Summary
Over the last ten years there have been major advances in documenting and understanding dynamic changes to DNA methylation, small RNAs, chromatin modifications, and chromatin structure that accompany reproductive development in flowering plants from germline specification to seed maturation. Here I highlight recent advances in the field, mostly made possible by microscopic analysis of epigenetic states or by the ability to isolate specific cell types or tissues and apply omics approaches. I consider in which contexts there is potentially reprogramming vs maintenance or reinforcement of epigenetic states.
Keywords: DNA methylation, embryo, endosperm, epigenetic reprogramming, female gametophyte, plant reproduction, pollen
I. Introduction
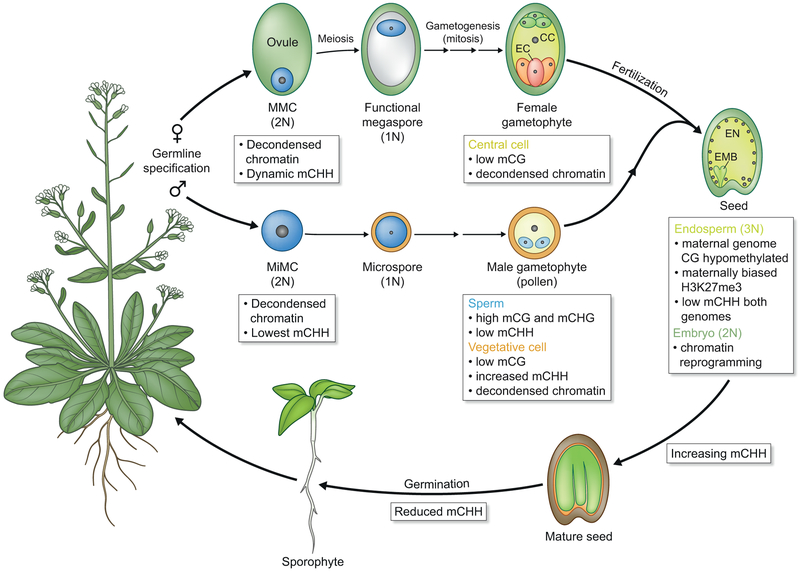
Reproduction is the most fundamental task of all organisms. In flowering plants, which alternate between haploid and diploid forms, successful reproduction occurs when egg and sperm unite to form a diploid embryo and, ultimately, a viable seed. Reproduction begins when cells in the diploid sporophyte undergo meiosis, creating haploid spores. Spores divide mitotically to form the haploid gametophyte phase of the life cycle. Gametes are specified in the male and female gametophytes (Fig. 1).
Figure 1: Epigenetic dynamics during flowering plant reproduction.
Summary of key steps in the flowering plant life cycle using Arabidopsis as an example. Associated epigenetic changes are described in the text. MMC, MiMC, functional megaspore, and microspore are the cells in dark blue. MMC, megaspore mother cell; MiMC, microspore mother cell, EC, egg cell; CC, central cell; VC, vegetative cell; EN, endosperm; EM, embryo; mCHH, CHH methylation; mCG, CG methylation; mCHG, CHG methylation.
Current evidence suggests that the most dynamic and consequential alterations to the epigenome occur during reproductive development (Fig. 1). This is sometimes referred to as epigenetic reprogramming. The term reprogramming is most frequently used in the context of animal stem cell biology, where it is taken to mean the erasure of information and the establishment of new information that allows cells to undergo a fate change over multiple cell divisions. For plants, this definition is perhaps useful with regards to regeneration from callus tissue, but appears less applicable to processes that occur during reproduction, in which differences in cell identity can be established over a single cell division. The key questions addressed here are 1) How much molecular reprogramming takes places during reproduction? 2) And what are the functional consequences of these dynamics?
Chromatin structure and modification, small non-coding RNAs, and DNA methylation will be considered here as reprogrammable substrates (Matzke and Mosher, 2014; Mozgova et al., 2015). These are not the only potential mediators of epigenetic inheritance, but they can be reasonably measured and the pathways that control them are understood genetically or biochemically. A major challenge in evaluating the extent of reproductive reprogramming is that the epigenetic status of the cells that become the next generation (the starting point for reprogramming) are few in number and inaccessible. All cells arise from meristems, with reproductive cells formed specifically from the L2 layer of apical meristems. The epigenetic state of the L2 cells is largely unknown, although features of meristematic regions are beginning to be probed (Sijacic et al., 2018; Gutzat et al., 2018). Thus, the molecular epigenetic state of reproductive cells is often compared to leaf or seedling tissue, and it is unknown how reflective these tissues are of the relevant cells in meristems.
II. Meiosis
In flowering plants, the cells that undergo meiosis are chosen based on their position within the developing flower. Lack of a segregated germline is a fundamental distinction between plant and animal reproduction. A sub-epidermal cell in the ovule primordium is chosen as the megaspore mother cell (MMC), which undergoes female meiosis. Both the MMC and its male analogue, the microspore mother cell (MiMC), experience large-scale chromatin changes during cell specification, including heterochromatin decondensation and increased nuclear volume, perhaps indicative of a highly active transcriptional state (She et al., 2013; She and Baroux, 2015). In maize and Arabidopsis, mutations in different AGO (ARGONAUTE) genes involved in small RNA-mediated heterochromatin silencing, and in the maize de novo DNA methyltransferases DMT102/103, either cause division of the MMC without meiosis (germ cells act like somatic cells) or result in the formation of additional cells that display physical and gene expression signatures similar to MMCs (somatic cells act like germ cells) (Garcia-Aguilar et al., 2010; Olmedo-Monfil et al., 2010; Singh et al., 2011). The AGO genes are expressed in the cells surrounding the MMC, indicating a cell non-autonomous effect on meiosis, which could be mediated by mobile small RNAs. The targets of these processes are unknown and the epigenetic state of the MMC has not been profiled at molecular resolution. Recently, fusions between DNA methylation-binding proteins and fluorescent reporters allowed visualization of methylation dynamics at a broad scale (Ingouff et al., 2017). CG methylation remains steady but CHH methylation transiently decreases and is then restored during MMC specification. Typically, CG and CHG methylation are maintained by maintenance methyltransferases MET1 and CMT3. CHH methylation is maintained by CMT2 or by the RNA-directed DNA methylation (RdDM) pathway, in which 24 nt small RNAs guide de novo methyltransferases to matching sites, usually in repetitive DNA like transposable elements (TEs) (Matzke and Mosher, 2014). Thus, low CHH methylation is often indicative of low de novo methylation activity. The cause and function of CHH methylation dynamics in the MMC is unknown.
DNA methylation has recently been profiled in the diploid Arabidopsis MiMC (Walker et al., 2018). The MiMC has high levels of CG and CHG methylation but low levels of CHH methylation. Interestingly, despite global CHH hypomethylation, Walker et al. (2018) identified several hundred genic regions of RdDM-dependent CHH hypermethylation compared to leaves. This is unusual because genes are not typical targets of RdDM. In some instances, this hypermethylation has functional consequences – for the MPS1 gene, loss of genic reproduction-specific RdDM led to incorrect splicing and a mild meiosis defect (Walker et al., 2018). Why and how RdDM is directed to genic regions should be an interesting area of future research.
III. Male gametogenesis
Mitosis of the haploid meiotic products generates the gametophytes. The ability to isolate pure populations of cells at different stages of male gametogenesis has illuminated developmental DNA methylation dynamics in Arabidopsis (Walker et al., 2018; Calarco et al., 2012; Ibarra et al., 2012). The male gametophyte, or pollen grain, consists of two gametes (sperm) and a vegetative cell, which functions in pollen tube growth. The vegetative cell and sperm are in distinct epigenetic states, despite a separation of only two mitotic divisions (Fig. 1). Sperm DNA is highly methylated in the CG and CHG context but has low CHH methylation in retrotransposons. By contrast, the vegetative cell has low CG and CHG methylation and high CHH methylation in heterochromatic TEs, chromatin is decondensed, and hypomethylated retrotransposons are mobile (Slotkin et al., 2009). How do these differences arise? CG and CHG methylation levels are quite similar from the MiMC, to the microspores, and then sperm (Walker et al., 2018). CHH methylation progressively increases, although it remains lower in sperm than in somatic cells (Walker et al., 2018; Calarco et al., 2012; Ibarra et al., 2012). The functional consequence of these dynamics are unknown. By contrast, in the vegetative cell RdDM restores CHH methylation from the lower levels present in the microspore. RdDM in heterochromatin might be important to counteract the loss of DDM1 expression in the vegetative cell because DDM1 is normally required to maintain heterochromatic DNA methylation (Slotkin et al., 2009; Schoft et al., 2009). Loss of CG methylation in the vegetative cell is at least partially due to active DNA demethylation by 5-methylcytosine DNA glycosylases (Ibarra et al., 2012; Calarco et al., 2012). There is increasing evidence that the epigenetic state of the vegetative cell might influence mRNA or translation profiles of the sperm (Slotkin et al., 2009; Grant-Downton et al., 2013; Martinez et al., 2016). In vegetative cells, hypomethylated, expressed retrotransposons give rise to abundant small RNAs that have been termed epigenetically activated siRNAs (easiRNAs). In a series of elegant experiments, Martinez et al. (2016) demonstrated that small RNAs expressed in the vegetative cell can target a miRNA site or an Athila retrotransposon siRNA site on reporter constructs in sperm. This process is dependent on AGO and Dicer genes typically involved in post-transcriptional gene silencing (Martinez et al., 2016). It has been speculated that small RNAs loaded into sperm from the vegetative cell might have functions after fertilization in the zygote or endosperm, but direct evidence is lacking (Slotkin et al., 2009; Calarco et al., 2012, Martinez et al., 2016).
IV. Female gametogenesis
The epigenetic dynamics associated with female gametogenesis are less clear, largely due to its relative inaccessibility. Female gametogenesis occurs when the haploid functional megaspore (the surviving product of meiosis) divides mitotically three times to yield the 8-celled female gametophyte, which is surrounded by maternal integument tissue (Fig. 1). As part of a recurring theme, the Argonaute AGO5 acts in a cell non-autonomous manner to promote the mitotic division of the functional megaspore (Tucker et al., 2012). Fluorescent reporters suggest that CG and CHH methylation remain fairly steady throughout female gametogenesis (Ingouff et al., 2017). The mature female gametophyte contains two gametes, a haploid egg cell and a diploid central cell. Fertilization of the egg by sperm makes the embryo, whereas fertilization of the central cell generates the triploid endosperm, which supports embryo growth. These female gametes, separated by 1-3 mitoses, are cytologically distinct, with the central cell exhibiting decondensed chromatin and reduced repressive histone modifications compared to the egg (Pillot et al., 2010). Like the pollen vegetative cell, the central cell expresses the 5-methylcytosine DNA glycosylase DME. It also has reduced expression of the maintenance and de novo methyltransferases (Jullien et al., 2012). Egg and central cells have been isolated from rice, Arabidopsis, and maize using manual microdissection or tagging approaches like INTACT and methylation analyzed. There is lower CG methylation in genes and TEs in Arabidopsis central cells compared to sperm, which is at least partly due to the expression of DME (Park et al., 2016). Like for the vegetative cell and sperm, it has been proposed that small RNAs move from the central cell to the egg to promote TE silencing (Ibarra et al., 2012). Direct evidence for such a process occurring is scarcer than for vegetative cell-sperm cell transmission of siRNAs. In some instances, passive movement of exogenously supplied small RNAs has been observed. In Torenia fournieri, 25 nt small RNAs injected into the central cell acted as morpholinos in the synergid cells, which flank the egg (Okuda et al., 2009). In Arabidopsis, labeled double-stranded 24 nt small RNAs injected into the central cell were later detected as both double and single-stranded RNAs in the egg cell, indicating movement (Erdmann et al., 2017a). Additionally, expression of an artificial miRNA against GFP from a central cell promoter reduced GFP fluorescence in the Arabidopsis egg cell (Ibarra et al., 2012).
V. Heritability and reinforcement of DNA methylation in the embryo
The frequency with which stable epialleles have been discovered in plants and direct measurements of methylation heritability between generations indicate that most DNA methylation is inherited through sexual reproduction (Schmitz et al., 2011; Becker et al., 2011). Existing methylation is reinforced during embryogenesis. When methylation is lost in hypomethylated recombinant inbred lines, it is partially replaced by RdDM by passage through reproduction (Texeira et al., 2009). Wild type sperm has low CHH methylation (Calarco et al., 2012), indicating that the paternal genome must gain methylation after fertilization. Recent studies from soybean and Arabidopsis have shown that CHH methylation increases during embryogenesis (Lin et al., 2017; Kawakatsu et al., 2017; Narsai et al., 2017). In mature Arabidopsis embryos, CHH methylation approaches 100% at individual cytosines, whereas in other tissues, including younger embryos, individual CHH sites are ~20% methylated (Bouyer et al., 2017). Methylation decreases upon seed germination, likely through a passive mechanism (Lin et al., 2017; Kawakatsu et al., 2017; Narsai et al., 2017; Bouyer et al., 2017). It is unknown whether the increased CHH methylation in developing embryos is functional – mutation of the de novo methyltransferases DRM2 and CMT2 prevents CHH methylation but without obvious effects on seed development or germination, although it is unclear how comprehensively phenotypes have been assessed.
VI. Chromatin-based epigenetic reprogramming in the embryo
Despite the heritability of DNA methylation between generations, there is clear evidence that chromatin-based epigenetic memory established during vegetative growth and development is erased during embryogenesis. In Arabidopsis, histone H3 from the egg and sperm are removed in the zygote and replaced by newly synthesized histones (Ingouff et al., 2010). The FLC locus represents a specific case of reprogramming. Levels of FLC expression control flowering time in vernalization-sensitive plants. Stable mitotic epigenetic silencing of FLC after prolonged cold stimulus is mediated by spreading of H3K27me3, mediated by PRC2 complexes (Yang et al., 2017). Vernalization memory is not transmitted to the next generation, indicating the occurrence of resetting. In vernalized plants, FLC is silenced in gametes but expression is reactivated during early embryogenesis (Sheldon et al., 2008; Choi et al., 2009; Tao et al., 2017). Two mechanisms have been indicated: activation by the NF-YB transcription factor LEC1 (Tao et al., 2017) and regulation by the histone demethylase ELF6 (Cervillen et al., 2014), which appears to play a more minor role. Reprogramming of chromatin by pioneer transcription factors (defined as transcription factors that can bind to their DNA motif even in the context of a nucleosome) during embryonic development is well-documented in animals (Zaret and Mango, 2016). It has been suggested that LEC1 acts as a pioneer factor, but this has not been specifically demonstrated biochemically (Tao et al., 2017).
VII. Modulation of epigenetic information in the endosperm
Endosperm is epigenetically divergent from other plant tissues, as has been reviewed extensively elsewhere (Satyaki and Gehring, 2017), with the added twist that some effects are specific to maternally or paternally inherited genomes. Consistent with DNA demethylation in the central cell, endosperm DNA is maternally hypomethylated. The endosperm maternal genome is even less CG methylated than the central cell, suggesting additional loss of DNA methylation occurs either passively or actively after fertilization (Park et al., 2016). CHH methylation also appears dynamic, with low expression of RdDM genes early in development (Jullien et al., 2012; Belmonte et al., 2013) and low CHH methylation at 4 DAP (Moreno-Romero et al., 2016). By 6-8 DAP when the endosperm has cellularized and differentiated, typical levels of CHH methylation are observed on the paternal endosperm genome (Ibarra et al., 2012; Pignatta et al., 2014). Histone modifications are also asymmetric between maternal and paternal genomes, with the PRC2 mark H3K27me3 favoring maternal alleles in euchromatin, particularly at regions that were demethylated in the central cell, and paternal alleles in pericentromeric heterochromatin (Moreno-Romero et al., 2016). Small RNAs have also been profiled in Arabidopsis, rice, and maize endosperm. In Arabidopsis, the endosperm 24 nt small RNA population is paternally-biased, consistent with higher methylation of the paternal genome (Erdmann et al., 2017b). Endosperm also has a population of genic small RNAs, another instance of increased genic small RNAs observed during reproduction. Discrete regions of maternal and paternal sRNA bias have been identified, but is unknown how small RNAs are preferentially produced from one allele or whether this is connected to other asymmetric epigenetic modifications.
There are several possible consequences of endosperm epigenetic dynamics. Methylation state can impact the expression of key epigenetic regulators, thus potentially creating cascading effects caused by the expression of alternative epigenetic pathways in tissues that undergo dynamic DNA methylation changes, like endosperm. Epigenetic regulators regulated by DNA methylation include the histone demethylase IBM1, the 5-methylcytosine DNA glycosylase ROS1, and the Dicer enzyme DCL4 (Rigal et al., 2012; Williams et al., 2015; Pumplin et al., 2016). For these genes, DNA methylation alters splicing, promotes expression, or alters transcriptional start site selection. Another consequence of endosperm methylation dynamics is the establishment of gene imprinting, the preferential expression of one allele of a gene dependent on its parent-of-origin. For example, the majority of paternally expressed imprinted genes are associated with methylated paternal alleles and repressed maternal alleles that are DNA hypomethylated, modified by H3K27me3, and associated with genic small RNAs (Pignatta et al., 2014; Moreno-Romero et al., 2016; Erdmann et al., 2017b, Satyaki and Gehring, 2017). Imprinting dynamics can intersect with natural epigenetic polymorphisms or environmentally induced methylation changes to generate diverse seed phenotypes (Pignatta et al. 2018; Iwasaki et al., 2019). For example, the maternally expressed imprinted gene ALN negatively regulates seed dormancy (Iwasaki et al., 2019). Seeds that develop at 10°C have increased DNA methylation at the ALN locus, reduced ALN expression, and higher levels of dormancy, consistent with an increase in seed dormancy associated with development in cold temperatures (Iwasaki et al., 2019). Differences in endosperm DNA methylation and gene expression programs induced by environmental conditions is largely unexplored (and exciting) territory.
VIII. Conclusions
Recent studies with improved tissue- or cell type-specificity have expanded understanding of the extent of epigenetic dynamics that occur during flowering plant reproductive development. Unlike in mammals, there is no genome-wide erasure and reestablishment of epigenetic information during reproduction. Rather, chromatin-based epigenetic information appears to be either passively lost or actively reprogrammed in the embryonic lineage, at least based on a limited number of examples, and DNA methylation-based epigenetic information is maintained, reinforced, and inherited. Although CHH methylation dynamics have been discovered at multiple stages of reproductive development, it is not yet clear whether any of these instances represent reprogramming or are important for change from one cell state to another. Integrating concepts and language from other disciplines is important for bridging scientific fields, especially those with many true commonalities, like the study of epigenetics in plant, animal, and fungal systems. When we talk about epigenetic reprogramming in plants, it is important to be clear where this concept might differ from other organisms, and how this is integrated with differences in development.
Acknowledgements
I thank members of my lab for discussions and Colette Picard for help with figure preparation. Research in my lab on plant epigenetics is supported by NSF MCB 1453459 and NIH R01GM112851 grants.
References
- Becker C, Hagmann J, Müller J, Koenig D, Stegle O, Borgwardt K, Weigel D. (2011) Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature 480: 245–9. [DOI] [PubMed] [Google Scholar]
- Belmonte MF, Kirkbride RC, Stone SL, Pelletier JM, Bui AQ, Yeung EC, Hashimoto M, Fei J, Harada CM, Munoz MD, Le BH, Drews GN, Brady SM, Goldberg RB, Harada JJ. (2013) Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed. Proc Natl Acad Sci U S A. 110: E435–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouyer D, Kramdi A, Kassam M, Heese M, Schnittger A, Roudier F, Colot V. (2017) DNA methylation dynamics during early plant life. Genome Biol. 18: 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarco JP, Borges F, Donoghue MT, Van Ex F, Jullien PE, Lopes T, Gardner R, Berger F, Feijó JA, Becker JD, Martienssen RA. (2012) Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell 151: 194–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crevillén P, Yang H, Cui X, Greeff C, Trick M, Qiu Q, Cao X, Dean C. (2014). Epigenetic reprogramming that prevents transgenerational inheritance of the vernalized state. Nature 515: 587–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Hyun Y, Kang MJ, In Yun H, Yun JY, Lister C, Dean C, Amasino RM, Noh B, Noh YS, Choi Y. (2009) Resetting and regulation of Flowering Locus C expression during Arabidopsis reproductive development. Plant J. 57:918–31. [DOI] [PubMed] [Google Scholar]
- Erdmann RM, Hoffmann A., Walter H-K, Wagenknecht H-A, Grob-Hardt R, Gehring M. (2017a) Molecular movement in the Arabidopsis thaliana female gametophyte. Plant Reprod 30: 141–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann RM, Satyaki PR, Klosinska M, Gehring M. (2017b) A small RNA pathway mediates allelic dosage in endosperm. Cell Rep. 21: 3364–3372. [DOI] [PubMed] [Google Scholar]
- Garcia-Aguilar M, Michaud C, Leblanc O, Grimanelli D. (2010) Inactivation of a DNA methylation pathway in maize reproductive organs results in apomixis-like phenotypes. Plant Cell 22:3249–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant-Downton R, Kourmpetli S, Hafidh S, Khatab H, Le Trionnaire G, Dickinson H, Twell D. (2013) Artificial microRNAs reveal cell-specific differences in small RNA activity in pollen. Curr Biol 23: R599–R601. [DOI] [PubMed] [Google Scholar]
- Gutzat R, Rembart K, Nussbaumer T, Pisupati R, Hofmann F, Bradamante G, Daubel N, Gaidora A, Lettner N, Dona M, Nordborg M, Nodine M, Mittelsten Scheid O. (2018) Stage-specific transcriptomes and DNA methylomes indicate an early and transient loss of transposon control in Arabidopsis shoot stem cells. bioRxiv 430447; doi: https://doi.org/10.110¼30447 [Google Scholar]
- Jullien PE, Susaki D, Yelagandula R, Higashiyama T, Berger F. (2012). DNA methylation dynamics during sexual reproduction in Arabidopsis thaliana. Curr Biol. 22: 1825–30. [DOI] [PubMed] [Google Scholar]
- Ibarra CA, Feng X, Schoft VK, Hsieh TF, Uzawa R, Rodrigues JA, Zemach A, Chumak N, Machlicova A, Nishimura T, Rojas D, Fischer RL, Tamaru H, Zilberman D. (2012) Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science. 337:1360–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingouff M, Rademacher S, Holec S, Soljić L, Xin N, Readshaw A, Foo SH, Lahouze B, Sprunck S, Berger F (2010). Zygotic resetting of the HISTONE 3 variant repertoire participates in epigenetic reprogramming in Arabidopsis. Curr Biol. 20: 2137–43. [DOI] [PubMed] [Google Scholar]
- Ingouff M, Selles B, Michaud C, Vu TM, Berger F, Schorn AJ, Autran D, Nowack MK, Martienssen RA, Grimanelli D. (2017) Live-cell analysis of DNA methylation during sexual reproduction in Arabidopsis reveals context and sex-specific dynamics controlled by noncanonical RdDM. Genes Dev 31: 72–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki M, Hyvärinen L, Piskurewicz U, Lopez-Molina L. (2019). Non-canonical RNA-directed DNA methylation participates in maternal and environmental control of seed dormancy. eLife 8: e37434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakatsu T, Nery JR, Castanon R, Ecker JR. (2017) Dynamic DNA methylation reconfiguration during seed development. Genome Biol. 18: 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JY, Le BH, Chen M, Henry KF, Hur J, Hsieh TF, Chen PY, Pelletier JM, Pellegrini M, Fischer RL, Harada JJ, Goldberg RB. (2017) Similarity between soybean and Arabidopsis seed methylomes and loss of non-CG methylation does not affect seed development. Proc Natl Acad Sci U S A. 114: E9730–E9739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez G, Panda K, Kohler C, Slotkin RK (2016). Silencing in the sperm cells is directed by RNA movement from the surrounding nurse cell. Nat Plants 2: 16030. [DOI] [PubMed] [Google Scholar]
- Matzke MA, Mosher RA (2014) RNA-directed DNA methylation: an epigenetic pathways of increasing complexity. Nat Rev Genet 15: 394–408. [DOI] [PubMed] [Google Scholar]
- Moreno-Romero J, Jiang H, Santos-Gonzalez J, Kohler C. (2016). Parental epigenetic asymmetry of PRC2-mediated histone modifications in the Arabidopsis endosperm. EMBO J. 35: 1298–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozgova I, Kohler C, Hennig L (2015) Keeping the gate closed: functions of the polycomb repressive complex PRC2 in development. Plant J 83: 121–132. [DOI] [PubMed] [Google Scholar]
- Narsai R, Gouil Q, Secco D, Srivastava A, Karpievitch YV, Liew LC, Lister R, Lewsey MG, Whelan J. (2017) Extensive transcriptomic and epigenomic remodeling occurs during Arabidopsis thaliana germination. Genome Biol. 18: 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda S, Tsutsui H, Shiina K, Sprunck S, Takeuchi H, Yui R, Kasahara RD, Hamamura Y, Mizukami A, Susaki D, Kawano N, Sakakibara T, Namiki S, Itoh K, Otsuka K, Matsuzaki M, Nozaki H, Kuroiwa T, Nakano A, Kanaoka MM, Dresselhaus T, Sasaki N, Higashiyama T. (2009) Defensin-like polypeptide LUREs are pollen tube attractants secreted from synergid cells. Nature 458: 357–361. [DOI] [PubMed] [Google Scholar]
- Olmedo-Monfil V, Durán-Figueroa N, Arteaga-Vázquez M, Demesa-Arévalo E, Autran D, Grimanelli D, Slotkin RK, Martienssen RA, Vielle-Calzada JP (2010). Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature. 464: 628–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park K, Kim MY, Vickers M, Park JS, Hyun Y, Okamoto T, Zilberman D, Fischer RL, Feng X, Choi Y, Scholten S. (2016) DNA demethylation is initiated in the central cells of Arabidopsis and rice. Proc Natl Acad Sci U S A. 113: 15138–15143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignatta D, Erdmann RM, Scheer E, Picard CL, Bell GW, Gehring M. (2014) Natural epigenetic polymorphisms lead to intraspecific variation in Arabidopsis gene imprinting. eLife 3: e03198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignatta D, Novitzky K, Satyaki PRV, Gehring M. (2018) A variably imprinted epiallele impacts seed development. PLoS Genet. 14: e1007469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillot M, Baroux C, Vazquez MA, Autran D, Leblanc O, Vielle-Calzada JP, Grossniklaus U, Grimanelli D. (2010) Embryo and endosperm inherit distinct chromatin and transcriptional states from the female gamete in Arabidopsis. Plant Cell 22: 307–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pumplin N, Sarazin A, Jullien PE, Bologna NG, Oberlin S, Voinnet O. (2016). DNA methylation influences the expression of DICER-LIKE4 isoforms, which encode proteins of alternative localization and function. Plant Cell 28: 2786–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigal M, Kevei Z, Pélissier T, Mathieu O. (2012) DNA methylation in an intron of the IBM1 histone demethylase gene stabilizes chromatin modification patterns. EMBO J. 31: 2981–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satyaki PR, Gehring M. (2017) DNA methylation and imprinting in plants: machinery and mechanisms. Crit Rev Biochem Mol Biol. 52: 163–175. [DOI] [PubMed] [Google Scholar]
- Schoft VK, Chumak N, Mosiolek M, Slusarz L, Komnenovic V, Brownfield L, Twell D, Kakutani T, Tamaru H. (2009). Induction of RNA-directed DNA methylation upon decondensation of constitutive heterochromatin. EMBO Rep. 10: 1015–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz RJ, Schultz MD, Lewsey MG, O'Malley RC, Urich MA, Libiger O, Schork NJ, Ecker JR. (2011) Transgenerational epigenetic instability is a source of novel methylation variants. Science 334: 369–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She W, Grimanelli D, Rutowicz K, Whitehead MW, Puzio M, Kotlinski M, Jerzmanowski A, Baroux C. (2013). Chromatin reprogramming during the somatic-to-reproductive cell fate transition in plants. Development 140: 4008–4019. [DOI] [PubMed] [Google Scholar]
- She W, Baroux C. (2015) Chromatin dynamics in pollen mother cells underpin a common scenario at the somatic-to-reproductive fate transition of both the male and female lineages in Arabidopsis. Front Plant Sci. 6: 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon CC, Hills MJ, Lister C, Dean C, Dennis ES, Peacock WJ. (2008) Resetting of FLOWERING LOCUS C expression after epigenetic repression by vernalization. Proc Natl Acad Sci U S A. 105: 2214–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijacic P, Bajic M, McKinney EC, Meagher RB, Deal RB (2018). Changes in chromatin accessibility between Arabidopsis stem cells and mesophyll cells illuminate cell type-specific transcription factor networks. Plant J. 94:215–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M, Goel S, Meeley RB, Dantec C, Parrinello H, Michaud C, Leblanc O, Grimanelli D. (2011) Production of viable gametes without meiosis in maize deficient for an ARGONAUTE protein. Plant Cell 23: 443–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin RK, Vaughn M, Borges F, Tanurdzić M, Becker JD, Feijó JA, Martienssen RA. (2009) Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136: 461–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Z, Shen L, Gu X, Wang Y, Yu H, He Y. (2017). Embryonic epigenetic reprogramming by a pioneer transcription factor in plants. Nature 551: 124–128. [DOI] [PubMed] [Google Scholar]
- Teixeira FK, Heredia F, Sarazin A, Roudier F, Boccara M, Ciaudo C, Cruaud C, Poulain J, Berdasco M, Fraga MF, Voinnet O, Wincker P, Esteller M, Colot V. (2009) A role for RNAi in the selective correction of DNA methylation defects. Science 323: 1600–1604. [DOI] [PubMed] [Google Scholar]
- Tucker MR, Okada T, Hu Y, Scholefield A, Taylor JM, Koltunow AM. (2012). Somatic small RNA pathways promote the mitotic events of megagametogenesis during female reproductive development in Arabidopsis. Development 139:1399–404. [DOI] [PubMed] [Google Scholar]
- Walker J, Gao H, Zhang J, Aldridge B, Vickers M, Higgins JD, Feng X. (2018). Sexual-lineage-specific DNA methylation regulates meiosis in Arabidopsis. Nat Genet. 50: 130–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BP, Pignatta D, Henikoff S, Gehring M. (2015) Methylation-sensitive expression of a DNA demethylase gene serves as an epigenetic rheostat. PLOS Genet. 11: e1005142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Berry S, Olsson TSG, Hartley M, Howard M, Dean C. (2017) Distinct phases of Polycomb silencing to hold epigenetic memory of cold in Arabidopsis. Science 357: 1142–1145 [DOI] [PubMed] [Google Scholar]
- Zaret KS, Mango SE. (2016) Pioneer transcription factors, chromatin dynamics, and cell fate control. Curr Opin Genet Dev 37: 76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]