Abstract
Background
Treatment for Cutaneous T Cell Lymphoma (CTCL) is generally not curative. Therefore, selecting therapy that is effective and tolerable is critical to clinical decision-making. Histone deacetylase inhibitors (HDACi), epigenetic modifier drugs, are commonly used but effective in only ~30% of patients. There are no predictive markers of HDACi response and the CTCL histone acetylation landscape remains unmapped. We sought to identify pre-treatment molecular markers of resistance in CTCL that progressed on HDACi therapy.
Methods
Purified T cells from 39 pre/post-treatment peripheral blood samples and skin biopsies from 20 patients were subjected to RNA-seq and ChIP-seq for histone acetylation marks (H3K14/9 ac, H3K27ac). We correlated significant differences in histone acetylation with gene expression in HDACi-resistant/sensitive CTCL. We extended these findings in additional CTCL patient cohorts (RNA-seq, microarray) and using ELISA in matched CTCL patient plasma.
Findings
Resistant CTCL exhibited high levels of histone acetylation, which correlated with increased expression of 338 genes (FDR < 0·05), including some novel to CTCL: BIRC5 (anti-apoptotic); RRM2 (cell cycle); TXNDC5, GSTM1 (redox); and CXCR4, LAIR2 (cell adhesion/migration). Several of these, including LAIR2, were elevated pre-treatment in HDACi-resistant CTCL. In CTCL patient plasma (n = 6), LAIR2 protein was also elevated (p < 0·01) compared to controls.
Interpretation
This study is the first to connect genome-wide differences in chromatin acetylation and gene expression to HDACi-resistance in primary CTCL. Our results identify novel markers with high pre-treatment expression, such as LAIR2, as potential prognostic and/or predictors of HDACi-resistance in CTCL.
Funding
NIH:CA156690, CA188286; NCATS: WU-ICTS UL1 TR000448; Siteman Cancer Center: CA091842.
Keywords: Lymphoma, Epigenetics, Therapeutic resistance, Predictive biomarker
Research in context.
Evidence before this study
Cutaneous T Cell Lymphomas are a heterogeneous group of which Mycosis Fungoides (MF) and Sèzary Syndrome (SS) are the most common subtypes; 5-year survival for advanced MF/SS is only 20%. Treatment for MF/SS is generally not curative; thus, selecting a therapy that is both effective and tolerable is critical to clinical decision-making. Histone deacetylase inhibitors (HDACi) have been used to treat CTCL for >10 years, yet epigenome-wide histone acetylation, the direct target of these drugs, has not previously been mapped. Essentially all previously reported mechanisms of HDACi resistance were defined in cell lines, not in primary patient samples. Moreover, only ~30% of patients respond to HDACi, and we lack predictive markers for response.
Added value of this study
Our study is the first to map epigenome-wide differences in chromatin acetylation (ChIP-seq) and link them to altered gene expression (RNA-seq) in HDACi-resistant CTCL (39 samples, 20 patients). We identified novel mechanisms of HDACi resistance in MF/SS patient samples and validated these findings in additional CTCL patient cohorts (RNA-seq, microarray). Elevated expression of several genes, including BIRC5, NRP2, and LAIR2, was detectable prior to HDACi therapy, suggesting their utility as predictive markers. Of these, LAIR2 was the most robust and is readily detected in patient plasma using routine clinical laboratory methods (ELISA).
Implications of all the available evidence
Our study demonstrates that elevated levels of histone acetylation and novel cell adhesion/migration pathways are likely mechanisms of HDACi resistance in CTCL patients. While further studies are necessary, LAIR2 and other adhesion proteins may promote inflammation and migration of malignant and benign immune cells in the CTCL microenvironment. The predictive markers we identified represent novel therapeutic targets and could inform therapeutic decision-making in CTCL.
Alt-text: Unlabelled Box
1. Introduction
Cutaneous T cell lymphomas (CTCLs) are a heterogeneous group of non-Hodgkin lymphomas thought to derive from mature CD4+ skin-homing T lymphocytes [1]. Mycosis Fungoides (MF) and its leukemic variant, Sèzary Syndrome (SS), are the most common subtypes of CTCL. Patients with MF/SS have a highly variable clinical course. While some with early stage disease do not progress beyond limited skin disease and do not have significant morbidity or mortality from MF/SS, others develop advanced stage disease characterized by more extensive skin involvement, skin tumors, and lymph node and peripheral blood involvement. Those with advanced stage MF/SS have a 5-year overall survival of only 20% [2,3]. Treatment for MF/SS is generally not curative, except for allogeneic stem cell transplant, which is a last resort. Therefore, selecting a therapy that is both effective and tolerable is a critical part of clinical decision making for treatment of MF/SS. Biologic agents, immunomodulators, targeted therapies, histone deacetylase inhibitors, and chemotherapy all have efficacy in some patients. However, except for CD30-targeted drugs, predictive biomarkers to guide choice of therapy do not exist for MF/SS [4].
Romidepsin and vorinostat, histone deacetylase inhibitors (HDACi), are two of the drugs FDA-approved for advanced MF/SS, underscoring the significance of histone modification in MF/SS pathogenesis and treatment. HDACs remove acetyl groups from lysine residues on histone proteins, causing condensation and decreased accessibility for transcription factors and other transcriptional machinery. HDACs also remove acetyl groups from non-histone proteins, including transcription factors. Thus, by broadly regulating transcription, HDACs modulate a wide range of cellular processes, including cell cycle and apoptosis, cell growth and survival, DNA repair, development and differentiation, each of which has been implicated in MF/SS pathogenesis when dysregulated [[5], [6], [7], [8]]. HDACi therefore may be effective in treating MF/SS because they disrupt many key cellular processes and may impact pathogenic pathways. However, the exact mechanism(s) of HDACi response and resistance remains largely unknown [6].
In the studies that led to the approval of romidepsin in MF/SS, the overall response rate on average was 34% with a median duration of response of 13·7–15 months [9,10]. Nevertheless, clinical, biologic, or molecular markers that predict sensitivity or resistance to HDACi therapy in MF/SS have not been defined [1,6]. Increases in bulk histone acetylation (measured by Western blot) have been reported with HDACi treatment of cell lines, primary normal cells, and primary MF/SS samples [11]. More recently, HDACi response in MF/SS was linked to global increases in accessible DNA and to enrichment in several transcription factor motifs as mapped by ATAC-seq. However, the differential ATAC-seq profiles could not be directly linked to changes in the gene expression because expression profiling was not performed in the same patient samples [12]. In other studies, genes and pathways have been associated with resistance to HDACi treatment, including ATP-binding cassette transporter, pro-apoptotic (Fas, Caspase), anti-apoptotic (BCL), JAK-STAT, MAPK/PI3K, NFKB, Redox, and TNF pathways [13,14]. However, the majority of these studies were performed in cell lines and it is not clear that the same processes drive resistance in patients with MF/SS.
To address these outstanding questions, we performed chromatin immunoprecipitation and sequencing (ChIP-seq) and transcriptome sequencing (RNA-seq) on purified malignant T cells from skin biopsies and peripheral blood from patients treated with HDACi. Our studies revealed significant differences in the histone acetylation of gene regulatory elements in HDACi-resistant versus -sensitive samples and we linked these to significant expression changes in apoptosis, cell cycle, cytokine/chemokine signaling, and cell migration pathways. We identified a number of genes not previously associated with MF/SS or with HDACi-resistance. Notably, some of these changes are detectable prior to HDACi therapy. One of these novel HDACi-resistance genes, LAIR2, encodes a secreted collagen receptor protein that is also significantly elevated in the plasma of patients with HDACi-resistant MF/SS. In summary, we report the first epigenome-wide map of chromatin acetylation in primary MF/SS and link significant differences in acetylation to gene expression in HDACi-resistant versus -sensitive samples. Our findings identify previously unrecognized mechanisms of HDACi resistance and define novel predictive markers as potential targets for therapeutic development.
2. Materials and methods
2.1. Sample collection
De-identified peripheral blood draws or skin punch biopsies were obtained from patients seen at the Washington University School of Medicine Cutaneous Lymphoma Clinic under IRB-approved protocols with patients providing informed consent. Peripheral blood was also drawn from healthy volunteers at WUSM with IRB approval.
2.2. PBMC isolation from primary skin samples
PBMCs were isolated from skin punches through mechanical separation and repeated flushing of the tissue with sort buffer (PBS, 1% FBS, 2 mM EDTA), followed by red blood cell lysis (155 mM NH4Cl, 10 mM KHCO3, 0.1 mM EDTA) for 10 min.
2.3. T cell isolation from peripheral blood samples
Primary peripheral blood samples were incubated with RosetteSep Human Monocyte (CD36) Depletion Cocktail for 15 min at room temperature, layered onto a Histopaque-1077 gradient, and centrifuged at 400g for 30 min with no brake. The interphase was collected and washed with 10 mL of sort buffer (PBS, 1% FBS, 2 mM EDTA), followed by red blood cell lysis (155 mM NH4Cl, 10 mM KHCO3, 0·1 mM EDTA) for 10 min. Cells were washed and resuspended in sort buffer. Malignant cell populations were isolated with the EasySep Human CD4 Positive Selection Kit according to the manufacturer's instructions or through FACS on a Sony iCyt Synergy SY3200 after staining with CD4 (Miltenyi Biotec Cat# 130–092-373, RRID:AB_871684), CD7 (Miltenyi Biotec Cat# 130–105-842, RRID:AB_2659107), and/or CD26 (Miltenyi Biotec Cat# 130–093-441, RRID:AB_1103210) fluorophore conjugated antibodies.
2.4. Cell culture
HH (ATCC Cat# CRL-2105, RRID:CVCL_1280), HUT78 (ATCC Cat# CRM-TIB-161, RRID:CVCL_0337), and Jurkat (ATCC Cat# TIB-152, RRID:CVCL_0367) cell lines were acquired from ATCC. HH and Jurkat cells were cultured in RPMI (Gibco) media supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. HUT78 cells were cultured in IMDM (Gibco) media supplemented with 20% fetal bovine serum and 1% penicillin-streptomycin. Primary T cells were cultured in IMDM media supplemented with 20% fetal bovine serum and 1% penicillin-streptomycin. All cells were grown at 37 °C with 5% CO2. All cell lines were collected for RNA and gDNA extraction within 10 passages after receipt from ATCC.
2.5. In vitro Romidepsin treatments
Cells cultured as above were treated with 2·5 nM Romidepsin for 4 h, media was replaced, and cells were collected 20 h later for RNA-seq and ChIP-seq.
2.6. Luciferase reporter assays
Putative regulatory regions were PCR amplified from HH, HUT78, or Jurkat cell line gDNA with Accuprime Pfx DNA polymerase (Invitrogen) using primers with a NheI or XhoI cut site added to their 5′ end (Table S1). PCR samples were gel-extracted (Qiagen cat. 28,706), digested with NheI and XhoI (New England Biolabs), gel-purified again, and ligated into the Promega pGL4·23 plasmid overnight at 23 °C with T4 DNA ligase (New England BioLabs, cat. M0202S). Sanger sequencing confirmed successful cloning. 1 μg of each luciferase plasmid and 15 ng of Promega's pNL1·1·TK vector (#N1501) were co-nucleofected into 1 × 106 HUT78 cells with an Amaxa Nucleofector 2b system using the X-001 program and a homemade nucleofection buffer (SM1) as described [15]. Cells were incubated for 24 h in IMDM (Gibco) supplemented with 20% FBS and 1% penicillin-streptomycin, centrifuged at 200g for 10 min, and resuspended in 200 μL media. 60 μL were added to each well of a 96 well flat-bottom, white, opaque plate. The Promega Nano-Glo system (#N1110) was used to read the firefly and renilla luciferase for each well according to the manufacturer's instructions in a BioTek Cytation5 platereader. All experiments were read in triplicate and performed at least twice. The average ratios between the firefly and renilla luciferase readings for each sample were compared to the average ratio for the empty pGL3-promoter vector to determine relative luciferase.
2.7. ChIP-seq
0.5–1.0 × 105 cells were snap-frozen for 15 min on dry ice and stored at −80 °C until use. Ultra-low-input chromatin immunoprecipitation for H3K9/K14 ac (EMD-Millipore 06–599) and H3K27ac (Abcam ab4729) was performed as described [16]. DNA was sequenced by the Washington University Genome Technology Access Center on an Illumina Hi-Seq 3500 to generate 50 bp single-end reads. Reads were aligned to hg19 with bowtie2 (v2·2·5) with default settings [17]. Reads in ENCODE blacklisted regions were removed with samtools (v1·3) [18,19]. Peaks were called with MACS2 (v2·1·0·20150420) with the settings –q 0·01 –m 10 50 ‐‐nomodel ‐‐shiftsize = 150 and input controls [20]. RPKM normalized genome browser tracks were created with deepTools' (v3·1·0) bamCoverage utility with settings ‐‐binSize 10 ‐‐extendReads 150 ‐‐normalizeUsing RPKM and visualized on the UCSC genome browser [21,22]. ChIPQC (v1·14·0) was used for quality control [23], and samples with fewer than 30% (H3ac) or 25% (H3K27ac) reads in peaks were removed from subsequent analyses. The DiffBind R package (v2·8·0) was used to derive consensus peak sets and determine differentially bound peaks between sample groups [24]. The chipSeeker R package (v1·16·1) was used to annotate peaks [25].
2.8. RNA-seq
0·5-2 × 106 cells from each sample were stored in 1 mL TRIzol reagent (Invitrogen cat. 15,596,026). RNA was isolated by the Washington University Tissue Procurement Center and sequenced by the Washington University Genome Technology Access Center on an Illumina Hi-Seq 3500 to generate 50 bp single-end reads. Reads were aligned to hg19 with UCSC annotations using STAR (v2·5·3a) [26]. RPKM normalized genome browser tracks were created with deepTools' (v3·1·0) bamCoverage utility and visualized on the UCSC genome browser. Read quantification was performed by Salmon (v0·11·0) using UCSC hg19 knownGene annotations [27], and differential gene expression analyses were done with the DESeq2 R package (v1·20·0) [28]. Genotify (v1·2·1) was used for manual gene curation [29]. All data analysis was done in SoS Notebook environments [30]. All gene ontology and pathway enrichments were performed on the Enrichr web server [31].
2.9. Quantitative real-time PCR
RNA was isolated from 0·5-2 × 106 cells stored in 1 mL TRIzol reagent following reagent instructions. DNA was removed with the TURBO DNA-free kit (Invitrogen cat. AM1907) and cDNA synthesized from 1 μg of RNA with a High-Capacity cDNA Reverse Transcription kit (Applied Biosystems cat. 4368814). PCR reactions were performed with SsoAdvanced Universal SYBR Green Supermix (Bio-Rad cat. 1725271) in a Bio-Rad CFX96 Connect. Target primers are available in Table S1. Relative expression to healthy CD4+ cells was calculated via the ΔΔCT method [32].
2.10. LAIR2 ELISA
Plasma was collected from primary MF/SS and healthy control peripheral whole blood by centrifuging for 15 min at 1500g within 30 min of collection in EDTA-coated tubes. Plasma was stored at −80 °C until use. The LAIR2 sandwich ELISA (LifeSpan Biosciences cat. LS-F6502) was performed according to manufacturer instructions with plasma samples diluted 1:2. All samples were read in duplicate, and a third-order polynomial regression line was fit to the standard curve to calculate concentration.
2.11. Statistical analyses
All statistical tests were performed with GraphPad Prism (v8·1·1).
2.12. Data accession
Sequencing data was deposited in GEO: GSE132053. External datasets used include GEO: GSE59307, GSE9479, and GSE113113.
3. Results
To define epigenetic changes that are associated with response or resistance to HDACi, we first sought to map the epigenome-wide histone acetylation landscape of primary patient CTCL samples. Patients being treated at the Cutaneous Lymphoma Clinic at the Washington University School of Medicine were consented for tissue banking. Patient demographics and clinical data are summarized in Table 1 and individual patient data is included in Table S2. For this study, 20 patients had specimens collected and subjected to epigenome and transcriptome analyses. Comparisons of HDACi sensitivity or resistance were performed for 17 MF/SS patients treated with romidepsin (n = 14) or vorinostat (n = 3). Skin biopsies and peripheral blood were collected at timepoints prior to the start of HDACi therapy, during the course of therapy, and/or at subsequent follow-up visits. For romidepsin patients, “pre” specimens were collected just prior to first infusion and “post” specimens were collected at the end of the infusion, one week later (prior to the second infusion), or at subsequent visits (Fig. 1A and Table S3). Thirty-nine specimens defined this “discovery set” for epigenome and/or transcriptome studies. We assigned these specimens to HDACi-sensitive or -resistant groups based on the patients' clinical responses to HDACi therapy, determined by blinded review of the medical record. Sensitive: specimens from patients who experienced partial or complete responses or stable disease with HDACi therapy; Resistant: specimens from patients who experienced progressive disease during treatment with HDACi (Tables 1 and Table S2, Table S3). Assignment to these groups was based on HDACi response during the course of therapy in which the specimen was collected. We also collected peripheral blood from healthy volunteers and purified control CD4+ T cells. From patient specimens, we purified malignant MF/SS cells (CD4+/CD7− or CD4+/CD26−, see Methods). Purity of malignant T cells from Resistant and Sensitive samples was similar (92.5% and >99%, respectively); median purity for all samples was >90%.
Table 1.
Summary of patient clinical characteristics.
| Characteristics | WUSM cohort (N = 20) |
|---|---|
| Age at diagnosis, median (range) | 65 (27–84) |
| Sex: F, M % | 40%, 60% |
| Ethnicity | |
| Caucasian % | 75% |
| African-American % | 25% |
| Deceased | 30% |
| Initial Stage - range | IA - IVA2 |
| Worst Stage - range | IIA - IVA2 |
| IIA | 5% |
| IIB | 20% |
| IIIA | 5% |
| IIIB | 10% |
| IVA1 | 40% |
| IVA2 | 15% |
| N/A | 5% |
| Treated with HDACi | N = 17 |
| Romidepsin | 82% |
| Vorinostat | 18% |
| HDACi sensitive/resistant | |
| Sensitive | 24% |
| Resistant | 76% |
| Previous therapy lines, n (%) | |
| 0 | 24% |
| 1 | 18% |
| 2 | 24% |
| ≥ 3 | 35% |
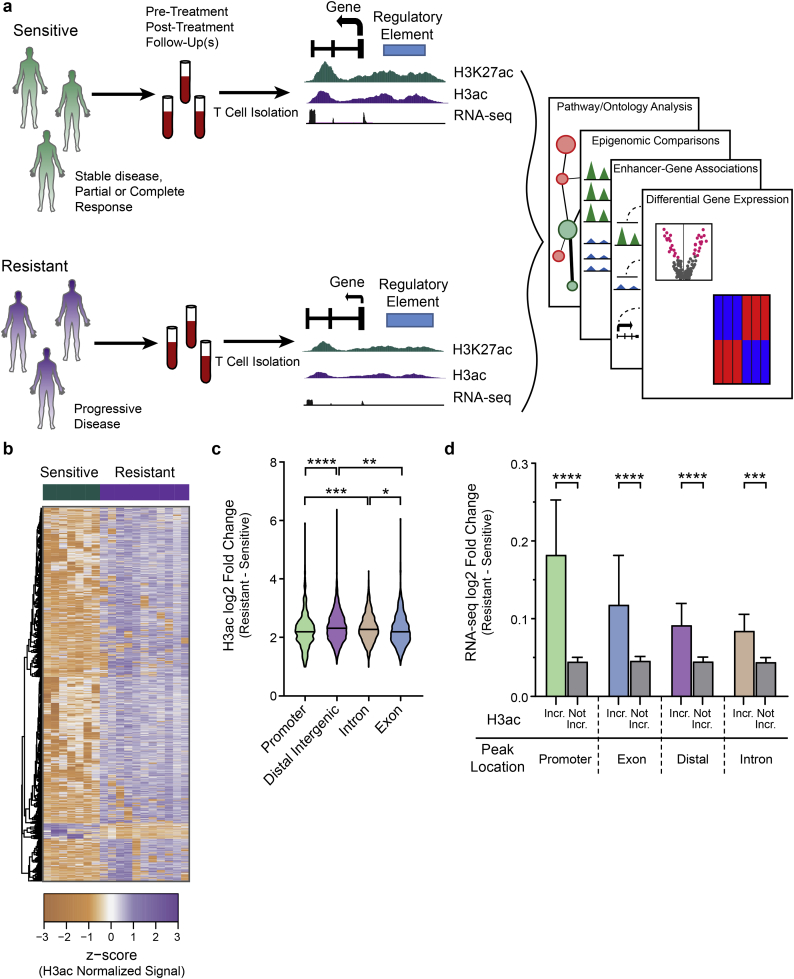
Fig. 1.
Epigenome-wide profiling identifies hyperacetylation of genes and regulatory elements in CTCL that responded or progressed on HDACi-therapy. a) Diagram of study design. b) Heatmap of differentially bound (FDR ≤ 0·05) H3ac peaks (n = 4868) Sensitive and Resistant groups. c) Violin plot showing differences in log2 fold changes for H3ac peaks enriched in Resistant samples categorized by functional genomic region (Kruskal-Wallis test; *, P ≤ 0·05; **, P ≤ 0·01; ***, P ≤ 0·001; ****, P ≤ 0·0001). d) Bar plot comparing RNA-seq log2 fold changes of genes associated with H3ac peaks enriched in Resistant samples (Incr.) to genes not associated with enriched peaks (Not Incr.). H3ac peaks are categorized by functional genomic region. (Mann-Whitney test; ***, P ≤ 0·001; ****, P ≤ 0·0001). Mean with 95% confidence interval shown.
HDAC inhibitors prevent the deacetylation of chromatin histone proteins, resulting in increased levels of acetylation. Histone acetylation is most often an activating modification that leads to increased expression of nearby genes. Two of the most prominent of these are H3K9/14 ac (H3ac), which mark active enhancers and promoters, and H3K27ac, which marks active enhancers. Because cellular yields were limiting in primary MF/SS samples, we optimized a method for performing histone acetylation profiling (H3K27ac, H3K9/14 ac) from small numbers of cells: ultra-low input chromatin immunoprecipitation and sequencing (ULI-ChIP-seq) [33]. For comparison, we treated healthy control CD4+ T lymphocytes with romidepsin using a dose and timing based on existing literature and to mimic, to the extent possible, the exposure of primary MF/SS tumors treated in vivo [34]. RNA-sequencing of the same samples was performed in parallel.
3.1. Altered acetylation corresponds to dysregulated gene expression in HDACi resistant MF/SS
To define epigenetic and linked gene expression changes associated with HDACi resistance or response, we updated and optimized an integrative informatic pipeline that we had created previously to analyze ULI-ChIP- and RNA-seq data from serial MF/SS samples and the in vitro treated T cells (see Methods). Briefly, we used MACS2 to call acetylation peaks with an FDR q value <0·01 [20]. We first derived consensus peak sets and then identified differentially bound peaks between sample groups using the DiffBind R package and an FDR q value <0·05 [24]. The heatmap in Fig. 1B shows the ~5000 acetylation peaks with significantly altered activity in resistant compared to sensitive sample groups (Table S3); the majority of these are increased in samples from patients with progressive disease on HDACi. Comparison of functional genomic regions (promoter, enhancer, exon, intron) revealed that the greatest number of significantly altered acetylation peaks are located within introns and distal intergenic regions (enhancers), compared to promoters and exons. Most of these peaks have increased acetylation in resistant samples (Fig. S1A). Indeed, we observed a greater than two-fold change (log2) in all functional genomic regions, with distal intergenic (enhancer) regions having the highest relative increase in acetylation (Fig. 1C). The small number of peaks with decreased acetylation in resistant samples similarly showed a greater than two-fold reduction in all functional regions (Fig. S1B). Because acetylation is an activating histone modification associated with increased transcription, we examined the expression of genes linked to genomic regions with significantly higher acetylation in resistant versus sensitive samples. As expected, genes with highly acetylated regulatory elements (distal enhancer regions and promoters), as well as exons and introns, had significantly higher expression compared to genes with unchanged or decreased acetylation at these elements (Fig. 1D). These data demonstrate that MF/SS tumors that progress during HDACi treatment have significantly increased histone acetylation at thousands of regulatory elements compared to HDACi-sensitive MF/SS tumors, and that their target genes exhibit significantly increased expression.
3.2. HDACi-resistant MF/SS exhibits high expression of anti-apoptotic, cell cycle, and cell adhesion/migration genes
We next directly evaluated the transcriptomes of primary patient MF/SS samples based on their response to HDACi. Using DESeq2, we identified 491 genes with significantly different expression in resistant versus sensitive samples (Fig. 2A–B and Table S3, Table S4) [28]. Similar to the histone acetylation changes we observed above (see Fig. 1), a majority of significantly altered genes have higher expression in the HDACi-resistant group (357 up versus 134 down). To validate these expression changes in additional MF/SS samples, we obtained publicly available RNA-seq data from a study of early to advanced stage MF (n = 49) samples and healthy control CD4+ T cells (n = 3) [35]. Genes upregulated in HDACi-resistance also exhibited higher expression in MF samples compared to control CD4+ T cells from the Querfeld et al. study (Fig. S2A) [35]. In contrast, genes that were down-regulated in HDACi-resistant samples were not significantly different across this independent dataset (Fig. S2B).
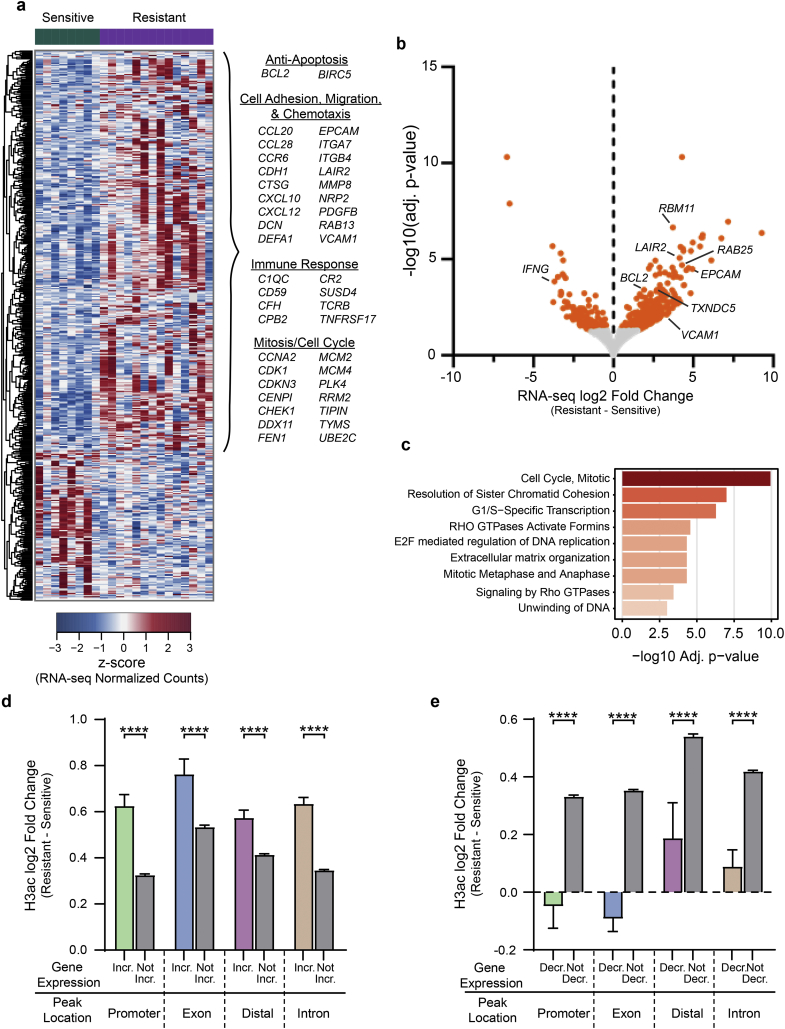
Fig. 2.
Genes in anti-apoptosis, cell cycle, cell adhesion and migration pathways are upregulated in Resistant samples. a) Heatmap showing mRNA expression of differentially expressed genes (adj. p-value ≤0·05) in Sensitive versus Resistant samples. Selected genes that are upregulated in Resistant samples are listed. b) Volcano plot of gene expression differences in Resistant versus Sensitive samples. DEGs (adj. p-value ≤0·05) are coloured orange. c) Select enriched (adj. p-value ≤0·05) Reactome terms for genes upregulated in Resistant samples (full list Table S4). d) Bar plot comparing log2 fold changes for H3ac peaks linked to genes with increased (Incr.) and unchanged/decreased (Not Incr.) expression in Resistant vs Sensitive samples. H3ac peaks are categorized by functional genomic region (Mann-Whitney test; ****, P ≤ 0·0001; mean with 95% confidence interval shown). e) Bar plot as in d, but for genes with decreased (Decr.) and unchanged/increased (Not Decr.) expression in Resistant vs Sensitive samples (Mann-Whitney test; ****, P ≤ 0·0001). Mean with 95% confidence interval shown.
Gene ontology and pathway analyses revealed several significantly enriched pathways for HDACi-resistance upregulated genes, but none for downregulated genes (Fig. 2C and Table S5). A number of the upregulated genes in these pathways have been previously associated with MF/SS or HDACi treatment, but many are novel to this analysis of HDACi response/resistance in MF/SS. These include anti-apoptotic, BCL2 (known) and BIRC5 (novel); cell cycle, CDK1 (known) and CDC6 (novel); and proliferation, RAB25 and RBM11 (both novel) [36,37]. We also observed a number of upregulated genes with inflammatory functions, such as TNFAIP3, which is known to be upregulated and/or mutated in B cell lymphoma [[38], [39], [40], [41]], and STAT4, which was reported to be downregulated in several MF/SS studies [42], but we find to be nearly two-fold upregulated (1·9 log2 fold change, p < 0·0001) in HDACi-resistant samples. Quite striking was the number of extra-cellular matrix, cell adhesion, and cell migration genes that were upregulated in the resistant group, including several not previously associated with MF/SS: CCR6 (known), CCL28 (novel), EPCAM (novel), VCAM1 (novel), and LAIR2 (novel) [[42], [43], [44], [45]]. Upregulation of these genes in the resistant group suggest that increased proliferation, anti-apoptotic signaling, and higher levels of chemotaxis and migration may represent functional mechanisms of HDACi resistance and/or MF/SS progression.
Alterations in histone acetylation likely contribute to the differences in gene expression we identified in HDACi-resistant or responsive MF/SS. Indeed, upregulated genes had significantly higher levels of histone acetylation at regulatory elements and gene bodies compared to unchanged/downregulated genes, with promoters exhibiting the greatest relative increase in acetylation (Fig. 2D). Similarly, downregulated genes had significantly lower chromatin acetylation levels compared to genes with unchanged/increased expression, with promoters and exons exhibiting the greatest decreases in acetylation (Fig. 2E). Taken with the data presented in Fig. 1, these results further support a role for differential histone acetylation in HDACi resistance and link acetylation to anti-apoptotic, cell cycle, and cell adhesion/migration pathways as potential mechanisms of resistance and MF/SS progression.
3.3. Primary MF/SS HDACi-resistance genes are distinct from those identified by in vitro studies
A number of studies have identified mechanisms that contribute to HDACi resistance, the vast majority of which were performed in vitro, generally in cancer cell lines. Approximately 30 genes and proteins reportedly contribute to HDACi resistance, including those in ABC transporter (MDR), pro-apoptotic (Fas, Caspase), anti-apoptotic (BCL), heat shock, HDAC, JAK-STAT, MAPK/PI3K, NFKB, Redox, and TNF pathways [13,14]. To evaluate these mechanisms in primary MF/SS samples, we examined the expression of genes in these pathways (n = 361) (Fig. S2C and Table S6). Of these, 14 were significantly altered in resistant versus sensitive groups. Notably, only 3/14 had been previously associated with HDACi resistance: anti-apoptotic gene BCL2 and PI3K genes PIK3C2A and PIK3C2B, and these latter two were significantly decreased in the resistant group while increases have been reported [13]. While other STATs were unchanged, we detected significantly increased expression of STAT4, which has not been previously connected to HDACi resistance. We also identified three genes involved in Redox pathways that had not previously been reported: TXNDC5, whose product catalyzes thiol-disulfide interchanges, and GSTM1 and GSTM3, which are glutathione S-transferases that detoxify drugs, toxins, and reactive oxygen species. Decreases in TNF-related apoptotic pathway proteins are also associated with resistance to HDACi treatment in vitro [13], but we detected increases in two of these genes, TNFRSF17 and TNFAIP3, both of which promote B cell survival, and no change in the rest of these genes (51/53). A caveat to this RNA-seq analysis is that it does not measure levels of protein or phosphoprotein, which may be different from mRNA levels. Nevertheless, we demonstrate that mRNA expression of most of the HDACi-resistance pathways previously identified in vitro are not significantly altered in primary MF/SS HDACi-resistant versus -sensitive groups. In addition, we identify STAT4, TNFRSF17, TNFAIP3, GSTM1, GSTM3, and TXNDC5 as previously unrecognized genes whose upregulated expression may contribute to HDACi-resistance in MF/SS disease.
3.4. Differences in the expression of key resistance-associated genes are detectable pre-HDACi treatment
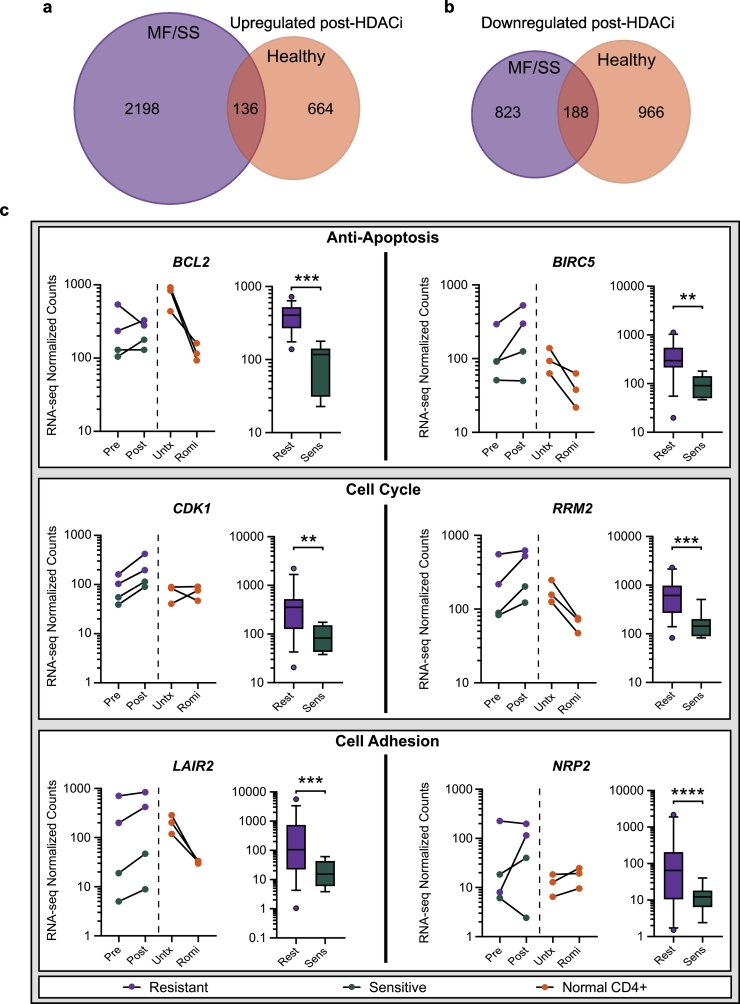
We next asked which of the gene expression differences in MF/SS samples collected pre- and post-one-week romidepsin therapy coincided with, or diverged from, expression changes induced by in vitro romidepsin treatment of normal CD4+ T cells. We therefore compared RNA-seq from healthy donor CD4+ T cells pre- and post-romidepsin treatment and paired MF/SS samples collected immediately pre-infusion and one-week post-infusion of romidepsin. We identified genes with increased expression (log2 fold change ≥1) in paired post- versus pre-treatment MF/SS samples (>2300) or healthy CD4+ T cells (800). Of these upregulated genes, only 136 were in common (5·8% of MF/SS, 17% of healthy CD4+ T cells) (Fig. 3A). The numbers of downregulated genes (log2 fold change ≤ −1) in each comparison were more similar: 1011 for MF/SS and 1154 for healthy CD4+ T cells. The 188 genes in common comprised 18·6% of MF/SS and 16·3% of healthy CD4+ T cell downregulated genes (Fig. 3B). As we observed with the MF/SS samples, genes with expression changes in HDACi-treated control T cells had corresponding changes in acetylation (Fig. S3). Thus, the MF/SS cells exhibited a substantially greater number of genes with increased expression after romidepsin treatment compared to healthy donor CD4+ T cells.
Fig. 3.
Resistant-associated expression changes are detectable pre-HDACi treatment. a) Venn diagram showing overlap of genes upregulated (log2 fold change ≥1) by romidepsin treatment in paired MF/SS samples (one-week post-treatment vs immediately pre-treatment, purple) and genes upregulated in healthy CD4+ T cells (24 h post-treatment vs untreated, orange). b) Venn diagram showing overlap of genes downregulated (log2 fold change ≤ −1) by romidepsin treatment in paired MF/SS samples (as in A, purple) and genes downregulated in healthy CD4+ T (as in A, orange). c) Paired dot plots (left) and box plots (right) for select differentially expressed genes. Paired MF/SS pre-HDACi and one-week post-HDACi treatment samples connected (purple = Resistant, green = Sensitive). Paired healthy CD4+ T cell pre- and 24 h post-romidepsin treatment samples connected (orange). (Wald test – DESeq2; **, P ≤ 0·01; ***, P ≤ 0·001; ****, P ≤ 0·0001). Whiskers show 10-90th percentile. Pre = pre-HDACi treatment; Post = one-week post HDACi treatment; Untx = Untreated; Romi = Romidepsin treated; Rest = HDACi Resistant; Sens = HDACi Sensitive. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Given that we observed significant differences in hundreds of genes in HDACi-resistant versus -sensitive groups (Fig. 1, Fig. 2), we asked whether these differences were also detectable in the post- versus pre-treatment analysis. Indeed, we found that the HDACi-resistant MF/SS samples showed consistently higher expression of these differentially expression genes, and surprisingly, many of these were significantly increased in pre-treatment samples (Fig. S4). Several examples are shown in Fig. 3C. These include anti-apoptotic genes BCL2 and BIRC5 (Survivin), which promote proliferation and prevent apoptosis in multiple cancer types, including B and T cell lymphomas [46]. We also identified genes in cell cycle pathways such as CDK1, a cyclin-dependent kinase that promotes mitosis, and RRM2, a ribonucleotide reductase enzyme involved in DNA replication/repair. Cell adhesion pathway genes also showed this pattern in HDACi-resistance, including NRP2, whose protein product binds semaphorins and VEGF and is involved in the migration of T cells and other immune cells [47], and LAIR2, which codes for a secreted collagen receptor protein expressed by T and NK cells [48]. In an independent study of CTCL that did not evaluate HDACi therapy [35], expression levels of RRM2, BIRC5, CDK1, CCR6, CXCR4, and LAIR2 are significantly higher in MF compared to control skin samples, with a trend toward higher expression in more advanced stages (Fig. S5). NRP2 expression showed a non-significant trend toward higher expression in MF samples, but BCL2 expression was not different from control skin (Fig. S5). The pattern of higher expression of these genes in advanced disease and HDACi-resistant samples, detectable pre- and post-HDACi, suggests that they may be involved in disease progression and/or mechanisms of HDACi resistance.
3.5. Highly acetylated regions demonstrate robust enhancer activity in reporter assays
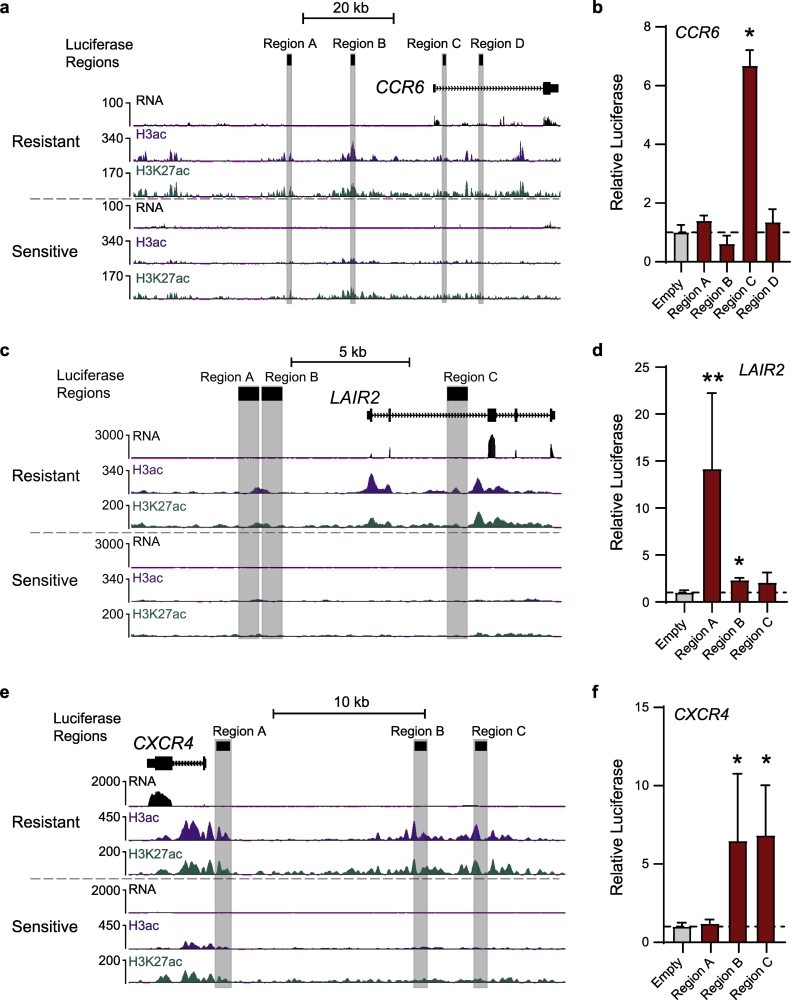
To confirm enhancer activity of regions with higher levels of acetylation in HDACi-resistant versus sensitive samples, we performed luciferase reporter assays in an MF/SS cell line (HUT78) by adapting previously described protocols [49]. We selected putative enhancer regions that met the following criteria: 1) higher acetylation in resistant samples, 2) likely to regulate expression of nearby genes based on GeneHancer interactions and ENCODE transcription factor binding data, 3) target gene(s) with significantly elevated expression in resistant samples, and 4) the regulated genes have known or potential roles in lymphoma progression and/or drug resistance. Three gene loci with enhancer regions meeting these criteria were subjected to luciferase reporter assays (Figs. 4 and S6). CCR6 is a beta chemokine receptor expressed by normal memory T cells and MF/SS that may regulate their migration and recruitment in inflammation and MF/SS metastasis [43]. Expression of CCR6 is significantly higher in resistant samples (Fig. S7), and the gene locus contains two distal intergenic elements upstream of the promoter/transcription start site (TSS) and two elements within the first intron that exhibit significantly higher levels of H3K27 and K3K9/14 acetylation in the resistant group (Figs. 4A and S6C). Luciferase assays confirmed strong enhancer activity in one of the intronic elements (6-fold higher activity compared to empty vector, Fig. 4B). In the LAIR2 locus, we tested three regions with higher acetylation in resistant samples, two distal intergenic elements upstream of the promoter/TSS and another element in the second intron of LAIR2 (Figs. 4C and S6D). Luciferase assays confirmed strong enhancer activity for the intronic element, with >10-fold greater activitya compared to the vector alone (Fig. 4D). The chemokine receptor CXCR4 binds CXCL12/SDF-1 and this signaling axis plays major roles in cell migration and immune response [50]. CXCR4 expression is higher in the resistant group (Fig. S7), and we identified three upstream intergenic regions with higher acetylation in resistant samples (Figs. 4E and S6E). Luciferase reporter assays demonstrated robust enhancer activity (>5-fold) for the two most distal of these elements (Fig. 4F). These results demonstrate that highly acetylated elements near upregulated genes in resistant samples harbor strong enhancer activity, suggesting that these elements may drive high expression of genes that promote HDACi resistance in MF/SS.
Fig. 4.
Luciferase reporter assays confirm enhancer activity for regulatory elements near HDACi-resistant upregulated genes. a) UCSC genome browser snapshot showing the CCR6 locus with representative Resistant and Sensitive samples. All signal tracks are normalized to reads per kilobase per million mapped reads (RPKM). Putative enhancer regions assayed by luciferase reporter are highlighted. b) Enhancer activity of putative CCR6 enhancer regions measured by luciferase reporter in HUT78 cells. All assays were performed at least twice and read in triplicate. Ratios are relative to empty vector. c) UCSC genome browser snapshot showing the LAIR2 locus with representative Resistant and Sensitive samples, as in a. d) Enhancer activity of putative LAIR2 enhancer regions (c) in HUT78 cells, performed as in b. e) Enhancer activity of putative CXCR4 enhancer regions (f) in HUT78 cells, performed as in b. (Mann-Whitney test *, p ≤ 0·05; **, p ≤ 0·01) Mean with standard deviation shown.
3.6. LAIR2 is a novel marker of HDACi-resistance in MF/SS
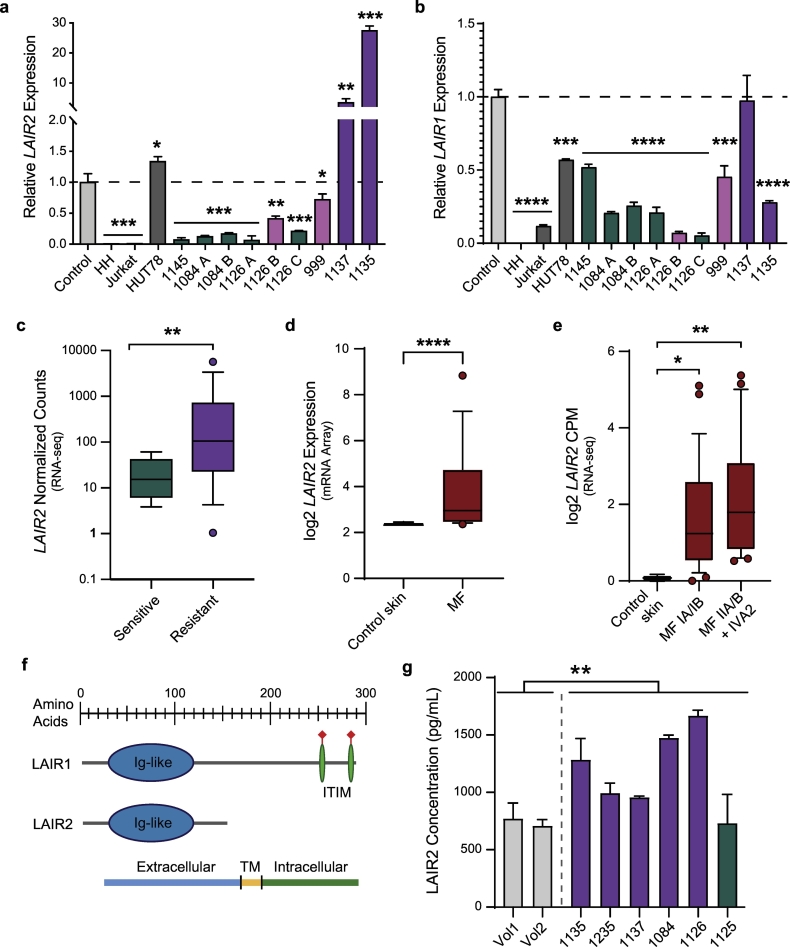
Among the genes with highly acetylated enhancers and significantly upregulated expression in the HDACi-resistant group, LAIR2 was particularly intriguing because of the degree of upregulation and its roles in T cell adhesion, migration, and activation [48,51]. We confirmed the RNA-seq results using qRT-PCR for LAIR2 in additional MF/SS samples from the same patients, including additional time points; in some cases, the additional samples were collected during treatment with non-HDACi therapies due to progression on HDACi. We also tested peripheral blood CD4+ T cells from healthy donors and T cell lines (Jurkat, leukemia; HH and HUT78, MF/SS). We found substantially higher levels of LAIR2 mRNA in samples from patients with MF/SS that were progressing on HDACi therapy compared to normal CD4+ T cells or to samples from patients with MF/SS that was responding to therapy (Fig. 5A). We noted that in two samples from two patients with mixed response to therapy (i.e., response in lymph nodes/blood and progression in skin, 1126 B, 999), LAIR2 expression was trending higher than samples from the same patient at different time points in complete response or compared to samples from other patients in complete response. LAIR1 is a highly homologous gene located near LAIR2 on chromosome 19q13·42. In contrast to LAIR2, LAIR1 expression was not significantly higher in MF/SS compared to healthy control CD4+ T cells, and HDACi-resistant samples were not different from -sensitive (Fig. 5B).
Fig. 5.
LAIR2 is upregulated in multiple independent MF/SS datasets. a) LAIR2 mRNA expression measured by qRT-PCR in healthy control CD4+ T cells, T cell lines, and primary MF/SS samples. Green bars: responding to therapy, Pink bars: mixed response to therapy; Purple bars: progressive disease. (Unpaired student's t-test compared to control; *, P ≤ 0·05; **, P ≤ 0·01; ***, P ≤ 0·001). Mean with standard deviation shown. b) LAIR1 mRNA expression measured as in A. (Unpaired student's t-test compared to control; *, P ≤ 0·05; **, P ≤ 0·01; ***, P ≤ 0·001; ***, P ≤ 0·0001). Mean with standard deviation shown. c) LAIR2 mRNA expression in CD4+ T cells purified from Sensitive and Resistant MF/SS samples (WUSM, RNAseq). d) LAIR2 mRNA expression in control skin (n = 8) and primary MF (n = 6) (microarray; Mann-Whitney test; Humme et al., 2015). e) LAIR2 mRNA expression in control skin (n = 3) and primary MF (n = 49) biopsies (RNAseq; Kruskall-Wallis test; Querfeld et al., 2018). c-e show mean with 95% CI. f) Diagram showing protein domains in LAIR2 and LAIR1. g) LAIR2 protein expression measured by ELISA in plasma from healthy controls and CTCL patients (colours as in A&B; unpaired Welch's t-test; **, P ≤ 0·01). Mean with standard deviation shown. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To evaluate LAIR2 expression in additional MF/SS patient cohorts, we obtained publicly available expression data from four studies in which mRNA was collected from MF or SS samples and from healthy control CD4+ T cells in 2 of 4 studies [35,[52], [53], [54]]. Similar to our findings in HDACi-resistant MF/SS samples measured by RNA-seq (Fig. 5C), LAIR2 expression quantified by microarray was higher in MF and SS samples compared to control CD4+ T cells, (Figs. 5D and S8A) [53]. In a larger study using RNA-seq (49 MF, 3 control), LAIR2 expression was significantly higher (two-fold on a log2 scale) in all stages of MF compared to controls [35]. More advanced stage disease (IIA and above) showed higher LAIR2 levels than early stage MF (IA/IB) in this study, as well as another CTCL study that performed microarray analysis (Figs. 5E and S8B) [52]. LAIR1 levels were not statistically different in purified MF/SS cells from resistant versus sensitive samples (Fig. S8C) or were slightly decreased in Sezary cells (Fig. S8D) compared to healthy control CD4+ T cells [54]. In unsorted MF samples, LAIR1 expression was increased relative to control samples (Fig. S8E) [35]. However unlike LAIR2, which is expressed only in T and NK cells, LAIR1 is expressed on nearly all immune cells, including monocytes, macrophages, T, B, and NK cells, dendritic cells, mast cells, and eosinophils, so the increased levels may reflect the inflammatory microenvironment of MF-involved skin [55]. Thus, we demonstrate using several different methods that LAIR2 expression is significantly higher in HDACi-resistant MF/SS disease.
The gene that encodes LAIR2 lies within the leukocyte receptor complex (LRC), a gene cluster on chromosome 19q13.4 that is rich in leukocyte immunoglobulin-like receptor (LIR) and Killer cell immunoglobulin-like receptors (KIRs) genes (Fig. S9). These genes encode transmembrane inhibitory and activating receptors expressed by T cells and/or NK cells, which are highly polymorphic; expression differences and genomic polymorphisms have been associated with auto-immune and infectious diseases [56,57]. LAIR2 is present in primates but absent in most other organisms, including mice (Fig. S9) [48]. In amino acid sequence, LAIR2 is highly homologous to the N-terminus of LAIR1, which includes the Ig-like domain, but lacks the transmembrane and intracellular domains that are present in LAIR1 (Fig. 5F). LAIR2 is a secreted protein and has been detected in the body fluids of patients with autoimmune disease [48,58]. Therefore, we collected plasma from MF/SS patients and healthy donors and performed ELISA using an anti-LAIR2 antibody. Consistent with mRNA levels, we detected significantly higher levels of LAIR2 protein in MF/SS plasma compared to healthy controls (Fig. 5G). Notably, the patient with the lowest plasma concentration of LAIR2 protein, which was equivalent to healthy controls, has had a sustained response to romidepsin over four years of treatment (Patient 1125). Taken together, these results demonstrate that LAIR2 mRNA and protein expression levels are significantly higher in both malignant cells and plasma from patients with HDACi-resistant MF/SS.
4. Discussion
An outstanding question in CTCL research is why only a fraction of patients respond to HDACi therapy [6], and the related question of how response and resistance could be predicted to inform therapeutic choice. Here we have identified significant differences in the histone acetylation of gene regulatory elements in samples from CTCL patients with HDACI-resistant versus -sensitive disease and linked them to significant expression changes in cell cycle, apoptosis, cytokine/chemokine signaling, and cell adhesion/migration pathways. We and others have previously shown that high levels of acetylation of enhancers and promoters can lead to overexpression of oncogenes that promote cancer pathogenesis [49,[59], [60], [61], [62]]. In this study, we demonstrated that enhancer elements near potential MF/SS oncogenes CCR6, CXCR4, and LAIR2 had significantly increased acetylation levels in HDACI-resistant samples also showed strong enhancer activity by luciferase reporter assay. These results suggest that highly acetylated elements may drive the high expression levels of target genes in resistant samples, promoting enhanced chemotaxis, cell migration, and inflammation that contribute to progressive disease.
We validated our findings in several independent expression profiling studies of MF/SS, showing that HDACi-resistance genes were generally elevated in MF/SS compared to normal CD4+ T cells and several genes exhibited higher levels in more advanced stages of MF [35,[52], [53], [54]]. One caveat is that these studies did not evaluate response to HDACi, however, there were no expression datasets with HDACi response data available for comparison to our study. Several HDACi-resistance genes had higher levels of acetylation and expression in pre-treatment samples, including BCL2, CDK1, CXCR4 and CCR6. Previous studies have shown that knock-down or inhibition of BCL2, or deficiency of CDK1 family member, CDKN1A (p21), increased the sensitivity of cells treated with HDACi in vitro [[63], [64], [65], [66], [67]]. Other inhibition and knock-down studies demonstrated that CXCR4 and CCR6 may play a role in CTCL pathogenesis, though synergy with HDACi remains to be demonstrated [[68], [69], [70], [71], [72]]. We also identified several genes that had not been previously associated with MF/SS or HDACi-resistance, including BIRC5, NRP2, RRM2, and LAIR2, whose role in CTCL and HDACi-resistance may be further elucidated by future studies. Taken together, these results suggest that elevated levels of histone acetylation and novel cell adhesion/migration pathways are likely mechanisms of HDACi resistance in CTCL patients.
Our analysis of primary samples from HDACi-treated MF/SS patients also revealed several novel genes upregulated in previously identified HDACi resistance pathways. These include three genes in Redox pathways, TXNDC5, GSTM1, and GSTM3, which process therapeutic drugs and reactive oxygen species. Cells with altered levels of other components of these pathways have decreased cell death when treated with HDACi in vitro [13], and these three genes may contribute to in vivo mechanisms of HDACi resistance. STAT4 expression was nearly two-fold in the resistant group, in contrast to previous reports of downregulation in Sèzary and upregulation in MF skin, though no association with HDACi resistance was previously reported [42]. Also, in contrast to published in vitro HDACi resistance studies, the resistant group exhibited higher expression of TNF pathway genes TNFRSF17 and TNFAIP3, though notably both of these are associated with the pathogenesis of B cell lymphomas [[38], [39], [40], [41],73]. In summary, we identify STAT4, TNFRSF17, TNFAIP3, GSTM1, GSTM3, and TXNDC5 as previously unrecognized genes whose upregulated expression may contribute to HDACi-resistance in primary MF/SS and may represent novel targets for therapeutic development.
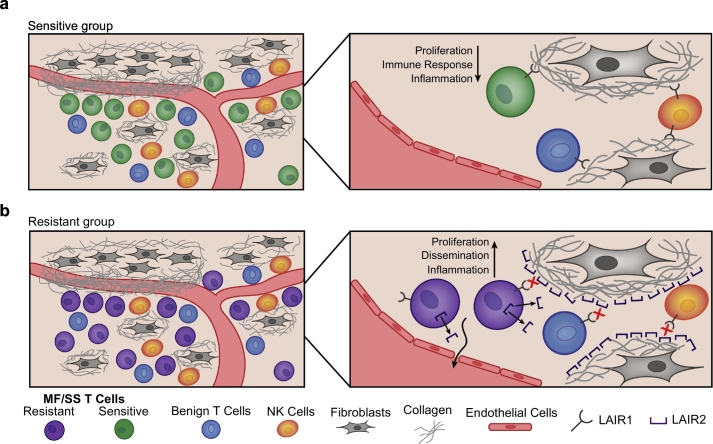
One of the most striking differences we detected in HDACI-resistant compared to sensitive groups was elevated expression of LAIR2, a gene that encodes a secreted collagen receptor protein [48]. LAIR2 mRNA was significantly higher in MF/SS skin biopsies and blood from patients with resistant disease, and the LAIR2 protein was elevated in the plasma of these patients. The LAIR2 gene may have arisen by duplication from a close homologue, LAIR1, which encodes a transmembrane inhibitory receptor expressed on the majority of immune cells, in contrast to LAIR2 expression only on T and NK cells [48]. The ligands of LAIR1 and LAIR2 receptors are collagen types I, III, and IV. When bound to collagens, LAIR1 transmits inhibitory signaling that reduces immune cell activation and proliferation (Fig. 6A) [51,74]. Binding of collagen by LAIR2 prevents binding of LAIR1, which may promote activation, proliferation, and migration of malignant T cells from skin sites into lymph nodes and peripheral blood (Fig. 6B) [75,76]. Downregulation of LAIR1-mediated inhibitory signaling may also contribute to inflammation in the MF/SS tumor microenvironment, in which benign immune cells are activated and contribute to morbidity of the disease [77]. In this way, MF/SS cells that produce LAIR2 might have a higher rate of proliferation and could more readily migrate through and out of tissue into lymphatics and/or the peripheral blood. LAIR2 could also promote a more inflammatory milieu by blocking collagen binding of LAIR1 on benign immune T and NK cells in the MF/SS microenvironment. Thus, LAIR2 represents a novel pathway for HDACi-resistance in MF/SS. It is also a potentially clinically useful marker for prognosis, HDACi resistance prediction, and monitoring disease burden and may represent a new therapeutic target for this challenging cancer.
Fig. 6.
Proposed model for the role of LAIR2 in MF/SS pathogenesis. a) MF/SS cells that produce low/no LAIR2, such as in the Sensitive group, would still bind collagen through LAIR1, causing inhibitory signaling through LAIR1 ITIM domains. LAIR1-mediated inhibitory signaling could decrease proliferation, immune response, and inflammation. b) MF/SS cells that produce high levels of LAIR2, such as in the Resistant group, would have decreased binding of LAIR1 to collagen, causing decreased inhibitory ITIM signaling. Loss of LAIR1-inhibitory signaling could promote migration and proliferation of MF/SS cells and increased inflammation in the tumor microenvironment through decreased inhibitory signaling in local benign T and NK cells.
In conclusion, these studies are the first to connect differences in epigenome-wide acetylation and gene expression to HDACi resistance in primary samples from CTCL patients. Many of the HDACi resistance genes are involved in cell adhesion/migration, suggesting they may play a role in the variable manifestations of MF/SS disease in skin, lymph node, and peripheral blood. Elevated expression of several of these significantly altered genes was detectable prior to HDACi therapy, suggesting their utility as prognostic and/or predictive markers. Of these, we characterized LAIR2 as the most robust, which may contribute to inflammation and migration of MF/SS and benign immune cells in the microenvironment. Further studies to define the role of LAIR2 in MF/SS pathogenesis and progression, as well as its potential as a predictive marker and therapeutic target in this challenging cancer, are already underway.
The following are the supplementary data related to this article.
Primers used for qRT-PCR and amplification of putative regulatory elements used in luciferase assays.
Additional patient clinical information. Romi – Romidepsin; Vori – Vorinostat; Pre/Post-Treatment – paired samples collected pre- and post-HDACi treatment.
Genes differentially expressed in resistant compared to sensitive samples and Romidepsin-treated versus untreated healthy CD4+ T cells.
Gene pathway and ontology enrichment analyses for genes upregulated (adj. p-value ≤0·05) in resistant compared to sensitive samples.
Genes in previously reported HDACi resistance pathways.
Supplementary material
Funding sources
This work was supported by NIH grants CA156690, CA188286 (J.E.P.), the Washington University Institute of Clinical and Translational Sciences grant UL1 TR000448 from the National Center for Advancing Translational Sciences (NCATS) (Pilot Award to J.E.P.), and Siteman Cancer Center (CA091842) (Pilot Award to J.E.P.) grants. Sequencing provided by the Genome Technology Access Center, which is partially supported by NCI Cancer Center Support Grant #P30 CA91842 to the Siteman Cancer Center and by ICTS/CTSA Grant# UL1 TR000448 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH), and NIH Roadmap for Medical Research. The ICTS is funded by the NIH's NCATS Clinical and Translational Science Award (CTSA) program grant #UL1 TR002345. This publication is solely the responsibility of the authors and does not necessarily represent the official view of NCRR or NIH. None of these sources had any role in data collection, analysis, interpretation, trial design, patient recruitment, writing the manuscript, the decision to submit the manuscript, or any other aspect pertinent to the study. None of the authors was paid to write the article by a company or any other agency. The corresponding author (Jacqueline Payton) had full access to all of the data in the study and had final responsibility for the decision to submit for publication.
Author contributions
Conception and design: J.E. Payton, J.M. Andrews, N. Mehta-Shah.
Development of methodology: J.M. Andrews, J.E. Payton, J.A. Schmidt.
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): J.M. Andrews, J.E. Payton, A.C. Musiek, N. Mehta-Shah, J.A. Schmidt, K.R. Carson.
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): J.M. Andrews, J.E. Payton.
Writing, review, and/or revision of the manuscript: J.M. Andrews, J.E. Payton, A.C. Musiek, N. Mehta-Shah, J.A. Schmidt, K.R. Carson.
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): J.M. Andrews, J.A. Schmidt, J.E. Payton.
Study supervision: N. Mehta-Shah, J.E. Payton.
Declaration of Competing Interest
N. Mehta-Shah reports research funding from Celgene, Verastem Pharmaceuticals, Roche/Genentech, and Bristol Myers Squibb and is a consultant for Kiowa Hakka Kirin. K. R. Carson is also employed by Flatiron Health. A. C. Musiek reports research funding from Pfizer, Helsinn, miRagen, Solgenix, Kyowa, Elorac, and Actelion and is also on the advisory boards for Actelion and Kyowa. No potential conflicts of interest were disclosed by the other authors.
Acknowledgements
We thank the Washington University School of Medicine Lymphoma Banking Program of the Division of Medical Oncology in the Department of Medicine for support of the biopsy and banking program. We are grateful to the patients, their caregivers and families, and the patient coordinators who participated in this study.
References
- 1.Jawed S.I., Myskowski P.L., Horwitz S., Moskowitz A., Querfeld C. Primary cutaneous T-cell lymphoma (mycosis fungoides and Sézary syndrome): Part I. Diagnosis: clinical and histopathologic features and new molecular and biologic markers. J Am Acad Dermatol. 2014;70(205) doi: 10.1016/j.jaad.2013.07.049. [e1–205.e16] [DOI] [PubMed] [Google Scholar]
- 2.Agar N.S., Wedgeworth E., Crichton S. Survival outcomes and prognostic factors in mycosis fungoides/Sézary syndrome: validation of the revised International Society for Cutaneous Lymphomas/European Organisation for Research and Treatment of Cancer staging proposal. J Clin Oncol Off J Am Soc Clin Oncol. 2010;28:4730–4739. doi: 10.1200/JCO.2009.27.7665. [DOI] [PubMed] [Google Scholar]
- 3.Kim Y.H., Jensen R.A., Watanabe G.L., Varghese A., Hoppe R.T. Clinical stage IA (limited patch and plaque) mycosis fungoides. A long-term outcome analysis. Arch Dermatol. 1996;132:1309–1313. [PubMed] [Google Scholar]
- 4.Wilcox R.A. Cutaneous T-cell lymphoma: 2017 update on diagnosis, risk-stratification, and management. Am J Hematol. 2017;92:1085–1102. doi: 10.1002/ajh.24876. [DOI] [PubMed] [Google Scholar]
- 5.Wilcox R.A. Cutaneous T-cell lymphoma: 2014 update on diagnosis, risk-stratification, and management. Am J Hematol. 2014;89:837–851. doi: 10.1002/ajh.23756. [DOI] [PubMed] [Google Scholar]
- 6.Lopez A.T., Bates S., Geskin L. Current status of HDAC inhibitors in cutaneous T-cell lymphoma. Am J Clin Dermatol. 2018;19:805–819. doi: 10.1007/s40257-018-0380-7. [DOI] [PubMed] [Google Scholar]
- 7.Haery L., Thompson R.C., Gilmore T.D. Histone acetyltransferases and histone deacetylases in B- and T-cell development, physiology and malignancy. Genes Cancer. 2015;6:184–213. doi: 10.18632/genesandcancer.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baylin S.B., Jones P.A. A decade of exploring the cancer epigenome — biological and translational implications. Nat Rev Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Piekarz R.L., Frye R., Turner M. Phase II multi-institutional trial of the histone deacetylase inhibitor romidepsin as monotherapy for patients with cutaneous T-cell lymphoma. J Clin Oncol. 2009;27:5410–5417. doi: 10.1200/JCO.2008.21.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whittaker S.J., Demierre M.-F., Kim E.J. Final results from a multicenter, international, pivotal study of romidepsin in refractory cutaneous T-cell lymphoma. J Clin Oncol. 2010;28:4485–4491. doi: 10.1200/JCO.2010.28.9066. [DOI] [PubMed] [Google Scholar]
- 11.Bates S.E., Zhan Z., Steadman K. Laboratory correlates for a phase II trial of romidepsin in cutaneous and peripheral T-cell lymphoma. Br J Haematol. 2010;148:256–267. doi: 10.1111/j.1365-2141.2009.07954.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qu K., Zaba L.C., Satpathy A.T. Chromatin accessibility landscape of cutaneous T Cell lymphoma and dynamic response to HDAC inhibitors. Cancer Cell. 2017;32:27–41. doi: 10.1016/j.ccell.2017.05.008. [e4] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robey R.W., Chakraborty A.R., Basseville A. Histone deacetylase inhibitors: emerging mechanisms of resistance. Mol Pharm. 2011;8:2021–2031. doi: 10.1021/mp200329f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee J.-H., Choy M.L., Marks P.A. Mechanisms of resistance to histone deacetylase inhibitors. Adv Cancer Res. 2012;116:39–86. doi: 10.1016/B978-0-12-394387-3.00002-1. [DOI] [PubMed] [Google Scholar]
- 15.Chicaybam L., Barcelos C., Peixoto B. An efficient electroporation protocol for the genetic modification of mammalian cells. Front Bioeng Biotechnol. 2017;4 doi: 10.3389/fbioe.2016.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brind'Amour J., Liu S., Hudson M., Chen C., Karimi M.M., Lorincz M.C. An ultra-low-input native ChIP-seq protocol for genome-wide profiling of rare cell populations. Nat Commun. 2015;6:6033. doi: 10.1038/ncomms7033. [DOI] [PubMed] [Google Scholar]
- 17.Langmead B., Salzberg S.L. Fast gapped-read alignment with bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Consortium T.E.P. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H., Handsaker B., Wysoker A. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng J., Liu T., Qin B., Zhang Y., Liu X.S. Identifying ChIP-seq enrichment using MACS. Nat Protoc. 2012;7 doi: 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramírez F., Ryan D.P., Grüning B. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Casper J., Zweig A.S., Villarreal C. The UCSC genome browser database: 2018 update. Nucleic Acids Res. 2018;46:D762–D769. doi: 10.1093/nar/gkx1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carroll T.S., Liang Z., Salama R., Stark R., de Santiago I. Impact of artifact removal on ChIP quality metrics in ChIP-seq and ChIP-exo data. Front Genet. 2014;5 doi: 10.3389/fgene.2014.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ross-Innes C.S., Stark R., Teschendorff A.E. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481:389–393. doi: 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu G., Wang L.-G., He Q.-Y. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015;31:2382–2383. doi: 10.1093/bioinformatics/btv145. [DOI] [PubMed] [Google Scholar]
- 26.Dobin A., Davis C.A., Schlesinger F. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andrews J.M., El-Alawi M., Payton J.E. Genotify: fast, lightweight gene lookup and summarization. J Open Source Softw. 2018;3 doi: 10.21105/joss.00885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng B., Wang G., Ma J. SoS notebook: an interactive multi-language data analysis environment. Bioinformatics. 2018 doi: 10.1093/bioinformatics/bty405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuleshov M.V., Jones M.R., Rouillard A.D. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schefe J.H., Lehmann K.E., Buschmann I.R., Unger T., Funke-Kaiser H. Quantitative real-time RT-PCR data analysis: current concepts and the novel “gene expression's CTdifference” formula. J Mol Med. 2006;84:901–910. doi: 10.1007/s00109-006-0097-6. [DOI] [PubMed] [Google Scholar]
- 33.Kaiko G.E., Ryu S.H., Koues O.I. The colonic crypt protects stem cells from microbiota-derived metabolites. Cell. 2016;165:1708–1720. doi: 10.1016/j.cell.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tiffon C., Adams J., van der Fits L. The histone deacetylase inhibitors vorinostat and romidepsin downmodulate IL-10 expression in cutaneous T-cell lymphoma cells. Br J Pharmacol. 2011;162:1590–1602. doi: 10.1111/j.1476-5381.2010.01188.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Querfeld C., Leung S., Myskowski P.L. Primary T cells from cutaneous T-cell lymphoma skin explants display an exhausted immune checkpoint profile. Cancer Immunol Res. 2018;6:900–909. doi: 10.1158/2326-6066.CIR-17-0270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mitra S., Cheng K.W., Mills G.B. Rab25 in cancer: a brief update. Biochem Soc Trans. 2012;40:1404–1408. doi: 10.1042/BST20120249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pavlyukov M.S., Yu H., Bastola S. Apoptotic cell-derived extracellular vesicles promote malignancy of glioblastoma via intercellular transfer of splicing factors. Cancer Cell. 2018;34:119–135. doi: 10.1016/j.ccell.2018.05.012. [e10] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weniger M.A., Küppers R. NF-κB deregulation in Hodgkin lymphoma. Semin Cancer Biol. 2016;39:32–39. doi: 10.1016/j.semcancer.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 39.Carbone A., Gloghini A., Kwong Y.-L., Younes A. Diffuse large B cell lymphoma: using pathologic and molecular biomarkers to define subgroups for novel therapy. Ann Hematol. 2014;93:1263–1277. doi: 10.1007/s00277-014-2116-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okosun J., Bödör C., Wang J. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat Genet. 2014;46:176–181. doi: 10.1038/ng.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Du M.-Q. MALT lymphoma: genetic abnormalities, immunological stimulation and molecular mechanism. Best Pract Res Clin Haematol. 2017;30:13–23. doi: 10.1016/j.beha.2016.09.002. [DOI] [PubMed] [Google Scholar]
- 42.Dulmage B.O., Geskin L.J. Lessons learned from gene expression profiling of cutaneous T-cell lymphoma. Br J Dermatol. 2013;169:1188–1197. doi: 10.1111/bjd.12578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abe F., Kitadate A., Ikeda S. Histone deacetylase inhibitors inhibit metastasis by restoring a tumor suppressive microRNA-150 in advanced cutaneous T-cell lymphoma. Oncotarget. 2016;8:7572–7585. doi: 10.18632/oncotarget.13810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choi J., Goh G., Walradt T. Genomic landscape of cutaneous T cell lymphoma. Nat Genet. 2015;47:1011–1019. doi: 10.1038/ng.3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Damsky W.E., Choi J. Genetics of cutaneous T cell lymphoma: from bench to bedside. Curr Treat Options Oncol. 2016;17:33. doi: 10.1007/s11864-016-0410-8. [DOI] [PubMed] [Google Scholar]
- 46.Altieri D.C. Survivin, cancer networks and pathway-directed drug discovery. Nat Rev Cancer. 2008;8:61–70. doi: 10.1038/nrc2293. [DOI] [PubMed] [Google Scholar]
- 47.Curreli S., Arany Z., Gerardy-Schahn R., Mann D., Stamatos N.M. Polysialylated neuropilin-2 is expressed on the surface of human dendritic cells and modulates dendritic cell-T lymphocyte interactions. J Biol Chem. 2007;282:30346–30356. doi: 10.1074/jbc.M702965200. [DOI] [PubMed] [Google Scholar]
- 48.Lebbink R.J., van den Berg M.C.W., de Ruiter T. The soluble leukocyte-associated Ig-like receptor (LAIR)-2 antagonizes the collagen/LAIR-1 inhibitory immune interaction. J Immunol. 2008;180:1662–1669. doi: 10.4049/jimmunol.180.3.1662. [DOI] [PubMed] [Google Scholar]
- 49.Koues O.I., Kowalewski R.A., Chang L.-W.W. Enhancer sequence variants and transcription-factor deregulation synergize to construct pathogenic regulatory circuits in B-cell lymphoma. Immunity. 2015;42:186–198. doi: 10.1016/j.immuni.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eckert F., Schilbach K., Klumpp L. Potential role of CXCR4 targeting in the context of radiotherapy and immunotherapy of cancer. Front Immunol. 2018;9 doi: 10.3389/fimmu.2018.03018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meyaard L. LAIR and collagens in immune regulation. Immunol Lett. 2010;128:26–28. doi: 10.1016/j.imlet.2009.09.014. [DOI] [PubMed] [Google Scholar]
- 52.Shin J., Monti S., Aires D.J. Lesional gene expression profiling in cutaneous T-cell lymphoma reveals natural clusters associated with disease outcome. Blood. 2007;110:3015–3027. doi: 10.1182/blood-2006-12-061507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Humme D., Haider A., Möbs M. Aurora kinase a is upregulated in cutaneous T-cell lymphoma and represents a potential therapeutic target. J Invest Dermatol. 2015;135:2292–2300. doi: 10.1038/jid.2015.139. [DOI] [PubMed] [Google Scholar]
- 54.Wang Y., Su M., Zhou L.L. Deficiency of SATB1 expression in Sezary cells causes apoptosis resistance by regulating FasL/CD95L transcription. Blood. 2011;117:3826–3835. doi: 10.1182/blood-2010-07-294819. [DOI] [PubMed] [Google Scholar]
- 55.Meyaard L., Adema G.J., Chang C. LAIR-1, a novel inhibitory receptor expressed on human mononuclear leukocytes. Immunity. 1997;7:283–290. doi: 10.1016/s1074-7613(00)80530-0. [DOI] [PubMed] [Google Scholar]
- 56.Rajalingam R. The impact of HLA class I-specific killer cell immunoglobulin-like receptors on antibody-dependent natural killer cell-mediated cytotoxicity and organ allograft rejection. Front Immunol. 2016;7 doi: 10.3389/fimmu.2016.00585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hirayasu K., Arase H. Functional and genetic diversity of leukocyte immunoglobulin-like receptor and implication for disease associations. J Hum Genet. 2015;60:703–708. doi: 10.1038/jhg.2015.64. [DOI] [PubMed] [Google Scholar]
- 58.Simone R., Pesce G., Antola P., Merlo D.F., Bagnasco M., Saverino D. Serum LAIR-2 is increased in autoimmune thyroid diseases. PLoS One. 2013;8 doi: 10.1371/journal.pone.0063282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jones P.A., Issa J.-P.J., Baylin S. Targeting the cancer epigenome for therapy. Nat Rev Genet. 2016;17:630–641. doi: 10.1038/nrg.2016.93. [DOI] [PubMed] [Google Scholar]
- 60.Luo H., Schmidt J.A., Lee Y.-S., Oltz E.M., Payton J.E. Targeted epigenetic repression of a lymphoma oncogene by sequence-specific histone modifiers induces apoptosis in DLBCL. Leuk Lymphoma. 2016;0:1–12. doi: 10.1080/10428194.2016.1190973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang Y., Koues O.I., Zhao J. Cis-regulatory circuits regulating NEK6 kinase overexpression in transformed B cells are super-enhancer independent. Cell Rep. 2017;18:2918–2931. doi: 10.1016/j.celrep.2017.02.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baylin S.B., Jones P.A. Epigenetic determinants of cancer. Cold Spring Harb Perspect Biol. 2016;8 doi: 10.1101/cshperspect.a019505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cyrenne B.M., Lewis J.M., Weed J.G. Synergy of BCL2 and histone deacetylase inhibition against leukemic cells from cutaneous T-cell lymphoma patients. Blood. 2017;130:2073–2083. doi: 10.1182/blood-2017-06-792150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Whitecross K.F., Alsop A.E., Cluse L.A. Defining the target specificity of ABT-737 and synergistic antitumor activities in combination with histone deacetylase inhibitors. Blood. 2009;113:1982–1991. doi: 10.1182/blood-2008-05-156851. [DOI] [PubMed] [Google Scholar]
- 65.Wei Y., Kadia T., Tong W. The combination of a histone deacetylase inhibitor with the Bcl-2 homology domain-3 mimetic GX15-070 has synergistic antileukemia activity by activating both apoptosis and autophagy. Clin Cancer Res. 2010;16:3923–3932. doi: 10.1158/1078-0432.CCR-10-0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yazbeck V.Y., Buglio D., Georgakis G.V. Temsirolimus downregulates p21 without altering cyclin D1 expression and induces autophagy and synergizes with vorinostat in mantle cell lymphoma. Exp Hematol. 2008;36:443–450. doi: 10.1016/j.exphem.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 67.Sandor V., Senderowicz A., Mertins S. P21-dependent g(1)arrest with downregulation of cyclin D1 and upregulation of cyclin E by the histone deacetylase inhibitor FR901228. Br J Cancer. 2000;83:817–825. doi: 10.1054/bjoc.2000.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ikeda S., Kitadate A., Ito M. Disruption of CCL20-CCR6 interaction inhibits metastasis of advanced cutaneous T-cell lymphoma. Oncotarget. 2016;7:13563–13574. doi: 10.18632/oncotarget.6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kremer K.N., Dinkel B.A., Sterner R.M., Osborne D.G., Jevremovic D., Hedin K.E. TCR-CXCR4 signaling stabilizes cytokine mRNA transcripts via a PREX1-Rac1 pathway: implications for CTCL. Blood. 2017;130:982–994. doi: 10.1182/blood-2017-03-770982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Narducci M.G., Scala E., Bresin A. Skin homing of Sézary cells involves SDF-1-CXCR4 signaling and down-regulation of CD26/dipeptidylpeptidase IV. Blood. 2006;107:1108–1115. doi: 10.1182/blood-2005-04-1492. [DOI] [PubMed] [Google Scholar]
- 71.Maj J., Jankowska-Konsur A.M., Hałoń A., Woźniak Z., Plomer-Niezgoda E., Reich A. Expression of CXCR4 and CXCL12 and their correlations to the cell proliferation and angiogenesis in mycosis fungoides. Adv Dermatol Allergol Dermatol Alergol. 2015;32:437–442. doi: 10.5114/pdia.2015.48034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ito M., Teshima K., Ikeda S. MicroRNA-150 inhibits tumor invasion and metastasis by targeting the chemokine receptor CCR6, in advanced cutaneous T-cell lymphoma. Blood. 2014;123:1499–1511. doi: 10.1182/blood-2013-09-527739. [DOI] [PubMed] [Google Scholar]
- 73.Cho S.-F., Anderson K.C., Tai Y.-T. Targeting B cell maturation antigen (BCMA) in multiple myeloma: potential uses of BCMA-based immunotherapy. Front Immunol. 2018;9:1821. doi: 10.3389/fimmu.2018.01821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lebbink R.J., de Ruiter T., Adelmeijer J. Collagens are functional, high affinity ligands for the inhibitory immune receptor LAIR-1. J Exp Med. 2006;203:1419–1425. doi: 10.1084/jem.20052554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lenting P.J., Westerlaken G.H.A., Denis C.V., Akkerman J.W., Meyaard L. Efficient inhibition of collagen-induced platelet activation and adhesion by LAIR-2, a soluble Ig-like receptor family member. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nordkamp M.J.M.O., van Roon J.A.G., Douwes M., de Ruiter T., Urbanus R.T., Meyaard L. Enhanced secretion of leukocyte-associated immunoglobulin-like receptor 2 (LAIR-2) and soluble LAIR-1 in rheumatoid arthritis: LAIR-2 is a more efficient antagonist of the LAIR-1–collagen inhibitory interaction than is soluble LAIR-1. Arthritis Rheum. 2011;63:3749–3757. doi: 10.1002/art.30612. [DOI] [PubMed] [Google Scholar]
- 77.Vieyra-Garcia P., Crouch J.D., O'Malley J.T. Benign T cells drive clinical skin inflammation in cutaneous T cell lymphoma. JCI Insight. 2019;4 doi: 10.1172/jci.insight.124233. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used for qRT-PCR and amplification of putative regulatory elements used in luciferase assays.
Additional patient clinical information. Romi – Romidepsin; Vori – Vorinostat; Pre/Post-Treatment – paired samples collected pre- and post-HDACi treatment.
Genes differentially expressed in resistant compared to sensitive samples and Romidepsin-treated versus untreated healthy CD4+ T cells.
Gene pathway and ontology enrichment analyses for genes upregulated (adj. p-value ≤0·05) in resistant compared to sensitive samples.
Genes in previously reported HDACi resistance pathways.
Supplementary material