Laboratory surveillance plays an important role in the detection and control of hepatitis A outbreaks and requires the application of rapid and accurate molecular diagnostic tools for hepatitis A virus (HAV) RNA detection, subgenotype identification, and sequence-based genotyping.
KEYWORDS: hepatitis A virus, genotyping, outbreak, public health, reverse transcription-PCR
ABSTRACT
Laboratory surveillance plays an important role in the detection and control of hepatitis A outbreaks and requires the application of rapid and accurate molecular diagnostic tools for hepatitis A virus (HAV) RNA detection, subgenotype identification, and sequence-based genotyping. We describe the development and validation of a triplex real-time, reverse transcription-PCR (triplex rRT-PCR) assay for the identification and discrimination of HAV subgenotypes IA, IB, and IIIA and a singleplex rRT-PCR assay designed to detect all HAV genotypes infecting humans. Overall, the accuracy, sensitivity, and specificity of the new assays were >97% for serum and plasma specimens collected during unrelated outbreaks of HAV in California and Michigan compared to a nested RT-PCR genotyping assay and the ISO 15216-1 rRT-PCR method for HAV detection. The new assays will permit the rapid detection of HAV RNA and discrimination among subgenotypes IA, IB, and IIIA in serum and plasma specimens, which will strengthen public health surveillance efforts for HAV outbreak detection and response.
INTRODUCTION
The incidence of hepatitis A virus (HAV) infections has declined significantly in the United States following the 1996 Advisory Committee on Immunization Practices recommendation for childhood hepatitis A vaccination (1). However, outbreaks of hepatitis A are still of public health concern, particularly among the susceptible adult population. In 2016 to 2019, 22 states reported outbreaks of hepatitis A, primarily among persons experiencing homelessness and/or users of illicit drugs. Based on preliminary data, the cumulative impact of these outbreaks will extend to more than 19,000 cases, more than 11,000 hospitalizations, and at least 189 deaths (2) (https://www.cdc.gov/hepatitis/outbreaks/2017March-HepatitisA.htm). The two earliest states to report cases, California and Michigan, experienced 708 and 909 outbreak-related confirmed cases, respectively, with each outbreak attributed to genetically distinct HAV subgenotype IB strains. The dramatic increase in the number of hepatitis A cases and outbreaks in the United States underscored the need for more rapid diagnostic tools and improved surveillance methods for hepatitis A in public health laboratory settings.
HAV, the causative agent of hepatitis A infections, is a quasi-enveloped, icosahedral virus. The sole human virus within the genus Hepatovirus of the family Picornaviridae, it has a single-stranded, positive-polarity RNA genome. Humans are the only known reservoir for virus propagation, and viral transmission occurs via a fecal-oral route. The incubation period for hepatitis A infections ranges from 15 to 50 days, and patients may shed virus before and after the onset of symptoms. Symptoms often go unrecognized until the onset of jaundice and may include fever, malaise, anorexia, nausea, abdominal pain, and dark urine (3). In young children, hepatitis A infection is typically asymptomatic, but in older children and adults, HAV tends to cause a self-limiting, acute disease, with jaundice present in >70% of patients (4). Illness tends to be more severe in adults than in children, and in patients with underlying medical conditions, infection may lead to more severe disease such as fulminant hepatitis. Risk groups for hepatitis A infections include international travelers, men who have sex with men, and users of illicit drugs (1). While infrequent, community-based and common source outbreaks in the United States have caused significant morbidity and mortality in recent years (5, 6) (https://www.cdc.gov/hepatitis/outbreaks/2016/hav-strawberries.htm).
Comprehensive laboratory surveillance for hepatitis A outbreak detection includes serodiagnostics, nucleic acid amplification testing (NAAT), and genotyping through subgenomic or whole-genome sequencing. Serologic testing can identify recent infections through the detection of IgM anti-HAV and is often used to help classify cases for public health reporting and surveillance purposes (https://wwwn.cdc.gov/nndss/conditions/hepatitis-a-acute/case-definition/2019/). Detection of HAV RNA by NAAT can confirm an active infection and trigger reflex testing to determine genotype by Sanger or next-generation sequencing (NGS). The ∼7.5-kb HAV genome has been fully sequenced and consists of a 5′ untranslated region (5′-UTR) of ∼735 nucleotides (nt), followed by structural genes (VP4, VP2, VP3, and VP1), nonstructural genes (P2A-C and P3A-D), and a 3′-UTR of ∼63 nt (7). The relatively well conserved 5′-UTR is often targeted in the design of rRT-PCR assays for detecting HAV RNA, whereas the highly variable VP1-P2A junction is often selected for HAV genotyping (8). HAV strains have been classified into six genotypes, genotypes I to VI, based on nucleotide sequence diversity. Human HAV strains are represented by genotypes I to III, whereas genotypes IV to VI are simian strains (9). Each human HAV genotype can be further subdivided into subgenotypes A and B. A third subgenotype, C, has been proposed for genotype I (10). Nucleotide sequence diversity in the VP1-P2A junction is >15% between genotypes and >7.5% between subgenotypes (11). Comparison of strain sequences through phylogenetic analysis can provide useful information about strain relatedness as well as the possible geographic origin of strains (12). Historically, subgenotype IA has represented >80% of the HAV strains detected in the United States (1). This dynamic has changed recently with 15%, 84%, and <1% of the strains genotyped as IA, IB, and IIIA, respectively (13). Combined with epidemiological data, genotyping can be a powerful public health tool. Understanding strain similarities and differences among case-patients can help public health investigators detect new outbreaks, determine the source of infections, track the modes and chains of transmission, and monitor control and prevention efforts.
In this report, we describe two new rRT-PCR assays for the detection of HAV RNA: a triplex rRT-PCR assay that provides subgenotype level identification of IA, IB, and IIIA strains and a complementary 5′-UTR rRT-PCR assay. This 5′-UTR assay reflects the expanding number of HAV sequences in GenBank and detects HAV genotypes I to III. The performance characteristics of these two assays were assessed against both a nested VP1-P2B RT-PCR genotyping assay and a standardized rRT-PCR assay using a panel of serum and plasma specimens collected during two unrelated and genetically distinct HAV outbreaks in the United States.
MATERIALS AND METHODS
Specimens.
Serum or plasma specimens were requested and submitted for genotyping as part of a statewide investigation of an HAV subgenotype IB outbreak among persons experiencing homelessness and/or using illicit drugs. Criteria for specimen acceptance included but were not restricted to serum or plasma specimens that were either IgM anti-HAV seropositive or from symptomatic patients with known risk factors for hepatitis A exposure. Two hundred specimens from California were included in this study. Additionally, 46 serum specimens from a concurrent, but unrelated HAV subgenotype IB outbreak were kindly provided by the Michigan Department of Health and Human Services. An exclusivity panel of sera was also assembled to assess assay specificity and included a hepatitis E seropositive specimen and 17 specimens in which the following viruses had been detected by NAAT: hepatitis B virus (n = 5), hepatitis C virus (n = 7), human immunodeficiency virus (n = 3), and enterovirus (n = 2). Samples were collected for public health surveillance and deidentified prior to analysis, and thus are considered exempt from human subject regulations by the California Health and Human Services Agency Committee for the Protection of Human Subjects.
Nucleic acid extraction.
Total nucleic acids were extracted and purified from 250 μl of specimen using the Nuclisens EasyMag system (bioMérieux, Cambridge, MA) and collected in an output volume of 80 μl. The nucleic acid extracts were either tested immediately or stored at −20°C until used.
Nested RT-PCR and Sanger sequencing.
Oligonucleotide primer sequences for nested RT-PCR and DNA sequencing of a 315-nt segment of VP1-P2B were previously described (14). The first round of RT-PCR utilized the OneStep RT PCR kit (Qiagen Inc., Valencia, CA) with 1× Q solution and primers 2870 and 3381 at a final concentration of 300 nM each. Twenty microliters of total nucleic acids was added to the reaction mix and amplified in a total reaction volume of 50 μl. Amplification was performed with an ABI 9700 thermal cycler (Thermo Fisher Scientific, Carlsbad, CA) using the following parameters: a 40-min hold at 50°C, a 15-min hold at 94°C, 45 cycles with 1 cycle consisting of 95°C for 15 s, 50°C for 20 s, and 72°C for 40 s, followed by a final extension step of 72°C for 10 min. Five microliters of the first-round product were then amplified in a second round of PCR. The second-round reaction mix consisted of 1× PerfeCTa SYBR green Supermix (Quantabio, Beverly, MA) and 300 nM concentration of primers 2897 and 3288 in a final volume of 50 μl. Amplification was performed with an ABI 9700 thermal cycler beginning with a 5-min hold at 95°C, followed by 35 cycles, with 1 cycle consisting of 95°C for 15 s, 50°C for 20 s, and 72°C for 30 s, and ending with a 10-min hold at 72°C. The final product was purified using the QIAamp PCR purification kit (Qiagen Inc.) and detected by agarose gel electrophoresis using 2% E-gels (Thermo Fisher Scientific). Bidirectional dye terminator cycle sequencing of the purified amplicons was performed by a commercial sequencing service (Sequetech, Mountain View, CA). Sequence assembly and phylogenetic analysis were performed using MEGA version 5.05. A second nested RT-PCR and Sanger sequencing assay that targets VP1-P2A was used in a limited capacity to resolve discrepant results (15). A unidirectional workflow with dedicated workstations was adhered to throughout the nested RT-PCR process.
rRT-PCR assays.
rRT-PCR assay designs were based on 115 complete HAV genomes downloaded from GenBank (database queried 21 November 2017) and aligned using MEGA version 5.05. The genomes included 56 IA sequences, 36 IB sequences, one IIA sequence, one IIB sequence, 17 IIIA sequences, and four IIIB sequences. The aligned sequences were manually screened for regions specific to IA, IB, or IIIA strains and a region that was broadly conserved among genotypes I to III. A triplex assay for the subgenotype-specific detection of IA, IB, or IIIA strains and a singleplex assay for genotype I to III detection were designed using Primer Quest (Integrated DNA Technologies Inc., Coralville, IA). For the triplex assay, oligonucleotide primer and probe sequences were selected to minimize heterologous interactions, and the probes were designed to allow discrimination of single nucleotide polymorphisms. Initial evaluation of the IA primer-probe set indicated that a minor variant (1 of 56 IA genome sequences) existed in our study population and required the addition of a second probe (HAVIA_P2; Table 1) to detect this sequence variant. For the 5′-UTR assay, a consensus primer and probe set was selected to minimize mismatches among the 115 aligned sequences. In silico primer pair specificity analysis was performed using NCBI Primer BLAST (16). Primer pair specificity was checked by searching selected organisms in the Refseq representative genome database or the nr database (Rickettsia rickettsii) for unintended targets using a primer mismatch threshold of ≤20% and a maximum target length of 1000 nt (17).
TABLE 1.
Oligonucleotide primer and probe sets for the triplex and 5′-UTR rRT-PCR assays
| Assay | Oligonucleotide | Reference sequence positiona | Oligonucleotide sequence and modificationsb | Final concn (nM) |
|---|---|---|---|---|
| Triplex IA | HAVIA_F | 2616−2639 | ATAGAGGACCAYTGGAYTTGACAA | 900 |
| HAVIA_P1 | 2648−2664 | FAM-ACAGGAGCCACTGATGT-BHQ1 | 100 | |
| HAVIA_P2 | 2647−2665 | FAM-TCACAGGAGCTACTGATGT-BHQ1 | 100 | |
| HAVIA_R | 2712−2733 | GCTGAYTYCTTTTCYACCCAAG | 900 | |
| Triplex IB | HAVIB_F | 5336−5355 | GTTTGGAGTTGGAGAGAAGA | 900 |
| HAVIB_P | 5430−5449 | VIC-AAATTTGAGAAAGATTATGA-MGBNFQ | 200 | |
| HAVIB_R | 5479−5502 | CAGCTGAAATTGAATAGTARGTTC | 900 | |
| Triplex IIIA | HAVIIIA_F | 4507−4530 | TGCATCTGATTATTGGGATGGATA | 300 |
| HAVIIIA_P | 4575−4596 | Q670-TCAGACCAATCCTCATCAGTTG-BHQ2 | 200 | |
| HAVIIIA_R | 4612−4632 | AAYCTCATTGGACATCCAGAC | 300 | |
| 5′-UTR | HAV5UTR_F | 249−265 | CTAGGCTCTGGCCGTTG | 800 |
| HAV5UTR_P | 296−322 | FAM-TCCCCAATT-NOVA-TAGACTCCTACAGCTCCA-BHQ1 | 150 | |
| HAV5UTR_R | 343−362 | GCCAAGTTAACACTGCAAGG | 800 |
Genome position relative to GenBank accession no. M14707 for the 5′-UTR and Triplex IB primer-probe set, GenBank accession no. AB020564 (HAVIA_F, HAVIA_R, and HAVIA_P1) and AB020568 accession no. (HAVIA_P2) for the Triplex IA primer-probe set, and GenBank accession no. AB279732 for the Triplex IIIA primer-probe set.
Oligonucleotide modifications: BHQ1, black hole quencher 1; MGBNFQ, minor grove binder nonfluorescent quencher; Q670, Quasar 670; BHQ2, black hole quencher 2; NOVA, BHQnova double-quenched probe.
The nucleotide sequence, genome location, oligonucleotide modifications, and final assay concentrations of the primer and probes used in the triplex and 5′-UTR assays are provided in Table 1. With the exception of HAV IB_P synthesis (Thermo Fisher Scientific), all other custom oligonucleotides were synthesized by Biosearch Technologies (Novato, CA). The 25-μl reaction mixtures consisted of 1× qScript XLT 1-Step RT-qPCR ToughMix L-Rox (Quantabio), 5 μl of total nucleic acids, and either the triplex primer probe sets (IA, IB, IIIA) or the 5′-UTR primer-probe set. The reactions were amplified using standard cycling conditions with the ABI 7500 Fast Real Time PCR system (Thermo Fisher Scientific) beginning with a 20- min hold at 50°C, followed by a 1-min hold at 95°C, and then 40 cycles, with 1 cycle consisting of 95°C for 10 s and 60°C for 30 s. Cycle threshold (CT) values were determined using a manual threshold setting option with each detector. A no-template control and low-copy-number positive controls were included in each thermal cycling run. The ISO 15216-1 rRT-PCR method for HAV detection was used as an additional reference test for the evaluation of the 5′-UTR assay performance (18).
Synthetic and in vitro-transcribed RNA controls.
A custom RNA synthesis service (Biosynthesis Inc., Lewisville, TX) was utilized to synthesize IA, IB, IIIA, and 5′-UTR RNA target sequences for use as amplification controls and determine the limit of assay detection. The reference genomes and region synthesized were GenBank accession no. AB020564 nucleotide positions 2613 to 2736 for IA1, GenBank accession no. AB020568 nucleotide positions 2614 to 2737 for IA2, GenBank accession no. M14707 nucleotide positions 5332 to 5505 for IB, GenBank accession no. AB279732 nucleotide positions 4504 to 4635 for IIIA, and GenBank accession no. M14707 nucleotide positions 246 to 365 for the 5′-UTR target.
IIA, IIB, and IIIB exclusivity controls for the triplex assay were constructed by concatenating the corresponding IA, IB, and IIIA target regions derived from IIA, IIB, and IIIB reference genomes. The concatenated regions consisted of GenBank accession no. AY644676 regions 2558 to 2698, 5270 to 5458, and 4466 to 4615 for the IIA construct, GenBank accession no. AY644670 regions 2557 to 2697, 5275 to 5463, and 4465 to 4614 for the IIB construct, and GenBank accession no. AB279735 regions 2588 to 2728, 5306 to 5494, and 4496 to 4645 for the IIIB construct. These constructs were designed to include the 5′-UTR target region as an amplification control when tested concurrently with the 5′-UTR assay. The constructs were synthesized initially as DNA, cloned into a plasmid vector, and transcribed into RNA by in vitro transcription (Biosynthesis Inc.).
RESULTS
We evaluated the test performance characteristics of a triplex rRT-PCR assay designed for the subgenotype-specific detection of IA, IB, and IIIA strains of HAV and a 5′-UTR rRT-PCR assay designed to detect all human HAV strains. Analytical sensitivity was assessed by testing in triplicate, serial 10-fold dilutions of synthetic RNA target sequences ranging from 105 to 0.1 copies per reaction (Table 2). The limit of detection (LOD) was defined as the lowest concentration of RNA detected in all replicates. A LOD of 1 RNA copy per reaction was obtained for the triplex IA and 5′-UTR analytes and 10 RNA copies per reaction for the triplex IB and IIIA analytes. No cross-reactivity was detected with the heterologous analytes at any of the concentrations tested. To determine whether the triplex assay format influenced the subgenotype target LOD, the subgenotyping primer-probe sets were also tested in singleplex. No difference in the LOD was observed for each subgenotype target when tested in the singleplex and triplex formats (Table 2).
TABLE 2.
Limit of detection titration for the hepatitis A virus IA, IB, IIIA (singleplex and triplex formats) and 5′-UTR rRT-PCR assays
| No. of copies/reaction | IA-1 RNA |
IA-2 RNA |
IB RNA |
IIIA RNA |
5′-UTR |
||||
|---|---|---|---|---|---|---|---|---|---|
| Singleplex CTa | Triplex CT | Singleplex CT | Triplex CT | Singleplex CT | Triplex CT | Singleplex CT | Triplex CT | Singleplex CT | |
| 100,000 | 19.96 (0.08) | 18.51 (0.02) | 20.76 (0.06) | 19.01 (0.10) | 23.03 (0.05) | 23.27 (0.08) | 20.69 (0.07) | 20.13 (0.04) | 17.86 (0.03) |
| 10,000 | 23.30 (0.07) | 21.82 (0.12) | 24.20 (0.09) | 22.59 (0.25) | 26.30 (0.22) | 26.56 (0.10) | 24.02 (0.03) | 23.58 (0.10) | 21.40 (0.30) |
| 1,000 | 26.41 (0.16) | 25.01 (0.19) | 27.45 (0.18) | 25.84 (0.39) | 29.54 (0.08) | 29.47 (0.17) | 27.29 (0.09) | 27.09 (0.06) | 24.40 (0.15) |
| 100 | 29.39 (0.20) | 27.61 (0.32) | 30.14 (0.17) | 28.52 (0.19) | 32.94 (0.66) | 32.52 (0.40) | 30.60 (0.15) | 30.17 (0.10) | 27.72 (0.22) |
| 10 | 32.18 (0.30) | 30.44 (0.33) | 32.98 (0.13) | 31.29 (0.68) | 36.65 (0.88) | 35.75 (0.33) | 34.19 (0.76) | 33.91 (0.56) | 31.02 (0.27) |
| 1 | 35.20 (0.18) | 34.33 (0.67) | 36.71 (0.26) | 35.27 (0.21) | NDb | ND | ND | 36.2c | 34.11 (0.64) |
| 0.1 | 38.26 (0.08)d | 38.12 (0.18)d | 39.2c | 38.61 (0.50)d | ND | ND | 36.5c | ND | 36.67 (0.56)d |
CT, average cycle threshold value with standard deviation shown in parentheses.
ND, not detected.
One of three replicates detected.
Two of three replicates detected.
To evaluate assay exclusivity, serum specimens that were nucleic acid amplification positive for hepatitis B virus, hepatitis C virus, human immunodeficiency virus, or enterovirus and a specimen that was seropositive for hepatitis E virus were tested by the triplex and 5′-UTR assays. In addition, in vitro-transcribed RNA constructs representing the concatenated homologous target sequences from subgenotypes IIA (reference sequence GenBank accession no. AY644676), IIB (reference sequence GenBank accession no. AY644670), and IIIB (reference sequence GenBank accession no. AB279735) were tested at 105 copies per reaction to further assess the specificity of the triplex assay. No cross-reactivity was observed with any of the serum specimens or the in vitro-transcribed RNA constructs representing the homologous IIA, IIB, and IIIB target sequences (data not shown). In silico exclusivity testing was performed using the primer pair specificity feature of Primer BLAST. No unintended targets were identified with the triplex and 5′-UTR assay primer sets for the following genome sequences: poliovirus, enterovirus B, enterovirus C, enterovirus D, encephalomyocarditis virus, Epstein-Barr virus, cytomegalovirus, herpes simplex viruses 1 and 2, varicella-zoster virus, yellow fever virus, dengue virus, measles virus, parvovirus B-19, Salmonella enterica serotype Typhi, Mycobacterium tuberculosis, Brucella melitensis, Coxiella burnetti, Leptospira interrogans, Borrelia burgdorferi sensu lato, Escherichia coli, Klebsiella pneumoniae, Rickettsia rickettsii, Candida albicans, and Plasmodium falciparum.
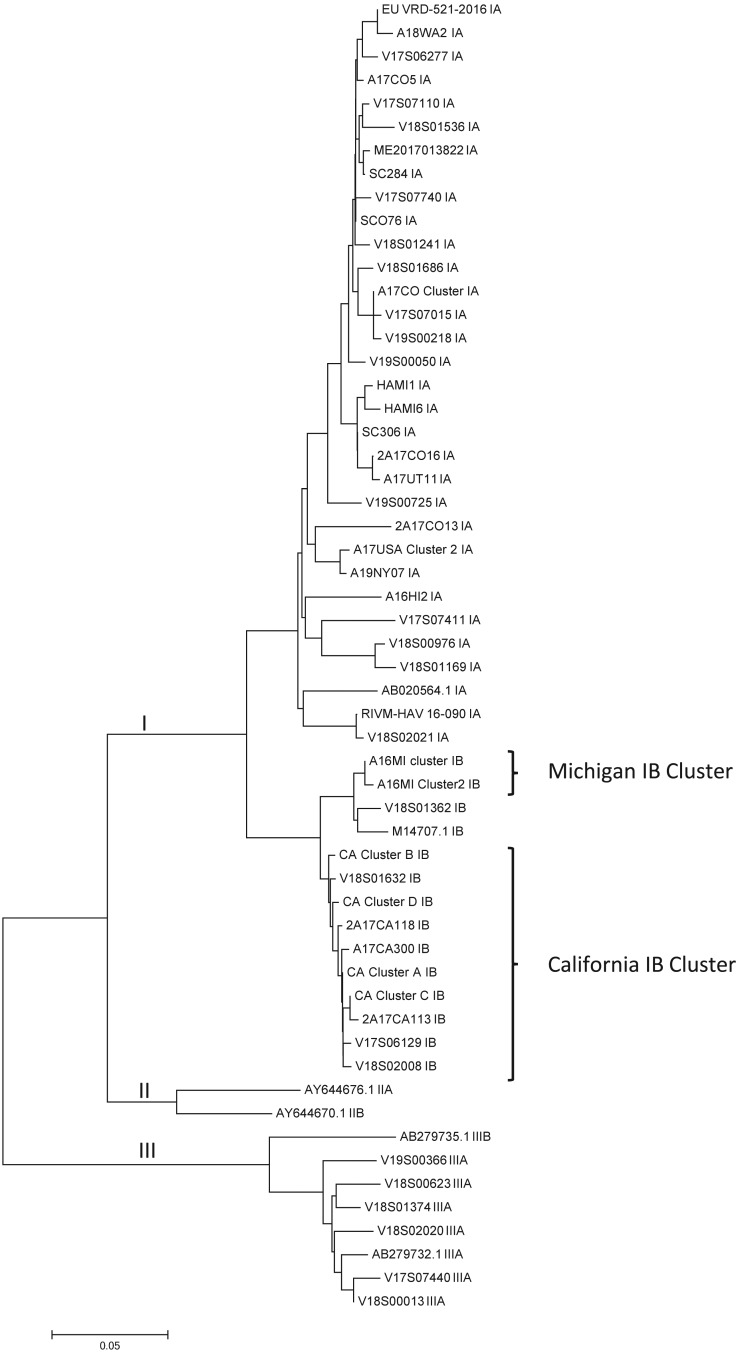
Assay performance characteristics were further defined by comparing test results from the triplex and 5′-UTR assays against genotyping results achieved with a nested VP1-P2B genotyping assay. The comparison was done using serum and plasma specimens from 200 suspected HAV cases submitted to our laboratory and 46 serum specimens from a concurrent, but unrelated, Michigan HAV outbreak. Using the nested VP1-P2B genotyping assay, sequencing results were obtained from 169 of these 246 samples (68.7%), with 71, 93, and 5 specimens classified as subgenotypes IA, IB, and IIIA, respectively. A total of 48 unique VP1-P2B sequences were identified, encompassing 31 IA sequence types, 13 IB sequence types, and 4 IIIA sequence types (Fig. 1).
FIG 1.
Phylogenetic relationship of 50 unique HAV VP1-P2B sequences detected in serum and plasma specimens. HAV reference sequences are indicated by GenBank accession numbers. Sequence alignments were performed with ClustalW, and the dendrogram was generated using the neighbor-joining algorithm and the maximum composite likelihood model. Evolutionary distance is indicated by the scale at the bottom of the dendrogram. Evolutionary branches corresponding to genotype lineage are labeled. The Michigan and California HAV outbreak strains are indicated with the brackets to the right of the dendrogram.
When tested with the triplex assay, 169 samples (68.7%) tested positive for HAV RNA. Four discrepancies were observed for the triplex assay, including two false-negative results with the IB analyte and two false-positive results with the IIIA analyte. Repeat testing of the discrepant samples with the triplex assay yielded late CT values (∼38 cycles) in only one of two replicates for both IB specimens and an early CT value (∼25 cycles) and a late CT value (∼35 cycles) for the IIIA specimens. The late sporadic CT values obtained upon repeat testing of the discrepant IB samples suggested that the viral load in these specimens was at or near the LOD of the triplex assay for the IB target. To resolve the discrepant IIIA results, the samples were tested with an alternate nested RT-PCR genotyping assay that utilizes degenerate amplification primer sequences for broader HAV coverage. Using this assay, both samples yielded distinct IIIA sequences (Fig. 1, sequences V18S01374 and V19S00366), confirming the result of the triplex assay. After discrepancy analysis, the accuracy, sensitivity, and specificity of the triplex assay were 100%, 100%, and 100% for the IA analyte, 99.2%, 97.8%, and 100% for the IB analyte, and 100%, 100%, and 100% for the IIIA analyte, respectively (Table 3).
TABLE 3.
Performance characteristics of hepatitis A virus triplex and 5′-UTR rRT-PCR assays
| Assay | Performance characteristic |
|||
|---|---|---|---|---|
| LOD (no. of copies/reaction) | Accuracy (%) | Sensitivity (%) | Specificity (%) | |
| Triplex IA | 1 | 100 | 100 | 100 |
| Triplex IB | 10 | 99.2 | 97.8 | 100 |
| Triplex IIIA | 10 | 100a | 100 | 100a |
| 5′-UTR | 1 | 98.4a | 98.8a | 97.3a |
Performance characteristic determined after discrepancy resolution.
Comparison of the 5′-UTR assay with the nested RT-PCR genotyping assay yielded five discrepant results: two false-negative results and three false-positive results. The false-negative samples included one of the two IB samples that was falsely negative in the triplex assay. Repeat testing with the 5′-UTR assay produced a CT value of 36 cycles in one of two replicates, reaffirming that the false-negative sample likely possessed a low viral load. The other false-negative sample yielded a late CT value for the IIIA analyte in the triplex assay that was confirmed using the alternate nested genotyping assay. Of the three false-positive 5′-UTR results, the IIIA discrepancy analysis resolved one of these false-positive results. Sufficient material for retesting was available for only one of the two remaining 5′-UTR false-positive samples. Retesting this sample produced a CT value of 36 for both replicates; however, no amplification was detected with the alternate nested RT-PCR genotyping assay. The accuracy, sensitivity, and specificity of the 5′-UTR assay was 98.4%, 98.8%, and 97.3%, respectively, following discrepancy analysis (Table 3).
The ISO 15216-1 rRT-PCR method for HAV detection was also used as a comparator test for the 5′-UTR assay using a subset of specimens. Agreement between the two assays was 100% for the 143 specimens (94 positive specimens and 49 negative specimens) tested. Two of these specimens had yielded discrepant results (a false-negative result for a IIIA specimen and a false-positive result) with the 5′-UTR assay and were also discrepant with the ISO 15216-1 HAV rRT-PCR assay compared to the nested RT-PCR genotyping assays.
DISCUSSION
A large outbreak of hepatitis A in California in 2017-2018 highlighted the need for better diagnostic tools to quickly confirm and prioritize patient specimens for investigation by molecular epidemiology (2). Laboratory confirmation of acute HAV infections for outbreak investigations has primarily relied upon IgM anti-HAV testing. While highly sensitive and specific when applied to persons meeting the clinical criteria for acute viral hepatitis, IgM testing for HAV can have a low predictive value when patients do not meet those criteria (19, 20). False-positive IgM anti-HAV results may occur through nonspecific IgM binding or cross-reactivity with other picornaviruses (21, 22). In addition, the IgM response to HAV may persist for long periods, making interpretation of recent infections problematic (23). NAAT offers an alternative and complementary tool for the laboratory confirmation of HAV infections and was recently added as an acceptable laboratory method for classifying acute hepatitis A cases by the U.S. Centers for Disease Control and Prevention and the Council of State and Territorial Epidemiologists (https://wwwn.cdc.gov/nndss/conditions/hepatitis-a-acute/case-definition/2019/). HAV RNA in serum may be detectable earlier than IgM anti-HAV during the course of infection and may be detected for weeks to months after symptom onset (24). NAAT results can also indicate the suitability of a sample for genotyping. However, a FDA-cleared nucleic acid amplification test is not available, and few commercial laboratories offer laboratory-developed tests for HAV RNA detection.
We have designed a triplex HAV subgenotyping rRT-PCR assay and a consensus 5′-UTR rRT-PCR assay based on 115 complete genome sequences downloaded from GenBank and have defined assay performance characteristics for detecting HAV RNA in serum and plasma specimens. The LOD for each analyte in the triplex assay was ≤10 copies per reaction and 1 copy per reaction in the 5′-UTR assay. The accuracy, sensitivity, and specificity of the assays were >97% for all analytes compared to a nested VP1-P2B RT-PCR genotyping assay. The triplex assay provides subgenotyping level identification of IA, IB, and IIIA strains. These three subgenotypes represent the majority of the strains detected in the United States (13). The multiplex format of the triplex assay offers economical and ease of use advantages over the set of singleplex subgenotyping assays described by Coudray-Meunier et al. (25). Moreover, the triplex assay represents a significant improvement in analytical sensitivity compared to these earlier assays. When performed in parallel with the triplex assay, the 5′-UTR assay design should permit detection of all genotypes, including the rarely encountered subgenotype IIA, IIB, and IIIB strains.
The relatively well-conserved 5′-UTR region of HAV has been a common target for RT-PCR design (26). However, several of these assay designs were based on a limited number of HAV sequences, and consequently, may not target regions that are highly conserved across HAV strains. A standardized method, ISO 15216-1, utilizes primer sequences described in 2006 and targets the region from 68 to 241 nt of HAV strain HM175 (27). The reverse primer for this assay, HAV240R, has a 3′-terminal G-U nucleotide mismatch with all genotype III sequences (27, 28). Mismatches within the 3′-end primer region may adversely impact the efficiency of PCR (29). The effect of the HAV240R 3′-terminal mismatch was recently investigated and found to underestimate IIIA genome concentration by a factor of 2.8 (28). This same study also noted that 54% of the HAV genome sequences had one or more mismatches with the ISO 15216-1 HAV primer and probe sequences. In contrast, the region targeted by our 5′-UTR assay is much more highly conserved, with fewer than 9% of the genome sequences having a single mismatch with the primer and probe sequences. The assays also differ significantly in run time: our 5′-UTR assay takes only 1.25 h compared with 3.25 h for the ISO 15216-1 rRT-PCR. Notably, we found perfect agreement between the two assays for the detection of HAV in serum and plasma specimens. Sequence conservation, shorter run time, and compatible amplification parameters with the triplex subgenotyping assay represent significant advantages to our 5′-UTR assay.
The 5′-UTR and triplex assays were designed to fulfill gaps in our ability to rapidly respond to and inform a public health response to a large HAV subgenotype IB outbreak. The ability to run both assays simultaneously provided a rapid and inexpensive method for confirming suspected HAV cases as well as a way to monitor disease trends through subgenotype identification. During the course of the outbreak, we found that rapid communication of subgenotype data helped in two ways. Epidemiologists were able to rule out case-patients with subgenotype IA or IIIA strains, which aided in focusing resource-intensive disease investigation and outbreak mitigation efforts on subgenotype IB case-patients and their contacts (30). Similarly, based on the initial subgenotype identification, sequence-based genotyping efforts were prioritized and expedited for IB cases. We consider these rRT-PCR assays as a complement to, and not a replacement for, sequence-based genotyping, with the added value of having actionable results within a shorter time frame. This capacity may be sufficient for laboratories that may not have the capacity to add the more-expensive and technically challenging sequence-based approach to HAV detection or that prefer to refer HAV samples for genotyping. While these assays cannot determine the actual strain in a given patient, the availability of these rapid, same-day tests strengthen public health surveillance efforts for HAV outbreak detection and response. Results from rRT-PCR can be available within a few hours of specimen receipt, whereas sequence-based genotyping may require several days, or longer if referred, to complete. For HAV reference laboratories where batching of specimens is a common practice for NGS, prior knowledge of the subgenotype may help prioritize specimens for sequence-based genotyping.
There are two notable limitations to our study. First, the specimens included in this study were collected during two large unrelated outbreaks of HAV subgenotype IB occurring in California and Michigan, and 54.4% of the positive specimens in our study are represented by outbreak strains or related variants. Therefore, the diversity of IB strains analyzed in our study is limited. Likewise, only seven subgenotype IIIA strains were detected in our study, reflecting the paucity of this subgenotype in the United States (13). Overall, our panel of specimens likely represents regional strain diversity at best and does not adequately capture the global diversity of HAV. Additional studies are needed to assess the performance of these assays against a global collection of HAV strains. Second, we have not assessed the suitability of testing specimen types other than sera and plasma with these assays. Stool specimens may also serve as an acceptable specimen type for the detection of HAV RNA, since HAV may be present at a higher titer and for a longer duration in stool specimens than in serum specimens (31). However, stool specimens may contain more substances that can inhibit PCR (32). Many of these inhibitory substances can be reduced or eliminated by the use of a magnetic bead-based nucleic acid extraction method coupled with an RT-PCR master mix that has been formulated to be less susceptible to common PCR inhibitors, similar to the methodology selected for this study.
The implementation of rapid and sensitive molecular diagnostic tools and assays has the potential to improve public health laboratory surveillance for HAV outbreak detection and characterization. Periodic updating of molecular diagnostic assays is necessary to account for RNA virus diversity and evolution (33). This process is facilitated by the increasing availability of whole-genome sequences in public databases. Using these resources, we have developed and validated both a triplex rRT-PCR assay for subgenotype IA, IB, and IIIA identification and discrimination, as well as an updated 5′-UTR rRT-PCR assay with the potential to efficiently detect all genotypes of HAV infecting humans. These two new HAV assays will enable rapid laboratory confirmation and subgenotype identification to help define and stratify case-patients in support of epidemiological investigations of hepatitis A outbreaks.
ACKNOWLEDGMENTS
This work was supported, in part, by the Epidemiological & Laboratory Capacity cooperative agreement 5NU50CK000410 funded by the Centers for Disease Control and Prevention.
The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the Centers for Disease Control and Prevention or the Department of Health and Human Services.
We are grateful to Marty Soehnlen of the Michigan Department of Health and Human Services, the San Diego County Public Health Laboratory, and the Contra Costa County Public Health Laboratory for providing serum specimens. We are also grateful for the epidemiological and data support provided by Jennifer Zipprich, Rosie Glenn-Finer, Cynthia Yen, and Maria Salas (CDPH).
REFERENCES
- 1.Hofmeister MG, Foster MA, Teshale EH. 2019. Epidemiology and transmission of hepatitis A virus and hepatitis E virus infections in the United States. Cold Spring Harb Perspect Med 9:a033431. doi: 10.1101/cshperspect.a033431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Foster M, Ramachandran S, Myatt K, Donovan D, Bohm S, Fiedler J, Barbeau B, Collins J, Thoroughman D, McDonald E, Ballard J, Eason J, Jorgensen C. 2018. Hepatitis A virus outbreaks associated with drug use and homelessness - California, Kentucky, Michigan, and Utah, 2017. MMWR Morb Mortal Wkly Rep 67:1208–1210. doi: 10.15585/mmwr.mm6743a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lemon SM, Ott JJ, Van Damme P, Shouval D. 2018. Type A viral hepatitis: a summary and update on the molecular virology, epidemiology, pathogenesis and prevention. J Hepatol 68:167–184. doi: 10.1016/j.jhep.2017.08.034. [DOI] [PubMed] [Google Scholar]
- 4.Lednar WM, Lemon SM, Kirkpatrick JW, Redfield RR, Fields ML, Kelley PW. 1985. Frequency of illness associated with epidemic hepatitis A virus infections in adults. Am J Epidemiol 122:226–233. doi: 10.1093/oxfordjournals.aje.a114093. [DOI] [PubMed] [Google Scholar]
- 5.Latash J, Dorsinville M, Del Rosso P, Antwi M, Reddy V, Waechter H, Lawler J, Boss H, Kurpiel P, Backenson PB, Gonzalez C, Rowe S, Poissant T, Lin Y, Xia GL, Balter S. 2017. Notes from the field: increase in reported hepatitis A infections among men who have sex with men - New York City, January-August 2017. MMWR Morb Mortal Wkly Rep 66:999–1000. doi: 10.15585/mmwr.mm6637a7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Viray MA, Hofmeister MG, Johnston DI, Krishnasamy VP, Nichols C, Foster MA, Balajadia R, Wise ME, Manuzak A, Lin Y, Xia G, Basler C, Nsubuga J, Woods J, Park SY. 2018. Public health investigation and response to a hepatitis A outbreak from imported scallops consumed raw-Hawaii, 2016. Epidemiol Infect 17:1–8. doi: 10.1017/S0950268818002844. [DOI] [PubMed] [Google Scholar]
- 7.Cohen JI, Ticehurst JR, Purcell RH, Buckler-White A, Baroudy BM. 1987. Complete nucleotide sequence of wild-type hepatitis A virus: comparison with different strains of hepatitis A virus and other picornaviruses. J Virol 61:50–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nainan OV, Xia G, Vaughan G, Margolis HS. 2006. Diagnosis of hepatitis A virus infection: a molecular approach. Clin Microbiol Rev 19:63–79. doi: 10.1128/CMR.19.1.63-79.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costa-Mattioli M, Di Napoli A, Ferré V, Billaudel S, Perez-Bercoff R, Cristina J. 2003. Genetic variability of hepatitis A virus. J Gen Virol 84:3191–3201. doi: 10.1099/vir.0.19532-0. [DOI] [PubMed] [Google Scholar]
- 10.Pérez-Sautu U, Costafreda MI, Lite J, Sala R, Barrabeig I, Bosch A, Pintó RM. 2011. Molecular epidemiology of hepatitis A virus infections in Catalonia, Spain, 2005-2009: circulation of newly emerging strains. J Clin Virol 52:98–102. doi: 10.1016/j.jcv.2011.06.011. [DOI] [PubMed] [Google Scholar]
- 11.Robertson BH, Jansen RW, Khanna B, Totsuka A, Nainan OV, Siegl G, Widell A, Margolis HS, Isomura S, Ito K, Ishizu T, Moritsugu Y, Lemon SM. 1992. Genetic relatedness of hepatitis A virus strains recovered from different geographical regions. J Gen Virol 73:1365–1377. doi: 10.1099/0022-1317-73-6-1365. [DOI] [PubMed] [Google Scholar]
- 12.Kroneman A, de Sousa R, Verhoef L, Koopmans MPG, Vennema H, HAVNet Network. 2018. Usability of the international HAVNet hepatitis A virus database for geographical annotation, backtracing and outbreak detection. Euro Surveill 23(37):pii=1700802 10.2807/1560-7917.ES.2018.23.37.1700802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foster MA, Hofmeister MG, Kupronis BA, Lin Y, Xia GL, Yin S, Teshale E. 2019. Increase in hepatitis A virus infections - United States, 2013-2018. MMWR Morb Mortal Wkly Rep 68:413–415. doi: 10.15585/mmwr.mm6818a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hutin YJ, Pool V, Cramer EH, Nainan OV, Weth J, Williams IT, Goldstein ST, Gensheimer KF, Bell BP, Shapiro CN, Alter MJ, Margolis HS. 1999. A multistate, foodborne outbreak of hepatitis A. National Hepatitis A Investigation Team. N Engl J Med 340:595–602. doi: 10.1056/NEJM199902253400802. [DOI] [PubMed] [Google Scholar]
- 15.Stene-Johansen K, Tjon G, Schreier E, Bremer V, Bruisten S, Ngui SL, King M, Pinto RM, Aragonès L, Mazick A, Corbet S, Sundqvist L, Blystad H, Norder H, Skaug K. 2007. Molecular epidemiological studies show that hepatitis A virus is endemic among active homosexual men in Europe. J Med Virol 79:356–365. doi: 10.1002/jmv.20781. [DOI] [PubMed] [Google Scholar]
- 16.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. 2012. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lefever S, Pattyn F, Hellemans J, Vandesompele J. 2013. Single-nucleotide polymorphisms and other mismatches reduce performance of quantitative PCR assays. Clin Chem 59:1470–1480. doi: 10.1373/clinchem.2013.203653. [DOI] [PubMed] [Google Scholar]
- 18.International Organization for Standardization. 2017. ISO 15216-1. Microbiology of the food chain: horizontal method for determination of hepatitis A virus and norovirus using real-time PCR, part 1: Method for quantification, 1st ed., 2017-03 ed International Organization for Standardization, Geneva, Switzerland. [Google Scholar]
- 19.Centers for Disease Control and Prevention. 2005. Positive test results for acute hepatitis A virus infection among persons with no recent history of acute hepatitis–United States, 2002-2004. MMWR Morb Mortal Wkly Rep 54:453–456. [PubMed] [Google Scholar]
- 20.Castrodale L, Fiore A, Schmidt T. 2005. Detection of immunoglobulin M antibody to hepatitis A virus in Alaska residents without other evidence of hepatitis. Clin Infect Dis 41:e86–e88. doi: 10.1086/497073. [DOI] [PubMed] [Google Scholar]
- 21.Landry ML. 2016. Immunoglobulin M for acute infection: true or false? Clin Vaccine Immunol 23:540–545. doi: 10.1128/CVI.00211-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wollersheim SK, Humphries RM, Cherry JD, Krogstad P. 2015. Serological misdiagnosis of acute liver failure associated with echovirus 25 due to immunological similarities to hepatitis A virus and prozone effect. J Clin Microbiol 53:309–310. doi: 10.1128/JCM.02724-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kao HW, Ashcavai M, Redeker AG. 1984. The persistence of hepatitis A IgM antibody after acute clinical hepatitis A. Hepatology 4:933–936. doi: 10.1002/hep.1840040525. [DOI] [PubMed] [Google Scholar]
- 24.Bower WA, Nainan OV, Han X, Margolis HS. 2000. Duration of viremia in hepatitis A virus infection. J Infect Dis 182:12–17. doi: 10.1086/315701. [DOI] [PubMed] [Google Scholar]
- 25.Coudray-Meunier C, Fraisse A, Mokhtari C, Martin-Latil S, Roque-Afonso AM, Perelle S. 2014. Hepatitis A virus subgenotyping based on RT-qPCR assays. BMC Microbiol 14:296. doi: 10.1186/s12866-014-0296-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sánchez G, Bosch A, Pintó RM. 2007. Hepatitis A virus detection in food: current and future prospects. Lett Appl Microbiol 45:1–5. doi: 10.1111/j.1472-765X.2007.02140.x. [DOI] [PubMed] [Google Scholar]
- 27.Costafreda MI, Bosch A, Pintó RM. 2006. Development, evaluation, and standardization of a real-time TaqMan reverse transcription-PCR assay for quantification of hepatitis A virus in clinical and shellfish samples. Appl Environ Microbiol 72:3846–3855. doi: 10.1128/AEM.02660-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Persson S, Karlsson M, Borsch-Reniers H, Ellström P, Eriksson R, Simonsson M. 2019. Missing the match might not cost you the game: primer-template mismatches studied in different hepatitis A virus variants. Food Environ Virol doi: 10.1007/s12560-019-09387-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stadhouders R, Pas SD, Anber J, Voermans J, Mes TH, Schutten M. 2010. The effect of primer-template mismatches on the detection and quantification of nucleic acids using the 5′ nuclease assay. J Mol Diagn 12:109–117. doi: 10.2353/jmoldx.2010.090035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Probert WS, Gonzalez C, Espinosa A, Hacker JK. 1 July 2019. Molecular genotyping of hepatitis A virus, California, USA, 2017–2018. Emerg Infect Dis doi: 10.3201/eid2508.181489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin A, Lemon SM. 2006. Hepatitis A virus: from discovery to vaccines. Hepatology 43:S164–S172. doi: 10.1002/hep.21052. [DOI] [PubMed] [Google Scholar]
- 32.Rådström P, Knutsson R, Wolffs P, Lövenklev M, Löfström C. 2004. Pre-PCR processing: strategies to generate PCR-compatible samples. Mol Biotechnol 26:133–146. doi: 10.1385/MB:26:2:133. [DOI] [PubMed] [Google Scholar]
- 33.Stellrecht KA. 2018. The drift in molecular testing for influenza: mutations affecting assay performance. J Clin Microbiol 56:e01531-17. doi: 10.1128/JCM.01531-17. [DOI] [PMC free article] [PubMed] [Google Scholar]