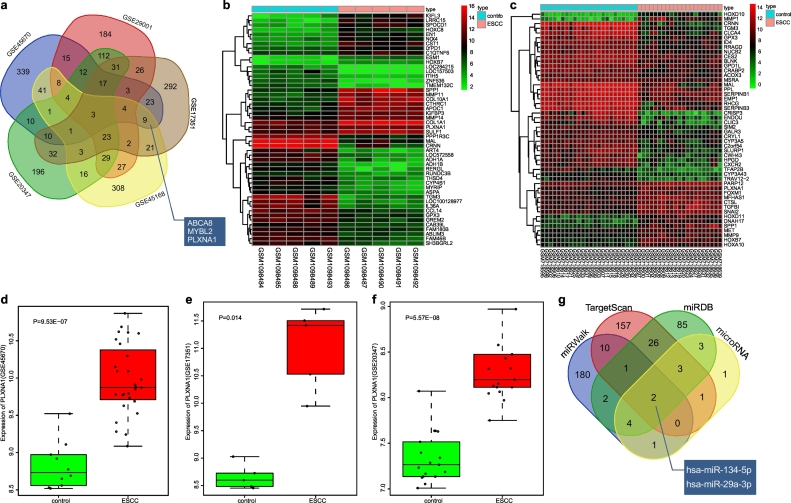
Fig. 1.
MiR-134 is predicted as a candidate miRNA that affects the progression of ESCC through regulation of PLXNA1. (a) the top 500 DEGs determined from GSE17351, GSE20347, GSE29001, GSE45168 and GSE45670 datasets; (b and c) a heat map depicting the top 50 DEGs from GSE45168 and GSE29001 datasets (the x-axis represents the sample number and the y-axis represents the DEGs; the upper right histogram represents the colour gradation; each rectangle in the graph corresponds to one sample's gene expression, red represents high expression and green represents low expression); (d) the expression of PLXNA1 in ESCC in the GSE45670 dataset; (e) the expression of PLXNA1 in ESCC in the GSE17351 dataset; (f) the expression of PLXNA1 in ESCC in the GSE20347 dataset; (g) the top 200 miRNAs predicted from four web-based resources, TargetScan, miRDB, miRWalk and microRNA. miR-134, microRNA-134; ESCC, esophageal squamous cell carcinoma; PLXNA1, plexin A1; MAPK, mitogen-activated protein kinase. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)