Abstract
The common marmoset, a New World (platyrrhine) monkey, is currently being fast-tracked as a nonhuman primate model species, especially for genetic modification but also as a general-purpose model for research on the brain and behavior bearing on the human condition. Compared to the currently dominant primate model, the catarrhine macaque monkey, marmosets are notable for certain evolutionary specializations, including their propensity for twin births, their very small size (a result of phyletic dwarfism), and features related to their small size (rapid development and relatively short lifespan), that result in these animals yielding experimental results more rapidly and at lower cost. Macaques, however, have their own advantages. Importantly, macaques are more closely related to humans (which are also catarrhine primates) than are marmosets, sharing approximately 20 million more years of common descent, and are demonstrably more similar to humans in a variety of genomic, molecular, and neurobiological characteristics. Furthermore, the very specializations of marmosets that make them attractive as experimental subjects, such as their rapid development and short lifespan, are ways in which marmosets differ from humans and in which macaques more closely resemble humans. These facts warrant careful consideration of the trade-offs between convenience and cost, on the one hand, and biological realism, on the other, in choosing between nonhuman primate models of human biology. Notwithstanding the advantages marmosets offer as models, prudence requires continued commitment to research on macaques and other primate species.
Keywords: brain evolution, gene editing, genetic modification, model organism, animal model, primates
Why Primates?
Historically, the most popular mammalian species for experimental neuroscientific research have been members of the rodent order, including rats (especially Rattus norwegicus) and mice (Mus musculus) [Manger et al. 2008], the latter being increasingly favored in this era of translational research owing to its tractability for genetic manipulation. Rodents have been favored because of their convenience, being easy to breed and maintain in captivity, and by the belief that the important features of mammalian biology (including neurobiology) are shared widely, if not universally, among mammals [Logan 2001, 2002].
Notwithstanding the convenience of rodents as research animals, there is increasing evidence of the inadequacy of rodents in translational research, at least for certain disorders [Check Hayden 2014; Kolata 2013]. Currently available genetically modified mouse models of human neurological disorders, especially age-related diseases such as Alzheimer’s disease, Parkinson’s disease, amyotrophic lateral sclerosis, and Huntington’s disease, have failed to reproduce important features of the human disease phenotype (e.g., Burns et al. [2015]; Levine et al. [2004]; Li and Li [2012]; Onos et al. [2016]; Perrin [2014]). Furthermore, drugs developed in rodents frequently do not produce comparable effects in humans (e.g. Hyman [2012]; Perlman [2016]; Rittirsch et al. [2006]; van Dyck [2018]). Sometimes, much higher drug doses in rodents are necessary to yield effects seen at lower doses in nonhuman primates and humans, as for example with the adrenergic a-2A agonist guanfacine (A. Arnsten, pers. comm.), which has proven useful for treatment of ADHD [Arnsten 2010] and PTSD [Arnsten et al. 2015]. In addition to the deficiencies of mouse models of neurological diseases, mouse models of human immunology, inflammation, and sepsis have also been found wanting (see Mestas and Hughes [2004], and citations therein), and the genetics of development in rodents and humans differ in important respects (e.g., Liao and Zhang [2008]).
In retrospect, it is not difficult to understand why rodent models of the human brain have important limitations: rather than sharing a common brain organization, comparative studies have revealed that mammals are remarkably diverse. For example, rodents and primates differ in the numbers of cortical areas, the fiber systems that link them, the numbers of neurons in a cortical column, the morphologies of pyramidal and nonpyramidal cells, the pattern of peptide expression by cortical neurons, the regional and laminar distribution of neurotransmitters and receptors, and the embryology of the cortex (for reviews, see Hof and Sherwood [2007]; Kaas [2012]; Molnar and Clowry [2012]; Preuss [2001]; Preuss [2007]; Preuss [2010]) — and that is just the anatomy of the cortex! Differences in neuroanatomy and other aspects of brain biology are only to be expected, given the substantial phylogenetic distance between primates and rodents (Figure 1).
Figure 1.

The ordinal and supraordinal phylogeny of the primates. The primates (black lines) are a monophyletic group, the closest relatives of which are the Scandentia (tree shrews) and Dermoptera (colugos or flying lemurs). Together, these clades constitute the Euarchonta. The sister group of the euarchontans is Glires, comprised of Order Rodentia and Order Glires (rabbits and pikas). Collectively, these groups constitute the Euarchontoglires. The primate phylogeny is from Fleagle [Fleagle 2013]. The phylogeny of Euarchontoglires is from Murphy et al. [Murphy et al. 2001].
Why Marmosets?
It is evident that rodent models of the human brain and human neurological diseases have shortcomings that, for many purposes, outweigh the benefits of convenience. The case for models more closely related to humans—nonhuman primate models, that is— has received new impetus. But in what nonhuman primate should effort and resources be concentrated? The most intensively studied nonhuman primates are species of the genus Macaca, most commonly Macaca mulatta, the rhesus macaque. Macaques have many advantages: we have had captive colonies for many decades, and know much about their rearing, housing needs, behavior, and neurobiology. Macaques, however, have liabilities as well: they are relatively large, mature slowly and have a low reproductive rate (compared to smaller primates), are quite aggressive (especially M. mulatta), and their saliva and other body fluids and tissues can harbor a virus (macacine herpes virus, also known as herpes B virus) that is potentially lethal to humans [Wisely et al. 2018], necessitating specialized engineering and administrative controls. Collectively, these factors make macaque colonies expensive to establish and maintain, with the result that the number of macaque facilities available to researchers is limited. Moreover, the diversity of primate species available for neuroscientific research has been markedly reduced in recent years.
Recently, support has been growing for increased use of the common marmoset, Callithrix jacchus, as a nonhuman primate model. In this role, marmosets have been placed at the center of the Japanese national brain initiative, the Brain/MINDS project [Cyranoski 2014; Okano et al. 2016], and the importance of marmosets has been highlighted in recent special issues of Neuroscience Research (2015) and Developmental Neurobiology (2017); see especially the essays by Mitchell and Leopold [2015], Burkart and Finkenwirth [2015], and Miller [2017]). Interest in marmosets as research animals is not new: there is, in fact, a long history of research on marmosets, particularly (although not exclusively) in the neurosciences, where the small size of the brain facilitates cortical mapping studies [Miller 2017]. What is different today is that marmosets have come to be seen as having important advantages compared to macaques. Among the favorable features of are their small size, rapid development, rapid reproduction, and a lifespan that is longer than that of mice, but shorter than that of macaques [Okano et al. 2012; Salmon 2016; Sasaki 2015; Tokuno et al. 2015] (Figure 2). In addition, marmosets are safer to work with, as they do not carry macacine herpes virus. These factors make them less expensive to rear and maintain than macaques, and the rapid development and short lifespan of marmosets means that studies of brain development and aging can be completed more quickly in marmosets than in macaques. Marmosets also possess features of pro-social behavior that, due to convergent evolution, mirror those of humans, including female-male bonding and aspects of cooperative breeding, such as infant carrying and food sharing by adult males, the infants’ siblings, and even non-kin [Burkart and Finkenwirth 2015; Erb and Porter 2017; Fernandez-Duque et al. 2009; Schiel and Souto 2017]. Marmosets are also sometimes said to be monogamous, although they have been observed in the wild to form monogamous, polygynous, and polyandrous groupings (reviewed by Schiel and Souto [2017]).
Figure 2.

Some characteristics of marmosets and macaques relevant to their utility as models of human neurobiology. Marmoset photograph courtesy of Texas Biomedical Research Institute and Kathy West Studios. The macaque photograph is courtesy of the Yerkes National Primate Research Center, Emory University.
Marmosets, then, are increasingly being viewed as a general nonhuman primate model with which to address issues of human behavioral and cognitive neuroscience across the lifespan. Nevertheless, there is no question that the greatest impetus for the current interest in marmosets stems from the opportunity they provide to apply techniques for genetic modification that have been developed in mice to a nonhuman primate, including the powerful CRISPR/Cas9 techniques (Kishi et al. [2014]; Okano et al. [2012]; Sasaki [2015]; see also Ledford [2016]). Compared to macaques, the high rate of reproduction and rapid maturation exhibited by marmosets could accelerate the process of creating and evaluating genetically modified models. American researchers, perceiving the need for marmosets to fill this role, have decried their present lack of availability in the United States [Servick 2018].
The question I want to address here is whether marmosets are really the best choice as a nonhuman primate stand-in for humans, paying particular attention to the advantages and disadvantages of marmosets compared to the currently dominant macaque model. My purpose is not to discourage research on one species or the other. In fact, I hold the view that our understanding of the fundamental principles underlying nervous system structure and function benefits from studying a diverse array of species [Preuss 2000; Preuss and Robert 2014; Striedter et al. 2014], as does the reconstruction of brain evolution. That said, the variety of primate species available for study by neuroscientists has shrunk dramatically over the past several decades. For this reason, and because of the concentration of resources that must be achieved to develop genetically modified animals, for practical purposes, we have been nearly reduced to having to choose between marmosets and macaques, or some combination of the two. While the choice of marmosets over macaques will seem to many to be an obvious one, given their many practical advantages, I think the case is less clear. To understand why, it is necessary to delve into the phylogeny and comparative biology of the primates.
What is a marmoset? A very short course in primate phylogeny and brain evolution
Primate diversity and taxonomy.
It is tempting to think of nonhuman primates as a homogenous entity-“the primate” or “the monkey.” Yet primates are a diverse group of mammals consisting of no fewer than 200 species in several major subgroups, varying markedly in ecology, behavior, and social organization [Fleagle 2013]. With a few exceptions, the composition and relationships among these major subgroups are now largely agreed upon, and in any event the areas of controversy do not affect the following presentation. The platyrrhine primates, which include the marmosets, have historically been one of those exceptions, but there is now broad consensus about the relationships of marmosets and their close relatives, the tamarins and Goeldi’s monkey, which collectively constitute the callitrichine subfamily of primates [Schneider and Sampaio 2015]. For the purposes of this paper, I base the taxonomic terms, phylogenies, and divergence dates mainly on the authoritative treatment of Fleagle [2013]. Additional valuable sources about primate evolution and anatomy include Ankel-Simons [2007], Gebo and Severson [2014], Martin [1990], Ravosa and Dagosto [2007], and Ross and Kay [2004], while Baum and Smith [2012] provide an excellent introduction to modern phylogenetic concepts and methods. Note, however, that while there is now considerable (if not universal) agreement about who’s related to whom among primates—that is, the branching order of primate tree—the same tree can be accommodated within different systems of taxonomic nomenclature. So, for example, the marmosets and their close relatives are classified by some workers as a subfamily (Callitrichinae) and by others as a family (Callitrichidae). Fleagle [2013] treats them as a subfamily and I have adopted that usage. To complicate matters further, while most authorities prefer their taxonomies to consist only of monophyletic groups, others retain older, non-monophyletic taxonomies even when they accept modern interpretations of relationships. So, while most authors accept that tarsiers are more closely related to anthropoid primates than they are to the lemurs and lorises, and so prefer a main division of the primates into Strepsirrhini and Haplorhini, others prefer a more traditional taxonomy, grouping tarsiers with lemurs and lorises in the Prosimii, as distinct from the Anthropoidea (also known as the Simiiformes). Finally, I make little distinction in this paper between common marmosets and other marmoset species or between rhesus macaques and other macaque species.
Primate phylogenetics and evolutionary specializations.
Groups of animals are defined by common ancestry; a natural group of animals (a “clade” in phyletic parlance) is the complete set of species descended from a single ancestral species, also known as a “monophyletic” group. The features (character states) of the last common ancestor (LCA) that distinguish a species or a clade from its close relatives (its outgroups) are referred to as its shared derived traits (informally: “specializations”). While comparative molecular data are increasingly used to identify clades and reconstruct their relationships, reconstructing the anatomy of ancestral species typically requires comparative anatomical data from living species, with additional data provided by fossils, when available.
What, then, were the shared, derived traits present in the last common ancestor of the primates? Answering this question requires an examination of the living primate groups, and comparison to their close relatives. The closest relatives of the Order Primates are the Orders Scandentia (tree shrews) and Dermoptera (flying lemurs or colugos), followed by Glires (the rodent-rabbit group) (Figure 1). The features primates share that distinguish them from these non-primate groups include, among other things, close-set, forward-facing eyes (orbital approximation and convergence, respectively) surrounded by complete bony rings, and grasping extremities with opposable first digits and broad terminal digits tipped with nails rather than claws [Cartmill 1974, 1992; Martin 1968; Sussman and Kinzey 1984] (Figure 3). The behavioral reconstructions presented in the papers cited above suggest that the primate LCA was nocturnally active and foraged for flowers, fruit, and insects in the terminal branches of trees (the “fine-branch niche”) using their nail-tipped digits to grasp branches too fine to be effectively gripped with claws.
Figure 3.

Relationships of the Callitrichinae, a subfamily of the platyrrhine family Cebidae. Based on Fleagle [2013].
We can conduct the same kind of analysis for each of the different primate subgroups. Today, the Order Primates is usually considered to consist of two infraorders, Strepsirrhini and Haplorihini. The former consists of two main groups, the lemurs of Madagascar and the loris-galago (bushbaby) group from Africa and Asia. The strepsirrhines are important for comparative analysis because they are in certain respects more conservative evolutionarily than the haplorines, retaining such ancestral mammalian characteristics as a dog-like wet, hairless nose (rhinarium), and a groove connecting the rhinarium to the vomeronasal organ (VNO), a chemosensory receptor epithelium that in turn sends neural projections to the accessory olfactory bulb (AOB). Moreover, many of the living strepsirrhines are nocturnal, consistent with reconstructions of the primate LCA, and strepsirrhines retain a number of ancestral features of the visual system (to be discussed below).
While strepsirrhines are the “wet-nosed” primates, haplorhines are the “dry-nosed” primates: they lack a wet rhinarium and associated median groove and typically have a shorter snout than the strepsirrhines. The eye is enclosed partially (tarsiers) or completely (anthropoids) posteriorly by a bony plate, and the orbits are set even more closely together than in strepsirrhines [Fleagle 2013; Ross 1996]. The haplorhines include the tiny, but huge-eyed, tarsiers and the anthropoids. The species-rich anthropoid group includes the platyrrhines and catarrhines—the New World and Old World anthropoids, respectively. The catarrhines consist of the hominoids (apes and humans) and the cercopithecoids (the Old World “monkeys”). The platyrrhines are usually referred to as the New World “monkeys.” Features of the visual system suggest that the haplorhine LCA was diurnal, although nocturnality evolved secondarily in tarsiers and in the platyrrhine owl monkeys.
Anthropoid primates are distinguished from other primates by a variety of modifications of the teeth and skull, as shown in Figure 5. Also, they are generally larger than other primates, and display a wide range of morphologies, social organizations, and behaviors. Among anthropoids, the platyrrhine and catarrhine groups are distinguished by the breadth of the external nose, differences in the sutural patterns of the skull, in the structure of the middle ear, and in the number of teeth in the dental rows (Figure 5).
Figure 5.
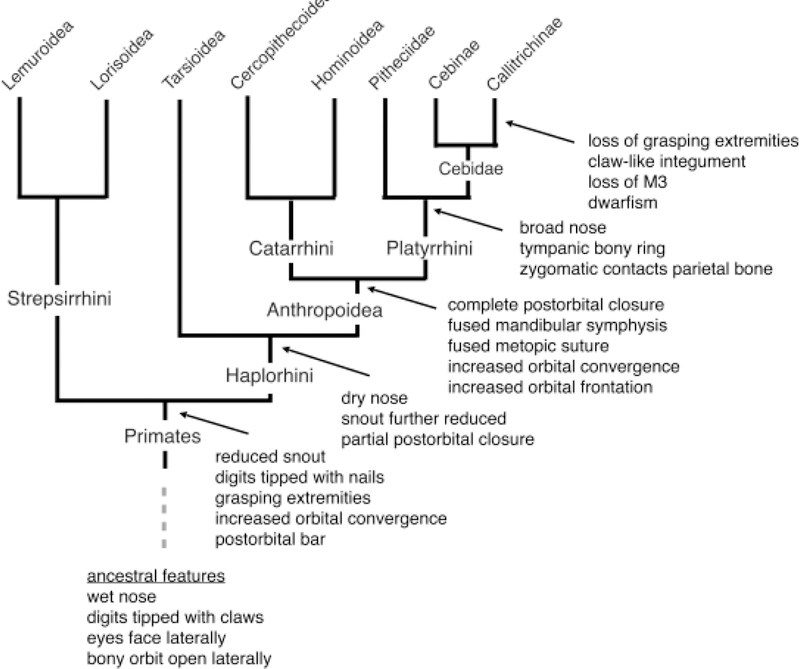
The evolutionary history of the anatomical characteristics of the callitrichines. See the text for citations.
“Monkey”—an aside.
I’ve put “monkey” is scare quotes above because the term is problematic, for at least two reasons. For one, it suggests that New World monkeys (platyrrhines) and Old World monkeys (catarrhines; cercopithecoids) are, collectively, a natural group, more closely related to each other than either is to apes and humans. In fact, Old World monkeys are more closely related to apes and humans than they are to New World monkeys (Figure 1). From a phylogenetic perspective, “monkey” is not a monophyletic group and therefore does not represent a valid biological category [Baum and Smith 2012].
The problem is amplified by the central place of “monkey” in an older, but highly influential, view of primate evolution, exemplified in the work of the pioneering primate anatomist W.E. Le Gros Clark, whose influential synthesis of primatology, The Antecedents of Man [Le Gros Clark 1959] went through multiple editions. Like most scientists working before the modern era of phylogenetics, Le Gros Clark viewed the primate order as approximating an ascending scale, with tree shrew, lemur, tarsier, monkey, and ape stages, each representing an adaptive grade that increasingly approximates the highest stage, namely humans (Figure 4). This view of primate evolution has been rejected by modern evolutionary biologists, who understand evolution in terms of diversification rather than ascent, a view better represented by the metaphor of a tree than by a scale [Baum and Smith 2012; Preuss and Robert 2014]. In primatology, this change has come about with the accumulation of knowledge about the differences and similarities displayed by the various primate clades. The idea that the New World platyrrhines and Old World cercopithecoids collectively represent a coherent monkey stage of primate evolution cannot be sustained today, nor can the idea that the living primates form a progressive series from simpler to more complex forms.
Figure 4.

Le Gros Clark’s conception of primate evolution as a series of grades, including a monkey grade, culminating in humans, based on Le Gros Clark [Le Gros Clark 1959]. This older view stands in contrast to the modern conception of primate evolution as being treelike, and in which Old World monkeys (cercopithecoids) are understood to be more closely related to humans than are New World monkeys (platyrrhines). Note that tree shrews are no longer considered primates (although they are close relatives), and that none of the living insectivore species are considered close relatives or primates.
The marmoset anatomical mosaic.
The branching nature of the evolutionary tree implies that any natural group of animals can be understood as displaying a mosaic of features: some shared with its relatives, by virtue of common ancestry, and some distinctive of that group—its derived or specialized features. It is instructive to consider a few of the morphological features that make up the marmoset mosaic (Figure 5). These include character states inherited from the common ancestor of the primate order, including relatively forward-facing, closely spaced bony orbits. As anthropoids, however, marmosets exhibit even greater convergence of the bony orbits and their eyes are enclosed within a bony cup. As platyrrhines, and in contrast to catarrhines, marmosets have widely spaced nasal apertures and a ring-like tympanic bone to which the tympanic membrane is attached (in contrast to catarrhines, in which the tympanic bone extends laterally to form a tube), and they retain three premolars from their haplorhine ancestors.
Marmosets also possess distinctive specializations. Marmosets and other callitrichines are remarkable for having lost the grasping hands and feet that characterize most primates, and for having transformed the digital nails into claw-like forms. These changes permit callitrichines to cling to tree trunks in order to exploit saps and gums as food sources. As callitrichines, marmosets also exhibit the loss of the most distal molar in the upper and lower tooth rows, which is retained in other platyrrhines and in catarrhines. This may be a consequence of the very small size of callitrichines, compared to other anthropoids. Callithrix jacchus adult body size averages approximately 320 grams, which is in the mid-range for callitrichines (approximately 110—620 grams), and as a group, the callitrichines show no overlap with the size range of other anthropoid primates [Martin 1992]. The very small size of callitrichines is evidently the result of phyletic dwarfism, a dramatic evolutionary reduction of size from ancestors that were larger [Ford 1980; Martin 1992; Montgomery and Mundy 2013; Rosenberger 1984; Sussman and Kinzey 1984].
Whereas callitrichines were once considered primitive anthropoids, recognition of the extensive and remarkable anatomical and behavioral specializations of callitrichines has led to their being characterized as the most specialized of all the platyrrhine primates. As Sussman and Kinsey put it, “[Callitrichines] have a suite of highly derived morphological features, and they can no longer be regarded as morphologically primitive New World primates” [Sussman and Kinzey 1984]. Similarly, Fleagle has concluded, “Callitrichines are the smallest and most morphologically derived New World anthropoids” [Fleagle 2013, p. 109].
The marmoset neuroanatomical mosaic.
One can carry out a similar exercise with callitrichine and marmoset neurobiological features (Figure 6). A disproportionate number of these features involve the visual system, likely reflecting the fact that this is the part of the nervous system that has received the most comparative study in primates. For example, callitrichines possess a pair of retinal cone photopigments, namely the short-wavelength sensitive (S) and medium-to-long wavelength sensitive (M/L) opsins [Jacobs 2008], which they inherited from their mammalian ancestors. However, the X-linked M/L opsin gene is polymorphic in callitrichines, as in many other platyrrhines, so that some females are trichromats, whereas the males are dichromats [Jacobs 2008].
Figure 6.
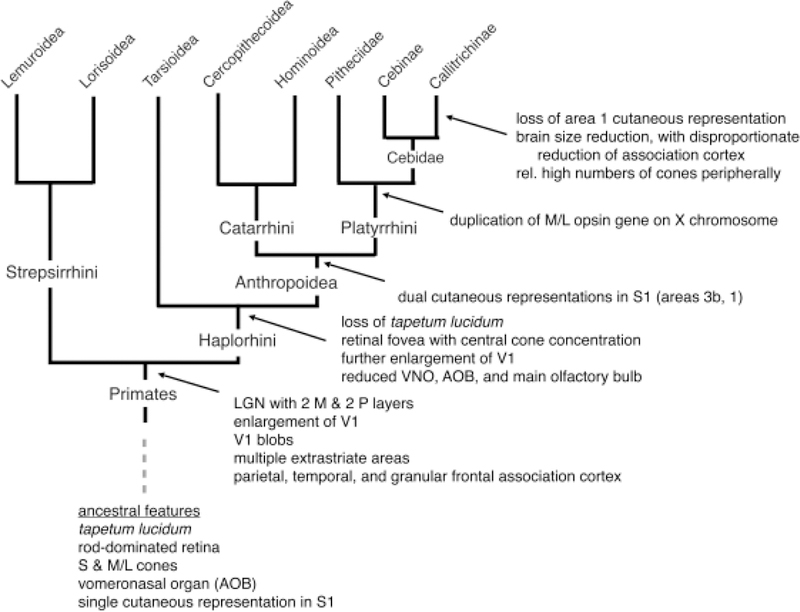
The evolutionary history of the neuroanatomical characteristics of the callitrichines. See the text for citations.
Callitrichines also possess a host of features inherited from early primates that appear to be primate specializations. These include an enlarged primary visual area (area V1) [Stephan et al. 1981], and a lateral geniculate nucleus (LGN) that has four main layers, two magnocellular and two parvocellular [Kaas et al. 1978; Le Gros Clark 1941; Solomon and Rosa 2014]. In some other anthropoids, such as macaques and humans (but not all other platyrrhines and catarrhines), the parvocellular layers have split and interdigitated, creating the appearance of a six-layered LGN [Kaas et al. 1978]. Marmosets also possess cytochrome oxidase-rich “blobs” in the primary visual area [Solomon 2002; Solomon and Rosa 2014], which receive projections from the koniocellular cells of the LGN, cells that reside outside the magnocellular and parvocellular layers. This is a specialization of primate area V1 [Horton 1984; Horton and Hubel 1981; Preuss and Kaas 1996], although similar features evolved convergently in carnivores [Murphy et al. 1995].
Callitrichines also share with haplorhines and with other anthropoids modifications of the visual system related to the change from ancestral primate nocturnality to diurnality [Ross 1996] (Figure 6). Among these are the loss of a reflecting tapetum lucidum, which enhances nocturnal visual sensitivity and is present in strepsirrhines and many other mammals [Martin 1990, pp. 298–300; Ollivier et al. 2004; Peichl 2005]. Also, like most haplorhines, but unlike most strepsirrhines [Collins et al. 2005; Ross 1996], callitrichines possess a retinal fovea, with a concentration of cones in and around the fovea [Finlay et al. 2008; Troilo et al. 1993]. As in other haplorhines and anthropoids, the primary visual area is enlarged compared to strepsirrhines [Stephan et al. 1981]. Furthermore, whereas area V1 in the catarrhines that have been examined show clear segregation of projections from the LGN representing the left and right eyes [Adams et al. 2007; Cheng et al. 2001; Florence and Kaas 1992; Horton 1984; Horton et al. 1990; LeVay et al. 1985], in callitrichines and several other platyrrhine taxa, the degree of segregation may be more variable or reduced [Hendrickson et al. 1978; Livingstone 1996; Roe et al. 2005; Sengpiel et al. 1996].
As in strepsirrhines and in other anthropoids that have been studied [Lyon 2006; Rosa and Tweedale 2005], marmosets possess a large number of extrastriate visual areas, divisible into dorsal and ventral systems, and including the higher-order outposts of the visual system in the posterior parietal cortex and inferotemporal cortex [Atapour et al. 2018; Paxinos et al. 2012; Rosa and Krubitzer 1999]. Marmosets possess additional regions of primate-specific, higher-order territories: These include the granular areas of dorsolateral prefrontal cortex [Burman et al. 2006; Burman et al. 2011], which are unique to primates [Passingham and Wise 2012; Preuss 1995]. Marmosets possess primate-specific limbic cortices [Preuss and Goldman-Rakic 1991b; Vogt et al. 2013; Vogt and Paxinos 2012], including posterior cingulate areas 23 and 31 [Armstrong 1985; Zilles et al. 1986] and posterior parahippocampal cortex [Palmer and Rosa 2005]. Finally, marmosets, like other strepsirrhine and anthropoid primates, possess a dorsal pulvinar (also known as medial pulvinar) nucleus [Brysch et al. 1990; Hackett et al. 1998; Roberts et al. 2007], a structure that appears to be unique to primates and is connected mainly with higher-order frontal, parietal, temporal, and limbic cortical areas [Preuss 2007].
In addition to these primate specializations, callitrichines share with other haplorhines a reduction of the main olfactory bulb and accessory olfactory bulb compared to early primates (Figure 6), which themselves had reduced these structures relative to their mammalian ancestors [Barton 2006; Stephan et al. 1981]. In catarrhines, the major elements of the accessory olfactory system (vomeronasal organ and accessory olfactory bulb) are absent or vestigial in adults [Bhatnagar and Smith 2007; Smith et al. 2014]. However, the VNO and AOB remain present in callitrichines and evidently in most other platyrrhines [Bhatnagar and Smith 2007; Smith et al. 2004; Smith et al. 2011], and consistent with this, vomeronasal receptor genes that appear to be functional are present in callitrichines [Moriya-Ito et al. 2018]. Thus, the accessory olfactory system, including VNO and AOB, is probably functional in callitrichines, but not in catarrhines.
Callitrichines also possess a number of neuroanatomical specializations that evolved after their separation from other platyrrhines (Figure 6). For one, marmosets and tamarins are reported to have higher densities of cones in the peripheral retina (particularly in the nasal retina) than do other platyrrhines or catarrhines [Finlay et al. 2008; Troilo et al. 1993]. In the somatosensory cortex, all anthropoid primates that have been examined, with the exception of callitrichines, have a pair of representations of the body surface’s cutaneous receptors, occupying areas 3b and 1 of the primary somatosensory region (S1) [Kaas 1983; Padberg et al. 2007]. The callitrichines, however, have only a single cutaneous representation, corresponding to area 3b [Carlson et al. 1986; Krubitzer and Kaas 1990]. Callitrichines also lack monosynaptic projections of corticospinal neurons onto the motor neurons of the spinal cord ventral horn [Kondo et al. 2015]. In this, callitrichines differ from Homo and Macaca, among catarrhines, and at least Cebus among platyrrhines—all animals with well-developed digital opposability and grasping abilities (reviewed by Padberg et al. [2007]). These features of the somatosensory and motor systems may be related to callitrichine modifications of gross anatomy and locomotor behavior: it seems plausible that with the anatomical transformation of flattened nails into claw-like forms, and the behavioral change from digital grasping to gripping with the claws, there was a reduction in the role of sensory feedback from the digital pads and nail beds to the corticospinal system.
Callitrichines also have very small brains, much smaller in absolute terms than those of any other anthropoids, and among the smallest relative to body size [Isler et al. 2008]. While data on the size of higher-order cortical regions relative to the rest of the cortex are not available for comparing marmosets to other primates, visual inspection of recent marmoset cortical maps [Atapour et al. 2018; Majka et al. 2016; Paxinos et al. 2012] and quantitative analysis of regional size change [Chaplin et al. 2013] suggest that portions of prefrontal cortex are smaller in marmosets than in other, larger-brained platyrrhines (Cebus [Cruz-Rizzolo et al. 2011]) and catarrhines (Macaca and Homo [Glasser et al. 2016; Markov et al. 2012; Neubert et al. 2014; Petrides and Pandya 1999; Preuss and Goldman-Rakic 1991a; Sallet et al. 2013]). Chaplin and colleagues identified temporoparietal junction cortex and anterior cingulate cortex as additional regions that are relatively larger in macaques and humans than in marmosets [Chaplin et al. 2013].
Marmosets Compared to Macaques.
It is useful to summarize some features shared by marmosets and macaques and those they don’t share (Figure 7). Marmosets and macaques share a number of features by virtue of being primates (reduced main olfactory bulbs, V1 blobs; multiple extrastriate areas divisible into dorsal and ventral streams, new higher-order association areas, addition of the dorsal pulvinar) and by virtue of being anthropoid primates (a fovea, with a central concentration of cones). In addition, there are features of ancestral primates and ancestral anthropoids that marmosets and macaques do not share, notably the accessory olfactory system (present in marmosets, absent or vestigial in macaques and other catarrhines). The visual systems of marmosets and macaques also differ in a number of respects that mainly reflect catarrhine specializations, involving opsin proteins, the LGN, and the primary visual area,. Finally, marmosets differ from macaques in ways that reflect callitrichine specializations, including differences in the retinal distribution of photoreceptors and in sensorimotor cortex, and a likely reduction of brain size that disproportionately affected higher-order association cortex.
Figure 7.

A comparison of neuroanatomical features of marmosets and macaques discussed in the text, indicating points of similarity (=) and difference (≠).
Despite the limitations of the comparative neuroanatomical data set, it is clear that marmosets are, in a variety of ways, more different from humans than are macaques. That is not to say that macaques do not also differ from humans in significant ways—they clearly do. Even in the visual system, commonly considered to differ only in minor, quantitative ways between humans and macaques, one can in fact identify numerous differences at all levels of the visual system [Preuss 2004; Preuss and Robert 2014]. Every clade has its own mosaic of features, including its own specializations, and macaques are no exception. Nevertheless, for the purpose of modeling the human condition, macaques (along with other catarrhines) have the advantage of being much more closely related to humans than are marmosets, sharing about 20 million years of additional common ancestry (Figure 1). Furthermore, human-macaque comparisons are uncomplicated by the platyrrhine and callitrichine specializations present in marmosets, and by the retention in marmosets of ancestral features lost in catarrhines (Figures 6 and 7).
Additional considerations
The foregoing discussion has focused on anatomical features of the nervous system, primarily because anatomy has received more comparative study than other dimensions of nervous system organization. As our knowledge of primate neurobiology grows, so will the number of different characters that can be evaluated, and, assuming we study a large enough variety of species, this knowledge will extend beyond anatomical features to include life history and behavior, on the one hand, and biochemistry, molecular biology, and genomics, on the other. Even with the limited information currently available, however, there is evidence for additional differences between callitrichines and catarrhines that likely bear on the utility of marmosets as stand-ins for humans in experimental studies.
I have noted above some differences in aspects of higher-order cortical organization between the small-brained marmosets and the larger-brained platyrrhines and catarrhines. The very small size of the marmoset brain makes it very likely that the functions of its cortical systems differ in important ways from those of larger-brained primates, if only because of the much more limited amount of neural machinery marmosets and other callitrichines have to work with. In this regard, it is significant that two recent meta-analyses concluded that absolute brain size is a better predictor of cognitive function than is brain size relative to body size [Deaner et al. 2007; Reader et al. 2011] and a third indicated that absolute brain size is the best predictor of self control [Maclean et al. 2014].
Given the small size and rapid development of marmosets, it is tempting to view marmoset life history as a condensed version of that of longer-lived primates. Yet there is evidence primates vary in patterns of postnatal growth and development. Bogin [2007] indicates that cercopithecoid and hominoid development includes an extended period of slow growth, defining a juvenile stage that has no counterpart in marmosets. This difference, and the specializations of human development recognized by Bogin—namely, the addition of childhood and adolescent stages—imply differences in the hormonal control of development [Bogin 2009]. It is noteworthy that comparative studies of humans, macaques, and marmosets have identified remarkable species differences in the production of the androgen hormone dehydroepiandrosterone (DHEA) [Abbott and Bird 2009], which is involved in sexual maturation. There is also a remarkable sex difference in marmosets, with adult males lacking a functional zona reticulata [Pattison et al. 2009], the component of the adrenal gland that produces most of the body’s DHEA.
Callitrichines, like humans, engage in prosocial behaviors that are unusual among primates. They include cooperative breeding, in which adult males as well as females participate in carrying and sharing food with offspring, and alloparenting, in which other individuals, both related to and unrelated to the mother, contribute to the support of her offspring [Erb and Porter 2017; Fernandez-Duque et al. 2009]. These have been cited as possible instances of convergent evolution in the human and callitrichine clades [Burkart and Finkenwirth 2015], and there are other behavioral and cognitive similarities between humans and marmosets [Miller et al. 2016] that could be viewed in the same light. In evolutionary biology, convergence is considered to provide information about the similarities in selection pressures that shape similar phenotypes [Arendt and Reznick 2008]. Convergence may not be what one is looking for in a human model, however, as convergently evolved features of social behavior or other phenotypes need not share homologous neural and genetic underpinnings [Arendt and Reznick 2008]. Moreover, the proximate mechanisms regulating social behavior in callitrichines likely differ from those of humans in some important respects, considering that callitrichines mark their substrates, and sometimes other group members, with a mixture of urine and circumgenital secretions, and these chemical cues have been postulated to regulate the reproductive physiology of group individuals [Abbott et al. 1997; Epple 1970; Lazaro-Perea et al. 1999; Smith et al. 2001]. As in many other mammalian groups, detection of such cues likely involves the accessory olfactory system, which is absent or vestigial in catarrhines, as discussed above.
Callitrichines are highly unusual among mammals in that their dizygotic twins exchange cell lines very early in development, resulting in genetically chimeric individuals [Benirschke et al. 1962; Gengozian et al. 1964]. Although the range of tissues that exhibit chimerism is controversial (compare Ross et al. [2007] and Sweeney et al. [2012]), it is clear that it involves hematopoietic tissues, such as blood and lymph cells, and possibly also germ-line tissues [Sweeney et al. 2012]. Chimerism could complicate the interpretation of genetic modification experiments because the presence of a modified gene in blood cells collected in routine assays would not necessarily indicate its presence in neural cells. In addition, the analysis of phenotypic responses in experiments involving brain damage, infection, inflammation, and aging could be complicated by the fact that macrophages and other components of the immune system that infiltrate brain tissue in these conditions are derived from hematopoietic cells.
There are numerous differences in the distribution of receptors in the cortex of marmosets compared to catarrhines (based mainly on studies of macaques and humans). These include differences in the regional and laminar distribution of adrenergic α1, muscarinic M1 and M2, and serotonergic 5-HT2 receptors in the hippocampus [Kraemer et al. 1995] and in the laminar distribution of GABAa and 5-HT1 receptors in primary visual cortex [Gebhard et al. 1993]. Given the lack of evidence about receptor distribution in platyrrhines other than marmosets, it is unclear whether these represent differences between platyrrhines and catarrhines or between marmosets and catarrhines or, most likely, some combination of both. There is evidence for additional molecular and biochemical differences as well, for example, in the distribution and concentrations of manganese and zinc in marmoset and human brains [Knauer et al. 2017], and in the neuroendocrinology of stress and reproduction between platyrrhines and catarrhines [Abbott et al. 2003; Bercovitch and Ziegler 2002].
The existence of differences such as these should not be surprising, given the longer period of shared ancestry between humans and macaques compared to humans and marmosets (Figure 1). The greater shared ancestry of humans and macaques is reflected in genomic similarities as well: The genome-wide identity of amino-acid coding nucleotide sequences is 94.0% for macaques and humans, compared to 91.7% for marmosets and humans (R. Alan Harris, Jeff Rogers, pers. comm.; see also Worley et al. [2014]). Given that chimpanzees and humans, which are even more similar genetically, exhibit a remarkable diversity of genomic, molecular, and biochemical differences (e.g., Bauernfeind et al. [2015; Konopka et al. [2012]; Li et al. [2017]; Mora-Bermúdez et al. [2016]; Reilly et al. [2015]), there is a great deal of room for molecular and biochemical differences between marmosets and humans, and corresponding differences in nervous system organization and function. In fact, a recent comparison of human-marmoset orthologs (R. Alan Harris, pers. comm.) provides evidence for selection acting on marmoset genes, identifying 49 potentially positively selected genes at p < 0.01 with a role in the brain, based on Gene Ontology Biological Process terms. (The analysis employed HyPhy aBSREL [Smith et al. 2015] to evaluate selection on orthologs identified in Ensembl v. 91 [Herrero et al. 2016].)
Conclusions
Given the variability of primate brain organization, how do we choose the model or models most relevant for understanding the human brain and behavior? In view of the decreasing diversity of primate species available for research, and the growing imperative to generate genetically modified primate models, we are likely faced with the choice of marmosets or macaques or some combination of both. Therefore, it is important to evaluate the relative merits of marmosets and macaques, as nonhuman primate models for understanding the human brain and behavior. From the standpoint of cost and convenience, marmosets have some clear advantages, such as their rapid rate of growth and reproduction, which provide the potential for the speedier development of genetically modified models and for the speedier completion of developmental and aging studies. From a purely biological standpoint, however, macaques have clear advantages over marmosets: they have more in common with us, owing to our closer phylogenetic relationship.
There are additional points in favor of macaques. Compared to marmosets, we have much more experience with macaque husbandry and with the assessment of macaque neurobiology and behavior, including changes across the lifespan. It should be remembered, too, that the very characteristics of marmosets that make them convenient experimentally for the purposes of genetic modification, studies of development and aging, and cortical mapping—their small size, rapid development, and short lifespan—all related to the callitrichine specialization of dwarfing—are ways in which marmosets differ from humans but in which macaques are more like humans (Figure 2). Finally, while it is clear that marmosets offer advantages for genetic modification, it is not clear that these advantages involve anything more than cost and speed of development: the same procedures appear to be feasible in macaques, and in fact China’s brain project, which includes gene editing approaches, focuses on macaques [Poo et al. 2016].
From a research-resources perspective, if the question is how best to understand the features of human brain organization that cannot be studied directly in humans, the best solution (in lieu of even broader studies) would be to study both macaques and marmosets. Studying multiple species is necessary for reconstructing evolutionary history, and in particular, for disambiguating features shared among larger groups from the specializations of subgroups. So, for example, features shared by both macaques and marmosets are likely to have been present in the immediate ancestry of catarrhines (including humans), whereas features present only in macaques, for example, might be shared derived features of macaques, or of cercopithecoids, or of catarrhines, or of anthropoids, and so forth down the phylogenetic tree—there’s no way to tell by studying only macaques. Analogous considerations apply to research limited to marmosets. Therefore, we should view marmoset and macaque research as complementary (see also Miller [2017] and Mitchell and Leopold [2015]). If we could only study one animal from which to extrapolate results to humans, the choice would necessarily be macaques, given their longer period of shared ancestry with humans—the likelihood of a given feature of macaques having a homologue in humans is greater than for a given feature of marmosets. But it would be extremely unfortunate if we were forced to make that choice, and inimical to good science.
If the question is, what animal is best for genetic modification, measured as the rate at which translatable results can be obtained, the answer is less clear, depending as it does on two unknown quantities: the rate at which models can be developed and the likelihood that a model will yield results that translate to humans. Marmosets probably have the edge with respect to model development, whereas macaques probably have the edge in translatability. We could perhaps narrow this knowledge gap by carrying out parallel pilot experiments in macaques and marmosets to try to estimate those rates. However, obtaining translatable results for a single gene in one species is no guarantee that other genes might not yield favorable results in the other species. Moreover, it is entirely possible that neither macaques nor marmosets will provide translatable results for a given gene of interest, but that some other primate (or even non-primate) species will. The most reasonable strategy would be to hedge our bets, devoting some resources to marmosets, while maintaining a robust investment in macaques. One hopes, too, that we will ultimately be able to expand the range of primate and non-primate species to which we can apply gene editing and other advanced research techniques.
Practitioners of genetic modification, hoping for a rapid transition from mice to primates and wishing to regard marmosets as “the new mouse,” might find these prescriptions unsatisfactory. But just as the mouse model, convenient as it is, has run afoul of the diversity of mammalian biology, so might the marmoset model run afoul of the diversity of primate biology. Be careful what you wish for.
Acknowledgements
The author is grateful for the comments and suggestions of Amy Arnsten, Anthony Chan, R. Alan Harris, Paul Johnson, Jeff Rogers, and Georg Striedter on early drafts of this manuscript, and to Mary Ann Cree and Jeromy Dooyema for their close reading of the manuscript. This project was supported in part by NIH ORIP/OD P51 0D011132.
Footnotes
The author has no conflicts of interest to declare.
References
- Abbott DH, Saltzman W, Schultz-Darken NJ, Smith TE (1997) Specific neuroendocrine mechanisms not involving generalized stress mediate social regulation of female reproduction in cooperatively breeding marmoset monkeys. Ann N Y Acad Sci 807:219–238. [DOI] [PubMed] [Google Scholar]
- Abbott DH, Barnett DK, Yamamoto ME, Schultz-Darken NJ (2003) Aspects of common marmoset basic biology and life history important for biomedical research. 53:339–350. [PubMed] [Google Scholar]
- Abbott DH, Bird IM (2009) Nonhuman primates as models for human adrenal androgen production: function and dysfunction. Rev Endocr Metab Disord 10:33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams DL, Sincich LC, Horton JC (2007) Complete pattern of ocular dominance columns in human primary visual cortex. J Neurosci 27:10391–10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ankel-Simons F (2007) Primate anatomy: an introduction. 3rd ed Amsterdam; Boston: Elsevier Academic Press. [Google Scholar]
- Arendt J, Reznick D (2008) Convergence and parallelism reconsidered: what have we learned about the genetics of adaptation? Trends Ecol Evol (Amst) 23:26–32. [DOI] [PubMed] [Google Scholar]
- Armstrong E (1985) Relative brain size in monkeys and prosimians. Am J Phys Anthropol 66:263–273. [DOI] [PubMed] [Google Scholar]
- Arnsten AF (2010) The use of α−2A adrenergic agonists for the treatment of attention-deficit/hyperactivity disorder. Expert Rev Neurother 10:1595–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnsten AF, Raskind MA, Taylor FB, Connor DF (2015) The effects of stress exposure on prefrontal cortex: Translating basic research into successful treatments for posttraumatic stress disorder. Neurobiol Stress 1:89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atapour N, Majka P, Wolkowicz IH, Malamanova D, Worthy KH, Rosa MGP (2018) Neuronal distribution across the cerebral cortex of the marmoset monkey (Callithrix jacchus). Cerebral Cortex. [DOI] [PubMed] [Google Scholar]
- Barton RA (2006) Olfactory evolution and behavioral ecology in primates. Am J Primatol 68:545–558. [DOI] [PubMed] [Google Scholar]
- Bauernfeind AL, Soderblom EJ, Turner ME, Moseley MA, Ely JJ, Hof PR, Sherwood CC, Wray GA, Babbitt CC (2015) Evolutionary divergence of gene and protein expression in the brains of humans and chimpanzees. Genome Biol Evol 7:2276–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum DA, Smith SD (2012) Tree thinking: an introduction to phylogenetic biology. Greenwood Village, Colo: Roberts. [Google Scholar]
- Benirschke K, Anderson JM, Brownhill LE (1962) Marrow chimerism in marmosets. Science 138:513–515. [DOI] [PubMed] [Google Scholar]
- Bercovitch FB, Ziegler TE (2002) Current topics in primate socioendocrinology. Annu Rev Anthropol 31:45–67. [Google Scholar]
- Bhatnagar KP, Smith TD (2007) The vomeronasal organ and its evolutionary loss in catarrhine primates In: Evolution of Nervous Systems: Vol 4 Primates (Kaas JH, Preuss TM, eds), pp 141–148. Amsterdam: Elsevier. [Google Scholar]
- Bogin B (2007) The evolution of human brain and body growth patterns In: Evolution of Nervous Systems: Vol 4 Primates (Kaas JH, Preuss TM, eds), pp 337–345. Amsterdam: Elsevier. [Google Scholar]
- Bogin B (2009) Childhood, adolescence, and longevity: A multilevel model of the evolution of reserve capacity in human life history. Am J Hum Biol 21:567–577. [DOI] [PubMed] [Google Scholar]
- Brysch W, Brysch I, Creutzfeldt OD, Schlingensiepen R, Schlingensiepen KH (1990) The topology of the thalamo-cortical projections in the marmoset monkey (Callithrix jacchus). Exp Brain Res 81:1–17. [DOI] [PubMed] [Google Scholar]
- Burkart JM, Finkenwirth C (2015) Marmosets as model species in neuroscience and evolutionary anthropology. Neurosci Res 93:8–19. [DOI] [PubMed] [Google Scholar]
- Burman KJ, Palmer SM, Gamberini M, Rosa MGP (2006) Cytoarchitectonic subdivisions of the dorsolateral frontal cortex of the marmoset monkey (Callithrix jacchus), and their projections to dorsal visual areas. J Comp Neurol 495:149–172. [DOI] [PubMed] [Google Scholar]
- Burman KJ, Reser DH, Yu H-H, Rosa MGP (2011) Cortical input to the frontal pole of the marmoset monkey. Cerebral Cortex 21:1712–1737. [DOI] [PubMed] [Google Scholar]
- Burns TC, Li MD, Mehta S, Awad AJ, Morgan AA (2015) Mouse models rarely mimic the transcriptome of human neurodegenerative diseases: A systematic bioinformatics-based critique of preclinical models. European Journal of Pharmacology 759:101–117. [DOI] [PubMed] [Google Scholar]
- Carlson M, Huerta MF, Cusick CG, Kaas JH (1986) Studies on the evolution of multiple somatosensory representations in primates: the organization of anterior parietal cortex in the New World callitrichid, Saguinus. J Comp Neurol 246:409–426. [DOI] [PubMed] [Google Scholar]
- Cartmill M (1974) Rethinking primate origins. Science, New Series 184:436–443. [DOI] [PubMed] [Google Scholar]
- Cartmill M (1992) New views on primate origins. Evol Anthropol 1:105–111. [DOI] [PubMed] [Google Scholar]
- Chaplin TA, Yu H-H, Soares JGM, Gattass R, Rosa MGP (2013) A conserved pattern of differential expansion of cortical areas in simian primates. J Neurosci 33:15120–15125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Check Hayden E (2014) Misleading mouse studies waste medical resources. Nature News http://wwwnaturecom/doifinder/101038/nature201414938. [Google Scholar]
- Cheng K, Waggoner RA, Tanaka K (2001) Human ocular dominance columns as revealed by high-field functional magnetic resonance imaging. Neuron 32:359–374. [DOI] [PubMed] [Google Scholar]
- Collins CE, Hendrickson A, Kaas JH (2005) Overview of the visual system of Tarsius. Anat Rec A Discov Mol Cell Evol Biol 287:1013–1025. [DOI] [PubMed] [Google Scholar]
- Cruz-Rizzolo RJ, De Lima MA, Ervolino E, de Oliveira JA, Casatti CA (2011) Cyto-, myelo-and chemoarchitecture of the prefrontal cortex of the Cebus monkey. BMC Neuroscience 12:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyranoski D (2014) Marmosets are stars of Japan’s ambitious brain project. Nature 514:151–152. [DOI] [PubMed] [Google Scholar]
- Deaner RO, Isler K, Burkart J, van Schaik C (2007) Overall brain size, and not encephalization quotient, best predicts cognitive ability across non-human primates. Brain Behav Evol 70:115–124. [DOI] [PubMed] [Google Scholar]
- Epple G (1970) Quantitative studies on scent marking in the marmoset (Callithrix jacchus). Folia Primatol 13:48–62. [DOI] [PubMed] [Google Scholar]
- Erb WM, Porter LM (2017) Mother’s little helpers: What we know (and don’t know) about cooperative infant care in callitrichines. Evol Anthropol 26:25–37. [DOI] [PubMed] [Google Scholar]
- Fernandez-Duque E, Valeggia CR, Mendoza SP (2009) The biology of paternal care in human and nonhuman primates. Annu Rev Anthropol 38:115–130. [Google Scholar]
- Finlay BL, Franco ECS, Yamada ES, Crowley JC, Parsons M, Muniz JAPC, Silveira LCL (2008) Number and topography of cones, rods and optic nerve axons in New and Old World primates. Vis Neurosci 25:289–299. [DOI] [PubMed] [Google Scholar]
- Fleagle JG (2013) Primate adaptation and evolution. 3rd ed San Diego, CA: Elsevier/Academic Press. [Google Scholar]
- Florence SL, Kaas JH (1992) Ocular dominance columns in area 17 of Old World macaque and talapoin monkeys: complete reconstructions and quantitative analyses. Vis Neurosci 8:449–462. [DOI] [PubMed] [Google Scholar]
- Ford SM (1980) Callitrichids as phyletic dwarfs, and the place of the Callitrichidae in Platyrrhini. Primates 21:31–43. [Google Scholar]
- Gebhard R, Zilles K, Schleicher A, Everitt BJ, Robbins TW, Divac I (1993) Distribution of seven major neurotransmitter receptors in the striate cortex of the New World monkey Callithrix jacchus. Neuroscience 56:877–885. [DOI] [PubMed] [Google Scholar]
- Gebo DL, Severson M (2014) Primate comparative anatomy. Baltimore: Johns Hopkins University Press. [Google Scholar]
- Gengozian N, Batson JS, Eide P (1964) Hematologic and cytogenetic evidence for hematopoietic chimerism in the marmoset, Tamarinus nigricollis. Cytogenet Genome Res 3:384–393. [DOI] [PubMed] [Google Scholar]
- Glasser MF, Coalson TS, Robinson EC, Hacker CD, Harwell J, Yacoub E, Ugurbil K, Andersson J, Beckmann CF, Jenkinson M, Smith SM, Van Essen DC (2016) A multi-modal parcellation of human cerebral cortex. Nature 536:171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett TA, Stepniewska I, Kaas JH (1998) Thalamocortical connections of the parabelt auditory cortex in macaque monkeys. J Comp Neurol 400:271–286. [DOI] [PubMed] [Google Scholar]
- Hendrickson AE, Wilson JR, Ogren MP (1978) The neuroanatomical organization of pathways between the dorsal lateral geniculate nucleus and visual cortex in Old World and New World primates. J Comp Neurol 182:123–136. [DOI] [PubMed] [Google Scholar]
- Herrero J, Muffato M, Beal K, Fitzgerald S, Gordon L, Pignatelli M, Vilella AJ, Searle SM, Amode R, Brent S, Spooner W, Kulesha E, Yates A, Flicek P (2016) Ensembl comparative genomics resources. Database (Oxford) 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hof PR, Sherwood CC (2007) The evolution of neuron classes in the neocortex of mammals In: Evolution of Nervous Systems Vol 3: Mammals, pp 113–124: Elsevier. [Google Scholar]
- Horton JC, Hubel DH (1981) Regular patchy distribution of cytochrome oxidase staining in primary visual cortex of macaque monkey. Nature 292:762–764. [DOI] [PubMed] [Google Scholar]
- Horton JC (1984) Cytochrome oxidase patches: a new cytoarchitectonic feature of monkey visual cortex. Philos Trans R Soc Lond, B, Biol Sci 304:199–253. [DOI] [PubMed] [Google Scholar]
- Horton JC, Dagi LR, McCrane EP, de Monasterio FM (1990) Arrangement of ocular dominance columns in human visual cortex. Arch Ophthalmol 108:1025–1031. [DOI] [PubMed] [Google Scholar]
- Hyman SE (2012) Revolution stalled. Science Translational Medicine 4:−155cm111. [DOI] [PubMed] [Google Scholar]
- Isler K, Christopher Kirk E, Miller JMA, Albrecht GA, Gelvin BR, Martin RD (2008)Endocranial volumes of primate species: scaling analyses using a comprehensive and reliable data set. Journal of Human Evolution 55:967–978. [DOI] [PubMed] [Google Scholar]
- Jacobs GH (2008) Primate color vision: a comparative perspective. Vis Neurosci 25:619–633. [DOI] [PubMed] [Google Scholar]
- Kaas JH, Huerta MF, Weber JT, Harting JK (1978) Patterns of retinal terminations and laminar organization of the lateral geniculate nucleus of primates. J Comp Neurol 182:517–553. [DOI] [PubMed] [Google Scholar]
- Kaas JH (1983) What, if anything, is SI? Organization of first somatosensory area of cortex. Physiol Rev 63:206–231. [DOI] [PubMed] [Google Scholar]
- Kaas JH (2012) The evolution of brains from early mammals to humans. Wiley Interdiscip Rev Cogn Sci 4:33–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi N, Sato K, Sasaki E, Okano H (2014) Common marmoset as a new model animal for neuroscience research and genome editing technology. Dev Growth Differ 56:53–62. [DOI] [PubMed] [Google Scholar]
- Knauer B, Majka P, Watkins KJ, Taylor AWR, Malamanova D, Paul B, Yu H-H, Bush AI, Hare DJ, Reser DH (2017) Whole-brain metallomic analysis of the common marmoset (Callithrixjacchus). Metallomics 9:411–423. [DOI] [PubMed] [Google Scholar]
- Kolata G (2013) Mice fall short as test subjects for human’s deadly ills. In: New York Times, February 11. [Google Scholar]
- Kondo T, Yoshihara Y, Yoshino-Saito K, Sekiguchi T, Kosugi A, Miyazaki Y, Nishimura Y, Okano HJ, Nakamura M, Okano H, Isa T, Ushiba J (2015) Histological and electrophysiological analysis of the corticospinal pathway to forelimb motoneurons in common marmosets. Neurosci Res 98:35–44. [DOI] [PubMed] [Google Scholar]
- Konopka G, Friedrich T, Davis-Turak J, Winden K, Oldham MC, Gao F, Chen L, Wang G-Z, Luo R, Preuss TM, Geschwind DH (2012) Human-specific transcriptional networks in the brain. Neuron 75:601–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer M, Zilles K, Schleicher A, Gebhard R, Robbins TW, Everitt BJ, Divac I (1995) Quantitative receptor autoradiography of 8 different transmitter-binding sites in the hippocampus of the common marmoset, Callithrix jacchus. Anat Embryol 191:213–225. [DOI] [PubMed] [Google Scholar]
- Krubitzer LA, Kaas JH (1990) The organization and connections of somatosensory cortex in marmosets. J Neurosci 10:952–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaro-Perea C, Snowdon CT, de Fatima Arruda M (1999) Scent-marking behavior in wild groups of common marmosets (Callithrix jacchus ). Behavioral Ecology and Sociobiology 46:313–324. [Google Scholar]
- Le Gros Clark WE (1941) The lateral geniculate body in the platyrrhine monkeys. J Anatomy 76:131. [PMC free article] [PubMed] [Google Scholar]
- Le Gros Clark WE (1959) The antecedents of man; an introduction to the evolution of the Primates. Edinburgh,: University Press. [Google Scholar]
- Ledford H (2016) CRISPR: gene editing is just the beginning. Nature 531:156–159. [DOI] [PubMed] [Google Scholar]
- LeVay S, Connolly M, Houde J, Van Essen DC (1985) The complete pattern of ocular dominance stripes in the striate cortex and visual field of the macaque monkey. J Neurosci 5:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine MS, Cepeda C, Hickey MA, Fleming SM, Chesselet M-F (2004) Genetic mouse models of Huntington’s and Parkinson’s diseases: illuminating but imperfect. Trends in Neurosciences 27:691–697. [DOI] [PubMed] [Google Scholar]
- Li Q, Bozek K, Xu C, Guo Y, Sun J, Paabo S, Sherwood CC, Hof PR, Ely JJ, Li Y, Willmitzer L, Giavalisco P, Khaitovich P (2017) Changes in lipidome composition during brain development in humans, chimpanzees, and macaque monkeys. Molecular Biology and Evolution 34:1155–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X-J, Li S (2012) Influence of species differences on the neuropathology of transgenic Huntington’s disease animal models. 39:239–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao B-Y, Zhang J (2008) Null mutations in human and mouse orthologs frequently result in different phenotypes. Proc Natl Acad Sci USA 105:6987–6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingstone MS (1996) Ocular dominance columns in New World monkeys. J Neurosci 16:2086–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan CA (2001) “[A]re Norway Rats … Things?”: Diversity versus generality in the use of albino rats in experiments on development and sexuality. J Hist Biol 34:287–314. [DOI] [PubMed] [Google Scholar]
- Logan CA (2002) Before there were standards: the role of test animals in the production of empirical generality in physiology. J Hist Biol 35:329–363. [DOI] [PubMed] [Google Scholar]
- Lyon DG (2006) The evolution of visual cortex and visual systems In: Evolution of Nervous Systems, Vol 3: Mammals, pp 1–40: Elsevier. [Google Scholar]
- Maclean EL, Hare B, Nunn CL, Addessi E, Amici F, Anderson RC, Aureli F, Baker JM, Bania AE, Barnard AM, Boogert NJ, Brannon EM, Bray EE, Bray J, Brent LJN, Burkart JM, Call J, Cantlon JF, Cheke LG, Clayton NS, Delgado MM, DiVincenti LJ, Fujita K, Herrmann E, Hiramatsu C, Jacobs LF, Jordan KE, Laude JR, Leimgruber KL, Messer EJE, de A Moura AC, Ostojic L, Picard A, Platt ML, Plotnik JM, Range F, Reader SM, Reddy RB, Sandel AA, Santos LR, Schumann K, Seed AM, Sewall KB, Shaw RC, Slocombe KE, Su Y, Takimoto A, Tan J, Tao R, van Schaik CP, Virànyi Z, Visalberghi E, Wade JC, Watanabe A, Widness J, Young JK, Zentall TR, Zhao Y (2014) The evolution of self-control. Proc Natl Acad Sci USA 111:E2140–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majka P, Chaplin TA, Yu HH, Tolpygo A, Mitra PP, Wojcik DK, Rosa MG (2016) Towards a comprehensive atlas of cortical connections in a primate brain: Mapping tracer injection studies of the common marmoset into a reference digital template. J Comp Neurol 524:2161–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manger PR, Cort J, Ebrahim N, Goodman A, Henning J, Karolia M, Rodrigues S-L, Strkalj G (2008) Is 21st century neuroscience too focussed on the rat/mouse model of brain function and dysfunction? Front Neuroanat 2:2–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markov NT, Ercsey-Ravasz MM, Ribeiro Gomes AR, Lamy C, Magrou L, Vezoli J, Misery P, Falchier A, Quilodran R, Gariel MA, Sallet J, Gamanut R, Huissoud C, Clavagnier S, Giroud P, Sappey-Marinier D, Barone P, Dehay C, Toroczkai Z, Knoblauch K, Van Essen DC, Kennedy H (2012) A weighted and directed interareal connectivity matrix for macaque cerebral cortex. Cerebral Cortex 24:17–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin RD (1968) Towards a new definition of primates. Man 3:377–401. [Google Scholar]
- Martin RD (1990) Primate origins and evolution: a phylogenetic reconstruction. Princeton, N.J: Princeton University Press. [Google Scholar]
- Martin RD (1992) Goeldi and the dwarfs: the evolutionary biology of the small New World monkeys. Journal of Human Evolution 22:367–393. [Google Scholar]
- Mestas J, Hughes CCW (2004) Of mice and not men: Differences between mouse and human immunology. J Immunol 172:2731–2738. [DOI] [PubMed] [Google Scholar]
- Miller CT, Freiwald WA, Leopold DA, Mitchell JF, Silva AC, Wang X (2016) Marmosets: A neuroscientific model of human social behavior. Neuron 90:219–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CT (2017) Why marmosets? Dev Neurobiol 77:237–243. [DOI] [PubMed] [Google Scholar]
- Mitchell JF, Leopold DA (2015) The marmoset monkey as a model for visual neuroscience. Neurosci Res 93:20–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnár Z, Clowry G (2012) Cerebral cortical development in rodents and primates. Prog Brain Res 195:45–70. [DOI] [PubMed] [Google Scholar]
- Montgomery SH, Mundy NI (2013) Parallel episodes of phyletic dwarfism in callitrichid and cheirogaleid primates. J Evol Biol 26:810–819. [DOI] [PubMed] [Google Scholar]
- Mora-Bermúdez F, Badsha F, Kanton S, Camp JG, Vernot B, Köhler K, Voigt B, Okita K, Maricic T, He Z, Lachmann R, Pääbo S, Treutlein B, Huttner WB (2016) Differences and similarities between human and chimpanzee neural progenitors during cerebral cortex development. Elife 5:e18683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya-Ito K, Hayakawa T, Suzuki H, Hagino-Yamagishi K, Nikaido M (2018) Evolution of vomeronasal receptor 1 (V1R) genes in the common marmoset (Callithrix jacchus). Gene 642:343–353. [DOI] [PubMed] [Google Scholar]
- Murphy KM, Jones DG, Van Sluyters RC (1995) Cytochrome-oxidase blobs in cat primary visual cortex. J Neurosci 15:4196–4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy WJ, Eizirik E, O'Brien SJ, Madsen O, Scally M, Douady CJ, Teeling E, Ryder OA, Stanhope MJ, de Jong WW, Springer MS (2001) Resolution of the early placental mammal radiation using Bayesian phylogenetics. Science 294:2348–2351. [DOI] [PubMed] [Google Scholar]
- Neubert F-X, Mars RB, Thomas AG, Sallet J, Rushworth MFS (2014) Comparison of human ventral frontal cortex areas for cognitive control and language with areas in monkey frontal cortex. Neuron 81:700–713. [DOI] [PubMed] [Google Scholar]
- Okano H, Hikishima K, Iriki A, Sasaki E (2012) The common marmoset as a novel animal model system for biomedical and neuroscience research applications. Semin Fetal Neonatal Med 17:336–340. [DOI] [PubMed] [Google Scholar]
- Okano H, Sasaki E, Yamamori T, Iriki A, Shimogori T, Yamaguchi Y, Kasai K, Miyawaki A (2016) Brain/MINDS: A Japanese national brain project for marmoset neuroscience. Neuron 92:582–590. [DOI] [PubMed] [Google Scholar]
- Ollivier FJ, Samuelson DA, Brooks DE, Lewis PA, Källberg ME, Komáromy AM (2004) Comparative morphology of the tapetum lucidum (among selected species). Vet Ophthalmol 7:11–22. [DOI] [PubMed] [Google Scholar]
- Onos KD, Rizzo SJS, Howell GR, Sasner M (2016) Toward more predictive genetic mouse models of Alzheimer’s disease. Brain Res Bull 122:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padberg J, Franca JG, Cooke DF, Soares JG, Rosa MG, Fiorani MJ, Gattass R, Krubitzer L (2007) Parallel evolution of cortical areas involved in skilled hand use. Journal of Neuroscience 27:10106–10115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer SM, Rosa MGP (2005) Quantitative analysis of the corticocortical projections to the middle temporal area in the marmoset monkey: Evolutionary and functional implications. Cerebral Cortex 16:1361–1375. [DOI] [PubMed] [Google Scholar]
- Passingham RE, Wise SP (2012) The neurobiology of the prefrontal cortex : anatomy, evolution, and the origin of insight. Oxford, United Kingdom: Oxford University Press. [Google Scholar]
- Pattison JC, Abbott DH, Saltzman W, Conley AJ, Bird IM (2009) Plasticity of the zona reticularis in the adult marmoset adrenal cortex: voyages of discovery in the New World. J Endocrinol 203:313–326. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Watson C, Petrides M, Rosa M, Tokuno H (2012) The Marmoset Brain in Stereotaxic Coordinates. London: Academic Press, Elsevier. [Google Scholar]
- Peichl L (2005) Diversity of mammalian photoreceptor properties: Adaptations to habitat and lifestyle? Anat Rec A Discov Mol Cell Evol Biol 287A:1001–1012. [DOI] [PubMed] [Google Scholar]
- Perlman RL (2016) Mouse models of human disease: An evolutionary perspective. Evol Med Public Health 2016:170–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin S (2014) Preclinical research: Make mouse studies work. Nature News 507:423–425. [DOI] [PubMed] [Google Scholar]
- Petrides M, Pandya DN (1999) Dorsolateral prefrontal cortex: comparative cytoarchitectonic analysis in the human and the macaque brain and corticocortical connection patterns. Eur J Neurosci 11:1011–1036. [DOI] [PubMed] [Google Scholar]
- Poo M-M, Du J-L, Ip NY, Xiong Z-Q, Xu B, Tan T (2016) China Brain Project: basic neuroscience, brain diseases, and brain-inspired computing. Neuron 92:591–596. [DOI] [PubMed] [Google Scholar]
- Preuss TM, Goldman-Rakic PS (1991a) Myelo-and cytoarchitecture of the granular frontal cortex and surrounding regions in the strepsirhine primate Galago and the anthropoid primate Macaca. J Comp Neurol 310:429–474. [DOI] [PubMed] [Google Scholar]
- Preuss TM, Goldman-Rakic PS (1991b) Architectonics of the parietal and temporal association cortex in the strepsirhine primate Galago compared to the anthropoid primate Macaca. J Comp Neurol 310:475–506. [DOI] [PubMed] [Google Scholar]
- Preuss TM (1995) Do rats have prefrontal cortex? The Rose-Woolsey-Akert program reconsidered. J Cogn Neurosci 7:1–24. [DOI] [PubMed] [Google Scholar]
- Preuss TM, Kaas JH (1996) Cytochrome oxidase “blobs” and other characteristics of primary visual cortex in a lemuroid primate, Cheirogaleus medius In: Brain Behav Evol, pp 103–112. Karger Publishers. [DOI] [PubMed] [Google Scholar]
- Preuss TM (2000) Taking the measure of diversity: comparative alternatives to the model-animal paradigm in cortical neuroscience. Brain Behav Evol 55:287–299. [DOI] [PubMed] [Google Scholar]
- Preuss TM (2001) The discovery of cerebral diversity: an unwelcome scientific revolution In: Evolutionary anatomy of the primate cerebral cortex (Falk D, Gibson KR, eds), pp 138–164. Cambridge: Cambridge University Press. [Google Scholar]
- Preuss TM (2004) Specializations of the human visual system: the monkey model meets human reality In: The Primate Visual System (Kaas JH, Collins CE, eds), pp 231–259. Boca Raton, FL: CRC Press. [Google Scholar]
- Preuss TM (2007) Evolutionary specializations of primate brain systems In: Primate Origins: Adaptations and Evolution (Ravosa MJ, Dagasto M, eds), pp 625–675. Boston, MA: Springer. [Google Scholar]
- Preuss TM (2010) Reinventing primate neuroscience for the twenty-first century. In: Neuroethology Primate (Platt ML, Ghazanfar A, eds), vol 1, pp 422–454. Oxford: Oxford University Press. [Google Scholar]
- Preuss TM, Robert JS (2014) Animals models of the human brain: repairing the paradigm In: The Cognitive Neurosciences Fifth Edition (Gazzaniga MS, Mangun GR, eds), pp 1–6. Cambridge, MA: MIT Press. [Google Scholar]
- Ravosa MJ, Dagosto M (eds) (2007) Primate origins adaptations and evolution. New York: Springer. [Google Scholar]
- Reader SM, Hager Y, Laland KN (2011) The evolution of primate general and cultural intelligence. Philosophical Transactions of the Royal Society B: Biological Sciences 366:1017–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly SK, Yin J, Ayoub AE, Emera D, Leng J, Cotney J, Sarro R, Rakic P, Noonan JP (2015) Evolutionary genomics. Evolutionary changes in promoter and enhancer activity during human corticogenesis. Science 347:1155–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittirsch D, Hoesel LM, Ward PA (2006) The disconnect between animal models of sepsis and human sepsis. Journal of Leukocyte Biology 81:137–143. [DOI] [PubMed] [Google Scholar]
- Roberts AC, Tomic DL, Parkinson CH, Roeling TA, Cutter DJ, Robbins TW, Everitt BJ (2007) Forebrain connectivity of the prefrontal cortex in the marmoset monkey (Callithrix jacchus): An anterograde and retrograde tract-tracing study. J Comp Neurol 502:86–112. [DOI] [PubMed] [Google Scholar]
- Roe AW, Fritsches K, Pettigrew JD (2005) Optical imaging of functional organization of V1 and V2 in marmoset visual cortex. Anat Rec A Discov Mol Cell Evol Biol 287:1213–1225. [DOI] [PubMed] [Google Scholar]
- Rosa MG, Krubitzer LA (1999) The evolution of visual cortex: where is V2? Trends in Neurosciences 22:242–248. [DOI] [PubMed] [Google Scholar]
- Rosa MGP, Tweedale R (2005) Brain maps, great and small: lessons from comparative studies of primate visual cortical organization. Philos Trans R Soc Lond, B, Biol Sci 360:665–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberger AL (1984) Aspects of the systematics and evolution of the marmosets In: A Primatoiogia no Brasil (Thiago de Melto M, ed), pp 159–180. Brasilia: Sociedade Brasileira de Primatologia. [Google Scholar]
- Ross C (1996) Adaptive explanation for the origins of the Anthropoidea (Primates). Am J Primatol 40:205–230. [DOI] [PubMed] [Google Scholar]
- Ross C, Kay RF (eds) (2004) Anthropoid origins: new visions. New York: Kluwer Academic/Plenum Publishers. [Google Scholar]
- Ross CN, French JA, Orti G (2007) Germ-line chimerism and paternal care in marmosets (Callithrix kuhlii). Proceedings of the National Academy of Sciences 104:6278–6282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallet J, Mars RB, Noonan MP, Neubert F-X, Jbabdi S, O’Reilly JX, Filippini N, Thomas AG, Rushworth MF (2013) The organization of dorsal frontal cortex in humans and macaques. Journal of Neuroscience 33:12255–12274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon AB (2016) Moving toward ‘common’ use of the marmoset as a non-human primate aging model. Pathobiol Aging Age Relat Dis 6:32758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki E (2015) Prospects for genetically modified non-human primate models, including the common marmoset. Neurosci Res 93:110–115. [DOI] [PubMed] [Google Scholar]
- Schiel N, Souto A (2017) The common marmoset: An overview of its natural history, ecology and behavior. Dev Neurobiol 77:244–262. [DOI] [PubMed] [Google Scholar]
- Schneider H, Sampaio I (2015) The systematics and evolution of New World primates -A review. Mol Phylogenet Evol 82:348–357. [DOI] [PubMed] [Google Scholar]
- Sengpiel F, Troilo D, Kind PC, Graham B, Blakemore C (1996) Functional architecture of area 17 in normal and monocularly deprived marmosets (Callithrix jacchus). Vis Neurosci 13:145–160. [DOI] [PubMed] [Google Scholar]
- Servick K (2018) US labs clamor for marmosets. Science 362:383–384. [DOI] [PubMed] [Google Scholar]
- Smith MD, Wertheim JO, Weaver S, Murrell B, Scheffler K, Kosakovsky Pond SL (2015) Less is more: an adaptive branch-site random effects model for efficient detection of episodic diversifying selection. Mol Biol Evol 32:1342–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TD, Dennis JC, Bhatnagar KP, Bonar CJ, Burrows AM, Morrison EE (2004) Ontogenetic observations on the vomeronasal organ in two species of tamarins using neuron-specific β-tubulin III. Anat Rec 278A:409–418. [DOI] [PubMed] [Google Scholar]
- Smith TD, Garrett EC, Bhatnagar KP, Bonar CJ, Bruening AE, Dennis JC, Kinznger JH, Johnson EW, Morrison EE (2011) The vomeronasal organ of New World monkeys (Platyrrhini). Anat Rec 294:2158–2178. [DOI] [PubMed] [Google Scholar]
- Smith TD, Laitman JT, Bhatnagar KP (2014) The shrinking anthropoid nose, the human vomeronasal organ, and the language of anatomical reduction. Anat Rec 297:2196–2204. [DOI] [PubMed] [Google Scholar]
- Smith TE, Tomlinson AJ, Mlotkiewicz JA, Abbott DH (2001) Female marmoset monkeys (Callithrix jacchus) can be identified from the chemical composition of their scent marks. Chem Senses 26:449–458. [DOI] [PubMed] [Google Scholar]
- Solomon SG (2002) Striate cortex in dichromatic and trichromatic marmosets: neurochemical compartmentalization and geniculate input. J Comp Neurol 450:366–381. [DOI] [PubMed] [Google Scholar]
- Solomon SG, Rosa MGP (2014) A simpler primate brain: the visual system of the marmoset monkey. Frontiers in Neural Circuits 8:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan H, Frahm H, Baron G (1981) New and revised data on volumes of brain structures in insectivores and primates. Folia Primatol 35:1–29. [DOI] [PubMed] [Google Scholar]
- Striedter GF, Belgard TG, Chen C-C, Davis FP, Finlay BL, Güntürkün O, Hale ME, Harris JA, Hecht EE, Hof PR, Hofmann HA, Holland LZ, Iwaniuk AN, Jarvis ED, Karten HJ, Katz PS, Kristan WB, Macagno ER, Mitra PP, Moroz LL, Preuss TM, Ragsdale CW, Sherwood CC, Stevens CF, Stüttgen MC, Tsumoto T, Wilczynski W (2014) NSF workshop report: discovering general principles of nervous system organization by comparing brain maps across species. Brain Behav Evol 83:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman RW, Kinzey WG (1984) The ecological role of the Callitrichidae: A review. Am J Phys Anthropol 64:419–449. [DOI] [PubMed] [Google Scholar]
- Sweeney CG, Curran E, Westmoreland SV, Mansfield KG, Vallender EJ (2012) Quantitative molecular assessment of chimerism across tissues in marmosets and tamarins. BMC genomics 13:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuno H, Watson C, Roberts A, Sasaki E, Okano H (2015) Marmoset neuroscience. Neurosci Res 93:1–2. [DOI] [PubMed] [Google Scholar]
- Troilo D, Howland HC, Judge SJ (1993) Visual optics and retinal cone topography in the common marmoset (Callithrix jacchus). Vision Res 33:1301–1310. [DOI] [PubMed] [Google Scholar]
- van Dyck CH (2018) Anti-amyloid-β monoclonal antibodies for Alzheimer’s disease: Pitfalls and promise. Biol Psychiatry 83:311–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt BA, Paxinos G (2012) Cytoarchitecture of mouse and rat cingulate cortex with human homologies. Brain Struct Funct 219:185–192. [DOI] [PubMed] [Google Scholar]
- Vogt BA, Hof PR, Zilles K, Vogt LJ, Herold C, Palomero-Gallagher N (2013) Cingulate area 32 homologies in mouse, rat, macaque and human: cytoarchitecture and receptor architecture. J Comp Neurol 521:4189–4204. [DOI] [PubMed] [Google Scholar]
- Wisely SM, Sayler KA, Anderson CJ, Boyce CL, Klegarth AR, Johnson SA (2018) Macacine Herpesvirus 1 Antibody Prevalence and DNA Shedding among Invasive Rhesus Macaques, Silver Springs State Park, Florida, USA. Emerg Infect Dis 24:345–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worley KC, Warren WC, Rogers J, Locke D, Muzny DM, Mardis ER, Weinstock GM, Tardif SD, Aagaard KM, Archidiacono N, Rayan NA, Batzer MA, Beal K, Brejova B, Capozzi O, Capuano SB, Casola C, Chandrabose MM, Cree A, Dao MD, de Jong PJ, del Rosario RC-H, Delehaunty KD, Dinh HH, Eichler EE, Fitzgerald S, Flicek P, Fontenot CC, Fowler RG, Fronick C, Fulton LA, Fulton RS, Gabisi RA, Gerlach D, Graves TA, Gunaratne PH, Hahn MW, Haig D, Han Y, Harris RA, Herrero J, Hillier LW, Hubley R, Hughes JF, Hume J, Jhangiani SN, Jorde LB, Joshi V, Karakor E, Konkel MK, Kosiol C, Kovar CL, Kriventseva EV, Lee SL, Lewis LR, Liu Y-s, Lopez J, Lopez-Otin C, Lorente-Galdos B, Mansfield KG, Marques-Bonet T, Minx P, Misceo D, Moncrieff JS, Morgan MB, Nazareth LV, Newsham I, Nguyen NB, Okwuonu G, Prabhakar S, Perales L, Pu L-L, Puente XS, Quesada V, Ranck MC, Raney BJ, Raveendran M, Deiros DR, Rocchi M, Rodriguez D, Ross C, Ruffier M, Ruiz SJ, Sajjadian S, Santibanez J, Schrider DR, Searle S, Skaletsky H, Soibam B, Smit AFA, Tennakoon JB, Tomaska L, Ullmer B, Vejnar CE, Ventura M, Vilella AJ, Vinar T, Vogel J-H, Walker JA, Wang Q, Warner CM, Wildman DE, Witherspoon DJ, Wright RA, Wu Y, Xiao W, Xing J, Zdobnov EM, Zhu B, Gibbs RA, Wilson RK, Co MGSA (2014) The common marmoset genome provides insight into primate biology and evolution. Nat Genet 46:850–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilles K, Armstrong E, Schlaug G, Schleicher A (1986) Quantitative cytoarchitectonics of the posterior cingulate cortex in primates. J Comp Neurol 253:514–524. [DOI] [PubMed] [Google Scholar]


