Abstract
A genetic basis for otitis media is established, however the role of rare variants in disease etiology is largely unknown. Previously a duplication variant within A2ML1 was identified as a significant risk factor for otitis media in an indigenous Filipino population and in US children. In this report exome and Sanger sequencing was performed using DNA samples from the indigenous Filipino population, Filipino cochlear implantees, US probands, and Finnish and Pakistani families with otitis media. Sixteen novel, damaging A2ML1 variants identified in otitis media patients were rare or low-frequency in population-matched controls. In the indigenous population, both gingivitis and A2ML1 variants including the known duplication variant and the novel splice variant c.4061+1G>C were independently associated with otitis media. Sequencing of salivary RNA samples from indigenous Filipinos demonstrated lower A2ML1 expression according to carriage of A2ML1 variants. Sequencing of additional salivary RNA samples from US patients with otitis media revealed differentially expressed genes that are highly correlated with A2ML1 expression levels. In particular RND3 is upregulated in both A2ML1 variant carriers and high-A2ML1-expressors. These findings support a role for A2ML1 in keratinocyte differentiation within the middle ear as part of otitis media pathology and the potential application of ROCK inhibition in otitis media.
Keywords: A2ML1, alpha-2-macroglobulin-like-1, exome sequencing, otitis media, RNA-sequencing
Introduction
Otitis media is a very common and costly disease in young children that can cause hearing loss and further lead to speech and reading difficulties (le Clerq et al. 2017; Khavarghazalani et al. 2016; Carroll et al. 2017; Cai & McPherson, 2017). Known risk factors for otitis media include young age, lack of breastfeeding, allergies, upper respiratory infection, second-hand smoke, low social status, day care attendance, multiple siblings and family history (Brennan-Jones et al. 2015; Zhang et al. 2014). In the US, otitis media incidence in children remains high at 6%, 23% and 46% at ages 3, 6 and 9 months, respectively (Chonmaitree et al. 2016). In pediatric and adult emergency departments, 2.2% and 6.8% of visits are due to ear complaints and nearly two-thirds of these complaints are diagnosed as otitis media (Kozin et al. 2015). Annual health care expenditures due to office visits, antibiotics, and surgeries for US children <30 months old is estimated to cost $5 billion (Casey & Pichichero, 2014).
The persistence of high incidence of otitis media in children despite maximization of public health interventions point to other risk factors including immune weaknesses and genetic predisposition. Heritability of otitis media ranges from 22–74% depending on otitis media type and cohort (Casselbrant et al. 1999; Hafrén et al. 2012). The identification of genetic risk factors and disease-related pathways is one area of otitis media study for which efficient tools are available but discovery remains very limited compared to other common complex, inflammatory, immune, or infectious disorders. While the most current catalog of genome-wide association studies (GWAS) lists >3,500 studies, only five studies (<0.15%) using common single nucleotide polymorphisms (SNP) identified significant loci for otitis media susceptibility, namely: intergenic rs10497394 on 2q31.1 (Allen et al. 2013); rs16974263 in the 19q13.2 region which is intronic to PRX (MIM 605725) encoding periaxin (Einarsdottir et al. 2016); FNDC1 (MIM 609991) at 6q25.3 (van Ingen et al. 2016); and rs76488276 at 16p12.3 which is ~94kb away from innate immune gene GP2 (MIM 602977; Li et al. 2017). In the largest GWAS to date including >120,000 European-descent individuals (Pickrell et al. 2016; Tian et al. 2017), 15 risk variants were identified, including four SNPs that were coding and/or intronic but in linkage disequilibrium with coding variants. However the heritability estimated to be due to these common variants is low at ~1% (Tian et al. 2017).
On the other hand, more studies have been done for the otitis media transcriptome, although these were mostly done using microarrays in rodent models and cultured human middle ear epithelial cells (HMEEC). In these studies an acute otitis media-like condition was induced with Streptococcus pneumoniae (Spn), non-typeable Haemophilus influenzae (ntHI), influenza A virus, TLR gene knockdown, particulate matter, or lipopolysaccharide (Li et al. 2003; Li-Korotky et al. 2004; Leichtle et al. 2009a, 2009b, 2012; Lee et al. 2011; Preciado et al. 2013; MacArthur et al. 2013; Kurabi et al. 2015; Hernandez et al. 2015). In ntHI-inoculated mice, top upregulated genes included inflammatory cytokines Cxcl1, Cxcl2 and IL-6 (Preciado et al. 2013; MacArthur et al. 2013; Hernandez et al. 2015). Differential expression of these genes were likewise detected in Tlr−/− mice, treatment with particulate matter, influenza infection and aging (Leichtle et al. 2012; Nielsen et al. 2016; Kim et al. 2016; Tong et al. 2004; Song et al. 2013). Gene ontology and network analyses identified genes involved in NFKB signaling, innate and immunoglobulin-mediated immune response, inflammatory response, complement activation and cytokine activity (MacArthur et al. 2013; Hernandez et al. 2015; Song et al. 2011). However the expression of these pro-inflammatory cytokines and enrichment of these pathways are not unique to middle ear but are also seen in various inflammatory processes in the nose, lung, and colon and in autoimmune diseases such as diabetes and rheumatoid arthritis (Bartling et al. 2009; Sadighi Akha et al. 2013; Ong et al. 2016; Chen et al. 2016; Vozarova et al. 2003; Kishimoto 1992). Nonetheless these studies increased our knowledge of multiple otitis media-related genes and pathways in a time- and context-dependent manner.
Pichichero et al. conducted two transcriptome studies using serum samples from children with culture-verified acute otitis media pre- and post-infection (Liu et al. 2012, 2013; Pichichero et al. 2016). Genes for host immune response such as complement activation, TLR, and cytokines were differentially expressed in Spn- and ntHI-infected children (Liu et al. 2012, 2013). Differential expression of genes for antimicrobial activity according to pathogen were suggested to correlate with less local inflammation and systemic illness during acute otitis media due to ntHI vs. Spn (Pichichero et al. 2016). Genes encoding lactotransferrin and peptidoglycan recognition protein were downregulated in Spn-infected children (Liu et al. 2012); both proteins are abundantly secreted in the apical air-liquid interface of mouse middle ear epithelium (Mulay et al. 2016). In ntHI-infected children, STAT1 (MIM 600555) and PTGS2 (MIM 600262) were downregulated (Liu et al. 2013), which was inconsistent with their upregulation in ntHI-treated mice and influenza-infected HMEECs (MacArthur et al. 2013; Tong et al. 2004), possibly in part due to the small sample size (n=4) per study (Liu et al. 2012, 2013; Pichichero et al. 2016)
Our previous discovery of A2ML1 (MIM 610627), which encodes alpha-2-macroglobulin-like-1, as an autosomal dominant gene for otitis media susceptibility suggested that rare variants play a role in otitis media pathology (Santos-Cortez et al. 2015). An indigenous Filipino population with a ~50% prevalence of otitis media was found to have an A2ML1 duplication variant as the strongest predictor for disease (Santos-Cortez et al. 2016b). The same duplication variant was also identified to be associated with otitis-prone status in US children (Santos-Cortez et al. 2015). The duplication variant is predicted to cause aberrant coding of alpha-2-macroglobulin-like-1, a middle-ear-localized protein that may play a role in mitigating mucosal damage during infection and bears close structural resemblance to alpha-2-macroglobulin (A2M), which is a known inflammatory marker in the middle ear and oral cavity (Santos-Cortez et al. 2015). Salivary A2M is increased during inflammatory conditions in the oral cavity, such as gingivitis and periodontal disease (Pederson et al. 1995). Furthermore in a microbiome study, indigenous Filipino carriers with the A2ML1 duplication and otitis media harbor bacterial pathogens that are commonly associated with dental and oropharyngeal infections e.g. Fusobacteria and Bacteroidetes (Santos-Cortez et al. 2016a), suggesting the possibility of A2ML1-related pathophysiologic processes in the oral cavity.
Here we report novel A2ML1 variants from exome and Sanger sequence data of Filipino, Finnish, Pakistani and US patients with otitis media. We further describe A2ML1 variants in relation to gene transcription and oral cavity conditions in indigenous Filipinos. Lastly using RNA-sequence analyses we demonstrate that upregulation of A2ML1 is correlated with differential expression of multiple genes, particularly genes within keratinocyte and epidermal cell differentiation pathways.
Methods
Study participants
Ethical approval of this study was obtained from: the University of the Philippines Manila Research Ethics Board; the National Commission on Indigenous Peoples, the Institutional Review Board (IRB) of the Helsinki University Hospital; IRB of the University of Maryland School of Medicine; IRB of the University of Texas Medical Branch (UTMB) Galveston; IRB of the Institute of Molecular Biology and Biotechnology, Bahauddin Zakariya University, Multan, Pakistan; and the Colorado Multiple IRB. For the indigenous Filipino population, community consent was obtained prior to study initiation. Individual informed consent was given by all adult participants and parents or guardians of children enrolled in the study.
The indigenous Filipino (Negrito) population is a relatively closed community resulting in extensive intermarriage within six founding families that can be traced genealogically by oral history to 6–7 generations ago. Few individuals who are from other Negrito tribes from adjacent islands married into the community. Due to their physical features of short stature, darkly pigmented skin, curly hair and flat noses, the community has suffered racial segregation from the general Filipino population, resulting in limited opportunities for education, economic advancement, socio-cultural assimilation and health care access. Their community is protected by the government, allowing access only to researchers who have fulfilled both the community’s and the government’s requirements for conducting research projects.
For the indigenous community, otitis media was diagnosed based on otoscopic findings at last examination. Chronic otitis media was diagnosed for eardrum perforations with smooth edges, usually with mucoid or mucopurulent discharge and thickened middle ear mucosa. Acute otitis media was diagnosed for hyperemic eardrums with or without perforation or discharge, while otitis media with effusion was identified if with dull non-hyperemic intact eardrums with poor mobility or visible fluid behind the eardrum. Healed otitis media was noted for previously diagnosed chronic, acute or effusive otitis media that has resolved on follow-up examination, or if with healed perforations or eardrum scarring. An individual with chronic, acute, effusive or healed otitis media was labeled as affected with otitis media. Of 135 individuals with DNA samples who were examined by otologists for otitis media, fifty agreed to be checked by dentists for gum disease and dental caries. For the dental exams, gingivitis is defined as gum inflammation with clinical signs and symptoms of bleeding and swelling, with probing depths at 1–3 mm. Extensive review of systems during medical history and physical examination of different parts of the body including skin ruled out additional features that may be part of a syndrome, immunodeficiency or other genetic disease.
From an indigenous community of ~200 individuals, 135 (67.5%) provided saliva samples for DNA isolation using Oragene DNA Collection Kits (DNAgenotek, Ottawa, Ontario, Canada). Of these 135 individuals with DNA samples, 124 (91.9%) have known relations that can be traced to a single pedigree. DNA samples were isolated from saliva using the manufacturer’s protocol. An additional 29 Filipino cochlear implantees provided DNA samples isolated from blood for a study on genetic variants for hearing impairment (Chiong et al. 2013, 2018; Truong et al. 2019).
The Finnish families (Hafrén et al. 2012; Einarsdottir et al. 2016) were ascertained from the Helsinki University Hospital upon referral of the proband for otitis media. Finnish patients were considered positive for otitis media if they had insertion of tympanostomy tubes, effusive otitis media for >2 months, or recurrent otitis media (i.e. >3 episodes in 6 months or >4 episodes in 12 months).
For Pakistani families with otitis media, detailed interviews were conducted with family members to gather information on pedigree structure, comorbidities, onset of disease and initial symptoms. The clinical diagnosis was based on ear discharge and air/bone conduction audiometry. The different groups of study participants are further described in Supp. Table S1.
Exome and Sanger sequencing
Six DNA samples from indigenous Filipinos with otitis media were submitted for exome sequencing at the University of Washington Center for Mendelian Genomics (UWCMG) on an Illumina HiSeq. Sequence capture was performed in solution with either the Roche NimbleGen SeqCap EZ Human Exome v.2.0 or the Big Exome 2011 Library. Fastq files were aligned to the hg19 human reference sequence using Burrows-Wheeler Aligner (BWA; Li & Durbin, 2009, 2010) to generate demultiplexed BAM files. Realignment of indel regions, recalibration of base qualities, and variant detection and calling were performed using the Genome Analysis Toolkit (GATK; McKenna et al. 2010) to produce VCF files. Annotation was performed with SeattleSeq. Two A2ML1 (RefSeq NM_144670.5) variants, a duplication and a splice variant, identified from exome data were Sanger-sequenced using the 135 DNA samples from indigenous Filipinos. Available exome sequence data from 29 cochlear implantees from the general Filipino population were also examined for A2ML1 variants (Chiong et al. 2018). Clinical data of A2ML1 variant carriers were then checked for otitis media diagnoses. For the Filipino population, identified A2ML1 variants were Sanger- sequenced using >180 DNA samples from unrelated individuals from the Cebu Longitudinal Health and Nutrition Survey cohort, which were not ascertained for otitis media (Adair et al. 2011).
DNA isolated from blood samples of 234 individuals with otitis media from 218 Finnish families were also submitted for exome sequencing at the University of Washington Northwest Genomics Center, and using the Roche NimbleGen SeqCap EZ Human Exome v.2.0 library, processed as described above. Identified A2ML1 variants were Sanger-sequenced in the probands and the rest of family members (Supp. Fig. S1).
From all participating family members of 16 Pakistani families with otitis media, peripheral blood samples were collected for DNA extraction. All coding exons of A2ML1 were Sanger-sequenced in two families. For 14 additional families, a DNA sample of an affected individual was submitted for exome sequencing. Genomic libraries were recovered for exome enrichment using the Agilent SureSelect Human Expanded All Exon V5 (62 Mb) kit. Libraries were sequenced on an Illumina HiSeq4000 with average 100× coverage. Alignment and variant calling were likewise performed using BWA and GATK, respectively. Sanger sequencing of the A2ML1 c.3676_3677delGC variant was performed for the rest of family members with DNA samples from two families PKOM-10 and PKOM-15 (Supp. Fig. S2).
Previously we Sanger-sequenced all 35 coding exons of A2ML1 using DNA samples from 123 otitis-prone children who were ascertained at UTMB (Patel et al. 2006; Santos-Cortez et al. 2015). These children were considered otitis-prone based on the following criteria: first episode of acute otitis media at <6 months; >3 episodes of acute otitis media within a 6-month period; >4 episodes of acute otitis media within a 12-month period; >6 episodes by 6 years old; or tympanostomy tube surgery for recurrent or persistent otitis media (Patel et al. 2006). In our previous publication, A2ML1 variants in these children were selected only if (1) it is the most deleterious variant even though there are multiple variants observed in the same child, (2) is absent in controls particularly if missense, and (3) if with scaled Combined Annotation Dependent Depletion (CADD) score >15 plus damaging prediction by at least two bioinformatics tools. For this report the Sanger sequence data from these otitis-prone children were reviewed for additional A2ML1 variants based on less stringent criteria. The decision to use less stringent criteria is based on our recent observations of common variants that are deemed polymorphisms due to higher MAF but are shown to be involved in otitis media susceptibility, and of otitis media patients carrying multiple variants from the same gene or multiple genes despite observation of autosomal dominant inheritance with reduced penetrance in families (Santos-Cortez et al. 2018).
For all identified variants whether previously published or novel, variants were classified as pathogenic/likely pathogenic or variant of unknown significance (VUS) based on current criteria from the American College of Medical Genetics (ACMG; Richards et al. 2015) using the Genetic Variant Interpretation Tool.
Bioinformatics, linkage and mixed model analyses
From exome or Sanger sequence data, variants were considered further if they have MAF less than 0.02 in the general population, have a scaled CADD score greater than 3, and are considered damaging by at least one additional bioinformatics tool (Table 1). For the Finnish, Pakistani and US populations, MAF was derived from the genome Aggregation Database (gnomAD) using Finnish, South Asian, non-Finnish European or Latino allele data, when appropriate.
Table 1.
A2ML1 Variants Identified in Multi-ethnic Families and Probands with Otitis Media
| Cohort-Patient ID |
hg19 chr12 Coordinate |
Variant (NM_144670.5)† | Control MAF‡ |
Protein Domain |
CADD | Mutation Taster | PolyPhen-2 HumDiv |
PROVEAN | SIFT | Mutation Assessor |
|---|---|---|---|---|---|---|---|---|---|---|
| I. Pathogenic/likely pathogenic | ||||||||||
| CIFIL-11 | 8975257 | c.10C>T (p.(Gln4*)) | 0 | MG1 | 35.0 | D(NMD) | -- | -- | -- | -- |
| UTMB-1039 | 8990070 | c.763C>T (p.(Gln255*)) | 0 | MG3 | 35.0 | D(NMD) | -- | -- | -- | -- |
| IPOM, UTMB-959/969/970 | 9004827 | c.2478_2485dupGGCTAAAT (p.(Ser829Trpfs*)) | 0 | MG7 | -- | D(NMD) | -- | -- | -- | -- |
| UTMB-1031 | 9009825 | c.2914G>T (p.(Glu972*)) | 0 | CUB/TED | 44.0 | D(NMD) | -- | -- | -- | -- |
| IPOM, CIFIL-21 | 9020954 | c.4061+1G>C | 0 | RBD | 25.7 | D | -- | -- | -- | -- |
| II. Variants of unknown significance | ||||||||||
| UTMB-1031 | 8975879 | c.164C>T (p.(Thr55Ile) | 0 | MG1 | 19.6 | P | PoD | D | D | M |
| UTMB-1178, UMN-123 | 8990963 | c.887T>C (p.(Val296Ala)) | 0.0009 | MG3 | 22.5 | P | B | D | D | M |
| UHF-269 | 8991701 | c.971–8C>T | 0.0006 | MG3 | 13.9 | D | -- | -- | -- | -- |
| UTMB-1017 | 8991805 | c.lO67C>G (p.(Pro356Arg)) | 0 | MG4 | 25.0 | P | PoD | D | D | M |
| UHF-101 | 8995789 | c.1308A>C (p.(Gln436His)) | 0.00005 | MG4 | 15.4 | P | B | N | T | M |
| UTMB-1026 | 8998818 | c.1683G>C (p.(Gln561His)) | 0 | MG6 | 33.0 | D | PoD | D | D | M |
| CIFIL-14 | 9001494 | c.2012T>C (p.(Leu671Pro)) | 0 | MG6/BRD | 13.3 | P | B | D | D/r | L |
| UTMB-998 | 9002825 | c.2189G>A (p.(Arg730His) | 0.00003 | MG6 | 21.3 | P | B | D | T/D | L |
| UHF-254/255 | 9002833 | c.2197T>C (p.(Phe733Leu)) | 0.004 | MG6 | 23.0 | D | B | D | T/D | L |
| UTMB-1027 | 9002864 | c.2228C>T (p.(Pro743Leu)) | 0 | MG6 | 23.4 | P | B | D | T | L |
| CIFIL-11 | 9004474 | c.2329G>A (p.(Gly777Arg)) | 0 | MG7 | 23.3 | D | PrD | D | D | H |
| UTMB-1031 | 9004573 | c.2428G>A (p.(Ala810Thr)) | 0 | MG7 | 25.2 | D | PrD | D | D | H |
| UTMB-1019 | 9004887 | c.2545G>T (p.(Asp849Tyr)) | 0 | MG7 | 15.6 | P | PoD | D | D | M |
| UTMB-1030 | 9006810 | c.2677C>T (p.(Arg893*)) | 0.00009 | MG7 | 34.0 | D(NMD) | -- | -- | -- | -- |
| 7 UHF families | 9007368 | c.2713–8C>A§ | 0.013 | MG7 | 4.8 | D | -- | -- | -- | -- |
| UTMB-1018 | 9009882 | c.2971G>C (p.(Ala991Pro)) | 0 | CUB/TED | 20.8 | P | B | N | D | L |
| UTMB-971 | 9009912 | c.3001C>T (p.(Arg1001Trp)) | 0 | CUB/TED | 24.5 | P | PrD | D | D | M |
| UTMB-959 | 9013882 | c.3491C>T (p.(Ala1164Val)) | 0 | CUB/TED | 27.5 | D | PrD | D | D | M |
| PKOM-10/15 | 9016563 | c.3676_3677delGC (p.(Ala1226Glnfs*34)) | 0.07 | CUB/TED | -- | P(NMD) | -- | -- | -- | -- |
| UTMB-1027 | 9027091 | c.4292C>T (p.(Ala1431Val)) | 0 | RBD | 25.6 | D | PrD | D | D | L |
Abbreviations: BRD, bait-region domain; CADD, Combined Annotation Dependent Depletion; CIFIL, Filipino cochlear implantee; CUB, complement protein subcomponent C1r/C1s, urchin embryonic growth factor and bone morphogenetic protein 1 domain; IPOM, indigenous Filipino cohort; MAF, minor allele frequency; MG, macroglobulin domain; RBD, receptor-binding domain; TED, thiol ester-containing domain; UHF, Finnish cohort; UMN, Minnesota cohort; UTMB, Texas cohort. MutationTaster: D, disease-causing; NMD, nonsense-mediated decay; P, polymorphism. PolyPhen-2: PrD, probably damaging; PoD, possibly damaging; B, benign. PROVEAN: D, deleterious; N, neutral. SIFT: D, deleterious; T, tolerated (“/” denotes multiple predictions depending on isoform). MutationAssessor: H, high; M, medium; L, low.
Novel variants are in bold font. Known variants were previously reported in Santos-Cortez et al. 2015.
For CIFIL and IPOM, control MAF is from the Cebu Longitudinal Health and Nutrition Survey. For UHF, UMN and PKOM, control MAF is from gnomAD Finnish, non-Finnish European and South Asian, respectively. For UTMB, control MAF is either from gnomAD non-Finnish European or Latino populations depending on self-reported ethnicity. UTMB IDs 959, 1030, 1031, 1039 and 1178 are Hispanic while the rest of UTMB IDs are non-Hispanic White. In some cases the gnomAD MAF in another population is higher, e.g. the c.2677C>T (p.(Arg893*)) variant has Latino MAF 0.00009 but has African and non-Finnish European MAF=0.0002.
Eight individuals with exome data from seven Finnish families carry the c.2713–8C>A variant (Supp. Fig. S1). From the exome data two common SNPs namely rs73037000 (chr12:8987285G>A) and rs1860967 (chr12:9013755C>T) flank the c.2713–8C>A variant, which comprise a short 26,470-bp haplotype found in all eight carriers. However four carriers have various common or low-frequency variants within the haplotype, suggesting that this haplotype is very old and multiple recombinations have occurred within the region.
For the two A2ML1 variants c.3676_3677delGC and c.4061+1G>C, two-point linkage analysis was performed using Superlink (Fishelson & Geiger, 2002). For the frameshift variant, linkage analysis was performed using variant MAF of 0.07, disease allele frequency of 0.01, and two modes of inheritance, namely: (a) affecteds-only with autosomal dominant inheritance, 90% penetrance and 5% phenocopy rate; and (b) autosomal recessive inheritance with full penetrance and no phenocopies (Supp. Fig. S2). For the A2ML1 c.4061+1G>C variant in the indigenous population, two-point linkage analysis was performed using an affecteds-only model with autosomal dominant inheritance, 90% penetrance, 5% phenocopy rate, disease allele frequency of 0.01 and variant MAF of 0.000001 (Supp. Fig. S3).
Fisher exact test was used to test associations between A2ML1 variants, otitis media and/or dental findings in the indigenous Filipino population. For mixed model analysis testing the association between otitis media and multiple variables including age, sex, carriage of at least one A2ML1 variant and gingivitis as fixed effects, grouping by family branch or household was used as a random effect variable.
RNA sequencing and analysis
For RNA studies, saliva samples were collected from nine indigenous Filipinos using the Oragene·RNA RE-100 kit. An additional 23 saliva samples were collected from otitis media patients undergoing surgery at the Children’s Hospital Colorado and the University of Colorado Hospital (Supp. Tables S1–S2) using Oragene·RNA kits. Salivary RNA was extracted according to the manufacturer’s protocol.
RNA samples were analyzed on an Agilent 2200 Tapestation and processed with the NuGen Trio RNA-Seq Kit at the University of Colorado Denver Genomics and Microarray Core. RIN values ranged from 4.5–7.7 (± S.D. 0.92). Sequencing libraries were sequenced on an Illumina HiSeq 4000 generating 50 bp single-end reads, with Filipino samples pooled at equimolar concentrations and sequenced in a single lane while Colorado samples were pooled and sequenced across three lanes. Reads were trimmed and adaptor sequences were removed using the FASTX-Toolkit (v0.0.13) prior to alignment to the hg38 human genome (GENCODE release 24) using STAR v2.5.3a (Dobin et al. 2013). Aligned reads were summarized at the gene level using featureCounts v1.5.2 with default parameters (Liao et al. 2014). For Filipino RNA samples, raw counts were analyzed according to variant carriage using Wilcoxon tests in R.
For the Colorado samples, count values from the technical replicates per sample were summed. Genes with an average count value of <3 were discarded resulting in ~12,000 remaining genes. The filtered count matrix was input to DESeq2 v1.20.0 (Love et al. 2014) in R v3.5.1 for differential expression analysis, while comparing high- vs. low-A2ML1-expressors. For correlation analysis, the normalized count matrix from DESeq2 was rlog-transformed, then correlation was performed for A2ML1 vs. all other genes using the Spearman method in R. Heatmap visualization of DESeq2 results was performed using the ‘pheatmap’ package in R. Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were identified using the Generally Applicable Gene-set Enrichment (GAGE v2.30.0) package in R (Luo et al. 2009). Log-2 fold changes calculated by DESeq2 were input to GAGE and processed using default parameters.
For validation of differentially expressed genes, cDNA was generated from 23 RNA samples from Colorado using the Invitrogen SuperScript IV protocol. Each cDNA sample was used for qPCR on a Bio-Rad machine (Hercules, CA, USA) in triplicate using the Applied Biosystems PowerUp SYBR green master mix and each set of primers for A2ML1 (NM_144670.5), differentially expressed genes AHNAK (NM_001620.2) and RND3 (NM_005168.4), and ACTB (NM_001101.3) as control. Fold change was determined according to ΔCT values.
Results
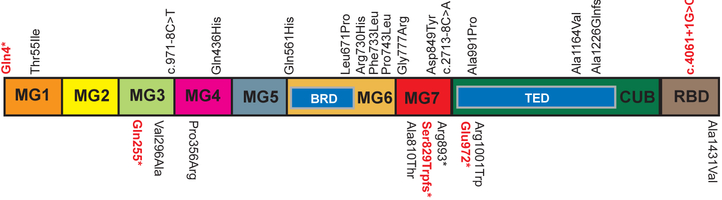
Sixteen novel, rare or low-frequency A2ML1 variants were identified from exome data of individuals with otitis media (Table 1). Seven of these variants were observed in UTMB otitis-prone probands. Notably two probands UTMB-959 and UTMB-1031 have 2–3 A2ML1 variants each with at least one as a novel variant (Table 1). Most of these variants found in UTMB probands did not pass previously set criteria due to lower CADD scores using the earlier software version and/or having only one damaging prediction, for example, c.2228C>T (p.(Pro743Leu)) in UTMB-1027 and c.2971G>C (p.(Ala991Pro)) in UTMB-1018. However due to lack of additional evidence, these novel variants along with some previously identified variants in UTMB probands were classified as VUS (Table 1; Fig. 1).
Fig. 1.
A2ML1 variants identified in families and probands with otitis media and their occurrence within protein domains. Domain names as in footnote to Table 1. Variants on top of the boxed representation of the A2ML1 protein are novel and are included in this report; below are previously published (Santos-Cortez et al. 2015). Variants in bold red font are pathogenic/likely pathogenic while the rest of variants are of unknown significance.
Four novel variants were identified as heterozygous in Finnish otitis media patients with exome data, including two missense and two non-canonical splice variants, all of which are predicted to be damaging (Table 1; Supp. Fig. S1). Two variants c.971–8C>T and c.1308A>C (p.(Gln436His)) occurred uniquely in a single Finnish proband. The c.2197T>C (p.(Phe733Leu)) was found in two individuals and has a MAF=0.004 in the Finnish population, while the low-frequency splice variant c.2713–8C>A was observed in seven Finnish families (Table 1). The Finnish probands carrying A2ML1 variants have no known syndromic features.
From the sequence data of 16 Pakistani families with otitis media, affected individuals from two families carried a frameshift variant c.3676_3677delGC (p.(Ala1226Glnfs*34)) that is predicted by MutationTaster (Schwarz et al. 2010) to be a polymorphism though leading to nonsense-mediated decay (Supp. Fig. S2). This variant has MAF=0.07 in gnomAD South Asian alleles and does not fully co-segregate with otitis media in the two Pakistani families using an autosomal dominant model or affecteds-only analysis (Supp. Fig. S2). Because three affected individuals of family PKOM-15 are homozygous for the frameshift variant and has otitis media from early childhood, the variant potentially co-segregates with autosomal recessive otitis media in branch 2 of family PKOM-15. However the LOD score is deflated compared to the maximum LOD score that is expected given pedigree branch structure and autosomal recessive inheritance with full penetrance (Supp. Fig. S2). All other A2ML1 variants identified in the sequence data of the Pakistani families were frequent (MAF>0.20) or deemed non-damaging to function. For these families, autosomal recessive variants that remain to be identified likely play a greater role in otitis media pathology.
Four A2ML1 variants were identified in three Filipino children who had cochlear implantation for congenital hearing impairment and who also have a history of otitis media. One child was heterozygous for two A2ML1 variants, namely c.10C>T (p.(Gln4*)) and c.2329G>A (p.(Gly777Arg)). Of these two variants, the stop variant is classified as pathogenic while the missense variant is a VUS (Table 1; Fig. 1). Unfortunately we have no parental DNA for testing if these two variants are compound heterozygous or in linkage disequilibrium. At age 8 months, a year prior to cochlear implantation, the child with these two variants had bilateral type C tympanograms. This patient is also homozygous for SLC26A4 (MIM 605646) c.706C>G (p.Leu236Val) which is the known cause for his profound hearing loss and enlarged vestibular aqueducts (Chiong et al. 2018). The second Filipino cochlear implantee is heterozygous for A2ML1 c.2012T>C (p.(Leu671Pro)) and had a flat tympanogram for the right implanted ear 14 months post-surgery at age 6 1/2 years. She is also homozygous for a splice variant c.2301+1G>T within OTOA (MIM 607038), likely the genetic cause of her congenital profound hearing loss (Truong et al. 2019). The third cochlear implantee is heterozygous for both a variant in hearing loss gene COL4A3 (MIM 120070) c.764C>T (p.(Thr255Met)) and A2ML1 c.4061+1G>C. All four A2ML1 variants identified in Filipino cochlear implantees were predicted to be damaging and were absent in the general Filipino population (Table 1).
Previously the A2ML1 c.2478_2485dupGGCTAAAT (p.Ser829Trpfs*9) variant was identified in the exome sequence data of two second-cousins from the indigenous Filipino population (Santos-Cortez et al. 2015). Out of 135 individuals (Table 2), this duplication variant is heterozygous in 62 and homozygous in eight indigenous Filipinos, and of these 70 variant carriers, 49 (70%) have otitis media. However, 33 individuals who were wildtype for the duplication variant also had otitis media. We submitted for exome sequencing DNA samples from four additional individuals who were wildtype for the duplication variant and come from different subpedigrees within the community. In two individuals with exome data, the A2ML1 splice variant c.4061+1G>C was identified. Of 135 indigenous individuals (Table 2), one is homozygous and 24 are heterozygous for the splice variant, including nine that are compound heterozygous for the two A2ML1 variants. Majority of those who carry the splice variant can be connected by three subpedigrees based on known relations (Supp. Fig. S3). Two-point linkage analysis for the splice variant resulted in a LOD score of 3.2 (θ=0; Supp. Fig. S3). In total, 86 (63.7%) of the screened population carry A2ML1 variants, and of these 60 (69.7%) have otitis media (Table 1). Conversely of 82 individuals with otitis media, 60 (73.2%) carry A2ML1 variants. Therefore among 135 indigenous Filipinos, the odds ratio for an A2ML1 variant carrier having otitis media is 2.8 (95%CI: 1.3, 6.2; p=0.006; Table 2).
Table 2.
A2ML1 Genotypes and Otitis Media Status for 135 Indigenous Filipinos
| A2ML1 genotype | Normal otoscopy | Otitis media† | Total |
|---|---|---|---|
| Wildtype | 27 | 22 | 49 |
| Heterozygous, duplication | 17 | 36 | 53 |
| Heterozygous, splice | 5 | 10 | 15 |
| Compound heterozygous | 1 | 8 | 9 |
| Homozygous, duplication | 3 | 5 | 8 |
| Homozygous, splice | 0 | 1 | 1 |
| Total with variant‡ | 26 | 60 | 86 |
| Overall total | 53 | 82 | 135 |
Of 60 A2ML1 variant carriers with otitis media, 22 (36.7%) have chronic otitis media, 23 (38.3%) with healed otitis media, 14 (23.3%) with effusive otitis media, and 1 with acute otitis media based on last exam. In contrast, among 22 wildtype individuals with otitis media, 9 (40.9%) have chronic otitis media, 5 (22.7%) with healed otitis media, another 5 (22.7%) with effusive otitis media and 3 (13.6%) with acute otitis media. Fifteen of the 22 wildtype individuals with otitis media carry the FUT2 c.604C>T (p.Arg202*) variant which also plays a role in otitis media susceptibility (Santos-Cortez et al. 2018; Supp. Fig. S3).
The Fisher exact odds ratio for an A2ML1 variant carrier having otitis media is 2.8 (95%CI: 1.3, 6.2; p=0.006).
Among 50 indigenous Filipinos with dental examinations, 28 (56%) had otitis media and 34 (68%) carried A2ML1 variants, which is comparable to the bigger cohort (Table 2). In addition, 52% were male with a mean age of 13.64 years (range 4 months - 47 years). Of these 50 individuals, 44 had dental caries, 24 had gum bleeding and 38 were diagnosed with gingivitis. None of these dental conditions were associated with A2ML1 variants, however gingivitis was associated with otitis media (OR=5.6, 95%CI: 1.1, 37.5; p=0.02). In mixed models analysis, in which family branch or household was analyzed as a random effect, otitis media was associated with both gingivitis (p=0.0007) and A2ML1 variants (p=0.048). Neither age nor sex was associated with otitis media, consistent with previous findings (Santos-Cortez et al. 2016b).
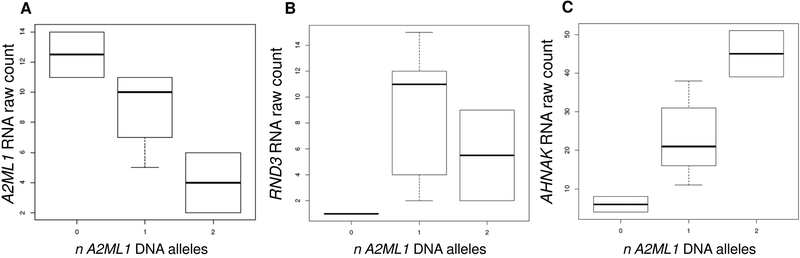
To further determine the effect of the A2ML1 duplication and splice variants in the indigenous Filipinos, RNA-sequencing was performed using salivary DNA from nine indigenous individuals, two of whom are wildtype, five are heterozygous, one compound heterozygous and one homozygous. RNA counts showed a decrease in salivary A2ML1 expression according to carriage of A2ML1 variants (Fig. 2A).
Fig. 2.
RNA counts from indigenous Filipinos for [A] A2ML1, [B] RND3 and [C] AHNAK. RND3 and AHNAK are upregulated in A2ML1-variant carriers (p=0.05) and high-A2ML1-expressors (RND3 2.7-log2foldΔ, adj-p=4.1×10−6; AHNAK 1.5-log2foldΔ, adj-p=0.003).
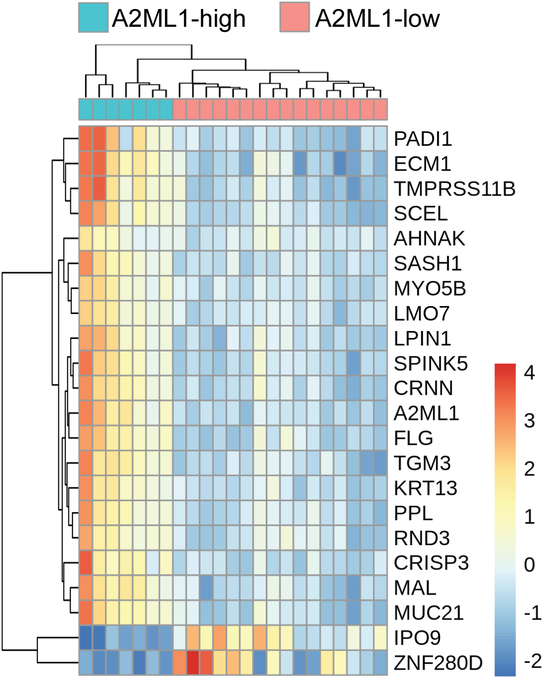
Because A2ML1 variants related to otitis media are rare, in order to study the effects of changes in A2ML1 expression in otitis media, salivary DNA samples from 23 otitis media patients from Colorado were also submitted for RNA-sequencing. For this analysis, based on the inflection of the count plot for A2ML1 in the Colorado samples (Supp. Fig. S4), differential expression analysis was initially performed using a threshold count of 33 for the Colorado samples, dividing the group into low- and high-A2ML1-expressors. Out of 12,107 post-filtered genes in salivary RNA, differential expression analysis using DESeq2 identified 745 (6.2%) genes at FDR-adjusted p<0.05, with 442 upregulated and 303 downregulated genes in high-A2ML1-expressors (Supp. Tables S3–S4). Because A2ML1 count values ranged from ~5–550 (±S.D.125), we also performed a genome-wide correlation analysis and A2ML1 values were compared against all other genes. A total of 41 genes were highly correlated with A2ML1 (r >±0.81, Bonferroni-corrected p<0.05; Table 3). Of note, 14 genes overlap among the top differentially expressed genes identified by both DESeq2 and correlation analysis, including: MUC21 (MIM 616991); PPL (MIM 602871); SPINK5 (MIM 605010); LPIN1 (MIM 605518); KRT13 (MIM 148065); SCEL (MIM 604112); CRNN (MIM 611312); TGM3 (MIM 600238); LMO7 (MIM 604362); MAL (MIM 188860); MYO5B (MIM 606540); SASH1 (MIM 607955); FLG (MIM 135940); and TMPRSS11B (Fig. 3; Table 3). Of the 41 correlated genes, AHNAK and RND3 were also found to be upregulated in indigenous Filipino A2ML1-variant carriers compared to wildtype (Fig. 2B–2C). Validation using qPCR on the Colorado RNA samples confirmed that RND3 is significantly enriched in high-A2ML1-expressors (Table 4).
Table 3.
Top 41 genes correlated with A2ML1 in Colorado patients and their known role in mucosal, epithelial or infectious traits
| Gene | Spearman R | p-value | Known Role in Mucosa/Infection/Epithelium | Reference |
|---|---|---|---|---|
| EMP1 | 0.919 | 3.30E-06 | Downregulated in nasal polyps | Yu et al. 2013 |
| SPRR3 | 0.903 | 3.28E-06 | Upregulated in chronic rhinosinusitis | Ramakrishnan et al. 2017 |
| RND3 | 0.896 | 3.19E-06 | Forms confluent layer of HMEECs | Mulay et al. 2016 |
| CSTB | 0.887 | 3.02E-06 | Upregulated in gingival epithelial cells after contact with Fusobacterium nucleatum | Yin & Dale 2007 |
| MUC21 | 0.886 | 3.00E-06 | Induced by RSV and hMPV | Banos-Lara et al. 2015 |
| RNR2 | 0.880 | 2.84E-06 | Expressed in olfactory mucosa | Bergstrom et al. 2007 |
| MTRNR2L8 | 0.876 | 2.73E-06 | -- | -- |
| KRT4 | 0.876 | 2.73E-06 | Upregulated in cholesteatoma | Britze et al. 2014 |
| PPL | 0.873 | 2.60E-06 | Autoimmune target in IPF | Taille et al. 2011 |
| SPINK5 | 0.870 | 2.50E-06 | Otitis media in Netherton syndrome | Renner et al. 2009 |
| TMPRSS11B | 0.869 | 2.47E-06 | Expressed in squamous epithelia of cervix, esophagus and oral cavity | Miller et al. 2014 |
| ECM1 | 0.867 | 2.40E-06 | Mucosal defect in lipoid proteinosis | Mirancea et al. 2007 |
| ERO1L | 0.867 | 2.40E-06 | High expression in gastric cancer with poor prognosis | Seol et al. 2016 |
| LPIN1 | 0.866 | 2.36E-06 | Downregulated due to high-fat diet along with changes in gastrointestinal mucosa | Zhou et al. 2016 |
| S100A10 | 0.862 | 2.23E-06 | Interaction with TRPV6 in airway and gut epithelia | Borthwick et al. 2008 |
| KRT13 | 0.858 | 2.09E-06 | Mutations lead to white sponge naevus in buccal mucosa | Liu et al. 2011 |
| TMPRSS11D | 0.858 | 2.09E-06 | Protease in airway epithelium | Menou et al. 2017 |
| AHNAK | 0.851 | 1.89E-06 | Upregulated in otitis media | This report |
| SPRR1B | 0.848 | 1.82E-06 | Upregulated in chronic rhinosinusitis | Ramakrishnan et al. 2017 |
| SCEL | 0.847 | 1.80E-06 | Forms cornified envelope in stratified squamous epithelium | Baden et al. 2005 |
| RNR1 | 0.845 | 1.76E-06 | -- | -- |
| CRNN | 0.843 | 1.73E-06 | Downregulated in esophageal cancer | Pawar et al. 2013 |
| CAST | 0.840 | 1.71E-06 | Decreases influenza A infection | Blanc et al. 2016 |
| PABPC1 | 0.837 | 1.71E-06 | Interaction with HSV-1 proteins | Dobrikova et al. 2010 |
| ADAM9 | 0.836 | 1.72E-06 | Increased in COPD bronchial cells | Wang et al. 2018 |
| ATF6B | −0.836 | 1.72E-06 | -- | -- |
| KRT78 | 0.834 | 1.74E-06 | Expressed in squamous epithelia | Langbein et al. 2016 |
| TGM3 | 0.833 | 1.76E-06 | Cross-links with HPV proteins | Brown et al. 2006 |
| LMO7 | 0.832 | 1.78E-06 | Upregulated by H.pylori in gastric cells | Lim et al. 2003 |
| AIM1 | 0.831 | 1.81E-06 | Expressed in EBV-infected cells | Cahir-McFarland et al. 2004 |
| ANXA1 | 0.831 | 1.81E-06 | Regulates intestinal mucosal injury, inflammation and repair | Babbin et al. 2008 |
| FAM129B | 0.831 | 1.81E-06 | Knockout results in delayed healing of wounded skin; keratinocyte-expressed | Oishi et al. 2012 |
| SEMA3F | −0.831 | 1.81E-06 | Regulates neurite growth in lingual epithelium | Vilbig et al. 2004 |
| ANXA2 | 0.830 | 1.84E-06 | Increased bacterial opsonization and clearance in otitis media | Renner et al. 2016 |
| MAL | 0.830 | 1.84E-06 | Methylated in dysplastic cervix | Mersakova et al. 2014 |
| EHF | 0.823 | 2.21E-06 | Regulates airway gene expression | Fossum et al. 2017 |
| ERGIC2 | 0.823 | 2.21E-06 | -- | -- |
| MYO5B | 0.820 | 2.47E-06 | Mutation causes intestinal microvillus defects | Muller et al. 2008 |
| SASH1 | 0.816 | 2.91E-06 | Mutation causes lentiginous skin phenotypes | Zhang et al. 2016 |
| FLG | 0.813 | 3.34E-06 | Lower expression in cholesteatoma | Nguyen et al. 2014 |
| KLK13 | 0.810 | 3.85E-06 | In periodontitis Porphyromonas gingivalis proteases degrade SPINK6 resulting in loss of KLK13 inhibition | Plaza et al. 2016 |
HMEEC, human middle ear epithelial cells; RSV, respiratory syncytial virus; hMPV, human metapneumovirus; IPF, idiopathic pulmonary fibrosis; HSV-1, herpes simplex virus-1; COPD, chronic obstructive pulmonary disease; HPV, human papilloma virus; EBV, Epstein-Barr virus.
Fig. 3.
Heatmap for top 20 differentially expressed genes (plus RND3 and AHNAK) according to A2ML1 expression in 23 otitis media patients from Colorado. For each data point, the row mean has been subtracted.
Table 4.
Fold Changes by Gene Based on qPCR ΔCT Values from Colorado patients
| Gene | High-Expressors | Low-Expressors | ΔΔCT | Fold Change |
|---|---|---|---|---|
| A2ML1 | 5.94 | 8.45 | −2.52 | 5.72* |
| AHNAK | 5.32 | 6.08 | −0.76 | 1.69 |
| RND3 | 4.05 | 6.53 | −2.48 | 5.58** |
p<0.05,
p<0.01
When genes with log-2-fold differential expression >2 were analyzed using the GAGE package, 88 KEGG pathways and gene ontology terms were enriched (p<0.05; Supp. Table S5). In particular, keratinocyte differentiation (p=2.9×10−6) and epidermal cell differentiation (p=3.2×10−5) were enriched in high-A2ML1-expressors (Supp. Table S5).
Because the mean age of low-A2ML1-expressors was noticeably higher than high-A2ML1-expressors (Supp. Table S2), we reanalyzed the Colorado dataset excluding samples with age >10 years (low-A2ML1-expressors mean 3.27 years vs. high-A2ML1-expressors mean 1.71 years). Differential expression analysis yielded similar results, albeit fewer significant genes were detected i.e. 371 upregulated and 157 downregulated genes in A2ML1-high-expressors (Supp. Table S6). Many of the same KEGG and GO terms were upregulated in high-A2ML1-expressors, however downregulated KEGG and GO terms were different (Supp. Table S6). We attribute these differences to the reduced number of significantly dysregulated genes. Regardless we conclude that the age range in the low-A2ML1expressors does not account for any of the dysregulated genes we observed.
Discussion
In this study we identified sixteen novel A2ML1 variants in otitis media patients, two of which are pathogenic based on ACMG criteria (Table 1; Fig. 1), further providing evidence to support a role for A2ML1 in middle ear mucosal pathology. Interestingly the variants that are deemed pathogenic are loss-of-function variants that are mostly predicted by MutationTaster to result in nonsense-mediated decay (Table 1). This is further supported by decreased transcription levels of A2ML1 due to the duplication and c.4061+1G>C variants (Fig. 2A). All the loss-of-function variants are also predicted to remove important domains, with at least the receptor-binding domain (RBD) being affected (Fig. 1). Based on the known crystallographic structure of homotetrameric A2M complex, after induction the RBD of one monomer protrudes into the circulation which results in the recognition of the tetramer by cell-surface receptors and leads to endocytosis, lysosomal degradation and clearance of the protease inhibitor complex along with the proteases it trapped (Marrero et al. 2012). Loss of the RBD is therefore predicted to result in lack of recognition of A2ML1 and failure of clearance of proteases for removal in order to prevent undue damage to mucosa. In our previous study, we showed that some missense variants are predicted to result in more subtle torsional changes in the macroglobulin (MG) or thiol-ester domains that interact to form the tetrameric structure (Santos-Cortez et al. 2015). In particular, most of the A2ML1 variants identified in otitis media patients occur within the MG6-MG7 domains, and these MG6-MG7 domains are required to close the superhelical structure in order to trap proteases that are baited by the bait-region domain (BRD) within MG6 (Marrero et al. 2012). Because most of the missense variants identified were seen in single probands and were classified as VUS, identification of additional otitis media patients with these specific variants will aid in establishing variant pathogenicity.
Three of the novel A2ML1 variants were recurrent: the splice variant c.4061+1G>C variant was identified in the indigenous Filipino population and in a cochlear implantee with otitis media from the general Filipino population; and a missense variant c.2197C>T (p.(Phe733Leu)) and a splice variant c.2713–8C>A in Finnish patients with otitis media. Here we also identified a frameshift variant in two Pakistani families, however this variant did not co-segregate with otitis media suggesting both intra-familial genetic heterogeneity (Rehman et al. 2015) and unidentified otitis media susceptibility variants with likely autosomal recessive inheritance (Supp. Fig. S2). In our previous study the A2ML1 duplication variant c.2478_2485dupGGCTAAAT that was initially identified to co-segregate with otitis media in the intermarried indigenous Filipino population was also genome-wide significant in three European-American and Hispanic-American children (Santos-Cortez et al. 2015; Table 1). Taken together, these findings suggest that A2ML1 variants conferring otitis media susceptibility are population-specific.
Although A2ML1 variants in general confer susceptibility to autosomal dominant nonsyndromic otitis media, the consanguineous Pakistani family PKOM-15 has additional cognitive, cranial and cardiac anomalies that do not co-segregate with the c.3676_3677delGC (p.(Ala1226Glnfs*34)) variant or otitis media. These additional phenotypes are not exactly the same but have some overlaps with the clinical features of intellectual disability, craniofacial and cardiac defects i.e. pulmonary valve stenosis in Noonan-like syndrome due to rare A2ML1 variants p.Arg592Leu, p.Arg802Leu and p.Arg802His (Vissers et al. 2015). Note that A2ml1-mutant zebrafish also had broad heads and failed cardiac looping (Vissers et al. 2015). A2ML1 variants have also been associated with hypertension, however the variants identified for hypertension do not overlap with otitis media-related variants, except for two variants p.Val296Ala and p.Arg893* which were observed separately in US probands with otitis media and in Europeans with hypertension (Table 1; Fig.1; Santos-Cortez et al. 2015; Surendran et al. 2016). This may suggest that allelic heterogeneity for A2ML1 contributes to phenotypic heterogeneity; for example, A2ML1 variants identified for otitis media cluster within the MG6 and MG7 domains while variants for hypertension tend to lie within the MG3 domain and the TED region of the CUB domain, alluding to differences in genotype-phenotype effects (Fig. 1; Surendran et al. 2016). On the other hand, it might also mean that A2ML1 variants confer susceptibility to otitis media in childhood and hypertension in later life.
While potential pleiotropic effects of A2ML1 remain to be fully resolved, our data showed that A2ML1 variants are not associated with oral disease. Previously A2M protein was found to be higher in gingival crevicular fluid of patients with gingivitis and chronic periodontitis, but whether A2M plays a role in pathogenesis or is merely a marker for inflammation is unknown (Ertugrul et al. 2013). We first hypothesized that the structural similarity between A2M and A2ML1 might translate to an increased risk for dental disease conferred by A2ML1 variants. However in this study A2ML1 variants were not associated with dental conditions including gum disease. Saliva possesses a vast array of enzymes, immunoglobulins, glycoproteins and cystatins that preserve the equilibrium of the microbiological flora in the oral cavity and are crucial in maintaining oral health (Amerongen & Veerman, 2002). This could account for why a defect in a single immunoregulatory protein in saliva would not necessarily lead to breakdown of protective barriers and consequently manifest as dental disease. It is also possible that in the head and neck region the mucosal protection conferred by A2ML1 is specific to the middle ear despite the upper airway being contiguous with the oral cavity (Santos-Cortez et al. 2015). Such compartmentalization of tissue-site expression of immune factors has been documented in other parts of the body with mucosal surfaces (Burgener et al. 2013).
On the other hand, both gingivitis and A2ML1 variants contribute independently to otitis media status. These significant effects are observed independent of age, and with correction for household or close familial relations as a random effect. When gum inflammation is present, retrograde movement of pathogens from the oral cavity into the nasopharynx and eventually the middle ear may contribute to otitis media pathogenesis. For example, clonal similarity between Fusobacterium nucleatum isolated from middle ear effusion and the oropharynx was documented previously (Topcuoglu et al. 2012). In middle ears of A2ML1 variant carriers, we observed a nominally significant increase in relative abundance of Fusobacteria and Bacteroidetes, which are more commonly identified as oral cavity or oropharyngeal pathogens (Santos-Cortez et al. 2016a; Suzuki et al. 2015; Yang et al. 2014). The additional finding of gingivitis as an independent risk factor for otitis media further supports the oral cavity as a potential source of otitis media pathogens in middle ears with weaker mucosal protection due to A2ML1 defects (Santos-Cortez et al. 2015, 2016a). It also suggests that prevention of gum disease may be an effective public health measure towards decreasing otitis media burden in the indigenous Filipino population.
Another significant finding is the co-upregulation of genes, including genes involved in keratinocyte and epidermal cell differentiation, when A2ML1 is upregulated. One of the byproducts of chronic otitis media is cholesteatoma, which is a collection of squamous debris encapsulated by keratinized epithelium in the middle ear (Maniu et al. 2014). In contrast to pseudostratified ciliated or simple squamous epithelium of healthy middle ear mucosa, the cholesteatoma capsule consists of keratinized stratified squamous epithelial matrix and a collagenous submatrix with inflammatory cells (Lim & Saunders, 1972). Given that many of the cellular, biochemical and regulatory factors favoring cholesteatoma growth, expansion and bony erosion remain unknown, the differentially expressed genes due to lower or higher A2ML1 expression may provide clues to the process of cholesteatoma formation.
Interestingly RND3 and AHNAK were shown to be upregulated in otitis media patients, whether in high-A2ML1-expressors or in A2ML1 variant carriers with lower A2ML1 expression. This may suggest that RND3 and AHNAK are involved in middle ear homeostasis in response to changes in A2ML1 expression. RND3 encodes Rho GTPase 3/RhoE which disorganizes the actin cytoskeleton by inhibiting ROCK-1, RhoA and Rac signaling while increasing cytokines NFKB and IRAK (Guasch et al. 2007). Interestingly Rac activates JNK, and in the infected mouse middle ear JNK inhibition resulted in decreased mucosal hyperplasia (Furukawa et al. 2007). In addition, injured astrocytes treated with Fasudil, a ROCK inhibitor, had widespread AHNAK labeling and downregulated protein degradation pathways, indicating a healthy state (O’Shea et al. 2015). Likewise treatment of the inflamed gut with Rnd3 reduced microvascular permeability (Breslin et al. 2016). ROCK inhibition is actually a common method for forming a confluent layer of HMEECs in culture studies (Mulay et al. 2016), however this application has not been considered for augmenting therapies for otitis media.
Overall our studies further support a role for A2ML1 in otitis media and reveal novel variants, genes and pathways related to otitis media pathology.
Supplementary Material
Acknowledgments:
We are deeply grateful to the patients who provided clinical data and samples and to the indigenous community for their graciousness and continued participation in this project. We thank C Brands, S Lu, V Ramakrishnan and S Kinnamon for general support and S Yousaf and R Ishaq for technical assistance.
Grant support: This work was funded by: the US National Institutes of Health (NIH) via grants from the National Human Genome Research Institute and the National Heart, Lung and Blood Institute UM1 HG006493 and U24 HG008956 (to D.A.N., M.J.B. and S.M.L.); the NIH -National Institute on Deafness and Other Communication Disorders R01 DC015004 (to R.L.P.S.C.) and R56 DC011803 (to S.R.); and by the Philippine Council for Health Research and Development - Department of Science and Technology via the Balik Scientist Program (to R.L.P.S.C.) and project no. FP 150010 (to C.M.C.).
Footnotes
Consortium:
Members of the University of Washington Center for Mendelian Genomics are listed in http://uwcmg.org/docs/Crediting_UW-CMG/UW_CMG_Banner.pdf
Disclosure: The authors declare no conflict of interest.
Web Resources
Burrows-Wheeler Aligner, bio-bwa.sourceforge.net
ClinVar, www.ncbi.nlm.nih.gov/clinvar (SCV000882552 - SCV000882559, SCV000900042 – SCV000900048)
Combined Annotation Dependent Depletion, cadd.gs.washington.edu
DESeq2 v1.20.0, bioconductor.org/packages/release/bioc/html/DESeq2.html
FastX-ToolKit v0.0.13, hannonlab.cshl.edu/fastx_toolkit
featureCounts v1.5.2, bioinf.wehi.edu.au/featureCounts
GAGE v2.30.0, bioconductor.org/packages/release/bioc/html/gage.html
GENCODE, www.gencodegenes.org
Gene Ontology Consortium, www.geneontology.org
Genetic Variant Interpretation Tool,
www.medschool.umaryland.edu/Genetic_Variant_Interpretation_Tool1.html/
Genome Aggregation Database, gnomad.broadinstitute.org
Genome Analysis Toolkit, software.broadinstitute.org/gatk/
GWAS Catalog, www.ebi.ac.uk/gwas/
KEGG, www.genome.jp/kegg/
MutationAssessor, mutationassessor.org/r3/
MutationTaster, www.mutationtaster.org
Online Mendelian Inheritance in Man, www.omim.org
Pheatmap v1.0.10, CRAN.R-project.org/package=pheatmap
PolyPhen-2, genetics.bwh.harvard.edu/pph2/
PROVEAN, provean.jcvi.org
R v3.5.1, www.r-project.org
SeattleSeq Annotation, snp.gs.washington.edu/SeattleSeqAnnotation151
STAR v2.5.3a, github.com/alexdobin/STAR
Superlink Online SNP 1.1, cbl-hap.cs.technion.ac.il/superlink-snp/
UCSC Genome Browser, genome.ucsc.edu
References
- Adair LS, Popkin BM, Akin JS, Guilkey DK, Gultiano S, Borja J, Perez L, Kuzawa CW, McDade T, & Hindin MJ (2011). Cohort profile: the Cebu Longitudinal Health and Nutrition Survey. International Journal of Epidemiology 40, 619–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen EK, Chen WM, Weeks DE, Chen F, Hou X, Mattos JL, Mychaleckyj JC, Segade F, Casselbrant ML, Mandel EM, Ferrell RE, Rich SS, Daly KA, & Sale MM (2013). A genome-wide association study of chronic otitis media with effusion and recurrent otitis media identified a novel susceptibility locus on chromosome 2. Journal of the Association for Research in Otolaryngology 14, 791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amerongen AN, & Veerman EC (2002). Saliva – the defender of the oral cavity. Oral Diseases 8, 12–22. [DOI] [PubMed] [Google Scholar]
- Babbin BA, Laukoetter MG, Nava P, Koch S, Lee WY, Capaldo CT, Peatman E, Severson EA, Flower RJ, Perretti M, Parkos CA, & Nusrat A (2008) Annexin A1 regulates intestinal mucosal injury, inflammation and repair. Journal of Immunology 181, 5035–5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden HP, Champliaud MF, Sundberg JP, & Viel A (2005). Targeted deletion of the sciellin gene resulted in normal development and maturation. Genesis 42, 219–228. [DOI] [PubMed] [Google Scholar]
- Banos-Lara MDR, Piao B, & Guerrero-Plata A (2015). Differential mucin expression by respiratory syncytial virus and human metapneumovirus infection in human epithelial cells. Mediators of Inflammation 2015, 347292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartling TR, & Drumm ML (2009). Oxidative stress causes IL8 promoter hyperacetylation in cystic fibrosis airway cell models. American Journal of Respiratory Cell and Molecular Biology 40, 58–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrom U Olsson JA, Hvidsten TR, Komorowski J, & Brandt I (2007). Differential gene expression in the olfactory bulb following exposure to the olfactory toxicant 2,6-dichlorophenyl methylsulphone and its 2,5-dichlorinated isomer in mice. Neurotoxicology 28, 1120–1128. [DOI] [PubMed] [Google Scholar]
- Blanc F, Furio L, Moisy D, Yen HL, Chignard M, Letavernier E, Naffakh N, Mok CK, & Si-Tahar M (2016). Targeting host calpain proteases decreases influenza A virus infection. American Journal of Physiology Lung Cellular and Molecular Physiology 310, L689–L699. [DOI] [PubMed] [Google Scholar]
- Borthwick LA, Neal A, Hobson L, Gerke V, Robson L, & Mulmo R (2008). The annexin 2-S100A10 complex and its association with TRPV6 is regulated by cAMP/PKA/CnA in airway and gut epithelia. Cell Calcium 44, 147–157. [DOI] [PubMed] [Google Scholar]
- Brennan-Jones CG, Whitehouse AJ, Park J, Hegarty M, Jacques A, Eikeboom RH, Swanepoel de W, White JD, & Jamieson SE (2015). Prevalence and risk factors for parent-reported recurrent otitis media during early childhood in the Western Australian Pregnancy Cohort (Raine) Study. Journal of Paediatrics and Child Health 51, 403–409. [DOI] [PubMed] [Google Scholar]
- Breslin JW, Daines DA, Doggett TM, Kurtz KH, Souza-Smith FM, Zhang XE, Wu MH, & Yuan SY (2016). Rnd3 as a novel target to ameliorate microvascular leakage. Journal of the American Heart Association 5, e003336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britze A, Birkler RI, Gregersen N, Ovesen T, & Palmfeldt J (2014). Large-scale proteomics differentiates cholesteatoma from surrounding tissues and identified novel proteins related to the pathogenesis. PLoS One 9, e104103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DR, Kitchin D, Qadadri B, Neptune N, Batteiger T, & Ermel A (2006). The human papillomavirus type 11 E1—E4 protein is a transglutaminase 3 substrate and induces abnormalities of the cornified cell envelope. Virology 345, 290–298. [DOI] [PubMed] [Google Scholar]
- Burgener A, Tjernlund A, Kaldensjo T, Abou M, McCorrister S, Westmacott GR, Mogk K, Ambrose E, Broliden K, & Ball B (2013). A systems biology examination of the human female genital tract shows compartmentalization of immune factor expression. Journal of Virology 87, 5141–5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahir-McFarland ED, Carter K, Rosenwald A, Giltnane JM, Henrickson SE, Staudt LM, & Kieff E (2004). Role of NF-kappa B in cell survival and transcription of latent membrane protein 1-expressing or Epstein-Barr virus latency III-infected cells. Journal of Virology 78, 4108–4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai T, & McPherson B (2017). Hearing loss in children with otitis media with effusion: a systematic review. International Journal of Audiology 56, 65–76. [DOI] [PubMed] [Google Scholar]
- Carroll JM, & Breadmore HL (2017). Not all phonological awareness deficits are created equal: evidence from a comparison between children with otitis media and poor readers. Developmental Science 21, e12588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey JR, & Pichichero ME (2014). Payment analysis of two diagnosis and management approaches of acute otitis media. Clinical Pediatrics 53, 865–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casselbrant ML, Mandel EM, Fall PA, Rockette HE, Kurs-Lasky M, Bluestone CD, & Ferrell RE (1999). The heritability of otitis media: a twin and triplet study. JAMA 282, 2125–2130. [DOI] [PubMed] [Google Scholar]
- Chen CY, Chang JT, Ho YF, & Shyu AB (2016). MiR-26 down-regulates TNF-α/NF-KB signalling and IL-6 expression by silencing HMGA1 and MALT1. Nucleic Acids Research 44, 3772–3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiong CM, Cutiongco-de la Paz EM, Reyes-Quintos MRT, Tobias CAM, Hernandez K, & Santos-Cortez RLP (2013). GJB2 variants and auditory outcomes among Filipino cochlear implantees. Audiology & Neurotology Extra 3, 1–8. [Google Scholar]
- Chiong CM, Reyes-Quintos MRT, Yarza TKL, Tobias-Grasso CAM, Acharya A, Leal SM, Mohlke KL, Mayol NL, Cutiongco-de la Paz EM, & Santos-Cortez RLP (2018). The SLC26A4 c.706C>G (p.Leu236Val) variant is a frequent cause of hearing impairment in Filipino cochlear implantees. Otology Neurotology 39, e726–e730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chonmaitree T, Trujillo R, Jennings K, Alvarez-Fernandez P, Patel JA, Loeffelholz MJ, Nokso-Koivisto J, Matalon R, Pyles RB, Miller AL, & McCormick DP (2016). Acute otitis media and other complications of viral respiratory infection. Pediatrics 137, e20153555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, & Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrikova E, Shveygert M, Walters R, & Gromeier M (2010). Herpes simplex virus proteins ICP27 and UL47 associate with polyadenylate-binding protein and control its subcellular distribution. Journal of Virology 84, 270–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einarsdottir E, Hafrén L, Leinonen E, Bhutta MF, Kentala E, Kere J, & Mattila PS (2016). Genome-wide association analysis reveals variants on chromosome 19 that contribute to childhood risk of chronic otitis media with effusion. Scientific Reports 6, 33240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertugrul AS, Sahin H, Dikilitas A, Alpaslan N, & Bozoglan A (2013). Evaluation of beta-2 microglobulin and alpha-2 macroglobulin levels in patients with different periodontal diseases. Australian Dental Journal 58, 170–175. [DOI] [PubMed] [Google Scholar]
- Fishelson M, & Geiger D (2002). Exact genetic linkage computations for general pedigrees. Bioinformatics 18 Suppl 1, S189–S198. [DOI] [PubMed] [Google Scholar]
- Fossum SL, Mutolo MJ, Tugores A, Ghosh S, Randell SH, Jones LC, Leir SH, & Harris A (2017). Ets homologous factor (EHF) has critical roles in epithelial dysfunction in airway disease. The Journal of Biological Chemistry 292, 10938–10949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa M, Ebmeyer J, Pak K, Austin DA, Melhus A, Webster NJ, & Ryan AF (2007). Jun N-terminal protein kinase enhances middle ear mucosal proliferation during bacterial otitis media. Infection and Immunity 75, 2562–2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guasch RM, Blanco AM, Perez-Arago A, Miñambres R, Talens-Visconti R, Peris B, & Guerri C (2007). RhoE participates in the stimulation of the inflammatory response induced by ethanol in astrocytes. Experimental Cell Research 313, 3779–3788. [DOI] [PubMed] [Google Scholar]
- Hafrén L, Kentala E, Einarsdottir E, Kere J, & Mattila PS (2012). Current knowledge of the genetics of otitis media. Current Allergy and Asthma Reports 12, 582–589. [DOI] [PubMed] [Google Scholar]
- Hernandez M, Leichtle A, Pak K, Webster NJ, Wasserman SI, & Ryan AF (2015). The transcriptome of a complete episode of acute otitis media. BMC Genomics 16, 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khavarghazalani B, Farahani F, Emadi M, & Hosseni Dastgerdi Z (2016). Auditory processing abilities in children with chronic otitis media with effusion. Acta Oto-Laryngologica 136, 456–459. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Kim SY, Kwon JY, Kim YJ, Hun Kang S, Jang WH, Lee JH, Seo MW, Song JJ, Seo YR, & Park MK (2016). Identification of potential novel biomarkers and signaling pathways related to otitis media induced by diesel exhaust particles using transcriptomic analysis in in an in vivo system. PLoS One 11, e0166044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto T (1992). Interleukin-6 and its receptor in autoimmunity. Journal of Autoimmunity 5 Suppl A, 123–132. [DOI] [PubMed] [Google Scholar]
- Kozin ED, Sethi RK, Remenschneider AK, Kaplan AB, Del Portal DA, Gray ST, Shrime MG, & Lee DJ (2015). Epidemiology of otologic diagnoses in United States emergency departments. Laryngoscope 125, 1926–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurabi A, Lee J, Wong C, Pak K, Hoffman HM, Ryan AF, & Wasserman SI (2015). The inflammasome adaptor ASC contributes to multiple innate immune processes in the resolution of otitis media. Innate Immunity 21, 203–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langbein L, Eckhart L, Fischer H, Rogers MA, Praetzel-Wunder S, Parry DA, & Kittstein W (2016). Localisation of keratin K78 in the basal layer and first suprabasal layers of stratified epithelia completes expression catalogue of type II keratins and provides new insights into sequential keratin expression. Cell and Tissue Research 363, 735–750. [DOI] [PubMed] [Google Scholar]
- le Clercq CMP, van Ingen G, Ruytjens L, Goedegebure A, Moll HA, Raat H, Jaddoe VWV, Baatenburg de Jong RJ, & van der Schroeff MP (2017). Prevalence of hearing loss among children 9 to 11 years old: the Generation R Study. JAMA Otolaryngology-Head & Neck Surgery 143, 928–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YW, Chung Y, Juhn SK, Kim Y, & Lin J (2011). Activation of the transforming growth factor beta pathway in bacterial otitis media. Annals of Otology, Rhinology & Laryngology 120, 204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leichtle A, Hernandez M, Lee J, Pak K, Webster NJ, Wollenberg B, Wasserman SI, Ryan AF (2012). The role of DNA sensing and innate immune receptor TLR9 in otitis media. Innate Immunity 18, 3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leichtle A, Hernandez M, Pak K, Webster NJ, Wasserman SI, Ryan AF (2009a). The toll-like receptor adaptor TRIF contributes to otitis media pathogenesis and recovery. BMC Immunology 10, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leichtle A, Hernandez M, Pak K, Yamasaki K, Cheng CF, Webster NJ, Ryan AF, & Wasserman SI (2009b). TLR4-mediated induction of TLR2 signaling is critical in the pathogenesis and resolution of otitis media. Innate Immunity 15, 205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, & Durbin R (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, & Durbin R (2010). Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HS, Doyle WJ, Swarts JD, Lo CY, & Hebda PA (2003). Mucosal expression of genes encoding possible upstream regulators of Na+ transport during pneumococcal otitis media. Acta Oto-Laryngologica 123, 575–582. [DOI] [PubMed] [Google Scholar]
- Li J, van Ingen G, Li YR, Goedegebure A, March ME, Jaddoe VWV, Mentch FD, Thomas K, Wei Z, Chang T, Uitterlinden AG, Moll HA, van Duijn CM, Rivadeneira F, Raat H, Baatenburg de Jong RJ, Sleiman PM, van der Schroeff MP, & Hakonarson H (2017). Genome-wide association study of otitis media in children (Abstract #1908). Presented at the 67th Annual Meeting of The American Society of Human Genetics, October 20, 2017, Orlando, Florida. [Google Scholar]
- Liao Y, Smyth GK, & Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Li-Korotky HS, Swarts JD, Hebda PA, & Doyle WJ (2004). Cathepsin gene expression profile in rat pneumococcal otitis media. Laryngoscope 114, 1032–1036. [DOI] [PubMed] [Google Scholar]
- Lim DJ, & Saunders WH (1972). Acquired cholesteatoma: light and electron microscopic observations. Annals of Otology, Rhinology & Laryngology 81, 1–11. [PubMed] [Google Scholar]
- Lim JW, Kim H, & Kim KH (2003). Cell adhesion-related gene expression by Helicobacter pylori in gastric epithelial AGS cells. The International Journal of Biochemistry & Cell Biology 35, 1284–1296. [DOI] [PubMed] [Google Scholar]
- Liu K, Chen L, Kaur R, & Pichichero ME (2012). Transcriptome signature in young children with acute otitis media due to Streptococcus pneumoniae. Microbes and Infection 14, 600–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Chen L, Kaur R, & Pichichero ME (2013). Transcriptome signature in young children with acute otitis media due to non-typeable Haemophilus influenzae. International Immunology 25, 353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Li Q, Gao Y, Song S, & Hua H (2011). Mutational analysis in familial and sporadic patients with white sponge naevus. British Journal of Dermatology 165, 448–451. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, & Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Friedman MS, Shedden K, Hankenson KD, & Woolf PJ (2009). GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics 10, 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur CJ, Hausman F, Kempton JB, Choi D, & Trune DR (2013). Otitis media impacts hundreds of mouse and middle and inner ear genes. PLoS One 8, e75213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniu AA, Harabagiu O, Perde Schrepler M, Catana A, Fanuta B, & Mogoanta CA (2014). Molecular biology of cholesteatoma. Romanian Journal of Morphology and Embryology 55, 7–13. [PubMed] [Google Scholar]
- Marrero A, Duquerroy S, Trapani S, Goulas T, Guevara T, Andersen GR, Navaza J, Sottrup-Jensen L, & Gomis-Rüth FX (2012). The crystal structure of human α2-macroglobulin reveals a unique molecular cage. Angewandte Chemie International Edition 51, 3340–3344. [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, & DePristo MA (2010). The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research 20, 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menou A, Duitman J, Flajolet P, Sallenave JM, Mailleux AA, & Crestani B (2017). Human airway trypsin-like protease, a serine protease involved in respiratory diseases. American Journal of Physiology Lung Cellular and Molecular Physiology 312, L657–L668. [DOI] [PubMed] [Google Scholar]
- Mersakova S, Visnovsky J, Holubekova V, Nachajova M, Kudela E, Danko J, & Lasabova Z (2014). Detection of methylation of the promoter region of the MAL and CADM1 genes by pyrosequencing in cervical carcinoma. Neuro Endocrinology Letters 35, 619–623. [PubMed] [Google Scholar]
- Miller GS, Zoratti GL, Murray AS, Bergum C, Tanabe LM, & List K (2014). HATL5: a cell surface serine protease differentially expressed in epithelial cancers. PLoS One 9, e87675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirancea N, Hausser I, Metze D, Stark HJ, Boukamp P, & Breitkreutz D (2007). Junctional basement membrane anomalies of skin and mucosa in lipoid proteinosis (hyalinosis cutis et mucosae). Journal of Dermatological Science 45, 175–185. [DOI] [PubMed] [Google Scholar]
- Mulay A, Akram KM, Williams D, Armes H, Russell C, Hood D, Armstrong S, Stewart JP, Brown SD, Bingle L, & Bingle CD (2016). An in vitro model of murine middle epithelium. Disease Models & Mechanisms 9, 1405–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller T, Hess MW, Schiefermeier N, Pfaller K, Ebner HL, Heinz-Erian P, Ponstingl H, Partsch J, Rollinghoff B, Kohler H, Berger T, Lenhartz H, Schlenck B, Houwen RJ, Taylor CJ, Zoller H, Lechner S, Goulet O, Utermann G, Ruemmele FM, Huber LA, & Janecke AR (2008). MYO5B mutations cause microvillus inclusion disease and disrupt epithelial cell polarity. Nature Genetics 40, 1163–1165. [DOI] [PubMed] [Google Scholar]
- Nguyen KH, Suzuki H, Ohbuchi T, Wakasugi T, Koizumi H, Hashida K, Baba R, Morimoto H, & Doi Y (2014). Possible participation of acidic pH in bone resorption in middle ear cholesteatoma. Laryngoscope 124, 245–250. [DOI] [PubMed] [Google Scholar]
- Nielsen MC, Friis M, Martin-Bertelsen T, Winther O, Friis-Hansen L, & Cayé-Thomasen P (2016). The middle ear immune defense changes with age. European Archives of Oto-Rhino-Laryngology 273, 81–86. [DOI] [PubMed] [Google Scholar]
- Oishi H, Itoh S, Matsumoto K, Ishitobi H, Suzuki R, Ema M, Kojima T, Uchida K, Kato M, Miyata T, & Takahashi S (2012). Delayed cutaneous wound healing in Fam129b/Minerva-deficient mice. Journal of Biochemistry 152, 549–555. [DOI] [PubMed] [Google Scholar]
- Ong CB, Kumagai K, Brooks PT, Brandenberger C, Lewandowski RP, Jackson-Humbles DN, Nault R, Zacharewski TR, Wagner JG, & Harkema JR (2016). Ozone-induced type 2 immunity in nasal airways: development and lymphoid cell dependence in mice. American Journal of Respiratory Cell and Molecular Biology 54, 331–340. [DOI] [PubMed] [Google Scholar]
- O’Shea RD, Lau CL, Zulaziz N, Maclean FL, Nisbet DR, Horne MK, & Beart PM (2015). Transcriptomic analysis and 3D bioengineering of astrocytes indicate ROCK inhibition produces cytotrophic astrogliosis. Frontiers in Neuroscience 9, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel JA, Nair S, Revai K, Grady J, Saeed K, Matalon R, Block S, & Chonmaitree T (2006). Association of proinflammatory cytokine gene polymorphisms with susceptibility to otitis media. Pediatrics 118, 2273–2279. [DOI] [PubMed] [Google Scholar]
- Pawar H, Maharudraiah J, Kashyap MK, Sharma J, Srikanth SM, Choudhary R, Chavan S, Sathe G, Manju HC, Kumar KV, Vijayakumar M, Sirdeshmukh R, Harsha HC, Prasad TS, Pandey A, & Kumar RV (2013). Downregulation of cornulin in esophageal squamous cell carcinoma. Acta Histochemica 115, 89–99. [DOI] [PubMed] [Google Scholar]
- Pederson ED, Stanke SR, Whitener SJ, Sebastiani PT, Lamberts BL, & Turner DW (1995). Salivary levels of alpha 2-macroglobulin, alpha 1-antitrypsin, C-reactive protein, cathepsin G and elastase in humans with or without destructive periodontal disease. Archives of Oral Biology 40, 1151–1155. [DOI] [PubMed] [Google Scholar]
- Pichichero ME (2016). Ten-year study of acute otitis media in Rochester, NY. Pediatric Infectious Disease Journal 35, 1027–1032. [DOI] [PubMed] [Google Scholar]
- Pickrell JK, Berisa T, Liu JZ, Ségurel L, Tung JY, & Hinds DA (2016). Detection and interpretation of shared genetic influences on 42 human traits. Nature Genetics 48, 709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaza K, Kalinska M, Bochenska O, Meyer-Hoffert U, Wu Z, Fischer J, Falkowski K, Sasiadek L, Bielecka E, Potempa B, Kozik A, Potempa J, & Kantyka T (2016). Gingipains of Porphyromonas gingivalis affect the stability and function of serine protease inhibitor of Kazal-type 6 (SPINK6), a tissue inhibitor of human kallikreins. Journal of Biological Chemistry 291, 18753–18764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preciado D, Burgett K, Ghimbovschi S, & Rose M (2013). NTHi induction of Cxcl2 and middle ear mucosal metaplasia in mice. Laryngoscope 123, E66–E71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan VR, Gonzalez JR, Cooper SE, Barham HP, Anderson CB, Larson ED, Cool CD, Diller JD, Jones K, & Kinnamon SC (2017). RNA sequencing and pathway analysis identify tumor necrosis factor alpha driven small proline-rich protein dysregulation in chronic rhinosinusitis. American Journal of Rhinology & Allergy 31, 283–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman AU, Santos-Cortez RL, Drummond MC, Shahzad M, Lee K, Morell RJ, Ansar M, Jan A, Wang X, Aziz A, Riazuddin S, Smith JD, Wang GT, Ahmed ZM, Gul K, Shearer AE, Smith RJ, Shendure J, Bamshad MJ, Nickerson DA, University of Washington Center for Mendelian Genomics, Hinnant J, Khan SN, Fisher RA, Ahmad W, Friderici KH, Riazuddin S, Friedman TB, Wilch ES, & Leal SM (2015). Challenges and solutions for gene identification in the presence of familial locus heterogeneity. European Journal of Human Genetics 23, 1207–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner B, Tong HH, Laskowski J, Jonscher K, Goetz L, Woolaver R, Hannan J, Li YX, Hourcade D, Pickering MC, Holers VM, & Thurman JM (2016). Annexin A2 enhances complement activation by inhibiting factor H. Journal of Immunology 196, 1355–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner ED, Hartl D, Rylaarsdam S, Young ML, Monaco-Shawver L, Kleiner G, Markert ML, Stiehm ER, Belohradsky BH, Upton MP, Torgerson TR, Orange JS, & Ochs HD (2009). Comel-Netherton syndrome defined as primary immunodeficiency. Journal of Allergy and Clinical Immunology 124, 536–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL, ACMG Laboratory Quality Assurance Committee. (2015). Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in Medicine 17, 405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadighi Akha AA, Theriot CM, Erb-Downward JR, McDermott AJ, Falkowski NR, Tyra HM, Rutkowski DT, Young VB, & Huffnagle GB (2013). Acute infection of mice with Clostridium difficile leads to eIF2α phosphorylation and pro-survival signaling as part of the mucosal inflammatory response. Immunology 140, 111–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Cortez RLP, Chiong CM, Frank DN, Ryan AF, Giese APJ, Bootpetch Roberts T, Daly KA, Steritz MJ, Szeremeta W, Pedro M, Pine H, Yarza TKL, Scholes MA, Llanes E.G.d.V., Yousaf S, Friedman N, Tantoco MLC, Wine TM, Labra PJ, Benoit J, Ruiz AG, de la Cruz RAR, Greenlee C, Yousaf A, Cardwell J, Nonato RMA, Ray D, Ong KMC, So E, Robertson CE, Dinwiddie J, Lagrana-Villagracia SM, University of Washington Center for Mendelian Genomics, Gubbels SP, Shaikh RS, Cass SP, Einarsdottir E, Lee NR, Schwartz DA, Gloria-Cruz TLI, Bamshad MJ, Yang IV, Kere J, Abes GT, Prager JD, Riazuddin S, Chan AL, Yoon PJ, Nickerson DA, Cutiongco-de la Paz EM, Streubel SO, Reyes-Quintos MRT, Jenkins HA, Mattila P, Chan KH, Mohlke KL, Leal SM, Hafrén L, Chonmaitree T, Sale MM, & Ahmed ZM (2018). FUT2 variants confer susceptibility to familial otitis media. American Journal of Human Genetics 103, 679–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Cortez RLP, Chiong CM, Reyes-Quintos MRT, Tantoco MLC, Wang X, Acharya A, Abbe I, Giese AP, Smith JD, Allen EK, Li B, Cutiongco-de la Paz EM, Garcia MC, Llanes E.G.d.V., Labra PJ, Gloria-Cruz TLI, Chan AL, Wang GT, Daly KA, Shendure J, Bamshad MJ, Nickerson DA, Patel JA, Riazuddin S, Sale MM, University of Washington Center for Mendelian Genomics, Chonmaitree T, Ahmed ZM, Abes GT, & Leal SM (2015). Rare A2ML1 variants confer susceptibility to otitis media. Nature Genetics 47, 917–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Cortez RLP, Hutchinson DS, Ajami NJ, Reyes-Quintos MRT, Tantoco MLC, Labra PJ, Lagrana SM, Pedro M, Llanes E.G.d.V., Gloria-Cruz TLI, Chan AL, Cutiongco-de la Paz EM, Belmont JW, Chonmaitree T, Abes GT, Petrosino JF, Leal SM, & Chiong CM (2016a). Middle ear microbiome differences in indigenous Filipinos with chronic otitis media due to a duplication in the A2ML1 gene. Infectious Diseases of Poverty 5, 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Cortez RLP, Reyes-Quintos MRT, Tantoco MLC, Abbe I, Llanes E.G.d.V., Ajami NJ, Hutchinson DS, Petrosino JF, Padilla CD, Villarta RL Jr., Gloria-Cruz TLI, Chan AL, Cutiongco-de la Paz EM, Chiong CM, Leal SM, & Abes GT (2016b). Genetic and environmental determinants of otitis media in an indigenous Filipino population. Otolaryngology-Head and Neck Surgery 155, 856–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz JM, Rödelsperger C, Schuelke M, & Seelow D (2010). MutationTaster evaluates disease-causing potential of sequence alterations. Nature Methods 7, 575–576. [DOI] [PubMed] [Google Scholar]
- Seol SY, Kim C, Lim JY, Yoon SO, Hong SW, Kim JW, Choi SH, & Cho JY (2016). Overexpression of endoplasmic reticulum oxidoreduction 1-α (ERO1L) is associated with poor prognosis of gastric cancer. Cancer Research and Treatment 48, 1196–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JJ, Kwon SK, Cho CG, Park SW, & Chae SW (2011). Microarray analysis of microRNA expression in LPS induced inflammation of human middle ear epithelial cells (HMEECs). International Journal of Pediatric Otorhinolaryngology 75, 648–651. [DOI] [PubMed] [Google Scholar]
- Song JJ, Kwon JY, Park MK, & Seo YR (2013). Microarray analysis of gene expression alteration in human middle ear epithelial cells induced by micro particle. International Journal of Pediatric Otorhinolaryngology 77, 1760–1764. [DOI] [PubMed] [Google Scholar]
- Surendran P, Drenos F, Young R, Warren H, Cook JP, Manning AK, Grarup N, Sim X, Barnes DR, Witkowska K, Staley JR, Tragante V, Tukiainen T, Yaghootkar H, Masca N, Freitag DF, Ferreira T, Giannakopoulou O, Tinker A, Harakalova M, Mihailov E, Liu C, Kraja AT, Fallgaard Nielsen S, Rasheed A, Samuel M, Zhao W, Bonnycastle LL, Jackson AU, Narisu N, Swift AJ, Southam L, Marten J, Huyghe JR, Stančáková A, Fava C, Ohlsson T, Matchan A, Stirrups KE, Bork-Jensen J, Gjesing AP, Kontto J, Perola M, Shaw-Hawkins S, Havulinna AS, Zhang H, Donnelly LA, Groves CJ, Rayner NW, Neville MJ, Robertson NR, Yiorkas AM, Herzig KH, Kajantie E, Zhang W, Willems SM, Lannfelt L, Malerba G, Soranzo N, Trabetti E, Verweij N, Evangelou E, Moayyeri A, Vergnaud AC, Nelson CP, Poveda A, Varga TV, Caslake M, de Craen AJ, Trompet S, Luan J, Scott RA, Harris SE, Liewald DC, Marioni R, Menni C, Farmaki AE, Hallmans G, Renström F, Huffman JE, Hassinen M, Burgess S, Vasan RS, Felix JF, CHARGE-Heart Failure Consortium, Uria-Nickelsen M, Malarstig A, Reily DF, Hoek M, Vogt T, Lin H, Lieb W, Traylor M, Markus HF, METASTROKE Consortium, Highland HM, Justice AE, Marouli E, GIANT Consortium, Lindström J, Uusitupa M, Komulainen P, Lakka TA, Rauramaa R, Polasek O, Rudan I, Rolandsson O, Franks PW, Dedoussis G, Spector TD, EPIC-InterAct Consortium, Jousilahti P, Männistö S, Deary IJ, Starr JM, Langenberg C, Wareham NJ, Brown MJ, Dominiczak AF, Connell JM, Jukema JW, Sattar N, Ford I, Packard CJ, Esko T, Mägi R, Metspalu A, de Boer RA, van der Meer P, van der Harst P, Lifelines Cohort Study, Gambaro G, Ingelsson E, Lind L, de Bakker PI, Numans ME, Brandslund I, Christensen C, Petersen ER, Korpi-Hyövälti E, Oksa H, Chambers JC, Kooner JS, Blakemore AI, Franks S, Jarvelin MR, Husemoen LL, Linneberg A, Skaaby T, Thuesen B, Karpe F, Tuomilehto J, Doney AS, Morris AD, Palmer CN, Holmen OL, Hveem K, Willer CJ, Tuomi T, Groop L, Käräjämäki A, Palotie A, Ripatti S, Salomaa V, Alam DS, Shafi Majumder AA, Di Angelantonio E, Chowdhury R, McCarthy MI, Poulter N, Stanton AV, Sever P, Amouyel P, Arveiler D, Blankenberg S, Ferrières J, Kee F, Kuulasmaa K, Müller-Nurasyid M, Veronesi G, Virtamo J, Deloukas P, Wellcome Trust Case Control Consortium, Elliott P, Understanding Society Scientific Group, Zeggini E, Kathiresan S, Melander O, Kuusisto J, Laakso M, Padmanabhan S, Porteous D, Hayward C, Scotland G, Collins FS, Mohlke KL, Hansen T, Pedersen O, Boehnke M, Stringham HM, EPIC-CVD Consortium, Frossard P, Newton-Cheh C, CHARGE+ Exome Chip Blood Pressure Consortium, Tobin MD, Nordestgaard BG, T2D-GENES Consortium, GoT2DGenes Consortium, ExomeBP Consortium, CHD Exome+ Consortium, Caulfield MJ, Mahajan A, Morris AP, Tomaszewski M, Samani NJ, Saleheen D, Asselbergs FW, Lindgren CM, Danesh J, Wain LV, Butterworth AS, Howson JM, & Munroe PB (2016). Trans-ancestry meta-analyses identify rare and common variants associated with blood pressure and hypertension. Nature Genetics 48, 1151–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Kurono Y, Ikeda K, Watanabe A, Iwamoto A, Totsuka K, Kaku M, Iwata S, Kadota J, & Hanaki H (2015). Nationwide surveillance of 6 otorhinolaryngological infectious diseases and antimicrobial susceptibility pattern in the isolated pathogens in Japan. Journal of Infection and Chemotherapy 21, 483–491. [DOI] [PubMed] [Google Scholar]
- Taille C, Grootenboer-Mignot S, Boursier C, Michel L, Debray MP, Fagart J, Barrientos L, Mailleux A, Cigna N, Tubach F, Marchal-Somme J, Soler P, Chollet-Martin S, & Crestani B (2011). Identification of periplakin as a new target for autoreactivity in idiopathic pulmonary fibrosis. American Journal of Respiratory and Critical Care Medicine 183, 759–766. [DOI] [PubMed] [Google Scholar]
- Tian C, Hromatka BS, Kiefer AK, Eriksson N, Noble SM, Tung JY, & Hinds DA (2017). Genome-wide association and HLA region fine-mapping studies identify susceptibility loci for multiple common infections. Nature Communications 8, 599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong HH, Long JP, Li D, & DeMaria TF (2004). Alteration of gene expression in human middle ear epithelial cells induced by influenza A virus and its implication for the pathogenesis of otitis media. Microbial Pathogenesis 37, 193–204. [DOI] [PubMed] [Google Scholar]
- Topcuoglu N, Keskin F, Ciftci S, Paltura C, Kulekci M, Ustek D, & Kulekci G (2012). Relationship between oral anaerobic bacteria and otitis media with effusion. International Journal of Medical Sciences 9, 256–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong BT, Yarza TKL, Bootpetch Roberts T, Roberts S, Xu J, Steritz MJ, Tobias-Grasso CAM, Azamian M, Lalani SR, Mohlke KL, Lee NR, Cutiongco-de la Paz EM, Reyes-Quintos MRT, Santos-Cortez RLP, & Chiong CM (2019). Exome sequencing reveals novel variants and unique allelic spectrum for hearing impairment in Filipino cochlear implantees. Clinical Genetics, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ingen G, Li J, Goedegebure A, Pandey R, Li YR, March ME, Jaddoe VW, Bakay M, Mentch FD, Thomas K, Wei Z, Chang X, Uitterlinden AG, Moll HA, van Duijn CM, Rivadeneira F, Raat H, Baatenburg de Jong RJ, Sleiman PM, van der Schroeff MP, & Hakonarson H (2016). Genome-wide association study for acute otitis media in children identifies FNDC1 as disease contributing gene. Nature Communications 7, 12792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilbig R, Cosmano J, Giger R, & Rochlin MW (2004). Distinct roles for Sema3A, Sema3F, and an unidentified trophic factor in controlling the advance of geniculate axons to gustatory lingual epithelium. Journal of Neurocytology 33, 591–606. [DOI] [PubMed] [Google Scholar]
- Vissers LE, Bonetti M, Paardekooper Overman J, Nillesen WM, Frints SG, de Ligt J, Zampino G, Justino A, Machado JC, Schepens M, Brunner HG, Veltman JA, Scheffer H, Gros P, Costa JL, Tartaglia M, van der Burgt I, Yntema HG, & den Hertog J (2015). Heterozygous germline mutations in A2ML1 are associated with a disorder clinically related to Noonan syndrome. European Journal of Human Genetics 23, 317–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vozarova B, Fernandez-Real JM, Knowler WC, Gallart L, Hanson RL, Gruber JD, Ricart W, Vendrell J, Richart C, Tataranni PA, & Wolford JK (2003). The interleukin-6 (−174) G/C promoter polymorphism is associated with type-2 diabetes mellitus in Native Americans and Caucasians. Human Genetics 112, 409–413. [DOI] [PubMed] [Google Scholar]
- Wang X, Polverion F, Rojas-Quintero J, Zhang D, Sanchez J, Yambayev I, Lindqvist E, Virtala R, Djukanovic R, Davies DE, Wilson S, O’Donnell R, Cunoosamy D, Hazon P, Higham A, Singh D, Olsson H, & Owen CA (2018). A disintegrin and a metalloproteinase-9 (ADAM9): a novel proteinase culprit with multifarious contributions to COPD. American Journal of Respiratory and Critical Care Medicine, doi: 10.1164/rccm.201711-2300OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang NY, Zhang Q, Li JL, Yang SH, & Shi Q (2014). Progression of periodontal inflammation in adolescents is associated with increased number of Porphyromonas gingivalis, Prevotella intermedia, Tannerella forsynthesis and Fusobacterium nucleatum. International Journal of Paediatric Dentistry 24, 226–233. [DOI] [PubMed] [Google Scholar]
- Yin L, & Dale BA (2007). Activation of protective responses in oral epithelial cells by Fusobacterium nucleatum and human β-defensin-2. Journal of Medical Microbiology 56, 976–987. [DOI] [PubMed] [Google Scholar]
- Yu XM, Li CW, Li YY, Liu J, Lin ZB, Li TY, Zhao L, Pan XL, Shi L, & Wang de Y (2013). Down-regulation of EMP1 is associated with epithelial hyperplasia and metaplasia in nasal polyps. Histopathology 63, 686–695. [DOI] [PubMed] [Google Scholar]
- Zhang J, Cheng R, Liang J, Ni C, Li M, & Yao Z (2016). Lentiginous phenotypes caused by diverse pathogenic genes (SASH1 and PTPN11): clinical and molecular discrimination. Clinical Genetics 90, 372–377. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Xu M, Zhang J, Zeng L, Wang Y, & Zheng QY (2014). Risk factors for chronic and recurrent otitis media-a meta-analysis. PLoS One 9, e86397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Wang Y, Jiang Y, Diao Y, Strappe P, Prenzler P, Ayton J, & Blanchard C (2016). Deep-fried oil consumption in rats impairs glycerolipid metabolism, gut histology and microbiota structure. Lipids in Health and Disease 15, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.