPII were used in the assay; Sal1, which was made in S2 cells as described above; PvAH, PvO and PvP alleles 44, which were a kind gift from Chetan Chitnis (Insitute Pasteur, France); and PvHMP013, which was sequenced from a vivax-infected patient in Australia 46. The expression of recombinant PvDBPII HMP013 protein has been previously described 19 and is summarised above. An anti-Ebolavirus glycoprotein (GP)-reactive human IgG1 mAb ‘EBL040’ 47 was used as a negative isotype control for mAb samples. Polyclonal human anti-PvDBPII serum from the clinical trial 19 was used as the positive control.
Dot blot assay
Briefly, 1.5 μL of each of the ten human anti-PvDBPII mAbs, a human anti-Zaire Ebolavirus GP IgG1 mAb, recombinant PvDBPII (all at 1 mg/mL) and PBS were spotted onto 0.2 μm nitrocellulose membrane and air-dried for 10 min. The membrane was then blocked in 3 % BSA + 3 % skimmed milk in PBS for 1 h and washed in PBS. It was then immersed in P. vivax culture supernatant for 1 h and washed again in PBS. This supernatant was taken from a short-term in vitro parasite viability assay using a frozen P. vivax blood-stage inoculum produced for controlled human malaria infection clinical trials (Minassian AM et al., in preparation; Clinicaltrials.gov NCT03797989). Bound PvDBPII was detected by incubating the membrane in anti-PvDBPII rabbit serum20 diluted 2000-fold in PBS followed by an alkaline phosphatase-conjugated anti-rabbit IgG mAb (clone RG-96, 403 Sigma-Aldrich) also diluted 1:2000, separated by two wash steps in PBS. After a final series of six PBS washes, the dot blot was developed with Sigmafast BCIP/NBT alkaline phosphatase substrate at 1 mg/mL (Sigma-Aldrich).
Genetic modification of Plasmodium knowlesi parasites
The genetic modification of Plasmodium knowlesi A1-H.1 strain 30 is described in detail elsewhere 31. Briefly, parasites were modified using a two-plasmid CRISPR-Cas9 system, comprised of a plasmid (pCas/sg) providing Cas9, sgRNA, and a hDHFR-yFCU (for positive and negative selection) to create a locus specific double strand break and a separate “donor” DNA plasmid (pDonor) to act as the repair template. To create the PkDBPαOR or PvDBPOR lines the PkDBPα locus (sgRNA: GCTGATCCAGGTTCTCAATC) was targeted using a pDonor plasmid containing full length recodonised PkDBPα or PvDBP (SalI) genes respectively, both flanked by 500 bp homology regions targeting the 5’UTR and 3’UTR of PkDBPα. To create the PkDBPαOR/Δβγ and PvDBPOR/Δβγ lines this process was repeated in an A1-H.1 line which had a natural deletion of PkDBPβ (due to a truncation of chromosome 14) and then a subsequent round of gene editing was used to delete the PkDBPγ locus (sgRNA: CATGCAACAATTTACACCCC) using a pDonor plasmid containing a spacer sequence flanked by 500 bp homology regions targeting the 5’UTR and 3’UTR of PkDBPγ.
Assays of growth inhibitory activity (GIA) with Plasmodium knowlesi lines
In vitro parasite culture and synchronisation
Human RBC-adapted parasites were maintained in culture as previously described 30. Briefly, parasites were grown at 2 % haematocrit in O+ human RBC, which were prepared twice monthly. Culture medium contained 10 % heat-inactivated pooled human serum mixed with RPMI 1640 supplemented with 25 mM HEPES, 35 μM hypoxanthine, 2 mM L-glutamine and 20 μg/mL gentamycin. Parasite cultures were synchronised at trophozoite/schizont stage by magnetic separation (MACS LS columns, Miltenyi Biotech).
In vitro assay of GIA
Methodology was adapted from the protocol of the International GIA Reference Centre at NIH, USA 48. Synchronised trophozoites were adjusted to 1.5 % parasitaemia, and 20 μL aliquots were pipetted into 96-well flat/half area tissue culture cluster plates (Appleton Woods). 20 μL test antibody or controls were added in duplicate or triplicate test wells over a concentration range (usually; 1, 0.5, 0.25, 0.125, 0.0625, 0.0312, 0.015 and 0.0075 mg/mL) and incubated for one erythrocytic parasite cycle (26-30 h). Parasitaemia was measured using the lactate dehydrogenase (pLDH) activity assay following standard protocols 49. Percentage GIA was calculated as below;
An anti-DARC VHH camelid nanobody 50, a kind gift from Dr Olivier Bertrand (INSERM, France) was included in the test plate as a positive control in every assay (at a final concentration of 6, 3 or 1.5 μg/mL) and an anti-Ebolavirus glycoprotein-reactive human IgG1 mAb as a negative isotype control for mAb samples. The assays were performed using five different lines of Plasmodium knowlesi parasites; the non-transgenic strain (PkA1-H.1); two transgenic lines containing the PvDBP gene (PvDBPOR and PvDBPOR/Δβγ) and two transgenic control lines (PkDBPαOR and PkDBPαOR/Δβγ) 31. In the mAb synergy assays (Figure 5) ‘Bliss additivity’ between two mAbs (mAb A and mAb B) was calculated using the formula below.
Ex-vivo Plasmodium vivax invasion assays
The assay methodology has been previously described 51,52. The major steps are summarised below.
Purification of reticulocytes from umbilical cord blood
20 mL aliquots of umbilical cord blood were collected from consenting volunteers (OXTREC 027-025 (Centre for Clinical Vaccinology and Tropical Medicine, University of Oxford, Oxford, United Kingdom) and MUTM 2008-215 from the Ethics Committee of the Faculty of Tropical Medicine (Mahidol University, Bangkok, Thailand)) in lithium heparin tubes and the white blood cells and platelets depleted using Non-woven filters™ (Antoshin). The RBC were then layered onto 70 % isotonic Percoll, centrifuged and the resulting band of enriched reticulocytes was incubated with anti-CD71 MicroBeads (Miltenyi) before being passed through a large selection (LS) column (Miltenyi) to obtain a CD71-depleted (negative) fraction and a CD71-rich fraction (positive). The purity levels of these CD71+ fractions were assessed with microscopy using new methylene blue staining (Sigma-Aldrich).
Purification of schizonts from ex-vivo Plasmodium vivax-infected blood samples
5 mL samples of whole blood were collected from consenting patients (OXTREC 027-025 (Centre for Clinical Vaccinology and Tropical Medicine, University of Oxford, Oxford, United Kingdom) and MUTM 2008-215 from the Ethics Committee of the Faculty of Tropical Medicine (Mahidol University, Bangkok, Thailand)) diagnosed with Plasmodium vivax malaria and rapidly transported at room temperature back to the laboratory. Samples were not collected from patients who had taken antimalarial or antimicrobial drugs within the previous month, or whose parasitaemia was < 0.1 % on smear microscopy. The samples were leukodepleted using Non-woven filters™ (Antoshin), and the parasites were cultured to schizont stage. The culture was then treated with trypsin, overlaid on a 45 % Percoll (isotonic) cushion and centrifuged to isolate a fine band of concentrated schizonts for use in the assay.
Invasion assay
The concentrated schizont preparation was mixed with the enriched reticulocyte fraction at a ratio of 1:6, giving a starting schizont parasitaemia of ~14 %. The mixture was diluted to 1.3 % haematocrit in 300 μL of complete McCoy 5A medium and cultured in 96-well cell culture plates in an atmosphere of 5 % O2 at 37.5 °C. Test antibodies and controls were buffer exchanged into Pv culture medium (McCoy 5A medium (Gibco) supplemented with 2.4 g/L D-glucose, 40 mg/mL gentamycin sulfate, and 20% heat-inactivated human AB serum) and added to the final invasion assay mixture at the desired concentration. In the case of the anti-PvDBPII mAbs this was 1 mg/mL. The anti-DARC nanobody, at 25 μg/mL, and the recombinant IgG1 anti-Ebolavirus mAb, at 1mg/mL, were added to all assays as positive and negative controls respectively. Maturation was obtained after incubation for an average of 24 h. At the end of the incubation period, thin smears (each made with 1 μL packed cells) were made, and stained with Giemsa (Sigma-Aldrich). The number of ring stages and trophozoites per 4000 erythrocytes was determined by examining the Giemsa thin film smears by light microscopy. For isolates #1-3, the invasion assays were performed in Singapore using cryopreserved Thai Plasmodium vivax isolates and reticulocyte invasion inhibition was quantified by flow cytometry as previously described 51.
Sequencing of the Pv DBPII gene from Thai isolates
Genomic DNA was extracted either from an aliquot (200 μL) of the initial blood sample or from a dried blood spot, using the DNeasy Blood and Tissue kit (QIAGEN). PCR amplification was performed on 1 μL purified genomic DNA in a total volume of 50 μL, containing 1x PCR buffer, 2.5 mM MgCl2, 0.2 mM of each dNTP, 2 μM each primer (forward 5’ – gtg act ggg cat gag gga aat tct cg and reverse 5’ – gcg tag aat ctc ctg gaa cct tct cc) and 1.25U of AmpliTaq Gold DNA polymerase (Applied Biosystems). The cycling conditions were: 95 °C for 5 min followed by 30 cycles at 94 °C for 1 min, 63 °C for 2 min, and 72 °C for 2 min. The product was purified using the QIAquick PCR purification kit (QIAGEN) and then sequenced (GATC biotech).
Structural Methods
Protein Cloning, Expression and Purification
PvDBPII coding region (residues 211-507; SalI sequence) was cloned into pET15b vector. This vector allowed expression of the protein with an N-terminal His6 tag and a TEV cleavage site, in E. coli (BL21-DE3 strain). The transformed bacteria were induced with 1 mM IPTG at an optical density of 0.8 at 600 nm. PvDBPII was expressed exclusively in inclusion bodies and was prepared as previously described 15. In short, the inclusion bodies were solubilised in 6 M guanidine hydrochloride, 20 mM Tris pH8, 300 mM NaCl, and refolded by flash-dilution into 400 mM L-arginine, 50 mM Tris pH8, 10 mM EDTA, 0.1 mM PMSF, 3 mM reduced glutathione, and 0.3 mM oxidised glutathione. Refolded proteins were buffer exchanged into 20 mM Tris pH8, 150 mM NaCl, 20 mM imidazole and then affinity purified on NiNTA resin. The His-tag was removed by TEV protease cleavage and the tagless PvDBPII was separated from the His-tag on NiNTA resin and purified by SEC (S75 16/60, GE Healthcare) into 20 mM Hepes pH7.5, 150 mM NaCl.
Subdomain3 (Sd3) (residues 211-508) was subcloned from the PvDBPII construct in the pET15b vector. Sd3 also expressed in E. coli BL21 (DE3) cells in the insoluble fraction and was solubilised in 6 M guanidine hydrochloride, 20 mM Tris (pH8), 150 mM NaCl, 20 mM imidazole. Sd3 was then refolded by binding to a NiNTA column and gradually decreasing the concentration of guanidine hydrochloride, until the protein was in 20 mM Tris (pH8), 150 mM NaCl, 20 mM imidazole. The refolded Sd3 protein was then eluted and further purified using SEC (S75 16/60 GE Healthcare) into 20 mM Hepes pH7.5, 150 mM NaCl.
Fab Generation and Purification
Fab fragments for DB9 were generated from IgG by incubating with immobilised papain (ThermoFisher Scientific) for 16 h at 37 °C. The Fab was separated from the un-cleaved DB9 and Fc regions using protein A resin (ThermoFisher Scientific). The Fab fragments were then purified by SEC into 20 mM Hepes pH7.5, 150 mM NaCl.
Protein Cystallisation and Data Collection
PvDBPII and DB9Fab were mixed in a 1.2:1 molar ratio and incubated at RT for 1 h. The complex was purified by SEC (S75, GE Healthcare) in 20 mM Hepes pH7.5, 150 mM NaCl. Broad crystallisation trials were set up by sitting-drop vapour diffusion in SwisSci 96-well plates by mixing 100 nl protein with 100 nl reservoir solution. Crystals grew in the reservoir solution of 45 % v/v polypropylene glycol 400, 10% v/v ethanol and were then cryo-cooled in liquid nitrogen. Data were collected on beamline I03 at Diamond Light Source at a wavelength of 0.97625Å and were indexed using XDS 53 and scaled using Scala 54 to a resolution of 3.04 Å.
Structure Solution
The PvDBPII:DB9Fab structure was solved by molecular replacement in Phaser 55 using the known structures of PvDBPII (PDB 4NUV) and a human mAb Fab fragment (PDB: 3DIF), separated into the variable and constant regions, as search models. This identified one copy of the PvDBPII:DB9 complex in the asymmetric unit. Refinement and rebuilding was completed using Buster 56 and Coot 57, respectively. The final Ramachandran plot showed 94.9% of residues in the favoured region, 5.1% in the allowed region and none in the disallowed region.
Statistical Analyses
Data were analysed using GraphPad Prism version 6.07 for Windows (GraphPad Software Inc.). In Figure 3A, a four-parameter sigmoidal dose-response curve was fitted to the relationship between log10 (antibody concentration) and percentage GIA for each dataset and used to interpolate EC30 values. In Figure 3B, the nonparametric Spearman’s rank correlation coefficient (ρ) was used to assess a correlation between the variables kon/koff/KD and GIA EC30. In all statistical tests the P values reported are two-tailed with P < 0.05 considered significant.
Supplementary Material
Acknowledgements
The authors are grateful for the assistance of Julie Furze, David Llewellyn, Drew Worth, Kim Johnson, Jordan Barrett and Sumi Biswas (Jenner Institute, University of Oxford); Patrick C. Wilson (University of Chicago) for expression plasmids; Oleg Fedorov (Structural Genomics Consortium, University of Oxford) for use of the Octet RED384; Amy Duckett and Carly Banner (University of Oxford) for arranging contracts; Yves Durocher for HEK293E cells (CNRC-NRC, Canada); A. Rushdi Shakri (ICGEB, India) and Chetan E. Chitnis (Institute Pasteur, France) for PvDBPII variant proteins; James McCarthy (QIMR, Australia) for HMP013 P. vivax strain PvDBPII sequence; Olivier Bertrand (INSERM, Paris) for VHH camelid anti-DARC nanobody; and the VAC051 PvDBPII vaccine clinical trial study volunteers and the staff and patients associated with the Shoklo Malaria Research Unit for Plasmodium vivax sample donation (SMRU, Thailand).
This work was supported in-part by funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement for MultiViVax [number 733073]. T.A.R. held a Wellcome Trust Research Training Fellowship [108734/Z/15/Z]; N.M.B. is funded by the Wellcome Trust D. Phil. program in Structural Cell Biology; J.S.C. held a Singapore International Graduate Award (SINGA); D.G.W.A. held a UK Medical Research Council (MRC) iCASE PhD Studentship [MR/K017632/1]; M.K.H. is a Wellcome Trust Investigator [101020/Z/13/Z]; and S.J.D. is a Jenner Investigator; a Lister Institute Research Prize Fellow and a Wellcome Trust Senior Fellow [106917/Z/15/Z]. SMRU is part of the Mahidol Oxford Research Unit supported by the Wellcome Trust. R.W.M and F.M are supported by a UK MRC Career Development Award [MR/M021157/1] jointly funded by the UK MRC and Department for International Development.
Footnotes
Data Availability
Crystallographic data is deposited in the Protein Data Bank with accession number 6R2S. DNA sequences are deposited in GenBank with accession numbers MK752505-24. The remaining data that support the findings of this study are available from the corresponding author on request.
Author Contributions
T.A.R., N.M.B., M.K.H. and S.J.D. conceived the study and wrote the manuscript.
T.A.R., N.M.B., F.M., J.S.C., V.K., S.F.G., D.G.W.A., G.M.L., S.C.E., S.E.S., D.Q., J.J., J.M.M., R.W.M. and M.K.H. performed experiments.
T.A.R., N.M.B., F.M., J.S.C., D.G.W.A., M.K.H. and S.J.D. performed data analysis and interpretation. R.O.P., A.M.M., J.J., R.W.M., B.R., L.R. and F.H.N. contributed reagents or materials and facilities.
Competing Interests Statement
D.G.W.A, M.K.H. and S.J.D. are named inventors on patent applications relating to malaria vaccines, mAbs and/or immunization regimens.
References
- 1.WHO. Control and elimination of plasmodium vivax malaria: a technical brief. 2015. ISBN 978 92 4 150924 4. [Google Scholar]
- 2.Gething PW, et al. A long neglected world malaria map: Plasmodium vivax endemicity in 2010. PLoS Negl Trop Dis. 2012;6:e1814. doi: 10.1371/journal.pntd.0001814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gruszczyk J, et al. Transferrin receptor 1 is a reticulocyte-specific receptor for Plasmodium vivax. Science. 2018;359:48–55. doi: 10.1126/science.aan1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malleret B, Renia L, Russell B. The unhealthy attraction of Plasmodium vivax to reticulocytes expressing transferrin receptor 1 (CD71) Int J Parasitol. 2017;47:379–383. doi: 10.1016/j.ijpara.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 5.Singh SK, Hora R, Belrhali H, Chitnis CE, Sharma A. Structural basis for Duffy recognition by the malaria parasite Duffy-binding-like domain. Nature. 2006;439:741–4. doi: 10.1038/nature04443. [DOI] [PubMed] [Google Scholar]
- 6.Chitnis CE, Sharma A. Targeting the Plasmodium vivax Duffy-binding protein. Trends Parasitol. 2008;24:29–34. doi: 10.1016/j.pt.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 7.Singh AP, et al. Targeted deletion of Plasmodium knowlesi Duffy binding protein confirms its role in junction formation during invasion. Mol Microbiol. 2005;55:1925–34. doi: 10.1111/j.1365-2958.2005.04523.x. [DOI] [PubMed] [Google Scholar]
- 8.Singh AP, Puri SK, Chitnis CE. Antibodies raised against receptor-binding domain of Plasmodium knowlesi Duffy binding protein inhibit erythrocyte invasion. Mol Biochem Parasitol. 2002;121:21–31. doi: 10.1016/s0166-6851(02)00017-8. [DOI] [PubMed] [Google Scholar]
- 9.Miller LH, Mason SJ, Clyde DF, McGinniss MH. The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy. N Engl J Med. 1976;295:302–4. doi: 10.1056/NEJM197608052950602. [DOI] [PubMed] [Google Scholar]
- 10.Howes RE, et al. Plasmodium vivax Transmission in Africa. PLoS Negl Trop Dis. 2015;9:e0004222. doi: 10.1371/journal.pntd.0004222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Culleton RL, et al. Failure to detect Plasmodium vivax in West and Central Africa by PCR species typing. Malar J. 2008;7:174. doi: 10.1186/1475-2875-7-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Draper SJ, et al. Malaria Vaccines: Recent Advances and New Horizons. Cell Host Microbe. 2018;24:43–56. doi: 10.1016/j.chom.2018.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Batchelor JD, Zahm JA, Tolia NH. Dimerization of Plasmodium vivax DBP is induced upon receptor binding and drives recognition of DARC. Nat Struct Mol Biol. 2011;18:908–14. doi: 10.1038/nsmb.2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choe H, et al. Sulphated tyrosines mediate association of chemokines and Plasmodium vivax Duffy binding protein with the Duffy antigen/receptor for chemokines (DARC) Mol Microbiol. 2005;55:1413–22. doi: 10.1111/j.1365-2958.2004.04478.x. [DOI] [PubMed] [Google Scholar]
- 15.Batchelor JD, et al. Red blood cell invasion by Plasmodium vivax: structural basis for DBP engagement of DARC. PLoS Pathog. 2014;10:e1003869. doi: 10.1371/journal.ppat.1003869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hans D, et al. Mapping binding residues in the Plasmodium vivax domain that binds Duffy antigen during red cell invasion. Mol Microbiol. 2005;55:1423–34. doi: 10.1111/j.1365-2958.2005.04484.x. [DOI] [PubMed] [Google Scholar]
- 17.VanBuskirk KM, Sevova E, Adams JH. Conserved residues in the Plasmodium vivax Duffy-binding protein ligand domain are critical for erythrocyte receptor recognition. Proc Natl Acad Sci U S A. 2004;101:15754–9. doi: 10.1073/pnas.0405421101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh K, et al. Malaria vaccine candidate based on Duffy-binding protein elicits strain transcending functional antibodies in a Phase I trial. NPJ Vaccines. 2018;3:48. doi: 10.1038/s41541-018-0083-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Payne RO, et al. Human vaccination against Plasmodium vivax Duffy-binding protein induces strain-transcending antibodies. JCI Insight. 2017;2 doi: 10.1172/jci.insight.93683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Cassan SC, et al. Preclinical Assessment of Viral Vectored and Protein Vaccines Targeting the Duffy-Binding Protein Region II of Plasmodium Vivax. Front Immunol. 2015;6:348. doi: 10.3389/fimmu.2015.00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moreno A, et al. Preclinical assessment of the receptor-binding domain of Plasmodium vivax Duffy-binding protein as a vaccine candidate in rhesus macaques. Vaccine. 2008;26:4338–44. doi: 10.1016/j.vaccine.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bermudez M, Moreno-Perez DA, Arevalo-Pinzon G, Curtidor H, Patarroyo MA. Plasmodium vivax in vitro continuous culture: the spoke in the wheel. Malar J. 2018;17:301. doi: 10.1186/s12936-018-2456-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cole-Tobian JL, et al. Strain-specific duffy binding protein antibodies correlate with protection against infection with homologous compared to heterologous plasmodium vivax strains in Papua New Guinean children. Infect Immun. 2009;77:4009–17. doi: 10.1128/IAI.00158-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.King CL, et al. Naturally acquired Duffy-binding protein-specific binding inhibitory antibodies confer protection from blood-stage Plasmodium vivax infection. Proc Natl Acad Sci U S A. 2008;105:8363–8. doi: 10.1073/pnas.0800371105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicolete VC, Frischmann S, Barbosa S, King CL, Ferreira MU. Naturally Acquired Binding-Inhibitory Antibodies to Plasmodium vivax Duffy Binding Protein and Clinical Immunity to Malaria in Rural Amazonians. J Infect Dis. 2016;214:1539–1546. doi: 10.1093/infdis/jiw407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen E, et al. Broadly neutralizing epitopes in the Plasmodium vivax vaccine candidate Duffy Binding Protein. Proc Natl Acad Sci U S A. 2016;113:6277–82. doi: 10.1073/pnas.1600488113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chootong P, et al. Mapping epitopes of the Plasmodium vivax Duffy binding protein with naturally acquired inhibitory antibodies. Infect Immun. 2010;78:1089–95. doi: 10.1128/IAI.01036-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ye J, Ma N, Madden TL, Ostell JM. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 2013;41:W34–40. doi: 10.1093/nar/gkt382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shakri AR, Rizvi MM, Chitnis CE. Development of quantitative receptor-ligand binding assay for use as a tool to estimate immune responses against Plasmodium vivax Duffy binding protein Region II. J Immunoassay Immunochem. 2012;33:403–13. doi: 10.1080/15321819.2012.659781. [DOI] [PubMed] [Google Scholar]
- 30.Moon RW, et al. Adaptation of the genetically tractable malaria pathogen Plasmodium knowlesi to continuous culture in human erythrocytes. Proc Natl Acad Sci U S A. 2013;110:531–6. doi: 10.1073/pnas.1216457110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mohring F, et al. Rapid and iterative genome editing in the zoonotic malaria parasite Plasmodium falciparum: New tools for P. vivax research. BioRxiv. 2019 doi: 10.7554/eLife.45829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adams JH, et al. The Duffy receptor family of Plasmodium knowlesi is located within the micronemes of invasive malaria merozoites. Cell. 1990;63:141–53. doi: 10.1016/0092-8674(90)90295-p. [DOI] [PubMed] [Google Scholar]
- 33.Gruring C, et al. Human red blood cell-adapted Plasmodium knowlesi parasites: a new model system for malaria research. Cell Microbiol. 2014;16:612–20. doi: 10.1111/cmi.12275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pain A, et al. The genome of the simian and human malaria parasite Plasmodium knowlesi. Nature. 2008;455:799–803. doi: 10.1038/nature07306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ord RL, et al. A malaria vaccine candidate based on an epitope of the Plasmodium falciparum RH5 protein. Malar J. 2014;13:326. doi: 10.1186/1475-2875-13-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Douglas AD, et al. Neutralization of Plasmodium falciparum merozoites by antibodies against PfRH5. J Immunol. 2014;192:245–58. doi: 10.4049/jimmunol.1302045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saul A. Kinetic constraints on the development of a malaria vaccine. Parasite Immunol. 1987;9:1–9. doi: 10.1111/j.1365-3024.1987.tb00483.x. [DOI] [PubMed] [Google Scholar]
- 38.Williams AR, et al. Enhancing blockade of Plasmodium falciparum erythrocyte invasion: assessing combinations of antibodies against PfRH5 and other merozoite antigens. PLoS Pathog. 2012;8:e1002991. doi: 10.1371/journal.ppat.1002991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nobrega de Sousa T, Carvalho LH, Alves de Brito CF. Worldwide genetic variability of the Duffy binding protein: insights into Plasmodium vivax vaccine development. PLoS One. 2011;6:e22944. doi: 10.1371/journal.pone.0022944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tiller T, et al. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J Immunol Methods. 2008;329:112–24. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quan J, Tian J. Circular polymerase extension cloning of complex gene libraries and pathways. PLoS One. 2009;4:e6441. doi: 10.1371/journal.pone.0006441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wrammert J, et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453:667–71. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jin J, et al. Accelerating the clinical development of protein-based vaccines for malaria by efficient purification using a four amino acid C-terminal 'C-tag'. Int J Parasitol. 2017;47:435–446. doi: 10.1016/j.ijpara.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ntumngia FB, et al. Conserved and variant epitopes of Plasmodium vivax Duffy binding protein as targets of inhibitory monoclonal antibodies. Infect Immun. 2012;80:1203–8. doi: 10.1128/IAI.05924-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hostetler JB, et al. A Library of Plasmodium vivax Recombinant Merozoite Proteins Reveals New Vaccine Candidates and Protein-Protein Interactions. PLoS Negl Trop Dis. 2015;9:e0004264. doi: 10.1371/journal.pntd.0004264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Payne RO, Griffin PM, McCarthy JS, Draper SJ. Plasmodium vivax Controlled Human Malaria Infection - Progress and Prospects. Trends Parasitol. 2017;33:141–150. doi: 10.1016/j.pt.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rijal P, et al. Therapeutic Monoclonal Antibodies for Ebola Virus Infection Derived from Vaccinated Humans. Cell Rep. 2019;27:172–186 e7. doi: 10.1016/j.celrep.2019.03.020. [DOI] [PubMed] [Google Scholar]
- 48.Miura K, et al. Anti-apical-membrane-antigen-1 antibody is more effective than anti-42-kilodalton-merozoite-surface-protein-1 antibody in inhibiting plasmodium falciparum growth, as determined by the in vitro growth inhibition assay. Clin Vaccine Immunol. 2009;16:963–8. doi: 10.1128/CVI.00042-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kennedy MC, et al. In vitro studies with recombinant Plasmodium falciparum apical membrane antigen 1 (AMA1): production and activity of an AMA1 vaccine and generation of a multiallelic response. Infect Immun. 2002;70:6948–60. doi: 10.1128/IAI.70.12.6948-6960.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smolarek D, et al. A recombinant dromedary antibody fragment (VHH or nanobody) directed against human Duffy antigen receptor for chemokines. Cell Mol Life Sci. 2010;67:3371–87. doi: 10.1007/s00018-010-0387-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cho JS, et al. Unambiguous determination of Plasmodium vivax reticulocyte invasion by flow cytometry. Int J Parasitol. 2016;46:31–9. doi: 10.1016/j.ijpara.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 52.Russell B, et al. A reliable ex vivo invasion assay of human reticulocytes by Plasmodium vivax. Blood. 2011;118:e74–81. doi: 10.1182/blood-2011-04-348748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–32. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 55.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bricogne. Buster version 2.10.2. Global Phasing Limited; Cambridge, United Kingdom: 2017. [Google Scholar]
- 57.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.