Abstract
Metastasis accounts for most of cancer-related deaths. Paracrine signaling between tumor cells and the stroma induces changes in the tumor microenvironment required for metastasis. Transcription factor c-Myb was associated with breast cancer (BC) progression but its role in metastasis remains unclear. Here we show that increased c-Myb expression in BC cells inhibits spontaneous lung metastasis through impaired tumor cell extravasation. On contrary, BC cells with increased lung metastatic capacity exhibited low c-Myb levels. We identified a specific inflammatory signature, including Ccl2 chemokine; that was expressed in lung metastatic cells but was suppressed in tumor cells with higher c-Myb levels. Tumor cell-derived Ccl2 expression facilitated lung metastasis and rescued trans-endothelial migration of c-Myb overexpressing cells. Clinical data show that the identified inflammatory signature, together with a MYB expression, predicts lung metastasis relapse in BC patients. These results demonstrate that the c-Myb-regulated transcriptional program in BCs results in a blunted inflammatory response and consequently suppresses lung metastasis.
Keywords: Myb, endothelium, inflammation, Ccl2
Introduction
Breast cancer (BC) 5 year disease-free survival rate is about 90% in patients with localized disease, but only 25% of patients with metastasis will survive 5 years from the date of diagnosis.1 While many therapies contribute to prolonged patients survival, the identification of metastatic mediators driving tissue-specific metastasis remains the major objective for further therapeutic development. Distribution of BC metastasis to the bones, brain, lungs and liver is a non-random process and each tumor type manifests a specific pattern of metastatic progression to distant organs.2,3 Metastasis comprises multiple steps including the tumor cell (TC) escape from primary location, survival in the circulation, seeding to distant sites and initiation of the growth at secondary sites. Circulating TCs engage with the endothelial cells in the vasculature, extravasate into distant organs and adopt the environment permissible for tumor growth. Tumor cells seeding and tissue colonization of distant organs are considered to be the rate-limiting step defining metastasis.
The c-Myb protein encoded by MYB/Myb gene is an essential transcriptional regulator for the maintenance of stem cells in bone marrow, colon epithelia, and neurogenic niches in an adult brain.4 Furthermore, normal hematopoiesis and cell lineage commitment are dependent on MYB function.5 The c-Myb binds to the specific sequence t/cAACt/gG, known as a Myb-binding site (MBS), within the control regions of target genes.4 The Myb-targeted genes are associated with cell proliferation, differentiation and apoptosis. However, c-Myb regulates different genes in distinct cell types, suggesting a context-specific regulation of transcription.6,7
The pioneering research on MYB focused on its oncogenic function in leukemia, but MYB expression has later been also linked to epithelial cancers, particularly breast and colon cancers.4 The presence of c-Myb is considered to be essential for the proliferation of ER-positive BC cells and also a prerequisite for mammary carcinogenesis in murine models.8,9 However, clinical data show that high MYB levels are associated with good prognosis for BC patients.10–12 One possibility to explain these contradictory findings is that c-Myb-driven proliferation of ER-positive BC tumors might be more responsive to cytotoxic drug treatment.13 Recently, we showed that c-Myb expression is inversely correlated with distant metastases in CRC patients and prevents murine mammary tumors to disseminate to lungs.14,15 However, the molecular mechanism how c-Myb contributes to metastasis remains unclear.
In this study, we used complementary strategies of c-Myb overexpression and in vivo selection of metastatic cells to evaluate transcriptional program regulated by c-Myb in BC cells. We identified an inflammatory signature required for pulmonary BC metastasis, which is suppressed by c-Myb; that may serve as a clinical predictor of tissue-specific relapse in BCs patients.
Results
Myb expression inhibits breast cancer lung metastasis
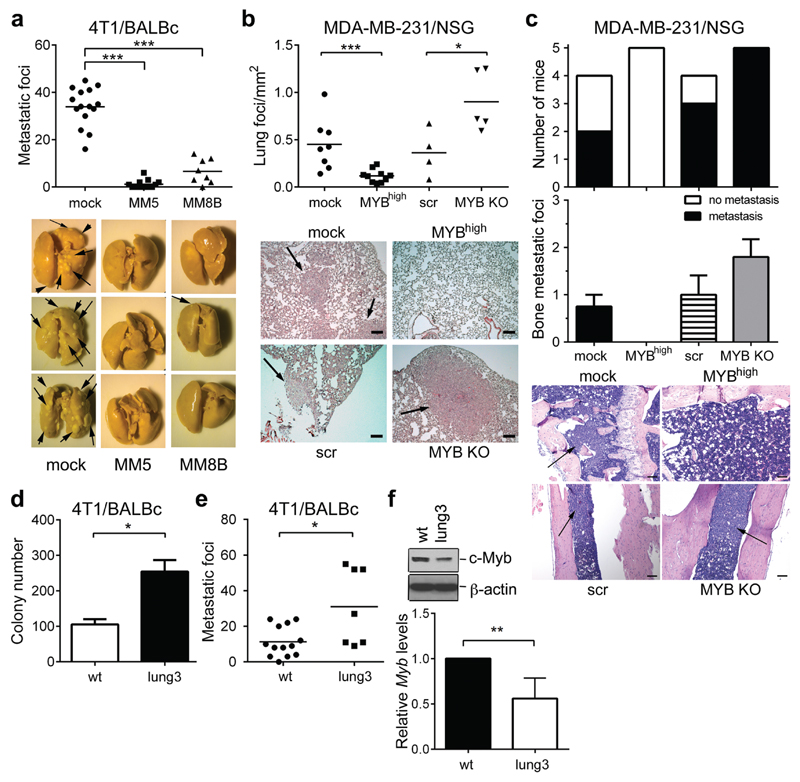
Overexpression of transcription factor (TF) Myb in murine mammary cancer cells 4T1 hinders spontaneous lung metastasis.15 To analyze the mechanism of the c-Myb activity, two independent clones (MM5 and MM8B) overexpressing c-Myb were injected into the mammary fat pads (m.f.p.) of BALB/c mice and metastasis were analyzed 24-28 days post injection (p.i.) (Supplementary Figure 1a). Mice bearing Myb-overexpressing tumors have significantly less metastatic foci in lungs compared to mock-transfected controls (Figure 1a), while liver metastasis remained unchanged (Supplementary Figure 1b). When we used 4T1 cells expressing luciferase, we observed particularly lung metastasis that was decreased in Myb-overexpressing tumors (Supplementary Figure 1c). To determine whether c-Myb affects metastatic dissemination, we assessed the lung seeding of 4T1 cells after intravenous injection (i.v.). The clonogenic growth of 4T1 cells isolated from lungs 24 h after i.v. injection revealed that MM5 cells produced less colonies compared to mock-transfected cells (Supplementary Figure 1d). To validate these findings we tested human breast cancer cell line MDA-MB-231, and prepared these cells with MYB overexpression (MYbhigh) and MYB deletion (MYB KO), respectively. Increased MYB expression resulted in significantly reduced lungs metastasis, but also decreased metastasis to the bone and to the liver (Figure 1b,c, Supplementary figure 1e). On contrary, MYB deletion caused overall increased metastasis in all three tissues. These data indicate that c-Myb expression in MDA-MB-231 breast cancer cells correlates with reduced metastasis.
Figure 1. c-Myb inhibits lung metastasis of BC cells.
(a) Number of metastatic foci in lungs of tumor-bearing BALB/c mice 28 days after m.f.p. injection of 4T1 cells: mock or Myb-overexpressing clones MM5 and MM8B (2 independent experiments). Representative images of lungs fixed in Bouin’s solution. (b-c) Metastasis of MDA-MB-231 cells upon m.f.p. injection into NSG mice and terminated at day 42, using MDA-MB-231 cells: transfected with empty vector (mock), expressing human MYB (MYBhigh), transfected with control gRNA (Scr); and deficient in MYB expression (MYB KO). (b) Quantification of lung metastasis with representative H&E stained lung sections; scale bar = 50μm. (c) Quantification of bone metastasis incidence and amounts with representative H&E stained bone sections; scale bar = 100μm. (d) Lung seeding of parental = wt, and lung3 cells. Number of colonies formed by 4T1 cells lodged in lungs 24 hours p.i (n=3). (e) Quantification of lung metastatic foci from BALB/c mice bearing 4T1 wt and lung3 tumors 28 days after m.f.p. injection (2 independent experiments). (f) Immunoblot analysis of c-Myb expression in parental (wt), and lung3 subline of 4T1 cells. Expression levels of Myb mRNA in the lung3 subline analyzed by qPCR and normalized to Gapdh, showed as relative values to parental cells (n=5). *; p<0.05; **; p<0.01; ***; p<0.001.
To corroborate the c-Myb involvement in metastasis, we passaged parental 4T1 cells in vivo and selected cells with high lung-seeding capacity (Supplementary Figure 1f). After three selection rounds the resulting cell line (named lung3) exhibited significantly increased lung seeding when compared to parental cells (wt) as determined by clonogenic assay (Figure 1d, Supplementary Figure 1g). In addition, m.f.p. injection of lung3 cells resulted in increased spontaneous lung metastasis (Figure 1e), while the tumor growth and liver metastasis was not affected (Supplementary Figure 1h-j). Interestingly, lung3 cells showed decreased Myb expression both on mRNA and protein levels (Figure 1f).
Low Myb expression correlates with increased expression of inflammatory response genes
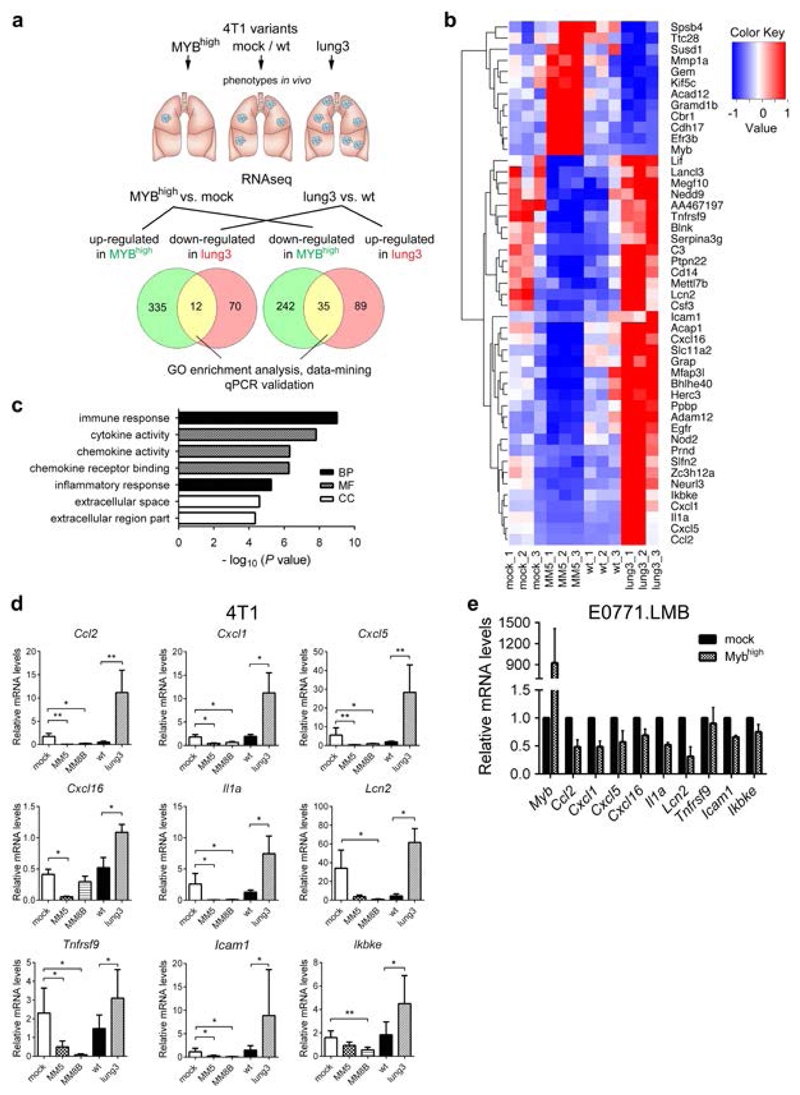
We tested the hypothesis that c-Myb drives transcriptional program preventing lung colonization by breast cancer cells, which is blunted in highly metastatic cells. To identify components of c-Myb-driven transcriptional program, we performed RNAseq of the complementary cell lines; Myb-overexpressing cells MM5 and lung3 cells with respective controls (Figure 2a). Genes that are down-regulated in MYBhigh cells and up-regulated in lung3 or vice versa were further analyzed. We identified a group of 12 transcripts up-regulated in MM5 and down-regulated in lung3 cells and a group of 35 transcripts down-regulated in MM5 but up-regulated in lung3 cells (Figure 2b). Gene ontology analysis revealed that the group with 35 genes upregulated in lung3 cells is significantly enriched in immune/inflammatory response genes (Figure 2c, Supplementary Figure 2, Supplementary Tables 4, 5). However, no significant GO enrichment was identified in the group of transcripts up-regulated in MM5 cells. Hence, c-Myb levels in 4T1 cells inversely correlate with specific inflammatory molecules. Nine genes of this inflammation-enriched set, Ccl2, Cxcl1, Cxcl5, Cxcl16, Il1a, Lcn2, Icam1, Tnfrsf9, Ikbke, were validated by qPCR in independent 4T1 clone overexpressing Myb MM8B (Figure 2d) and another mammary carcinoma cell line with basal-like characteristics, E0771.LMB. Transient Myb overexpression in E0771.LMB cells resulted in down-regulation of Ccl2, Cxcl1, Cxcl5, Cxcl16, Il1a, Lcn2, Icam1, Ikbke mRNA levels (Figure 2e). Taken together, these data indicate that c-Myb regulates lung metastasis through suppression of an inflammatory signature of tumor cells.
Figure 2. c-Myb represses immune/inflammatory response genes in BC cells.
(a) Strategy of RNAseq analysis to identify overlapping genes up/down-regulated in 4T1 MYBhigh and lung3 cells. (b) Heat map of 47 genes identified as induced/repressed in 4T1 MYBhigh and lung3 cells (fold change>1.5, p<0.01). (c) DAVID analysis software was used to find the Gene Ontology (GO) terms that were significantly enriched (FDR<0.05) in a 35 gene set highly expressed in lung3 and suppressed in MYBhigh cells (BP, biological process; MF, molecular function; CC, cellular compartment). (d) Expression levels of selected mRNAs in 4T1 MYBhigh clones and the lung3 subline were analyzed by qPCR, normalized to Gapdh and displayed as relative values to parental cells (n=3). *; p<0.05; **; p<0.01. (e) Expression levels of selected mRNAs after transient (48 hours) Myb overexpression in E0771.LMB cells by qPCR, normalized to Gapdh and displayed as relative values to mock-transfected E0771.LMB cells (n=3).
c-Myb suppresses Ccl2 expression in tumor cells and thereby inhibits lung metastasis
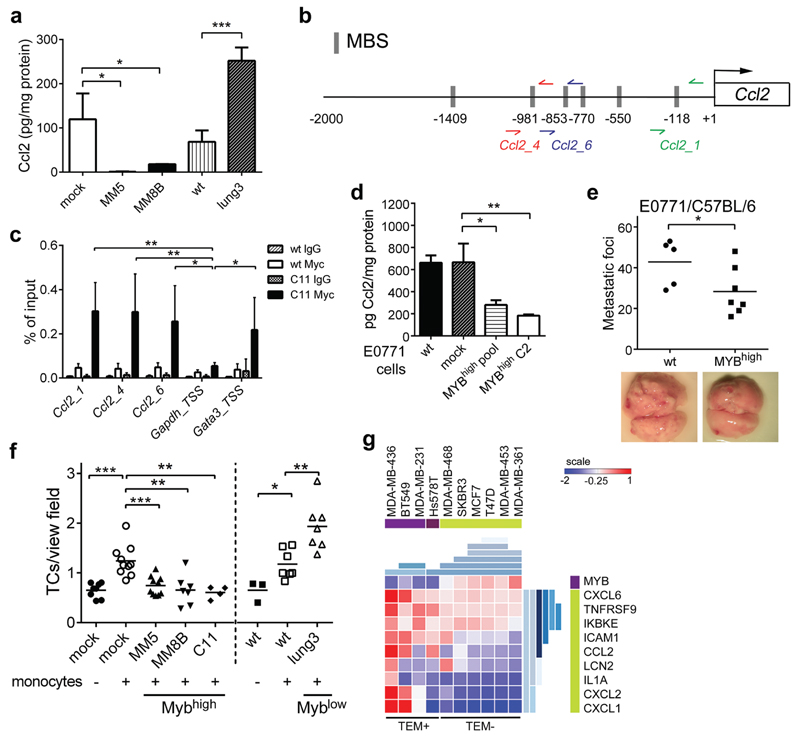
To validate the effect of Myb expression on inflammatory signature, we selected Ccl2 chemokine, which has been previously shown to promote metastasis in various cancers.16–19 MM5 and MM8B cells showed significantly reduced while lung3 cells significantly increased amounts of Ccl2 in the conditioned media when compared to controls (Figure 3a). Of note, Ccl2 expression was attenuated also upon transient Myb induction in 4T1 cells (Supplementary Figure 3a). To test whether c-Myb is directly responsible for suppression of the Ccl2 expression, we analyzed the regulatory regions of the Ccl2 gene for potential Myb-binding sites (MBSs). Six MBSs were identified in proximal region 2000 bp upstream of the transcription starting site (Figure 3b), and ChIP assays were performed to examine the c-Myb occupancy of these sites. We prepared 4T1-derived clone C11 overproducing c-Myb with C-terminal Myc-tag for the ChIP assay (Supplementary Figure 3b). Three different non-overlapping amplicons showed a c-Myb binding to the endogenous Ccl2 promoter (Figure 3c and Supplementary Figure 3c). In addition, ChIP assay in MDA-MB-231 cells showed binding of both endogenous and exogenous c-Myb to human CCL2 promoter. Anti-Myb antibody precipitated a sequence spanning two MBSs (hCCL2) upstream TSS of human CCL2, but not a control region without MBSs (hCCL2_neg) in wt and MYBhigh (M5) cells (Supplementary Figure 3d). These data indicate that c-Myb directly modulates the expression of Ccl2.
Figure 3. c-Myb suppresses Ccl2 expression and inhibits tumor cell metastasis and TEM.
(a) Ccl2 protein levels in the tumor cell conditioned media were normalized to total protein content (n=3). (b) Schematic representation of the promoter region of murine Ccl2 gene on chromosome 11. The potential c-Myb binding sites (MBSs, identified by TFSEARCH and ConSite, http://consite.genereg.net), the regions amplified after ChIP (primer pairs: red, blue, and green arrows) are depicted. (c) ChIP assays were performed with anti-Myc-Tag antibody or non-specific mouse IgG in 4T1 cells transfected with Myc-tagged-Myb (C11) and wt cells (n=3). Three primer sets spanning different MBSs were used for qPCR amplification and data are shown as percentage of input, unpaired t-test. Gapdh was used a negative control; Gata3 as a positive control. (d) Ccl2 protein levels in the E0771.LMB tumor cell conditioned media: wt, mock-transfected controls, MYBhigh pool and MYBhigh C2 normalized to total protein content (n=3, unpaired t-test). (e) Quantification of lung metastasis in mice i.v. injected with E0771.LMB cells 17 days p.i. (2 independent experiments) (f) Transmigrated 4T1 cells through primary lung ECs in the absence or presence of monocytes after 16 hours of co-culture. (3 independent experiments) (g) Expression-based clustering of BC cell lines (GSE44552) according to the TEM activity. Signature genes expression levels as determined by microarrays are visualized in a heat-map. Hierarchical clustering by Gitools is shown in shades of blue. *; p<0.05; **; p<0.01; ***; p<0.001.
To validate that Myb expression affects Ccl2 production and metastasis, we generated stable transfectants of E0771.LMB cells expressing exogenous c-Myb (MYBhigh) (Supplementary Figure 3e) and these cells produced lower amounts of Ccl2 compared to controls (Figure 3d). Intravenous injection of MYBhigh E0771.LMB resulted in attenuated lung metastasis when compared to parental cells (Figure 3e). Of note, c-Myb overexpression in two independent clones of MDA-MB-231 cells also resulted in reduced secretion of CCL2 (Supplementary Figure 3f). Next, we tested whether down-regulation of Ccl2 affects lung colonization of 4T1 cells. Stable Ccl2 knock-down in 4T1 cells (shCcl2) resulted in reduced Ccl2 production (Supplementary Figure 3g). Significantly less lung metastasis were observed upon m.f.p. injection of 4T1 shCcl2 cells when compared to sh-scrambled control cells (scRNA) (Supplementary Figure 3h).
The reduced capacity of Myb-expressing tumor cells to form lung metastasis can be linked to impaired tumor cell extravasation, which is known to be dependent on Ccl2.17,20 Therefore, we tested the capacity of tumor cells to migrate through endothelial cells in vitro. Trans-endothelial migration (TEM) of mock 4T1 cells through endothelial cell monolayers was enhanced by monocytes as observed previously. However; the migration of all 3 Myb overexpressing 4T1 clones MM5, MM8B, and C11 was impaired, while lung3 cells expressing lower levels of Myb showed enhanced TEM capacity (Figure 3f). Next, we assessed whether c-Myb interferes with efficient TEM by data-mining in microarray databases (GSE44552). Human BC cell lines known to be active in TEM21 express rather low endogenous MYB RNA levels compared to TEM-negative cell lines (Figure 3g). Further analysis of inflammatory signature genes in this dataset: CCL2, CXCL1, CXCL2 (human orthologue of murine Cxcl1), CXCL6 (human orthologue of murine Cxcl5), IL1A, ICAM1, IKBKE, LCN2, TNFRSF9; showed a reduced expression in Mybhigh TEM-negative cell lines, while in TEM-positive BC cells an induction of a specific inflammatory signature was detected. These data support the inverse correlation between c-Myb and Ccl2 expression and its effect on lung metastasis.
c-Myb inhibits tumor cell extravasation in an endothelial Ccr2-dependent manner
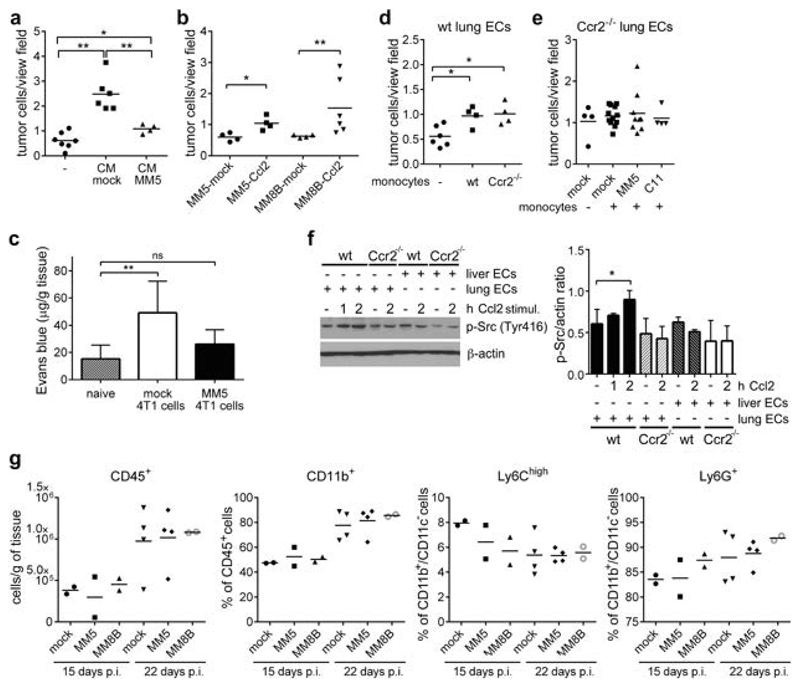
To test whether Ccl2 is sufficient to rescue TEM of Myb-overexpressing 4T1 cells we applied conditioned medium (CM) from mock or MM5 cells. Mock CM significantly promoted TEM of MM5 cells; while the CM from MM5 cells had a little effect (Figure 4a). Furthermore, exogenous Ccl2 expression in Myb-overexpressing 4T1 cells also rescued TEM (Figure 4b, Supplementary Figure 4a), indicating that c-Myb inhibits trans-endothelial migration of tumor cells through down-regulation of Ccl2. To test whether the reduced capacity of Myb-overexpressing tumor cells to migrate through the endothelial cells can be observed also in vivo, we intravenously injected MM5 cells and analyzed lung vascular permeability. Indeed, MM5 cells failed to induce pulmonary vessel permeability when compared to mock-transfected controls (Figure 4c, Supplementary Figure 4b). Since the failure of tumor cells to induce vascular permeability results in impaired cancer cell extravasation20, these findings indicate that c-Myb expression impairs the tumor cell extravasation and thereby lung colonization.
Figure 4. TEM of tumor cells is dependent on lung endothelial Ccr2 expression.
(a) TEM of MM5 cells untreated (-) or stimulated with conditioned medium (CM) from mock or MM5 cells, in the presence of monocytes after 16 hours. (2 independent experiments) (b) TEM of 4T1 MYBhigh cells overexpressing Ccl2 through lung ECs in the presence of monocytes. (2 independent experiments) (c) Quantification of Evans blue extracted from lungs of mice: untreated (naïve) or 24 hours p.i. with 4T1 mock and MM5 cells (n=4-6). (d) Tumor cells (4T1 mock) transmigration through lung ECs derived from wt C57BL/6 mice in the presence of wt and Ccr2-/- monocytes. (2 independent experiments) (e) TEM of mock 4T1, MM5 and C11 cells through Ccr2-/- lung ECs in the presence/absence of monocytes. (2 independent experiments) (f) Immunoblot analysis of p-Src in Ccl2-stimulated (100ng/ml) ECs derived from wt and Ccr2-/- mice, left. Densitometry quantification of p-Src signal (n=5), right. (g) Flow cytometry analysis of total leukocytes (CD45+), myeloid cells (CD45+CD11b+), granulocytes (CD45+CD11b+CD11c-Ly6G+), and inflammatory monocytes (CD45+CD11b+CD11c-Ly6G-Ly6Chi) recruited to the lungs 15 and 22 days p.i. *; p<0.05; **; p<0.01; ns; not significant.
To identify Ccl2-responsive cells involved in TEM, we first determined Ccr2 expression in 4T1 cells, lung and liver ECs and monocytes. Ccr2 was not expressed in 4T1 cells (Supplementary Figure 4c); that is in agreement with previous reports.22 Monocytes isolated either from wt or Ccr2-/- mice promoted TEM of 4T1 cells to the same level (Figure 4d). However, there was no monocyte-induced TEM observed on lung ECs derived from Ccr2-/- mice (Figure 4e). Inflammatory mediators are known to disrupt endothelial integrity by the Src kinase activation.23,24 To test whether endothelial cells respond directly to Ccl2, we stimulated ECs derived from lungs and liver. A time-dependent increase in p-Src (Y316) was observed in wt lung ECs, but not in Ccr2-/- ECs (Figure 4f), while no changes were observed in liver ECs irrespective of Ccr2 expression.
To test whether c-Myb modulation in the primary tumor affects the formation of lung and liver metastatic niche, we analyzed leukocyte recruitment to the metastatic sites in mice m.f.p.-injected with 4T1 cells at 15 and 22 days p.i. We observed an increased recruitment of leukocytes (CD45+) at day 22, but no significant difference between mock and Myb-overexpressing (MM5, MM8B) tumors (Figure 4g). The composition of leukocytes recruited to the lungs and liver, including myeloid cells (CD11b+), inflammatory monocytes (Ly6Chi), granulocytes (Ly6G+), macrophages (CD64+), was not different (Figure 4g, Supplementary Figure 4d).
Low MYB/high inflammatory signature is associated with poor prognosis and increased risk of BC lung relapse
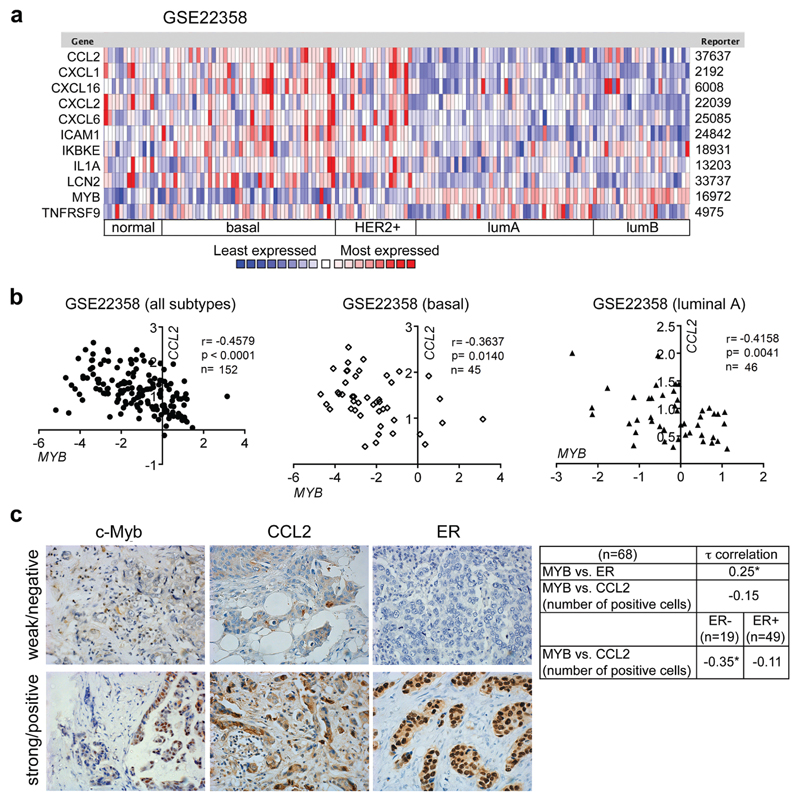
High MYB expression is associated with good prognosis for BC patients.10,11 Our meta-analyses using KM-Plotter confirmed that high MYB levels are linked with longer overall survival and lower risk of relapse or occurrence of distant metastasis (Supplementary Figure 5a). High MYB levels are generally found in less aggressive luminal subtypes (Figure 5a, Supplementary Figure 5b) that tend to metastasize to the bone rather than to the lungs or brain.25–27 To determine whether MYB modulates metastasis, we first calculated correlations between the expression of MYB and inflammatory signature genes within and across molecular subtypes of BCs in several independent publically available datasets (Figure 5b, Supplementary Table 3). Significant inverse correlations of MYB with CCL2, CXCL1, CXCL2, CXCL6, LCN2, were found in all six cohorts and in 5 cohorts additionally for ICAM1 across subtypes. Importantly, CCL2 mRNA levels were inversely correlated with MYB expression also within basal and luminal subtype from 3 datasets. To verify these data we evaluated c-Myb and Ccl2 protein levels in a cohort of 68 BCs patients by IHC. Estrogen receptor (ER) expression was significantly associated with c-Myb expression, while there was a trend towards negative association with a number of Ccl2-positive cells (Figure 5c). When the patient samples were analyzed according to the ER status, a significant inverse correlation between MYB and CCL2 was found within ER- group (n=19).
Figure 5. Inversed MYB-inflammatory signature expression in human BCs.
(a) Heat map showing expression levels of MYB and identified signature genes in human BCs (GSE22358). Red and blue indicate high and low mRNA expression levels, respectively. (b) Significant correlations between mRNA expression of MYB and CCL2 in the same dataset of BCs samples as in A and stratified according to the molecular subtype (basal, luminal A). (c) Representative IHC stainings of human BCs for c-Myb, Ccl2, ER (left). Kendall’s Tau correlation between ER, CCL2 and c-Myb detection analyzed in a group of 68 BC patients, and in ER+ and ER- patients. *; p<0.05.
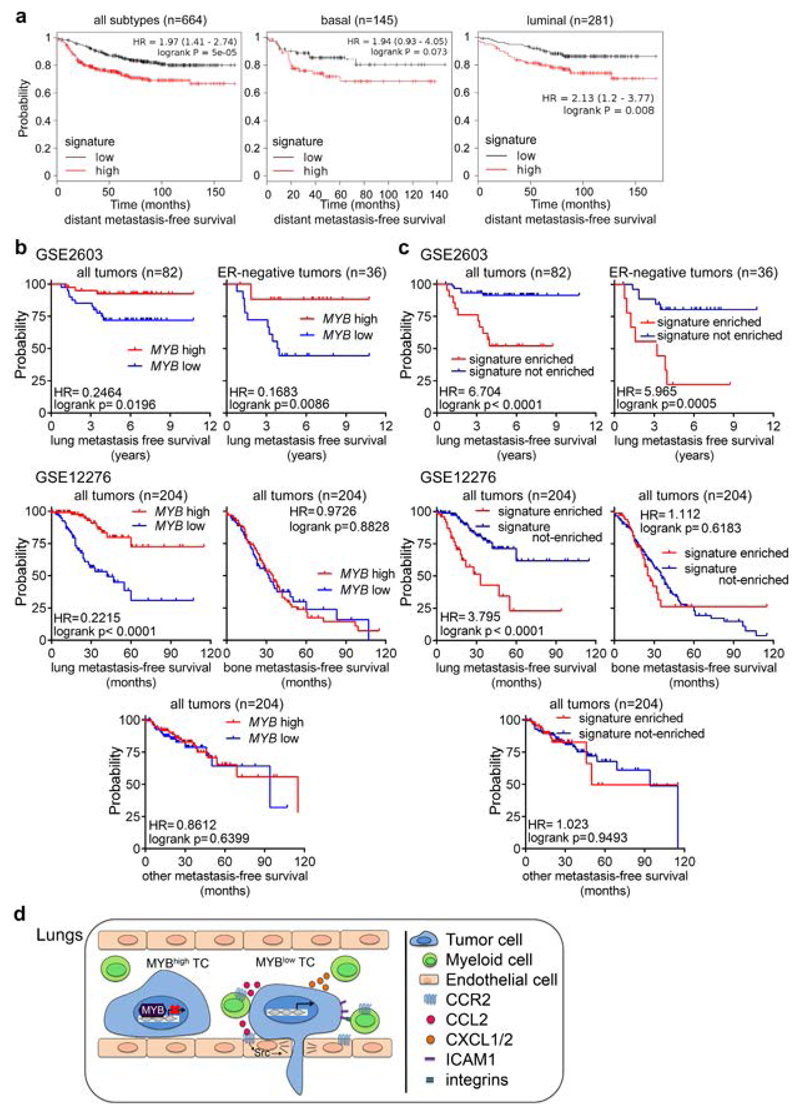
Next, we performed survival meta-analysis to test the probability of distant metastasis-free survival (DMFS), relapse-free survival (RFS) and overall survival (OS) in BCs stratified according to the expression status of the signature genes (CCL2, CXCL1, CXCL2, CXCL6, CXCL16, ICAM1, IL1A, TNFRSF9, LCN2, IKBKE) along with MYB (inversed). A significant correlation was observed between the signature expression and shorter survival outcome (Figure 6a, Supplementary Figure 6a). Importantly, the risk of distant metastasis remained increased in patients within basal (HR=1.94, p=0.073) or luminal group (HR=2.13, p=0.008) with a low MYB/high inflammatory signature status. Next, we analyzed datasets GSE2603 and GSE12276 for lung metastasis-free survival based on MYB expression using cohorts of primary tumors from BC patients with known site of relapse. We found a significant association between high MYB expression and longer lung-relapse free survival (HR=0.2215, p<0.0001; and HR=0.2464, p=0.0196) that remained significant also in ER-negative tumors (HR=0.1683, p=0.0086) (Figure 6b).
Figure 6. Inflammatory signaling that is suppressed by MYB is required for lung metastasis.
(a) Meta-analyses of BCs patients available on KMplot.com representing the probability of distant metastasis free survival in BCs stratified according to the expression status of the signature genes (CCL2, CXCL1, CXCL2, CXCL6, CXCL16, ICAM1, IL1A, TNFRSF9, LCN2, IKBKE and inversed MYB). The log-rank test P value reflects the significance of the correlation between inflammatory gene signature high/MYB low and shorter survival outcome. b-c) Kaplan–Meier plots showing lung metastasis-free, bone metastasis-free and other metastasis-free (liver/LN) survival of BCs patients from datasets: GSE260351 and GSE1227652, respectively. Survival analysis based on MYB expression (b) and based on signature enrichment (defined in A) as determined by z-score (c). Signature enriched = samples with significant up-regulation of the signature; signature not-enriched = the rest of samples (d) Model of the Myb-repressed inflammatory circuit during pulmonary metastasis.
To determine whether c-Myb-repressed inflammatory signature correlates with lung relapse we used Sample Level Enrichment Analysis (SLEA). 28 We observed a significant correlation between inflammatory signature genes and the risk of lung relapse (HR=3.795, p<0.0001; and HR=6.704, p<0.0001) even within the ER-negative group (HR=5.965, p=0.0005) (Figure 6c). Similar results were obtained with other independent cohorts (Fig. S6B). The analysis of the GSE12276 dataset for site-specific distant metastasis relapse showed that neither high MYB levels, nor inflammatory signature was associated with the risk of bone or liver/lymph node metastases (Figure 6c). When we compared MYB expression in primary BCs (GSE2603 and GSE12276 datasets) with known site of relapse and in BCs metastases (GSE11078), we observed lower MYB expression in tumors metastasizing to the lungs when compared to metastasizing tumors to other tissues (Supplementary Figure 6c). Overall these data show that the identified inflammatory signature, together with the MYB expression, predicts lung metastasis relapse in BC patients independently of the ER status.
Discussion
TF c-Myb was described to have oncogenic and tumor suppressor activities in BCs.8–11,29 Clinical data have unanimously associated MYB with a good prognosis for BC patients 10–12 that is agreement with our findings. There is only one recent study indicating the opposite tendency in one dataset;30 however, we could not reproduce these results by analysis of the same dataset. Here we showed that MYB expression in human BC tumor cells predicts a low risk of lung relapse. Low MYB expression is commonly associated with ER-negative BCs 25 and these tumors are known to preferentially colonize lung tissue when compared to ER-positive BCs.26,27 Consequently, high MYB marks BCs with longer lung-metastasis-free survival. Importantly, our data show that the association between MYB and the lung-metastasis-free survival is sustained within ER-negative cohort of BC patients, emphasizing c-Myb as an ER-independent risk factor for lung metastasis. We postulate that c-Myb expression, albeit potentially important for proliferation and early tumorigenesis8,9,29, has a negative impact on lung colonization of BCs.
TF c-Myb is a transcription regulator that is involved in regulation of cell growth and survival acts in a context-dependent manner.4,6,7 In our study the modulation of Myb expression did not alter cell proliferation or the cell cycle (Supplementary Figure 7a,b). Interestingly, higher Myb expression makes cells to be more resistant to chemotherapy (Supplementary Figure 7c), as previously observed.8,31 Myb expression was recently also linked to metastatic progression in pancreatic and breast cancer models.30,32 While c-Myb may act differently depending on the tissue origin32, there is discrepancy in breast cancer between our data and the previously published data.30 This may be due to a different subline (MDA-MB-231 BM) used in the report of Li et al30, while we were using the parental MDA-MB-231 cell line. Several genes previously identified as Myb-responsive11 were also confirmed in this work (Supplementary Figure 8). To define the transcriptional module responsible for lung metastasis suppression in mammary tumors, we compared transcriptomes of two complementary cell populations of lung metastasis-proficient and -deficient 4T1 cells. Increased c-Myb expression in lung-deficient cells suppressed genes involved in inflammation and immune-responses. Inflammatory mediators, including cytokines, were shown to directly promote metastasis through: increased vascular permeability and tumor cell survival in the circulation, mediating the immune escape and outgrowth at secondary sites, particularly in the lungs.17,18,33,34 Although causal interdependence between inflammation and cancer progression has long been established, unveiling the molecular mechanisms from a TF to a specific inflammatory circuit that modulates metastatic seeding is highly relevant for prospective therapy application.
Despite a variation in MYB expression among BCs subtypes, several inflammatory genes within the c-Myb-repressed module are associated with ER-negative status of BCs: e.g. LCN2 is highly expressed in ER-negative BCs 35, ICAM1 is overexpressed in human triple-negative BCs 36, CXCL1, CXCL3, CXCL5, CXCL6, and IL-8 are highly present in ER-negative tumors.37 Of note, a unique subgroup of ER+ BCs that express high levels of c-Myb and low level of innate inflammatory genes has been identified.11 Our findings indicate that c-Myb-initiated transcriptional program in ER-negative BCs results in blunted inflammatory response thereby limiting lungs metastasis while enabling metastasis in other organs.
TF c-Myb can repress expression of approximately half of its direct targets, despite being usually considered as a trans-activator.38 However, the repressing function of c-Myb remains poorly understood and is likely target-specific.6,7 Here we provide evidence that c-Myb expression suppresses lung metastasis in two mammary mouse models. Thus we postulate that both direct binding of c-Myb to the regulatory elements of the inflammatory genes, demonstrated for Ccl2, together with additional indirect effects contribute to the c-Myb-repressed transcriptional program in a gene-specific fashion. How the c-Myb represses all inflammatory genes in the identified signature in vivo remains to be defined.
Ccl2 is one of the inflammatory mediators repressed by c-Myb, which has been identified as a prominent tumor- and metastasis-promoting factor in BCs.18,19,39,40 Previously we demonstrated that endothelial Ccr2 expression in the lungs is required for tumor cell extravasation and E-selectin-dependent monocyte recruitment required for metastasis.17,20 In line with these results our current data show that suppression of tumor-derived Ccl2 by c-Myb impairs TEM of BC cells due to missing endothelial engagement while the recruitment of myeloid-derived cells remained unaffected. These findings suggest that enhanced c-Myb levels diminish the ability of circulating BC cells to activate pulmonary endothelium, thereby inhibits lung metastasis (Figure 6d).
Extravasation into the pulmonary tissue requires specific functions for breaching the tight capillary walls, while liver is relatively compliant to TCs extravasation owing to the fenestrated endothelium.41,42 Interestingly, we found that the c-Myb-dependent inhibition of TEM and metastasis is tissue-specific. Liver is permissive for Myb-overexpressing TCs that is in contrast to the lungs. We observed that liver ECs do not respond to recombinant Ccl2 stimulation by Src activation when compared to lung ECs. This data indicate that tumor-derived factors may elicit tissue-dependent responses promoting metastasis.
In conclusion, the inflammatory response controlled by c-Myb determines the metastatic competency of BCs cells. The identified inflammatory signature improves the identification of high-risk patients for lung relapse in majority of analyzed datasets compared to a single MYB assessment. Importantly, these markers predict lung relapses in BC patients irrespective of ER expression. Inhibition of the c-Myb-driven paracrine inflammatory axis may improve targeting of tumor cells that are inaccessible to standard anti-proliferative therapy and thereby lower the incidence of metastasis.
Material and Methods
Cell culture
Cell lines 4T1 and MDA-MB-231 were purchased from American Type Culture Collection and were cultured RPMI-1640/10% FCS. 4T1 and MDA-MB-231 cells with constitutive c-Myb overexpression (MYBhigh) were previously described.15 E0771.LMB cells were kindly provided by Dr. Robin Anderson (University of Melbourne, Australia)43 and were cultured in DMEM/10% FCS. E0771.LMB cells overexpressing Myb were prepared by transfection with pcDNA3.Myc-tag-Myb using Lipofecamine® LTX (Life Technologies) and selected with 500μg/ml G418. Stable knock-down clones of Ccl2 in 4T1 cells were generated by lentiviral infection with a two MISSION shRNAs targeting Ccl2 in pLKO.1 or non-mammalian shRNA control vector (both Sigma) and selection with 1.5μg/ml puromycin. 4T1 lung3 cells were prepared by three rounds of in vivo passage of wt cells, where lung tissues were harvested 24 hours later, and tumor cells grew under selection of 6-thioguanine (60μM).
Generation of MDA-MB-231 MYB knock-out cell line with CRISPR/Cas9
Guide RNA (gRNA) sequences for CRISPR/Cas9 were designed at CRISPR design web site (http://crispr.mit.edu/). The 25-bp forward and reverse oligonucleotides (Table S1) comprising 20 bp MYB-target sequence and Esp3I sticky ends were annealed and inserted into the lentiCRISPRv2 plasmid (Addgene 52961, Cambridge, MA) digested with Esp3I enzyme. Similarly, oligonucleotides comprising GFP-target sequence (Table S1) were used for derivation of a control plasmid. All plasmids were sequenced. MDA-MB-231 cells were transfected with lentiCRISPRv2 plasmids using Lipofectamine LTX, selected under puromycin (1μg/ml) for 2 weeks. Single-cell colonies were expanded for DNA extraction, protein extraction, and cryopreservation. PCR primers spanning potential sites of mutation were designed (Table S1), mutations were confirmed by Sanger sequencing and the efficiency of MYB knock-out was verified by immunoblotting.
Mice
Animal experiments were performed according to the guidelines of the Swiss Animal Protection Law, and approved by Veterinary Office of Kanton Zurich. Female mice (6-9 weeks old) BALB/c, NOD scid gamma (NSG), C57BL/6, and Ccr2 deficient mice (Ccr2-/-) were purchased from The Jackson Laboratory.
Metastatic assay and lung seeding experiments
4T1 cells (100’000 cells) in HBSS:Matrigel (1:1) were orthotopically injected into a mammary fat pad (m.f.p.) of a BALB/c mice and metastasis was analyzed after 24-28 days. MDA-MB-231 cells (2’000’000) were injected in m.f.p. of NSG mice and metastases were evaluated after 42 days. E0771.LMB (600’000) cells were intravenously injected (i.v.) in the tail vein of C57BL/6 mice and lung metastatic foci were analyzed after 17 days. For lung seeding experiments, 300’000 cells were i.v. injected and perfused lungs after 24 hours were digested with collagenase A/D (Sigma), passed through 40μm cell strainers, and cells were grown in presence of 6-thioguanine (60μM) for 10 days. Clonogenic growth was assessed after fixation with 4% PFA and 0.04% crystal violet staining.
Flow cytometry analysis of lung and liver tissues
The lungs of 4T1-tumor bearing mice were perfused, minced and digested in Collagenase D and A (2 mg each, Roche) for 1 hour at 37°C. A single-cell suspension using 40μm cell strainers was prepared, erythrocytes lysed. Cells were resuspended in staining buffer with Fc block (eBioscience), incubated on ice for 10 min, followed by staining with following antibodies: CD45-PB, CD11b-BV510, CD64-PE, Ly6C-APC, Ly6G-PerCP-Cy5.5 (all from BD) for 30 min. Data were collected with CountBright absolute counting beads (Life Technologies) using a FACSCanto (BD) and analyzed by FlowJo software (FlowJo, LCC).
Vascular permeability assay
Lung vascular permeability was determined with Evans blue assay 17.
RNA isolation and quantitative real-time PCR
Total RNA was isolated with RNeasy Mini kit (Qiagen) and cDNA was prepared with Omniscript RT Kit (Qiagen). Real-time PCR was performed with KAPA SYBR Fast Master mix (KAPA Biosystems) with intron-spanning primers (Table S1) using the LightCycler 480 (Roche) and data were normalized to Gapdh.15
Cytokine assay
Supernatants of cells cultured for 48 hours were collected and the amount of Ccl2 was assessed using the Cytometric Bead Array kit (BD Biosciences).
Isolation and stimulation of primary endothelial cells
Pulmonary and liver endothelial cells (ECs) were isolated using a positive immuno-magnetic selection as described previously.17 Seven days after isolation, cells were stimulated with murine recombinant (mr) IL1β (40 ng/ml, R&D Systems) for 2 hours, followed by treatment with mrCcl2 (100 ng/ml, R&D Systems) for indicated time. ECs were then lysed and proteins analyzed by immunoblotting.
Trans-endothelial migration assay
Trans-endothelial migration assay was performed as described previously.17 Alternatively, tumor cells were seeded in the upper chamber in conditioned media.
Immunoblotting
Tumor cell lysis and Western blot analysis were performed as described.15 Primary ECs were lysed with lysis buffer (20mM Tris, 150mM NaCl, 1mM EDTA, 1% Triton X-100, protease/phosphatase inhibitors) for 30 min on ice and supernatants were resolved by SDS-PAGE. Following antibodies were used: Myb (05-175, Millipore), p-Src (Tyr416) (6943S, Cell Signaling), Myc-Tag (9B11, Cell Signaling) and FLAG (F3165, Sigma). The membranes were probed with β-actin (A5060, Sigma) or α-tubulin (T9026, Sigma) antibodies for sample loading controls.
Chromatin immunoprecipitation (ChIP) analysis
Forty millions of 4T1 cells overexpressing Myc-tagged Myb (C11) and wt cells were cross-linked with 1% methanol-free formaldehyde for 10 min, followed by neutralization with glycine (final conc. 0.125M). Cells were lysed and chromatin was sheared to ∼400 bp fragments using the Bioruptor Plus sonicator (Diagenode). ChIP assays were performed using Imprint Chromatin immunoprecipitation kit (Sigma) with anti-Myc-Tag antibody (9B11, Cell Signaling) and isotype control (mouse IgG). DNA was analyzed by qPCR using three primer pairs flanking different Myb binding sites (MBSs; 118 bp, 853 bp, and 981 bp upstream transcription start site, TSS) within the Ccl2 promoter region. MBSs were identified using TFSEARCH and ConSite (http://consite.genereg.net). TSS of the Gapdh gene (Gapdh_TSS) was used as a negative control. TSS of a known c-Myb target gene Gata3 was used as positive control.44 Immunoprecipitated chromatin fragments from C11 and wt cells were used as templates for conventional PCR. The regions in the Ccl2 promoter harboring the MBSs were PCR amplified. Non-immunoprecipitated chromatin was used as an input. ChIP assay with human MDA-MB-231 cells (wt and MYB-overexpressing M5) was performed similarly with anti-Myb antibody (ab45150, Abcam) and rabbit IgG. Primers flanking two MBSs (677 bp and 723 bp upstream TSS) and negative control region without MBSs were used. All primer sequences are in listed the Table S1.
Immunohistochemistry
The study group included 68 BC patients undergoing surgical treatment at Lviv Regional Oncological Center between 2013-2015. Immunohistochemical (IHC) detection of c-Myb, CCL2, and ER was performed on formalin fixed paraffin embedded tissues of primary tumors using the following antibodies: c-Myb (EP769Y, Abcam), CCL2 (PA5-34505) and ER (SP1; MA5-14501; both from Thermo Fisher).
RNA-sequencing and functional enrichment analysis
RNA quality control, library preparation (TruSeq RNA Kit), sequencing on an Illumina HiSeq™ 2500, and data pre-processing were performed at the Functional Genomics Center Zurich (FGCZ). NGS data analysis system developed by the FGCZ bioinformatics, supporting selected open source, NGS data analysis packages, along with SUSHI data analysis was used.45 Reads were aligned using the STAR aligner 46 of at least 30bp matching; and acceptance at most 5 mismatches, and at most 5% of mismatches was used. Read alignments were only reported for reads with less than 50 valid alignments (Table S2). The mouse genome build and annotation from Ensembl (GRCm38) were used as the reference. Expression counts were computed using the Bioconductor package Genomic Ranges.47 Differential expression was computed using the Edger package.48 Only transcripts with a fold change >1.5 and an uncorrected p-value <0.01 between mock and MYBhigh cell lines, and between wt and lung3 cell lines were considered. We subtracted RNAs differentially expressed in mock-transfected and parental (wt) cells, and analyzed overlapped transcripts between subsets upregulated in lung3 and downregulated in MYBhigh, and vice versa. The set of enriched genes (35 RNAs) was analyzed using DAVID Bioinformatics Resources for functional classification.49 GO terms with FDR<0.05 were reported.
Correlation analysis
We used Oncomine (www.oncomine.org), Medisapiens (ist.medisapiens.com) and NCBI GEO databases to assess the differential expression of MYB, CCL2, CXCL1, CXCL2, CXCL6, CXCL16, ICAM1, IL1A, TNFRSF9, LCN2, IKBKE mRNA in human BCs. Pearson correlations were calculated with the GraphPad Prism software (version 6.07). Six independent datasets were used (accession numbers: EGAS00000000083, GSE25066, GSE22358, GSE22226, GSE12276, GSE20685), results in Table S3.
Survival analysis
Kaplan–Meier plots representing the probability of distant metastasis free survival (DMFS), relapse-free survival (RFS) and overall survival (OS) in BCs stratified according to the expression status of the signature genes (CCL2, CXCL1, CXCL2, CXCL6, CXCL16, ICAM1, IL1A, TNFRSF9, LCN2, IKBKE) and MYB (inversed) were calculated with KM plotter kmplot.com50. Patients were stratified according to median expression of indicated genes. The log-rank test was used to assess the significance of the correlation between gene(s) expression and shorter survival outcome.
Kaplan–Meier curves for site-specific distant metastasis-free survival based on MYB expression were calculated with the GraphPad Prism (version 6.07) using cohorts of GSE260351 and GSE1227652 of BC patients with known site of relapse.
The correlation between c-Myb-repressed inflammatory signature and lung relapse was analyzed using Sample Level Enrichment Analysis (SLEA) with datasets GSE2603 and GSE1227628. Gitools (www.gitools.org) were used to calculate the Z-score indicating the transcriptional status of the gene set in each sample. Analysis of the site specific-relapse free survival of patients was performed using the GraphPad Prism (version 6.07).
Statistics
Statistical analysis was performed with the GraphPad Prism software (version 6.07). All data are presented as mean ±SD and were analyzed with Mann-Whitney test unless stated otherwise.
Supplementary Material
Acknowledgements
This work was funded by SCOPES/SNF grant IZ73Z0-152361 (L. Borsig, L. Knopfova and N. Volodko). Further by Czech Science Foundation grant 17-08985Y (L. Knopfova), National Program of Sustainability II LQ1605-MEYS CR; and ICRC-ERA-HumanBridge/no. 316345 and by the MUNI/0877/2016 project of Grant Agency of Masaryk University. This work was also supported by the SNF grant #310030-152901 (L. Borsig). The authors acknowledge the assistance of the Center for Microscopy and Image Analysis and the Functional Genomic Center at University of Zurich.
Footnotes
Disclosures
The authors report no conflicts of interest.
References
- 1.Jones SE. Metastatic breast cancer: the treatment challenge. Clin Breast Cancer. 2008;8:224–233. doi: 10.3816/CBC.2008.n.025. [DOI] [PubMed] [Google Scholar]
- 2.Obenauf AC, Massague J. Surviving at a distance: organ specific metastasis. Trends Cancer. 2015;1:76–91. doi: 10.1016/j.trecan.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu X, Kang Y. Organotropism of breast cancer metastasis. J Mammary Gland Biol Neoplasia. 2007;12:153–162. doi: 10.1007/s10911-007-9047-3. [DOI] [PubMed] [Google Scholar]
- 4.Ramsay RG, Gonda TJ. MYB function in normal and cancer cells. Nat Rev Cancer. 2008;8:523–534. doi: 10.1038/nrc2439. [DOI] [PubMed] [Google Scholar]
- 5.Mucenski ML, McLain K, Kier AB, Swerdlow SH, Schreiner CM, Miller TA, et al. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell. 1991;65:677–689. doi: 10.1016/0092-8674(91)90099-k. [DOI] [PubMed] [Google Scholar]
- 6.Bengtsen M, Klepper K, Gundersen S, Cuervo I, Drablos F, Hovig E, et al. c-Myb Binding Sites in Haematopoietic Chromatin Landscapes. PLoS One. 2015;10:e0133280. doi: 10.1371/journal.pone.0133280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ness SA. Myb protein specificity: evidence of a context-specific transcription factor code. Blood Cells Mol Dis. 2003;31:192–200. doi: 10.1016/s1079-9796(03)00151-7. [DOI] [PubMed] [Google Scholar]
- 8.Drabsch Y, Robert RG, Gonda TJ. MYB suppresses differentiation and apoptosis of human breast cancer cells. Breast Cancer Res. 2010;12:R55. doi: 10.1186/bcr2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miao RY, Drabsch Y, Cross RS, Cheasley D, Carpinteri S, Pereira L, et al. MYB is essential for mammary tumorigenesis. Cancer Res. 2011;71:7029–7037. doi: 10.1158/0008-5472.CAN-11-1015. [DOI] [PubMed] [Google Scholar]
- 10.Thorner AR, Parker JS, Hoadley KA, Perou CM. Potential tumor suppressor role for the c-Myb oncogene in luminal breast cancer. PLoS One. 2010;5:e13073. doi: 10.1371/journal.pone.0013073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nicolau M, Levine AJ, Carlsson G. Topology based data analysis identifies a subgroup of breast cancers with a unique mutational profile and excellent survival. Proc Natl Acad Sci U S A. 2011;108:7265–7270. doi: 10.1073/pnas.1102826108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu LY, Chang LY, Kuo WH, Hwa HL, Chang KJ, Hsieh FJ. A supervised network analysis on gene expression profiles of breast tumors predicts a 41-gene prognostic signature of the transcription factor MYB across molecular subtypes. Comput Math Methods Med. 2014;2014:813067. doi: 10.1155/2014/813067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hugo HJ, Saunders C, Ramsay RG, Thompson EW. New Insights on COX-2 in Chronic Inflammation Driving Breast Cancer Growth and Metastasis. J Mammary Gland Biol Neoplasia. 2015;20:109–119. doi: 10.1007/s10911-015-9333-4. [DOI] [PubMed] [Google Scholar]
- 14.Tichy M, Knopfova L, Jarkovsky J, Pekarcikova L, Veverkova L, Vlcek P, et al. Overexpression of c-Myb is associated with suppression of distant metastases in colorectal carcinoma. Tumour Biol. 2016;37:10723–10729. doi: 10.1007/s13277-016-4956-7. [DOI] [PubMed] [Google Scholar]
- 15.Knopfova L, Benes P, Pekarcikova L, Hermanova M, Masarik M, Pernicova Z, et al. c-Myb regulates matrix metalloproteinases 1/9, and cathepsin D: implications for matrix-dependent breast cancer cell invasion and metastasis. Mol Cancer. 2012;11:15. doi: 10.1186/1476-4598-11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borsig L, Wolf MJ, Roblek M, Lorentzen A, Heikenwalder M. Inflammatory chemokines and metastasis-tracing the accessory. Oncogene. 2014;33:3217–3224. doi: 10.1038/onc.2013.272. [DOI] [PubMed] [Google Scholar]
- 17.Wolf MJ, Hoos A, Bauer J, Boettcher S, Knust M, Weber A, et al. Endothelial CCR2 signaling induced by colon carcinoma cells enables extravasation via the JAK2-Stat5 and p38MAPK pathway. Cancer Cell. 2012;22:91–105. doi: 10.1016/j.ccr.2012.05.023. [DOI] [PubMed] [Google Scholar]
- 18.Kitamura T, Qian BZ, Soong D, Cassetta L, Noy R, Sugano G, et al. CCL2-induced chemokine cascade promotes breast cancer metastasis by enhancing retention of metastasis-associated macrophages. J Exp Med. 2015;212:1043–1059. doi: 10.1084/jem.20141836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soria G, Ben-Baruch A. The inflammatory chemokines CCL2 and CCL5 in breast cancer. Cancer Lett. 2008;267:271–285. doi: 10.1016/j.canlet.2008.03.018. [DOI] [PubMed] [Google Scholar]
- 20.Hauselmann I, Roblek M, Protsyuk D, Huck V, Knopfova L, Grassle S, et al. Monocyte Induction of E-Selectin-Mediated Endothelial Activation Releases VE-Cadherin Junctions to Promote Tumor Cell Extravasation in the Metastasis Cascade. Cancer Res. 2016;76:5302–5312. doi: 10.1158/0008-5472.CAN-16-0784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bauer K, Mierke C, Behrens J. Expression profiling reveals genes associated with transendothelial migration of tumor cells: a functional role for alphavbeta3 integrin. Int J Cancer. 2007;121:1910–1918. doi: 10.1002/ijc.22879. [DOI] [PubMed] [Google Scholar]
- 22.Yoshimura T, Howard OM, Ito T, Kuwabara M, Matsukawa A, Chen K, et al. Monocyte Chemoattractant Protein-1/CCL2 Produced by Stromal Cells Promotes Lung Metastasis of 4T1 Murine Breast Cancer Cells. PLoS One. 2013;8:e58791. doi: 10.1371/journal.pone.0058791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mucha DR, Myers CL, Schaeffer RC., Jr Endothelial contraction and monolayer hyperpermeability are regulated by Src kinase. Am J Physiol Heart Circ Physiol. 2003;284:H994–H1002. doi: 10.1152/ajpheart.00862.2002. [DOI] [PubMed] [Google Scholar]
- 24.Roberts TK, Eugenin EA, Lopez L, Romero IA, Weksler BB, Couraud PO, et al. CCL2 disrupts the adherens junction: implications for neuroinflammation. Lab Invest. 2012;92:1213–1233. doi: 10.1038/labinvest.2012.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonda TJ, Leo P, Ramsay RG. Estrogen and MYB in breast cancer: potential for new therapies. Expert Opin Biol Ther. 2008;8:713–717. doi: 10.1517/14712598.8.6.713. [DOI] [PubMed] [Google Scholar]
- 26.Smid M, Wang Y, Zhang Y, Sieuwerts AM, Yu J, Klijn JG, et al. Subtypes of breast cancer show preferential site of relapse. Cancer Res. 2008;68:3108–3114. doi: 10.1158/0008-5472.CAN-07-5644. [DOI] [PubMed] [Google Scholar]
- 27.Harrell JC, Prat A, Parker JS, Fan C, He X, Carey L, et al. Genomic analysis identifies unique signatures predictive of brain, lung, and liver relapse. Breast Cancer Res Treat. 2012;132:523–535. doi: 10.1007/s10549-011-1619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gundem G, Lopez-Bigas N. Sample-level enrichment analysis unravels shared stress phenotypes among multiple cancer types. Genome Med. 2012;4:28. doi: 10.1186/gm327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hugo HJ, Pereira L, Suryadinata R, Drabsch Y, Gonda TJ, Gunasinghe NP, et al. Direct repression of MYB by ZEB1 suppresses proliferation and epithelial gene expression during epithelial-to-mesenchymal transition of breast cancer cells. Breast Cancer Res. 2013;15:R113. doi: 10.1186/bcr3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y, Jin K, van Pelt GW, van Dam H, Yu X, Mesker WE, et al. c-Myb Enhances Breast Cancer Invasion and Metastasis through the Wnt/beta-Catenin/Axin2 Pathway. Cancer Res. 2016;76:3364–3375. doi: 10.1158/0008-5472.CAN-15-2302. [DOI] [PubMed] [Google Scholar]
- 31.Pekarcikova L, Knopfova L, Benes P, Smarda J. c-Myb regulates NOX1/p38 to control survival of colorectal carcinoma cells. Cell Signal. 2016;28:924–936. doi: 10.1016/j.cellsig.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 32.Srivastava SK, Bhardwaj A, Arora S, Singh S, Azim S, Tyagi N, et al. MYB is a novel regulator of pancreatic tumour growth and metastasis. Br J Cancer. 2015;113:1694–1703. doi: 10.1038/bjc.2015.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hiratsuka S, Ishibashi S, Tomita T, Watanabe A, Akashi-Takamura S, Murakami M, et al. Primary tumours modulate innate immune signalling to create pre-metastatic vascular hyperpermeability foci. Nat Commun. 2013;4:1853. doi: 10.1038/ncomms2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gruvberger S, Ringner M, Chen Y, Panavally S, Saal LH, Borg A, et al. Estrogen receptor status in breast cancer is associated with remarkably distinct gene expression patterns. Cancer Res. 2001;61:5979–5984. [PubMed] [Google Scholar]
- 36.Guo P, Huang J, Wang L, Jia D, Yang J, Dillon DA, et al. ICAM-1 as a molecular target for triple negative breast cancer. Proc Natl Acad Sci U S A. 2014;111:14710–14715. doi: 10.1073/pnas.1408556111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bieche I, Chavey C, Andrieu C, Busson M, Vacher S, Le Corre L, et al. CXC chemokines located in the 4q21 region are up-regulated in breast cancer. Endocr Relat Cancer. 2007;14:1039–1052. doi: 10.1677/erc.1.01301. [DOI] [PubMed] [Google Scholar]
- 38.Zhao L, Glazov EA, Pattabiraman DR, Al-Owaidi F, Zhang P, Brown MA, et al. Integrated genome-wide chromatin occupancy and expression analyses identify key myeloid pro-differentiation transcription factors repressed by Myb. Nucleic Acids Res. 2011;39:4664–4679. doi: 10.1093/nar/gkr024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fang WB, Yao M, Brummer G, Acevedo D, Alhakamy N, Berkland C, et al. Targeted gene silencing of CCL2 inhibits triple negative breast cancer progression by blocking cancer stem cell renewal and M2 macrophage recruitment. Oncotarget. 2016 doi: 10.18632/oncotarget.9885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Svensson S, Abrahamsson A, Rodriguez GV, Olsson AK, Jensen L, Cao Y, et al. CCL2 and CCL5 Are Novel Therapeutic Targets for Estrogen-Dependent Breast Cancer. Clin Cancer Res. 2015;21:3794–3805. doi: 10.1158/1078-0432.CCR-15-0204. [DOI] [PubMed] [Google Scholar]
- 41.Sevenich L, Bowman RL, Mason SD, Quail DF, Rapaport F, Elie BT, et al. Analysis of tumour- and stroma-supplied proteolytic networks reveals a brain-metastasis-promoting role for cathepsin S. Nat Cell Biol. 2014;16:876–888. doi: 10.1038/ncb3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aird WC. Phenotypic heterogeneity of the endothelium: I. Structure, function, and mechanisms. Circ Res. 2007;100:158–173. doi: 10.1161/01.RES.0000255691.76142.4a. [DOI] [PubMed] [Google Scholar]
- 43.Johnstone CN, Smith YE, Cao Y, Burrows AD, Cross RS, Ling X, et al. Functional and molecular characterisation of EO771.LMB tumours, a new C57BL/6-mouse-derived model of spontaneously metastatic mammary cancer. Dis Model Mech. 2015;8:237–251. doi: 10.1242/dmm.017830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maurice D, Hooper J, Lang G, Weston K. c-Myb regulates lineage choice in developing thymocytes via its target gene Gata3. EMBO J. 2007;26:3629–3640. doi: 10.1038/sj.emboj.7601801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hatakeyama M, Opitz L, Russo G, Qi W, Schlapbach R, Rehrauer H. SUSHI: an exquisite recipe for fully documented, reproducible and reusable NGS data analysis. BMC Bioinformatics. 2016;17:228. doi: 10.1186/s12859-016-1104-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lawrence M, Huber W, Pages H, Aboyoun P, Carlson M, Gentleman R, et al. Software for computing and annotating genomic ranges. PLoS Comput Biol. 2013;9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 50.Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 51.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bos PD, Zhang XH, Nadal C, Shu W, Gomis RR, Nguyen DX, et al. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459:1005–1009. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.