Abstract
Background
Twist-related protein 1 (TWIST1) plays an essential role in the carcinogenesis and metastasis of NSCLC. Our aims were to identify the molecule at the downstream of TWIST1 and to evaluate its potential as a diagnostic and a prognostic marker in NSCLC.
Methods
The functional genes at the downstream of TWIST1 were obtained via microarray gene expression analyses in the NSCLC cell line. The expression levels of synaptotagmin 7 (SYT7) in a cohort of patients with NSCLC (n = 154) were examined using immunohistochemistry staining and assessed by Kaplan-Meier survival analysis and Cox regression analysis. The effects of SYT7 on the tumorigenesis and metastasis of NSCLC were measured in NSCLC cells. In vivo xenograft lung cancer models were used to study the tumorigenesis role of SYT7.
Findings
We discovered that SYT7 is significantly altered by TWIST1 expression. We further confirmed that SYT7 protein was significantly higher in NSCLC than that in the adjacent normal lung tissue, and higher SYT7 expression was associated with poor survival of NSCLC patients. The protein level of SYT7 was positively correlated with TWIST1 in NSCLC tissue. Functional experiments indicated that SYT7 promoted proliferation, invasion, and metastasis and inhibited cell apoptosis of NSCLC cells in vitro. In vivo experiments showed that shSYT7 inhibited the xenograft tumor growth of NSCLC cells. Knocking down of SYT7 increased the expression of E-cadherin and decreased the level of N-cadherin and Vimentin in cultured cells.
Interpretation
Our data indicate that SYT7 is an important promoter for EMT and tumor progression in NSCLC.
Fund
This project was supported by grants from the Major Scientific and Technological Innovation Project of Shandong Province (2018CXGC1212), Science and Technology Foundation of Shandong Province (2014GSF118084, 2016GSF121043), Medical and Health Technology Innovation Plan of Jinan City (201805002) and the National Natural Science Foundation of China (81372333).
Keywords: SYT7, NSCLC, TWIST1, Tumorigenesis, Metastasis
Research in context.
Evidence before this study
Metastasis remains a serious problem in the management of non-small cell lung cancer (NSCLC). TWIST1 is associated with tumorigenesis and metastasis in NSCLC, while the mechanism of how TWIST1 regulates the occurrence and development of NSCLC remains unclear.
Added value of this study
Findings from this study indicate that SYT7 is a critical downstream gene of TWIST1. SYT7 promoted the proliferation, invasion, and metastasis and inhibited cell apoptosis of NSCLC cells through promoting EMT. The repression of SYT7 resulted in the dramatic suppression of tumor growth in vivo. SYT7 might be an independent prognostic marker for the prognosis of NSCLC patients.
Implications of all the available evidence
This study provide evidence that SYT7 is a novel oncogene and a diagnostic and a prognostic marker of NSCLC.
Alt-text: Unlabelled Box
1. Introduction
Lung cancer is the leading cause of cancer-related mortality worldwide, and non-small cell lung cancer (NSCLC) accounts for ~85% of all lung cancers [1]. Despite improvements in the chemotherapeutic drugs used over time, the 5-year survival rate of NSCLC patients is only 18% [1]. Although the response rates in subsets of NSCLC with tyrosine kinase receptors (mutant EGFR, ALK, and ROS1) were high, drug resistance has been a major challenge [[2], [3], [4]]. Hence, identification of novel drug targets related to tumor metastasis and their mechanisms of action remains an unmet medical need for the treatment and management of NSCLC.
Epithelial-mesenchymal transition (EMT) is an important process that acts as an indispensable driver for invasion and metastasis of a variety of cancer cells. Important factors driving EMT include many transcription factors, such as the TWIST, SNAIL, ZEB, and PREP families [[5], [6], [7]]. EMT is characterized by decreased expression of epithelial cell markers (for example, E-cadherin and α- and γ-catenin), and an increase in mesenchymal cell markers (for example, vimentin and N-cadherin) [8]. Loss of E-cadherin expression is considered as a key event during EMT [[9], [10], [11]]. EMT-inducing transcription factors possess the ability to repress E-cadherin directly or indirectly [12], as well as to promote the expression of matrix metalloproteases to activate the remodeling of the basement membrane and invasion into surrounding tissues [13].
TWIST1, a crucial EMT-inducing transcription factor, plays an essential role in driving NSCLC tumorigenesis, metastasis, and drug resistance [[14], [15], [16]]. Several studies have reported potential mechanisms of TWIST1 in the EMT process. TWIST1 participates in EMT through regulation by the upstream regulatory molecules such as the AKT families [17,18], Nkx2.8, and VGF [19,20], or regulating the downstream molecules such as p53 and Bmi1 [21,22]. TWIST1 is also involved in oncogenesis, metastasis, drug resistance, and tumor stem cell maintenance in various tumors including head and neck, breast, and prostate cancers [[23], [24], [25], [26]].
While TWIST1 is attracting considerable interest, the precise mechanism of how TWIST1 regulates the occurrence and development of NSCLC remains unclear. Elucidating the downstream target genes or signal pathways by which TWIST1 mediates EMT is critical for understanding the tumor progression process. In the present study, we identified SYT7 as an important downstream effector for TWIST1 function, initially through gene expression analyses, followed by functional studies. Our data may provide an effective therapeutic target for NSCLC.
2. Materials and methods
2.1. Cell lines and culture conditions
The human NSCLC cell lines H1299, H1975, H358, H125, A549 and human embryonic kidney 293T cells were obtained from Cell Bank, Institute of Life Sciences, Chinese Academy of Sciences Cell Bank (Shanghai, China). These NSCLC cells were cultured in RPMI 1640 medium with 10% fetal bovine serum (FBS) under a humidified atmosphere of 37 °C and 5% of CO2.
2.2. Plasmid construction and lentivirus transfection experiment
Lentiviral vectors encoding the human TWIST1 and SYT7 gene were purchased from GeneChem (Shanghai, China) for gain-of-function studies. Silencing of target genes was achieved via lentiviral transduction with the following specific shRNA vectors obtained from GeneChem: DUSP5, TNIP1, TMEM154, CCBE1, CYB5R2, MYEOV, NUAK2, RPS6KL1, PPIF, RIMS2, SYT7, ABHD5, CEP85, LST1, NFKBIE, HYI, POLR3G, SNHG12, UPP1, PXK, TWIST1, and the negative control duplex with a scramble sequence. The transduction processes and the establishment of stable cell lines were executed according to the manufacturer's instructions. The expressions of the vectors were proved by RT-qPCR and Western blotting. Sequences of the interference targets are listed in Supplementary Table S1.
2.3. Cell migration and invasion assays
The upper chambers of Transwell migration chambers (3422, Corning, NY, USA) were placed at 37 °C for 1 h with serum-free medium. After the removal of the medium, the upper chambers were seeded with serum-starved cells suspensions (1 × 105 cells/well). The 10% FBS was added into the lower chambers. As a reference of migration, cells suspensions were also seeded into MTS 96-well plate (G3580, Promega, Madison, USA) and OD570 was measured immediately. The cells from the two groups (control group and experimental group) in chambers were incubated at 37 °C for 24 h. After incubation, the medium and the cells remaining on the upper surface of the membrane were removed with cotton swabs. The cells on the lower surface of the membrane were fixed and stained with Giemsa (32884, Sigma-Aldrich, Taufkirchen, Germany), and then photographed under an inverted microscope. Three visual fields were chosen at random and counted per chamber, and the results were analyzed with Image J and calculated by t-test to determine the statistical significance.
The serum-free medium was placed in the lower and upper chambers of invasion chambers (354,480, Corning). After the removal of the medium, the upper chambers were seeded with serum-starved cells suspensions (1 × 105 cells/well). The 10% FBS was added into the lower chambers. As a reference of invasion, cells suspensions were also seeded into MTS plate (Promega) and OD570 was measured immediately. The cells were incubated at 37 °C for 24 h. Cells that had invaded through Matrigel were visualized with Giemsa (Sigma-Aldrich). Three microscopic fields were photographed and counted per chamber, and the data were analyzed via t-test to determine the statistical significance.
2.4. High-Content Screening (HCS) scratch assay
HCS is based on the Cellomics automatic image acquisition and data analysis system. The images were formed by the fluorescence stimulation of targets and the collection of emitted light. The system analyzed the images to acquire the biological events associated with the optical information, including the coordinate position, signal intensity, temporal information, and their combinations. The biological phenomena such as cell quantity, angiogenesis, and apoptosis were then assessed.
Cell culture and transfection were performed as previously described. Cell suspensions of each group were seeded into triplicate wells of 96-well plates. After scratching using a scratch tester, the plates were washed with serum-free medium and incubated in 0.5% FBS. The cells were photographed at 0 h and then incubated at 37 °C and 5% of CO2. According to the degree of healing, the plates were scanned and the migration distances were analyzed by Cellomics (ArrayScan VT1, Thermo Fisher Scientific).
2.5. Microarray and computational analysis
Total RNA was extracted and purified as described in Supplementary Materials and Methods. All RNAs were labeled using a GeneChip 3′IVT Expression Kit and Hybridization Wash and Stain Kit (Affymetrix, Santa Clara, CA, USA) and analyzed using Affymetrix GeneChip Human Genome U133 plus 2.0 arrays (Affymetrix). The gene expression levels were normalized as log 2 values using GeneSpring software (Agilent Technologies, Palo Alto, CA, USA). Genes that were up- or down-regulated with >1.3 or 2-Fold Change (FC), and a significant difference of P < .05 was further subjected to computational simulation by Ingenuity Pathway Analysis (IPA; QIAGEN, Valencia, CA, USA) online tools to perform an enrichment analysis.
2.6. Dual-luciferase reporter assays
Luciferase reporters and expression plasmids were constructed using GeneChem. Cell culture and resuspension of 293T cells were performed as previously described. The cells were seeded into 24-well plates (1 × 105 cells/well) and assigned to 4 groups: TWIST1 -Normal Control (NC) + luc-NC; TWIST1-NC + luc-SYT7; TWIST1-OE + luc-NC and TWIST1-OE + luc-SYT7. Plasmids were co-transfected using X-tremegene HP DNA transfection reagents (06366236001, Roche, Basel, Switzerland). To normalize each transfection reaction, a Renilla reporter plasmid (0.02 μg) was co-transfected with the luciferase reporters (1 μg). The cells were harvested at 48 h post-transfection and lysed, and promoter activity was assayed using the Dual-Luciferase Reporter Assay System (E2910, Promega) on a GloMax 20/20 Luminometer (E5311, Promega) according to the manufacturer's recommendations.
2.7. Immunohistochemistry
Immunohistochemistry (IHC) was conducted on paraffin-embedded sections. Before staining, paraffin was removed from the slides by incubation at 65 °C for 60 min and treatment with dimethylbenzene two times for 10 min. The tissue sections were then rehydrated by immersion in ethanol-series with descending concentrations (100%, 100%, 95%, 80% and 70%) for 5 min each and in deionized water for 2 min, followed by microwave antigen retrieval using citrate. Then endogeneous peroxidase was blocked in 3% H2O2 for 15 min and in 5% BSA for 45 min. Afterwards, the slides were incubated overnight at 4 °C with primary rabbit anti-SYT7 antibody (1:100 dilution in PBS, ab121383, polyclonal; Abcam, Cambridge, UK) and anti-TWIST1 antibody (1:200 dilution in PBS, ab50581, polyclonal; Abcam, Cambridge, UK), respectively. After incubation with the second antibody and SABC of HRP conjugated anti-Rabbit IgG SABC Kit (SA1022, BosterBio, Pleasanton, CA, USA) for 30 min each, and chromogen development was performed using a DAB Chromogenic Substrate Kit (AR1022, BosterBio). The slides were stained with hematoxylin and covered with coverslips using neutral balsam. Negative controls were treated with PBS instead of the primary antibody; the other steps remained the same. The paraffin-embedded prostate cancer tissues were used as positive control and treated with the same steps as the NSCLC SYT7 staining.
An intensity score of 0 to 3 was allocated to the intensity of tumor cells (0, none; 1, weak; 2, moderate; and 3, strong). A proportional score was obtained by the estimated proportion of positive tumor cells in percentage. To assess the average degree of staining within a slide, multiple regions were randomly selected and analyzed, and at least 100 tumor cells were assessed. The cytoplasmic expression levels were assessed by the H-score system using the following formula: H-score = ∑(Pi×I) = (percentage of cells of weak intensity ×1) + (percentage of cells of moderate intensity ×2) + (percentage of cells of strong intensity ×3). In the formula, Pi represents the percentage of stained tumor cells and I represents the intensity of staining, producing cytoplasm or nuclear scores ranging from 0 to 300. The H-score above or below the median classified the tumor samples into a high expression group or a low expression group. The scoring was independently assessed by two investigators who were unaware of the clinical outcomes.
2.8. Clinical information and tissue samples
This study was conducted with the approval of the ethical committees of Qilu Hospital of Shandong University (Jinan, China). The patients or their relatives were informed that paraffin sections of resected specimens were used for scientific research. A total of 154 cancer tissue and 136 paracancerous tissue from patients with NSCLC who underwent pneumonectomy were used for the detection of SYT7 and TWIST1 expression by IHC. Clinical information including basic information, pathological type, tumor location, tumor node metastasis (TNM) stage (according to the eighth edition), and prognosis were collected. The overall survival (OS) time was calculated from the surgery date to the end of follow-up or the date of death.
2.9. In vivo tail vein xenograft lung cancer models
All of the surgical procedures and care given to the animals were in accordance with the institutional guidelines. Female BALB/c-nude mice (5 weeks of age) were purchased from Lingchang Biotech (Shanghai, China). A total of 20 nude mice were randomly assigned to a control group and an experimental group, with 10 mice in each group. Luciferase-labeled A549 shControl and A549 shSYT7 cells (2 × 106/per mouse) were injected into the tail vein of each nude mouse in two groups, respectively. An in-vivo imaging system (Lumina LT, Perkin Elmer) was used once a week to observe cell vaccination and metastasis in the mice. The tumor-bearing mice were sacrificed 38 days after inoculation. Tumor volume was calculated as follows: V (volume) = (length × width2)/2. The tumors were harvested and frozen at −80 °C at the end of the experiments for our next studies. All of the animal procedures were approved by the Ethics Committee of Qilu Hospital of Shandong University (KYLL-2013-097; February 25, 2014).
2.10. Bioinformatics analysis
RNA-Seq microarray gene expressions of TWIST1 and SYT7 in 21 NSCLC cell lines (LK2, NCIH1155, NCIH1755, NCIH2106, NCIH1693, NCIH522, SCLC21H, A427, NCIH520, NCIH23, NCIH1650, CORL47, EPLC272H, NCIH2444, NCIH2009, HCC95, NCIH2085, RERFLCSQ1, NCIH322, NCIH1573, HCC1171) were downloaded from Cancer Cell Line Encyclopedia (CCLE) [27]. Robust Multi-array Average (RMA) normalization was performed. Correlation between TWIST1 and SYT7 expression was analyzed by Spearman's rank correlation test.
The differential expression of SYT7 between lung adenocarcinoma (LUAD) and normal lung tissues, as well as lung squamous cell carcinoma (LUSC) and normal lung tissues, were verified using TCGA data by GEPIA online analysis tool (http://gepia.cancer-pku.cn/) [28]. The following settings were used for the expression analysis: ‘Boxplot’; ‘Gene = SYT7’; ‘|Log2FC| Cutoff = 1’; ‘P-value Cutoff = 0.01’; ‘Datasets = LUSC, LUAD’; ‘Log Scale = Yes’; ‘Jitter Size = 0.4’; and ‘Match TCGA normal data’. The relationship of SYT7 and TWIST1 mRNA expression in NSCLC was also analyzed using GEPIA by Pearson's correlation analyses. The following settings were used for the correlation analyses: ‘Correlation’; ‘Gene A = SYT7’; ‘Gene B = TWIST1’; ‘Correlation Coefficient = Spearman’; ‘Datasets = TCGA Tumor, TCGA Normal’.
The correlation of individual SYT7 mRNA expression with OS was analyzed using an online database [29] that was established using gene expression data and survival information of lung cancer patients and downloaded from the Gene Expression Omnibus (GEO). SYT7 was entered into the database called the Kaplan-Meier (K-M) Plotter (http://kmplot.com/analysis/index.php?p=service&cancer=lung) to obtain KM survival plots. The mRNA expression of SYT7 above or below the median classified the cases into a high expression group and a low expression group, respectively. These cohorts were compared with a Kaplan-Meier survival plot. Hazard ratios (HR), 95% confidence intervals (CIs), and log-rank P values were determined and displayed on the webpage.
2.11. Statistical analysis
The quantitative data are shown as the mean ± standard deviation (SD). The significance of a difference between the groups was tested using Student's t-test for the data from the cell and animal experiments. Mann-Whitney U test was used for comparison between two groups not normally distributed having quantitative variables. Correlation between the TWIST1 and SYT7 protein levels in NSCLC patient tissue was analyzed by chi-square (χ2) test, with R representing the correlation coefficient. The clinical variables between the groups were compared using the χ2 test. OS was calculated from the data from the lung cancer diagnosis to death from any cause or was censored at the last follow-up data. The OS rate was analyzed using Kaplan-Meier method with the log-rank test. The Univariate Cox regression proportional hazards model was performed to estimate the effect on OS. The variables with a P-value < .1 in the univariate analysis were further analyzed by a multivariate analysis of the prognostic factors. All of the analyses were performed using SPSS 21.0 software (IBM Corporation, Armonk, NY, USA). Differences with a P-value < .05 were considered statistically significant. More information about the methods, including the real-time quantitative reverse transcription PCR (RT-qPCR), Western blotting, scratch assay, and the cell proliferation and apoptosis assays is provided in the Supplementary Materials and Methods.
3. Results
3.1. Establishment of TWIST1-OE H1975 cells
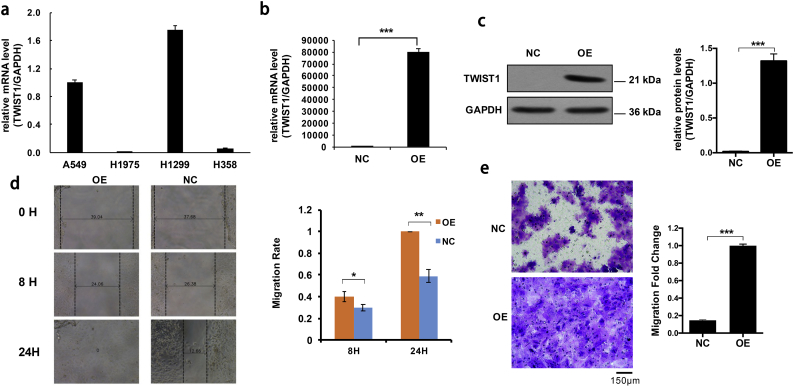
With H1975 cells showing a low TWIST1 transcript level in comparison with three other cell lines, (H1299, A549, and H358) (Fig. 1a), we ectopically expressed TWIST1 in H1975 cells using the lentivirus expression system. >80% of the cells had fluorescent protein expression 72 h after virus infection. We confirmed higher levels of TWIST1 transcript and TWIST1 protein in TWIST1-OE H1975 cells (Fig. 1b and c). Next, we performed cell migration experiments on the TWIST1-OE H1975 cells, and found them to be more aggressive than the cells expressing only the empty vector (Fig. 1d). Similarly, the TWIST1-OE H1975 cells were more invasive than the control cells in a transwell migration assay (Fig. 1e).
Fig. 1.
The expression and function of TWIST1 in NSCLC cell lines. a, The mRNA expression levels of TWIST1 in four NSCLC cell lines A549, H1975, H1299, and H358. b and c, The mRNA and protein expression of TWIST1 in control cells and TWIST1-OE H1975 cells. d and e, Scratch test and transwell migration test in control cells and TWIST1-OE H1975 cells. All of the data are expressed as mean values ± SEM (n = 3); ⁎P < .05, ⁎⁎P < .01, ⁎⁎⁎P < .001 (Student's t-test).
3.2. Expression of SYT7 was significantly altered by TWIST1 in the microarray analysis and HCS experiments
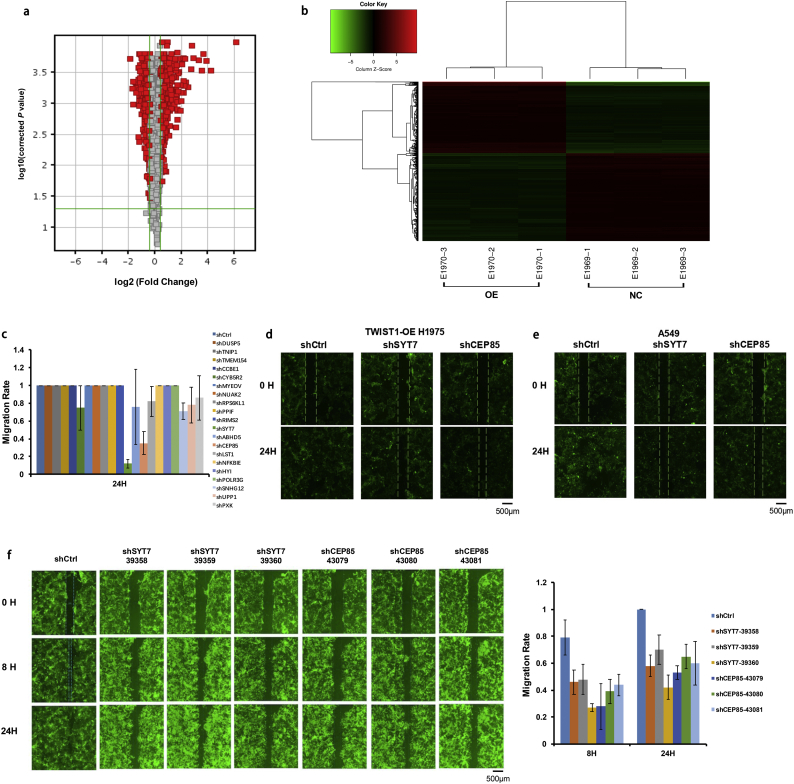
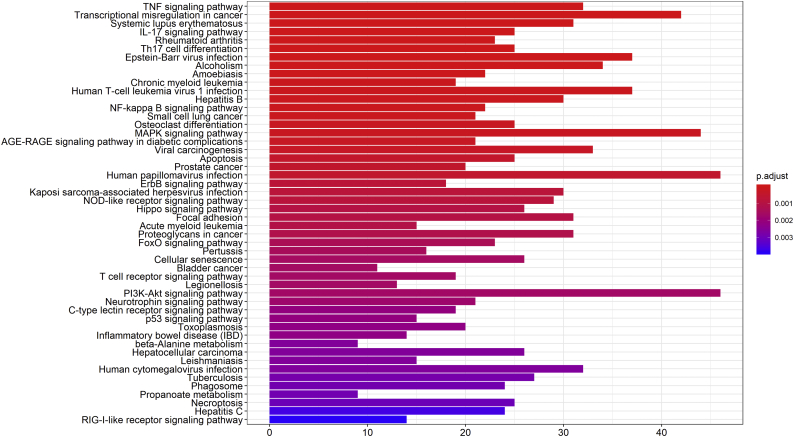
To explore the downstream genes of TWIST1 in NSCLC cells, we used the TWIST1-OE H1975 cells and control-H1975 cells for a whole genome expression profiling microarray analysis. A total of 1297 differentially expressed genes (DEGs) were identified, with 589 DEGs upregulated and 708 DEGs downregulated (Fig. 2a–b, Table S2). Pathway enrichment analysis showed that these DEGs were primarily involved in the following signaling pathways: TNF pathway, NF-κB pathway, MAPK pathway, PI3K/AKT pathway, p53 pathway, Hippo pathway, JAK/STAT regulatory pathway, and so on (Table S3 and Fig. S1). We selected 28 significant DEGs through a literature search and pathway analysis. They were relatively novel molecules and had the potential to regulate lung cancer. We validated these 28 DEGs using RT-qPCR, and found a total of 20 highly expressed DEGs in TWIST1-OE H1975 cells (Table S4).
Fig. 2.
The selection of TWIST1-downstream genes. a and b, Volcano Plots and Cluster Diagram of differentially expressed genes between TWIST1-OE H1975 cells and control cells. c and d, The HCS experiment and the migration distance ratio of 20 genes in TWIST1-OE H1975 cells. e, The suppression of the migration abilities of A549 cells by SYT7 and CEP85 knockdown. f, Representative photographs of the SYT7 and CEP85 groups and the control group and the comparison of the migration distances between the SYT7 or CEP85 groups with the control group. All of the data are expressed as mean values ± SEM (n = 3).
Next, we constructed 3 RNA interference (RNAi) targets for each gene and used a mixture of 3 plasmids for each gene in equal ratios for lentiviral packaging. As shown in Fig. 2c–d, HCS experiments in TWIST1-OE H1975 cells showed that the most significant disruption of cell migration was from SYT7 gene knockdown (0.12, P < .05 in comparison with the control group, and Student's t-test) and CEP85 gene knockdown (0.35, P < .05 in comparison with the control group, and Student's t-test). The suppression of cell migration by SYT7 and CEP85 gene knockdown was also demonstrated in A549 cells (0.28, P < .05; 0.57, P < .05, respectively; Student's t-test, Fig. 2e). We further validated the effects using the TWIST1-OE H1975 cells expressing a single shRNA for each gene (with three separate shRNAs for each gene). The results showed that each shRNA was sufficient to inhibit cell migration, further confirming that SYT7 and CEP85 promote cell migration in the TWIST1-OE H1975 cells (Fig. 2f). Of the two genes, the knockdown of SYT7 inhibited cell migration more strongly, so we chose SYT7 for the following functional study.
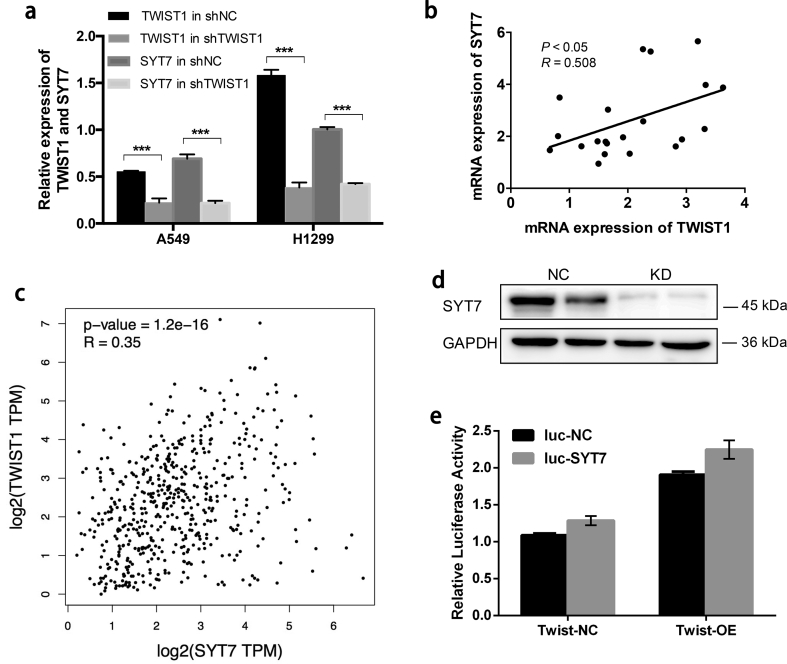
To further confirm whether TWIST1 can regulate the mRNA expression of SYT7, knockdown of TWIST1 was performed via lentiviral transduction with shTWIST1 vectors. In A549 and H1299 cells, we analyzed the relative mRNA expression of SYT7 in the shTWIST1 and shCtrl cells, respectively. The results showed that SYT7 was significantly downregulated after the knockdown of TWIST1 (P < .001, Student's t-test, Fig. S2a).
3.3. Association of SYT7 expression with TWIST1 and the clinicopathological parameters of NSCLC
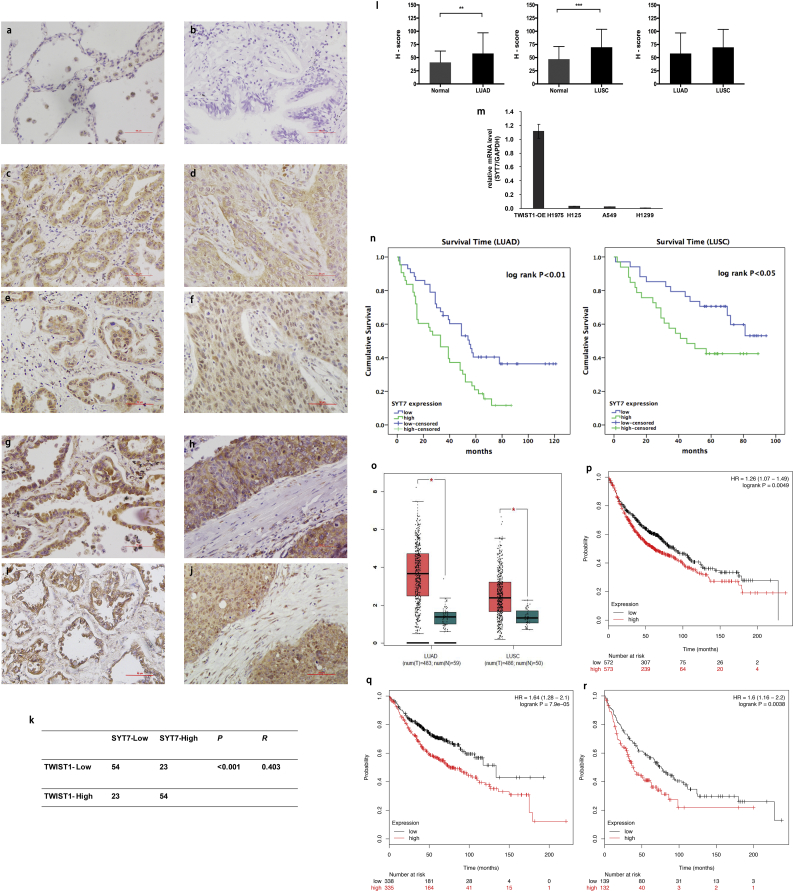
The relevance of our studies was evaluated in NSCLC lung tissues form human specimens. We detected the expressions and subcellular localization of SYT7 and TWIST1 protein by IHC in NSCLC tissues (N = 154) and adjacent normal tissues (N = 136) of patients (Fig. 3a–j). Positive SYT7 staining was mainly detected in cellular membrane and cytoplasm in NSCLC tissues (Fig. 3c–d and 3g–h). The expressions of SYT7 protein in the tumor tissues of LUAD (n = 86) and LUSC (n = 68) patients were significantly higher than in the adjacent normal tissues, respectively (P < .01, Mann-Whitney U test, Fig. 3l). To confirm the association between SYT7 and TWIST1 protein expression in NSCLC patients, we also performed IHC staining of TWIST1 in those 154 NSCLC tissues. Positive TWIST1 staining was mainly detected in the cell nucleus and cytoplasm in NSCLC (Fig. 3e–f, i–j). The correlation between the SYT7 and TWIST1 protein levels was significantly positive (P < .001, R = 0.403, Pearson's χ2 test). Overall, 70.1% (54 of 77) of the tumor samples exhibited the same staining intensity of TWIST1 and SYT7 protein (Fig. 3k).
Fig. 3.
The expression and correlation of SYT7 and TWIST1 in NSCLC tissue determined by IHC and survival analyses and the expression of SYT7 in NSCLC cell lines. a and b, Negative SYT7 protein expression in the normal tissues adjacent to the non-cancerous tissue samples. c, Low expression of SYT7 protein in the adenocarcinoma tissues. d, Low expression of SYT7 protein in the squamous cell carcinoma tissues. e and f, Expression of TWIST1 protein in the tumor tissue shown in c and d, respectively. g, High expression of SYT7 protein in the adenocarcinoma tissues. h, High expression of SYT7 protein in the squamous cell carcinoma tissues. i and j, Expression of TWIST1 protein in the tumor tissue shown in g and h, respectively; a-j were ×400 magnification (scale bar = 50 μm). k, Correlation between TWIST1 and SYT7 protein levels in NSCLC (P < .001, R = 0.403, Pearson's χ2 test). l, Semiquantitative analyses of the SYT7 protein expression in the NSCLC and normal tissues adjacent to the non-cancerous tissue samples (⁎⁎P < .01, ⁎⁎⁎P < .001, Mann-Whitney U test). m, The mRNA expression levels of SYT7 in four NSCLC cell lines TWIST1-OE H1975, H125, A549, and H1299. n, Kaplan-Meier survival curves indicated that the protein expression of SYT7 was significantly associated with the OS rate of the clinical LUAD and LUSC patients (Kaplan-Meier method with log-rank test). o, SYT7 was highly expressed in LUAD and LUSC compared with normal lung tissue in the TCGA database (⁎P < .05, one-way ANOVA test). p, The OS was lower in the patients with a high expression of SYT7 than in those with a low expression of SYT7 in all of the NSCLC patients in the K-M Plotter database (Kaplan-Meier method with log-rank test). q and r, OS was lower in the patients with a high expression of SYT7 than in those with low a expression of SYT7 in LUAD and LUSC patients in the K-M Plotter database (Kaplan-Meier method with log-rank test). The data in i and n are expressed as mean values ± SEM (n = 3).
To validate the correlation between TWIST1 and SYT7 mRNA expression, we performed Spearman's correlation analysis of TWIST1 and SYT7 mRNA expression (RNASeq) in 21 NSCLC cell lines on CCLE. There was a significantly positive correlation between TWIST1 and SYT7 mRNA expression (P < .05, R = 0.508, Spearman's rank correlation test, Fig. S2b). Correlation analyses in clinical LUAD and LUSC of TCGA were also analyzed, showing a positive correlation between SYT7 and TWIST1 expression in the LUSC tumor and normal samples (P < .001, R = 0.35, Spearman's rank correlation test, Fig. S2c).
The mRNA expression of SYT7 in four cell lines (A549, H1299, TWIST1-OE H1975 and H125) were detected by RT-qPCR. As shown in Fig. 3m, SYT7 was highly expressed in the TWIST-OE H1975 cells and moderately expressed in the other three lung cancer cells. The association between SYT7 protein expression and clinicopathological variables of the LUAD and LUSC patients is summarized in Table 1. In the LUAD patients, the high expression of SYT7 was significantly associated with gender (P = .03, Pearson's χ2 test), differentiation grade (P = .033, Pearson's χ2 test), and TNM stage (P = .03, Pearson's χ2 test). In the LUSC patients, there was no association between SYT7 and the clinicopathological variables (P > .05, respectively, Pearson's χ2 test).
Table 1.
Correlation of SYT7 expression in the tumorous tissues with clinicopathologic characteristics in the LUAD and LUSC patients.
| Clinicopathologic characteristic | SYT7 expression |
|||||
|---|---|---|---|---|---|---|
| n | Low expression | High expression | Pearson χ2 | P | ||
| LUAD | Gender | 4.715 | 0.030⁎ | |||
| Male | 48 | 19 (39.6) | 29 (60.4) | |||
| Female | 38 | 24 (63.2) | 14 (36.8) | |||
| Age at diagnosis (years) | 0.189 | 0.664 | ||||
| ≤60 | 38 | 20 (52.6) | 18 (47.4) | |||
| >60 | 48 | 23 (47.9) | 25 (52.1) | |||
| Smoking status | 0.422 | 0.516 | ||||
| No smoking | 39 | 18 (46.2) | 21 (53.8) | |||
| Smoking | 47 | 25 (53.2) | 22 (46.8) | |||
| Primary tumor location | 0.048 | 0.826 | ||||
| Left lung | 35 | 17 (48.6) | 18 (51.4) | |||
| Right lung | 51 | 26 (51.0) | 25 (49.0) | |||
| Differentiation grade | 4.568 | 0.033⁎ | ||||
| High and Middle | 61 | 35 (57.4) | 26 (42.6) | |||
| Low | 25 | 8 (32.0) | 17 (68.0) | |||
| Primary tumor size | 10.676 | 0.005# | ||||
| T1 | 18 | 13 (72.2) | 5 (27.8) | |||
| T2 | 47 | 16 (34.0) | 31 (66.0) | |||
| T3 + T4 | 28 | 21 (66.7) | 7 (33.3) | |||
| Lymph node metastasis | 1.675 | 0.196 | ||||
| N0 | 42 | 24 (57.1) | 18 (42.9) | |||
| N1-3 | 44 | 19 (43.2) | 25 (56.8) | |||
| Stage Grouping with TNM | 4.715 | 0.030⁎ | ||||
| Stage I + II | 48 | 29 (60.4) | 19 (39.6) | |||
| Stage III + IV | 38 | 14 (36.8) | 24 (63.2) | |||
| LUSC | Gender | 0.249 | 0.618 | |||
| Male | 42 | 20 (47.6) | 22 (52.4) | |||
| Female | 26 | 14 (53.8) | 12 (46.2) | |||
| Age at diagnosis (years) | 0.073 | 0.787 | ||||
| ≤60 | 19 | 9 (47.4) | 10 (52.6) | |||
| >60 | 49 | 25 (51.0) | 24 (49.0) | |||
| Smoking status | 0.971 | 0.324 | ||||
| No smoking | 28 | 12 (42.9) | 16 (57.1) | |||
| Smoking | 40 | 22 (55.0) | 18 (45.0) | |||
| Primary tumor location | 0.553 | 0.457 | ||||
| Left lung | 27 | 15 (55.6) | 12 (44.4) | |||
| Right lung | 41 | 19 (46.3) | 22 (53.7) | |||
| Differentiation grade | 0.302 | 0.582 | ||||
| High and Middle | 50 | 26 (52.0) | 24 (48.0) | |||
| Low | 18 | 8 (44.4) | 10 (55.6) | |||
| Primary tumor size | 0.657 | 0.417 | ||||
| T1 + T2 | 49 | 23 (46.9) | 26 (53.1) | |||
| T3 + T4 | 19 | 11 (57.9) | 8 (42.1) | |||
| Lymph node metastasis | 3.010 | 0.083 | ||||
| N0 | 41 | 24 (28.5) | 17 (41.5) | |||
| N1-3 | 27 | 10 (37.0) | 17 (63.0) | |||
| Stage Grouping with TNM | 0.078 | 0.779 | ||||
| Stage I + II | 51 | 26 (51.0) | 25 (49.0) | |||
| Stage III + IV | 17 | 8 (47.1) | 9 (52.9) | |||
LUAD: lung adenocarcinoma; LUSC: lung squamous cell carcinoma; TNM: tumor node metastasis.
P < .05, χ2 test.
A difference in expression was only seen in T2 tumor while T1 and T3–T4 had the same percentage of staining.
The univariate and multivariate analyses of prognostic variables for the survival rate of LUAD and LUSC are shown in Table 2. In the LUAD patients, univariate Cox regression analyses revealed that the overexpression of SYT7 (HR 1.920, 95% CI 1.158–3.186, P = .012) was a significant negative prognostic factor for OS. Multivariate analysis showed that SYT7 overexpression remained an independent prognosis factor (HR 1.823, 95% CI 1.009–3.292, P = .046) of OS after adjusting for TNM stage, lymph node metastasis, and gender. The Kaplan–Meier survival curves confirmed that OS was negatively associated with high SYT7 protein expression (P < .01, log-rank test, Fig. 3n).
Table 2.
Univariate and multivariate analyses of the prognostic factors of overall survival in the LUAD and LUSC patients.
| Characteristic | Univariate analysis |
Multivariate analysis |
|||||
|---|---|---|---|---|---|---|---|
| HR | P | 95% CI | HR | P | 95% CI | ||
| LUAD | SYT7 expression | 1.920 | 0.012⁎ | 1.158–3.186 | 1.823 | 0.046⁎ | 1.009–3.292 |
| Low vs High | |||||||
| Gender | 0.639 | 0.082 | 0.386–1.058 | ||||
| Male vs Female | |||||||
| Age at diagnosis (years) | 1.042 | 0.871 | 0.534–1.713 | ||||
| ≤60 vs >60 | |||||||
| Smoking status | 1.598 | 0.774 | 1.023–1.984 | ||||
| No smoking vs Smoking | |||||||
| Primary tumor location | 1.077 | 0.772 | 0.653–1.776 | ||||
| Left lung vs Right lung | |||||||
| Differentiation grade | 0.871 | 0.622 | 0.664–1.985 | ||||
| High and Middle vs Low | |||||||
| Primary tumor size | 1.987 2.455 |
0.070 0.032 |
0.946–4.173 1.082–5.571 |
||||
| T1 vs T2 vs T3 + T4 | |||||||
| Lymph node metastasis | 2.483 | 0.001⁎⁎ | 1.483–4.157 | ||||
| N0 vs N1–3 | |||||||
| Stage Grouping with TNM | 3.423 | <0.001⁎⁎⁎ | 1.860–6.299 | 3.202 | <0.001⁎⁎⁎ | 1.735–5.909 | |
| I + II vs III + IV | |||||||
| LUSC | SYT7 expression | 2.067 | 0.044⁎ | 1.020–4.188 | 2.329 | 0.022⁎ | 1.132–4.788 |
| Low vs High | |||||||
| Gender | 0.975 | 0.942 | 0.448–1.946 | ||||
| Male vs Female | |||||||
| Age at diagnosis (years) | 2.397 | 0.072 | 0.924–6.217 | 2.736 | 0.040⁎ | 1.047–7.153 | |
| ≤60 vs >60 | |||||||
| Smoking status | 0.684 | 0.296 | 0.335–1.395 | ||||
| No smoking vs Smoking | |||||||
| Primary tumor location | 0.803 | 0.533 | 0.403–1.602 | ||||
| Left lung vs Right lung | |||||||
| Differentiation grade | 1.410 | 0.365 | 0.670–2.966 | ||||
| High and Middle vs Low | |||||||
| Primary tumor size | 1.333 | 0.436 | 0.646–2.751 | ||||
| T1 + T2 vs T3 + T4 | |||||||
| Lymph node metastasis | 0.989 | 0.984 | 0.334–2.927 | ||||
| N0 vs N1–3 | |||||||
| Stage Grouping with TNM | 1.950 | 0.074 | 1.064–4.685 | ||||
| I + II vs III + IV | |||||||
LUAD: lung adenocarcinoma; LUSC: lung squamous cell carcinoma; HR: hazard ratio; CI: confidence interval; TNM: tumor node metastasis.
⁎P < .05, ⁎⁎P < .01, ⁎⁎⁎P < .001, the univariate and multivariate Cox regression analysis.
In the LUSC patients, univariate Cox regression analyses suggested that the overexpression of SYT7 (HR 2.067, 95% CI 1.020–4.188, P = .044) was a significant negative prognostic factor for OS. Multivariate analysis showed that SYT7 overexpression remained the independent prognosis factor (HR 2.329, 95% CI 1.132–4.788, P = .022) after adjusting age at diagnosis and TNM stage. The Kaplan–Meier survival curves also confirmed that the OS of the LUSC patients gradually declined with the increasing SYT7 expression (P < .05, log-rank test, Fig. 3n).
3.4. Differential expression of SYT7 mRNA in the TCGA database and survival analysis in the K-M Plotter database
The data of SYT7 mRNA expression, which contained 483 cases of LUAD, 486 cases of LUSC, and 59 cases of normal lung tissues, were obtained from the TCGA database for analyses using the GEPIA online tool. As shown in Fig. 3o, SYT7 was highly expressed in both LUAD and LUSC compared with normal lung tissue, respectively (P < .05, one-way ANOVA).
The association of SYT7 mRNA expression with prognosis was evaluated using the K-M Plotter database. From a total of 1926 NSCLC patients, poor OS was associated high expression of SYT7 (HR = 1.26, P < .01, Kaplan-Meier, and the log-rank test; Fig. 3p). For the LUAD patients (n = 720), poor OS was also associated with high expression of SYT7 (HR = 1.64, P < .001, Kaplan-Meier, and the log-rank test; Fig. 3q). A similar result was observed in the LUSC patients (n = 524) (HR = 1.6, P < .01, Kaplan-Meier and log-rank test; Fig. 3r). These data suggest that the high expression of SYT7 indicated a poor prognosis for the NSCLC patients, which is in accordance with our clinical prognosis analyses.
3.5. Knockdown of SYT7 inhibited metastasis, invasion, proliferation, and markers of EMT and increased apoptosis in vitro
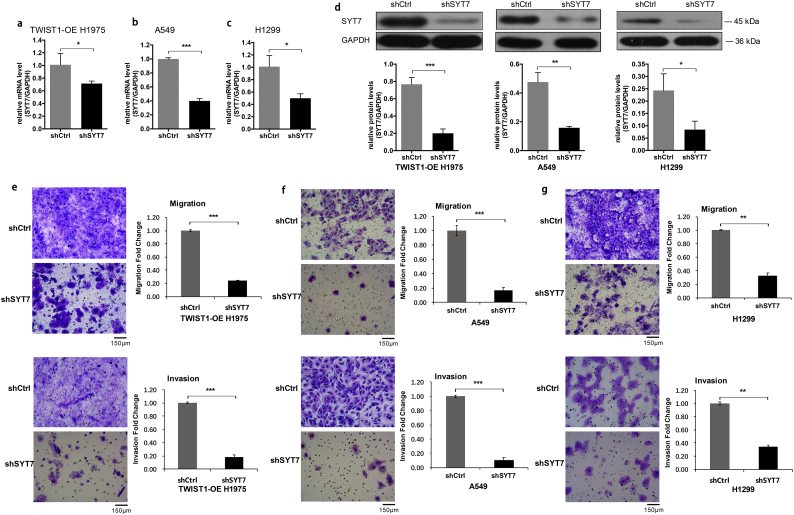
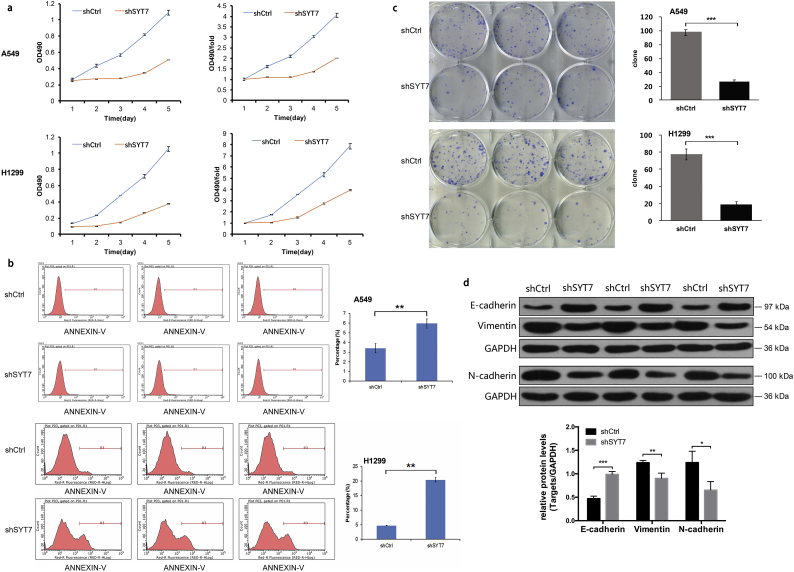
To evaluate the impact of SYT7 on the invasion and metastasis of the NSCLC cell lines, the TWIST1-OE H1975 cells, H1299 cells, and A549 cells were infected with shCtrl and shSYT7 lentivirus, respectively. After shSYT7 lentivirus infection, the expression levels of SYT7 mRNA were significantly decreased in the shSYT7 group of TWIST1-OE H1975 cells, H1299 cells, and A549 cells. The knockdown efficiency reached 38%, 50.1%, and 59.9%, respectively (all P < .05, Student's t-test; Fig. 4a–c). The expressions of SYT7 protein also significantly decreased in these three cell lines (Fig. 4d).
Fig. 4.
The repression of SYT7 in regulating metastasis and invasion in vitro. a-d, The expression level of SYT7 mRNA and protein were significantly decreased in the shSYT7 group of the TWIST1-OE H1975, H1299, and A549 cells. e, Knockdown of SYT7 inhibited the metastasis and invasion abilities of the TWIST1-OE H1975 cells. f and g, The metastasis and invasion abilities in the shSYT7 group of A549 and H1299 cells were significantly inhibited in the transwell migration and invasion experiments. All of the data are expressed as mean values ± SEM (n = 3); ⁎P < .05, ⁎⁎P < .01, ⁎⁎⁎P < .001 (Student's t-test).
In the TWIST1-OE H1975 cells, the transwell migration and invasion experiments showed that shSYT7 significantly inhibited migration and invasion (P < .001, Student's t-test; Fig. 4e). Similar results were also observed in A549 and H1299 cells following shSYT7 expression (P < .001 and P < .01, respectively, Student's t-test; Fig. 4f and g). To evaluate the effect of SYT7 on the proliferation and apoptosis of NSCLC, an MTT proliferation assay and FACS apoptosis assay were performed in A549 and H1299 cells. We found that shSYT7 expression also reduced cell proliferation (to obtain the exact number in the results plus the P-values) (P < .001, Student's t-test; Fig. 5a), suggesting that SYT7 had a significant effect on the proliferation of A549 and H1299 cells. When compared with the shCtrl group, the apoptotic rates of A549 and H1299 cells in the shSYT7 group significantly increased (P < .01, Student's t-test; Fig. 5b), suggesting that the knockout of SYT7 significantly increased apoptosis in A549 and H1299 cells. Using a colony formation assay, the numbers of colony cells in the shSYT7 group significantly declined compared with the shCtrl group (P < .01, Student's t-test; Fig. 5c).
Fig. 5.
Repression of SYT7 in regulating proliferation, apoptosis, and EMT markers in NSCLC cell lines. a, The proliferation ability of the shSYT7 group was significantly inhibited in A549 and H1299 cells. b, The apoptotic rates in the shSYT7 groups significantly increased. c, The numbers of colony cells in the shSYT7 groups significantly decreased. d, The EMT-related markers in H1299 cells. All of the data are expressed as mean values ± SEM (n = 3); ⁎P < .05, ⁎⁎P < .01, ⁎⁎⁎P < .001 (Student's t-test).
We further detected the expression of EMT-related markers in the shCtrl and shSYT7 group of the H1299 cells (Fig. 5d). The results showed that the expression of E-cadherin was significantly increased after the knockdown of SYT7. Meanwhile, the expressions of N-cadherin and Vimentin were significantly decreased. These results indicated that the knockdown of SYT7 significantly inhibited EMT of H1299 cells.
3.6. Overexpression of SYT7 promoted cell proliferation, migration and invasion, and inhibited apoptosis in vitro
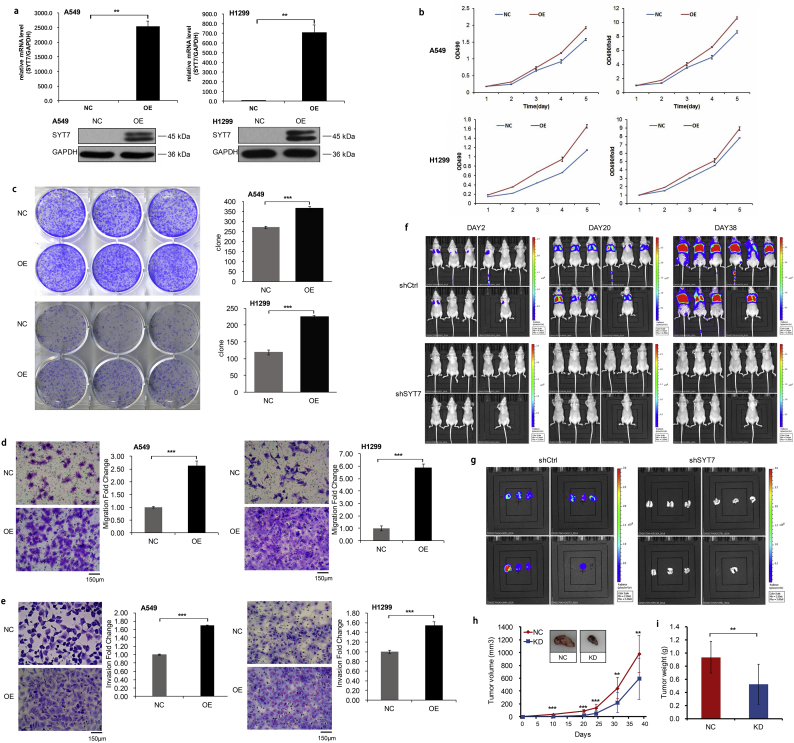
To confirm our data from the gene knockdown, we performed SYT7 overexpression in A549 and H1299 cells by lentivirus infection, and the overexpression efficiency was then confirmed by RT-qPCR and Western blotting (Fig. 6a). Compared with the negative control cells, the SYT7-OE groups had higher levels of cell proliferation at different time points by MTT assay in both A549 and H1299 cells (P < .001, Student's t-test; Fig. 6b). Using the colony formation assay, the numbers of colony cells in the SYT7-OE groups significantly increased compared with the control cells (P < .001, Student's t-test; Fig. 6c). We further explored the effects of the SYT7 overexpression on the invasion and migration of A549 and H1299 cells. The migration assay revealed that the cell motility was significantly increased in the SYT7-OE cells (P < .001, Student's t-test; Fig. 6d). Similarly, the invasion assay revealed that the invasiveness was significantly elevated in the SYT7-OE groups of A549 and H1299 cells (P < .001, Student's t-test; Fig. 6e).
Fig. 6.
Overexpression of SYT7 in vitro and repression of SYT7 in vivo. a, Overexpression efficiency in A549 and H1299 cells confirmed by RT-qPCR and Western blotting. b, MTT assay in the SYT7-OE groups and control groups. c, Colony formation assay. d, Transwell migration assay. e, Invasion assay. f and g, The amount of fluorescence expression in the nude mice model and their lung tissues. h and i, The volume and weight of the tumors formed by the SYT7 knockdown A549 cells and NC A549 cells. All of the data are expressed as mean values ± SEM; ⁎⁎P < .01, ⁎⁎⁎P < .001 (Student's t-test).
3.7. Knockdown of SYT7 inhibited tumorigenesis in vivo
To determine the effect of SYT7 on NSCLC cells tumorigenesis in vivo, we performed an experimental model of nude mice tail vein inoculation tumorigenicity. At 2 days, 10 days, 20 days, 24 days, 31 days, and 38 days after inoculation of A549 cells, the lung colonization of these cells in the A549-shCtrl and A549-shSYT7 groups was monitored and quantitatively measured using a non-invasive bioluminescence system. The results showed that the amount of fluorescence expression in the shSYT7 group significantly reduced compared with the shCtrl group (P < .05, Student's t-test; Fig. 6f). The lung tissues of the sacrificed mice then underwent in vivo imaging, and the fluorescent expression in the lungs of the shSYT7-mice was also significantly reduced compared with the shCtrl group (P < .05, Student's t-test; Fig. 6g). The size and weight of the tumors formed by the A549-shSYT7 cells significantly decreased in comparison with the tumors formed by the A549-shCtrl cells (Fig. 6h-Hi). The dissected lung tissues of the nude mice were further observed. In the shCtrl group, there were signs of metastasis in the lungs of 10 mice, and 155 metastases were found. Meanwhile, only 1 lung cancer cell metastasis in 1 mouse of in the shSYT7 group was found. We performed Western blotting to detect the protein expression of SYT7 in the lung tumor tissues, showing significantly decreased SYT7 protein levels in the shSYT7 group compared with the shCtrl group (Fig. S2d). Taken together, these results indicated that the knockdown of SYT7 significantly inhibited the tumor growth of the lung cancer cells in vivo.
3.8. The regulation function of TWIST1 on SYT7
To confirm whether TWIST1 acts as a transcription factor to directly regulate the expression of SYT7, we performed a Dual-luciferase reporter gene assay. As shown in Fig. S2e, the TWIST1-OE cells that were transfected with SYT7 recombinant promoter plasmid showed no significant difference in fluorescence signal intensity compared with the negative control group (P > .05, Student's t-test;). This suggested that TWIST1 and SYT7 promoter could not directly bind, indicating that the regulation of SYT7 expression by TWIST1 may be not through direct transcriptional regulation.
4. Discussion
In this study, we identified SYT7 as the downstream gene of TWIST1 in NSCLC cells. The knockdown of SYT7 weakened the metastasis and invasion abilities of the TWIST1-OE H1975 cells. SYT7 was highly expressed in both NSCLC cell lines and clinical tissues and demonstrated to be correlated with a poor OS rate in NSCLC patients. The protein level of SYT7 was positively correlated with TWIST1 in the clinical NSCLC tissues. Moreover, loss-of-function and gain-of-function analyses revealed that SYT7 promoted the proliferation, EMT, invasion, and metastasis and inhibits cell apoptosis of the NSCLC cells. The knockdown of SYT7 significantly inhibited tumor promotion role of lung cancer cells in vivo. These results emphasize the potential tumor promotion role of SYT7 in NSCLC.
TWIST1 belongs to a family of basic helix-loop-helix proteins that function as transcription factors [30]. It has been reported that TWIST1 mediates tumorigenesis, metastasis, EMT, and drug resistance in NSCLC, demonstrating its potential as a drug target [[14], [15], [16],[31], [32], [33], [34], [35]]. Our results showed that the overexpression of TWIST1 enhanced the invasion and metastasis abilities of the NSCLC cells, which was in accordance with previous studies reporting that TWIST1 was recognized as a critical regulator of metastasis in NSCLC [32]. To further seek novel genes related to the invasion and metastasis of NSCLC, we found that SYT7 and CEP85 were the functional genes at the downstream of TWIST1 through gene chip and HCS method. Functional experiments of SYT7 and CEP85 were then conducted in sequence, and the SYT7 results were exhibited in this study.
SYT7, a member of the synaptic binding protein gene family, is required for facilitation of the central synapses as the specialized Ca2+ sensor [[36], [37], [38], [39]]. SYT7 regulates the chemokine-induced fusion of lysosomes with the cell membrane in migrating cells, and the SYT7-deficient leukocytes showed less migration in vitro and in vivo [40]. As the calcium sensor involved in the lysosome-mediated repair mechanism of plasma membrane, SYT7 transcription is induced by prostaglandin E(2) [41]. A recent study indicated that SYT7 is associated with hepatic metastasis formation of gastric cancer cells [42]. It was also reported that SYT7 was overexpressed in colorectal cancer and regulated the proliferation colorectal cancer cells [43]. However, the detailed function and underlying mechanism of SYT7 in lung cancer tumorigenicity and metastasis remain unknown. Our study found the involvement of SYT7 in the proliferation and metastasis of NSCLC, which is consistent with its function in gastrointestinal tumors research. The knockdown and overexpression experiments demonstrated the promotion function of SYT7 in NSCLC cells proliferation, migration, and invasion in vitro. Furthermore, the repression of SYT7 inhibited the tumorigenicity and tumor growth of the NSCLC cells in vivo. However, the underlying causes of tumorigenic and promoting metastasis effects of SYT7 have not been reported. Hence, we explored the molecular mechanism of SYT7 in NSCLC.
We demonstrated that SYT7 may promote tumorigenesis and metastasis through activating EMT, an essential process during cancer metastasis and progression. After the knockdown of SYT7, the expression of E-cadherin significantly increased, and the expression of N-cadherin and Vimentin significantly reduced. We also demonstrated the upregulation of SYT7 protein in the tumor tissue of the NSCLC patients. The protein expression of SYT7 was positively correlated with TWIST1 in the NSCLC patients. In both the LUAD and LUSC tissues, higher SYT7 expression was significantly associated with inferior survival independently of conventional prognostic factors, which further confirmed that the upregulation of SYT7 promoted NSCLC development. Survival outcomes vary among NSCLC patients, even within groups that present with the same stage at the time of diagnosis and are treated with similar strategies [44,45]. Hence, it is necessary to develop accurate prognostic factors. In recent studies, emerging novel prognostic biomarkers were reported and showed promising efficacy in NSCLC patients, such as estrogen receptor β [46], regulatory T cells [47], and tripartite motif-containing 37 [48]. In our study, the upregulation of SYT7 demonstrated the potential to predict poor prognosis of NSCLC patients.
Some limitations to our study must to be acknowledged. First, we investigated the functional mechanism between TWIST1 and SYT7. However, the dual-luciferase reporter assay result suggested that the regulation of SYT7 expression by TWIST1 is not through the direct transcriptional regulation. The molecular mechanism of the interaction between SYT7 and TWIST1 has not been clarified. Second, the in-depth mechanism of SYT7 involved in the development and progression of NSCLC must be revealed. In the future, large-scale prospective researches are necessary to further assess the values of SYT7 in the diagnosis and predicting the prognosis of NSCLC, and the underlying biological mechanism of SYT7 will be illustrated in our future research.
In summary, our study suggested that SYT7, as the downstream gene of TWIST1, plays a potential tumor promotion role in NSCLC. We observed that SYT7 protein was highly expressed in NSCLC tissue, and was an independent prognostic marker for patient survival. The results of the knockdown and overexpression experiments revealed that SYT7 promotes the proliferation, invasion, and metastasis and inhibited cell apoptosis of NSCLC in vitro. These functions may be generated through promoting EMT. In vivo, the repression of SYT7 resulted in the dramatic suppression of tumor growth, which could be a vigorous therapeutic strategy for future NSCLC treatment.
The following are the supplementary data related to this article.
Sequences of interfering targets.
List of 1297 DEGs.
KEGG pathway enrichment analysis of 1297 DEGs.
Primers used in qRT-PCR and the expression abundance in TWIST1-OE H1975 cells.
Supplementary Fig. 1.
KEGG pathway enrichment analysis of 1297 DEGs shown as barplot.
Supplementary Fig. 2.
Association of SYT7 expression with TWIST1 and the regulation function of TWIST1 on SYT7.
Supplementary material
Funding sources
This project was supported by grants from the Major Scientific and Technological Innovation Project of Shandong Province (2018CXGC1212), Science and Technology Foundation of Shandong Province (2014GSF118084, 2016GSF121043), Medical and Health Technology Innovation Plan of Jinan City (201805002) and the National Natural Science Foundation of China (81372333).
Authors' contributions
Yiqing Qu and Xiao Liu designed this study; Xiao Liu and Chunyu Li performed most experiments; Yie Yang, Xiaoxia Liu, Rui Li, Mengyu Zhang, Yunhong Yin helped to perform experiments and collected the data; Xiao Liu and Chunyu Li analyzed the data; Xiao Liu and Yiqing Qu wrote and revised the manuscript. Yiqing Qu supervised the entire study.
Declaration of Competing Interest
None.
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Rotow J., Bivona T.G. Understanding and targeting resistance mechanisms in NSCLC. Nat Rev Cancer. 2017;17(11):637–658. doi: 10.1038/nrc.2017.84. [DOI] [PubMed] [Google Scholar]
- 3.McCoach C.E., Le A.T., Gowan K., Jones K., Schubert L., Doak A. Resistance mechanisms to targeted therapies in ROS1(+) and ALK(+) non-small cell lung cancer. Clin Cancer Res. 2018;24(14):3334–3347. doi: 10.1158/1078-0432.CCR-17-2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vaishnavi A., Schubert L., Rix U., Marek L.A., Le A.T., Keysar S.B. EGFR mediates responses to small-molecule drugs targeting oncogenic fusion kinases. Cancer Res. 2017;77(13):3551–3563. doi: 10.1158/0008-5472.CAN-17-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Puisieux A., Brabletz T., Caramel J. Oncogenic roles of EMT-inducing transcription factors. Nat Cell Biol. 2014;16(6):488–494. doi: 10.1038/ncb2976. [DOI] [PubMed] [Google Scholar]
- 6.Vandewalle C., Van Roy F., Berx G. The role of the ZEB family of transcription factors in development and disease. Cell Mol Life Sci. 2009;66(5):773–787. doi: 10.1007/s00018-008-8465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Risolino M., Mandia N., Iavarone F., Dardaei L., Longobardi E., Fernandez S. Transcription factor PREP1 induces EMT and metastasis by controlling the TGF-beta-SMAD3 pathway in non-small cell lung adenocarcinoma. Proc Natl Acad Sci U S A. 2014;111(36):E3775–E3784. doi: 10.1073/pnas.1407074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thiery J.P. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2(6):442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 9.Onder T.T., Gupta P.B., Mani S.A., Yang J., Lander E.S., Weinberg R.A. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 2008;68(10):3645–3654. doi: 10.1158/0008-5472.CAN-07-2938. [DOI] [PubMed] [Google Scholar]
- 10.Daugaard I., Sanders K.J., Idica A., Vittayarukskul K., Hamdorf M., Krog J.D. miR-151a induces partial EMT by regulating E-cadherin in NSCLC cells. Oncogenesis. 2017;6(7):e366. doi: 10.1038/oncsis.2017.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He Y., Northey J.J., Pelletier A., Kos Z., Meunier L., Haibe-Kains B. The Cdc42/Rac1 regulator CdGAP is a novel E-cadherin transcriptional co-repressor with Zeb2 in breast cancer. Oncogene. 2017;36(24):3490–3503. doi: 10.1038/onc.2016.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thiery J.P., Acloque H., Huang R.Y., Nieto M.A. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139(5):871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 13.Tsai J.H., Donaher J.L., Murphy D.A., Chau S., Yang J. Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell. 2012;22(6):725–736. doi: 10.1016/j.ccr.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yochum Z.A., Cades J., Mazzacurati L., Neumann N.M., Khetarpal S.K., Chatterjee S. A first-in-class TWIST1 inhibitor with activity in oncogene-driven lung cancer. Mol Cancer Res. 2017;15(12):1764–1776. doi: 10.1158/1541-7786.MCR-17-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yochum Z.A., Cades J., Wang H., Chatterjee S., Simons B.W., O'Brien J.P. Targeting the EMT transcription factor TWIST1 overcomes resistance to EGFR inhibitors in EGFR-mutant non-small-cell lung cancer. Oncogene. 2019;38(5):656–670. doi: 10.1038/s41388-018-0482-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gou W., Zhou X., Liu Z., Wang L., Shen J., Xu X. CD74-ROS1 G2032R mutation transcriptionally up-regulates Twist1 in non-small cell lung cancer cells leading to increased migration, invasion, and resistance to crizotinib. Cancer Lett. 2018;422:19–28. doi: 10.1016/j.canlet.2018.02.032. [DOI] [PubMed] [Google Scholar]
- 17.Li C.W., Xia W., Lim S.O., Hsu J.L., Huo L., Wu Y. AKT1 inhibits epithelial-to-mesenchymal transition in breast cancer through phosphorylation-dependent Twist1 degradation. Cancer Res. 2016;76(6):1451–1462. doi: 10.1158/0008-5472.CAN-15-1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vichalkovski A., Gresko E., Hess D., Restuccia D.F., Hemmings B.A. PKB/AKT phosphorylation of the transcription factor Twist-1 at Ser42 inhibits p53 activity in response to DNA damage. Oncogene. 2010;29(24):3554–3565. doi: 10.1038/onc.2010.115. [DOI] [PubMed] [Google Scholar]
- 19.Yu C., Liu Z., Chen Q., Li Y., Jiang L., Zhang Z. Nkx2.8 inhibits epithelial-mesenchymal transition in bladder urothelial carcinoma via transcriptional repression of Twist1. Cancer Res. 2018;78(5):1241–1252. doi: 10.1158/0008-5472.CAN-17-1545. [DOI] [PubMed] [Google Scholar]
- 20.Hwang W., Chiu Y.F., Kuo M.H., Lee K.L., Lee A.C., Yu C.C. Expression of neuroendocrine factor VGF in lung cancer cells confers resistance to EGFR kinase inhibitors and triggers epithelial-to-mesenchymal transition. Cancer Res. 2017;77(11):3013–3026. doi: 10.1158/0008-5472.CAN-16-3168. [DOI] [PubMed] [Google Scholar]
- 21.Piccinin S., Tonin E., Sessa S., Demontis S., Rossi S., Pecciarini L. A "twist box" code of p53 inactivation: twist box: p53 interaction promotes p53 degradation. Cancer Cell. 2012;22(3):404–415. doi: 10.1016/j.ccr.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 22.Yang M.H., Hsu D.S., Wang H.W., Wang H.J., Lan H.Y., Yang W.H. Bmi1 is essential in Twist1-induced epithelial-mesenchymal transition. Nat Cell Biol. 2010;12(10):982–992. doi: 10.1038/ncb2099. [DOI] [PubMed] [Google Scholar]
- 23.Xu Y., Qin L., Sun T., Wu H., He T., Yang Z. Twist1 promotes breast cancer invasion and metastasis by silencing Foxa1 expression. Oncogene. 2017;36(8):1157–1166. doi: 10.1038/onc.2016.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gajula R.P., Chettiar S.T., Williams R.D., Nugent K., Kato Y., Wang H. Structure-function studies of the bHLH phosphorylation domain of TWIST1 in prostate cancer cells. Neoplasia. 2015;17(1):16–31. doi: 10.1016/j.neo.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei S.C., Fattet L., Tsai J.H., Guo Y., Pai V.H., Majeski H.E. Matrix stiffness drives epithelial–mesenchymal transition and tumour metastasis through a TWIST1–G3BP2 mechanotransduction pathway. Nat Cell Biol. 2015;17:678. doi: 10.1038/ncb3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen H.F., Huang C.H., Liu C.J., Hung J.J., Hsu C.C., Teng S.C. Twist1 induces endothelial differentiation of tumour cells through the Jagged1-KLF4 axis. Nat Commun. 2014;5:4697. doi: 10.1038/ncomms5697. [DOI] [PubMed] [Google Scholar]
- 27.Ghandi M., Huang F.W., Jane-Valbuena J., Kryukov G.V., Lo C.C., McDonald E.R., III Next-generation characterization of the cancer cell line encyclopedia. Nature. 2019;569(7757):503–508. doi: 10.1038/s41586-019-1186-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang Z., Li C., Kang B., Gao G., Li C., Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gyorffy B., Surowiak P., Budczies J., Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8(12) doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang J., Mani S.A., Donaher J.L., Ramaswamy S., Itzykson R.A., Come C. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117(7):927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 31.Avasarala S., Van Scoyk M., Karuppusamy Rathinam M.K., Zerayesus S., Zhao X., Zhang W. PRMT1 is a novel regulator of epithelial-mesenchymal-transition in non-small cell lung cancer. J Biol Chem. 2015;290(21):13479–13489. doi: 10.1074/jbc.M114.636050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burns T.F., Dobromilskaya I., Murphy S.C., Gajula R.P., Thiyagarajan S., Chatley S.N. Inhibition of TWIST1 leads to activation of oncogene-induced senescence in oncogene-driven non-small cell lung cancer. Mol Cancer Res. 2013;11(4):329–338. doi: 10.1158/1541-7786.MCR-12-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin H.O., Hong S.E., Woo S.H., Lee J.H., Choe T.B., Kim E.K. Silencing of Twist1 sensitizes NSCLC cells to cisplatin via AMPK-activated mTOR inhibition. Cell Death Dis. 2012;3:e319. doi: 10.1038/cddis.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakashima H., Hashimoto N., Aoyama D., Kohnoh T., Sakamoto K., Kusunose M. Involvement of the transcription factor twist in phenotype alteration through epithelial-mesenchymal transition in lung cancer cells. Mol Carcinog. 2012;51(5):400–410. doi: 10.1002/mc.20802. [DOI] [PubMed] [Google Scholar]
- 35.Liu Y., Mayo M.W., Xiao A., Hall E.H., Amin E.B., Kadota K. Loss of BRMS1 promotes a mesenchymal phenotype through NF-kappaB-dependent regulation of Twist1. Mol Cell Biol. 2015;35(1):303–317. doi: 10.1128/MCB.00869-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jackman S.L., Turecek J., Belinsky J.E., Regehr W.G. The calcium sensor synaptotagmin 7 is required for synaptic facilitation. Nature. 2016;529(7584):88–91. doi: 10.1038/nature16507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu D., Bacaj T., Morishita W., Goswami D., Arendt K.L., Xu W. Postsynaptic synaptotagmins mediate AMPA receptor exocytosis during LTP. Nature. 2017;544(7650):316–321. doi: 10.1038/nature21720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo F., Sudhof T.C. Synaptotagmin-7-mediated asynchronous release boosts high-fidelity synchronous transmission at a central synapse. Neuron. 2017;94(4):826–839. doi: 10.1016/j.neuron.2017.04.020. [e3] [DOI] [PubMed] [Google Scholar]
- 39.Chen C., Satterfield R., Young S.M., Jr., Jonas P. Triple function of synaptotagmin 7 ensures efficiency of high-frequency transmission at central GABAergic synapses. Cell Rep. 2017;21(8):2082–2089. doi: 10.1016/j.celrep.2017.10.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Colvin R.A., Means T.K., Diefenbach T.J., Moita L.F., Friday R.P., Sever S. Synaptotagmin-mediated vesicle fusion regulates cell migration. Nat Immunol. 2010;11(6):495–502. doi: 10.1038/ni.1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Divangahi M., Chen M., Gan H., Desjardins D., Hickman T.T., Lee D.M. Mycobacterium tuberculosis evades macrophage defenses by inhibiting plasma membrane repair. Nat Immunol. 2009;10(8):899–906. doi: 10.1038/ni.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kanda M., Tanaka H., Shimizu D., Miwa T., Umeda S., Tanaka C. SYT7 acts as a driver of hepatic metastasis formation of gastric cancer cells. Oncogene. 2018;37(39):5355–5366. doi: 10.1038/s41388-018-0335-8. [DOI] [PubMed] [Google Scholar]
- 43.Wang K., Xiao H., Zhang J., Zhu D. Synaptotagmin7 is overexpressed in colorectal cancer and regulates colorectal cancer cell proliferation. J Cancer. 2018;9(13):2349–2356. doi: 10.7150/jca.25098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Molina J.R., Yang P., Cassivi S.D., Schild S.E., Adjei A.A. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;83(5):584–594. doi: 10.4065/83.5.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wouters M.W., Siesling S., Jansen-Landheer M.L., Elferink M.A., Belderbos J., Coebergh J.W. Variation in treatment and outcome in patients with non-small cell lung cancer by region, hospital type and volume in the Netherlands. Eur J Surg Oncol. 2010;36(Suppl. 1):S83–S92. doi: 10.1016/j.ejso.2010.06.020. [DOI] [PubMed] [Google Scholar]
- 46.Ma L., Zhan P., Liu Y., Zhou Z., Zhu Q., Miu Y. Prognostic value of the expression of estrogen receptor beta in patients with non-small cell lung cancer: a meta-analysis. Transl Lung Cancer Res. 2016;5(2):202–207. doi: 10.21037/tlcr.2016.04.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barua S., Fang P., Sharma A., Fujimoto J., Wistuba I., Rao A.U.K. Spatial interaction of tumor cells and regulatory T cells correlates with survival in non-small cell lung cancer. Lung Cancer. 2018;117:73–79. doi: 10.1016/j.lungcan.2018.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Y., Deng L., Zhao X., Li B., Ren D., Yu L. Tripartite motif-containing 37 (TRIM37) promotes the aggressiveness of non-small-cell lung cancer cells by activating the NF-kappaB pathway. J Pathol. 2018;246(3):366–378. doi: 10.1002/path.5144. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequences of interfering targets.
List of 1297 DEGs.
KEGG pathway enrichment analysis of 1297 DEGs.
Primers used in qRT-PCR and the expression abundance in TWIST1-OE H1975 cells.
Supplementary material