Abstract
In 1901 T.H. Morgan proposed in “Regeneration” that pattern formation in amphibian limb regeneration is a stepwise process. Since, biologist have continued to piece together the molecular components of this process to better understand the “patterning code” responsible for regenerate formation. Within this context, several different models have been proposed; however, all are based on one of two underlying hypotheses. The first is the “morphogen hypothesis” that dictates that pattern emerges from localized expression of signaling molecules, which produce differing position-specific cellular responses in receptive cells depending on the intensity of the signal. The second hypothesis is that cells in the remaining tissues retain memory of their patterning information, and use this information to generate new cells with the missing positional identities. A growing body of evidence supports the possibility that these two mechanisms are not mutually exclusive. Here, we propose our theory of hierarchical pattern formation, which consists of 4 basic steps. The first is the existence of cells with positional memory. The second is the communication of positional information through cell-cell interactions in a regeneration-permissive environment. The third step is the induction of molecular signaling centers. And the last step is the interpretation of these signals by specialized cell types to ultimately restore the limb in its entirety. Biological codes are intertwined throughout this model, and we will discuss their multiple roles and mechanisms.
Keywords: Patterning Hierarchy code, Positional information, axolotl, pattern formation, limb regeneration, biological codes
1). Biological codes
Although no singular definition exists, a code can be defined as a system whereby information is stored in one form, such as the sequence or pattern of letters or numbers, and then interpreted such that output is generated in a different form. Code biology is the study of the origin, refinement, function, evolution and conservation of life codes, broadly range from organic codes, which contribute to the phenomena such as of cellular hereditary and metabolism, to cultural codes, such as human language (as reviewed by (Barbieri, 2019, 2018, 2008)). At an organic level, examples of code-based systems are plentiful. One of the most classical illustrations being that of the genetic code. This phenomenon involves the conversion (via transcription and translation) of a sequence of nucleotides (a gene and informational entity), constituting a piece of a DNA strand, into a specific amino acid sequence (a protein and functional entity) (Crick et al., 1961). Specific rules govern this system to ensure not only the accuracy of the conversion itself but also when and where this conversion of information occurs. Regulation occurs at several levels, including the use of the histone code (Strahl and Allis, 2000). This epigenetic phenomenon involves specific modifications, such as methylation and acetylation, being added to histone proteins. These marks are read like a language, dynamically regulating the transcriptional output of the associated DNA molecule (Rothbart and Strahl, 2014; Strahl and Allis, 2000). The histone code and genetic code, although distinct, work together in a hierarchal fashion to facilitate the ability of a cell to interpret its genetic information in the context of its environment and give rise to the appropriate expressional fate.
The use of biological codes extends beyond gene regulation. A cell within a multicellular organism responds to a dynamic environment composed of a variety of other cells. Each cell must behave within this environment in an appropriate, cohesive manner, at both spatial and temporal levels, to form the complex structures that compose a fully functioning organism. A single fertilized egg cell must undergo profound changes in order to generate a three dimensional organism. This process requires fidelity, and a patterning code would provide reproducible outcomes during development. Furthermore, this code could be reemployed post-embryonically in cases where lost pattern must be replaced.
Pattern formation, whether in a developmental or regenerative context, is the process whereby the cells of an organism are organized spatially and temporally within a three dimensional framework. Within a developmental setting, the establishment of anterior-posterior (A/P) and dorsal-ventral (D/V) axes are required for the formation of the primary body field. The development of the primary body plan is a progressive, segmented process, with the head being established earlier than the more posterior structures such as the trunk and tail. The secondary body fields are subsequently initiated at locations dictated by the pattern of the primary field. These secondary fields are responsible for the patterning of limbs or wings de novo.
Within a regenerative context, the primary and secondary body patterns have already been established; therefore, any lost pattern must be re-specified in a manner that takes into account the pattern of the remaining stump. Hence, the deployment of any patterning code, used during development, must not only recognized pre-established, stump pattern but also ensure seamless integration of the new and old tissue. Evidence supports the idea that the mechanism by which this process occurs is hierarchical in nature, maintaining reproducibility in structure and function. Here we present a new model for pattern formation during limb regeneration called the Patterning Hierarchy Model. We will review the experimental evidence from a variety of regenerating and developing systems that contributed to the conception of this model. We will also discuss the potential roles of different biological codes in this new model.
2). Previous models describing pattern formation during regeneration
Despite the functional output of pattern formation being easily characterized by morphological and histological studies, there is much to learn about the language and regulation of the contributing codes facilitating this phenomenon. Many models have been developed to explain how pattern formation occurs during limb regeneration. Irrespective of the developmental or regenerative context, all models that describe patterning utilize a fundamental term - positional information. Positional information is the attribute of a cell, whereby the cell has a defined spatial position relative to one or more points. The interpretation of this information subsequently determines the specialization of the cell and, in so doing, generates pattern through spatial-specific differentiation (Wolpert, 1969).
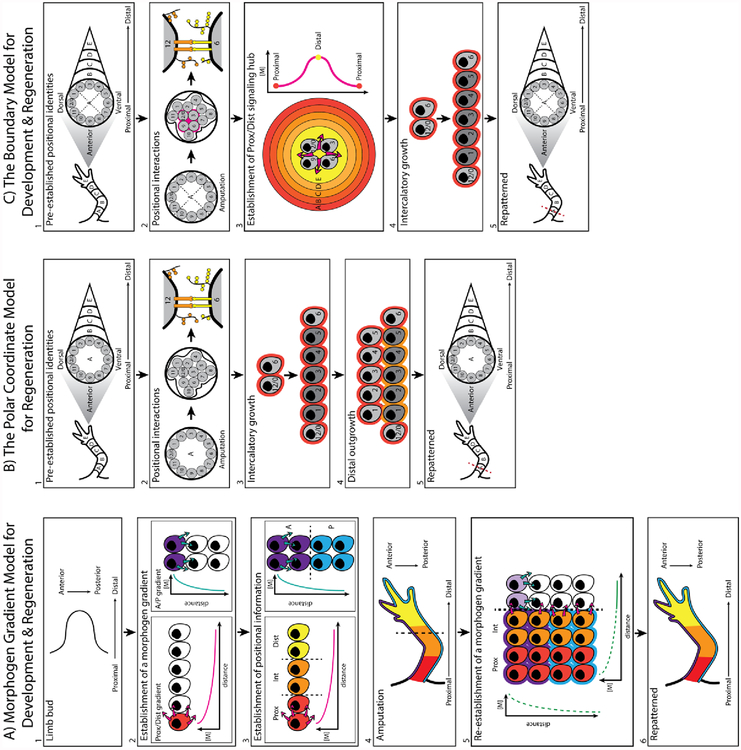
One of the earliest models describing the mechanism by which positional information is generated during development, involves a morphogen gradient in a sheet of cells (Figure 1A). Within a population of cells with uniform potential, a morphogen (a diffusible signaling molecule) is produced by a signaling center. In order to achieve a linear concentration gradient across the sheet, a “sink” may exist on the opposite end of the source. This sink will limit diffusion of the morphogen beyond a specific location, establishing a set end point for the gradient with a finite concentration. Therefore, the signaling hub and sink act as the reference points by which relative positional identities are established. A cell will then interpret the concentration of the morphogen to which it is exposed, at its location along the gradient, and act accordingly (Wolpert, 1969).
Figure 1:
Previously proposed patterning models.
A) Morphogen gradient model: During development cells exists in a naïve state, with uniform positional potential (white cells) (1). A morphogen generating signaling hub is established at one end of the cell sheet, in this model on the most proximal end (red cell) as well as the most anterior (purple cell) end of the sheet. Diffusion of each signaling molecule establishes an independent concentration gradient (2). The local concentration of the morphogen is interpreted by the surrounding cells and in turn specifies positional information, such as proximal to distal and anterior to posterior identities (3), which will result in the patterning of the complete limb (4). In the context of regeneration, amputation through the limb (5) would result in the generation of new signaling hubs along both the P/D and A/P axes (5). The newly established local concentrations of the morphogens are then interpreted by the new, positionally naïve cells and in turn specifies positional information (6).
B) The polar coordinate model: Prior to amputation identities pre-exist within the limb, established during development, along the P/D (arbitrarily denoted as A-E identities) and circumferential axes (arbitrarily denoted as 0 – 12). Pattern forming cells are demarcated in grey (1). In response to amputation at plane A, wound healing and local cell-cell communication will facilitate the establishment of a positional disparity (2). This disparity is resolved through the processes of intercalation, where new cells with the intermediate positional identities are generated (3), and distal outgrowth, where the new identities that are intercalated can assume the next distal identity (4). Repeated iteration of these two phenomena results in the reestablishment of complete limb pattern (5).
C) The boundary model: Positional identities also pre-exist within the limb along the P/D (arbitrarily denoted as A-E identities) and circumferential axes (arbitrarily denoted as 0 – 12). Pattern forming cells are demarcated in grey. Essential to this model is the established anterior, posterior, dorsal and ventral boundaries within the limb (dotted lines). In response to amputation at plane A, wound healing will juxtapose at least three limb boundaries, and the opposing cells communicate with one another locally (2). A signaling hub is established at the boundary intercept, which generates a circular morphogen gradient (3). Distal outgrowth of the regenerating organ stimulates higher morphogen levels to be produced, which then progressively specifies more distal identities. This progressive distal specification by increasing morphogen concentrations has been termed bootstrapping. Subsequently, any disparity between circumferential identities, within a single plane, are then resolved through intercalation (4). Through the process of bootstrapping and intercalation, the missing structure is re-patterned (5).
One in vivo example that contributed to this model is in the developing fly embryo, whereby a Bicoid gradient establishes different A/P positional identities in the syncytial embryo. Depending on its location in the embryo, each nuclei will receive differing abundances of Bicoid, which in turn regulates the transcription of Hunchback and Kruppel in specific A/P locations in the syncytium (Driever and Nüsslein-Volhard, 1988). The gradient model also explains the phenomenological effects documented in response to altered sonic hedgehog (Shh) expression in the developing limb bud. Shh is posteriorly expressed in the developing chick limb bud, and its graded distribution facilitates the distinct A/P digit pattern of the wing (Riddle et al., 1993; Tickle et al., 1975). Adding a new Shh signaling center to the anterior margin on the limb bud distorts A/P pattern by generating a mirror image duplication of the autopod. The most posterior digits are patterned closest to each of the Shh hubs and the most anterior digits centered furthest away, in the middle of autopod (Riddle et al., 1993; Tickle et al., 1975).
Within a regeneration context, this model predicts the re-establishment of developmental morphogen gradients in response to amputation which will facilitate the formation of new pattern (Figure 1A) (Wolpert, 1996). It is possible that either the original signaling centers, established during development, are responsible for generating the needed morphogen gradients or new signaling centers are created at the site of injury. The latter would be the case for Shh expression during axolotl limb regeneration as Shh expression disappears once limb development has completed and then re-appears during limb regeneration (Torok et al., 1999). However, there are several shortcomings of the gradient model in terms of regeneration. Firstly, if a morphogen gradient is re-established by a developmental signaling center, how is regeneration achieved if these centers are removed during amputation? Secondly, if a new signaling center is generated at the site of injury, such as for Shh in the case of amphibian limb regeneration (Torok et al., 1999), what mechanism ensures that a location-appropriate and -specific concentration of each of the necessary morphogens is produced to recapitulate the developmental gradients from that point? In order for this to be achieved, at least in part, cells must stably retain positional information long after the completion of development. Such a phenomenon is not described in the gradient model but is presented in a different model of patterning known as the polar coordinate model for regeneration.
The polar coordinate model was conceived from observations on regenerating amphibian and insect models to explain how new pattern emerges during regeneration, and it compensates for some of the short falls of the gradient model. In this model positional information is described in a polar coordinate system (Figure 1B) (Bryant et al., 1981; French et al., 1976). One dimension of information corresponds to a cells position on the circumference of the limb, which have been arbitrarily assigned identities 0 through 12. Another dimension corresponds to the location of the cells on the proximal/distal (P/D) limb axis, which have been arbitrarily assigned identities A through E. For example, a human thumb would roughly be ascribed the value 9E in this coordinate system. In this model, uninjured cells are in contact with ‘normal’ neighbors (example identities 2 and 3 in Figure 1B1) and are separate from or opposing ‘non-normal’ neighbors (example identities 3 and 9 in Figure 1B1). In response to amputation, wound healing brings ‘non-normal’ neighbors together which proliferate to restore the missing positional identities between them via a process called intercalation. This system has two fundamental rules. Firstly, intercalation takes place by the shortest route around the limb circumference as possible. Therefore, if identities 2 and 4 are brought together; identity 3 will be generated, rather than 5,6,7,8,9,10,11,12,1 (Bryant et al., 1981; French et al., 1976). Secondly, when new cells are intercalated, they can also acquire a more distal identity, thus resulting in distal outgrowth (Bryant et al., 1981). When identities 9A and 11A are brought together, identity 10A will be generated; if identity 10 already exists in plane A the new cell will assume a more distal identity (10B). The repeated iteration of intercalation and distal outgrowth, which depend solely on local cell-cell interactions, leads to regeneration.
Although the polar coordinate model provides a conceptual framework to understand pattern formation, little is known about how this model might work at the molecular level. Additionally, this model focuses on local interactions between cells that retain positional information; however, as will be discussed in section 3, not all cells retain positional memory. These cells, which lack positional information, can be considered as positionally naïve and also participate in the formation of the regenerate and therefore require location-specific instruction by some mechanism as well. Evidence for the importance of local cell interactions has since emerged in patterning systems. Mesoderm induction, for example, in the developing Xenopus embryo is reduced with a transfilter apparatus that physically separates the vegetal (inducing) and animal (responding) cells, suggesting that direct, local cell-cell interactions contribute to this developmental process (Slack, 1991). Glycosaminoglycans mediated communication appears to be one mechanism by which these localized interaction are facilitated, as mesoderm induction was abolished in the transfilter system with heparin treatment. This suggests that heparin is able to bind to signaling factors in the liquid junction between the different cell populations (Slack, 1991). Furthermore, pattern formation during regeneration is altered by heparin sulfate proteoglycan manipulations (McCusker et al., 2016; Phan et al., 2015).
Despite the absence of molecular mechanisms, the polar coordinate model has emerged as a powerful predicative tool for the pattern that is generated when confronting cells from different limb positions in a regenerative permissive environment. This model accurately predicts that in a regeneration permissive environment when completely opposing identities, such anterior and posterior, are brought together, a complete circumference of positional identities can be generated and, in turn, a new, complete limb will be patterned (Bryant and Iten, 1976; Endo et al., 2004; Iten and Bryant, 1975; McCusker et al., 2014). Alternatively, smaller disparities in positional information will fail to re-establish all the circumferential identities required for patterning, thus generating distally incomplete structures. Phenomenological examples of such distally incomplete patterns have been generated by 1) the amputation of double posterior limbs in Urodeles (Holder et al., 1980) and 2) the heat or radiation induced generation of small positional disparities in the imaginal discs of the fly (Girton, 1981; Girton and Russell, 1980).
Unfortunately, not all experimental data aligns with the polar coordinate model. Firstly, this model predicts that proximal patterning information is generated prior to distal identities. However, in situ hybridization (ISH) of blastema tissue (a collection of regeneration competent cells responsible for patterning the regenerate) reveals that both proximal and distal Hox genes (HoxA9 and HoxA13) are expressed in the early stages of limb regeneration, irrespective of location along the P/D axis. As regeneration commences the expression patterns of these genes become spatially restricted (Gardiner et al., 1995). Based on this evidence, the “distal-first” hypothesis was presented as a modification to the polar coordinate model. It was proposed that at the early stages of regeneration the regenerate has the most distal identity, while the remaining stump tissue has the most proximal identity. Since these are “non-normal” neighbors, this would drive an intercalary response to generate the intermediate limb identities along the P/D axis (Gardiner et al., 1995). However, there is some controversy regarding this modification to the model. Analysis of limb specific Hox expression using antibodies directed against HoxA9, HoxA11 and HoxA13 indicates a progressive, sequential segmentation of positional identities from proximal to distal (Roensch et al., 2013). The discrepancy between this data and that documented from ISH is uncertain and, therefore, the validity of the distal first hypothesis remains to be determined. Although Hox genes are well established P/D markers within the limb, evaluation of a larger pool of additional distal and proximal markers would likely improve our understanding in this context. Although antibody availability in the axolotl is limited, the genome has now been fully sequences and thus should aid in allowing for a variety of positional markers to be used in future.
Secondly, there are several regeneration associated patterning responses that do not perfectly align with the predictions of the polar coordinate model. For example, if double-half limbs are generated and then amputated, according to the polar coordinate model, only a limited number of identities will be present in the stump. Depending on how these identities interact locally, a variety of patterning outcomes could be generated - no growth, symmetrical and distally incomplete, or branched and distally complete outgrowths (Bryant et al., 1981). Furthermore, both anterior and posterior double half limbs would be predicted to have the same potential in this context. However, amputation of double anterior limbs in Urodeles results in no new pattern while double posterior limbs can (Holder et al., 1980). Additionally, amputating and rotating a portion of the limb by 90° relative to the stump and then reattaching it would result in only a small positional disparity, such as anterior meeting dorsal. The polar coordinate model predicts incomplete distal outgrowth but in vivo distally complete, supernumerary limbs are generated (Maden and Turner, 1978).
These deviations from the patterning predictions of the polar coordinate model could be accounted for, at least in part, by an uneven distribution of positional identities, both circumferentially and linearly, within the limb. The ability of periosteum tissue to induce supernumerary structures when implanted into distinct locations of the limb prior to amputation, through the host site, revealed that the connective tissue of the humerus and radius encoded only anterior and/or dorsal identities, while the ulna encoded the full complement of information (Gardiner and Bryant, 1989). Therefore, each half of the double posterior half limbs would contain more than half of the circumferential identities and therefore elicit more complex intercalatory growth relative to anterior half limbs, where each half has low positional diversity (Gardiner and Bryant, 1989). The same principle would apply to 90° rotations of the limb relative to the stump, where large disparities could be generated internally by small shifts between axes.
In an attempt to resolve the above mentioned phenomena, the boundary model of pattern formation was presented to provide more insight into the molecular basis of how new pattern emerges during embryogenesis and regeneration (Meinhardt, 1982, 1983a). The boundary model was originally derived utilizing experimental observations in developing and regenerating insect imaginal discs and limbs and was based on the idea, similar to the polar coordinate model, that cells have positional information and use this information to generate new pattern during regeneration. This model involved only cells with positional information during regeneration, like the polar coordinate model. It additionally incorporated the concept of a morphogen gradient (Figure 1C)(Meinhardt, 1982, 1983a).
According to the boundary model, secondary embryonic fields such as the limb are established by at least three specified boundaries in the primary body axis that are brought into direct contact – anterior (9), dorsal (12/0), posterior (3) and ventral (6) (Figure 1C2). At the site of this juxtaposition, a local morphogen gradient is induced that will establish rudimentary identities in the secondary field (Meinhardt, 1983b, 1983a). The morphogen would be strongest at the sight of juxtaposition and correspond to the most distal identity in the field. Moving away from this center, the morphogen would become more dilute and specify more proximal identities, in a process termed bootstrapping (Meinhardt 1982; Meinhardt 1983a) (Figure 1C3). Local cell-cell interactions within a particular P/D plane would then lead to intercalation and the establishment of all the outstanding circumferential identities (Meinhardt, 1982). In response to amputation and wound healing a new boundary intercept would be established that would then drive this process again, restoring the missing structure (Meinhardt, 1983c).
Part of the in vivo evidence that lead to the conception of the boundary model was derived from the two dimensional environment of the developing and regenerating imaginal discs of heat sensitive Drosophila mutants. In this system, exposure to elevated temperatures, or surgical manipulations, resulted in some cells within the secondary field changing their boundary specification. This generated boundary juxtapositions and, in turn supernumerary limbs. The location of the different boundaries relative to each other were able to accurately predict the completeness of the ectopic outgrowths (Girton, 1981; Girton and Russell, 1980; Meinhardt, 1983a; Russell and Girton, 1981). However, this phenomenon is not reproduced in the regenerating amphibian limb. The accessory limb model (ALM), is a surgical manipulation whereby an ectopic limb can be generated on existing, patterned limb field. In this “gain of function” regenerative assay the brachial nerve is deviated to a lateral wound site, which is implanted with a piece of full thickness skin from the opposite side of the limb axis (Endo et al., 2004). This surgery is sufficient to generate a complete limb (starting at the elbow joint), suggesting that only two opposing boundaries, instead of three, are sufficient to generate complete pattern (Bryant et al., 1981; French et al., 1976).
More recently, Nacu et al., (2016) was able to generate pattern from innervated limb wound sites treated with exogenous Shh or Fibroblastic growth factor (FGF) 8 instead of a skin graft. The ability of these signaling molecules to generate distal outgrowth correlates to the process of bootstrapping presented in the boundary model. However, these structures were symmetrical along the A/P axis and incomplete along the P/D axis. This suggests that morphogen gradients alone are insufficient to generate the finer features of pattern, which in the boundary model are induced by intercalation. As will be discussed further in this review, morphogen gradients appear sufficient to generate pattern, albeit incomplete, but fail to elicit the establishment of new positional identities.
The morphogen gradient, polar coordinate, and boundary models each provide valuable insight into the possible mechanisms by which pattern formation occurs during regeneration. However, as described above, each captures only a few aspects of a mutli-faceted biological process, and none completely characterizes the activity of both pattern forming and following cells during regeneration or the molecular underpinnings thereof. Despite each model being independent; there is considerable overlap between them. Therefore, we utilized each of the above-presented patterning models as well as the experimental evidence that support them and generated a hierarchical model to describe pattern formation during regeneration (Figure 2).
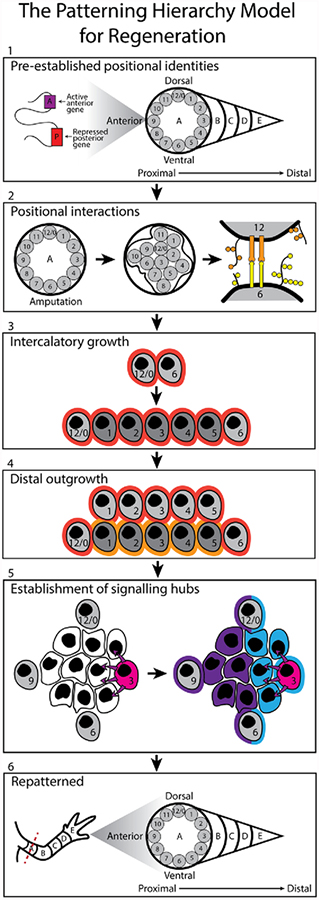
Figure 2:
Patterning Hierarchy Model: As in the polar coordinate model and boundary model, positional identities pre-exist within the limb prior to amputation. Epigenetic mechanisms, such as histone modifications, DNA methylation and DNA looping, ensure the stable retention of this information within pattern forming cells (grey cells). In response to amputation at plane A, wound healing and local cell-cell communication will facilitate the establishment of a positional disparity (2). This disparity is resolved through the processes of intercalation (3) and distal outgrowth (4). Pattern forming cells then establish location appropriate signaling centers (pink cell) for the production of morphogens and/or signaling molecules (5). Morphogen gradients generated by these centers are then interpreted by pattern following cells, which respond in a location appropriate manner. Pattern forming cells can also respond to these morphogen cues, in a location appropriate manner (5). This process, along with intercalation and distal outgrowth, ensures complete regeneration of the missing limb structure (6).
3). The Patterning Hierarchy Model of Regeneration:
Within the patterning hierarchy model two types of cells are defined, 1) pattern forming cells and 2) pattern following cells. Pattern forming cells acquire positional identity during development, and stably retain this information during the life of the organism. For example, fibroblasts appear to be a pattern forming cell type. Fibroblasts harvested from different sites within the human body have demonstrated a cell-autonomous and epigenetically-regulated retention of location specific information, which resembles that documented within the developing embryo (Rinn et al., 2008, 2007, 2006). Dermal fibroblasts contribute to the amphibian limb regenerate, and are able to induce new pattern to form when confronted with tissue with opposing positional information (Endo 2004). In contrast, pattern following cells do not retain this positional memory and can be viewed as being positionally naïve. These cells depend on informational cues from pattern forming cells in order behave in a location appropriate manner. For example, positionally naïve epithelial cells will undergo palmoplantar specialization when co-cultured with foot derived fibroblasts but not with leg fibroblasts (Rinn et al., 2008).
At the highest level of the hierarchy model, positional information is pre-established and stably maintained in pattern forming cells by epigenetic mechanisms (Figure 2.1) (Rinn et al., 2008, 2007). In uninjured tissues, these cells likely provide positional cues to the surrounding pattern following cells to maintain tissue homeostasis. However, in response to amputation, nerve-dependent signals generate a regeneration permissive environment in which pattern forming cells can communicate their positional information with one another and/or respond to this information appropriately (Endo et al., 2004; McCusker and Gardiner, 2013).
At this second level of the hierarchy, communication could be mediated through local cell-cell interactions (Figure 2.2)(Bryant and Gardiner, 1992; McCusker et al., 2016; Phan et al., 2015). Consistent with the polar coordinate model of regeneration, cells with opposing positional information are brought together by wound healing. The pattern forming cells that are juxtaposed to ‘non-normal’ neighbors will generate a positional disparity between them.
Pattern forming cells then interpret this discrepancy, and undergo a process of intercalation and distal outgrowth to resolve it (Figure 2.3 and 2.4)(Bryant et al., 1981; French et al., 1976). These two processes, proposed in the polar coordinate model, will restore both the circumferential and P/D positional identities through the generation of new pattern-forming cells with the missing identities. It is also plausible that some of the pattern-forming may be directly reprogrammed, in the absence of proliferation, and assume the required, outstanding identities.
At the third level of the hierarchy, once the missing positional identities have been restored, pattern forming cells generate one or more signaling centers (Figure 2.5). Pattern forming cells interpret their positional information to ensure the establishment of these signaling centers in a location-appropriate manner. The resultant signaling molecules are required to provide position-specific signaling cues to the pattern following cells in the regenerating environment.
The fourth and final level involves both pattern forming and following cells. The signaling molecules, generated at the third level of the hierarchy, are interpreted by both of these cell populations (Figure 2.5)(Rinn et al., 2008), such that overall pattern is achieved in all tissues (Figure 2.6).
Each level of this model, proceeding from pre-amputation to regeneration, will now be discussed in detail. Experimental evidence validating each hierarchical level as well as the mechanisms and biological codes assumed to be involved will be presented.
4). Level 1: The code of positional information
Since the conception of the field of developmental biology, embryologists have been fascinated by the complexity that arises from a single fertilized egg, sufficient to pattern a multicellular organism. Though there are many aspects involved in the development of a multicellular organism; ultimately, they either relate to the specification of distinct cell types, or the arrangement of these cell types according to a specific blueprint, or pattern. Patterning in the embryo starts early. For example, the location of sperm entry specifies the dorsal/ventral axis in amphibian embryo. Here, molecules which promote Wnt signaling are sequestered to the side of the embryo opposing the sperm penetration site and establish a dorsal fate (Elinson, 1975; Pierce and Kimelman, 1995; Schneidera et al., 1996; Vincent et al., 1986). In other organisms, patterning commences when multiple daughter cells have been generated, and cells begin to learn where they are relative to their neighbors (as reviewed by (Bedzhov et al., 2014)). At these early stages, this pattern information is still somewhat plastic, and can still be manipulated (as reviewed by (Bedzhov et al., 2014)). As the embryo continues to develop, additional layers of patterning complexity are added. The establishment of the A/P primary body axis, and subsequent axes of secondary fields such as limbs, will be formed. Hox genes have a well-established role in the patterning of the primary and secondary embryonic fields, and the position-specific regulation of these genes is often regarded as the Hox code.
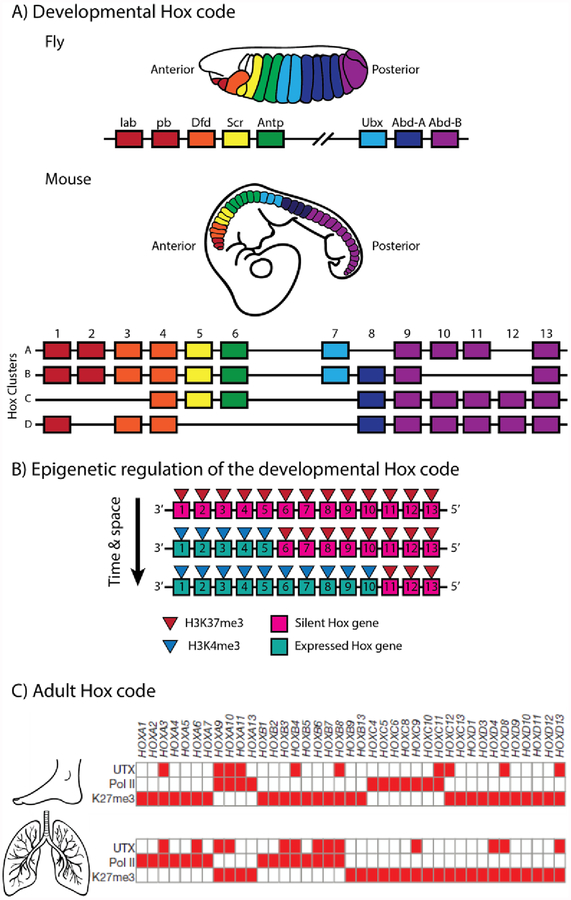
Hox genes, which encode a family of transcriptional regulators, were originally discovered in Drosophilia. These genes, encoded as two separate gene clusters (the Antennapedia and Bithorax complexes), are expressed in a spatial co-linear manner resulting in the coordinated and progressive segmentation of the developing fly embryo along the A/P axis (Figure 3A) (Kaufman et al., 1980; Lewis, 1978; Lewis et al., 1980). Subsequent research demonstrated that this gene family exhibits a remarkable degree of conservation during evolution, being documented in vertebrates, invertebrates and even plants (as reviewed by (Holland, 2013)). Genome duplications during evolution resulted in the duplication of the Hox genes and formation of four distinct Hox clusters within most vertebrates, designated A through to D (Figure 3A). Genes located at corresponding positions within each cluster and sharing sequence similarity are referred to as paralogous genes. The genes of each cluster are still expressed in a collinear fashion, both spatially and temporally, in a 3’ to 5’ direction during development and specify the positional information of the primary and secondary body axes (as reviewed by (Kmita and Duboule, 2003)).
Figure 3:
The Hox code and positional memory. A) The Hox genes are encoded as two separate gene clusters in the fly and share a high degree of conservation with the four clusters, arising through the process of genome duplication, in most vertebrates. The gene clusters are expressed in a spatial co-linear manner resulting in the coordinated and progressive segmentation of the developing embryo. B) The spatial and temporal co-linear expression pattern of the Hox genes during development is regulated, in part, through activating (H3K4me3) and repressive (H3K27me3) chromatin marks. C) Fibroblast populations derived from the adult lung and foot show distinct, location specific differences in transcriptional machinery (RNA polymerase II) and repressive epigenetic marks (H3K27me3) associated with the Hox genes (image modified with permission from (Lan et al., 2007)).
Based on studies in embryonic stem cells, the most primitive stage of mammalian development is assumed to lack Hox gene expression. The clusters are enriched with an epigenetic mark correlated with transcriptional inactivation – Histone 3 Lysine 27 trimethylation (H3K27me3) (Soshnikova and Duboule, 2009) (Figure 3B). As development proceeds, embryonically derived cells demonstrate a progressive loss in H3K27me3 marks in a 3’ to 5’ direction along the Hox clusters, corresponding with a progression from the anterior to posterior axis. This coordinated expression pattern corresponds with a directional accumulation of a transcriptional activation mark – Histone 3 Lysine 4 trimethylation (H3K4me3) (Soshnikova and Duboule, 2009) (Figure 3B). Although not completely understood, the polycomb repressive complexes (PRC) 2 and 1 appear to be responsible for the addition and retention of the H3K27me3 marks on the Hox clusters (as reviewed by (Mallo and Alonso, 2013)). This epigenetic mechanism, at least in part, ensure the establishment and maintenance of the Hox code during embryonic development.
Experimental observations indicate that the Hox code is retained after the completion of development as well. Fibroblasts harvested from different sites within the human body have demonstrated cell autonomous, distinct Hox gene expression profiles, which are reminiscent of developmental Hox expression profiles (Rinn et al., 2006), and maintained by a variety of epigenetic mechanisms including histone modifications and non-coding RNA (Rinn et al., 2008, 2007) (Figure 3C). This code appears sufficient to discern anatomical location, both globally and locally. At a global level the differential expression of Hox genes, as well as other genes, is sufficient to correctly cluster fibroblast populations according to body axis location – such as A/P or P/D locations (Rinn et al., 2006), suggesting a degree of continuity in information in structures located on the same end of an axis. At local level, however, there appears to be differences in positional information within organs and structures. Within the zeuogopod of the axolotl for example, the radius and ulna show distinct differences in circumferential positional information – the radius encoded only anterior and/or dorsal identities, while the ulna encoded the full complement of information (Gardiner and Bryant, 1989).
The stable retention of this location-specific Hox code is hypothesized to be a source of positional memory for homeostasis and regeneration (Rinn et al., 2008). To maintain homeostasis, fibroblasts populations are hypothesized to communicate positional cues to pattern following cells to ensure location appropriate specialization. For example, the positional cue Wnt5a ensures palmoplantar specialization of positionally naïve epithelium. This signal is produced by foot derived fibroblasts in a HoxA13-dependent manner (Rinn et al., 2008) (Figure 4A).
Figure 4:
Pattern forming and following cells facilitate tissue homeostasis and regeneration in adult organisms. A) Pattern following epithelium cells respond in a location appropriate manner when co-cultured with pattern forming cells, fibroblasts, from distinct locations (foot and thigh) along the human hind limb. Wnt5a, expressed by foot derived fibroblasts, resulted in the expression of keratin 9 in co-cultured epithelial (a marker of distal specialization) (Rinn et al., 2008). B) Pattern forming cells, such as dermis, retains memory of its location. Posterior dermis can be implanted into an innervated anterior wound site in order to induce a positional disparity sufficient to generate a new patterned limb on the existing limb field (Endo et al., 2004). Alternatively, pattern-following cells, such a Schwann cells, do not retain positional memory. Distal Schwann cells implanted into a proximal amputation site failed to hone to the distal region of the regenerate; instead the graft contributed along the entire length (Kragl et al., 2009) (Live and fluorescent images modified with permission from (Endo et al., 2004) and (Kragl et al., 2009))
In the context of regeneration, new pattern must be generated in cells that have existing pattern. Therefore, it could be assumed that a memory of already established pattern at the stump would play an essential role in the formation of the pattern of the missing structures. The ALM has demonstrated that a wound epithelium and nerve-specific signaling culminates in the formation of an ectopic blastema. This blastema regresses unless provided with a positional disparity. This disparity is generally achieved by generating a wound in the anterior of the limb and grafting a piece of posterior, full thickness skin into the wound site (Endo et al., 2004), allowing anterior cells to be juxtaposed to ‘non-normal’ neighbors (Figure 4B). Thus, in the ALM only two completely opposite identities are sufficient to generate the pattern of a distally complete limb. This observation aligns with our model as well as the polar coordinate model but not the boundary model, where at least three distinct positional boundaries are required (Bryant et al., 1981; French et al., 1976; Meinhardt, 1982).
It is important to note that not all cell types from the posterior side of the limb are sufficient to stimulate a positional disparity. For instance, only cells of connective tissue origin (dermal and periosteum cells), by retaining a memory of their location relative to the three dimensional body axis (Figure 4B), are able to elicit a regenerative response (Endo et al., 2004; McCusker et al., 2016; McCusker and Gardiner, 2013). These are cell types known to stably retain a HOX code in humans (Rinn et al., 2008, 2006). H3K27me3 levels are known to stabilize positional identity in pattern forming cells and the alteration of H3K27me3 in the regenerating cricket and flour beetle leg leads to defected pattern formation (Chou et al., 2019; Hamada et al., 2015). This observation further complements the idea that epigenetic mechanisms are involved in appropriate pattern formation during regeneration.
In summary, the first level of the hierarchal model of pattern formation involves patterning forming cells with epigenetically stabilized positional memory. These pattern forming cells are required to specify the existing pattern of the limb and will initiate the second level of the model when placed in contact with “non-normal” neighbors in a regeneration permissive environment.
5). Level 2: Local cell-cell interactions and intercalation by pattern forming cells
Wound closure, after limb amputation, is a rapid process and brings “non-normal”, pattern forming neighbors into contact with one another. This is exhibited by the migration of labeled connective tissue cells, such as dermal fibroblasts, into the early staged blastema (Endo et al., 2004; Muneoka et al., 1986; Vieira et al., 2019). It has been estimated that up to 80% of the early regenerate is of connective tissue origin (Muneoka et al., 1986). As epigenetic mechanisms ensure the retention of positional information within this cell population, a positional disparity is generated in response to these local cell-cell interactions, which stimulates the establishment of cells with the missing positional values, within the regeneration permissive environment. Achieving this requires two distinct and sequential biological phenomena – 1) communication of positional information between pattern-forming cells; 2) competence of pattern-forming cells to interpret, respond to and resolve (potentially by intercalation and distal outgrowth) a disparity in positional information. Successful resolution is achieved by the reprogramming, and subsequent stabilization of the positional identity of pattern forming cells during intercalation (McCusker and Gardiner, 2013). The use of local positional information will ensure that the structure regenerated is appropriate for the specific anatomical location. Therefore, the hierarchy model presented here could be applied to any regenerating structure; as the same step-wise process would occur with the resulting pattern dictated by the local positional interactions at the site of injury.
5.1). Communication of positional information between pattern-forming cells
The mechanisms by which positional information is communicated between cells within living organisms is slowly being elucidated. The growing evidence regarding the communication between pattern following and forming cells will be discussed in section 6 and 7 of this review; within this section we will focus on the evidence, albeit limited, which suggests that positional cues are mediated by cell-cell interactions between pattern forming cells.
As discussed earlier, mesoderm induction requires physical contact between vegetal (inducing) and animal (responding) cells, as separation of these two populations with a transfilter reduces the induction process in the developing Xenopus embryo (Slack, 1991). Patterning within the imaginal discs of the developing fly is also dependent on local cell-cell contacts. Decapentaplegic (Dpp), a morphogen expressed in the imaginal wing disc of Drosophila, is present as a gradient within this tissue. However, this gradient is not mediated by the secretion of Dpp into the extracellular matrix, but rather by the movement of this molecule from one cell to another by direct cell-cell contract. Dpp travels through specialized signaling filopodia (cytonemes), extending from Dpp receiving cells (which expressed the Dpp receptor Thickviens) to Dpp producing cells (Roy et al., 2014). Alternatively, cytonemes with Breathless, the Drosophila Fgf receptor, contact cells producing Branchless, the Drosophila Fgf (Roy et al., 2011; Sato and Kornberg, 2002), and facilitate directed migration of tracheoblast cells (Sato and Kornberg, 2002).
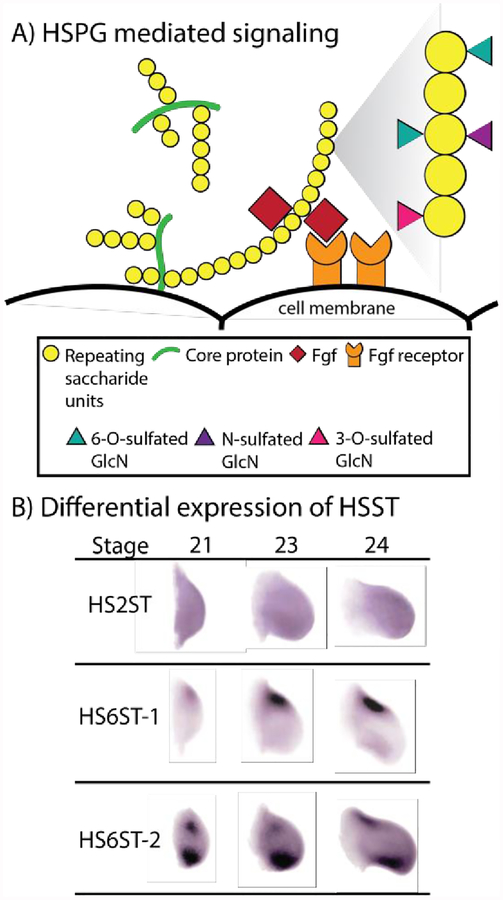
There is a growing body of evidence that indicates that the heparin sulfate proteoglycan (HSPG) code of the extracellular matrix (ECM) is used to communicate positional information between patterning forming cells during regeneration. HSPGs are an evolutionary ancient group of biomolecules, being identified in a wide variety of vertebrate and invertebrate phyla (as reviewed by (Yamada et al., 2011)). This glycoprotein is constituted by long, linear glycosaminoglycan heparan sulfate (HS) chains covalently linked to a core protein (Figure 5A). These molecules can either be anchored to the cell surface, through a transmembrane domain or glycosylphosphatidylinositol anchor, or released into ECM, either directly via secretion or enzymatic cleavage of the anchor. The HS chains are generated by alternating glucuronic acid and N-acetylglucosamine residues, which can undergo enzyme directed deacetylation, epimerization and sulfation. This diversity of modifications, which can occur multiple times on a single HS chain in a variety of combinations, are not evenly distributed along the length of the chain and, thus, generate subdomains (as reviewed by (Kreuger and Kjellén, 2012)). These modifications allow HS chains to act as epitopes for protein interactions. Structural motifs, the type of sulfation, and the negative charge density of these subdomains facilitates distinct protein binding, and allows for these biological molecules to modulate a variety of signaling pathways, by acting as receptors, co-receptors, recruiters, and/or ligand sinks (Lin, 2004; Poulain and Yost, 2015). HSPGs have been shown to be essential in the signaling of a variety of growth factors and morphogens that play fundamental roles in development and pattern formation, including Fgfs, Wnts, and Hedgehogs (as reviewed by (Lin, 2004)).
Figure 5:
The HSPG code. A) HSPGs, found either on the cell surface or free within the ECM, are composed of long, linear glycosaminoglycan HS chains covalently linked to a core protein. The saccharides of the HS chains can undergo a variety of enzyme directed modifications, including sulfation, which generates subdomains sufficient to participate in distinct protein binding. For example, as depicted, HSPGs mediate Fgf signaling by recruiting Fgf ligand and directing it towards the Fgf receptor. B) Within the developing chick wing bud, HS2ST (heparan sulfate 2-O-sulfotransferase), HS6ST-1 (heparan sulfate 6-Osulfotransferase isoform 1) and HS6ST-2 (heparan sulfate 6-Osulfotransferase isoform 2) have distinct differential expression patterns along the P/D and A/P axes. Proximal – left, Distal – right, Anterior – top, Posterior – bottom (image modified, with permission, from (Nogami et al., 2004)).
With this in mind, and based on their work in the developing nervous system of C. elegans, Bülow and Hobert (2002) proposed the HS code. They hypothesized that the combinatorial and differential expression of HS-modifying enzymes and core proteins could generate discrete HSPGs in a region specific manner that would affect local signaling cues and thus dictate developmental processes (Bülow and Hobert, 2004). Holt and Dickson, 2005, placed greater emphasis on HS modification, due to the limited number of core proteins and the diversity in HS side chain modifications documented in biological systems (Holt and Dickson, 2005). Supporting this idea was the observation that 1) different organs exhibit a distinct HS profile (Ledin et al., 2004); 2) during embryonic development dynamic alterations in HS composition, in a tissue specific manner, regulates Fgf signaling (Allen and Rapraeger, 2003); and 3) there is distinct differential expression patterns of HS modify enzymes along the different body axes during embryogenesis (Nogami et al., 2004) (Figure 5B). In the case of the latter observation, heparan sulfate sulfotransferases (HSST) are a type of HS modifying enzyme that display distinct temporal and spatial expression patterns in the developing chick wing. These patterns correlated with, and as a result are suspected to be responsible for, location- and stage-specific differences in the HS structure within the developing wing bud (Nogami et al., 2004).
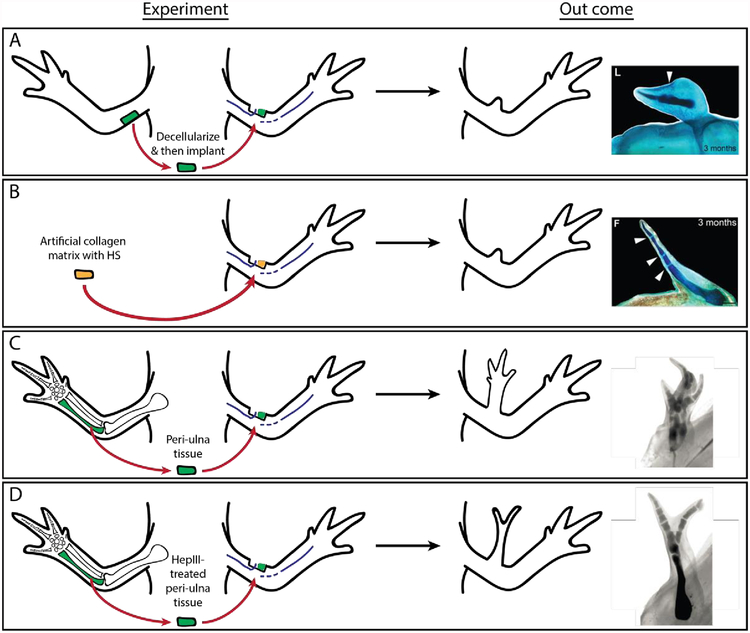
Limited observations within literature suggests that HSPGs contribute, at least in part, to pattern formation during Urodele limb regeneration (Figure 6). HSSTs, enzymes responsible for sulfation of the HS chains, exhibit differential expression between anterior and posterior blastema cells, and may therefore contribute towards an axis specific HS code (Phan et al., 2015). To test this, using the principles of the ALM, posterior de-cellularized ECM was grafted into an innervated, anterior wound site and was found to induce minimal pattern formation - what resembled a distal digit tip was generated (Phan et al., 2015) (Figure 6A). Artificial ECM, constituted by type I collagen and HS, induced more complicated pattern (jointed elements) relative to de-cellularized posterior ECM when grafted into an anterior, innervated wound site in a concentration dependent manner, suggesting specificity (Figure 6B) (Phan et al., 2015).
Figure 6:
HS mediated communication of positional information during regeneration. A) Posterior de-cellularized ECM stimulates the formation of minimally patterned outgrowths, when grafted into an innervated, anterior wound sites. B) Artificial, HS-containing ECM elicits the formation of more complicated patterning relative to de-cellularized posterior ECM, when grafted into an anterior, innervated wound site. C) A complete limb can be patterned when ulna-derived periosteum tissue is grafted into an innervated wound site; however D) heparinase-III treatment of the periosteum reduces the complexity of the resultant pattern. (Images modified, with permission, from (McCusker et al., 2016; Phan et al., 2015))
Further support for a patterning HSPG code during regeneration was identified using the ALM assay to test the pattern forming capabilities of periosteal tissue (Figure 6C and D). Ulna-derived periosteum induces the formation of a completely patterned ectopic limb when grafted into an innervated wound site. Enzymatic cleavage of the HS chains of this periosteum tissue by heparinase-III prior to grafting reduced the complexity of the pattern observed in the ectopic structures (McCusker et al., 2016). As a loss of HS chains did not abolish pattern formation, nor did exogenous addition recapitulate complete limb formation using the ALM system, HSPGs cannot be solely responsible for the communication of all patterning information.
Other cell-cell specific interactions, such as those facilitated by cadherins, ephrines and the like, may play a role in the communication of positional information; several of which show differential expression patterns in regenerating tissues. Relative to the stump, transcription of N-cadherin (Sader et al., 2019) and desmoglein 4 preprotein, a cadherin family member (Campbell et al., 2011), is upregulated in blastema tissue (Sader et al., 2019). The integrin proteins change dynamically as well, with a decrease in α1 and β1 integrin protein levels being coupled with an increase in α3, α6, αv and β3 integrins (Makanae and Satoh, 2012; Satoh et al., 2011; Tsonis et al., 1997). Although, there is no direct evidence to link any of these interactions with positional information, selective adhesion has been documented within regenerating tissues. Late bud stage blastema tissue, derived from different locations along the length of the axolotl forelimb, will migrate back to its relative position along the P/D axis of a regenerate if autografted into a more proximal, regenerating site within the hind limb (Crawford and Stocum, 1988). Prod-1, a CD59-like molecule that is GPI-anchored to the cell membrane of Notophtalmus viridescens, was thought to be a factor in the selective adhesion gradient in the regenerating limb (Da Silva et al., 2002). Prod-1 is differentially expressed in blastemas along the P/D limb axis, with highest expression being most proximally (Da Silva et al., 2002). Altering the expression of Prod-1 in blastema cells will result in the relocation of these cells to a new P/D location in the axolotl blastema (Echeverri and Tanaka, 2005). However, it was recently shown that Prod-1 is not anchored to the membrane of axolotl cells (Blassberg et al., 2010), which also exhibit selective adhesion properties. Thus, either multiple cell surface molecules play a role in this property, or species-specific differences are at play. Regardless, further research is required to determine if the selective adhesive properties of blastema cells could facilitate the communication of positional information.
In addition to surface molecules, other signals have been identified to convey positional information between cells within developing and regenerating systems. For example, more than 80 years ago, dramatic changes in electric potential were document within the amphibian embryo as it progressed through different developmental stages (Burr and Hovland, 1937), and it was proposed that pattern formation within living organisms is established by “electrodynamic” fields (Burr and Northrop, 1935). Although a paucity exists in our understanding of bioelectric signaling in biological systems; the regenerating tail of the axolotl demonstrates dependency on appropriate bioelectric signaling, as the use of specific ion channel antagonists inhibited regeneration of this structure (Franklin et al., 2017), and electric potential has been shown to control facial patterning (Vandenberg et al., 2011) as well as melanocyte migration, proliferation and morphology (Blackiston et al., 2011) during embryogenesis in Xenopus. In the case of the melanocytes, their biology was affected by the depolarization of cells expressing glycine receptor chloride channels, termed instructor cells. These instructor cells are scattered as small clusters throughout the embryo, and are able to affect the behavior of melanocytes at a distance (Blackiston et al., 2011). Whether instructor-like cells equate to pattern-forming cells, or even a portion of the population, is unknown, and requires further investigation.
Gap junctions, which can also modulate the electric potential of a cell by regulating ion movement, facilitate intercellular communication; and mutations in the proteins that constitute these structures impair regeneration and pattern formation. RNAi against smedinx-11, an innexin gap junction channel gene, inhibited regeneration and impaired stem cell maintenance in planarian tissue (Oviedo and Levin, 2007), while in developing and regenerating zebrafish tissue, mutations in the gap junction protein Connexin43 caused abnormal skeletal patterning (Hoptak-Solga et al., 2008; Misu et al., 2016). Intriguingly, hapln1a (hyaluronan and proteoglycan link protein 1a) is functionally downstream of Connexin43, and links hyaluronic acid with proteoglycans in the extracellular matrix (Govindan and Iovine, 2014). Therefore, gap junctions may communicate positional information by dictating the axis-specific proteoglycan profiles discussed above.
Neuronal polarity is also a possible mechanism by which positional information may be communicated during regeneration. A/P patterning in regenerating planarian tissue depends on neuronal polarity, whereby the direction in which signal molecules are transported along the neuronal axons ultimately dictates the direction of the patterning morphogen gradients in the regenerating tissue (Pietak et al., 2019). Although nerve signaling is vital for successful regeneration (Singer, 1974, 1946), it is unknown if neuronal polarity has similar effects in the regenerating amphibian limb.
5.2). Competence of patterning forming cells to interpret and respond to a disparity in positional information
Irrespective of the mechanism by which positional information is communicated, blastema cells must be competent to interpret and respond to this information. This competency is established within the injured environment through nerve derived signaling, a necessary requirement for limb regeneration (Endo et al., 2004; Singer, 1946). Makanae et al. demonstrated that a cocktail of Fgfs and Bone morphogenetic proteins (Bmps) can substitute for nerve signaling using the ALM; although, not all the combinations were equivalent (Lehrberg and Gardiner, 2015; Makanae et al., 2014). Of the eight different cocktail combinations, all demonstrated the capacity to stimulate initial blastema formation; however, only a combination of Bmp2, Fgf2 and Fgf8, or Bmp7, Fgf2 and Fgf8 made the blastema cells capable of forming new pattern when cells from the opposing side of the limb were grafted into the wound site. Therefore, a positional disparity in a wounded environment on its own is insufficient to facilitate pattern formation unless the cells are able to interpret this information and/or respond to it.
In the context of regeneration, a new limb structure requires new positional information to be generated. The blastema is constituted by cells derived from the underlying stump tissue. Therefore, the pattern forming cells that contribute to the blastemas must either assume or generate new cells with more distal identities to form the missing portion of the limb. The polar coordinate model calls the mechanism by which these new identities arise the process of intercalation (Bryant et al., 1981; French et al., 1976). Intercalation is defined as a growth response that is elicited between cells with disparaging positional information (in a regeneration permissive environment), where the newly generated cells have the identities of the intermediate, or missing, positional values. If the identity generated already exists in that particular proximal-to-distal plane, the cell assumes a more distal identity in a process referred to as distal outgrowth (Bryant et al., 1981).
Pattern forming cells must therefore exhibit positional plasticity, so to allow for the stable acquisition of new positional information. For example, when early stage blastemas, which are largely composed of dedifferentiated cells of connective tissue origin, are grafted into an innervated limb wound site they fail to elicit a patterning response and simply integrate with the surrounding tissue (McCusker and Gardiner, 2013). This suggests that this tissue is in a state of positional plasticity (McCusker and Gardiner, 2013). Nerve signaling is required to maintain plasticity in the early blastema cells because denervated early blastemas are capable of inducting limb structures in innervated wound sites (McCusker and Gardiner 2013). The highly proliferative apical end of the late stage blastema also exhibits this positional plasticity (Mccusker et al., 2015). However, the basal region of the late stage blastema, where differentiation and proliferation rates are more comparable to stump tissue, elicits a patterning response when grafted into an innervated limb wound (Mccusker et al., 2015). These basal cells therefore have stabilized positional identity sufficient to generate a positional disparity. Thus, cell proliferation and lack of differentiation are positively correlated with positional reprogramming.
Evidence supports the hypothesis that epigenetic modifications to pattering genes plays an important role in reprogramming positional information during pattern formation in the regenerate. At a transcriptional level, early bud and apical late bud blastema tissue are enriched for epigenetic modifiers including jarid2, zym2, habp4, uhrf1, lmnb2, parp1, thymopoietin, Prmt1, Smarcd1, Banf1, Set, Setd8, Hmgb3, Histone H3f3b, Smarcd1 (Baf60a), Prmt1, Smarcd4 and Smarca5 (Knapp et al., 2013; Mccusker et al., 2015; Monaghan et al., 2009). A re-expression of patterning genes has also been documented in teleost (Rabinowitz et al., 2017; Stewart et al., 2009) and Urodele blastema tissue (Carlson et al., 2001; Gardiner et al., 1995; Khan et al., 1999; Knapp et al., 2013; Torok et al., 1998). Irrespective of the location (jaw, caudal fin, tail or limb), location specific and non-specific patterning genes are re-expressed in blastema tissue, further supporting the notion that positional reprogramming occurs within this population (as reviewed by (Vieira and Mccusker, 2018)). In zebrafish, a loss in repressive epigenetic marks (H3K27me3) and gain of activating transcription marks (H3K4me3) was noted in the promoter regions for most, but not all, re-expressed genes considered (Stewart et al., 2009). Complementary to these observations, significant quantitative (Sosnik et al., 2017) and qualitative (Hay and Fischman, 1961) changes in nuclear architecture have been observed in early stage amphibian blastema tissue, relative to mature dermal tissue. The blastema cells exhibit a significant reduction in chromatin compaction. This large scale DNA remodeling event is reminiscent of somatic cell nuclear reprogramming, an inducible process where somatic cell assumes a more pluripotent state (as reviewed in (Gaspar‑Maia et al., 2011)). These previous observations were made on the blastema mesenchyme, which is composed mostly of cells of connective tissue origin (Muneoka et al., 1986). Therefore, pattern-forming cells would be expected to exhibit these properties during regeneration.
Within the limb, limb pattern forming cells with positional memory exhibit the capacity to respond to a positional disparity and generate appropriate pattern. However, cells with positional memory from different anatomical locations do not demonstrate this same capacity in the limb. Replacement of the limb dermis and epidermis with that derived from the head of the axolotl results in the inhibition of limb regeneration after amputation through the tissue graft site (Satoh et al., 2007; Tank, 1987). Additionally, head dermal cells, implanted into regenerating limb tissue, fail to participate in the regenerative response, demonstrating only a minimal capacity to contribute to a small number of connective tissue cells located around the graft itself (Satoh et al., 2007). Full thickness skin from the flank of the axolotl, which is closer in anatomical position to the limb relative to the head, however does demonstrate some capacity to participate in limb regeneration (Tank, 1987). Despite the inability of head derived cells to participate in limb regeneration, head structures, such as the jaw (as reviewed by (Vieira and Mccusker, 2018)), do exhibit regenerative capacity. Therefore, we hypothesize that the ability of mature, non-limb cells to participate in limb regeneration depends on their position in the body. When the disparity in positional information, in relation to the limb, is too large (such as from the head), it cannot be resolved, leading to a failure in intercalation and thus regeneration (Mccusker and Gardiner, 2014). Juxtaposition of limb and flank pattern-forming cells generates a disparity that can be resolved, although the pattern is frequently abnormal in some way (Tank, 1987). This failure in patterning pattern may relate to the inability of the non-limb cells to communicate their positional information and/or respond to positional cues in the limb environment. More research is required to determine the mechanisms by which positional information is communicated and interpreted by pattern forming cells and what regulates the responses of these cells when transplanted into new, distant anatomical locations.
In summary, level two of the hierarchy involves the communication of positional memory and the establishment of a positional disparity between pattern forming cells in response to amputation. Within a regeneration competent environment, established by nerve-derived signaling, pattern-forming cells participate in repeated iterations of intercalation and distal outgrowth in order to replace the missing identities. In order for new identities to be assumed, this population of cells must become positionally plastic, be re-patterned, and then stably retain the new positional information. This will ultimately lead to complete pattern formation as well as ensuring homeostasis and regenerative capabilities within the regenerate.
6). Level 3: Induction of signaling centers that confer positional cues
Once the pattern of the regenerate has been generated, this information must be communicated to pattern following cells to appropriately integrate all of the different tissue types into a functional structure. Interestingly, early staged regenerates, where pattern formation is presumably expected to occur, are mostly composed of cells of connective tissue origin. At later time points, other patterning following cell types contribute to the regenerate (Currie et al., 2016; Muneoka et al., 1986). Thus, it appears that the pattern forming cells must provide some sort of signal to recruit these pattern following cells to the blastema. In order for this to be achieved, we hypothesize that once pattern forming cells have generated the missing pattern through an intercalary response, they establish signaling centers to communicate this pattern information to the pattern-following cells.
The nature of these signaling centers would depend on the structure being regenerated. Signaling centers are well characterized during limb development (Figure 7). The apical ectodermal ridge (AER) is established at the most distal portion of the developing bud (Saunders, 1948). This specialized signaling hub drives the P/D patterning of underlying mesenchymal cells; although the precise mechanism by which this occurs is still controversial (as reviewed by (Zuniga, 2015)). Regardless, the AER acts as a signaling center for Fgf, Bmps and Wnt production; which dictates P/D patterning (Barrow et al., 2003; Lewandoski et al., 2000; Mariani et al., 2008; Mcqueeney et al., 2002; Parr et al., 1993; Pizette et al., 2001). An opposing interplay between Fgf8 and Retinoic acid (RA) also appears to contribute to P/D pattern formation. RA, which is produced in the lateral plate mesoderm by retinaldehyde dehydrogenase-2 (Mic et al., 2004), in the flank, has maximum concentration in the proximal end of the limb bud and induces the expression of Meis1 and Meis2, proximal determinate genes (Mercader et al., 2000). Moving to the distal end of the bud, RA is diminished, through the action of CYP26B1 (Yashiro et al., 2004) and Fgf represses Meis1 and Meis2 expression by inhibiting RA signaling in this region (Mercader et al., 2000).
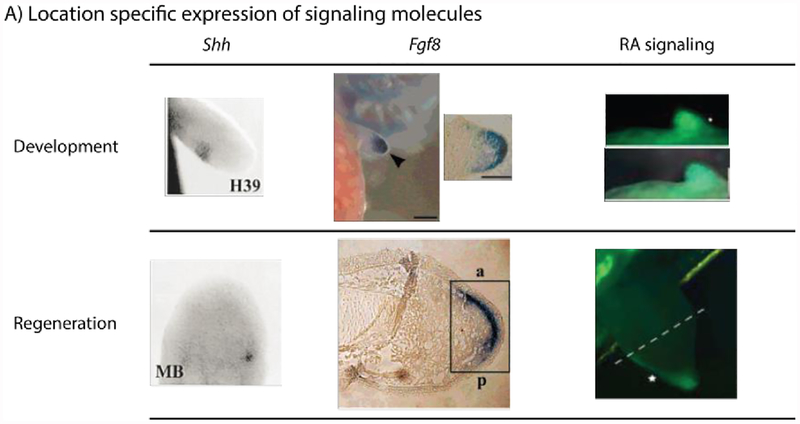
Figure 7:
Signaling molecules and their role in regeneration. Shh and Fgf8 are expressed in signaling centers in both the developing and regenerating axolotl limb. H39 – 39 days post hatching; MB – Medium-bud blastema stage; a – anterior; p – posterior. RA signaling, visualized in a RARE-GFP reporter transgenic axolotl, diminishes from the proximal to distal end of the developing limb bud of the axolotl. Unlike Shh and Fgf8, the RA signaling pattern is not recapitulated during regeneration; instead it is restricted to the posterior of the wound epithelium. (Images modified, with permission, from (Han et al., 2001; Monaghan and Maden, 2012; Torok et al., 1999).
Within the vertebrate embryo, the A/P limb axis is initially defined by mutual antagonism between two transcription factors, Hand2 and Gli3 (Galli et al., 2010; Zúñiga and Zeller, 1999). Hand2 becomes restricted to the posterior end of the bud, and, along with other transcription factors, generates a signaling center – the zone of polarizing activity (ZPA) (Galli et al., 2010). The ZPA is responsible for the production of the signaling molecule Shh, which regulates subsequent A/P pattern formation (Riddle et al., 1993). This signaling hub functions as part of a feedback loop with the AER as well to control limb outgrowth along the P/D axis (Laufer et al., 1994; Niswander et al., 1994).
The D/V axis is governed by ectodermal-derived factors. Wnt7a, which also contributes in part to A/P specification, is expressed within the dorsal ectoderm and dictates dorsal identity (Parr and Mcmahon, 1995). Ventral Bmp production induces the expression of Engrailed, which limits the expression of Wnt7a, and its downstream target Lmx1, to the dorsal side of the limb bud (Pizette et al., 2001).
Based on the importance of Shh, Fgf, and RA signaling in limb bud development, it is not surprising that these three signaling molecules participate in limb regeneration as well (Figure 7). A small zone of Shh expressing mesenchymal cells are formed on the posterior margin of the mid-bud blastema under the apical epithelial cap (AEC) in axolotl (Ambystoma mexicanum) and newt (Cynops pyrrhogaster), retained until late bud stage, and are completely lost by advanced palette stage, when patterning has been completed (Imokawa and Yoshizato, 2002; Torok et al., 1999). When anterior cells are surgically confronted with posterior cells within a regeneration permissive environment a small Shh signaling center is induced within a few days by the posterior population (Nacu et al., 2016). Therefore, a positional disparity is required for the induction of a Shh signaling center in these wounds.
Regeneration also induces the expression of Fgf8. The mRNA for this gene was detected in the basal layer of the AEC and as a thin layer in the underlying mesenchymal tissue of the mid to late bud stage blastema of the axolotl. The expression pattern demonstrated slight polarization to the anterior side of the tissue (Han et al., 2001). Thus, like Shh, Fgf8 recapitulates its developmental expression pattern.
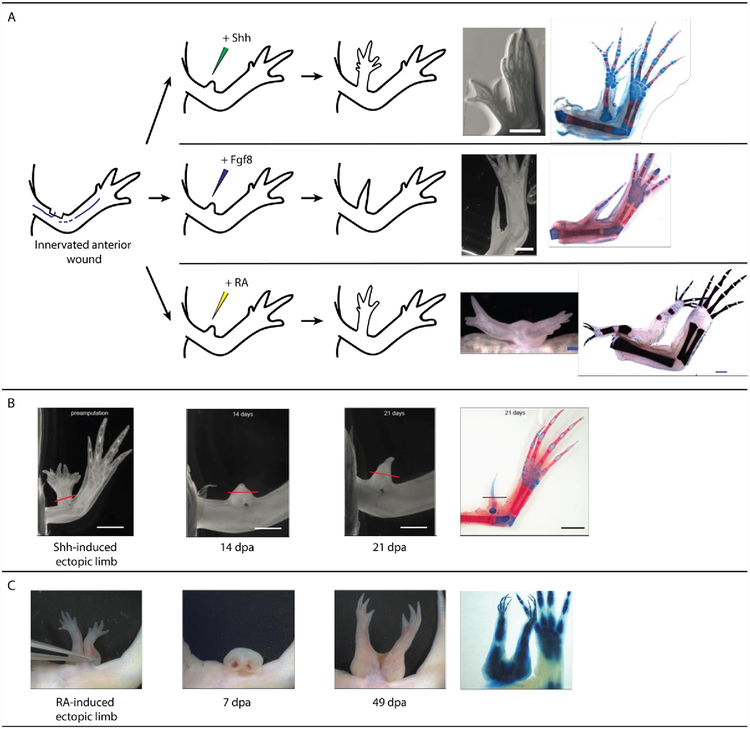
Work using the ALM regeneration assay has demonstrated the role of these two signaling molecules in pattern formation during regeneration (Nacu et al., 2016). Exogenous, overexpression of Shh and Fgf8 are sufficient to induce pattern from an ALM when acting as a surrogate for a contralateral tissue graft. Fgf8 is sufficient, in a shh dependent manner, to elicit pattern formation from both anterior and posterior innervated limb wounds (Nacu et al., 2016). Shh is only capable of inducing ectopic pattern from anterior innervated wound sites and this induction is Fgf8 dependent (Nacu et al., 2016). However, the pattern produced by both of these signaling molecules is abnormal – P/D structures are missing with incomplete, symmetrical A/P patterning (Nacu et al., 2016) (Figure 8). Additionally, although the application of these signaling molecules can induce ectopic pattern from an innervated wound site, the positional memory in these cells remains unchanged (has not been reprogramed). Upon amputation these structures fail to regenerate, suggesting an absence of positional disparity in these signaling molecule-induced regenerates (Figure 8) (Nacu et al., 2016). Furthermore, full thickness skin from these anterior located ectopic limbs can induce new pattern only when grafted into posterior, but not anterior, innervated wounds. Thus they retain only anterior positional information (Nacu et al., 2016). Therefore, despite their ability to facilitate pattern formation in a regeneration permissive environment, these signaling molecules are unable to establish (via intercalation) new positional identities.
Figure 8:
Signaling molecules and pattern formation during regeneration. A) A non-regenerative blastema, formed at an anterior innervated wound site, can generate pattern if treated with Shh, Fgf8 or RA. The RA-, but not Shh- and Fgf8-, induced structures have the capacity to be correctly patterned along the P/D and A/P limb axes. (B) Shh-induced ectopic limbs fail to regenerate in response to amputation, indicating a lack of positional diversity in these structures. (C) RA induced structures demonstrate regenerative capacity, indicating the establishment and stabilization of new positional identities within the RA-induced outgrowths. (Images modified, with permission, from (McCusker et al., 2014; Nacu et al., 2016; Vieira et al., 2019))
In contrast to Fgf and Shh, exogenous RA appears to reprogram the positional memory of blastema cells. RA, an active metabolite of vitamin A, has profound effects within developmental (as reviewed by (Cunningham and Duester, 2015; Rhinn and Dolle, 2012)) and regenerating systems (as reviewed by (Maden and Hind, 2003)). Within the developing embryo, RA is synthesized in the trunk and enters the limb bud. A gradient is established through AER-Fgf directed inactivation of this morphogen at the distal end of the bud (Mercader et al., 2000). Despite controversy related to the relevance and mechanism of action of endogenous RA in the patterning of developing limb buds (as reviewed by (Cunningham and Duester, 2015)), it is clear that exogenous RA disrupts P/D (Kochhar, 1973) and A/P (Tickle et al., 1982) pattern in this context.
RA appears to be present within the blastema as well. With the use of high performance liquid chromatography (HPLC), the posterior of the blastema was found to have a five-fold higher level of RA compared to the anterior (Scadding and Maden, 1994). Using an axolotl retinoic acid response element green fluorescent protein (RARE-GFP) reporter line, RA signaling appears localized distally in the apical epithelial cap of the early blastema, expands into the posterior region of the structure as regeneration progresses, and then disappears by the early stages of differentiation. Although, very little signal was detected in the blastema mesenchyme during normal regeneration; RA treatment induced reporter signaling within this population. This suggested that the mesenchymal cells are responsive to RA signaling (Monaghan and Maden, 2012). As the wound epithelium was not removed by Scadding and Maden (1994) during their analysis, the RA detected may have been derived from the AER and not the mesenchyme of the blastema itself during their analysis of regenerating amphibian limb tissue. HPLC analysis also revealed a higher level of RA in proximal blastema tissue relative to distal counterparts, suggesting RA levels within the blastema are dictated by P/D location (Scadding and Maden, 1994). Therefore, RA may play a role, like in the developing limb, in pattern formation during regeneration.
In axolotl, newt, and frog, exogenous treatment with vitamin A metabolites, RA being the best characterized in this context, affects the pattern of the regenerate in a reproducible manner. It is proposed that RA reprograms the positional information of pattern forming cells to a posterior-ventral-proximal limb identity (Bryant and Gardiner, 1992). The following observations support this hypothesis.
Firstly, after amputation, RA treatment induces the formation of a regenerate that has pattern more proximal than the amputation site (Kim and Stocum, 1986; Maden, 1983, 1982; Niazi and Saxena, 1978; Thoms and Stocum, 1984). This P/D duplication is concentration dependent, with the higher the RA concentration the more proximal the pattern formed (Kim and Stocum, 1986; Maden, 1983, 1982). The interpretation of these observations is that RA specifies P/D positional information within the regenerating limb (Maden, 1983).
Secondly, tissue grafting (Crawford and Stocum, 1988; Ludolph et al., 1990) and ALM surgeries (McCusker et al., 2014) have shown that RA also re-specifies positional information along the A/P and D/V axes of the regenerating limb, allowing blastema cells to assume more posterior and ventral identities. In the study by McCusker et al. 2014, RA was able to induce ectopic limb formation from anterior and dorsal innervated wounds – both having the capacity to generate complete, correctly patterned limbs. Ventral and posterior wound sites very rarely gave pattern, and if they did it was minimal, when RA treatment was used in place of a contralateral skin graft (McCusker et al., 2014). Additionally, RA, in combination with Fgf2, Fgf8 and Bmp-2, which supplement for nerve signaling, can elicit a regenerative response from a simple anterior lateral wound. These growth factor engineered limbs also demonstrate complete limb patterning (Vieira et al., 2019). A combination of Fgf2, Fgf8 and Bmp-2 fail to induce pattern from anterior lateral wounds in the absence of RA or a skin graft (Makanae et al., 2014; Vieira et al., 2019), therefore RA is sufficient to reprogram positional information in the regeneration permissive environment such that a positional disparity is generated. All of the above mentioned RA-related observations are in contrast to the incompletely patterned limbs induced in the ALM by Fgf8 and Shh (McCusker et al., 2014; Nacu et al., 2016) (Figure 8).
Third, transcriptional analysis of blastema tissue treated with RA, reveals that RA changes the positional identity of these cells on a molecular level. Gene microarray analysis of RA-treated blastema tissue derived from distal limb amputation sites reveals increased expression of proximal patterning genes (such as Meis1 and Meis2) and a decrease in distal genes (such as HoxA13 and Msx2). This suggests a proximalization of the treated tissue (Nguyen et al., 2017). Molecular analysis of anterior and posterior located ALM blastemas also indicates that RA reprograms A/P information. For example, untreated, anterior located ALM blastema tissue abundantly expresses the anterior marker Alx4, while expressing little of the posterior marker Shh. RA treatment results in the reversal of these expression patterns such that they mimic the patterns in blastema tissue from the posterior side of the limb (Vieira et al., 2019). Interestingly, it appears that mature limb tissue is refractory to being positionally reprogrammed by RA because the expression of A/P markers is unaffected in these tissues following treatment (Vieira et al., 2019).
Last, we have recently shown that the RA-induced ectopic limb structures generated in anterior located ALMs are capable of regenerating (Vieira et al., 2019). Additionally, full thickness skin from these RA-induced ectopic structures induce the formation of new ectopic structures when grafted into anterior innervated wound sites. These data indicate that RA treatment has permanently altered the positional memory in the ectopic limb structures and that these structures have both anterior and posterior positional information (Vieira et al., 2019).
In summary, since RA both reprograms the positional memory in regenerating limb cells and is upstream of the expression of Shh, we hypothesize that this signaling pathway plays a role on the second level of the hierarchy. Alternatively, Shh and Fgf signaling provide positional cues that pattern forming cells respond to, yet do not alter the positional memory in the regenerating cells. Thus, these latter signaling molecules are likely part of the third level of the patterning hierarchy (Figure 2.5).
7). Level 4: Cellular response by pattern following cells to positional cues
Limbs, and perhaps all organs, are constituted by both pattern forming and following cells (Mccusker and Gardiner, 2014; Rinn et al., 2008; Vieira and McCusker, 2018). During regeneration, a variety of different underlying stump tissues contribute towards the blastema, including pattern following lineages such as nerve, epidermis, and muscle cells (Hay and Fischman, 1961; Kragl et al., 2009; Tank, 1987). As we have explained previously, different cell types contribute to the blastema mesenchyme at different stages of regeneration. The pattern forming cells invade the blastema first and after pattern formation has commenced the pattern following cells immigrate. Thus, the last level of the patterning hierarchy is constituted by the response of pattern following cells to the positional cues generated by signaling centers (level three), which results in the arrangement and specialization of these cell types in a positionally appropriate manner.
Pattern forming cells, such as fibroblasts, have been demonstrated to communicate with pattern following cells, such as keratinocytes, though specific signaling molecules. These interactions regulate cellular activities such as proliferation and differentiation (Szabowski et al., 2000), and position specific specialization (Rinn et al., 2008; Yamaguchi et al., 2004). For example, plantar fibroblasts produce Wnt5a, a distal morphogen, which induces the expression of epidermal keratin 9 (K9), a palmoplantar differentiation marker, in co-cultured keratinocytes (Rinn et al 2008). This Wnt5a production is location specific and controlled by HoxA13, which demarcates the fibroblast population as distal. Silencing of HoxA13 by siRNA abrogates Wnt5a expression, and subsequently epidermal K9 expression (Rinn et al., 2008), indicating the importance of the positional HOX code in the ultimate specification of patterning output in pattern following cells. Palmoplantar fibroblasts are not only able to support the specialization of cells, but also suppress it. This cell population is able to reduce the growth and pigmentation of melanocytes, relative to non-palmoplantar fibroblasts, through the expression of Dickkopf 1 (DDK1), an inhibitor of canonical Wnt signaling. This location specific production of DDK1 allows for pigmentation differences between palmoplantar and non-palmoplantar regions of the body (Yamaguchi et al., 2004). In addition to Wnt5a and DDK1, Rinn et al (2006) identified a variety of other signaling molecules, including Wnts and Bmps that are produced by adult human fibroblasts in a location specific manner. It is likely that these position specific signals, in conjunction with those that are communicated from signaling centers (ZPA and AEC) are also playing an essential role in the position-appropriate behavior in the pattern-following cells during limb regeneration.
Despite the large paucity in understanding that exists regarding the precise mechanism by which position specific information is communicated, evidence indicates that it is a crucial step for final patterning output during regeneration. Further investigations are required to determine the nature and mechanisms of such signaling molecules in the context of limb regeneration.
8). Application of the pattern hierarchy model to mammalian system
The model described within this review can be used as means to assess regenerative capacity in other systems as well. For example, unlike the axolotl, mammals demonstrate poor regenerative abilities and, therefore, they must be deficient in one or more levels of this patterning hierarchy. However, mammalian system exhibit many of the biological codes described in the model. For example, the Hox (Rinn et al., 2008, 2007, 2006) and HSPG codes (Allen and Rapraeger, 2003; Ledin et al., 2004; Nogami et al., 2004) have been identified in mammalian systems; and communication between pattern forming and following cells has been documented and found to contribute to homeostasis (Rinn et al., 2008). Furthermore, transplantation of de-cellularized, posterior, murine ECM into an axolotl anterior ALM was sufficient to generate simple pattern, suggesting that there was sufficient information within the graft to communicate and establish a positional disparity (Phan et al., 2015). How mammalian cells would respond in the axolotl ALM is unknown as differences in thermal tolerances between the cells of these two species inhibit cross-species tissue grafting. To date, the ability of mammalian cells to assume new positional information is not well characterized, but seems to vary depending on the anatomical location of the cell population considered. HoxA11 expression is maintained in the murine tibia, specifically in periosteum and osteocytes, after development. In response to local injury and repair the expression of this gene is retained in the tissue (Leucht et al., 2008). Mature tibia periosteum also retains this expression pattern when implanted into a distinct, HoxA11-negative location, the mandible; however, the grafted tissue fails to integrate with the mandibular environment, instead forming a cartilaginous callus (Leucht et al., 2008). Surprisingly, HoxA11-negative mandible-derived periosteum can be successfully implanted into the Hox-positive tibia, where the grafted cells assume HoxA11 expression and integrating with the surrounding tissue. Although the hypothesis is limited by the consideration of only a single positional gene, the absence of positional plasticity appears to impede healing. Further investigations are required to determine the validity of this hypothesis; and, if true, why some mammalian cell populations exhibit positional plasticity while others do not. Never-the-less, based on all of these observations and the patterning hierarchy model, it is likely that regenerative failure in mammalian systems is due to the absence of a regeneration permissive environment and/or the inability of pattern forming cells to respond appropriately to a positional disparity. Research is therefore required to address these ideas.
9). Conclusion
In this review we have presented a new model of pattern formation during limb regeneration that we have called the Patterning Hierarchy Model. Throughout this review we have discussed the role of different biological codes that contribute to each level of the hierarchy. At the top of the hierarchy are the pattern forming cells with different positional identities. The Hox code appears to play an essential role in specifying these identities, and the epigenetic code ensures the retention of this information as positional memory. At the second level of the hierarchy are the molecular mechanisms by which these pattern forming cells communicate their positional identity to other pattern forming cells. We have discussed evidence that supports the idea that HSPGs, and thus the HSPG code, are among the cell surface molecules that communicates this information. Additionally, since positional plasticity appears to play a role in the establishment of new positional information in these cells, it is likely that the epigenetic code also contributes to this step. The third level of the hierarchy is the induction of signaling centers by the pattern forming cells, and the forth level is the cellular response to these signals by the pattern following cells. While the presence of biological codes is not yet evident at these last levels of the hierarchy, there is still a lot that can be learned about the molecular mechanisms at play. However, it is clear that pattern formation in the regenerating limb requires the integration of multiple biological codes.
Although more research is required to expand and more clearly characterize our model at a molecular level, the model provides a framework in which this can be achieved. Furthermore it provides a means to consider the regenerative potential of non-regenerative systems and to investigate where deficiencies may potentially lie and possibly be mitigated. Previously, opposing schools of thought have suggested that either morphogen gradients or local cell interactions alone are sufficient to dictate pattern. Although all previous models have provided crucial insight into the mechanisms of pattern formation during regeneration, they are limited. We believe that the model presented here, assembled from scientific observations and the valuable insight of the previously described patterning models, provides a mechanistic patterning code.
Funding
This work was supported by the National Institutes of Health [grant number 1R15HD092180–01A1].
Abbreviations
- A/P
anterior-posterior
- AEC
Apical epithelial cap
- AER
apical epithelial ridge
- ALM
accessory limb model
- BMP
Bone morphogenetic proteins
- D/V
dorsal-ventral
- Dpp
Decapentaplegic
- ECM
extracellular matrix
- Fgf
fibroblastic growth factor
- H3K37me3
histone 3 Lysine 27 trimethylation
- H3K4me3
Histone 3 lysine 4 trimethylation
- HPLC
high performance liquid chromatography
- HS
Heparan sulfate
- HSPG
heparin sulfate proteoglycan
- HSST
heparan sulfate sulfotransferase
- ISH
in situ hybridization
- K9
Keratin 9; P/D - proximal/distal
- RA
Retinoic acid
- Shh
Sonic Hedgehog
- ZPA
zone of polarizing activity
10) References
- Allen BL, Rapraeger AC, 2003. Spatial and temporal expression of heparan sulfate in mouse development regulates FGF and FGF receptor assembly. J. Cell Biol 163, 637–648. 10.1083/jcb.200307053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri M, 2019. A general model on the origin of biological codes. BioSystems 181, 11–19. 10.1016/j.biosystems.2019.04.010 [DOI] [PubMed] [Google Scholar]
- Barbieri M, 2018. What is code biology? BioSystems 164, 1–10. 10.1016/j.biosystems.2017.10.005 [DOI] [PubMed] [Google Scholar]
- Barbieri M, 2008. Biosemiotics: A new understanding of life. Naturwissenschaften 95, 577–599. 10.1007/s00114-008-0368-x [DOI] [PubMed] [Google Scholar]
- Barrow JR, Thomas KR, Boussadia-zahui O, Moore R, Kemler R, Capecchi MR, Mcmahon AP, 2003. Ectodermal Wnt3/B-catenin signaling is required for the establishment and maintenance of the apical ectodermal ridge. Genes Dev 17, 394–409. 10.1101/gad.1044903.ing [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedzhov I, Graham SJL, Leung CY, Zernicka-Goetz M, 2014. Developmental plasticity, cell fate specification and morphogenesis in the early mouse embryo. Philos. Trans. R. Soc. B Biol. Sci 369, 20130538 10.1098/rstb.2013.0538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackiston D, Adams DS, Lemire JM, Lobikin M, Levin M, 2011. Transmembrane potential of GlyCl-expressing instructor cells induces a neoplastic-like conversion of melanocytes via a serotonergic pathway. Dis. Model. Mech 4, 67–85. 10.1242/dmm.005561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blassberg RA, Garza-Garcia A, Janmohamed A, Gates PB, Brockes JP, 2010. Functional convergence of signalling by GPI-anchored and anchorless forms of a salamander protein implicated in limb regeneration. J. Cell Sci 124, 47–56. 10.1242/jcs.076331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant SV, French V, Bryant PJ, 1981. Distal regeneration and symmetry. Science (80-. ) 212, 993–1002. 10.1126/science.212.4498.993 [DOI] [PubMed] [Google Scholar]
- Bryant SV, Gardiner DM, 1992. Retinoic acid, local cell-cell interactions, and pattern formation in vertebrate limbs. Dev. Biol 152, 1–25. 10.1016/0012-1606(92)90152-7 [DOI] [PubMed] [Google Scholar]
- Bryant SV, Iten LE, 1976. Supernumerary limbs in amphibians: Experimental production in Notophthalmus viridescens and a new interpretation of their formation. Dev. Biol 50, 212–234. 10.1016/0012-1606(76)90079-8 [DOI] [PubMed] [Google Scholar]
- Bülow HE, Hobert O, 2004. Differential sulfations and epimerization define heparan sulfate specificity in nervous system development. Neuron 41, 723–736. [DOI] [PubMed] [Google Scholar]
- Burr HS, Hovland CI, 1937. Bio-Electric Correlates of Development in Amblystoma. Yale J. Biol. Med 9, 540–9. [PMC free article] [PubMed] [Google Scholar]
- Burr HS, Northrop FSC, 1935. The Electro-Dynamic Theory of Life. Quaterly Rev. Biol 10, 322–333. [Google Scholar]
- Campbell LJ, Suárez-castillo EC, Ortiz-zuazaga H, Knapp D, Tanaka EM, Crews CM, 2011. Gene Expression Profile of the Regeneration Epithelium during Axolotl Limb Regeneration. Dev. Dyn 240, 1826–1840. 10.1002/dvdy.22669.Gene [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson MRJ, Komine Y, Bryant SV, Gardiner DM, 2001. Expression of Hoxb13 and Hoxc10 in Developing and Regenerating Axolotl Limbs and Tails. Dev. Biol 229, 396–406. 10.1006/dbio.2000.0104 [DOI] [PubMed] [Google Scholar]
- Chou J, Ferris AC, Chen T, Seok R, Yoon D, Suzuki Y, 2019. Roles of Polycomb group proteins Enhancer of zeste (E(z)) and Polycomb (Pc) during metamorphosis and larval leg regeneration in the flour beetle Tribolium castaneum. Dev. Biol in press 10.1016/j.ydbio.2019.03.002 [DOI] [PubMed] [Google Scholar]
- Crawford K, Stocum DL, 1988. Retinoic acid coordinately proximalizes regenerate pattern and blastema differential affinity in axolotl limbs. Development 102, 687–98. [DOI] [PubMed] [Google Scholar]
- Crick FH., Barnett L, Brenner S, Watts-Tobin RJ, 1961. General nature of the genetic code for proteins. Nature 192, 1227–1231. [DOI] [PubMed] [Google Scholar]
- Cunningham TJ, Duester G, 2015. Mechanisms of retinoic acid signalling and its roles in organ and limb development. Nat. Rev. Mol. Cell Biol 16, 110–123. 10.1038/nrm3932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie JD, Kawaguchi A, Traspas RM, Schuez M, Chara O, Tanaka EM, 2016. Live Imaging of Axolotl Digit Regeneration Reveals Spatiotemporal Choreography of Diverse Article Live Imaging of Axolotl Digit Regeneration Reveals Spatiotemporal Choreography of Diverse Connective Tissue Progenitor Pools. Dev. Cell 39, 411–423. 10.1016/j.devcel.2016.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Silva SM, Gates PB, Brockes JP, 2002. The newt ortholog of CD59 is implicated in proximodistal identity during amphibian limb regeneration. Dev. Cell 3, 547–555. 10.1016/S1534-5807(02)00288-5 [DOI] [PubMed] [Google Scholar]
- Driever W, Nüsslein-Volhard C, 1988. The bicoid protein determines position in the Drosophila embryo in a concentration-dependent manner. Cell 54, 95–104. 10.1016/0092-8674(88)90183-3 [DOI] [PubMed] [Google Scholar]
- Echeverri K, Tanaka EM, 2005. Proximodistal patterning during limb regeneration. Dev. Biol 279, 391–401. 10.1016/j.ydbio.2004.12.029 [DOI] [PubMed] [Google Scholar]
- Elinson RP, 1975. Site of sperm entry and a cortical contraction associated with egg activation in the frog Rana pipens. Dev. Biol 47, 257–268. [DOI] [PubMed] [Google Scholar]
- Endo T, Bryant SV, Gardiner DM, 2004. A stepwise model system for limb regeneration. Dev. Biol 270, 135–145. 10.1016/j.ydbio.2004.02.016 [DOI] [PubMed] [Google Scholar]
- Franklin BM, Voss SR, Osborn JL, 2017. Ion channel signaling influences cellular proliferation and phagocyte activity during axolotl tail regeneration. Mech. Dev 146, 42–54. 10.1016/j.mod.2017.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French V, Bryant PJ, Bryant SV, 1976. Pattern Regulation in Epimorphic Fields. Science (80-. ) 193, 969–981. [DOI] [PubMed] [Google Scholar]
- Galli A, Robay D, Osterwalder M, Bao X, Be J, Tariq M, Paro R, Mackem S, Zeller R, 2010. Distinct Roles of Hand2 in Initiating Polarity and Posterior Shh Expression during the Onset of Mouse Limb Bud Development. PLoS Genet 6, e1000901 10.1371/journal.pgen.1000901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner DM, Blumberg B, Komine Y, Bryant SV, 1995. Regulation of HoxA expression in developing and regenerating axolotl limbs., in: Development pp. 1731–41. 10.1006/dbio.1998.8956 [DOI] [PubMed] [Google Scholar]
- Gardiner DM, Bryant SV, 1989. Organization of Positional Information in the Axolotl Limb. J. Exp. Zool 251, 47–55. [DOI] [PubMed] [Google Scholar]
- Gaspar‑Maia A, Alajem A, Meshorer E, Santos MR, 2011. Open chromatin in pluripotency and reprogramming. Nat. Publ. Gr 12, 36–47. 10.1038/nrm3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girton JR, 1981. Pattern Triplications Produced by a Cell-Lethal Mutation in Drosophila. Dev. Biol 84, 164–172. [DOI] [PubMed] [Google Scholar]
- Girton JR, Russell MA, 1980. A clonal analysis of pattern duplication in a temperature-sensitive cell- lethal mutant ofDrosophila melanogaster. Dev. Biol 77, 1–21. [DOI] [PubMed] [Google Scholar]
- Govindan J, Iovine MK, 2014. Hapln1a Is Required for Connexin43-Dependent Growth and Patterning in the Regenerating Fin Skeleton. PLoS One 9, e88574 10.1371/journal.pone.0088574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada Y, Bando T, Nakamura T, Ishimaru Y, Mito T, Noji S, Tomioka K, Ohuchi H, 2015. Leg regeneration is epigenetically regulated by histone H3K27 methylation in the cricket Gryllus bimaculatus. Development 142, 2916–2927. 10.1242/dev.122598 [DOI] [PubMed] [Google Scholar]
- Han MJ, An JY, Kim WS, 2001. Expression patterns of Fgf-8 during development and limb regeneration of the axolotl. Dev. Dyn 220, 40–48. [DOI] [PubMed] [Google Scholar]
- Hay ED, Fischman DA, 1961. Origin of the blastema in regenerating limbs of the newt Triturus viridescens: An autoradiographic study using tritiated thymidine to follow cell proliferation and migration. Dev. Biol 3, 26–59. [DOI] [PubMed] [Google Scholar]
- Holder N, Tank PW, Bryant SV, 1980. Regeneration of Symmetrical Forelimbs in Axolotl, Ambystoma mexicanum. Dev. Biol 74, 302–314. [DOI] [PubMed] [Google Scholar]
- Holland PWH, 2013. Evolution of homeobox genes. Wiley Interdiscip. Rev. Dev. Biol 2, 31–45. 10.1002/wdev.78 [DOI] [PubMed] [Google Scholar]
- Holt CE, Dickson BJ, 2005. Sugar codes for axons? Neuron 46, 169–172. 10.1016/j.neuron.2005.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoptak-Solga AD, Nielsen S, Jain I, Thummel R, Hyde DR, Iovine MK, 2008. Connexin43 (GJA1) is required in the population of dividing cells during fin regeneration. Dev. Biol 317, 541–548. 10.1016/j.ydbio.2008.02.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imokawa Y, Yoshizato K, 2002. Expression of Sonic hedgehog gene in regenerating newt limb blastemas recapitulates that in developing limb buds. Proc. Natl. Acad. Sci 94, 9159–9164. 10.1073/pnas.94.17.9159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iten LE, Bryant SV, 1975. The interaction between the blastema and stump in the establishment of the anterior-posterior and proximal-distal organization of the limb regenerate. Dev. Biol 44, 119–147. 10.1016/0012-1606(75)90381-4 [DOI] [PubMed] [Google Scholar]
- Kaufman TC, Lewis R, Wakimoto B, 1980. Cytogenetic Analysis of Chromosome 3 in DROSOPHILA MELANOGASTER: The Homoeotic Gene Complex in Polytene Chromosome Interval 84a-B. Genetics 94, 115–33. 10.1021/acsaem.8b01430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan PA, Tsilfidis C, Liversage RA, 1999. Hox C6 expression during development and regeneration of forelimbs in larval Notophthalmus viridescens. Dev. Genes Evol 209, 323–329. [DOI] [PubMed] [Google Scholar]
- Kim WS, Stocum DL, 1986. Effects of retinoids on regenerating limbs: comparison of retinoic acid and arotinoid at different amputation levels. Roux’s Arch. Dev. Biol 195, 455–463. 10.1007/BF00375749 [DOI] [PubMed] [Google Scholar]
- Kmita M, Duboule D, 2003. Organizing axes in time and space; 25 years of colinear tinkering. Science (80-. ) 301, 331–333. 10.1126/science.1085753 [DOI] [PubMed] [Google Scholar]
- Knapp D, Schulz H, Rascon CA, Volkmer M, Scholz J, Nacu E, Le M, Novozhilov S, Tazaki A, Protze S, Jacob T, Hubner N, Habermann B, Tanaka EM, 2013. Comparative Transcriptional Profiling of the Axolotl Limb Identifies a Tripartite Regeneration-Specific Gene Program. PLoS Genet 8, e61352 10.1371/journal.pone.0061352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochhar DM, 1973. Limb development in mouse embryos. I. Analysis of teratogenic effects of retinoic acid. Exp. Teratol 7, 289–298. [DOI] [PubMed] [Google Scholar]
- Kragl M, Knapp D, Nacu E, Khattak S, Maden M, Epperlein HH, Tanaka EM, 2009. Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature 460, 60–65. 10.1038/nature08152 [DOI] [PubMed] [Google Scholar]
- Kreuger J, Kjellén L, 2012. Heparan Sulfate Biosynthesis: Regulation and Variability. J. Histochem. Cytochem 60, 898–907. 10.1369/0022155412464972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, Chen S, Iwase S, Alpatov R, Issaeva I, Canaani E, Roberts TM, Chang HY, Shi Y, 2007. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature 449, 689–694. 10.1038/nature06192 [DOI] [PubMed] [Google Scholar]
- Laufer E, Nelson CE, Johnson RL, Morgan BA, Tabin C, 1994. Sonic hedgehog and Fgf-4 act through a signaling cascade and feedback loop to integrate growth and patterning of the developing limb bud. Cell 79, 993–1003. 10.1016/0092-8674(94)90030-2 [DOI] [PubMed] [Google Scholar]
- Ledin J, Staatz W, Li J-P, Götte M, Selleck S, Kjellén L, Spillmann D, 2004. Heparan Sulfate Structure in Mice with Genetically Modified Heparan Sulfate Production. J. Biol. Chem 279, 42732–42741. 10.1074/jbc.m405382200 [DOI] [PubMed] [Google Scholar]
- Lehrberg J, Gardiner DM, 2015. Regulation of axolotl (Ambystoma mexicanum) limb blastema cell proliferation by nerves and BMP2 in organotypic slice culture. PLoS One 10, e0123186 10.1371/journal.pone.0123186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leucht P, Kim J-B, Amasha R, James AW, Girod S, Helms JA, 2008. Embryonic origin and Hox status determine progenitor cell fate during adult bone regeneration. Development 135, 2845–2854. 10.1242/dev.023788 [DOI] [PubMed] [Google Scholar]
- Lewandoski M, Sun X, Martin GR, 2000. Fgf8 signalling from the AER is essential for normal limb development. Nat. Genet 26, 460–463. [DOI] [PubMed] [Google Scholar]
- Lewis EB, 1978. A gene complex controlling segmentation in Drosophila. Nature 276, 565–570. 10.1038/276565a0 [DOI] [PubMed] [Google Scholar]
- Lewis R, Wakimoto B, Denell RE, Kaufman TC, 1980. Genetic analysis of the Antennapedia gene complex (Ant-C) and adjacent chromosomal regions of Drosophila melanogaster. II. Polytene chromosome segments 84A- 84B1,2. Genetics 95, 383–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, 2004. Functions of heparan sulfate proteoglycans in cell signaling during development. Development 131, 6009–6021. 10.1242/dev.01522 [DOI] [PubMed] [Google Scholar]
- Ludolph D, Cameron JA, Stocum DL, 1990. The effect of retinoic acid on positional memory in the dorsoventral axis of regenerating axolotl limbs. Dev. Biol 140, 41–52. [DOI] [PubMed] [Google Scholar]
- Maden M, 1983. The effect of vitamin A on the regenerating axolotl limb. J. Embryol. Exp. Morphol 77, 273–95. [PubMed] [Google Scholar]
- Maden M, 1982. The effect of vitamin A on limb regeneration in Rana temporaria. Dev. Biol 98, 409–416. [DOI] [PubMed] [Google Scholar]
- Maden M, Hind M, 2003. Retinoic acid, a regeneration-inducing molecule. Dev. Dyn 226, 237–244. 10.1002/dvdy.10222 [DOI] [PubMed] [Google Scholar]
- Maden M, Turner RN, 1978. Supernumerary limbs in the axolotl. Nature 273, 232–235. [DOI] [PubMed] [Google Scholar]
- Makanae A, Mitogawa K, Satoh A, 2014. Co-operative Bmp- and Fgf-signaling inputs convert skin wound healing to limb formation in urodele amphibians. Dev. Biol 396, 57–66. 10.1016/j.ydbio.2014.09.021 [DOI] [PubMed] [Google Scholar]
- Makanae AKI, Satoh A, 2012. Early Regulation of Axolotl Limb Regeneration. Anat. Rec 295, 1566–1574. 10.1002/ar.22529 [DOI] [PubMed] [Google Scholar]
- Mallo M, Alonso CR, 2013. The regulation of Hox gene expression during animal development. Development 140, 3951–3963. 10.1242/dev.068346 [DOI] [PubMed] [Google Scholar]
- Mariani FV, Ahn CP, Martin GR, 2008. Genetic evidence that FGFs have an instructive role in limb proximal – distal patterning. Nature 453, 401–406. 10.1038/nature06876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCusker C, Lehrberg J, Gardiner D, 2014. Position-specific induction of ectopic limbs in non-regenerating blastemas on axolotl forelimbs. Regeneration 1, 27–34. 10.1002/reg2.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mccusker CD, Athippozhy A, Diaz-castillo C, Fowlkes C, Gardiner DM, Voss SR, 2015. Positional plasticity in regenerating Amybstoma mexicanum limbs is associated with cell proliferation and pathways of cellular differentiation. BMC Dev. Biol 1–17. 10.1186/s12861-015-0095-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCusker CD, Diaz-Castillo C, Sosnik J, Q. Phan A,,Gardiner DM, 2016. Cartilage and bone cells do not participate in skeletal regeneration in Ambystoma mexicanum limbs. Dev. Biol 416, 26–33. 10.1016/j.ydbio.2016.05.032 [DOI] [PubMed] [Google Scholar]
- Mccusker CD, Gardiner DM, 2014. Understanding positional cues in salamander limb regeneration : implications for optimizing cell-based regenerative therapies. Co. Biol 7, 593–599. 10.1242/dmm.013359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCusker CD, Gardiner DM, 2013. Positional Information Is Reprogrammed in Blastema Cells of the Regenerating Limb of the Axolotl (Ambystoma mexicanum). PLoS One 8, 1–14. 10.1371/journal.pone.0077064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcqueeney K, Soufer R, Dealy CN, 2002. B-Catenin-dependent Wnt signaling in apical ectodermal riMcqueeney K, Soufer R and Dealy CN (2002) ‘B-Catenin-dependent Wnt signaling in apical ectodermal ridge induction and FGF8 expression in normal and limbless mutant chick limbs’, Develop. Gro. Dev. Growth Differ 44, 315–325. [DOI] [PubMed] [Google Scholar]
- Meinhardt H, 1983a. Cell Detemination Boundries as Organizing Regions for Secondary Embryonic Fields. Dev. Genes Evol 96, 375–385. [DOI] [PubMed] [Google Scholar]
- Meinhardt H, 1983b. A bootstrap model for the proximodistal pattern formation in vertebrate limbs. J. Embryol. exp. Morph 76, 139–146. [PubMed] [Google Scholar]
- Meinhardt H, 1983c. A boundary model for pattern formation in vertebrate limbs. J. Embryol. exp. Morph 76, 115–137. [PubMed] [Google Scholar]
- Meinhardt H, 1982. The role of compartmentalization in the activation of particular control genes and in the generation of proximo-distal positional information in appendages. Am. Zool 22, 209–220. 10.1093/icb/22.1.209 [DOI] [Google Scholar]
- Mercader N, Leonardo E, Piedra ME, Martínez-A C, Ros MA, Torres M, 2000. Opposing RA and FGF signals control proximodistal vertebrate limb development through regulation of Meis genes. Development 127, 3961–70. [DOI] [PubMed] [Google Scholar]
- Mic FA, Sirbu IO, Duester G, 2004. Retinoic Acid Synthesis Controlled by Raldh2 Is Required Early for Limb Bud Initiation and Then Later as a Proximodistal Signal during Apical Ectodermal Ridge Formation *. J. Bioloical Chem 279, 26698–26706. 10.1074/jbc.M401920200 [DOI] [PubMed] [Google Scholar]
- Misu A, Yamanaka H, Aramaki T, Kondo S, Skerrett IM, Iovine MK, Watanabe M, 2016. Two different functions of Connexin43 confer two different bone phenotypes in zebrafish. J. Biol. Chem 291, 12601–12611. 10.1074/jbc.M116.720110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan JR, Epp LG, Putta S, Page RB, Walker JA, Beachy CK, Zhu W, Pao GM, Verma IM, Hunter T, Bryant SV, Gardiner DM, Harkins TT, Voss SR, 2009. Microarray and cDNA sequence analysis of transcription during nerve-dependent limb regeneration. BMC Biol 7, 10.1186/1741-7007-7-1. 10.1186/1741-7007-7-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan JR, Maden M, 2012. Visualization of retinoic acid signaling in transgenic axolotls during limb development and regeneration. Dev. Biol 368, 63–75. 10.1016/j.ydbio.2012.05.015.Visualization [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muneoka K, Fox WF, Bryant SV, 1986. Cellular Contribution from Dermis and Cartilage to the Regenerating Limb Blastema in Axolotls. Dev. Biol 116, 256–260. [DOI] [PubMed] [Google Scholar]
- Nacu E, Gromberg E, Oliveira CR, Drechsel D, Tanaka EM, 2016. FGF8 and SHH substitute for anterior-posterior tissue interactions to induce limb regeneration. Nature 533, 407–410. 10.1038/nature17972 [DOI] [PubMed] [Google Scholar]
- Nguyen M, Singhal P, Piet JW, Shefelbine SJ, Maden M, Voss SR, Monaghan JR, 2017. Retinoic acid receptor regulation of epimorphic and homeostatic regeneration in the axolotl. Development 144, 601–611. 10.1242/dev.139873 [DOI] [PubMed] [Google Scholar]
- Niazi IA, Saxena S, 1978. Abnormal hind limb regeneration in tadpoles of the toad, Bufo andersoni, exposed to excess vitamin A. Folia Biol 26, 3–8. [PubMed] [Google Scholar]
- Niswander L, Jeffrey S, Martin GR, Tickie C, 1994. A positive feedback loop coordinates the patterning and limb development. Nature 371, 609–612. [DOI] [PubMed] [Google Scholar]
- Nogami K, Suzuki H, Habuchi H, Ishiguro N, Iwata H, Kimata K, 2004. Distinctive Expression Patterns of Heparan Sulfate O-Sulfotransferases and Regional Differences in Heparan Sulfate Structure in Chick Limb Bluds. J. Biol. Chem 279, 8219–8229. 10.1074/jbc.M307304200 [DOI] [PubMed] [Google Scholar]
- Oviedo NJ, Levin M, 2007. smedinx-11 is a planarian stem cell gap junction gene required for regeneration and homeostasis. Development 134, 3121–3131. 10.1242/dev.006635 [DOI] [PubMed] [Google Scholar]
- Parr BA, Mcmahon AP, 1995. Dorsalizing signal Wnt-7a required for normal polarity of D-V and A-P axes of mouse limb. Nature 374, 350–353. [DOI] [PubMed] [Google Scholar]
- Parr BA, Shea MJ, Vassileva G, Mcmahon AP, 1993. Mouse Wnt genes exhibit discrete domains of expression in the early embryonic CNS and limb buds. Development 119, 247–261. [DOI] [PubMed] [Google Scholar]
- Phan AQ, Lee J, Oei M, Flath C, Hwe C, Mariano R, Vu T, Shu C, Dinh A, Simkin J, Muneoka K, Bryant SV, Gardiner DM, 2015. Positional information in axolotl and mouse limb extracellular matrix is mediated via heparan sulfate and fibroblast growth factor during limb regeneration in the axolotl (Ambystoma mexicanum). Regeneration 2, 182–201. 10.1002/reg2.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce SB, Kimelman D, 1995. Regulation of Spemann organizer formation by the intracellular kinase Xgsk-3. Development 121, 755–765. [DOI] [PubMed] [Google Scholar]
- Pietak A, Bischof J, Lapalme J, Morokuma J, Levin M, 2019. Neural control of body-plan axis in regenerating planaria. PLoS Comput. Biol 15, e1006904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizette S, Abate-shen C, Niswander L, 2001. BMP controls proximodistal outgrowth, via induction of the apical ectodermal ridge, and dorsoventral patterning in the vertebrate limb. Development 128, 4463–4474. [DOI] [PubMed] [Google Scholar]
- Poulain FE, Yost HJ, 2015. Heparan sulfate proteoglycans: a sugar code for vertebrate development? Development 142, 3456–3467. 10.1242/dev.098178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz JS, Robitaille AM, Wang Y, Ray CA, Thummel R, Gu H, Djukovic D, Raftery D, Berndt JD, Moon RT, 2017. Transcriptomic, proteomic, and metabolomic landscape of positional memory in the caudal fin of zebrafish. PNAS 114, E717–E726. 10.1073/pnas.1620755114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhinn M, Dolle P, 2012. Retinoic acid signalling during development. Development 139, 843–858. 10.1242/dev.065938 [DOI] [PubMed] [Google Scholar]
- Riddle RD, Johnson RL, Tabin C, Laufer E, 1993. Sonic hedgehog Mediates the Polarizing Activity of the ZPA. Cell 75, 1401–1416. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Bondre C, Gladstone HB, Brown PO, Chang HY, 2006. Anatomic demarcation by positional variation in fibroblast gene expression programs. PLoS Genet 2, 1084–1096. 10.1371/journal.pgen.0020119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY, 2007. Functional Demarcation of Active and Silent Chromatin Domains in Human HOX Loci by Noncoding RNAs. Cell 129, 1311–1323. 10.1016/j.cell.2007.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Wang JK, Allen N, Brugmann SA, Mikels AJ, Liu H, Ridky TW, Stadler HS, Nusse R, Helms JA, Chang HY, 2008. A dermal HOX transcriptional program regulates site-specific epidermal fate. Genes Dev 22, 303–307. 10.1101/gad.1610508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roensch K, Tazaki A, Chara O, Tanaka EM, 2013. Progressive Specification Rather than Intercalation of Segments During Limb Regeneration. Scienc 342, 225–240. 10.1016/b978-0-408-01434-2.50020-6 [DOI] [PubMed] [Google Scholar]
- Rothbart SB, Strahl BD, 2014. Interpreting the language of histone and DNA modifications. Biochim. Biophys. Acta - Gene Regul. Mech 1839, 627–643. 10.1016/j.bbagrm.2014.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Hsiung F, Kornberg TB, 2011. Specificity of Drosophila Cytonemes. Science (80-. ) 332, 354–358. 10.1126/science.1198949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Huang H, Liu S, Kornberg TB, 2014. Cytoneme-mediated contact-dependent transport of the Drosophila decapentaplegic signaling protein. Science (80-. ) 343, 1244624 10.1126/science.1244624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell MA, Girton JR, 1981. An analysis of compartmentalization in pattern duplications induced by a cell-lethal mutation in Drosophila. Dev. Biol 85, 55–64. [DOI] [PubMed] [Google Scholar]
- Sader F, Denis J, Laref H, Roy S, 2019. Epithelial to mesenchymal transition is mediated by both TGF- β canonical and non-canonical signaling during axolotl limb regeneration. Sci. Rep 9, 1144 10.1038/s41598-018-38171-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Kornberg TB, 2002. FGF is an essential mitogen and chemoattractant for the air sacs of the Drosophila tracheal system. Dev. Cell 3, 195–207. 10.1016/S1534-5807(02)00202-2 [DOI] [PubMed] [Google Scholar]
- Satoh A, Gardiner DM, Bryant SV, Endo T, 2007. Nerve-induced ectopic limb blastemas in the axolotl are equivalent to amputation-induced blastemas. Dev. Biol 312, 231–244. 10.1016/j.ydbio.2007.09.021 [DOI] [PubMed] [Google Scholar]
- Satoh A, Hirata A, Satou Y, 2011. Blastema induction in aneurogenic state and Prrx-1 regulation by MMPs and FGFs in Ambystoma mexicanum limb regeneration. Dev. Biol 355, 263–274. 10.1016/j.ydbio.2011.04.017 [DOI] [PubMed] [Google Scholar]
- Saunders JW, 1948. The proximo-distal sequence of origin of the parts of the chicken wing and the role of the ectoderm. J. Exp. Zool 108, 363–403. [DOI] [PubMed] [Google Scholar]
- Scadding SR, Maden M, 1994. Retinoic Acid Gradients during Limb Regeneration. Dev. Biol 162, 608–617. [DOI] [PubMed] [Google Scholar]
- Schneidera S, Steinbeissera H, Wargab RM, Hausen P, 1996. B-catenin translocation into nuclei demarcates the dorsalizing centers in frog and fish embryos. Mech. Dev 57, 191–198. [DOI] [PubMed] [Google Scholar]
- Singer M, 1974. Trophic functions of the neuron. VI. Other trophic systems. Neurotrophic control of limb regeneration in the newt. Ann. N. Y. Acad. Sci 228, 308–322. [DOI] [PubMed] [Google Scholar]
- Singer M, 1946. The nervous system and regeneration of the forelimb of adult Triturus. V. The influence of number of nerve fibers, including a quantitative study of limb innervation. J. Exp. Zool 101, 299–377. [DOI] [PubMed] [Google Scholar]
- Slack JM, 1991. The nature of the mesoderm-inducing signal in Xenopus: a transfilter induction study. Development 113, 661–669. [DOI] [PubMed] [Google Scholar]
- Soshnikova N, Duboule D, 2009. Epigenetic Temporal Control of Mouse Hox Genes in Vivo. Science (80-. ) 324, 1320–1323. [DOI] [PubMed] [Google Scholar]
- Sosnik J, Vieira WA, Webster KA, Siegfried KR, McCusker CD, 2017. A new and improved algorithm for the quantification of chromatin condensation from microscopic data shows decreased chromatin condensation in regenerating axolotl limb cells. PLoS One 12, e0185292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart S, Tsun Z, Carlos J, Belmonte I, 2009. A histone demethylase is necessary for regeneration in zebrafish. PNAS 106, 19889–19894 DEVELOPMENTAL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD, 2000. The language of covalent histone modications. Nature 403, 41–45. 10.1038/47412 [DOI] [PubMed] [Google Scholar]
- Szabowski A, Maas-Szabowski N, Andrecht S, Kolbus A, Schorpp-Kistner M, Fusenig NE, Angel P, 2000. c-Jun and JunB antagonistically control cytokine-regulated mesenchymal-epidermal interaction in skin. Cell 103, 745–755. 10.1016/S0092-8674(00)00178-1 [DOI] [PubMed] [Google Scholar]
- Tank PW, 1987. The Effect of Nonlimb Tissues on Forelimb Regeneration in the Axolotl, Ambystoma mexicanum. J. Exp. Zool 244, 409–423. [Google Scholar]
- Thoms SD, Stocum DL, 1984. Retinoic acid-induced pattern duplication in regenerating urodele limbs. Dev. Biol 10.1016/0012-1606(84)90320-8 [DOI] [PubMed] [Google Scholar]
- Tickle C, Alberts B, Wolpert L, Lee J, 1982. Local application of retinoic acid to the limb bond mimics the action of the polarizing region. Nature 296, 564–566. 10.1038/296564a0 [DOI] [PubMed] [Google Scholar]
- Tickle C, Summerbell D, Wolpert L, 1975. Positional signalling and specification of digits in chick limb morphogenesis. Nature 254, 199–202. [DOI] [PubMed] [Google Scholar]
- Torok MA, Gardiner DM, Izpisúa-Belmonte J-C, Bryant SV, 1999. Sonic Hedgehog (SHH) expression in developing and regenerating axolotl limbs. J. Exp. Zool 284, 197–206. [DOI] [PubMed] [Google Scholar]
- Torok MA, Gardiner DM, Shubin NH, Bryant SV, 1998. Expression of HoxD Genes in Developing and Regenerating Axolotl Limbs. Dev. Biol 233, 225–233. [DOI] [PubMed] [Google Scholar]
- Tsonis PA, Doane K, Del Rio-Tsonis K, 1997. Expression of integrins during axolotl limb regeneration. Dev. Growth Differ 39, 9–14. [DOI] [PubMed] [Google Scholar]
- Vandenberg LN, Morrie RD, Adams DS, 2011. V-ATPase-dependent ectodermal voltage and Ph regionalization are required for craniofacial morphogenesis. Dev. Dyn 240, 1889–1904. 10.1002/dvdy.22685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira WA, Mccusker CD, 2018. Regenerative Models for the Integration and Regeneration of Head Skeletal Tissues. Int. J. Mol. Sci 19, E3752 10.3390/ijms19123752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira WA, McCusker CD, 2018. Developmental plasticity and tissue integration, in: Gardiner DM (Ed.), Regenerative Engineering and Developmental Biology CRC Press, Boca Raton, pp. 411–429. [Google Scholar]
- Vieira WA, Wells KM, Raymond MJ, Souza L De, Garcia E, Mccusker CD, 2019. FGF, BMP, and RA signaling are sufficient for the induction of complete limb regeneration from non-regenerating wounds on Ambystoma mexicanum limbs. Dev. Biol 451, 146–157. 10.1016/j.ydbio.2019.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent JP, Oster GF, Gerhart JC, 1986. Kinematics of gray crescent formation in Xenopus eggs: The displacement of subcortical cytoplasm relative to the egg surface. Dev. Biol 113, 484–500. 10.1016/0012-1606(86)90184-3 [DOI] [PubMed] [Google Scholar]
- Wolpert L, 1996. One hundred years of positional information. Trends Genet. TIG 12, 359–364. 10.1017/S0025727300065728 [DOI] [PubMed] [Google Scholar]
- Wolpert L, 1969. Positional Information and the Spatial Pattern of Cellular Differentiationt 25, 1–47. [DOI] [PubMed] [Google Scholar]
- Yamada S, Sugahara K, Özbek S, 2011. Evolution of glycosaminoglycans. Commun. Integr. Biol 4, 150–158. 10.4161/cib.4.2.14547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Itami S, Watabe H, Yasumoto KI, Abdel-Malek ZA, Kubo T, Rouzaud F, Tanemura A, Yoshikawa K, Hearing VJ, 2004. Mesenchymal-epithelial interactions in the skin: Increased expression of dickkopf1 by palmoplantar fibroblasts inhibits melanocyte growth and differentiation. J. Cell Biol 165, 275–285. 10.1083/jcb.200311122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yashiro K, Zhao X, Uehara M, Yamashita K, Nishijima M, Nishino J, Saijoh Y, Sakai Y, Hamada H, 2004. Regulation of Retinoic Acid Distribution Is Required for Proximodistal Patterning and Outgrowth of the Developing Mouse Limb. Dev. Cell 6, 411–422. [DOI] [PubMed] [Google Scholar]
- Zuniga A, 2015. Next generation limb development and evolution : old questions, new perspectives. Development 142, 3810–3820. 10.1242/dev.125757 [DOI] [PubMed] [Google Scholar]
- Zúñiga A, Zeller R, 1999. Gli3 (Xt) and formin (ld) participate in the positioning of the polarising region and control of posterior limb-bud identity. Development 126, 13–21. [DOI] [PubMed] [Google Scholar]