Abstract
The intestinal epithelium has a high rate of cell turn over and is an excellent system to study stem cell-mediated tissue homeostasis. The Misshapen subfamily of the Ste20 kinases in mammals consists of MINK1, MAP4K4 and TNIK. Recent reports suggest that this subfamily has a novel function equal to the Hippo/MST subfamily as upstream kinases for Warts/LATS to suppress tissue growth. To study the in vivo functions of Mink1, Map4k4 and Tnik, we generated a compound knockout of these three genes in the mouse intestinal epithelium. The intestinal epithelia of the mutant animals were phenotypically normal up to approximately 12 months. The older animals then exhibited mildly increased proliferation throughout the lower GI tract. We also observed that the normally spatially organized Paneth cells in the crypt base became dispersed. The expression of one of the YAP pathway target genes Sox9 was increased while other target genes including CTGF did not show a significant change. Therefore, the Misshapen and Hippo subfamilies may have highly redundant functions to regulate growth in the intestinal epithelium, as illustrated in recent tissue culture models.
Keywords: Hippo, intestine, MAP4K4, MINK1, Misshapen, Ste20 kinases, TNIK
Introduction
The mammalian intestinal epithelium has a strong capacity of growth adaptation due to the balancing acts of continuous cell loss and intestinal stem cell (ISC)-mediated regeneration (Beumer and Clevers, 2016; Chin et al., 2017; Demitrack and Samuelson, 2016; Rubin and Levin, 2016; Tan and Barker, 2014; Tetteh et al., 2015). Multiple stem cell populations have been identified that are resided in the intestinal crypts, and the transit amplifying cells located in the more distal positions further provide a strong growth capacity (Bankaitis et al., 2018; Tetteh et al., 2016; Yan et al., 2012; Yousefi et al., 2017). Some of the interesting questions being pursued are how the intrinsic stemness, the surrounding niche signals and the lumenal content affect stem cell and proliferative activity (Degirmenci et al., 2018; Demitrack and Samuelson, 2016; Mana et al., 2017; Tan and Barker, 2014).
The Hippo (Hpo) pathway has a broad role in growth regulation under many physiological and pathological conditions (Harvey and Hariharan, 2012; Ma et al., 2018; Misra and Irvine, 2018; Pan, 2010). The central components of the pathway include a series of kinases, starting with Hpo in Drosophila and MST½ (or called STK¾) in mammals. This Hpo/MST subfamily of Ste20 kinases phosphorylate and positively regulate the functions of Warts/LATS of the NRD family of kinases. Warts/LATS then phosphorylate and suppress the transcriptional co-activators Yorkie/YAP. When the upstream kinases are inactive, Yorkie/YAP phosphorylation is reduced, which allows Yorkie/YAP to accumulate in the nucleus and collaborate with the transcription factors Scallop/TEAD to regulate growth and apoptosis related genes (Koontz et al., 2013; Yu and Pan, 2018). While the core kinases and transcriptional components have been well-defined, the physiological regulation of Hpo/MST is complex and many of the upstream components include membrane associated proteins, which in turn are possible mediators of mechanosensing, extracellular stimulation or intracellular cytoskeleton stretching (Deng et al., 2015; Fletcher et al., 2018; Han et al., 2018; Mao et al., 2017; Meng et al., 2018; Su et al., 2017; Sun et al., 2015)
Recent studies have expanded the Hpo/MST pathway to include other related kinases that can also regulate Warts/LATS and Yorkie/YAP under different biological contexts (Li et al., 2014; Li et al., 2018; Li et al., 2015; Meng et al., 2015; Meng et al., 2018; Poon et al., 2018; Zheng et al., 2015). The Ste20 kinases can be divided into 8 subfamilies, each contains the highly conserved kinase domain, but outside of that are quite different sequences and structural motifs. All these subfamilies are comprised of multiple proteins in mammals, but only one orthologous protein in Drosophila, making it potentially easier to study their functions in model organisms (Li et al., 2014; Li et al., 2015; Meng et al., 2015; Zheng et al., 2015). Our recent reports demonstrate that in differentiating enteroblasts of the adult Drosophila midgut, Misshapen (Msn) binds and phosphorylates Wts, thereby suppressing the activity of Yki and the expression of the JAK-STAT pathway ligand Upd3 to restrict intestinal growth (Li et al., 2014; Li et al., 2018). Furthermore, the mammalian Msn homolog MAP4K4 (or called NIK, HGK) also interacts with LATS to suppress YAP (Li et al., 2014; Li et al., 2018). Additional reports have similarly illustrated that the Ste20 kinases Hpo, Msn, and Happyhour (Hppy) of Drosophila, and their mammalian homologs Mst½, MAP4K4/6/7 and MAP4K½/3/5, respectively, have overlapping functions in regulating Wts/LATS and Yki/YAP in various assays (Li et al., 2015; Meng et al., 2015; Meng et al., 2018; Poon et al., 2018; Zheng et al., 2015). Therefore, the Hpo pathway has been expanded to include the Hpo, Msn and Hppy subfamilies of Ste20 kinases, by virtue of their common functions to regulate Wts/LATS.
An important question worth pursuing is whether distinct upstream mechanisms are employed to control the activity and specificity of these different subfamilies of Ste20 kinases in various biological processes (Li et al., 2018; Meng et al., 2018). Moreover, how the frequently shown overlapping functions of these kinases in different contexts play out their functions in vivo requires further investigation. In this report, we present results to show that compound conditional knockout of the three Msn subfamily kinases, which includes MINK1 (or called MAP4K6), MAP4K4 and TNIK (or called MAP4K7), in the mouse intestine causes a moderate increase of proliferation in aging animals. While the result reflects a functional role of these kinases in regulating intestinal growth, it also suggests possible extensive overlapping functions of Ste20 kinases in the intestinal epithelium.
Results
Loss of function analysis of Mink1, Map4k4 and Tnik
Previous reports have shown that Map4k4 is essential for embryonic development in mice. Animals that have lost Map4k4 function die around E10, with defects in mesodermal and endodermal cell migration, resulting in somite and hindgut developmental defects (Xue et al., 2001). Additional tissue specific knockouts demonstrate an essential function of Map4k4 in endothelial cells of blood and lymphatic vessels (Roth Flach et al., 2016; Roth Flach et al., 2015; Vitorino et al., 2015). Tnik loss of function mice are viable, and functional analyses suggest that Tnik has a role in regulating dendrite development and neuronal function (Anazi et al., 2016; Burette et al., 2015; Coba et al., 2012; Wang et al., 2016). Meanwhile, a knockout mouse model of Mink1 targeting the deletion of exon 3 and 4 has been performed in platelets and impaired thrombus formation has been observed (Yue et al., 2016).
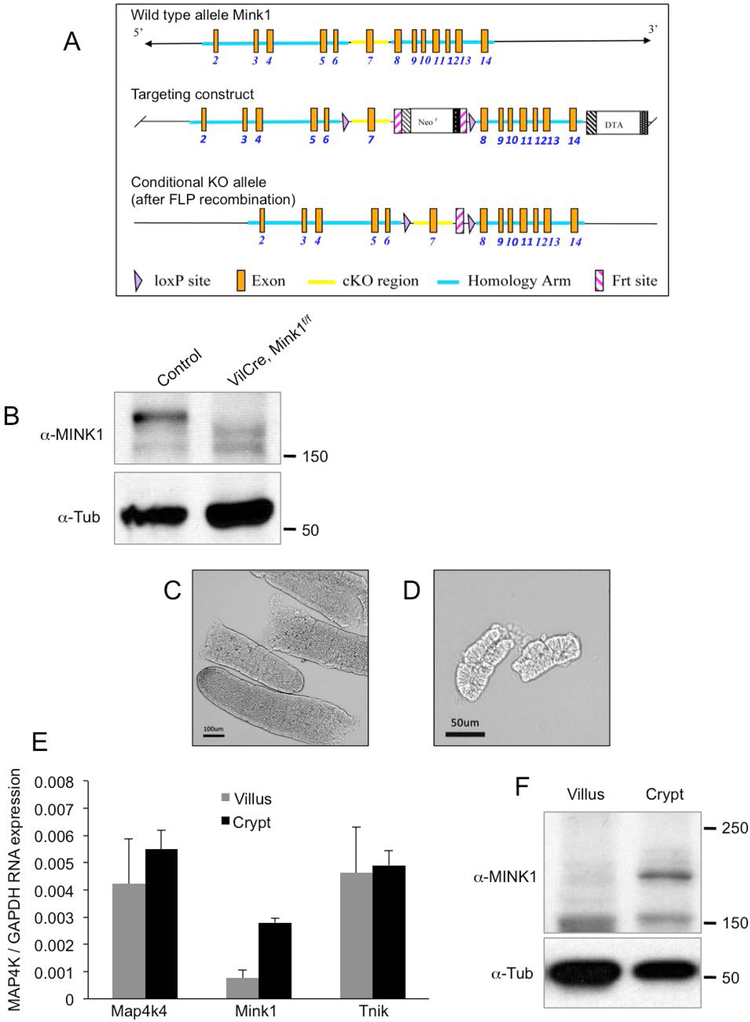
We independently generated a conditional allele of Mink1, aiming at deleting exon 7 of the gene and thereby part of the highly conserved kinase domain (Fig. 1A). This is an identical strategy as the Map4k4 and Tnik mutant alleles that we used in this report (Burette et al., 2015; Roth Flach et al., 2015). This Mink1f/f condition allele, when the sequences are properly deleted, should also produce a stop codon that causes premature termination of Mink1 translation close to the deletion. Western blot analysis by using intestinal tissues of this Mink1 conditional allele crossed with VillinCre (VilCre) showed that there was no detectable protein when comparing to that of the control tissues (Fig. 1B), consistent with the idea that this is a protein null mutation.
Figure 1.
The generation of Mink1f/f conditional knockout mouse. (A) The genomic locus of Mink1 is shown in the top panel. The Mink1 conditional knockout mouse strategy, targeting the deletion of exon 7, and the various sites are shown in the panels as indicated. (B) The Western blot showing the endogenous MINK1 protein in intestinal epithelial extracts, and the signal disappeared in the extracts from VilCre;Mink1f/f animals. The anti-Tubulin blot was used as an internal loading control. (C) Phase contrast microscope images of isolated small intestinal villi, which were used in the following quantitative real-time PCR and Western blot. (D) Phase contrast microscope images of similarly isolated small intestinal crypts. (E) Relative mRNA expression for Map4k4, Mink1 and Tnik in the separated villi and crypts. The expression level was normalized to that of GAPDH in parallel qPCR assays. (F) Endogenous protein expression of MINK1 is enriched in intestinal crypts. Extract lysates of indicated tissues were used for Western blot using the antibodies as indicated.
When crossed with the Sox2Cre strain to induce whole body knockout or VilCre for intestine specific knockout, the Mink1f/f conditional knockout allele produced viable homozygous progenies. No reduction in Mendelian ratio was observed. Therefore, Mink1 is probably not an essential gene for development and viability in mice.
Because the Map4k4 knockout alone would cause embryonic lethality, we performed a series of experiments to prepare for tissue specific compound knockout experiments of all three Msn subfamily of kinases in the intestine. We examined by quantitative PCR the RNA expression of these three kinase genes. RNA samples were isolated from the separated crypts and villi of intestinal tissues from normal, 1 month old animals by increased EDTA treatment (Fig. 1C, D). We could detect the expression of each of these three genes, with higher expression of Mink1 RNA in the crypt (Fig. 1E). Protein blot experimental results also corroborated the RNA expression, with a higher expression of MINK1 protein in the crypt preparations (Fig. 1F). Overall, the three genes have expression and therefore possible functions in intestine and the crypts.
Increased crypt proliferation after loss of Mink1, Map4k4 and Tnik
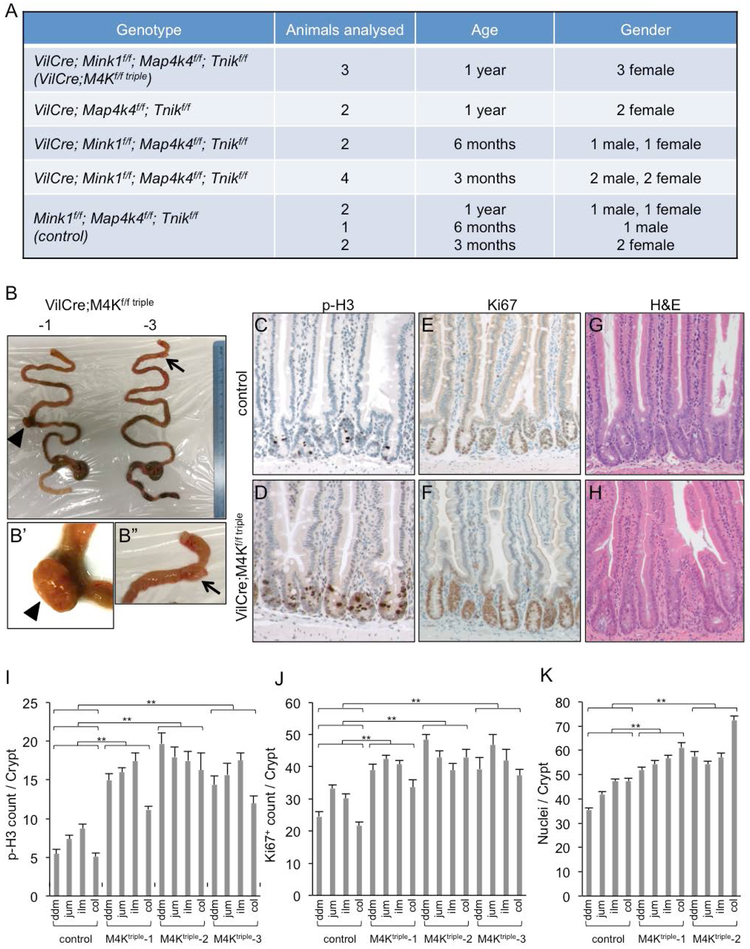
To examine the functional requirement in the intestinal epithelium, the three conditional knockout alleles were crossed together and then with the VilCre driver. The number of animals with different combinations of the mutant alleles and at different ages that were examined are listed in Fig. 2A. We did not detect observable phenotypic changes in control animals (homozygous for all three Mink1f/f; Map4k4f/f; Tnikf/f alleles without VilCre, here after referred to as control). Other single or multiple mutant combinations together with the VilCre we obtained at different time during the 1-year period similarly did not show lethality or observable phenotypic changes. The only group that showed phenotypic changes was the 1 year old age group containing all Mink1f/f; Map4k4f/f; Tnikf/f homozygous alleles together with VilCre (here after referred to as VilCre;M4Kf/f triple). All 3 animals we obtained showed some degree of abnormality in the intestine as detailed below. Furthermore, 1 more animal of this group died around the age of 1 year before we performed an examination. These results suggest that the three kinases are mostly dispensable in the postnatal intestinal epithelium and phenotypic changes occur around 1 year old.
Figure 2.
Loss of Mink1, Map4k4 and Tnik leads to increased proliferation in the intestines of aging animals. (A) A list of animals with the indicated genotype, gender and age used for quantification of intestinal phenotypes. The triple knockout animals, abbreviated as VilCre;M4Kf/f triple, showed the proliferation increase only after 1 year. The animals of the same triple knockout genotype at 3 or 6 months, or other combinations, did not show an increase. They had the same proliferation marker measurement as the control animals, which were the triple-floxed littermates without VilCre. (B) Photographs of whole intestines from two VilCre;M4Kf/f triple mice of 1 year old. The small tissue outgrowth of the small intestines are indicated by the arrowhead and arrow, and shown as the enlarged images in B’ and B”. (C-H) Images of immunohistochemical staining in small intestines of 1 year old control and VilCre;M4Kf/f triple mice. Phosphorylated histone 3 (p-H3) stains for mitotic chromosomes. Ki67 is a cell proliferating marker. Hematoxylin and eosin (H&E) shows the morphology to aid the counting of cell number in the intestinal crypts; this staining is included in all these panels, and is shown as double staining in panels C-F and as single staining in panels G-H. (I-K) Quantification of p-H3+, Ki67+ and cell number in control and VilCre;M4Kf/f triple mouse intestines. The counting of positive cell staining per crypt was performed separately in the different regions of the lower GI tract. ddm is duodenum, jum is jejunum, ilm is ileum, col is colon. Three different VilCre;M4Kf/f triple knockout animals (−1, −2, −3) are shown separately. p value indicates the comparison of the data sets in the specific region of the mutant to the same region of the control. All the individual comparisons show statistical significance. Experiments were performed at least 3 times independently. The error bar is standard error of the mean, and p-value is from Student’s T-test: * is p<0.05, ** is p<0.01, NS is no significance with p>0.05.
Two out of the three triple knockout intestinal epithelia each exhibited a lump of benign tissue outgrowth (indicated by arrow and arrowhead in Fig. 2B, C, and enlargements). We sectioned these lumps and found mostly mesenchymal tissue with probably infiltration of lymphocytes, but not apparent epithelial tumors. Therefore, we sectioned other parts of the intestinal epithelium and performed staining for growth markers. The phosphorylated histone 3 (p-H3) staining revealed cells in mitosis with condensed chromosomes. All three triple knockout tissues had more p-H3 staining in crypt epithelia, which is the proliferation compartment containing stem cells and transit amplifying cells. Staining for another proliferation marker Ki67 revealed a similar result of increased staining in crypts. We also counted the number of nuclei in crypts after H&E staining and revealed more cells in this compartment. We quantified the staining in the control and mutant animals along the lower intestinal tract for these markers. All 4 regions that is duodenum, jujunum, ileum and colon all showed significantly increased proliferation. Therefore, while the tissue growth had not caused tumor formation, the loss of the three kinases did result in significant increase of proliferation in intestinal crypts.
Increased proliferation leads to organizational changes in the mutant crypts
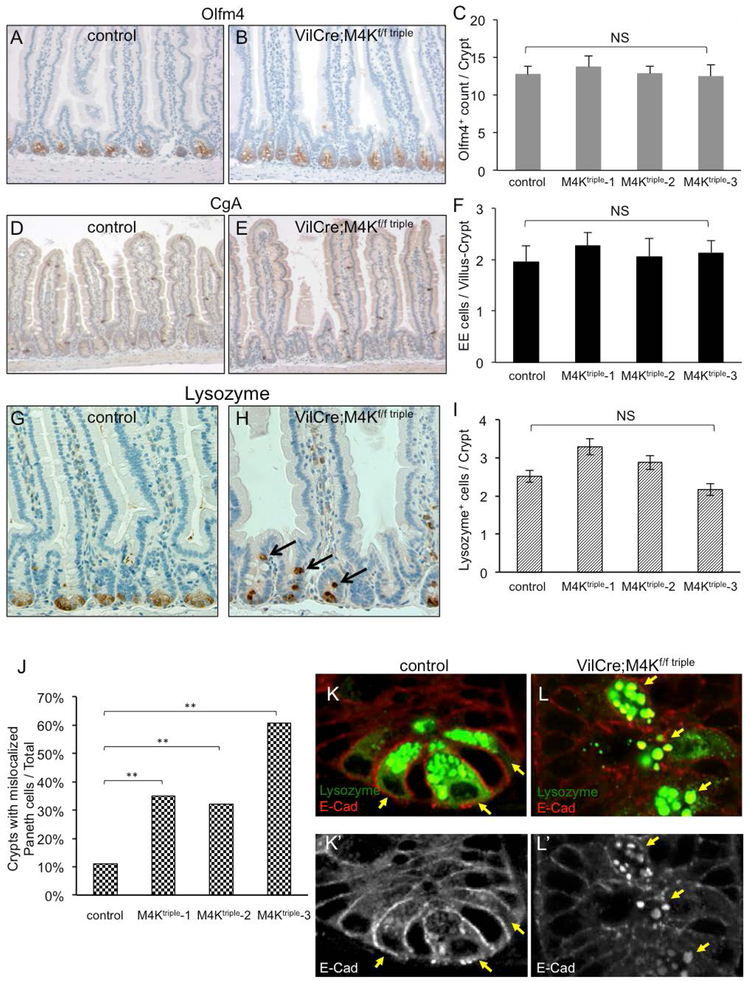
To investigate other possible phenotypic changes, we examined various cellular markers. The Olfm4 staining should reveal the crypt base stem cells and the quantification showed that there is no increase in Olfm4 staining (Fig. 3A-C), suggesting that the stem cell number was not changed. The staining for mature enteroendocrine cells by using the Chromogranin A (CgA) antibody also showed that the number remained the same in mutant intestines as in control intestines (D-F). Meanwhile, the staining for mature Paneth cells by using the Lysozyme antibody showed abnormality in spatial distribution, while the quantification of the Paneth cell number revealed no significant change (G-I). Together with visually normal villi, we conclude that the differentiation into various cell types in the intestine remains normal in the triple mutant animals.
Figure 3.
The MAP4K triple mutant intestines have normal cell fate but mislocalized paneth cells. (A-B) Immunohistochemical staining of the stem cell marker Olfm4, shown as brown staining at the crypt base, together with H&E staining to illustrate the tissue and cell morphology. A control and a triple mutant intestinal tissue are shown. (C) The quantification of the Olfm4+ cell staining in control and mutant intestines is shown. NS is no significance, with P value >0.05. (D-E) The images show immunohistochemical staining of the enteroendocrine cell marker Chromogranin A (CgA) in a control and a triple mutant intestines. (F) The quantification of the CgA+ cell staining in control and mutant intestines is shown. (G-H) The images show immunohistochemical staining of the Paneth cells marker Lysozyme in a control and a triple mutant intestine. (I) The quantification of the Lysozyme+ cell staining in control and mutant intestines is shown. (J) The percentage of crypts with mislocalized Paneth cells from control and triple mutant intestines is shown. For each crypt we examined, if there was one or more Lysozyme+ cell located above the crypt base, as those shown in panel H indicated by arrows, that crypt would be counted as positive for mislocalization. p-values are calculated by applying χ2-test. (K-L) Images of immunofluorescence staining of Lysozyme in green and E-Cadherin (E-Cad) in red, in small intestinal crypts of a control and a triple mutant. The bottom panels show single channel of E-Cad staining of the same images in white. The arrows point to Paneth cells, marked with high level of Lysozyme staining. These cells are localized at the crypt base of control, but are mislocalized into upper part of the crypt of mutant. The E-Cad staining in Paneth cells of the mutant (panel L’) shows lower membrane staining near cell-cell junctions but higher cytoplasmic staining that colocalizes with Lysozyme staining.
Paneth cells normally are located at the base of the crypts and interdigitally with the crypt base stem cells. In the triple mutant crypts, even though the number of Panel cells remained similar, the spatial distribution of these cells was clearly displaced (Fig. 3H, arrows). Many of them were located in the upper part of the crypt. We quantified this displacement phenotype, and counted that crypt as positively changed if there was 1 or more clearly visible Lysozyme staining in the upper part of the crypt. The result revealed a significant number of crypts containing mislocalized Paneth cells, increased from 10% in control intestines to 32%-60% in mutant intestines (Fig. 3J).
The staining of E-cadherin showed that in wild type tissues this adhesion protein is present mostly near the cytoplasmic membrane in both Paneth cells and neighboring crypt base stem cells (Fig. 3K). There is also visible E-cadherin co-staining with Lysozyme staining in Paneth cells (Fig. 3K’, arrows). However, in the mutant intestine the E-cadherin protein had reduced association with the cytoplasmic membrane but had stronger staining that co-localized with the Lysozyme staining. This result suggests that either the trafficking of E-cadherin in or the maturation of Paneth cells was abnormal. This abnormality may be caused by an intrinsic defect within Paneth cells after loss of the MAP4Ks, or by increased proliferation of other cells in the crypt as described above, which leads to impeded downward movement of Paneth cells and their maturation process.
Assessment of YAP activity after loss of Mink1, Map4k4 and Tnik
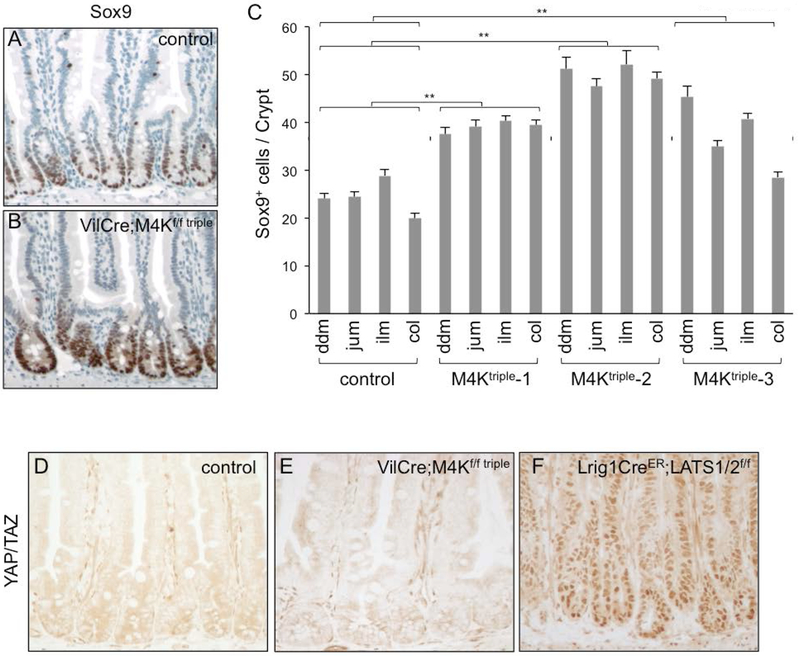
Each of MINK1, MAP4K4 and TNIK may have individual function but nonetheless all have been shown to activate LATS and thereby repress YAP to control tissue proliferation (Li et al., 2014; Li et al., 2018; Meng et al., 2015; Meng et al., 2018; Zheng et al., 2015). To assess the activity of this pathway, we examined some of the known target genes of YAP. Even though qPCR assays did not reveal an increased expression of two widely used reporter genes CTGF or ANKRD, the staining for the protein expression of another target gene SOX9 showed a substantial signal in many crypt nuclei (Fig. 4A, B. The quantification revealed similar results in intestines of the three mutant animals (Fig. 4C). Therefore, there may be a subtle increase of the pathway activity after loss of the three kinases.
Figure 4.
Assessment of YAP activity after loss of Mink1, Map4k4 and Tnik. (A-C) The images are immunohistochemical staining using an anti-Sox9 antibody, together with H&E staining. Sox9 nuclear staining shown in brown is strong in crypt nuclei, and appears to be stronger in the triple mutant. Panel C is quantification of the number of cells that are Sox9+ in control and mutant crypts. The increase of Sox9+ cell number in the mutants may result from a combination of increased cell number and increased staining. ** is P value <0.01. (D-F) Images of immunohistochemical staining using an anti-YAP/TAZ antibody in control and mutant intestines. There is no significant nuclear staining even after loss of the three kinases. Meanwhile, the nuclear staining is clear using this antibody after loss of LATS½.
We stained for YAP protein and found that in both control and triple mutant intestinal tissue sections the staining remained cytoplasmic (Fig. 4D, E). To illustrate the fidelity this antibody, nuclear staining of YAP/TAZ by was clearly detectable in the LATS½ knockout intestinal tissue (Fig. 4F) (Li et al, manuscript submitted) and therefore demonstrating the expected regulation. The lack of detectable nuclear YAP/TAZ staining in the VilCre;M4Kf/f triple mutants may represent further redundant regulation by other Ste20 kinases such as MST½. Tissue culture experiments have shown that compound knockout of 5 to 7 of these kinases could increase the misregulation of YAP and further increase of downstream events (Meng et al., 2015).
Regulation of MINK1, MAP4K4 and TNIK activities
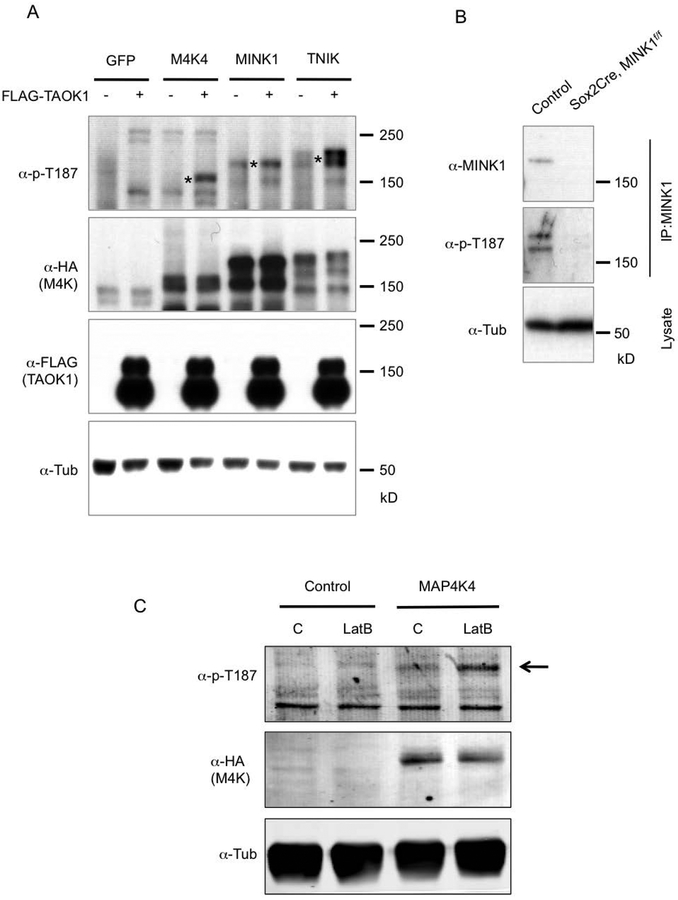
To examine possible biological regulations of MINK1, MAP4K4 and TNIK, we performed a series of biochemical experiments. In previous reports, we have demonstrated that a phosphorylation site within the highly conserved kinase domain of the Msn subfamily is essential for their activities (Li et al., 2018; Wang et al., 2016). This site is the threonine 194 (T194) in Msn and T187 in MINK1/MAP4K4/TNIK. Mutating this threonine residue to alanine abolish the activity towards the phosphorylation of the downstream target Warts/LATS (Li et al., 2018). In Drosophila, we have also shown that another Ste20 kinase Tao is the upstream kinase that can phosphorylate T194 of Msn (Li et al., 2018). Here we show that by co-transfection experiments, the mammalian homolog TAOK1 could also phosphorylate T187 of MINK1, MAP4K4 and TNIK (Fig. 5A). Therefore, the regulation of the activities of these kinases through the T194/T187 site is conserved. Furthermore, we could detect the phosphorylation of this site on MINK1 in intestinal tissue extracts (Fig. 5B). The Mink1 whole body knockout animals lost this signal in the intestinal tissue, demonstrating that this signal represents phosphorylation of MINK1 in vivo. We also tested whether the change of cytoskeleton tension, which is a widely used assay for mechanosensing (Meng et al., 2018), could also regulate the T187 phosphorylation. LatrunculinB treatment blocks actin polymerization and reduces tension. This treatment clearly led to increased MAP4K4 phosphorylation (Fig. 5C), which is consistent with our previous observation of Msn phosphorylation in response to reduced tension after gut starvation (Li et al., 2018).
Figure 5.
Regulation of MINK1, MAP4K4 and TNIK activities. (A) Western blots using extracts of transfected HEK293 cells. The plasmids containing cDNA of the genes indicated at the top of the panels were transfected into the cells. After 48 hours the cells were used for extract preparation, and the proteins resolved on SDS PAGE and analysed by Western blots using the antibodies indicated to the left of the panels. The asterisks in the top panel indicate the bands matching the predicted sizes of the full length proteins expressed. The MAP4K proteins are tagged with HA at the N-termini. (B) Western blot analysis of endogenous MINK1 protein phosphorylation. Protein extracts were prepared from mouse small intestinal epithelia of control (Mink1f/f without Cre) and mutant, which was 2 months old Sox2Cre; MINK1f/f. An anti-TNIK/MINK1 (BD Transduction Laboratories) antibody was used for IP, and then a rabbit anti-MINK1 (Bethyl) or the anti-p-T187 was used for blots. (C) Western blot analysis for the response of MAP4K4 to change of mechanical property. HEK293 cells were transfected with GFP cDNA as control or MAP4K4 cDNA. After 48 hours, the cells were treated with actin polymerization inhibitor Latrunculin B (LatB) for 1 hour. The cells were then used immediately for lysate preparation. The lysates were used for SDS PAGE and for blots using the indicated antibodies.
Discussion
We have demonstrated that the three Msn subfamily of kinases have functions in controlling proliferation of the mouse intestinal epithelium in aging animals. The proliferation increase is significant but relatively mild in the triple knockout animals, when compared to that of loss of function of LATS in the intestine (Li et al, manuscript submitted). Warts in Drosophila and LATS in mammals are central integrators of upstream signaling events from Hpo/MST, Msn/MAP4K4,6,7 and Hppy/MAP4K1-5. Warts/LATS in turn directly regulate Yorkie/YAP activity for transcriptional output (Ma et al., 2018; Misra and Irvine, 2018). Presumably, the Msn subfamily of kinases should only modulate part of the LATS function. The proliferation phenotype in the intestine is however still lower than expected, because the loss of Msn in Drosophila causes high proliferation in the midgut. It suggests that there is much higher redundancy with other Ste20 kinases in the mammalian intestine to regulate the LATS and YAP pathway. Alternatively, additional mechanisms remain to be uncovered, or that in the mouse intestine the Msn subfamily of kinases regulate pathways in addition to LATS and lead to control of proliferation. This is consistent with the redundant function of MINK1, MAP4K4 and TNIK in stress-induced mammalian neuronal degeneration involving the DLK and JNK pathway (Larhammar et al., 2017). Furthermore, the three Msn subfamily of kinases regulate other substrates such as SMAD (Fu et al., 2017; Kaneko et al., 2011). Nonetheless, biological assays after food ingestion or tissue damaged may help to unveil yet to be understood in vivo function of the Msn subfamily. Meanwhile, the mild proliferation phenotype suggests that these three kinases represent reasonable drug targets to improve regenerative proliferation perhaps without strong side effects. For example, total parenteral nutrition, which is feeding only via intravenous infusion, in human patients and mammalian models causes intestinal atrophy (Brinkman et al., 2012; Rubin and Levin, 2016). The stimulation of intestinal proliferation by inhibiting the Msn subfamily of kinases is a logical approach to improve patient recovery from intestinal atrophy.
Materials and Methods
Mouse models
All animals use protocols were reviewed and approved by The University of Massachusetts Medical School Institutional Animal Care and Use Committee. VillinCre (Madison et al., 2002) and Sox2Cre (Hayashi et al., 2002) mice were obtained from the Jackson laboratory. Map4k4flox was as reported previously (Roth Flach et al., 2015). Tnikflox was as reported previously (Burette et al., 2015).
To generate the Mink1flox allele, ES cells containing Mink1 exon 7 flanked by loxP sites were generated as shown in Fig. 1A. The ES cell lines bearing the targeted allele were injected to blastocysts to generate the mouse line. Animals bearing germline transmission were backcrossed to C57Bl6/J mice for 6 generations before crossing to Cre alleles for tissue knockout experiments. The generation of this Mink1flox strain was provided as a commercial service by Cyagen US Inc. (Santa Clara, CA, USA).
Compound knockout animals were generated by crossing repeatedly the Mink1flox, Map4k4flox and Tnikflox alleles and homozygous animals of the three flox alleles together were obtained based on genotyping.
Tissue collection and staining
Following euthanasia, intestines were dissected from adult mice and fixed in 10% Neutral Buffered Formalin (NBF) at 4°C overnight. For paraffin sections, tissue was dehydrated, embedded in paraffin, and sectioned at 6 μm thickness. The paraffin sections were stained using standard hematoxylin & eosin (H&E) reagents. For immunohistochemistry (IHC) and immunofluorescence (IF) staining, tissue sections were deparaffinized and rehydrated before undergoing heat-induced antigen retrieval in 10 mM sodium citrate buffer (pH 6.0) for 1 hour. Slides were blocked for endogenous peroxidase for 15 minutes, then blocked for 1 hour in 5% BSA, 1% goat serum, in PBST with 0.1% Tween-20, and incubated overnight at 4°C in primary antibody diluted in the blocking buffer. Slides were incubated in biotinylated secondary antibody for 1 hour at room temperature and signal was detected using the Vectastain Elite ABC kit (Vector Laboratories). Primary antibodies used for IHC/IF were: Ki67 (1:10,000, Abcam), phosphor-histone H3 (1:3000, Abcam), Olfm4 (1:200, Cell Signaling), Lysozyme (1:1000, Novus Biologicals), Sox9 (1:200, Abcam), Chromogranin A (CgA, 1:1000, Thermo Fisher, RB9003) and E-Cadherin (1:400, R&D Systems). Confocal images were obtained by using Nikon Spinning Disk confocal microscope (UMass Medical School Imaging Core Facility) and processed by Metaphor.
Isolation of crypts and villi from small intestine
The protocol used to isolate and separate the crypts and villus from mouse small intestine was as published previously (O’Rourke et al., 2016). In brief, 15 cm of small intestine was dissected, flushed, opened and washed in cold PBS. The tissue was cut into 5 mm pieces and placed in ice-cold 5 mM EDTA-PBS and gently mixed in a bench roller at 4°C for 10 minutes, during which the tube was inverted 10-15 times by hand. This step will release small portion of the villi into the supernatant. The pieces of intestinal tissue were transferred into a new tube containing ice-cold 5 mM EDTA-PBS and gently mixed at 4°C for 30 min, and then repeated another time. These supernatant collections were combined as villi extracts. For the remaining tissue, new cold PBS was added to the tissue, followed by 10 times vigorous trituration using a 10 ml pipette, then the supernatant was removed. This step was repeated two more times and the batches of supernatants were combined. This supernatant containing the crypts was then mixed with Basal Media (Advanced DMEM F/12 with L-Glutamine, HEPES, N-Acetylcysteine) and filtered through 100 μm and 70 μm filters, which should remove most of the villi from the crypts fractions. The isolated villi and crypts were spun down and used for protein and RNA extraction for Western blot and quantitative real-time PCR.
Tissue Culture, Transfection, and Plasmid Constructs
HEK293T cells were cultured in DMEM supplemented with 10% FBS and transfected with Lipofect-amine 2000 (Invitrogen) in Opti MEM Medium. Constructs that expressed the HA-tagged MAP4K4, MINK1 and TNIK, and the FLAG-tagged TAOK1 were generated in pMIPZ vector as described previously (Kaneko et al., 2011; Li et al., 2018).
Western blot and immunoprecipitation
Western blots were performed as described (Kaneko et al., 2011; Li et al., 2018) and the primary antibodies used were as following: anti-MINK1 (Bethyl), anti-phospho-MAP4KsT194/T187 (Li et al., 2018; Wang et al., 2016), anti-β-tubulin (Thermo Fisher), anti-HA (Roche) and anti-FLAG (Sigma). The endogenous MINK1 in mouse small intestine was first immunoprecipitated by purified mouse anti-TNIK/MINK1 (BD Biosciences) antibody and then blotted by anti-MINK1 antibody (Bethyl). The protocol for immunoprecipitation was as described (Kaneko et al., 2011; Li et al., 2018).
Real-Time Quantitative PCR
Total RNA of animal tissues was isolated using Trizol reagent (Invitrogen). Quantitative PCR was performed using iQ5 System (Bio-Rad). The mouse GAPDH gene was used as the internal control for normalization of cycle number. The sequences of forward (F) and reverse (R) primers used are as following:
MINK1-F: catctgcagggagattctcagg
MINK1-R: ctgtagtcataggtggcatcgg
MAP4K4-F: ggtccatcacagaccttgtgaag
MAP4K4-R: tctgtagtcgtaagtggcgtctg
TNIK-F: GGCCTGAGTCACCTGCACCAGC
TNIK-R: GGGCACCTTCTGCCATCTCA
GAPDH-F: aggccggtgctgagtatgtc
GAPDH-R: tgcctgcttcaccaccttct
Statistical Analysis
Experiments for which we showed representative qPCR results and the quantification of cells number counting were performed at least 3 times independently. For all statistics in this report, the error bar is standard error of the mean, and p-value is from Student’s T-test: * is p<0.05, ** is p<0.01, NS is no significance with p>0.05. For Figure 3J, p-values are calculated by applying χ2-test.
Acknowledgments
YTI is supported by NIH grants (DK083450, GM107457), is a member of the UMass DERC (DK32520), and a member of the UMass Center for Clinical and Translational Science (UL1TR000161). YTI, WW and BZ are members of the Guangdong Innovative Research Team Program (No. 201001Y0104789252) and members of the Science and Technology Program of Guangzhou (No. 201704030044). JM is supported by an NIH grant (DK099510). MPC is funded for this work by NIH Grant DK030898.
Funding Information
National Institutes of Health, Grant numbers:DK083450, DK099510, DK030898, GM107457.
Footnotes
No competing financial interests.
Data availability statement
The data that support the findings of this study are openly available in figshare:https://figshare.com/s/d9c2750dc024adbb28e7
References
- Anazi S, Shamseldin HE, AlNaqeb D, Abouelhoda M, Monies D, Salih MA, Al-Rubeaan K, and Alkuraya FS. 2016. A null mutation in TNIK defines a novel locus for intellectual disability. Hum Genet. 135:773–778. [DOI] [PubMed] [Google Scholar]
- Bankaitis ED, Ha A, Kuo CJ, and Magness ST. 2018. Reserve Stem Cells in Intestinal Homeostasis and Injury. Gastroenterology. 155:1348–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beumer J, and Clevers H. 2016. Regulation and plasticity of intestinal stem cells during homeostasis and regeneration. Development. 143:3639–3649. [DOI] [PubMed] [Google Scholar]
- Brinkman AS, Murali SG, Hitt S, Solverson PM, Holst JJ, and Ney DM. 2012. Enteral nutrients potentiate glucagon-like peptide-2 action and reduce dependence on parenteral nutrition in a rat model of human intestinal failure. Am J Physiol Gastrointest Liver Physiol. 303:G610–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burette AC, Phend KD, Burette S, Lin Q, Liang M, Foltz G, Taylor N, Wang Q, Brandon NJ, Bates B, Ehlers MD, and Weinberg RJ. 2015. Organization of TNIK in dendritic spines. J Comp Neurol. 523:1913–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin AM, Hill DR, Aurora M, and Spence JR. 2017. Morphogenesis and maturation of the embryonic and postnatal intestine. Semin Cell Dev Biol. 66:81–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coba MP, Komiyama NH, Nithianantharajah J, Kopanitsa MV, Indersmitten T, Skene NG, Tuck EJ, Fricker DG, Elsegood KA, Stanford LE, Afinowi NO, Saksida LM, Bussey TJ, O’Dell TJ, and Grant SG. 2012. TNiK is required for postsynaptic and nuclear signaling pathways and cognitive function. J Neurosci. 32:13987–13999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degirmenci B, Hausmann G, Valenta T, and Basler K. 2018. Wnt Ligands as a Part of the Stem Cell Niche in the Intestine and the Liver. Prog Mol Biol Transl Sci. 153:1–19. [DOI] [PubMed] [Google Scholar]
- Demitrack ES, and Samuelson LC. 2016. Notch regulation of gastrointestinal stem cells. J Physiol. 594:4791–4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H, Wang W, Yu J, Zheng Y, Qing Y, and Pan D. 2015. Spectrin regulates Hippo signaling by modulating cortical actomyosin activity. Elife. 4:e06567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher GC, Diaz-de-la-Loza MD, Borreguero-Munoz N, Holder M, Aguilar-Aragon M, and Thompson BJ. 2018. Mechanical strain regulates the Hippo pathway in Drosophila. Development. 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu G, Xu Q, Qiu Y, Jin X, Xu T, Dong S, Wang J, Ke Y, Hu H, Cao X, Wang D, Cantor H, Gao X, and Lu L. 2017. Suppression of Th17 cell differentiation by misshapen/NIK-related kinase MINK1. J Exp Med. 214:1453–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H, Qi R, Zhou JJ, Ta AP, Yang B, Nakaoka HJ, Seo G, Guan KL, Luo R, and Wang W. 2018. Regulation of the Hippo Pathway by Phosphatidic Acid-Mediated Lipid-Protein Interaction. Mol Cell. 72:328–340 e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey KF, and Hariharan IK. 2012. The hippo pathway. Cold Spring Harb Perspect Biol. 4:a011288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S, Lewis P, Pevny L, and McMahon AP. 2002. Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Gene Expr Patterns. 2:93–97. [DOI] [PubMed] [Google Scholar]
- Kaneko S, Chen X, Lu P, Yao X, Wright TG, Rajurkar M, Kariya K, Mao J, Ip YT, and Xu L. 2011. Smad inhibition by the Ste20 kinase Misshapen. Proc Natl Acad Sci U S A. 108:11127–11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koontz LM, Liu-Chittenden Y, Yin F, Zheng Y, Yu J, Huang B, Chen Q, Wu S, and Pan D. 2013. The Hippo effector Yorkie controls normal tissue growth by antagonizing scalloped-mediated default repression. Dev Cell. 25:388–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larhammar M, Huntwork-Rodriguez S, Rudhard Y, Sengupta-Ghosh A, and Lewcock JW. 2017. The Ste20 Family Kinases MAP4K4, MINK1, and TNIK Converge to Regulate Stress-Induced JNK Signaling in Neurons. J Neurosci. 37:11074–11084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Li S, Mana-Capelli S, Roth Flach RJ, Danai LV, Amcheslavsky A, Nie Y, Kaneko S, Yao X, Chen X, Cotton JL, Mao J, McCollum D, Jiang J, Czech MP, Xu L, and Ip YT. 2014. The conserved misshapen-warts-Yorkie pathway acts in enteroblasts to regulate intestinal stem cells in Drosophila. Dev Cell. 31:291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Nirala NK, Nie Y, Chen HJ, Ostroff G, Mao J, Wang Q, Xu L, and Ip YT. 2018. Ingestion of Food Particles Regulates the Mechanosensing Misshapen-Yorkie Pathway in Drosophila Intestinal Growth. Dev Cell. 45:433–449 e436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Cho YS, Yue T, Ip YT, and Jiang J. 2015. Overlapping functions of the MAP4K family kinases Hppy and Msn in Hippo signaling. Cell Discov. 1:15038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Meng Z, Chen R, and Guan KL. 2018. The Hippo Pathway: Biology and Pathophysiology. Annu Rev Biochem. [DOI] [PubMed] [Google Scholar]
- Madison BB, Dunbar L, Qiao XT, Braunstein K, Braunstein E, and Gumucio DL. 2002. Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J Biol Chem. 277:33275–33283. [DOI] [PubMed] [Google Scholar]
- Mana MD, Kuo EY, and Yilmaz OH. 2017. Dietary Regulation of Adult Stem Cells. Curr Stem Cell Rep. 3:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X, Li P, Wang Y, Liang Z, Liu J, Li J, Jiang Y, Bao G, Li L, Zhu B, Ren Y, Zhao X, Zhang J, Liu Y, Yang J, and Liu P. 2017. CRB3 regulates contact inhibition by activating the Hippo pathway in mammary epithelial cells. Cell Death Dis. 8:e2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Z, Moroishi T, Mottier-Pavie V, Plouffe SW, Hansen CG, Hong AW, Park HW, Mo JS, Lu W, Lu S, Flores F, Yu FX, Halder G, and Guan KL. 2015. MAP4K family kinases act in parallel to MST½ to activate LATS½ in the Hippo pathway. Nat Commun. 6:8357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Z, Qiu Y, Lin KC, Kumar A, Placone JK, Fang C, Wang KC, Lu S, Pan M, Hong AW, Moroishi T, Luo M, Plouffe SW, Diao Y, Ye Z, Park HW, Wang X, Yu FX, Chien S, Wang CY, Ren B, Engler AJ, and Guan KL. 2018. RAP2 mediates mechanoresponses of the Hippo pathway. Nature. 560:655–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra JR, and Irvine KD. 2018. The Hippo Signaling Network and Its Biological Functions. Annu Rev Genet. 52:65–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke KP, Ackerman S, Dow LE, and Lowe SW. 2016. Isolation, Culture, and Maintenance of Mouse Intestinal Stem Cells. Bio Protoc. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D 2010. The hippo signaling pathway in development and cancer. Dev Cell. 19:491–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon CLC, Liu W, Song Y, Gomez M, Kulaberoglu Y, Zhang X, Xu W, Veraksa A, Hergovich A, Ghabrial A, and Harvey KF. 2018. A Hippo-like Signaling Pathway Controls Tracheal Morphogenesis in Drosophila melanogaster. Dev Cell. 47:564–575 e565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth Flach RJ, Guo CA, Danai LV, Yawe JC, Gujja S, Edwards YJ, and Czech MP. 2016. Endothelial Mitogen-Activated Protein Kinase Kinase Kinase Kinase 4 Is Critical for Lymphatic Vascular Development and Function. Mol Cell Biol. 36:1740–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth Flach RJ, Skoura A, Matevossian A, Danai LV, Zheng W, Cortes C, Bhattacharya SK, Aouadi M, Hagan N, Yawe JC, Vangala P, Menendez LG, Cooper MP, Fitzgibbons TP, Buckbinder L, and Czech MP. 2015. Endothelial protein kinase MAP4K4 promotes vascular inflammation and atherosclerosis. Nat Commun. 6:8995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin DC, and Levin MS. 2016. Mechanisms of intestinal adaptation. Best Pract Res Clin Gastroenterol. 30:237–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su T, Ludwig MZ, Xu J, and Fehon RG. 2017. Kibra and Merlin Activate the Hippo Pathway Spatially Distinct from and Independent of Expanded. Dev Cell. 40:478–490 e473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Reddy BV, and Irvine KD. 2015. Localization of Hippo signalling complexes and Warts activation in vivo. Nat Commun. 6:8402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan DW, and Barker N. 2014. Intestinal stem cells and their defining niche. Curr Top Dev Biol. 107:77–107. [DOI] [PubMed] [Google Scholar]
- Tetteh PW, Basak O, Farin HF, Wiebrands K, Kretzschmar K, Begthel H, van den Born M, Korving J, de Sauvage F, van Es JH, van Oudenaarden A, and Clevers H. 2016. Replacement of Lost Lgr5-Positive Stem Cells through Plasticity of Their Enterocyte-Lineage Daughters. Cell Stem Cell. 18:203–213. [DOI] [PubMed] [Google Scholar]
- Tetteh PW, Farin HF, and Clevers H. 2015. Plasticity within stem cell hierarchies in mammalian epithelia. Trends Cell Biol. 25:100–108. [DOI] [PubMed] [Google Scholar]
- Vitorino P, Yeung S, Crow A, Bakke J, Smyczek T, West K, McNamara E, Eastham-Anderson J, Gould S, Harris SF, Ndubaku C, and Ye W. 2015. MAP4K4 regulates integrin-FERM binding to control endothelial cell motility. Nature. 519:425–430. [DOI] [PubMed] [Google Scholar]
- Wang Q, Amato SP, Rubitski DM, Hayward MM, Kormos BL, Verhoest PR, Xu L, Brandon NJ, and Ehlers MD. 2016. Identification of Phosphorylation Consensus Sequences and Endogenous Neuronal Substrates of the Psychiatric Risk Kinase TNIK. J Pharmacol Exp Ther. 356:410–423. [DOI] [PubMed] [Google Scholar]
- Xue Y, Wang X, Li Z, Gotoh N, Chapman D, and Skolnik EY. 2001. Mesodermal patterning defect in mice lacking the Ste20 NCK interacting kinase (NIK). Development. 128:1559–1572. [DOI] [PubMed] [Google Scholar]
- Yan KS, Chia LA, Li X, Ootani A, Su J, Lee JY, Su N, Luo Y, Heilshorn SC, Amieva MR, Sangiorgi E, Capecchi MR, and Kuo CJ. 2012. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc Natl Acad Sci U S A. 109:466–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousefi M, Li L, and Lengner CJ. 2017. Hierarchy and Plasticity in the Intestinal Stem Cell Compartment. Trends Cell Biol. 27:753–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, and Pan D. 2018. Validating upstream regulators of Yorkie activity in Hippo signaling through scalloped-based genetic epistasis. Development. 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue M, Luo D, Yu S, Liu P, Zhou Q, Hu M, Liu Y, Wang S, Huang Q, Niu Y, Lu L, and Hu H. 2016. Misshapen/NIK-related kinase (MINK1) is involved in platelet function, hemostasis, and thrombus formation. Blood. 127:927–937. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Wang W, Liu B, Deng H, Uster E, and Pan D. 2015. Identification of Happyhour/MAP4K as Alternative Hpo/Mst-like Kinases in the Hippo Kinase Cascade. Dev Cell. 34:642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]