Abstract
Cancer cell lines are critical models to study tumor progression and response to therapy. In 2008, we showed that ~50% of thyroid cancer cell lines were redundant or not of thyroid cancer origin. We therefore generated new authenticated thyroid cancer cell lines and patient derived xenograft (PDX) models using in vitro and feeder cell approaches, and characterized these models in vitro and in vivo. We developed four thyroid cancer cell lines, two derived from two different patients with papillary thyroid cancer (PTC) pleural effusions, CUTC5 and CUTC48; one derived from a patient with anaplastic thyroid cancer (ATC), CUTC60; and one derived from a follicular thyroid cancer patient (FTC), CUTC61. One PDX model (CUTC60-PDX) was also developed. Short tandem repeat genotyping showed that each cell line and PDX is unique and match the original patient tissue. The CUTC5 and CUTC60 cells harbor the BRAF (V600E) mutation, the CUTC48 cell line expresses the RET/PTC1 rearrangement, and the CUTC61 cells have the HRAS (Q61R) mutation. Moderate to high levels of PAX8 and variable levels of NKX2.1 were detected in each cell line and PDX. The CUTC5 and CUTC60 cell lines form tumors in orthotopic and flank xenograft mouse models.
Keywords: thyroid cancer, BRAF, RAS, RET/PTC1, cell line establishment
Introduction
Differentiated thyroid cancer is a major public health problem with 53,990 new patients predicted to have thyroid cancer diagnosed in United States in 2018 (1). Papillary thyroid cancer (PTC) is the most common thyroid tumor type, which comprises 90% of thyroid tumors, and follicular thyroid cancer (FTC) accounts for 5–10%. While many patients with PTC have a good prognosis, with a 95% survival at 5 years, a significant number of patients with radioiodine-refractory, progressive differentiated thyroid cancer have a poor prognosis, with a 10-year overall survival rate of approximately 10% (2). Widely invasive FTCs with distant metastasis have a poorer prognosis and occur in up to 46% of patients (3). Anaplastic thyroid cancer (ATC) is rare, but remains one of the most lethal human cancers, with a median survival of 6–12 months (4).
The mitogen activated protein kinase (MAPK) pathway is activated in approximately 70% of PTCs due to activating mutations in BRAF, RAS, or fusions in RET (5,6). BRAF (V600E) is the most commonly mutated oncogene in PTC and ATC with an average prevalence of 41 to 45% (5–8). Presence of the BRAF (V600E) mutation has been associated with more aggressive features in PTC, including recurrence, mortality, and resistance to radioactive iodine therapy. While BRAF and RAS are predominant drivers of aggressive thyroid cancer, additional genetic alterations including mutations in TP53, TERT, and PI3K pathway components are thought to be important for tumor progression and occur at increasing frequencies in PDTC and ATC (5,7,9,10). Mutations in RAS are commonly found in FTCs, with a prevalence of ~66% (7). Rearrangement of RET/PTC occurs commonly in PTC (~7–20% prevalence), with a lower prevalence in poorly differentiated thyroid cancer (PDTC; 13–17%) (6,7,11). RET/PTC rearrangements are more common in radiation-induced PTC than sporadic PTC. RET/PTC1 and RET/PTC3 are the most common, in which the RET tyrosine kinase domain is fused to the gene partner, CCDC6 or NCOA4, respectively. RET/PTC fusions activate the MAPK and PI3K pathways, resulting in increased proliferation and tumor progression.
Although there is much excitement regarding the development of targeted therapies, there is still much to be learned about how to effectively target these deregulated pathways in thyroid cancer. Human cancer-derived cell lines are critical models to study the biology of cancer and for preclinical testing of new therapeutic strategies. For thyroid cancer, the development of new therapies has been hampered by the lack of thyroid cancer cell lines in the widely used NCI-60 panel, which has been used to screen more than 100,000 drugs in other tumor types, as well as having a limited presence in more recent drug screening and functional genomics efforts (12,13). In addition, in 2008, we discovered that 17 out of 40 of widely used thyroid cancer cell lines were redundant or misidentified with cell lines from other tumor types (14). In response to this, we have generated and characterized a new set of authenticated thyroid cancer cell lines harboring the RET/PTC1 rearrangement (CUTC48), BRAF (V600E) mutation (CUTC5; CUTC60), or HRAS (Q61R) (CUTC61) in order to accurately study thyroid cancer pathogenesis and the efficacy of new therapies.
Materials and Methods
Patient tumors
All patient tissue samples were collected under an approved Institutional Review Board protocol, with written informed consent from the patients, at the University of Colorado Anschutz Medical Campus. CUTC5 cells were derived from a 73 year-old woman with a malignant pleural effusion (PTC). She was originally diagnosed with 4 cm left thyroid follicular carcinoma with focal Hurthle cell morphology, and a 2 mm follicular variant papillary thyroid carcinoma was also found on the right during surgery. Cytologic examination of pleural fluid showed cells positive for pan-cytokeratin, KRT8/KRT18, KRT7, and NKX2-1 and negative for KRT20, estrogen receptor, progesterone receptor, mammaglobin, GCFDP, MOC31, WT1, and calretinin.
CUTC48 cells were derived from a 68-year-old female with metastatic PTC to the lung (recurrent pleural effusions), bone, brain, and subcutaneous nodules. The patient had progressive disease requiring therapeutic thoracenteses (source of the cell line). The progressive cancer was unresponsive to radioiodine. Sorafenib was tried, but the patient did not tolerate this medication. Pleural effusion and blood were collected, and the patient was subsequently given 2 cycles of carboplatin and paclitaxel. Therapy was discontinued due to side effects.
The CUTC60 cell line was derived from a 59 year-old female with ATC. The patient was diagnosed with T2NXM0 stage II PTC in 2005, which was treated with total thyroidectomy and 100 mCi of I-131. After initial treatment, her thyroglobulin became undetectable and neck US showed stable 3 mm hypoechoic nodule in right zone 6. She presented in August 2015 with a rapidly growing painful anterior neck mass. Biopsy of this mass showed malignant cells consistent with ATC. The tumor was markedly positive for pan-cytokeratin, PAX8, and TP53 and negative for SOX10, thyroglobulin, and NKX2-1 on immunohistochemical stains. The patient underwent excision of the central neck mass and surgical pathology examination confirmed the diagnosis of thyroid carcinoma (90% anaplastic and 10% well-differentiated) with invasion into soft tissues, skeletal muscle, and sternum. Four out of 37 neck lymph nodes removed during the surgery were positive for metastatic ATC. The specimen for cell culture was obtained from the resected central neck mass. The patient received chemotherapy with paclitaxel and external beam radiation to the neck. Despite treatment, she developed pulmonary metastases complicated with recurrent pleural effusion and died from pneumonia and respiratory failure 5 months after the diagnosis of ATC.
The CUTC61 cell line was derived from the primary tumor of a 72 year-old female with metastatic FTC. The patient had a history of breast cancer treated with lumpectomy, adjuvant chemotherapy with docetaxel and cyclophosphamide, and adjuvant endocrine therapy with letrozole. Five years after the initial breast cancer treatment, she developed right shoulder pain and imaging with MRI and CT showed a 4.3 cm mass in the humerus, a 4.4 cm left rib mass, a 3.7 cm lung nodule, left calvarium lesions as well as multiple thyroid nodules. The biopsy of thyroid nodules and rib mass established the diagnosis of advanced thyroid cancer. Immunostaining was positive for KRT7, NKX2-1, and thyroglobulin, and negative for KRT20, GATA3, napsin A, chromogranin, and synaptophysin. In October 2015, the patient underwent total thyroidectomy with central neck dissection and a specimen for cell culture was obtained. Surgical pathology examination was consistent with FTC. The patient received additional treatment with external beam radiation therapy to bone metastases and two doses of I-131 (151 and 290 mCi), which resulted in a partial response to therapy.
Establishment of cell lines from tumor samples
Cell line generation method:
Pleural effusions (PEs) from two different patients were collected under sterile conditions. For the CUTC5 cell line, cells were lysed with red blood cell lysis buffer (Biolegend 420301), and washed and resuspended in RPMI 1640. Cells were under laid with Optiprep, and after centrifugation, the top (epithelial) layer of cells was collected and plated in RPMI/10% FBS supplemented with an antibiotic-antimycotic solution (Gibco by Life Technologies 15240096). Samples were diluted 1:1 with Hank’s Balanced Salt Solution (HBSS Gibco 24020117) or PBS (Life Technologies 14190–144), rinsed, and plated in RPMI supplemented with 10% FBS and an antibiotic-antimycotic solution (Gibco Life Technologies). For the CUTC48 cell line, pelleted cells were plated in Copland media (15) supplemented with an antibiotic-antimycotic solution (Gibco Life Technologies). When the cell lines reached confluency, attached and detached cells and colonies were passaged with 0.25% trypsin at a 1:3 ratio (Life Technologies-25200072). Cells were grown at 37° C in 5% CO2 and medium was changed 2 times per week.
The CUTC60 and CUTC61 cell lines were established from solid tumor tissue using the ROCK inhibitor-feeder cell approach (16). Tumor samples were minced into 2 mm3 pieces, and digested with 50 μg/ml liberase DL (Roche 05401160001) and 10 U/ml DNAse I (Roche 10104159001) in HBSS for 45 min at 37° C. After digestion, cells were plated on a layer of irradiated feeder cells in DMEM-HG (Gibco Life Technologies11965092) supplemented with heat inactivated bovine calf serum (Hyclone SH30073.03). Thyroid cancer cells proliferated and gradually replaced most of the feeder cells. Cells were passaged when 70% confluent using 0.25% trypsin (Gibco Life Technologies). The ability of thyroid cancer cells to grow independently of feeder cells and ROCK inhibitor was tested every 2–4 passages. All cell lines were routinely monitored for Mycoplasma contamination using the Lonza Mycoalert system (Lonza Walkersville, Inc. LT07–318), according to the manufacturer’s directions. Cell lines were considered permanent after > 6 months in culture, > 20 passages, and the ability to freeze down and recover. Cell lines have been deposited in the University of Colorado Cancer Center Tissue Culture Shared Resource.
Short tandem repeat profiling
Genomic DNA was extracted using the ZR genomic DNA kit (D3024, Zymo Research) from cultured cells, the patient blood sample (CUTC48), cells isolated directly from the PE (CUTC5), or tumor (CUTC60; CUTC61). Short tandem repeat (STR) profiling was conducted by either the University of Colorado Cancer Center DNA Sequencing & Analysis Core or the Barbara Davis Center BioResources Core Facility Molecular Biology Unit, at the University of Colorado, using the Identifiler kit (Thermo Fisher Scientific #4322288) or the Promega Powerplex® 16 HS kit (#DC2101). The STR electropherograms were analyzed and alleles called using GeneMapper™ version 4.0 software (Thermo Fisher #4440915) and the results were confirmed by visual inspection. Matches to the resulting genotypes were searched for by comparing them individually to a collection of 6,678 STR genotypes consisting of in-house data and data from the consolidation of STR genotypes from DSMZ, ATCC, RIKEN, and JCRB tissue culture repositories (available at the DSMZ website https://www.dsmz.de/services/services-human-and-animal-cell-lines/online-str-analysis.html; downloaded on July 10, 2017); and published STR data (17–25). Degree of matches were calculated by a modification of the Tanabe et al. formula (26):
This formula was applied to allelic data from shared STR loci. Single (monoallellic) peaks were counted only once, the results for the amelogenin locus amplicons from the X and Y chromosomes, were not included in the % Match calculation, as these alleles are not short tandem repeats. The results for the 13 shared CODIS loci (CSF1PO, D3S1358, D5S818, D7S820, D8S1179, D13S317, D16S539, D18S51, D21S11, FGA, THO1, TPOX, and vWA) were used to calculate the % Match. Most of the older external data (e.g., DSMZ consolidated, (19,20)) were produced with different STR genotyping kits with fewer STR loci and the % Match was calculated using only the results for shared loci.
RNA isolation and qRT-PCR
RNA was isolated using the PerfectPure RNA extraction kit (5 Prime FP2302340). mRNAs for human PAX8 and NKX2-1 were measured by real-time quantitative RT-PCR using ABI Prism 7700 Sequence detector, using the following primer/probe sequences, as previously described (14): PAX8 (F) 5’- GGACTACAAACGCCAGAACCC-3’, PAX8 (R) 5’- GGGCACAGTGTCATTGT CACA-3’, PAX8 (Probe) 6FAM- ACCATGTTTGCCTGGGA GATCCGA-TAMRA, NKX2-1 (F) 5’- ACGACT CCGTTCTCAGTGTCTG-3’, TTF-1 (R) 5’- GCCCTCCA TGCCCACTTT-3’, NKX2-1 (probe) 6FAM- CATCTTGAGTCCCCTGGAGGAAAGC TACA-TAMRA. Quantities of PAX8 and NKX2-1 were normalized to the corresponding 18s rRNA (PE ABI, P/N 4308310).
Detection of RET/PTC1
cDNA was used as a template to screen for RET/PTC rearrangements by quantitative PCR. First, the following primers were used to evaluate unbalanced expression of exons 12–13 versus exons 10–11 of RET, as previously described (14,27): Exons 10–11 (F) 5’-CACCTGCAACTGCTTCCCTGAGGA and (R) 5’-CGGCACAGCTCGTC-3’; and Exons 12–13 (F) 5’-GCAACGGCCTTCCATCTGAA-3’ and (R) 5’-ACTCGGGGAGGCGTTCTCTTT-3’. Samples with greater expression of exons 12–13 versus 10–11 of RET, which flank the rearrangement site in intron 11, were then screened for specific RET rearrangements using primers flanking the fusion points of RET/PTC1, RET/PTC2, and RET/PTC3, as previously described (27–29). Positive controls included cDNA from the TPC1 cells, which express the RET/PTC1 rearrangement, pCCL3 cells expressing RET/PTC2, and a PTC sample expressing RET/PTC3. mRNA was normalized to 18S rRNA, as described above. PCR products for RET/PTC1, 2, and 3 were analyzed on 2% agarose gels.
Mutational analysis
Mutational analysis was performed using the MSK-IMPACT targeted sequencing of 341 cancer genes (CUTC5, CUTC48), or a new panel covering 69 additional genes (410 genes total; CUTC60, CUTC61), as described (25,30).
Cell growth assays
Cells (2 × 104) were plated in triplicate, and counted after 24, 48, 72, and 96 hours using an automated ViCell counter (Beckman). Doubling time was calculated using an online calculator (http://www.doubling-time.com/compute.php).
Western blot analysis
Cells were harvested in CHAPS lysis buffer (10 mmol/L CHAPs, 50mmol/L Tris (pH 8.0), 150 mmol/L NaCl, and 2 mmol/L EDTA with 10 μmol/L Na3VO4 and 1x protease inhibitor cocktail (Roche)). Total protein concentrations were determined using the Bio-Rad DC kit (#5000111). Protein extracts were resolved on 8% or 10% PAGE-SDS gels and transferred to Immobilon-P membranes (Millipore IPFL00010), as previously described (31,32). Membranes were incubated at 4°C overnight with the following antibodies: RET (Abcam #134100), ppERK1/2, total ERK2 (Santa Cruz SC16982-R, Cell Signaling 9107), or α-tubulin (CalBiochem CP06) diluted in 5% nonfat dry milk or 5% BSA in 20 mmol/L Tris, pH 7.4, 138 mmol/L NaCl, 0.1% Tween (TBST). Blots were incubated with secondary goat anti-rabbit or goat anti-mouse horseradish peroxidase-conjugated antibodies (GE Healthcare NA934V) and proteins were detected by enhanced chemiluminescence (ECL) detection (Pierce 34080) (31,32).
Kinase inhibitor sensitivity
The sensitivity of CUTC cells lines to the indicated kinase inhibitors was assessed using CellTiterGlo 2.0 cell viability assay (Promega #G9243). All liquid handling operations were done using the JANUS Automated Workstation (Perkin Elmer) at the University of Colorado High Throughput Screening Core. Briefly, 400 cells in 25 µl of media were seeded into each well of a 384 well plate, and 24 hours later 1 μl of drug solution prepared in 12.5%/87.5% DMSO/RPMI 1640 cell culture media was added to each well. After 72 hour incubation with the drugs, 25 µl of CellTiterGlo 2.0 assay reagent was added to each well, and luminescence was read using the EnVision Multilabel Plate Reader (Perkin Elmer). Eight concentrations of drug, ranging from 0.64 to 40000 nM, were tested in quadruplicate. Drug responses were normalized to the negative control (DMSO). The final DMSO concentration was 0.5%, similar to the DMSO concentration (0.4%) used by the Cancer Cell Line Encyclopedia study (13). The DMSO concentration was the same for all wells in the assay. Each screening plate had 16 negative control assays (100% viability) and 16 positive control assays (0% viability, 1 μM staurosporine). To ensure quality, Z’ factor was calculated for each plate and Z’ ≥= 0.7 was considered acceptable. IC50 values (the concentration at which the drug response reached an absolute inhibition of 50%) were estimated using a 4-parameter log-logistic model in R.
Soft agar assays
Cells (104 – 4 × 104) were suspended in 0.3% agar with complete RPMI or Copland media, and plated on a base layer of 0.64% agar (Difco Agar Noble, BD 214220) in 6-well dishes, as previously described (33). Media was replenished every 3–4 days for 16 days. Colonies were stained with nitroblue tetrazolium chloride (VWR 0329), and incubated overnight at 37° C to develop the stain. Colonies were counted using Image J software.
Orthotopic mouse model
All animal studies were conducted in accordance with the animal protocol procedures approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Colorado Denver (Aurora, CO). Athymic nude mice (Hsd:Athymic Nude-Foxn1nu (069) from Envigo or in-house breeding for the CUTC60 and CUTC61 cells), SCID/Beige mice (C.B-17/IcrHsd-PrkdcscidLystbg-J from Envigo), or NRG mice (NOD.Cg-Rag1tm1MomIl2rgtm1Wjl/SzJ (#007799) from The Jackson Laboratory) were anesthetized with tribromoethanol (250 mg/kg), and the indicated thyroid cancer cells (5 × 105 in a 5 μL cell suspension) were injected into the right thyroid gland with the aid of a dissecting microscope (Nikon SMZ645), and the skin closed with staples, as previously described (33–35). Growth was monitored for the indicated days. Final thyroid tumor size for excised tumors was measured with calipers and volume was calculated using the following formula: tumor volume = (length x width x height)*0.5236.
Subcutaneous flank model
CUTC48 cells (107) were injected into the flanks of 5 athymic nu/nu mice (Hsd:Athymic Nude-Foxn1nu (069) from Envigo), 5 SCID/Beige mice (C.B-17/IcrHsd-PrkdcscidLystbg-J from Envigo), or 3 NOD/RAGKO/IL-2RgammaKO mice (NOD.Cg-Rag1tm1MomIl2rgtm1Wjl/SzJ (#007799) from Jackson Laboratory) using high concentration Matrigel (Corning #354248) 1:1 with RPMI 1640 (#11875119; Gibco Life Technologies), as previously described (36). Growth was monitored over 97 days (nude and SCID mice) or 6 months (NOD/RAGKO/IL-2RgammaKO). CUTC5 cells (5 × 106) were injected into the flanks of 3 NSG mice (NOD.Cg-Rag1tm1MomIl2rgtm1Wjl/SzJ (#007799) from Jackson Laboratory) using high concentration Matrigel 1:1 with RPMI 1640. Tumor growth was monitored, and caliper measurements taken over 42 days. CUTC60 cells (106) were injected into the flanks of 5 athymic nude mice (Hsd:Athymic Nude-Foxn1nu (069) from Envigo) using high concentration Matrigel 1:1 with RPMI 1640. Tumor growth was monitored, and caliper measurements taken over 42 days. Final tumor volumes were calculated as described above using measurements taken from excised tumors.
Establishment of patient-derived xenograft (PDX) models
Tumor tissue obtained from patient 60 was collected and placed in HBSS (Gibco) on ice for transport. Samples were then cut into pieces approximately 3mm3 with surgical scissors under sterile conditions. Tumor pieces were then transferred to high concentration Matrigel (Corning #354248) and placed on ice until injection. Female NSG mice (NOD.Cg-Rag1tm1MomIl2rgtm1Wjl/SzJ (#007799) from Jackson Laboratory) were anesthetized with 5% inhaled isoflurane in 95% oxygen. Tumors were subcutaneously engrafted into both hind flanks using a 12g trocar needle, and passaged into nude mice at mouse passage 3 (mF3). Tumor growth in vivo was monitored weekly by caliper measurements, with volume calculated using the equation: .
Results
Short Tandem Repeat genotyping
Short tandem repeat (STR) genotyping was performed on the CUTC5, CUTC48, CUTC60, and CUTC61 cell lines and their respective patient tissue samples and the CUTC60 patient derived xenograft (PDX) model. Specifically, for CUTC5, the patient’s PE tumor cells were profiled, for CUTC48, the patient’s blood was used for profiling; and for the CUTC60 and CUTC61, the patient tumors were used for profiling (Table 1). All profiles consisted of sharp peaks with negligible background and were not contaminated with DNA from other human samples (Figure S1). As shown in Table 1, the CUTC5 cell line STR profile matched that of the patient at all 13 STR loci. The STR profiles of the CUTC48 cells and patient sample were identical at the loci tested in common, except the CUTC48 cells exhibited loss of heterozygosity (LOH) at D16S539 (Table 1). The CUTC60 cell line matched their respective patient tissue sample, except with LOH at D13S317, D18S51, D21S11, and FGA (Table 1). The CUTC60 PDX exhibited LOH at FGA, TPOX, and vWA. The CUTC61 cell line matched its respective patient tissue sample at all STR loci tested (Table 1). As expected, since all of the cell lines were derived from female patients, they carry only the X-linked chromosomal Amelogenin locus (Table 1).
Table 1.
STR profiling of CUTC cell lines, PDX model, and corresponding patient tissue
| CUTC Sample Information | STR Locus | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample Name |
Sample Type |
Passage or PDX Generation Number |
Donor Gender |
Amelogenin | CSF1PO | D3S1358 | D5S818 | D7S820 | D8S1179 | D13S317 | D16S539 | D18S51 | D21S11 | FGA | THO1 | TPOX | vWA | % Identity Match 1 |
| Patient 5 | Tumor | NA | Female | X | 11 12 | 14 18 | 11 12 | 10 | 12 13 | 12 14 | 9 12 | 12 13 | 27 31.2 | 22.2 26 | 6 8 | 9 11 | 17 | Reference |
| CUTC5 | Cell Line | 16 | Female | X | 11 12 | 14 18 | 11 12 | 10 | 12 13 | 12 14 | 9 12 | 12 13 | 27 31.2 | 22.2 26 | 6 8 | 9 11 | 17 | 100% |
| Patient 48 | Patient Blood | NA | Female | X | 9 12 | 14 15 | 11 12 | 7 12 | 10 13 | 11 12 | 10 11 | 15 19 | 30 32.2 | 22 23 | 6 9.3 | 8 11 | 15 | Reference |
| CUTC48 | Cell Line | 20 | Female | X | 9 12 | 14 15 | 11 12 | 7 12 | 10 13 | 11 12 | 11 | 15 19 | 30 32.2 | 22 23 | 6 9.3 | 8 11 | 15 | 98% |
| Patient 60 | Tumor | NA | Female | X | 10 12 | 17 18 | 10 12 | 8 | 14 16 | 9 10 | 9 10 | 15 18 | 31 32.2 | 20 25 | 6 8 | 8 11 | 16 18 | Reference |
| CUTC60 | PDX | 0 | Female | X | 10 12 | 17 18 | 10 12 | 8 | 14 16 | 9 10 | 9 10 | 15 18 | 31 32.2 | 25 | 6 8 | 11 | 18 | 94% |
| CUTC60 | Cell Line | 44 | Female | X | 10 12 | 17 18 | 10 12 | 8 | 14 16 | 9 | 9 10 | 15 | 32.2 | 20 | 6 8 | 8 11 | 16 18 | 91% |
| Patient 61 | Tumor | NA | Female | X | 10 11 | 14 15 | 10 12 | 8 10 | 12 15 | 11 12 | 11 13 | 14 19 | 28 30 | 24 25 | 8 9 | 8 | 17 18 | Reference |
| CUTC61 | Cell Line | 35 | Female | X | 10 11 | 14 15 | 10 12 | 8 10 | 12 15 | 11 12 | 11 13 | 14 19 | 28 30 | 24 25 | 8 9 | 8 | 17 18 | 100% |
- % Identity Match calculated relative to donor sample using Tanabe et al. (1999) algorithm for 13 autosomal CODIS loci and not including the amelogenin results.
Characterization of thyroid-specific gene expression
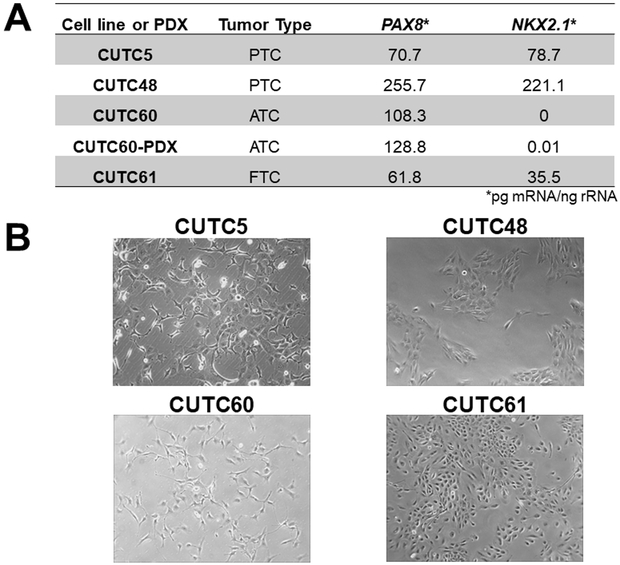
We next evaluated expression of the thyroid specific genes, PAX8 and NKX2-1, in the CUTC5, CUTC48, CUTC60, and CUTC61 cell lines, along with CUTC60-PDX tumor using qRT-PCR. Figure 1A shows that all cell lines and PDX tumor express moderate levels of PAX8 (61.8 to 255.7 pg mRNA/ng rRNA). Levels of NKX2-1 were more variable and ranged from 0 pg mRNA/ng rRNA in the CUTC60 cells to 221.1 pg mRNA/ng rRNA in the CUTC48 cells. Overall, PAX8 and NKX2-1 levels in cell lines are lower than expression levels in normal thyroid or tumor tissue, which exhibit PAX8 and NKX2-1 levels > 1000 pg mRNA/ng rRNA (14), but are detectable and similar to expression levels of many of the established thyroid cancer cell lines previously evaluated by our group (14), therefore supporting the thyroid origin of these cell lines.
Figure 1. Thyroid specific gene expression, morphologic and growth characteristics of the CUTC cell lines.
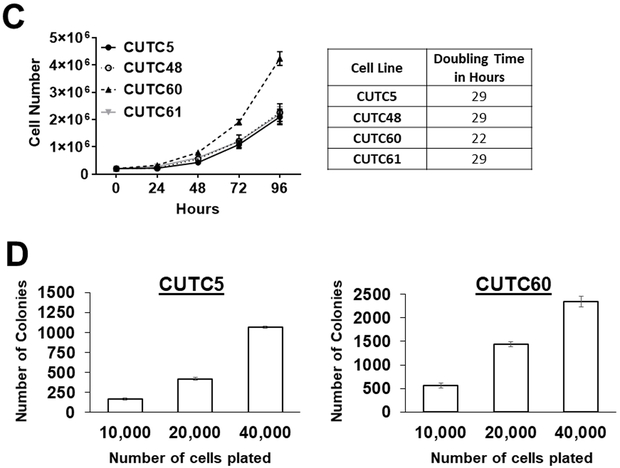
A) mRNA expression of PAX8 and NKX2-1 was measured by qRT-PCR. mRNA expression (pg) of PAX8 or NKX2-1 normalized to 18s rRNA (ng) is shown. B) Bright field images of the CUTC cell lines at 10X magnification. CUTC5 p17, CUTC48 p14, CUTC60 p28, and CUTC61 p20 are shown. C) The growth of the CUTC cell lines was determined by daily ViCell counting every 24 hours for 96 hours. Data shown are the mean +/− SEM of 3 independent experiments performed in triplicate. Doubling time was determined based on the growth data in 1C. D) Growth of the CUTC5 and CUTC60 cell lines in soft agar. The number of cells plated is indicated. Colony formation was quantified after 16 days using Image J. Results shown are the mean +/− SD of one experiment performed in 3 to 4 technical triplicates. The CUTC48 and CUTC61 cells did not form colonies in soft agar.
Morphologic and phenotypic characterization of the CUTC cell lines
We next assessed the morphology of the cell lines. The CUTC5, CUTC48, and CUTC61 cells have a polygonal-like shape, whereas the CUTC60 cells are more spindle-like (Fig. 1B). All cell lines grow as adherent cultures. The growth rates of the CUTC cell lines were evaluated using automated cell counting over a period of 96 hours, and showed the CUTC5, CUTC48, and CUTC61 cells have similar doubling times of 29 hours, while the CUTC60 cells have a doubling time of 22 hours (Fig. 1C). We further assessed the ability of the CUTC5, CUTC48, CUTC60, and CUTC61 cell lines to form colonies in soft agar assays. Figure 1D shows that the BRAF-mutant CUTC5 and CUTC60 cells readily form colonies in soft agar, whereas the RET/PTC1 expressing CUTC48 and CUTC61 cells did not form colonies in soft agar after 16 days of growth, and plating up to 40,000 cells (data not shown).
Mutational profiling
Mutations were identified previously using MSK-IMPACT targeted sequencing, and are summarized in Table 2 (25). The CUTC5 cells harbor mutations in BRAF c.T1799A (p.V600E) (allelic frequency 0.44) and TP53 c.C405G (p.C135W) (allelic frequency 0.99). For the CUTC48 cell line, no point mutations were detected in the coding regions of BRAF, RAS, or PIK3CA, but a TERT (telomerase reverse transcriptase) promoter mutation (c.−124C>T) was identified (allelic frequency 0.5). The CUTC60 cells and CUTC60-PDX harbor the BRAF T1799A (p.V600E) mutation with similar allelic frequencies (CUTC60 allelic frequency 0.67; CUTC60-PDX 0.59), a TP53 c.A578G (p.H193R) mutation (allelic frequency 1; CUTC60-PDX 0.98), and TERT promoter mutation (c.−124C>T; allelic frequency 0.49; CUTC60-PDX 0.58). The CUTC61 cells have an HRAS c.A182G (p.Q61R) mutation (allelic frequency 0.63), a splicing TP53 mutation (c.375+1G>A), and TERT promoter mutation (c.−124C>T; allelic frequency 0.43).
Table 2.
Mutational Analysis of the CUTC cell lines and PDX model.
| BRAF | HRAS | TERT | TP53 | |
|---|---|---|---|---|
| CUTC5 | c.T1799TA [p.V600E] |
WT | WT | c.C405G [p.C135W] |
| CUTC48 | WT | WT | c.−124C>T | WT |
| CUTC60 | c.T1799A [p.V600E] |
WT | c.−124C>T | c.A578G [p.H193R] |
| CUTC60-PDX | c.T1799TA [p.V600E] |
WT | c.−124C>T | c.A578G [p.H193R] |
| CUTC61 | WT | c.A182G [p.Q61R] |
c.−124C>T | Splicing c.375+1G>A |
Identification of the RET/PTC1 rearrangement in the CUTC48 cells
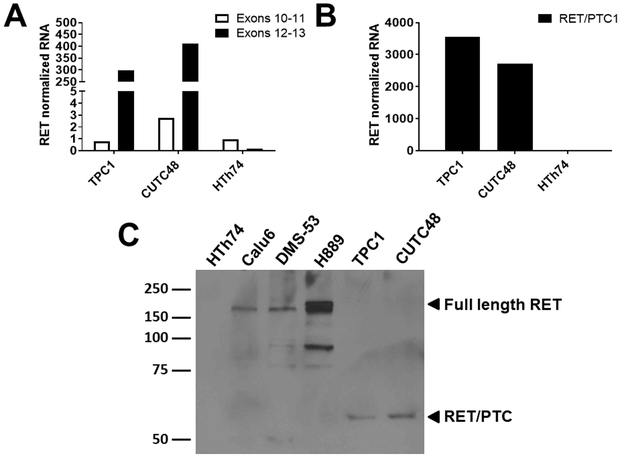
Given the lack of driver mutations identified in the CUTC48 cells, we screened the CUTC48 cell line for RET/PTC rearrangements using qRT-PCR and cDNA as a template. We first evaluated unbalanced expression of exons 10–11 relative to 12–13 of RET, which flanks the rearrangement site in intron 11. Figure 2A shows expression of exons 12–13 is higher than expression of exons 10–11 in the CUTC48 cells (150-fold), and is similar to the RET/PTC1-expressing TPC1 cells, which were used as a positive control. As expected, neither set of exons is expressed in the HTh74 cells, which was used as a negative control. To identify the specific RET/PTC rearrangement present, the CUTC48 cells were screened using primers bracketing the fusion point of RET/PTC1, as previously described (27–29). Using this approach, expression of RET/PTC1 was detected in the CUTC48 cells at similar levels of RET/PTC1 expression in the TPC1 cells (Fig 2B). These results are consistent with our Affymetrix gene expression data, showing expression of RET in the CUTC48 cells, as well as next generation sequencing (25). We next performed Western blot analysis to evaluate RET/PTC1 protein expression in the CUTC48 cells, using the TPC1 cells as a positive control for RET/PTC1 expression, the HTh74 cells as a negative control, and the lung cancer cell lines Calu6, DMS-53, and H889, to monitor full length RET expression. Figure 2C shows that similar to the TPC1 cells, the CUTC48 cells expression a 57 kDa protein, consistent with the expected size of the RET/PTC1 rearranged protein, compared to full length RET expression of 140–170 kDa.
Figure 2. Expression of RET/PTC1 in the CUTC48 cells.
A) qRT-PCR was used to screen for the presence of RET/PTC rearrangements. Expression of exons 12-13 versus exons 10-11 of RET are shown. Greater expression of exons 12-13 versus 10-11 indicates expression of RET/PTC1. The TPC1 cells were used as a positive control and the HTh74 cells were used as a negative control. Values shown are pg RET mRNA expression normalized to ng rRNA. B) RET/PTC1 expression was evaluated by qRT-PCR using primers flanking the fusion site. mRNA levels were normalized as in 2A. C) RET/PTC1 protein expression was evaluated by Western blotting using the TPC1 cells as a positive control, the HTh74 cells as a negative control, and the lung cancer cell lines, Calu6, DMS053, and H889 to compare full-length RET expression.
The sensitivity of CUTC cell lines to kinase inhibitors
The responses of CUTC cell lines to kinase inhibitors targeting their respective oncogenic drivers are summarized in Supplementary Table 1. Compared to other CUTC cell lines, the CUTC48 cells were sensitive to cediranib (IC50 = 667 nM), foretinib (IC50 = 111 nM), and lenvatinib (IC50 = 1525 nM), at concentrations known to inhibit RET (37,38), which likely explains their sensitivity. Consistent with the presence of BRAF V600E mutation, CUTC5 cell line was sensitive to BRAF inhibitors (IC50, dabrafenib, 8 nM; vemurafenib, 859 nM) and MEK inhibitors (IC50; selumetinib, 195 nM; PD-0325901, 18 nM; trametinib, 148 nM), at concentrations selective for their respective targets. Interestingly, the CUTC60 cell line was resistant to BRAF (dabrafenib and vemurafenib IC50s > 10 μM) and MEK1/2 (selumetinib, PD-0325901, and trametinib IC50s > 9 μM) inhibitors despite the presence of BRAF V600E mutation (Table S1). We have recently reported that the TCGA-derived BRAFV600E-RAS score (BRS) is a better predictor of MAPK pathway dependence than BRAF mutational status (6,25). Consistent with this, the CUTC5 cells have a lower BRS than the CUTC60 cells (CUTC5 BRS = −0.341 vs CUTC60 BRS = 0.089), indicating the CUTC60 cells are less reliant on the MAPK pathway and thus resistant to MEK1/2 or BRAF inhibition. Interestingly, the CUTC60 and CUTC61 cell lines did not show outlier drug sensitivities. Finally, we evaluated the sensitivity of the CUTC cells to the multi-kinase inhibitor, sorafenib, which is FDA-approved for differentiated thyroid cancer. The BRAF-mutant CUTC5 and CUTC60 cells exhibited IC50 values of < 2 μM to sorafenib, while the CUTC48 and CUTC61 cells were more resistant with IC50 values > 8 μM (Table S1), all of which are lower than achievable plasma levels in patients (39).
Growth of CUTC cell lines in preclinical mouse models
CUTC5 tumor establishment
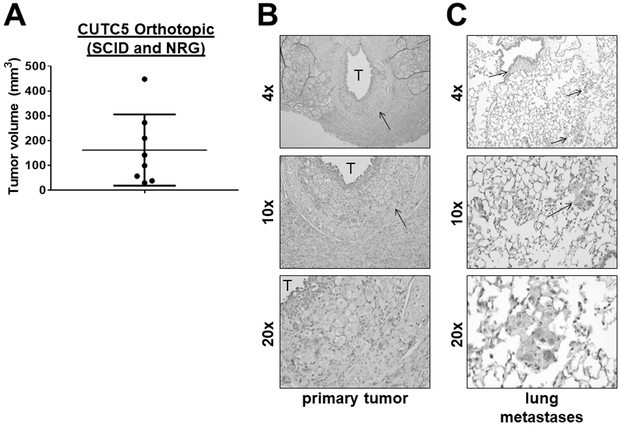
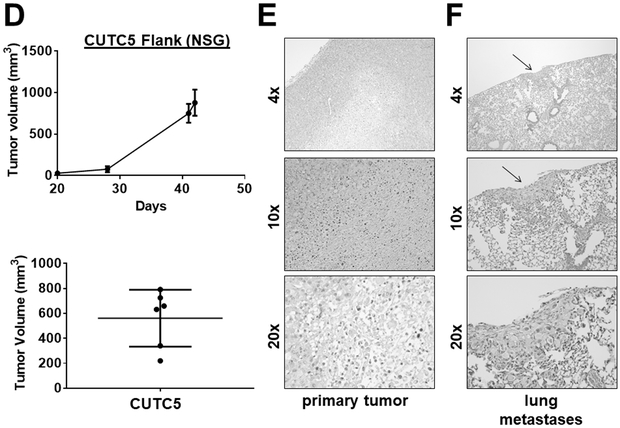
The ability of the CUTC cell lines to form tumors in vivo was evaluated using the orthotopic and flank models. Using the orthotopic model, to better recapitulate the microenviroment, where where 5 × 105 cells are injected into the right thyroid glands of mice, the CUTC5 cells formed small orthotopic tumors in nude mice after 48 days (79 mm3; not shown). In SCID and NRG mice, the CUTC5 orthotopic tumors reached an average tumor volume of 162 mm3 after 43 days (Fig. 3A; Table S2). H&E staining of a representative primary orthotopic tumor showing invasion into the muscle is shown in Figure 3B. Pulmonary metastases were also observed, with multiple foci in 6/8 mice and small foci in 2/8 mice (Fig. 3C; representative image). We also evaluated the ability of the CUTC5 cell lines to grow subcutaneously as flank tumors, as a robust and easily accessible model to study tumor growth and response to treatment. The CUTC5 cells established tumors in NSG mice after 20 days (6/6 mice; average ~25 mm3), reaching an average final tumor volume of 562 mm3 at 42 days (Fig. 3D & 3E; Table S2). Pulmonary metastases were observed in 3/3 of the CUTC5 mice evaluated, and representative images of the primary tumor and lung metastases are shown in Figure 3.
Figure 3. CUTC5 in vivo tumor growth.
A) CUTC5 cells (500,000) were injected into the right thyroid glands of SCID and NRG mice, and tumor volume was evaluated after 43 days. Results shown are the mean +/− SD of final tumor volumes from tumors grown in SCID and NRG mice. B) A representative image of an H&E-stained an orthotopic primary tumor is shown. The trachea is indicated by (T) and the arrow indicates invasion. C) Histologic analysis by H&E staining of lung metastases (indicated by arrows) is shown. Images are shown at 4, 10, or 20X, as indicated. D) CUTC5 cells were injected into the flanks of NSG mice and tumor growth was measured over 42 days, and final tumor volumes were calculated. Representative images of the primary tumor (E) and (F) lung metastases are shown. Metastases were observed in 3/3 mice.
CUTC60 tumor establishment
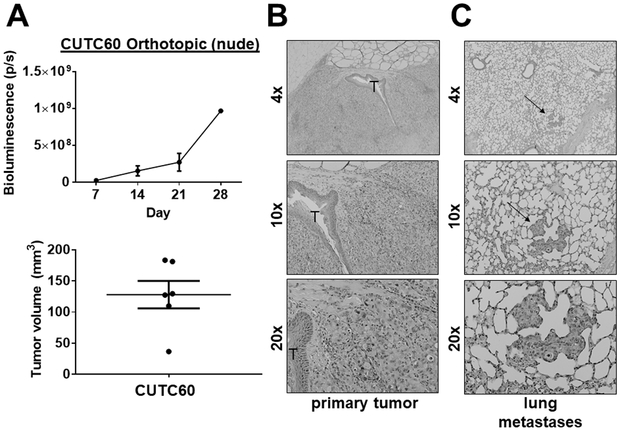
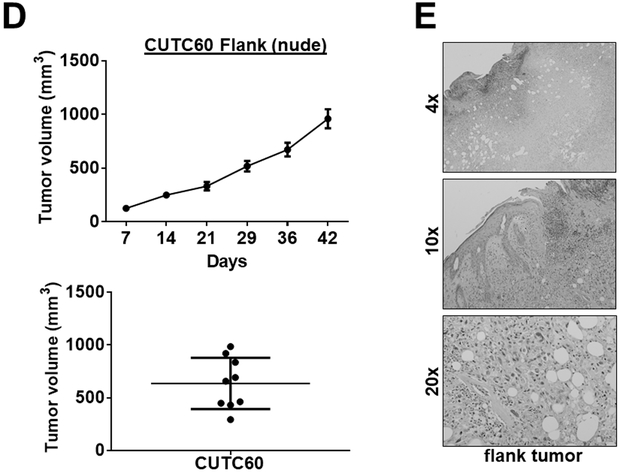
The CUTC60 cell line had an orthotopic take rate of 100% (6/6) in nude mice with an average tumor size of 128 mm3 after 28 days (Fig. 4A; Table S2). The CUTC60 cells formed invasive tumors, along with lung metastases with small foci in 3/6 mice. Representative images of the orthotopic primary tumors and lungs are shown in Fig. 4B and 4C. In the flank model, the CUTC60 cells established tumors in nude mice by day 7 (9/10 mice; average 125 mm3; Fig. 4D; Table S2), reaching an average final tumor volume of 636 mm3 at day 42 (Fig. 4D). Representative images of the flank tumors are show in Figure 4E. Lung metastases were not observed in the CUTC60 flank model.
Figure 4. CUTC60 tumor growth in vivo.
CUTC60 cells (500,000) were orthotopically injected into the right thyroid glands of nude mice. A) Tumor growth was monitored by weekly bioluminescence imaging over 28 days, and final tumor volumes were calculated. Histologic analysis was performed and representative H&E staining of the primary tumor (B) and lung metastases (indicated by the arrow) (C) are shown. The trachea (T) is also indicated. D) CUTC60 cells were injected into the flanks of nude mice and tumor growth was monitored by weekly caliper measurements for 42 days. Final tumor volumes are shown. E) A representative H&E stained image of the flank tumor is shown. Lung metastases were not observed in the CUTC60 flank model.
CUTC48 and CUTC61 tumor establishment
The CUTC48 or CUTC61 cells did not establish tumors in the orthotopic model using nude mice. The CUTC48 cells did not form flank tumors in SCID (0/10), or NOD/RAGKO/IL-2RgammaKO mice (0/6) even with 107 cells injected in the flank (Table S2). The CUTC61 cells were not tested in the flank model.
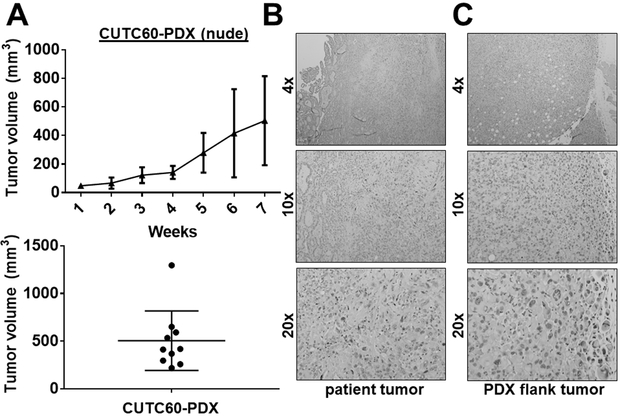
Patient-derived xenograft (PDX) models
The CUTC60-PDX model was generated directly from tumor tissue from the patient tumor, which was directly implanted into NSG mice, and subsequently passaged into nude mice. The growth of the CUTC60 PDX model (generation mF4) is shown in Figure 5A where tumor chunks (3 mm3) were injected subcutaneously into each flank of athymic nude mice. CUTC60-PDX tumors were invasive and reached an average tumor volume of 504 mm3 after 7 weeks (Fig. 5). Histology of the patient tumor and PDX flank tumor are shown in Fig. 5B and 5C. PDX models have been shown to accurately recapitulate the original patient’s tumor and therapeutic responses, thus the CUTC60-PDX model provides a new, clinically relevant thyroid cancer model to study BRAF-driven tumorigenesis and response to therapy, along with a matched cell line to study mechanisms in vitro (40).
Figure 5. Generation of the CUTC60 patient-derived xenograft (PDX) model.
Tumor tissue (3 mm3) from patient 60 was directly implanted into the flanks of NSG mice by trocar injection. Tumors were passaged into the flanks of nude mice at generation mF3. A) The growth of the CUTC60-PDX was measured weekly for 7 weeks. Final tumor volumes are the mean +/− SD of 10 tumors. Representative images of the patient tumor (B) and PDX flank tumor (C) are shown.
Discussion
Cancer cell lines remain a major tool to study cancer signaling mechanisms and response to therapy. In thyroid cancer, we previously reported the comprehensive characterization of a panel of 40 cell lines, and identified 23 unique thyroid cancer cell lines (14). While these cell lines represent valuable models to study thyroid cancer biology, the majority of these models are decades old, and have not been genetically linked to their corresponding patient tissue samples. Thus, there is a need to develop and characterize novel cell lines that closely recapitulate thyroid cancer pathogenesis. In support of this, new thyroid cancer cell lines have recently been established, including cell lines derived from ATC, PTC, and FTC (15,24,41,42). Here, we report the generation of two new PTC-derived cell lines isolated from pleural effusions (CUTC5, CUTC48), one cell line derived from the primary tumor of an ATC patient (CUTC60), and a fourth cell line derived from the primary tumor of a patient with metastatic FTC (CUTC61). The CUTC5 and CUTC60 cell lines and CUTC60-PDX harbor the BRAF V600E mutation, thus providing new authenticated models representing a common oncogenic driver mutation in PTC and ATC. The CUTC61 cell line harbors a HRAS Q61R mutation, to our knowledge, providing the first model of FTC to express this mutation. Finally, the CUTC48 cells express the RET/PTC1 rearrangement, representing another common genetic alteration in PTC, and importantly, providing the second RET/PTC thyroid cancer cell line model in existence.
Importantly, we have authenticated this panel of cell lines and PDX model using STR genotyping, and have genetically linked the cell lines and PDX models to their corresponding patient tissue samples. Of the 60 unique thyroid cancer cell lines recently characterized by our groups, approximately 20 of these cell lines (including the four cell lines reported here) have been linked to their corresponding patient tissue samples (15,24,25,41), thus providing a critical panel of thyroid cancer cell lines to accurately study thyroid cancer biology. In addition, these STR profiles provide an important resource for maintaining cell line integrity (19,43,44). For the CUTC cell lines, we have further shown that these cell lines cell lines express the thyroid specific transcription factors, PAX8 and/or NKX2-1, supporting the thyroid origin of these cell lines. We also evaluated mRNA expression of NIS; however, as with most preclinical thyroid cancer cell lines, we were unable to detect expression (data not shown). Overall, these results are consistent with the dedifferentiated nature of thyroid cancer cell lines in vitro, regardless of their derivation (25).
We further performed a mutational analysis of the CUTC cell lines using by MSK-IMPACT targeted sequencing (25). This analysis identified BRAF V600E and TP53 mutations in the CUTC5 cells. The CUTC60 cells harbor BRAF V600E, TP53, and a TERT promoter mutation, and the CUTC61 cells have HRAS Q61R, a TP53 splicing mutation, and a TERT promoter mutation. Of note, the TP53 alterations in the CUTC60 and CUTC61 cell lines were enriched compared to their corresponding tumor samples, as previously described (25). Recent studies have reported mutations in BRAF and TERT correlate with more aggressive disease (10), which is consistent with the aggressive nature of the tumors from which these cells were isolated. As expected, the BRAF-mutant CUTC5 cell line was highly sensitive to BRAF and MEK inhibitors, and the CUTC48 cell line, expressing RET/PTC1, was highly sensitive to drugs targeting RET. Interestingly, the BRAF-mutant CUTC60 cell line was resistant to BRAF and MEK inhibitors. This lack of sensitivity is consistent with a higher BRAFV600E-RAS score (BRS), indicating that despite the presence of the BRAF V600E mutation, the CUTC60 cells are not dependent on the MAP kinase pathway (25). The HRAS-mutant CUTC61 cell line was also resistant to MEK inhibitors, suggesting other pathways are important for the growth and survival of these cells. Together, these models will be valuable to further study why thyroid cancers are more resistant to MAPK-directed therapies than other tumor types with similar activating mutations (e.g. melanoma). Finally, all cell lines were responsive to the multi-kinase inhibitor, sorafenib, which is FDA-approved for differentiated thyroid cancer, at concentrations achievable in patients (39).
Notably, our mutational analysis of the CUTC48 cell line did not identify oncogenic mutations in BRAF, RAS, PIK3CA, or TP53. We noted expression of RET using Affymetrix gene expression profiling (25), which prompted us to evaluate expression of RET/PTC using RT-PCR approaches with primers flanking the rearrangement site of RET/PTC1. Accordingly, we found greater expression of exons 12–13 versus 10–11 of RET, indicating expression of the RET/PTC1 rearrangement in the CUTC48 cells. Thus, to our knowledge, the CUTC48 cells represent the second thyroid cancer cell line in the world to study RET/PTC1 biology, and the first RET/PTC1 thyroid cancer cell line that has been genetically linked to the original tumor. In addition, the CUTC48 cells harbor a TERT promoter mutation.
The ability to establish tumors in mouse models is an important characteristic of cancer cell lines in order to investigate the effects of the microenvironment and response to therapy in an in vivo model. We found that the CUTC5 cells form orthotopic tumors in SCID and NRG mice (8/8 take rate, 150 mm3 average tumor volume), with the development of lung metastases. The CUTC5 cells also form tumors in a subcutaneous tumor model in NRG mice, which also has lung metastases. The CUTC60 cells readily formed orthotopic and flank tumors in nude mice, demonstrating lung metastases in the orthotopic model. Interestingly, the CUTC48 cells did not form tumors in any mouse model that we tested, including the NRG mice, which is consistent with the take rate we and others have observed with the RET/PTC1-expressing TPC1 cells (34,45,46). Our previous studies have indicated that thyroid cancer cell lines with the highest take rate harbor mutations in BRAF V600E (34), thus indicating that the RET/PTC1-expressing cells may exhibit an overall less aggressive phenotype.
PDX models are thought to better resemble human tumors and have quickly become a valuable translational model to study cancer biology, response to therapy, and the development of biomarkers (40,47). Recently, 10 new thyroid cancer PDX models were reported by Marlow et al (24), and one other ATC PDX model has been published, which was used for drug screening (48). The models developed by Marlow et al (24) are of various thyroid subtypes expressing mutations in BRAF or RAS, and importantly, include three PDX models with matched cell lines. Here, we have developed an additional PDX model derived from ATC (CUTC60-PDX) with a matched cell line (CUTC60), which harbors a clinically relevant BRAF V600E mutation. Thus, providing another critical translational model to study oncogenic BRAF signaling in thyroid cancer and the ability to study mechanisms and responses in vitro and in vivo.
In summary, we have developed four new thyroid cancer cell lines and one new PDX model. The two PTC cell lines (CUTC5 and CUTC48) were each derived from pleural effusions, the CUTC60 cell line was derived from the primary tumor of an ATC patient, and the CUTC61 was derived from a the primary tumor of a patient with metastatic FTC. These cell lines and PDX model have been authenticated by STR profiling, and match their original patient tumor samples. In order to maintain cell line integrity, these models will be distributed through the University of Colorado Cancer Center Tissue Culture Shared Resource. The initial characterization of these models provides important information on growth rate, key mutations, thyroid-specific gene expression, kinase dependency, and tumor growth in vivo, thus providing an important resource to study thyroid cancer biology and BRAF, RAS, and RET/PTC signaling. Future studies will continue to focus on developing cell lines and PDX models from advanced forms of thyroid cancer.
Supplementary Material
Implications: We have developed the second RET/PTC1-expressing PTC-derived cell line in existence, which is a major advance in studying RET signaling. We have further linked all cell lines to the originating patients, providing a set of novel, authenticated thyroid cancer cell lines and PDX models to study advanced thyroid cancer.
Acknowledgements
The authors thank Randall Wong at the University of Colorado B. Davis Center BioResources Core Facility for STR profiling of the cell lines. Cell lines were also STR profiled at the UCCC DNA Sequencing & Analysis Core (supported by NCI Cancer Center support grant, P30CA046934). We thank Lori Sherman, Michelle Randolph, and Dr. Steve Anderson for assistance with depositing the cell lines in the UCCC Tissue Culture Shared Resource (supported by NCI Cancer Center support grant, P30CA046934, and the MSKCC Integrated Genomics and Bioinformatics Cores.
This work was supported by NCI grant 1RC1CA147371-01 (to B.R. Haugen, J.A. Fagin, J.A. Knauf, R.E. Schweppe), NCI grant K12-CA086913 (to R.E. Schweppe), American Cancer Society RSG-13-060-01-TBE (to R.E. Schweppe), Amercan Thyroid Association Research Grant (to N. Pozdeyev), Colorado Cancer Center Paul R. Ohara II Seed Grant (to N. Pozdeyev), NIH P50CA058187, NIH NCI Cancer Center grant P30CA046934, as well as P50CA172012, R01CA72597, and the Linn Family Fund (to J.A. Fagin), and the institutional support grant P30CA008748.
Footnotes
The authors declare no potential conflicts of interest.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA: a cancer journal for clinicians 2018;68(1):7–30 doi 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Durante C, Haddy N, Baudin E, Leboulleux S, Hartl D, Travagli JP, et al. Long-term outcome of 444 patients with distant metastases from papillary and follicular thyroid carcinoma: benefits and limits of radioiodine therapy. J Clin Endocrinol Metab 2006;91(8):2892–9 doi 10.1210/jc.2005-2838. [DOI] [PubMed] [Google Scholar]
- 3.Lamartina L, Grani G, Durante C, Borget I, Filetti S, Schlumberger M. Follow-up of differentiated thyroid cancer - what should (and what should not) be done. Nature reviews Endocrinology 2018;14(9):538–51 doi 10.1038/s41574-018-0068-3. [DOI] [PubMed] [Google Scholar]
- 4.Kebebew E, Greenspan FS, Clark OH, Woeber KA, McMillan A. Anaplastic thyroid carcinoma. Treatment outcome and prognostic factors. Cancer 2005;103(7):1330–5 doi 10.1002/cncr.20936. [DOI] [PubMed] [Google Scholar]
- 5.Fagin JA, Wells SA Jr. Biologic and Clinical Perspectives on Thyroid Cancer. The New England journal of medicine 2016;375(11):1054–67 doi 10.1056/NEJMra1501993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Research N. Integrated genomic characterization of papillary thyroid carcinoma. Cell 2014;159(3):676–90 doi 10.1016/j.cell.2014.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pozdeyev N, Gay LM, Sokol ES, Hartmaier R, Deaver KE, Davis S, et al. Genetic Analysis of 779 Advanced Differentiated and Anaplastic Thyroid Cancers. Clin Cancer Res 2018;24(13):3059–68 doi 10.1158/1078-0432.CCR-18-0373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Landa I, Ibrahimpasic T, Boucai L, Sinha R, Knauf JA, Shah RH, et al. Genomic and transcriptomic hallmarks of poorly differentiated and anaplastic thyroid cancers. J Clin Invest 2016;126(3):1052–66 doi 10.1172/JCI85271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu B, Ghossein R. Genomic landscape of poorly differentiated and anaplastic thyroid carcinoma. Endocr Pathol 2016;27(3):205–12 doi 10.1007/s12022-016-9445-4. [DOI] [PubMed] [Google Scholar]
- 10.Xing M, Liu R, Liu X, Murugan AK, Zhu G, Zeiger MA, et al. BRAF V600E and TERT promoter mutations cooperatively identify the most aggressive papillary thyroid cancer with highest recurrence. J Clin Oncol 2014;32(25):2718–26 doi 10.1200/JCO.2014.55.5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Santoro M, Papotti M, Chiappetta G, Garcia-Rostan G, Volante M, Johnson C, et al. RET activation and clinicopathologic features in poorly differentiated thyroid tumors. J Clin Endocrinol Metab 2002;87(1):370–9 doi 10.1210/jcem.87.1.8174. [DOI] [PubMed] [Google Scholar]
- 12.Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 2012;483(7391):570–5 doi 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012;483(7391):603–7 doi 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schweppe RE, Klopper JP, Korch C, Pugazhenthi U, Benezra M, Knauf JA, et al. Deoxyribonucleic acid profiling analysis of 40 human thyroid cancer cell lines reveals cross-contamination resulting in cell line redundancy and misidentification. J Clin Endocrinol Metab 2008;93:4331–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marlow LA, D’Innocenzi J, Zhang Y, Rohl SD, Cooper SJ, Sebo T, et al. Detailed molecular fingerprinting of four new anaplastic thyroid carcinoma cell lines and their use for verification of RhoB as a molecular therapeutic target. J Clin Endocrinol Metab 2010;95(12):5338–47 doi 10.1210/jc.2010-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu X, Ory V, Chapman S, Yuan H, Albanese C, Kallakury B, et al. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am J Pathol 2012;180(2):599–607 doi 10.1016/j.ajpath.2011.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu M, Selvaraj SK, Liang-Chu MM, Aghajani S, Busse M, Yuan J, et al. A resource for cell line authentication, annotation and quality control. Nature 2015;520(7547):307–11 doi 10.1038/nature14397. [DOI] [PubMed] [Google Scholar]
- 18.Lorenzi PL, Reinhold WC, Varma S, Hutchinson AA, Pommier Y, Chanock SJ, et al. DNA fingerprinting of the NCI-60 cell line panel. Mol Cancer Ther 2009;8(4):713–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masters JR, Thomson JA, Daly-Burns B, Reid YA, Dirks WG, Packer P, et al. Short tandem repeat profiling provides an international reference standard for human cell lines. Proc Natl Acad Sci U S A 2001;98(14):8012–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gazdar AF, Girard L, Lockwood WW, Lam WL, Minna JD. Lung cancer cell lines as tools for biomedical discovery and research. J Natl Cancer Inst 2010;102(17):1310–21 doi 10.1093/jnci/djq279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Staveren WC, Solis DW, Delys L, Duprez L, Andry G, Franc B, et al. Human thyroid tumor cell lines derived from different tumor types present a common dedifferentiated phenotype. Cancer Res 2007;67(17):8113–20. [DOI] [PubMed] [Google Scholar]
- 22.Chan SY, Choy KW, Tsao SW, Tao Q, Tang T, Chung GT, et al. Authentication of nasopharyngeal carcinoma tumor lines. Int J Cancer 2008;122(9):2169–71 doi 10.1002/ijc.23374. [DOI] [PubMed] [Google Scholar]
- 23.Griewank KG, Yu X, Khalili J, Sozen MM, Stempke-Hale K, Bernatchez C, et al. Genetic and molecular characterization of uveal melanoma cell lines. Pigment Cell Melanoma Res 2012;25(2):182–7 doi 10.1111/j.1755-148X.2012.00971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marlow LA, Rohl SD, Miller JL, Knauf JA, Fagin JA, Ryder M, et al. Methodology, criteria and characterization of patient-matched thyroid cell lines and patient-derived tumor xenografts. J Clin Endocrinol Metab 2018. doi 10.1210/jc.2017-01845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Landa I, Pozdeyev N, Korch C, Marlow L, Smallridge RC, Copland JA, et al. Comprehensive genetic characterization of human thyroid cancer cell lines: a validated panel for preclinical studies. Clin Cancer Res In Revision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanabe H, Takada Y, Minegishi D, Kurematsu M, Masui T, Mizusawa H. Cell line individualization by STR multiplex system in the cell bank found cross-contaminationi between ECV304 and EJ-1/T24. Tiss Cul Res Commun 1999;18:329–38. [Google Scholar]
- 27.Ricarte-Filho JC, Ryder M, Chitale DA, Rivera M, Heguy A, Ladanyi M, et al. Mutational profile of advanced primary and metastatic radioactive iodine-refractory thyroid cancers reveals distinct pathogenetic roles for BRAF, PIK3CA, and AKT1. Cancer Res 2009;69(11):4885–93 doi 10.1158/0008-5472.CAN-09-0727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Imkamp F, von Wasielewski R, Musholt TJ, Musholt PB. Rearrangement analysis in archival thyroid tissues: punching microdissection and artificial RET/PTC 1–12 transcripts. J Surg Res 2007;143(2):350–63. [DOI] [PubMed] [Google Scholar]
- 29.Vitagliano D, Carlomagno F, Motti ML, Viglietto G, Nikiforov YE, Nikiforova MN, et al. Regulation of p27Kip1 protein levels contributes to mitogenic effects of the RET/PTC kinase in thyroid carcinoma cells. Cancer Res 2004;64(11):3823–9. [DOI] [PubMed] [Google Scholar]
- 30.Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, et al. Memorial Sloan Kettering-integrated mutation profiling of actionable cancer targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. The Journal of molecular diagnostics : JMD 2015;17(3):251–64 doi 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schweppe R, Kerege A, Sharma V, Poczobutt J, Gutierrez-Hartmann A, Grzywa R, et al. Distinct Genetic Alterations in the MAPK Pathway Dictate Sensitivity of Thyroid Cancer Cells to MKK1/2 Inhibition. Thyroid 2009;19:825–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schweppe RE, Kerege AA, French JD, Sharma V, Grzywa RL, Haugen BR. Inhibition of Src with AZD0530 reveals the Src-Focal Adhesion Kinase complex as a novel therapeutic target in papillary and anaplastic thyroid cancer. J Clin Endocr Metab 2009;94(6):2199–203 doi 10.1210/jc.2008-2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chan CM, Jing X, Pike LA, Zhou Q, Lim DJ, Sams SB, et al. Targeted inhibition of SRC kinase with dasatinib blocks thyroid cancer growth and metastasis. Clin Cancer Res 2012;18(13):3580–91 doi 10.1158/1078-0432.CCR-11-3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morrison JA, Pike LA, Lund G, Zhou Q, Kessler BE, Bauerle KT, et al. Characterization of thyroid cancer cell lines in murine orthotopic and intracardiac metastasis models. Hormones & cancer 2015;6(2–3):87–99 doi 10.1007/s12672-015-0219-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kessler BE, Sharma V, Zhou Q, Jing X, Pike LA, Kerege AA, et al. FAK expression, not kinase activity, is a key mediator of thyroid tumorigenesis and protumorigenic processes. Mol Cancer Res 2016;14(9):869–82 doi 10.1158/1541-7786.MCR-16-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beadnell TC, Mishall KM, Zhou Q, Riffert SM, Wuensch KE, Kessler BE, et al. The Mitogen-Activated Protein Kinase Pathway Facilitates Resistance to the Src Inhibitor Dasatinib in Thyroid Cancer. Mol Cancer Ther 2016;15(8):1952–63 doi 10.1158/1535-7163.MCT-15-0702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roskoski R, Jr., Sadeghi-Nejad A. Role of RET protein-tyrosine kinase inhibitors in the treatment RET-driven thyroid and lung cancers. Pharmacol Res 2018;128:1–17 doi 10.1016/j.phrs.2017.12.021. [DOI] [PubMed] [Google Scholar]
- 38.Watson AJ, Hopkins GV, Hitchin S, Begum H, Jones S, Jordan A, et al. Identification of selective inhibitors of RET and comparison with current clinical candidates through development and validation of a robust screening cascade. F1000Research 2016;5:1005 doi 10.12688/f1000research.8724.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strumberg D, Richly H, Hilger RA, Schleucher N, Korfee S, Tewes M, et al. Phase I clinical and pharmacokinetic study of the Novel Raf kinase and vascular endothelial growth factor receptor inhibitor BAY 43–9006 in patients with advanced refractory solid tumors. J Clin Oncol 2005;23(5):965–72 doi 10.1200/JCO.2005.06.124. [DOI] [PubMed] [Google Scholar]
- 40.Tentler JJ, Tan AC, Weekes CD, Jimeno A, Leong S, Pitts TM, et al. Patient-derived tumour xenografts as models for oncology drug development. Nature reviews Clinical oncology 2012;9(6):338–50 doi 10.1038/nrclinonc.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Henderson YC, Ahn SH, Ryu J, Chen Y, Williams MD, El-Naggar AK, et al. Development and characterization of six new human papillary thyroid carcinoma cell lines. J Clin Endocrinol Metab 2015;100(2):E243–52 doi 10.1210/jc.2014-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Onoda N, Nakamura M, Aomatsu N, Noda S, Kashiwagi S, Hirakawa K. Establishment, characterization and comparison of seven authentic anaplastic thyroid cancer cell lines retaining clinical features of the original tumors. World J Surg 2014;38(3):688–95 doi 10.1007/s00268-013-2409-7. [DOI] [PubMed] [Google Scholar]
- 43.Barallon R, Bauer SR, Butler J, Capes-Davis A, Dirks WG, Elmore E, et al. Recommendation of short tandem repeat profiling for authenticating human cell lines, stem cells, and tissues. In Vitro Cell Dev Biol Anim 2010;46(9):727–32 doi 10.1007/s11626-010-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schweppe RE, Korch C. Challenges and advances in the development of cell lines and xenografts. Adv Mol Path 2018;1(1):239–51 doi 10.1016/j.yamp.2018.07.004. [DOI] [Google Scholar]
- 45.Bauerle KT, Schweppe RE, Kotnis G, Gagan D, Agarwal R, Haugen BR. NF kappa B-dependent regulation of proliferation and angiogenesis in an orthotopic model of thyroid cancer is associated with interleukin-8 secretion J Clin Endocrinol Metab 2014;99(8):1436–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gunda V, Bucur O, Varnau J, Vanden Borre P, Bernasconi MJ, Khosravi-Far R, et al. Blocks to thyroid cancer cell apoptosis can be overcome by inhibition of the MAPK and PI3K/AKT pathways. Cell death & disease 2014;5:e1104 doi 10.1038/cddis.2014.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lai Y, Wei X, Lin S, Qin L, Cheng L, Li P. Current status and perspectives of patient-derived xenograft models in cancer research. J Hematol Oncol 2017;10(1):106 doi 10.1186/s13045-017-0470-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wunderlich A, Khoruzhyk M, Roth S, Ramaswamy A, Greene BH, Doll D, et al. Pretherapeutic drug evaluation by tumor xenografting in anaplastic thyroid cancer. J Surg Res 2013;185(2):676–83 doi 10.1016/j.jss.2013.06.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.