Summary
BIR1 is a receptor-like kinase that functions as a negative regulator of basal immunity and cell death in Arabidopsis.
Using Arabidopsis thaliana and Tobacco rattle virus (TRV), we investigate the antiviral role of BIR1, the molecular mechanisms of BIR1 gene expression regulation during viral infections, and the effects of BIR1 overexpression on plant immunity and development.
We found that SA acts as a signal molecule for BIR1 activation during infection. Inactivating mutations of BIR1 cause strong antiviral resistance that is not due to constitutive cell death or SA defense priming in the bir1-1 mutant. RNA-directed DNA methylation (RdDM) and post-transcriptional silencing are both required to negatively regulate BIR1 expression upon viral induction. BIR1 overexpression causes severe developmental defects, cell death and premature death that correlate with the constitutive activation of plant immune responses.
Our findings suggest that BIR1 acts as a negative regulator of antiviral defense in plants, and indicate that RNA silencing contributes, alone or in conjunction with other regulatory mechanisms, to define a threshold expression for proper BIR1 function beyond which an autoimmune response may occur. This work provides novel mechanistic insights into the regulation of BIR1 homeostasis that may be common for other plant immune components.
Keywords: Antiviral defense, BAK1, BIR1, plant innate immunity, plant viruses, post-transcriptional silencing, RNA-directed DNA methylation, SOBIR1
Introduction
To defend themselves against invaders, plants have evolved potent inducible immune responses (Dangl & Jones, 2001). The frontline of active defense relies on the recognition of conserved microbial components named Pathogen-Associated Molecular Patterns (PAMPs) by membrane-localized receptor-like kinases (RLK) and receptor-like proteins (RLP) to induce PAMP-Triggered Immunity (PTI) (Boller & Felix, 2009; Tena et al., 2011). PTI prevents colonization by pathogens such as bacteria, fungi and oomycetes and includes activation of mitogen-activated protein kinases (MAPK), production of reactive oxygen species (ROS), generation of the signal molecule salicylic acid (SA), differential expression of genes, callose deposition and stomatal closure (Dodds & Rathjen, 2010). Pathogens hit back by producing effectors that suppress different steps of PTI, resulting in Effector-Triggered Susceptibility (ETS) (Jones & Dangl, 2006). As a counter-counter defense strategy, plants possess a repertoire of polymorphic disease resistance (R) proteins containing nucleotide-binding (NB) and leucine-rich repeat (LRR) domains (Martin et al., 2003; Meyers et al., 2003). These R immune receptors can sense effectors directly or indirectly and establish Effector-Triggered-Immunity (ETI). ETI responses significantly overlap with PTI signaling cascades, albeit with a stronger amplitude, and often result in a form of programmed cell death at the infection sites that restricts pathogen progression (Coll et al., 2011).
Recent studies show that RNA silencing is a key regulatory checkpoint modulating both PTI and ETI responses in plants (Zvereva & Pooggin, 2012; Boccara et al., 2014). Growing evidence illustrates the role of PAMP-responsive microRNAs (miRNAs) and small interfering RNAs (siRNAs) in plant innate immunity against microbial pathogens (Katiyar-Agarwal et al., 2006; Navarro et al., 2006; Katiyar-Agarwal et al., 2007; Navarro et al., 2008; Li et al., 2010; Zhang et al., 2011; Campo et al., 2013; Boccara et al., 2014; Li et al., 2014; Ouyang et al., 2014), and it is well documented how small RNA regulatory networks exert extensive post-transcriptional control of disease resistance genes to prevent undesirable R-mediated autoimmunity in unchallenged plants (Yi & Richards, 2007; Zhai et al., 2011; Boccara et al., 2014). Furthermore, RNA-directed DNA methylation (RdDM) provides epigenetic control of plant defenses by targeting transposable elements and their adjacent defense genes (Dowen et al., 2012; Yu et al., 2013; Lopez Sanchez et al., 2016). Immune responses against viruses are thought to rely mostly on ETI upon recognition of virus-specific effectors by intracellular immune-R receptors (Zvereva & Pooggin, 2012). In this line, interesting connections between RNA silencing-mediated regulation of R genes and viral infections have been made. For instance, Brassica miR1885 is induced specifically by Turnip mosaic virus (TuMV) infection, and targets NB–LRR class disease-resistant transcripts for cleavage (He et al., 2008). Also, members of the miR482/2118 superfamily mediate silencing of multiple NB-LRR disease resistance genes in tomato, which includes production of RNA-dependent RNA polymerase 6 (RDR6)-dependent secondary siRNAs (Shivaprasad et al., 2012). Interestingly, the miR482-mediated silencing cascade is suppressed in plants infected with viruses or bacteria allowing pathogen-inducible expression of NB-LRR targets (Shivaprasad et al., 2012). In another study, two miRNAs (miR6019 and miR6020) guide cleavage and production of functional secondary siRNAs from transcripts of the NB-LRR immune receptor N from tobacco that confers resistance to Tobacco mosaic virus (TMV) (Li et al., 2012). Overexpression of both miRNAs attenuates N-mediated resistance to TMV, demonstrating that miRNAs and secondary siRNAs have a functional role in regulating resistance to TMV.
Although in plants, apparently, there are no equivalent PAMPs derived from viruses, several studies have suggested a role of PTI in antiviral defense (Korner et al., 2013; Gouveia et al., 2016; Nicaise & Candresse, 2017). For instance, a recent report shows that Arabidopsis mutants deficient in the PTI master regulator BRASSINOSTEROID INSENSITIVE1 (BRI1)-ASSOCIATED RECEPTOR KINASE1 (BAK1) exhibit increased susceptibility to different RNA viruses (Korner et al., 2013). BAK1 interacts in vivo with the RLK BAK1-INTERACTING RECEPTOR-LIKE KINASE 1 (BIR1), a negative regulator of PTI responses and cell death pathways in Arabidopsis (Gao et al., 2009). It has been suggested that BIR1 sequesters BAK1 to prevent unwanted interactions with ligand-binding receptors in the absence of pathogens (Gao et al., 2009; Ma et al., 2017). Here, we study the role of BIR1 during viral infections and the molecular mechanisms whereby BIR1 is regulated. We further show that BIR1 regulation is critical to avoid constitutive activation of plant defense responses, which drastically impairs plant fitness and growth.
Materials and Methods
Plant material
Nicotiana benthamiana and Arabidopsis thaliana plants were grown in controlled environmental chambers under long day conditions (16h day/8h night) at 25°C and 22°C, respectively. Arabidopsis lines used in this study were derived from the Columbia-0 (Col-0) ecotype. Mutants for bir1-1 and sobir1-12 and bir1-1/BIR1 lines were donated by Yuelin Zhang (University of British Columbia, Canada). The Arabidopsis ago1-27, ago1-25, ago2-1 and mutant combinations involving the alleles rdr1-1, rdr2-1, rdr6-15, dcl2-1, dcl3-1 and dcl4-2 were donated by James C. Carrington (The Donald Danforth Plant Center, MO, USA). Arabidopsis mutant cmt3 and ddc were supplied by Steve Jacobsen (UCLA-HHMI, USA). The Arabidopsis nrpe1 (nrpd1b-11) was donated by Craig Pikaard (Indiana University, USA). The Arabidopsis mutant drm2-2 was supplied by Eric Richards (Boyce Thompson Institute, Cornell University, USA). The Arabidopsis npr1-1 and NPR1ox seeds were supplied by Xinniang Dong (Duke University, NC, USA).
Construction of a recombinant TRV-BIR1 vector and viral inoculation
Tobacco rattle virus (TRV) derivatives were created from an infectious TRV clone (Liu et al., 2002). TRV-GFP contained the soluble modified green fluorescence protein (GFP) under the promoter region of the Pea early browning virus (PEBV) replicase (Fernandez-Calvino et al., 2016a). TRV-BIR1 contained the Arabidopsis BIR1 coding region under the PEBV promoter. Briefly, the BIR1 cDNA containing its 5’ UTR was amplified by RT-PCR, cloned into the Gateway pDONR207 vector, and shuffled into the binary destination vector pGWB14. The HA-tagged BIR1 sequence was then PCR amplified, and cloned into pTRV2. The recombinant clones were screened by restriction enzyme digestion and sequencing. TuMV-GFP derived from an infectious clone of TuMV strain UK1 (Lellis et al., 2002). All primers used in this study are listed in Table S1.
N. benthamiana plants were inoculated at approximately 21 days after germination by infiltration of agrocultures containing TRV or TuMV (Johansen & Carrington, 2001; Liu et al., 2002). Three-week olds Arabidopsis plants were inoculated using sap extracts from virus-infected N. benthamiana leaves as described (Fernandez-Calvino et al., 2014). Arabidopsis plants inoculated with sap from non-infiltrated N. benthamiana were used as controls (mock). Additionally, experiments were paralleled using naïve Arabidopsis plants to discard potential side effects due to wounding caused by abrasion used during mechanical inoculation of sap extracts.
Construction of BIR1 transgenic plants
Arabidopsis Col-0 transgenic plants expressing the GFP:GUS dual reporter gene under the BIR1 promoter were generated using the Gateway compatible pBGWFS7 binary vector. A genomic DNA fragment of 3,297 bp containing the BIR1 promoter was cloned upstream to the fusion reporter gene as described (Xiao et al., 2010). Arabidopsis Col-0 transgenic plants expressing BIR1 were obtained using a glucocorticoid (DEX)-inducible gene expression system (Marques-Bueno et al., 2016). Briefly, the GVG::ter::6×UAS/pDONR221 contained the GVG cassette cloned into pDONR221. mCherry was added to this vector to generate GVG::ter::6×UAS::mCherry/ pDONR221. pDONR221-BIR1 contained the full-length BIR1 protein coding gene as described above. Final destination vectors were obtained by three-fragment recombination using the pH7m34GW destination vector. All the constructs were transformed into wild type Col-0 plants according to standard floral dipping (Clough & Bent, 1998). Independent homozygous lines harboring a single transgene insertion were selected in T4 and used for subsequent experiments.
Methylation analyses
Chop-PCR was carried out as described (Bohmdorfer et al., 2014) using genomic DNA (100 ng) from 3-week-old Arabidopsis rosette leaves and the methylation-sensitive restriction enzymes DdeI and NlaIII. Chop qPCR was done using Maxima Hot Start Taq DNA Polymerase (Thermo Scientific) and 25× SYBR Green (Invitrogen) diluted at 1:400.
Bisulfite sequencing was done as described (He et al., 2009). Briefly, genomic DNA from 3-week-old rosette leaves was extracted using DNeasy Plant Mini Kit (QIAGEN). Bisulfite conversion was done using EZ DNA Methylation Startup kit (Zymo Research). PCR was done using Maxima Hot Start Taq DNA Polymerase (Thermo Scientific), and amplification products were cloned into TOPO TA plasmids (Invitrogen). At least 30 clones per sample were sequenced. A non-methylated region at coordinates 19,573,407 to 19,573,671 in chromosome 4 was included as bisulfite conversion control. Primers for bisulfite were designed as described (Patterson et al., 2011) and listed in Table S1.
RNA analysis
Total RNA was extracted with TRIzol reagen (Invitrogen). One-step qRT-PCR was carried out using Brilliant III Ultra-Fast SYBR Green QRT-PCR Master Mix (Agilent Technologies) in a Rotor-Gene 6000/Rotor-Gene Q real-time PCR machine (Corbett/Qiagen) (Fernandez-Calvino et al., 2016a). Relative gene expression was determined using the Delta-delta cycle threshold method and Rotor-Gene 6000 Series Software (Corbett). Constitutively expressed CBP20 (At5g44200) or Actin2 (At3g18780) transcripts were used for normalization because of its similar level of expression in mock-inoculated and virus-infected leaves. A standard curve of known concentration of in vitro synthesized TRV transcripts was used to determine the TRV concentration as the number of viral copies per nanogram of total RNA (Fernandez-Calvino et al., 2016a). Significant differences between two or among several samples were compared by t-student test or one-way analysis of variance (ANOVA) followed by Duncan’s test, respectively, using Statgraphics Plus, version 5.1 (Statistical Graphics Corp.). Unless otherwise indicated, each Arabidopsis sample used for qRT-PCR analysis consisted in RNA extracted from a pool of rosette leaves from five plants (three leaves per plant, all leaves at identical positions).
Protein analysis
Protein extracts were prepared and analyzed by immunoblot assay after SDS-PAGE (Fernandez-Calvino et al., 2016b). Blotted proteins were detected using commercial horseradish peroxide (HRP)-conjugated secondary antibodies and a chemiluminescent substrate (LiteAblot Plus). Relative protein accumulation was measured by densitometry of protein blots exposed to autoradiographic films using the Image J Software.
Small RNA sequencing, construction of degradome libraries and 5’ RACE
Young rosette leaves from virus-infected plants and the corresponding mock-inoculated plants were pooled (10-12 plants) at 8 dpi (TRV) or 14 dpi (TuMV), and used for degradome or sRNA sequencing. Systemically infected inflorescences from TRV-infected or mock-inoculated Arabidopsis were pooled (10-15 plants) at 16 dpi, and used for degradome sequencing. Total RNA was extracted using TRIzol reagen (Invitrogen) or Plant RNeasy Kit (QIAGEN) and tested through Agilent 2100 bioanalyzer system to guarantee RNA quality. sRNA libraries were prepared and sequenced on an Illumina Genome Analyzer (HiSeq2000, 1×50bp, single-end run) by Ascidea Computational Biology Solutions (www.ascidea.com).
Parallel analysis of RNA ends (PARE) degradome libraries were done as described (German et al., 2009) and sequenced on an Illumina Genome Analyzer (HiSeq2000, 1×50bp, single-end run) by Fasteris (www.fasteris.com) and IGA technology (www.igatechnology.com). Sequencing data was then analyzed using CleaveLand4 (Addo-Quaye et al., 2009). Briefly, all degradome sequence reads with exact matches to structural RNA were removed and filtered dataset was mapped against the Arabidopsis cDNA sequence transcriptome (TAIR10) using Bowtie. For each exact match, 13-nt long sequences upstream and downstream of the location of the 5’-end of the matching degradome sequence was extracted to create a 26-nt long ‘query’ mRNA subsequence. Query sequences were then aligned to each sRNA sequence in our sRNA datasets or to miRNA reported in miRBase using GSTAr (Addo-Quaye et al., 2009). A modified 5’-RACE was used for mapping internal cleavage sites as described (Donaire et al., 2011).
SA application and determination of SA content
Three-week old plants grown on soil were sprayed with SA (1 mM) as described (Takahashi et al., 2007). To test the effect of SA on TRV accumulation, plants were TRV- or mock-inoculated 24h after the first SA application and then plants were treated for eight consecutive days by spraying the solution once at intervals of 24h (Exp #1) or 48h (Exp #2). To assess SA content in the plant tissue, rosette leaves were harvested at the same leaf position in order to minimize variations in the hormone content throughout the plant. SA was extracted and derivatized as described (Vallarino & Osorio, 2016). The samples were analyzed by using gas chromatography coupled to time-of-flight mass spectrometry (GC-TOF-MS) (Pegasus III, Leco), and quantified using an internal standard ([2H4]-SA; OlChemIm Ltd, Olomouc, Czech Republic).
Accession numbers
DNA methylation data (GSE39901) were used from (Stroud et al., 2013). Degradome sequencing data from naïve Col-0 inflorescences (GSM280226) was reported previously (German et al., 2008). Sequence data from this article can be found in the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession numbers: GSM3019138, GSM3019139, GSM3019140 (deep sequencing of degradome tags); GSM2808011, GSM2808012, GSM3019141, GSM3019142 (deep sequencing of sRNAs).
Results
Inactivating mutations in the immune repressor BIR1 triggers resistance to TRV
To gain insight into the role of Arabidopsis BIR1 (At5g48380) in the infectious process we monitored BIR1 expression during infection with TRV in a time-course experiment. We found that BIR1 transcripts were significantly induced in leaves of TRV-infected plants at 5 and 8 days post-inoculation (dpi) compared to mock-inoculated controls (Fig. 1a). BIR1 was also up regulated in response to the unrelated TuMV (Fig. S1a). Using an Arabidopsis bir1-1 mutant, we found that depletion of BIR1 led to strong antiviral resistance against TRV (Fig. 1b). However, TRV levels were reverted back to wild type plants, or even higher, in bir1-1 complemented lines (bir1-1/BIR1-HA) expressing a HA-tagged wild-type BIR1 coding gene (Fig. 1b). This result confirmed that the resistance phenotype observed in bir1-1 was caused by mutation in BIR1. Western blot assay using anti-HA antibody also revealed a significant induction of BIR1 protein in bir1-1/BIR1-HA lines after TRV infection, indicating that elevated BIR1 transcript levels reflected protein levels in systemically infected leaves (Fig. 1c). The bir1-1 mutant is known to constitutively activate cell death and defense responses that are partially dependent on the SA-dependent resistance pathway (Gao et al., 2009; Liu et al., 2016). Accordingly, we found that transcription of the defense marker genes PR1, PR4, PAD3 and WRKY29 remained similarly reactivated in TRV-infected bir1-1 mutants, indicating that virus infection does not impair the activation of defense when BIR1 is genetically suppressed (Fig. 1d and S1b). The autoimmune phenotypes in bir1-1 mutants are partially dependent on SUPPRESSOR OF BIR1-1 1 (SOBIR1), which promotes cell death and defense in conjunction with BAK1 (Chinchilla et al., 2007; Gao et al., 2009; Liu et al., 2016). Interestingly, we found a significant induction of SOBIR1 transcripts in Arabidopsis leaves at early time points of TRV or TuMV infection compared to mock-inoculated plants (Fig. 1e and Fig. S1a,c). In contrast, BAK1 transcripts decreased significantly after infection with TRV or TuMV (Fig. 1f and Fig. S1a,c). In our assay, the bak1-5 mutant, which is strongly impaired in PTI signaling (Schwessinger et al., 2011), was more susceptible to TRV accumulation (Fig. 1g), whereas TRV levels were moderately diminished in sobir1-12 mutants (Fig. 1h). Importantly, TRV RNA levels were also drastically reduced in a sobir1-1 bir1-1 double mutant, in which cell death and SA-dependent defense responses are significantly reduced by the sobir1-1 mutation (Gao et al., 2009). This result suggested that TRV resistance associated to loss of BIR1 function in the bir1-1 mutant was unrelated to constitutive cell death or SA defense priming (Fig. 1i). Consistently with this notion, we showed that exogenous application of SA triggered accumulation of PR1 transcripts in the plant tissue but was not sufficient to prime plant defense against TRV (Fig. 1i). Collectively, our results indicated that TRV triggers an immune response in which BIR1 likely functions as a negative regulator of antiviral defenses.
Figure 1.
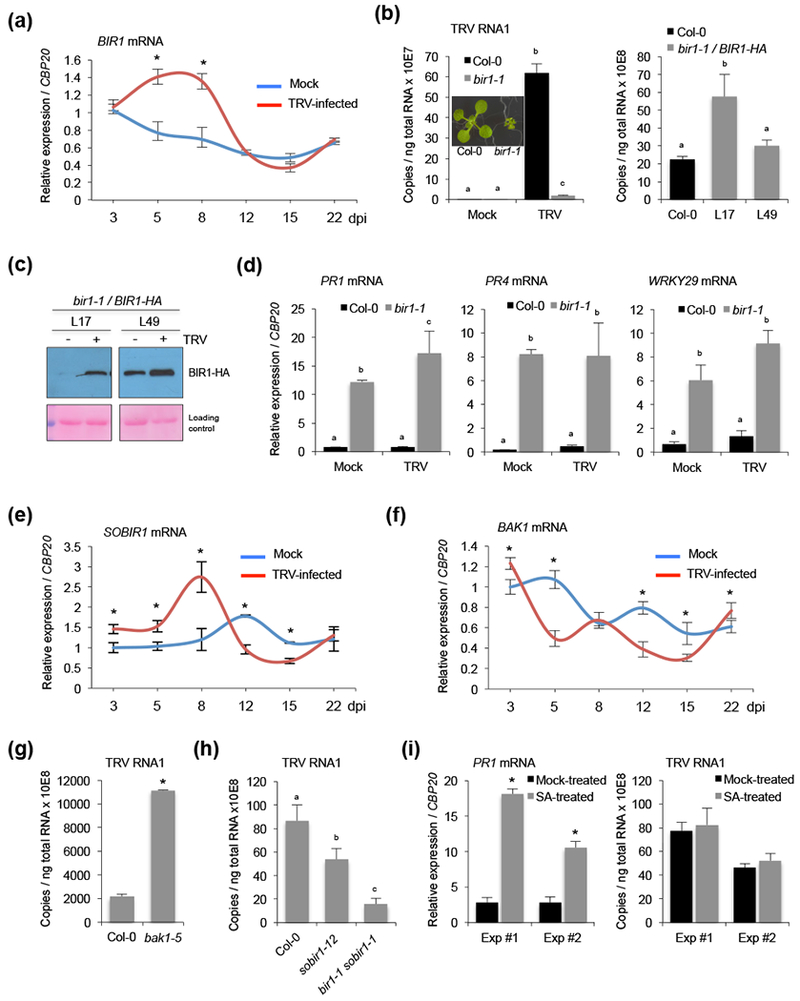
Expression of BIR1, SOBIR1 and BAK1 during TRV infection in Arabidopsis and effect of their loss-of-function mutations on TRV accumulation. (a) Time-course accumulation of BIR1 transcripts in mock-inoculated and TRV-infected leaves. (b) Accumulation of TRV genomic RNA in TRV-infected rosette leaves of Arabidopsis wild type (Col-0), bir1-1 mutants (lelf) and two bir1-1/BIR1-HA complemented lines (L17 and L49) (right) at 8 days post-inoculation (dpi). Mock-inoculated controls were included in the left panel to discriminate background amplification. The phenotype of wild type and bir1-1 plants grown on MS medium at 21° C is shown. (c) Western blot analysis of BIR1 proteins in extracts from leaves of mock-inoculated (−) or TRV-infected (+) bir1-1/BIR1-HA complemented lines (L17 and L49) at 8 dpi. Ponceau staining was used as a protein loading control. (d) Accumulation of defense-related PR1, PR4, and WRKY29 transcripts in mock-inoculated or TRV-infected leaves of Arabidopsis wild type and bir1-1 mutants at 8 dpi. (e) Time-course accumulation of SOBIR1 transcripts in mock-inoculated and TRV-infected leaves. (f) Time-course accumulation of BAK1 transcripts in TRV-infected and mock-inoculated leaves. (g) Accumulation of TRV genomic RNA in rosette leaves of wild type and bak1-5 mutants at 8 dpi. (h) Accumulation of TRV genomic RNA in rosette leaves of wild type, sobir1-12 and sobir1 bir1 mutants at 8 dpi. (i) Accumulation of PR1 transcripts (left) and TRV genomic RNA (right) in rosette leaves of wild type plants treated with or without (mock) salicylic acid (SA). Exp #1 and #2 are described in Materials and methods. Relative expression levels were determined by qRT-PCR and normalized to the CBP20 internal control. Error bars represent SD from three independent PCR measurements. Values in (a), (e) and (f) are related to the mock-inoculated sample at 3 dpi that was arbitrarily assigned to 1. Asterisks (Student’s t test) or different letters (one-way ANOVA) were used to indicate significant differences (P < 0.001). The experiments were repeated at least three times with similar results and one representative biological replicate is shown.
RdDM imparts transcriptional control of BIR1
Inspection of Arabidopsis small RNA sequencing datasets generated in our lab revealed the profuse accumulation of siRNAs upstream of the BIR1 transcription start site, the vast majority of which corresponded to the 24-nt class (Fig. 2a and Fig. S1d). Since 24-nt siRNAs guide methylation in the canonical RdDM pathway (Xie & Yu, 2015) we investigated if siRNA-dependent RdDM controls BIR1 expression. First, BIR1 transcripts were significantly more abundant in the RdDM mutants drm2, drm1 drm2 cmt3 (herein ddc), nrpe1 and ago4 mutants compared to wild type plants (Fig. 2b). BIR1 levels were unaffected in the single cmt3 mutant, likely due to redundancy between methyltransferases DRM2 and CMT3 in maintaining non-CG DNA methylation (Fig. 2b) (Cao & Jacobsen, 2002). Then, we used qRT-PCR to detect RNA products at the intergenic region containing the predicted BIR1 promoter. Interestingly, transcripts were amplified in wild type Col-0 plants but not in nrpe1 mutants, indicating that Pol V was required for their production (Fig. 2c). The accumulation of Pol V-dependent transcripts derived from INTERGENIC LOCUS 22 (IGN22) was used as a positive control (Rowley et al., 2011) (Fig. S2a).
Figure 2.
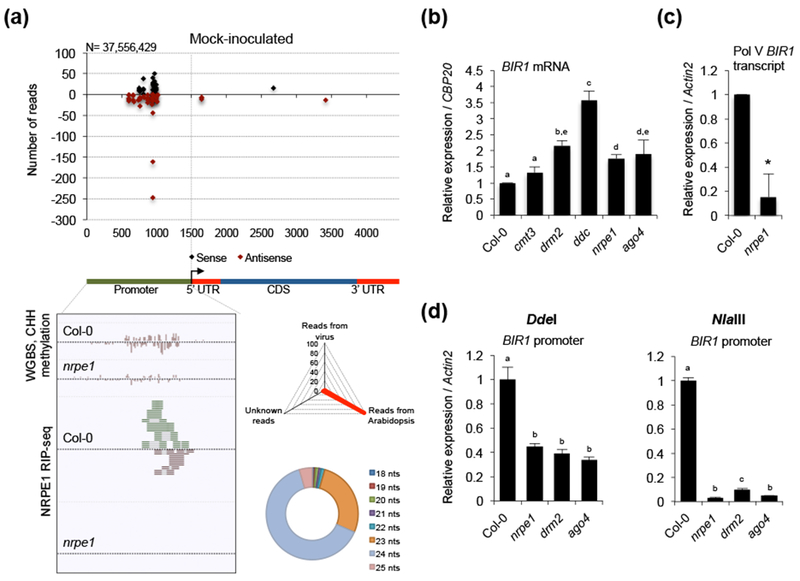
RdDM-mediated transcriptional regulation of BIR1. (a) Distribution of BIR1-derived siRNAs in rosette leaves of mock-inoculated Arabidopsis plants (upper diagram). Sense (black dots) and antisense (red dots) siRNA species are represented as positive and negative values on the y-axis, respectively. The triangle graph represents the genomic distribution (percentage) of sRNAs in the sequenced set. N denotes the total number of filtered sequenced reads. The circle graph represents the size distribution of BIR1-derived siRNAs. Genome browser screenshot of CHH methylation and Pol V transcripts at the BIR1 promoter in wild type (Col-0) and nrpe1 mutants using WGBS and Pol V (NRPE1) RIP-seq datasets is shown (Wierzbicki et al., 2012; Bohmdorfer et al., 2016) (lower diagram). (b) Accumulation of BIR1 transcripts in rosette leaves of wild type and RdDM mutants (cmt3, drm2, ddc, nrpe1 and ago4). (c) Accumulation of Pol V-dependent BIR1 promoter transcripts in rosette leaves of wild type and nrpe1 mutants. (d) Extent of asymmetric (CHH) cytosine methylation at the BIR1 promoter determined by chop-qPCR in rosette leaves of wild type and RdDM mutants (nrpe1, drm2 and ago4). PCR-amplified regions contain recognition sites of the methylation-sensitive DdeI and NlaIII endonucleases. Relative expression levels were determined by qRT-PCR and normalized to the CBP20 or Actin2 internal control as indicated. Error bars represent SD from three independent PCR measurements. Asterisks (Student’s t test) or different letters (one-way ANOVA) were used to indicate significant differences (P < 0.001). The experiments were repeated at least three times with similar results and one representative biological replicate is shown.
If BIR1 were an RdDM target, DNA methylation levels at this locus should be reduced in RdDM mutants. To test this idea, we performed methylation-specific Chop-PCR to examine DNA methylation at the BIR1 promoter region in wild type and several DNA methylation mutants. Genomic DNA was digested with the CHH methylation-sensitive restriction endonucleases DdeI and NlaIII prior to PCR amplification using flanking primers (Bohmdorfer et al., 2014). We found amplification products in DNA samples treated with either DdeI or NlaIII in the wild type background, indicative of active cytosine methylation (Fig. S2b). In contrast, low levels of amplification were reported in the RdDM mutants nrpe1, drm2 or ago4 (Fig. S2b). Similar results were obtained for At1g49490 and IGN36, used as positive RdDM controls for DdeI and NlaIII digestions, respectively (Bohmdorfer et al., 2014) (Fig. S2b). Parallel amplification of DNA sequences without restriction sites (At1g55535 and At2g36490) from the same digested DNA samples, used as internal digestion controls, produced amplification bands in all genetic backgrounds (Fig. S2b). Quantification of the difference in DNA methylation levels by Chop-qPCR indicated that CHH methylation levels at both the BIR1 promoter and the At1g49490 and IGN36 positive controls, but not the negative control, were reduced to a similar extent in all mutants tested (Fig. 2d and S2c). Finally, whole-genome bisulfite sequencing (WGBS) reported by (Wierzbicki et al., 2012) revealed extensive symmetrical and asymmetrical DNA methylation in the BIR1 promoter, whereas methylation was drastically diminished in nrpe1 compared to wild type plants (Fig. 2a and S3). Furthermore, published Pol V RIP-seq data (Bohmdorfer et al., 2016) revealed that Pol V-associated RNA accumulated in Col-0 wild type, but not in nrpe1 mutants, confirming that RNA reads originated at the BIR1 promoter were associated with Pol V (Fig. 2a). Collectively, our data demonstrated that BIR1 was an RdDM target under normal growing conditions.
SA mediates transcriptional activation of BIR1 during TRV infection
We wondered whether higher accumulation of BIR1 transcripts in infected tissues could reflect the transcriptional activation of the BIR1 locus in response to the virus. To test this idea, Arabidopsis plants expressing a GFP:GUS fusion protein under the control of the BIR1 promoter were challenged with TRV. GUS activity was strongly and consistently induced in rosette leaves and aerial tissues of TRV-infected transgenic plants when compared to the mock-inoculated ones (Fig. 3a). The spatial pattern of GUS induction suggested that BIR1 responded ubiquitously to TRV infection. Furthermore, northern blot revealed higher levels of GFP:GUS fusion transcripts in the presence of TRV confirming that TRV triggered transcriptional activation of BIR1 (Fig. 3a).
Figure 3.
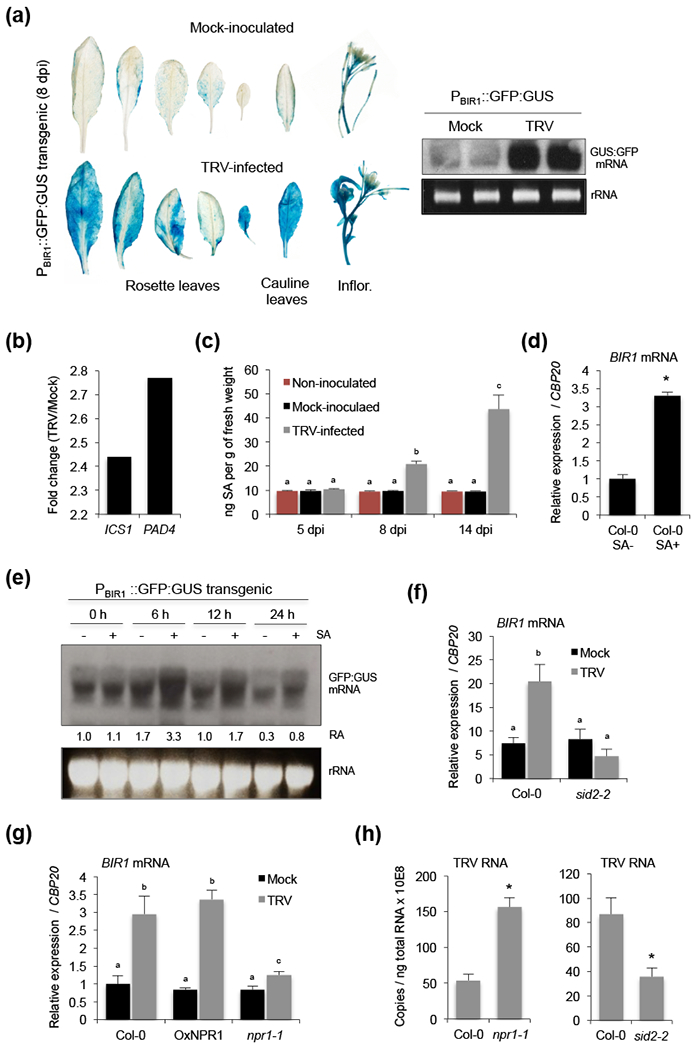
Salicylic acid (SA)-mediated transcriptional activation of BIR1 during viral infection. (a) Histochemical localization of GUS expression in mock-inoculated and TRV-infected transgenic Arabidopsis plants expressing a GFP:GUS fusion protein under the control of the BIR1 promoter (left panel). Northern blot analysis was used to monitor the expression of GFP:GUS mRNA using a GFP-specific radiolabeled probe (right panel). Ethidium-bromide stained RNA (prior to transfer) is shown as loading control. (b) Differential expression of SA biosynthetic genes ICS1 and PAD4. Fold-change (log2) in TRV-infected plants relative to mock-inoculated ones detected using a CATMA microarray (GSE15557) (Fernandez-Calvino et al., 2014). (c) Time-course accumulation of SA determined by GC-TOF-MS in leaves from non-inoculated, mock-inoculated and TRV-infected Arabidopsis. Error bars represent SD from five independent biological replicates. (d) Accumulation of BIR1 transcripts in rosette leaves of wild type (Col-0) plants treated with (+) or without (−) SA as indicated. (e) Northern blot analysis of GFP:GUS mRNA in extracts from transgenic leaves treated with (+) or without (−) SA as indicated. Samples were collected at 0, 6, 12 and 24 h post-treatment and blots were hybridized with a GFP-specific DNA radiolabeled probe. Ethidium-bromide stained RNA (prior to transfer) is shown as loading control. The relative accumulation (RA) level for each sample is indicated (level in mock-treated plants at 0 h was arbitrarily set at 1.0). (f) Accumulation of BIR1 transcripts in mock-inoculated and TRV-infected rosette leaves of wild type and sid2-2 mutants at 8 days post-inoculation (dpi). (g) Accumulation of BIR1 transcripts in mock-inoculated and TRV-infected rosette leaves of wild type, NPR1 overexpressor and nrp1-1 mutants at 8 dpi. (h) Accumulation of TRV genomic RNA in rosette leaves of wild type, npr1-1 and sid2-2 mutants at 8 dpi. Relative expression levels were determined by qRT-PCR and normalized to the CBP20 internal control. Unless otherwise indicated, error bars represent SD from three independent PCR measurements. Asterisks (Student’s t test) or different letters (one-way ANOVA) were used to indicate significant differences (P < 0.001). The experiments were repeated at least twice with similar results and one representative biological replicate is shown.
Inspection of transcriptomic data revealed that two key SA biosynthetic genes, ICS1 and PAD4 (Chen et al., 2009), were significantly up regulated in leaves of TRV-infected plants (Fig. 3b) (Fernandez-Calvino et al., 2014). We thus wondered if SA levels influence BIR1 expression in the infected tissue. To test this possibility, we first determined the levels of SA in the leaves of soil-grown plants using GC-TOF-MS. SA levels gradually increased from 5 to 14 dpi in TRV-infected plants, whereas they remained constant in both non-inoculated and mock-inoculated plants (Fig. 3c). We found that BIR1 transcripts were markedly enhanced in wild type Arabidopsis at 6 h after SA application compared to mock-treated controls (Fig. 3d). Furthermore, we observed increasing levels of GFP:GUS transcripts in Arabidopsis plants expressing a GFP:GUS reporter under the BIR1 promoter at 6, 12 and 24 h after SA treatment, indicating that SA efficiently promotes transcriptional activation of BIR1 (Fig. 3e). Importantly, SA-activation of BIR1 during TRV infection was largely inhibited in the Arabidopsis sid2-2 mutant, which has disrupted the pathogen-inducible ICS1 gene and reduced SA accumulation (Wildermuth et al., 2001) (Fig. 3f). We also found that induction of BIR1 in virus-infected plants was compromised in npr1-1 Arabidopsis mutants, which lack NPR1 receptor-dependent SA-signaling (Cao et al., 1997; Wu et al., 2012), compared to wild type or npr1 complemented transgenic lines (OxNPR1) (Fig. 3g). These findings indicated that SA acts as a signal molecule for BIR1 activation during TRV infection, and that TRV promotes BIR1 expression by increasing the levels of SA in infected cells. Interestingly, TRV levels in the SA-deficient sid2-2 mutants were lower than in wild type plants, whereas plants with the npr1-1 mutation display enhanced susceptibility to TRV (Fig. 3h). Our results supported the idea that SA lacks direct antiviral functions against TRV, and suggest a SA-independent role for NPR1 in the control of TRV infection.
TRV activates BIR1 without affecting its methylation status
We next asked if BIR1 induction in infected plants was due to changes in the methylation status of its promoter. We found that siRNAs of 24 nts produced upstream of the BIR1 transcription start were as much abundant in TRV-infected plants as in mock-inoculated controls, suggesting that epigenetic silencing of BIR1 was not compromised by TRV (Fig. 4a). Chop-qPCR experiments revealed comparable levels of CHH methylation at the BIR1 promoter in mock-inoculated and TRV-infected samples after digestion with NlaIII, whereas the relative levels of amplified DNA were slightly reduced in infected samples digested with DdeI, possibly due to star activity of the enzyme (Fig. 4b). No significant changes in the CHH methylation of the RdDM targets At1g49490 and IGN36, used as methylation controls, were observed in plants exposed to TRV infection relative to the mock-inoculated ones (Fig. S2d). BIR1 was induced by TRV to a similar extent in all RdDM mutants (except drm2), suggesting that TRV supported BIR1 transcription regardless of its methylation status (Fig. 4c). Importantly, BIR1 transcripts were elevated in TRV-infected ddc, nrpe1 or ago4 mutants compared to wild type plants, indicating that RdDM was important to contain BIR1 expression during infection (Fig. 4c). Finally, similar patterns of methylation at the BIR1 promoter were observed in healthy, mock-inoculated and virus-infected plants when methylation was analyzed using locus-specific bisulfite sequencing (Fig. 4d and Fig. S4).
Figure 4.
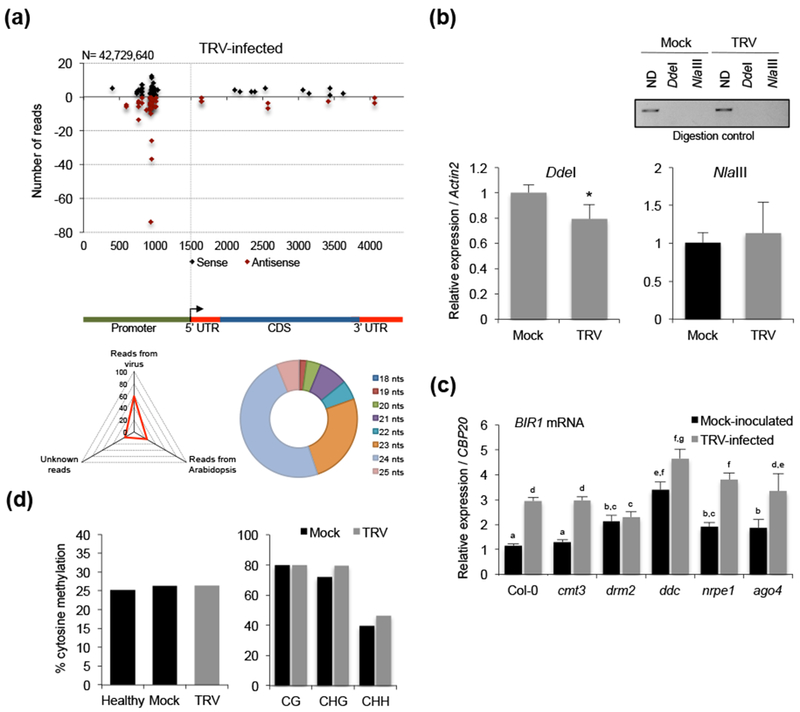
BIR1 methylation status in TRV-infected Arabidopsis. (a) Distribution of BIR1-derived siRNAs in rosette leaves of TRV-infected Arabidopsis plants. Sense (black dots) and antisense (red dots) siRNA species are represented as positive and negative values on the y-axis, respectively. The triangle graph represents the genomic distribution (percentage) of sRNAs in the sequenced set. N denotes the total number of filtered sequenced reads. The circle graph represents the size distribution of BIR1-derived siRNAs in TRV-infected plants. (b) Extent of asymmetric cytosine methylation at the BIR1 promoter determined by chop-qPCR in rosette leaves of mock-inoculated and TRV-infected plants at 8 days post-inoculation (dpi). The genomic DNA was digested with methylation-sensitive enzymes DdeI and NlaIII and qPCR amplified. Non-digested (ND) plants were used as control. Values were normalized to the Actin2 internal control. Error bars represent SD from three independent biological replicates. (c) Accumulation of BIR1 transcripts in rosette leaves of mock-inoculated and TRV-infected plants of wild type (Col-0) and RdDM mutants (cmt3, drm2, ddc, nrpe1 and ago4) at 8 dpi. Relative values were determined by qRT-PCR and normalized to the CBP20 internal control. Error bars represent SD from three independent PCR measurements. (d) Percentage of total cytosine methylation (left) and CG, CHG and CHH methylation (right) determined by in-house bisulfite sequencing at the BIR1 promoter in healthy (non-inoculated), mock-inoculated and TRV-infected Arabidopsis at 8 dpi. H represents A, T or C. Asterisks (Student’s t test) or different letters (one-way ANOVA) were used to indicate significant differences (P < 0.001). The experiments were repeated at least three times with similar results and one representative biological replicate is shown.
We next investigated if SA altered the DNA methylation pattern of the BIR1 promoter. We found low levels of DNA amplification diagnostic of loss of asymmetric methylation in nrpe1, drm2 or ago4 mutants compared to wild type Col-0 plants after 6 or 12 h of SA treatment (Fig. S5a,b). DNA methylation at the At1g49490 and IGN36 controls diminished in RdDM mutants regardless of SA treatments (Fig. S5a). BIR1 transcripts increased after SA treatment in wild type plants and in nrpe1, drm2 or ago4 mutants, indicating that loss of DNA methylation did not compromise SA-mediated induction of BIR1 (Fig. S5c). Finally, transcription at the BIR1 promoter was strongly reduced in the Pol V-defective npre1 mutants in leaves of both mock-treated plants and SA-sprayed plants (Fig. S5d). Collectively, our data proved that SA activates transcription of BIR1 during virus infections without interfering with its epigenetic regulation.
BIR1 is regulated by post-transcriptional RNA silencing
The analysis of our sRNA sequences revealed that siRNAs matching the BIR1 protein-coding region were abundant in plants systemically infected with TRV or TuMV, but not in mock-inoculated ones, suggesting that BIR1 is a target of post-transcriptional silencing during infections (Figs. 2a, 4a,e, and S1d,f). To test this possibility, we first monitored BIR1 transcripts in non-infected Arabidopsis silencing mutants. Although data between independent repeats showed slight variations, a subtle increment of BIR1 transcripts in some mutants involving dysfunctional DCL2, DCL3 or DCL4 as well as in mutants with genetic defects in RDR1, RDR2 or RDR6 suggested that BIR1 may undergo conditional post-transciptional silencing under non-challenging conditions (Fig. 5a and S6a).
Figure 5.
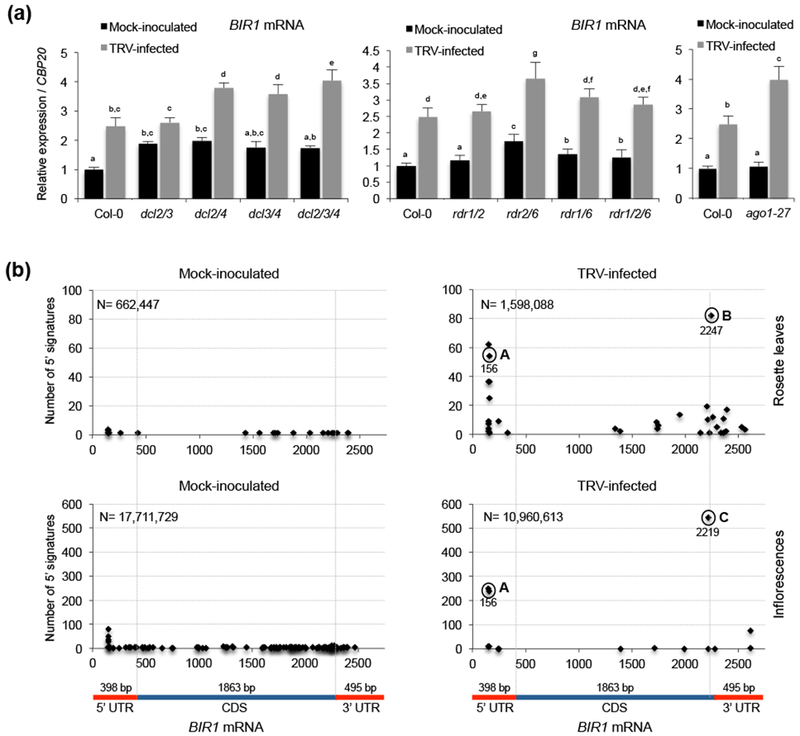
BIR1 mRNA accumulation in RNA silencing mutants and parallel-analysis of cDNA Ends (PARE)-based identification of preferential cleavage sites within the BIR1 mRNA. (a) Accumulation of BIR1 transcripts in mock-inoculated and TRV-infected Arabidopsis rosette leaves of wild type (Col-0) and mutants impaired in siRNA biogenesis [dcl2 dcl3 (dcl2/3), dcl2 dcl4 (dcl2/4), dcl3 dcl4 (dcl3/4) or dcl2 dcl3 dcl4 (dcl2/3/4)], secondary siRNA biogenesis [rdr1 rdr2 (rdr1/2), rdr2 rdr6 (rdr2/6), rdr1 rdr6 (rdrl1/6) or rdr1 rdr2 rdr6 (rdr½/6)], and AGO1 function (ago1). Relative expression levels were determined at 8 days post-inoculation (dpi) by qRT-PCR and normalized to the CBP20 internal control. Error bars represent SD from three independent PCR measurements. Different letters indicate significant differences according to one-way ANOVA and Duncan test (P < 0.001). The experiments were repeated at least three times with similar results and one representative biological replicate is shown. (b) Target plots showing 5’ signature abundance throughout the BIR1 mRNA identified through degradome sequencing. Circles in the t-plots denote highly abundant signatures at the indicated positions (referred to as A, B and C) identified in TRV-infected plants but not in mock-inoculated controls. Samples from rosette leaves and inflorescences were analyzed. N denotes the total number of filtered sequenced reads.
When BIR1 transcripts were measured in TRV-infected plants, we found that BIR1 was induced in the double dcl2 dcl3 mutants as much as the wild type (Fig. 5a). In contrast, BIR1 transcripts were significanlty more abundant in dcl2 dcl4, dcl3 dcl4 or dcl2 dcl3 dcl4 mutants compared to control plants, indicating that DCL4 was important to prevent excessive BIR1 accumulation in the infected tissue (Fig. 5a). Similarly, BIR1 transcripts were, in general, far more abundant in rdr2 rdr6 and, to a lower extent, in rdr1 rdr6 and rdr1 rdr2 rdr6 defective mutants than in wild type infected plants (Fig. 5a). Finally, BIR1 transcripts were similar in mock-inoculated wild type and ago1 mutants, whereas BIR1 transcripts were more abundant in ago1 when they were infected (Fig. 5a). Similar results were observed in plants systemically infected with TuMV, suggesting that post-transcriptional RNA silencing was accentuated in response to viral infections (Fig. S1e).
To support our findings, we examined BIR1 mRNA degradation via degradome sequencing. By plotting the abundance of 5’ signatures matching the BIR1 transcript we found that TRV infection correlated with the massive accumulation of degradome 5’ signatures at nucleotide positions 156, 2,219 and 2,247 (Fig. 5b). These cleavage site sequences were clearly discerned from a background of low abundant, non-specific degradation products at other positions (Fig. 5b). Cleavage at position 156 was reproducibly found with high abundance in all degradome libraries prepared from leaves or inflorescences of TRV-infected plants. Although this precise 5’ signature was not found in mock-inoculated controls, degradome tags diagnostic of sequential cleavage were identified at nearby nucleotide positions in all samples tested, suggesting that this region was particularly prone to RNA degradation (Fig. 5b and S6b). When we applied the CleaveLand4 computational pipeline to match BIR1-derived degradome 5’ signatures against the miRBase, we were unable to identify validated miRNAs as potentially responsible for cleavage at these positions, suggesting that BIR1-derived siRNAs may guide cis-cleavage events. Collectively, our data proved that BIR1 transcripts were exposed to selective post-transcriptional degradation in response to infection.
BIR1 overexpression causes extreme morphological defects and up regulation of plant defense in TRV-infected Arabidopsis
To further explore the relevance of BIR1 regulation in infected plants, we investigated the consequences of BIR1 overexpression during TRV infection in Arabidopsis. To do this, we used TRV as a viral expression vector to overproduce BIR1 in infected plants. We cloned a HA-tagged version of the Arabidopsis BIR1 into pTRV2 and introduced it along with pTRV1 in N. benthamiana by Agrobacterium-mediated infiltration (Fig. 6a). Western blot assay using anti-HA antibody detected BIR1 protein in systemically infected leaves (Fig. 6a). Interestingly, TRV-BIR1 RNA accumulated in upper non-infiltrated leaves to the same levels as the TRV-GFP control, suggesting that overexpression of BIR1 had negligible effects on TRV accumulation in N. benthamiana cells (Fig. 6a and S6c).
Figure 6.
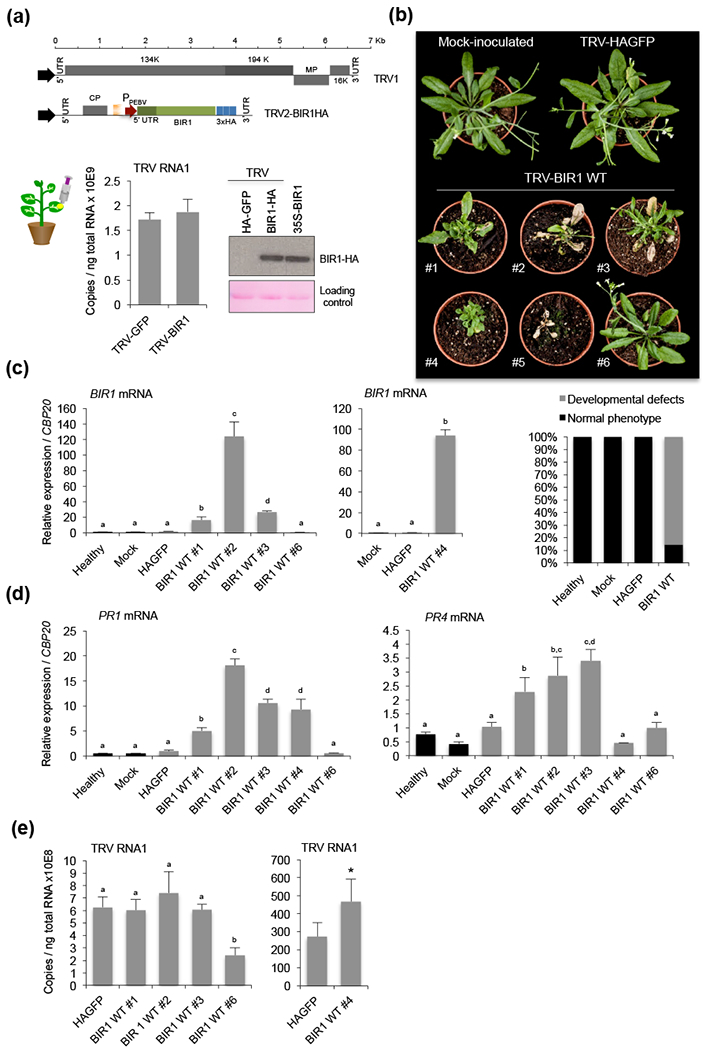
Phenotypes of TRV-BIR1-infected Arabidopsis. (a) TRV-derived constructs for HA-tagged expression of BIR1. The 5’UTR-contaninig BIR1 coding sequence was inserted adjacent to the PEBV replicase promoter in pTRV2. pTRV1 and pTRV2-BIR1 constructs were agroinjected in N. benthamiana. Accumulation of TRV genomic RNA in upper leaves of TRV-BIR1-infected plants at 5 days post-inoculation (dpi) is shown (left). Western blot analysis of HA-tagged BIR1 proteins in extracts from leaves infiltrated with TRV-BIR1 is shown (right). TRV-GFP and 35S-BIR1-HA were used as controls. Ponceau staining was used as a protein loading control. (b) Morphological phenotypes of plants mock-inoculated, systemically infected with TRV-GFP or infected with TRV-BIR1 WT (referred to as #1 to #6). Plants were grown on soil and photographed at 14 dpi. Percentage of plants displaying normal vs morphological phenotypes after inoculation with TRV-derivatives is indicated. Non-inoculated (healthy) and mock-inoculated plants were used as controls. TRV-GFP was used as control. (c) Accumulation of BIR1 transcripts in TRV-BIR1-infected individual plants shown in (b). Samples from non-inoculated (healthy), mock-inoculated or TRV-GFP-infected plants were included as controls. (d) Accumulation of defense-related PR1 and PR4 transcripts in TRV-BIR1-infected individual plants shown in (b). TRV-GFP was used as control. (e) Accumulation of TRV genomic RNA in TRV-BIR1-infected individual plants shown in (b). Relative expression levels were determined by qRT-PCR and normalized to the CBP20 internal control. Error bars represent SD from three independent PCR measurements. Asterisks (Student’s t test) or different letters (one-way ANOVA) were used to indicate significant differences (P < 0.001). The experiments were repeated at least three times with similar results and one representative biological replicate is shown.
Inoculation of three-week-old Arabidopsis plants with TRV-BIR1 revealed the appearance of a range of morphological defects at approximately 14 dpi, affecting more than 80% of the inoculated plants (Fig. 6b). Symptoms were more severe at later stages post-infection and included stunted morphology, abnormal leaf shape, extensive leaf necrosis, loss of apical dominance during bolting (bushy phenotype) and premature death (Fig. 6b). In contrast, plants infected with TRV-GFP, used as control, developed normally like non-inoculated or mock-inoculated plants (Fig. 6b). Interestingly, morphological phenotypes of TRV-BIR1-infected individual plants coincided with extremely high levels of BIR1 transcripts (Fig. 6c). Conversely, TRV-BIR1-infected plants that developed free of symptoms accumulated less BIR1 transcripts, similar to the TRV-GFP-infected control plants (Fig. 6c).
Growth arrest and cell death are reminiscent of plants that show constitute activation of defense responses (Lorrain et al., 2003). To gain insight into the effects of BIR1 overexpression in TRV-infected tissues, we measured relative transcript levels of defense genes PR1 and PR4. Despite BIR1 being a repressor of plant immunity, the expression of PR1 and PR4 was markedly up regulated in infected plants producing high amounts of BIR1 transcripts (Fig. 6d). In contrast, PR1 and PR4 accumulated to normal levels in symptomless plants producing low amounts of BIR1 transcripts (Fig. 6d). PR1 and PR4 were poorly induced in plants infected with TRV-GFP, confirming that defense activation was linked to BIR1 overexpression rather than virus infection (Fig. 6d). These experiments suggested that BIR1 overexpression induces constitutive immunity in Arabidopsis. Interestingly, TRV levels in TRV-BIR1-infected plants exhibited a marked variability between individuals and experimental replicates (Fig. 6e), and no correlation between BIR1 transcript levels and viral accumulation was found (Bilateral Spearman correlation, ρ= 0,48, p=0,84). We concluded that BIR1 overdosage had no direct effects on viral susceptibility in Arabidopsis.
Inducible BIR1 overexpression in transgenic Arabidopsis causes phenotypical defects and triggers the activation of plant defense
It is possible that the morphological phenotypes associated to high BIR1 doses in TRV-BIR1-infected cells were due to the combined effect of BIR1 overexpression and viral infection. To further investigate this possibility, we employed a dexamethasone (DEX)-inducible system to generate independent Arabidopsis homozygous lines that overexpress mCherry-tagged BIR1 proteins (Fig. S7a,b,c,d). DEX treatment had no apparent effects on wild type Col-0 seedlings, and BIR1 transgenics treated with water exhibited normal phenotypes (Fig. 7a and S8a,b). Conversely, more than 80% of DEX-treated BIR1 transgenics displayed stunting, abnormal leaf shape, leaf necrosis, bushy phenotype and cell death that resembled the morphological phenotypes observed in plants infected with TRV-BIR1 (Fig. 7a and S8a,b). As predicted, DEX-treated plants showing strong phenotypes accumulated over two orders of magnitude more BIR1 transcripts than control plants (Fig. 7b). Water-treated transgenic lines, wild type (non-transgenic) plants treated with DEX, and DEX-treated transgenics that exhibited normal growing phenotypes produced equivalent low amounts of BIR1 transcripts (Fig. 7b). Similarly, BIR1-mCherry fusion proteins were detected at much higher intensities in plants with morphological defects than in the above controls (Fig. 7c).
Figure 7.
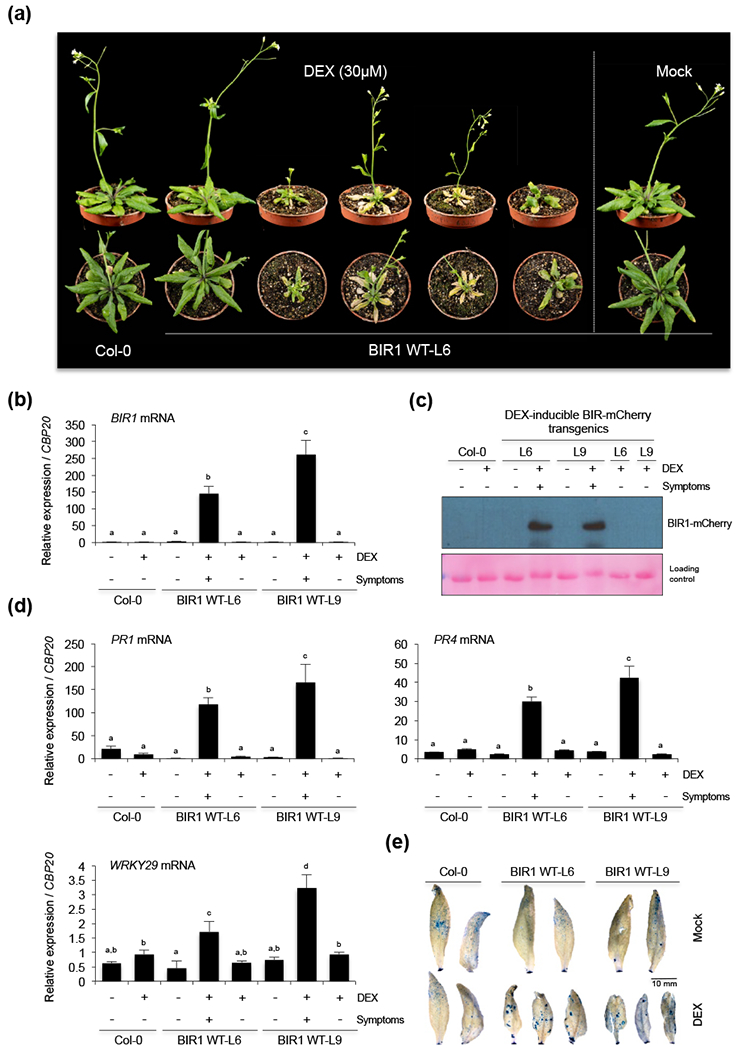
Phenotypes of BIR1 overexpressing transgenic Arabidopsis. (a) Morphological phenotypes of BIR1 transgenic plants after DEX treatment. Arabidopsis plants from transgenic line 6 (BIR1 WT L6) were grown for three weeks on soil and treated with 30 μM DEX or mock-treated for 6 consecutive days by spraying the solution (1 ml per plant) once at 24 h intervals. DEX-treated wild type (Col-0) plants are shown as controls. Plants were photographed at 7 days after the first DEX application. Morphological phenotypes of plants from transgenic line 9 (L9) are shown in Supporting Information Fig. S8(a). (b) Accumulation of BIR1 transcripts in plants from BIR1 overexpressor lines L6 and L9. Wild type plants are shown as controls. Plants were sprayed with DEX (+) or water (−). Plants showing wild type (−) or aberrant (+) phenotypes were analyzed. (c) Western blot analysis of BIR1 proteins in extracts from leaves of lines L6 and L9. Plants were sprayed with DEX (+) or water (−). Plants showing wild type (−) or aberrant (+) phenotypes were analyzed. Ponceau staining was used as a protein loading control. (d) Accumulation of defense-related PR1, PR4, and PAD3 transcripts in plants from lines L6 and L9. (e) Trypan blue staining of leaves of wild type and BIR1 overpression lines (L6 and L9). Leaves from DEX-treated and mock-treated plants grown on soil were stained with lactophenol trypan blue as described (Diaz-Tielas et al., 2012). Relative expression levels were determined by qRT-PCR and normalized to the CBP20 internal control. Error bars represent SD from three independent PCR measurements. Different letters indicate significant differences according to one-way ANOVA and Duncan test (P < 0.001). The experiments were repeated at least three times with similar results and one representative biological replicate is shown.
When the accumulation of defense gene markers was tested, high amounts of PR1, PR4, PAD3 or WRKY29 transcripts accumulated in plants overexpressing BIR1 as opposed to wild type or non-expressing transgenic plants (Fig. 7d and S8c). As predicted, none of the above markers were up regulated in asymptomatic BIR1 transgenics (Fig. 7d and S8c). We further demonstrated that overexpression of BIR1 triggered localized cell death in DEX-treated transgenic leaves, as deduced by trypan blue staining (Fig. 7e). These observations indicated that DEX-induced overexpression of BIR1 stimulated an autoimmune response in an infection-free cell environment.
Discussion
BIR1 is a negative regulator of several resistance pathways in which BAK1 and SOBIR1 have concerted roles (Gao et al., 2009; Dominguez-Ferreras et al., 2015; Liu et al., 2016). Here we provide compelling evidence that BIR1 transcription is positively regulated by SA and propose that TRV triggers NPR1-dependent expression of BIR1 during the infection by increasing SA levels in the infected tissue. We show that loss of BIR1 function in the bir1-1 mutant severely compromises TRV accumulation, likely due to constitutive activation of plant defenses in this mutant. A previous study reported that the bir1-1 mutation leads to extensive cell death, elevated levels of SA and SA-dependent gene expression (Gao et al., 2009). Based on this observation, it is possible that the SA defense pathway could prime an immune response against TRV in bir1-1 mutants. In some compatible plant–virus interactions, SA treatment or overexpression of SA biosynthetic genes can potentiate antiviral responses by affecting virus replication, coat protein accumulation and systemic virus movement (Chivasa et al., 1997; Mayers et al., 2005; Ishihara et al., 2008; Qi et al., 2018). However, we found that exogenous application of SA activated the SA defense pathway but was unable to antagonize the virus. Furthermore, a phenotype of strong resistance against TRV was also observed in the double bir1-1 sobir1-1 mutant, in which cell death and constitutive expression of SA-dependent defense genes are strongly reduced by the sobir1-1 mutation (Gao et al., 2009). These findings prove that enhanced TRV resistance in bir1-1 plants was not due to constitutive SA defense priming (Gao et al., 2009). On the contrary, we observed that loss of ICS1 function in the sid2-2 mutants correlated with reduced TRV proliferation, suggesting that SA may be important to support TRV infection. Importantly, altered susceptibility was not observed in plants expressing high levels of BIR1, even tough cell death and SA-mediated defense signaling pathway were substantially enhanced in BIR1 overexpressor plants. These results suggest that defense responses that were concomitant to both low and high expression of BIR1 may have a minor role in controlling viral proliferation in Arabidopsis. BAK1 is also required for activation of cell death and defense responses in the bir1-1 mutant (Liu et al., 2016). We show that BAK1 transcripts were diminished in infected plants, and bak1-5 mutants, which are impaired in PTI but not in BR signaling (Chinchilla et al., 2007; Heese et al., 2007; Schwessinger et al., 2011), were more susceptible to infection with TRV and other viruses (Korner et al., 2013). These findings suggest that BAK1, and likely SOBIR1, contribute to modulate viral proliferation, but their relationships with BIR1 and their potential interdependence during the antiviral response remain to be investigated. Furthermore, the role of NDR1-, PAD4- and EDS1-resistance pathways that are triggered in the bir1-1 mutant needs to be investigated to elucidate their contribution to antiviral resistance (Gao et al., 2009).
In our study, we prove that both transcriptional and post-transcriptional RNA silencing contribute, at least partly, to BIR1 homeostasis. We found that RdDM constitutively regulates BIR1. Under non-challenging conditions, our results suggests that post-transcriptional silencing may be mobilized to perform conditional fine-tune regulation of BIR1 expression. However, during viral infection, post-transcriptional silencing strongly reinforces the action of epigenetic silencing by removing the excess of BIR1 transcripts produced upon BIR1 transcriptional activation. This idea also emerges from our analysis of degradome according to which BIR1 gives rise to high amounts of discrete cleaved 3’ mRNA products in infected plants compared to mock-inoculated plants. The genetic requirement for RNA silencing components in the control of BIR1 is consistent with the widespread accumulation of BIR1-derived siRNAs of sense and antisense polarities in infected plants, but not in mock-inoculated ones. BIR1 siRNAs resemble viral-associated siRNAs (vasiRNAs) that are produced from multiple host genes during activation of antiviral silencing (Cao et al., 2014). vasiRNAs are competent in directing silencing of the host target genes in line with the idea that BIR1 siRNAs guide autosilencing of BIR1 transcripts. The requirement for BIR1 siRNA biogenesis and function seems to differ however from the predicted genetic pathway of vasiRNAs, which are mostly dependent on DCL4, RDR1 and AGO2 (Cao et al., 2014). From our data, it is possible that several complementary pathways that include RDR6 and AGO1 also contribute to vasiRNA biogenesis and function during viral infections.
We found that the strong overexpression of BIR1 triggers autoimmune phenotypes similar to those observed in bir1-1 mutants (Gao et al., 2009), indicating that a well-calibrated regulation of BIR1 guarantees a proper control of immune signaling pathways. Given that BIR1 is an active RLKs, overexpression of BIR1 may interfere with other closely related RLKs causing miscoordination of cellular signaling pathways, including plant defense or development. For instance, high levels of BIR1 may hinder BAK1-mediated regulation of SOBIR1-independent cell death (Liu et al., 2016). Although BIR1 represses immune responses in normal growing conditions, we demonstrated that BIR1 triggers plant defenses when expressed at a high dose, even in the absence of virus. As a plausible explanation, overproduction of BIR1 may either affect BIR1-dependent negative regulation of (co)receptor partners or, alternatively, promote inappropriate interactions with other immune (co)receptor proteins that result in the activation of resistance (Prelich, 2012; Rodriguez et al., 2016).
We saw that Arabidopsis mutants with defects in RdDM or siRNA biogenesis/function produce BIR1 at levels that barely compromise normal plant development. This finding has two important implications. First, one could argue that RNA silencing plays a secondary role in controlling BIR1 expression and that other yet unknown mechanisms provide additional layers of regulation that ultimately confine BIR1 below detrimental levels for plant fitness. This is a reasonable possibility, however, loss of function of one or several silencing genes does not necessarily imply a complete inhibition of the pathway (Bouche et al., 2006). And importantly, mutants tested in this study were affected either in the RdDM pathway or in the post-transcriptional silencing pathway, but not both. As a result, it is likely that residual RNA silencing activities in these mutants could yet exert effective BIR1 control preventing BIR1 from reaching deleterious expression levels upon virus or pathogen (SA-mediated) induction. The second implication is that phenotypes associated to BIR1 induction are likely dose-dependent. In our experiments, plants infected with TRV-BIR1 or DEX-treated transgenic plants showing developmental defects produced more than two orders of magnitude BIR1 transcripts than control plants. Conversely, we observed that seedlings of the same transgenic lines developed normally when they were grown on MS-DEX plates (Fig. S9a). In these experimental conditions transgenic plants accumulated only ten to 20 times more BIR1 transcripts than the wild type plants (Fig. S9b). This represented at least an order of magnitude less expression than that observed in DEX-treated, soil-grown plants. Furthermore, accumulation of defense genes was not substantially altered in transgenic seedlings (lines 5 and 6) grown on plates (Fig. S9c). Only, transgenic line 9 produced BIR1 transcripts at levels that triggered a modest induction of PR1, PR4 and PAD3, but they were insufficient to perturb normal development (Fig. S9c). A dose-dependent mechanism would explain why silencing mutants, in which increments in BIR1 expression were only mild, display normal phenotypes. Interestingly, ddc mutants show a suite of developmental abnormalities (Chan et al., 2006) and activation of defense genes (Fig. S9d) (Dowen et al., 2012), but morphological phenotypes in these plants are likely due to a broad misregulation of developmental genes that are normally controlled by non-CG methylation (Chan et al., 2006). BIR1 belongs to the BIR family, with four members of which BIR2 and BIR3 also function as negative regulators of BAK1-mediated immunity (Halter et al., 2014; Imkampe et al., 2017). Transgenic overexpression of BIR3 in Arabidopsis also leads to dwarf phenotypes that were dosage-dependent (Imkampe et al., 2017). From our experiments we conclude that regulation of BIR1 is critical for plant viability, and propose that the proper BIR1 functioning requires a threshold expression, and once BIR1 exceeds or falls behind such a threshold, misregulation of plant immunity takes place. Interestingly, in a previous study BIR1 transgenic Arabidopsis under a 35S promoter exhibited wild type morphology, and PTI responses were not apparently affected in these plants, suggesting that the BIR1 transgene was expressed at non-detrimental levels in their experimental conditions (Liu et al., 2016).
In conclusion, our results demonstrate that plant viruses initiate a basal immune response that involves SA-dependent activation of the immune repressor BIR1. We propose that BIR1 acts as a negative regulator of antiviral defense in Arabidopsis. Regulation of BIR1 gene expression is important to avoid constitutive defense responses that negatively impact plant development and fitness. In this scenario, RNA silencing provides two complementary layers of transcriptional and post-transcriptional regulation that prevent, alone or in conjunction with other regulatory mechanisms, BIR1 from reaching deleterious expression levels when BIR1 is transcriptionally activated (Fig. S10a,b). Our work provides novel mechanistic insights into the regulation of BIR1 homeostasis that may be common for other plant immune components.
Supplementary Material
Figure S1. Effect of RNA silencing on BIR1 expression in plants infected with TuMV.
Figure S2. Epigenetic regulation of BIR1 and RdDM-methylation controls.
Figure S3. Methylation status of the BIR1 promoter using whole-genome bisulfite sequencing (WGBS) data in Arabidopsis.
Figure S4. Methylation status of the BIR1 promoter using in-house bisulfite sequencing in Arabidopsis.
Figure S5. Epigenetic regulation of BIR1 and RdDM-methylation controls in salicylic acid (SA)-treated plants.
Figure S6. BIR1 mRNA accumulation in RNA silencing mutants, cleavage mapping at the 5’ UTR of BIR1 mRNA and viral accumulation in N. benthamiana leaves expressing BIR1.
Figure S7. DEX-inducible system for overexpression of BIR1 in Arabidopsis plants.
Figure S8. Phenotypes of BIR1 overexpressing transgenic Arabidopsis.
Figure S9. Phenotypes of BIR1 overexpressing transgenic seedlings grown in axenic conditions.
Figure S10. Model of BIR1 regulation
Table S1. List of primers.
Acknowledgements
This work has been supported by FPI fellowships (BES-2013-063138 and EEBB-I-16-10815) to IG-B, by a Ramon y Cajal grant (RyC-2011-07006) to VR-F and by National Research grants (BIO2012-39973 and BIO2015-70752-R) to CL from Ministerio de Economía y Competitividad (MINECO/FEDER), Spain, and by a National Institutes of Health (USA) grant R01GM108722 to ATW. We thank Yuelin Zhang, James Carrington, Steve Jacobsen, Craig Pikaard, Xinniang Dong and Eric Richards for providing seeds, and Ignacio Hamada, Jan Kuciński, Shriya Sethuraman and M. Hafiz Rothi for technical assistance.
REFERENCES
- Addo-Quaye C, Miller W, Axtell MJ. 2009. CleaveLand: a pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics 25(1): 130–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccara M, Sarazin A, Thiebeauld O, Jay F, Voinnet O, Navarro L, Colot V. 2014. The Arabidopsis miR472-RDR6 silencing pathway modulates PAMP- and effector-triggered immunity through the post-transcriptional control of disease resistance genes. PLoS Pathog 10(1): e1003883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohmdorfer G, Rowley MJ, Kucinski J, Zhu Y, Amies I, Wierzbicki AT. 2014. RNA-directed DNA methylation requires stepwise binding of silencing factors to long non-coding RNA. Plant J 79(2): 181–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohmdorfer G, Sethuraman S, Rowley MJ, Krzyszton M, Rothi MH, Bouzit L, Wierzbicki AT. 2016. Long non-coding RNA produced by RNA polymerase V determines boundaries of heterochromatin. eLife 5: e19092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T, Felix G. 2009. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60: 379–406. [DOI] [PubMed] [Google Scholar]
- Bouche N, Lauressergues D, Gasciolli V, Vaucheret H. 2006. An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J 25(14): 3347–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campo S, Peris-Peris C, Sire C, Moreno AB, Donaire L, Zytnicki M, Notredame C, Llave C, San Segundo B. 2013. Identification of a novel microRNA (miRNA) from rice that targets an alternatively spliced transcript of the Nramp6 (Natural resistance-associated macrophage protein 6) gene involved in pathogen resistance. New Phytol 199(1): 212–227. [DOI] [PubMed] [Google Scholar]
- Cao H, Glazebrook J, Clarke JD, Volko S, Dong X. 1997. The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88(1): 57–63. [DOI] [PubMed] [Google Scholar]
- Cao M, Du P, Wang X, Yu YQ, Qiu YH, Li W, Gal-On A, Zhou C, Li Y, Ding SW. 2014. Virus infection triggers widespread silencing of host genes by a distinct class of endogenous siRNAs in Arabidopsis. Proc Natl Acad Sci U S A 111(40): 14613–14618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Jacobsen SE. 2002. Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc Natl Acad Sci U S A 99 Suppl 4: 16491–16498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SW, Henderson IR, Zhang X, Shah G, Chien JS, Jacobsen SE. 2006. RNAi, DRD1, and histone methylation actively target developmentally important non-CG DNA methylation in arabidopsis. PLoS Genet 2(6): e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Zheng Z, Huang J, Lai Z, Fan B. 2009. Biosynthesis of salicylic acid in plants. Plant Signal Behav 4(6): 493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nurnberger T, Jones JD, Felix G, Boller T. 2007. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448(7152): 497–500. [DOI] [PubMed] [Google Scholar]
- Chivasa S, Murphy AM, Naylor M, Carr JP. 1997. Salicylic acid interferes with Tobacco Mosaic Virus replication via a novel salicylhydroxamic acid-sensitive mechanism. Plant Cell 9(4): 547–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16(6): 735–743. [DOI] [PubMed] [Google Scholar]
- Coll NS, Epple P, Dangl JL. 2011. Programmed cell death in the plant immune system. Cell Death Differ 18(8): 1247–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl JL, Jones JD. 2001. Plant pathogens and integrated defence responses to infection. Nature 411(6839): 826–833. [DOI] [PubMed] [Google Scholar]
- Diaz-Tielas C, Grana E, Sotelo T, Reigosa MJ, Sanchez-Moreiras AM. 2012. The natural compound trans-chalcone induces programmed cell death in Arabidopsis thaliana roots. Plant Cell Environ 35(8): 1500–1517. [DOI] [PubMed] [Google Scholar]
- Dodds PN, Rathjen JP. 2010. Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet 11(8): 539–548. [DOI] [PubMed] [Google Scholar]
- Dominguez-Ferreras A, Kiss-Papp M, Jehle AK, Felix G, Chinchilla D. 2015. An overdose of the Arabidopsis coreceptor BRASSINOSTEROID INSENSITIVE1-ASSOCIATED RECEPTOR KINASE1 or its ectodomain causes autoimmunity in a SUPPRESSOR OF BIR1–1-dependent manner. Plant Physiol 168(3): 1106–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaire L, Pedrola L, de la Rosa R, Llave C. 2011. High-throughput sequencing of RNA silencing-associated small RNAs in olive (Olea europaea L.). PLoS ONE 6(11): e27916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowen RH, Pelizzola M, Schmitz RJ, Lister R, Dowen JM, Nery JR, Dixon JE, Ecker JR. 2012. Widespread dynamic DNA methylation in response to biotic stress. Proc Natl Acad Sci U S A 109(32): E2183–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Calvino L, Guzman-Benito I, Del Toro FJ, Donaire L, Castro-Sanz AB, Ruiz-Ferrer V, Llave C. 2016a. Activation of senescence-associated Dark-inducible (DIN) genes during infection contributes to enhanced susceptibility to plant viruses. Mol Plant Pathol 17(1): 3–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Calvino L, Martinez-Priego L, Szabo EZ, Guzman-Benito I, Gonzalez I, Canto T, Lakatos L, Llave C. 2016b. Tobacco rattle virus 16K silencing suppressor binds ARGONAUTE 4 and inhibits formation of RNA silencing complexes. J Gen Virol 97(1): 246–257. [DOI] [PubMed] [Google Scholar]
- Fernandez-Calvino L, Osorio S, Hernandez ML, Hamada IB, Del Toro FJ, Donaire L, Yu A, Bustos R, Fernie AR, Martinez-Rivas JM, et al. 2014. Virus-induced alterations in primary metabolism modulate susceptibility to Tobacco rattle virus in Arabidopsis. Plant Physiol 166(4): 1821–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Wang X, Wang D, Xu F, Ding X, Zhang Z, Bi D, Cheng YT, Chen S, Li X, et al. 2009. Regulation of cell death and innate immunity by two receptor-like kinases in Arabidopsis. Cell Host Microbe 6(1): 34–44. [DOI] [PubMed] [Google Scholar]
- German MA, Luo S, Schroth G, Meyers BC, Green PJ. 2009. Construction of Parallel Analysis of RNA Ends (PARE) libraries for the study of cleaved miRNA targets and the RNA degradome. Nat Protoc 4(3): 356–362. [DOI] [PubMed] [Google Scholar]
- German MA, Pillay M, Jeong DH, Hetawal A, Luo S, Janardhanan P, Kannan V, Rymarquis LA, Nobuta K, German R, et al. 2008. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat Biotechnol 26(8): 941–946. [DOI] [PubMed] [Google Scholar]
- Gouveia BC, Calil IP, Machado JP, Santos AA, Fontes EP. 2016. Immune receptors and co-receptors in antiviral innate immunity in plants. Front Microbiol 7: 2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halter T, Imkampe J, Mazzotta S, Wierzba M, Postel S, Bucherl C, Kiefer C, Stahl M, Chinchilla D, Wang X, et al. 2014. The leucine-rich repeat receptor kinase BIR2 is a negative regulator of BAK1 in plant immunity. Curr Biol 24(2): 134–143. [DOI] [PubMed] [Google Scholar]
- He XF, Fang YY, Feng L, Guo HS. 2008. Characterization of conserved and novel microRNAs and their targets, including a TuMV-induced TIR-NBS-LRR class R gene-derived novel miRNA in Brassica. FEBS Lett 582(16): 2445–2452. [DOI] [PubMed] [Google Scholar]
- He XJ, Hsu YF, Pontes O, Zhu J, Lu J, Bressan RA, Pikaard C, Wang CS, Zhu JK. 2009. NRPD4, a protein related to the RPB4 subunit of RNA polymerase II, is a component of RNA polymerases IV and V and is required for RNA-directed DNA methylation. Genes Dev 23(3): 318–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heese A, Hann DR, Gimenez-Ibanez S, Jones AM, He K, Li J, Schroeder JI, Peck SC, Rathjen JP. 2007. The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci U S A 104(29): 12217–12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imkampe J, Halter T, Huang S, Schulze S, Mazzotta S, Schmidt N, Manstretta R, Postel S, Wierzba M, Yang Y, et al. 2017. The Arabidopsis leucine-rich repeat receptor kinase BIR3 negatively regulates BAK1 receptor complex formation and stabilizes BAK1. Plant Cell 29(9): 2285–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara T, Sekine KT, Hase S, Kanayama Y, Seo S, Ohashi Y, Kusano T, Shibata D, Shah J, Takahashi H. 2008. Overexpression of the Arabidopsis thaliana EDS5 gene enhances resistance to viruses. Plant Biol (Stuttg) 10(4): 451–461. [DOI] [PubMed] [Google Scholar]
- Johansen LK, Carrington JC. 2001. Silencing on the spot. Induction and suppression of RNA silencing in the Agrobacterium-mediated transient expression system. Plant Physiol 126(3): 930–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. 2006. The plant immune system. Nature 444(7117): 323–329. [DOI] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Gao S, Vivian-Smith A, Jin H. 2007. A novel class of bacteria-induced small RNAs in Arabidopsis. Genes Dev 21(23): 3123–3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Morgan R, Dahlbeck D, Borsani O, Villegas A Jr., Zhu JK, Staskawicz BJ, Jin H 2006. A pathogen-inducible endogenous siRNA in plant immunity. Proc Natl Acad Sci U S A 103(47): 18002–18007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korner CJ, Klauser D, Niehl A, Dominguez-Ferreras A, Chinchilla D, Boller T, Heinlein M, Hann DR. 2013. The immunity regulator BAK1 contributes to resistance against diverse RNA viruses. Mol Plant Microbe Interact 26(11): 1271–1280. [DOI] [PubMed] [Google Scholar]
- Lellis AD, Kasschau KD, Whitham SA, Carrington JC. 2002. Loss-of-susceptibility mutants of Arabidopsis thaliana reveal an essential role for eIF(iso)4E during potyvirus infection. Curr Biol 12(12): 1046–1051. [DOI] [PubMed] [Google Scholar]
- Li F, Pignatta D, Bendix C, Brunkard JO, Cohn MM, Tung J, Sun H, Kumar P, Baker B. 2012. MicroRNA regulation of plant innate immune receptors. Proc Natl Acad Sci U S A 109(5): 1790–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Lu YG, Shi Y, Wu L, Xu YJ, Huang F, Guo XY, Zhang Y, Fan J, Zhao JQ, et al. 2014. Multiple rice microRNAs are involved in immunity against the blast fungus Magnaporthe oryzae. Plant Physiol 164(2): 1077–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhang Q, Zhang J, Wu L, Qi Y, Zhou JM. 2010. Identification of microRNAs involved in pathogen-associated molecular pattern-triggered plant innate immunity. Plant Physiol 152(4): 2222–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Huang X, Li M, He P, Zhang Y. 2016. Loss-of-function of Arabidopsis receptor-like kinase BIR1 activates cell death and defense responses mediated by BAK1 and SOBIR1. New Phytol 212(3): 637–645. [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Marathe R, Dinesh-Kumar SP. 2002. Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to Tobacco mosaic virus. Plant J 30(4): 415–429. [DOI] [PubMed] [Google Scholar]
- Lopez Sanchez A, Stassen JH, Furci L, Smith LM, Ton J. 2016. The role of DNA (de)methylation in immune responsiveness of Arabidopsis. Plant J 88(3): 361–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S, Vailleau F, Balague C, Roby D 2003. Lesion mimic mutants: keys for deciphering cell death and defense pathways in plants? Trends Plant Sci 8(6): 263–271. [DOI] [PubMed] [Google Scholar]
- Ma C, Liu Y, Bai B, Han Z, Tang J, Zhang H, Yaghmaiean H, Zhang Y, Chai J. 2017. Structural basis for BIR1-mediated negative regulation of plant immunity. Cell Res 27(12): 1521–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques-Bueno MDM, Morao AK, Cayrel A, Platre MP, Barberon M, Caillieux E, Colot V, Jaillais Y, Roudier F, Vert G. 2016. A versatile Multisite Gateway-compatible promoter and transgenic line collection for cell type-specific functional genomics in Arabidopsis. Plant J 85(2): 320–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GB, Bogdanove AJ, Sessa G. 2003. Understanding the functions of plant disease resistance proteins. Annu Rev Plant Biol 54: 23–61. [DOI] [PubMed] [Google Scholar]
- Mayers CN, Lee KC, Moore CA, Wong SM, Carr JP. 2005. Salicylic acid-induced resistance to Cucumber mosaic virus in squash and Arabidopsis thaliana: contrasting mechanisms of induction and antiviral action. Mol Plant Microbe Interact 18(5): 428–434. [DOI] [PubMed] [Google Scholar]
- Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW. 2003. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15(4): 809–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JD. 2006. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312(5772): 436–439. [DOI] [PubMed] [Google Scholar]
- Navarro L, Jay F, Nomura K, He SY, Voinnet O. 2008. Suppression of the microRNA pathway by bacterial effector proteins. Science 321(5891): 964–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicaise V, Candresse T. 2017. Plum pox virus capsid protein suppresses plant pathogen-associated molecular pattern (PAMP)-triggered immunity. Mol Plant Pathol 18(6): 878–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang S, Park G, Atamian HS, Han CS, Stajich JE, Kaloshian I, Borkovich KA. 2014. MicroRNAs suppress NB domain genes in tomato that confer resistance to Fusarium oxysporum. PLoS Pathog 10(10): e1004464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson K, Molloy L, Qu W, Clark S. 2011. DNA methylation: bisulphite modification and analysis. J Vis Exp 56: 3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prelich G 2012. Gene overexpression: uses, mechanisms, and interpretation. Genetics 190(3): 841–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi G, Chen J, Chang M, Chen H, Hall K, Korin J, Liu F, Wang D, Fu ZQ. 2018. Pandemonium breaks out: Disruption of salicylic acid-mediated defense by plant pathogens. Mol Plant 11(12): 1427–1439. [DOI] [PubMed] [Google Scholar]
- Rodriguez E, El Ghoul H, Mundy J, Petersen M. 2016. Making sense of plant autoimmunity and ‘negative regulators’. FEBS J 283(8): 1385–1391. [DOI] [PubMed] [Google Scholar]
- Rowley MJ, Avrutsky MI, Sifuentes CJ, Pereira L, Wierzbicki AT. 2011. Independent chromatin binding of ARGONAUTE4 and SPT5L/KTF1 mediates transcriptional gene silencing. PLoS Genet 7(6): e1002120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwessinger B, Roux M, Kadota Y, Ntoukakis V, Sklenar J, Jones A, Zipfel C. 2011. Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. PLoS Genet 7(4): e1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaprasad PV, Chen HM, Patel K, Bond DM, Santos BA, Baulcombe DC. 2012. A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell 24(3): 859–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H, Greenberg MV, Feng S, Bernatavichute YV, Jacobsen SE. 2013. Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 152(1–2): 352–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Nasir KH, Ito A, Kanzaki H, Matsumura H, Saitoh H, Fujisawa S, Kamoun S, Terauchi R. 2007. A high-throughput screen of cell-death-inducing factors in Nicotiana benthamiana identifies a novel MAPKK that mediates INF1-induced cell death signaling and non-host resistance to Pseudomonas cichorii. Plant J 49(6): 1030–1040. [DOI] [PubMed] [Google Scholar]
- Tena G, Boudsocq M, Sheen J. 2011. Protein kinase signaling networks in plant innate immunity. Curr Opin Plant Biol 14(5): 519–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallarino JG, Osorio S. 2016. Simultaneous determination of plant hormones by GC-TOF-MS. Methods Mol Biol 1363: 229–237. [DOI] [PubMed] [Google Scholar]
- Wierzbicki AT, Cocklin R, Mayampurath A, Lister R, Rowley MJ, Gregory BD, Ecker JR, Tang H, Pikaard CS. 2012. Spatial and functional relationships among Pol V-associated loci, Pol IV-dependent siRNAs, and cytosine methylation in the Arabidopsis epigenome. Genes Dev 26(16): 1825–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM. 2001. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414(6863): 562–565. [DOI] [PubMed] [Google Scholar]
- Wu Y, Zhang D, Chu JY, Boyle P, Wang Y, Brindle ID, De Luca V, Despres C. 2012. The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep 1(6): 639–647. [DOI] [PubMed] [Google Scholar]
- Xiao YL, Redman JC, Monaghan EL, Zhuang J, Underwood BA, Moskal WA, Wang W, Wu HC, Town CD. 2010. High throughput generation of promoter reporter (GFP) transgenic lines of low expressing genes in Arabidopsis and analysis of their expression patterns. Plant Methods 6: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Yu B. 2015. siRNA-directed DNA methylation in plants. Curr Genomics 16(1): 23–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi H, Richards EJ. 2007. A cluster of disease resistance genes in Arabidopsis is coordinately regulated by transcriptional activation and RNA silencing. Plant Cell 19(9): 2929–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A, Lepere G, Jay F, Wang J, Bapaume L, Wang Y, Abraham AL, Penterman J, Fischer RL, Voinnet O, et al. 2013. Dynamics and biological relevance of DNA demethylation in Arabidopsis antibacterial defense. Proc Natl Acad Sci U S A 110(6): 2389–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J, Jeong DH, De Paoli E, Park S, Rosen BD, Li Y, Gonzalez AJ, Yan Z, Kitto SL, Grusak MA, et al. 2011. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev 25(23): 2540–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhao H, Gao S, Wang WC, Katiyar-Agarwal S, Huang HD, Raikhel N, Jin H. 2011. Arabidopsis Argonaute 2 regulates innate immunity via miRNA393(*)-mediated silencing of a Golgi-localized SNARE gene, MEMB12. Mol Cell 42(3): 356–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zvereva AS, Pooggin MM. 2012. Silencing and innate immunity in plant defense against viral and non-viral pathogens. Viruses 4(11): 2578–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Effect of RNA silencing on BIR1 expression in plants infected with TuMV.
Figure S2. Epigenetic regulation of BIR1 and RdDM-methylation controls.
Figure S3. Methylation status of the BIR1 promoter using whole-genome bisulfite sequencing (WGBS) data in Arabidopsis.
Figure S4. Methylation status of the BIR1 promoter using in-house bisulfite sequencing in Arabidopsis.
Figure S5. Epigenetic regulation of BIR1 and RdDM-methylation controls in salicylic acid (SA)-treated plants.
Figure S6. BIR1 mRNA accumulation in RNA silencing mutants, cleavage mapping at the 5’ UTR of BIR1 mRNA and viral accumulation in N. benthamiana leaves expressing BIR1.
Figure S7. DEX-inducible system for overexpression of BIR1 in Arabidopsis plants.
Figure S8. Phenotypes of BIR1 overexpressing transgenic Arabidopsis.
Figure S9. Phenotypes of BIR1 overexpressing transgenic seedlings grown in axenic conditions.
Figure S10. Model of BIR1 regulation
Table S1. List of primers.


